Summary:
We examined how baseline CD4+ T cell repertoire and precursor states impact responses to pathogen infection in humans using primary immunization with yellow fever virus (YFV) vaccine. YFV-specific T cells in unexposed individuals were identified by peptide-MHC tetramer staining and tracked pre- and post-vaccination by tetramers and TCR sequencing. A substantial number of YFV-reactive T cells expressed memory phenotype markers and contained expanded clones in the absence of exposure to YFV. After vaccination, pre-existing YFV-specific T cell populations with low clonal diversity underwent limited expansion, but rare populations with a reservoir of unexpanded TCRs generated robust responses. These altered dynamics reorganized the immunodominance hierarchy and resulted in an overall increase in higher avidity T cells. Thus, instead of further increasing the representation of dominant clones, YFV vaccination recruits rare and more responsive T cells. Our findings illustrate the impact of vaccines in prioritizing T cell responses and reveal repertoire reorganization as a key component of effective vaccination.
Graphical Abstract

dTOC blurb
Pan et al. examine how baseline CD4+ T cell repertoire and precursor states impact responses to pathogen infection in humans using primary immunization with yellow fever virus (YFV) vaccine. They find that instead of increasing the representation of dominant pre-existing YFV-reactive clones, vaccination reorganizes the immunodominance hierarchy by recruiting rare and more responsive T cells.
Introduction
The development of a functional memory response is required for protection against subsequent infections (Kathryn et al., 2013; Martin et al., 2012a). Decades of research inspired by questions on immunological memory have led to a wealth of knowledge into T cell responses and the differentiation process that governs memory cell selection (Malherbe et al., 2004; Williams et al., 2008; Zehn et al., 2009). However, as the majority of studies have used transferred transgenic T cells in laboratory mice, these data have been constrained by cells expressing a limited number of TCR specificities and experimental designs that relied on transferred cells. Using a sensitive peptide-MHC (pMHC) tetramer enrichment protocol that enabled the detection of rare antigen-specific T cells, the endogenous murine responses to antigen challenge were tracked by Moon et al and Obar et al (Moon et al., 2007; Obar et al., 2008). These seminal studies identified precursor frequency as a key determinant of response kinetics, immunodominance, and memory differentiation. Given the many differences between human and mice, it remains unclear if the same rules apply to human T cell responses to a novel pathogen. This question has gained a sense of urgency with the emergence of SARS-CoV-2. Understanding how the composition of the pre-immune repertoire governs post-challenge dynamics will be necessary for optimizing human T cell responses to existing microbes and future emerging pathogens.
The differentiation state of precursor T cells is of considerable interest following the identification of SARS-CoV-2-specific memory cells in unexposed individuals (Bacher et al., 2020; Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Nelde et al., 2020; Peng et al., 2020; Weiskopf et al., 2020). While some memory T cells recognizing SARS-CoV-2 exhibit cross-reactivity to common cold coronaviral strains (Braun et al., 2020; Mateus et al., 2020), immune memory to unexposed antigens is a general phenomenon and is found in other virus-specific T cells from uninfected adults (Kwok et al., 2012; Su et al., 2013). Pre-existing memory T cells to the human immunodeficiency virus (HIV) express the typical memory markers, are capable of rapid cytokine production in vitro, and exhibit evidence of clonal expansion by T cell receptor (TCR) sequencing. Furthermore, HIV-reactive T cell clones from unexposed individuals cross-recognize unrelated microbial peptides, suggesting that pre-existing memory cells may have developed as a cross-reactive response to a broad range of environmental antigens (Su et al., 2013). In mice, a free-living environment promotes the accumulation of memory T cells and protection against infectious pathogens (Beura et al., 2016; Le Bert et al., 2020; Rosshart et al., 2017). Along with the prevailing paradigm that memory cells provide faster and more robust immune response (Kathryn et al., 2013; Pihlgren et al., 1996; Rogers et al., 2000; Veiga-Fernandes et al., 2000), these findings suggest that pre-existing memory T cells in an individual could be a mechanism for protection against unencountered pathogens.
In this study, we examined the precursor CD4+ T cell repertoire and addressed how precursor states are related to post-vaccination T cell dynamics in humans. We used a highly effective live attenuated yellow fever virus (YFV) vaccine as a model for eliciting primary CD4+ T cell response to a novel pathogen challenge. Because YFV does not circulate in the United States and elicits long-lasting antibody responses, individuals naive to YFV were identified by the absence of exposure and confirmed by a negative serologic test. Longitudinal tracking of CD4+ T cell response to YFV using direct ex vivo pMHC tetramer staining and including responses to multiple YFV antigens in the same individual revealed heterogeneous pre-vaccine repertoires in healthy adults. A substantial number of YFV-specific T cells displayed a memory phenotype, indicating that pre-existing memory T cells can form in the absence of closely related circulating viruses. By following the same T cell specificities over time, we identified differences in T cell kinetics that depended on the pre-vaccination precursor frequencies and initial differentiation states. The clonal composition of YFV-specific T cells before and after vaccination was further assessed by single-cell TCR sequencing and revealed recruitment and predominant expansion of rare TCRs after vaccination. Our data link T cell precursor states to post-exposure responses and suggest that vaccines re-prioritize the immune repertoire to enlist most relevant T cells for responses against novel pathogens.
Results
The baseline repertoire of virus-specific CD4+ T cells
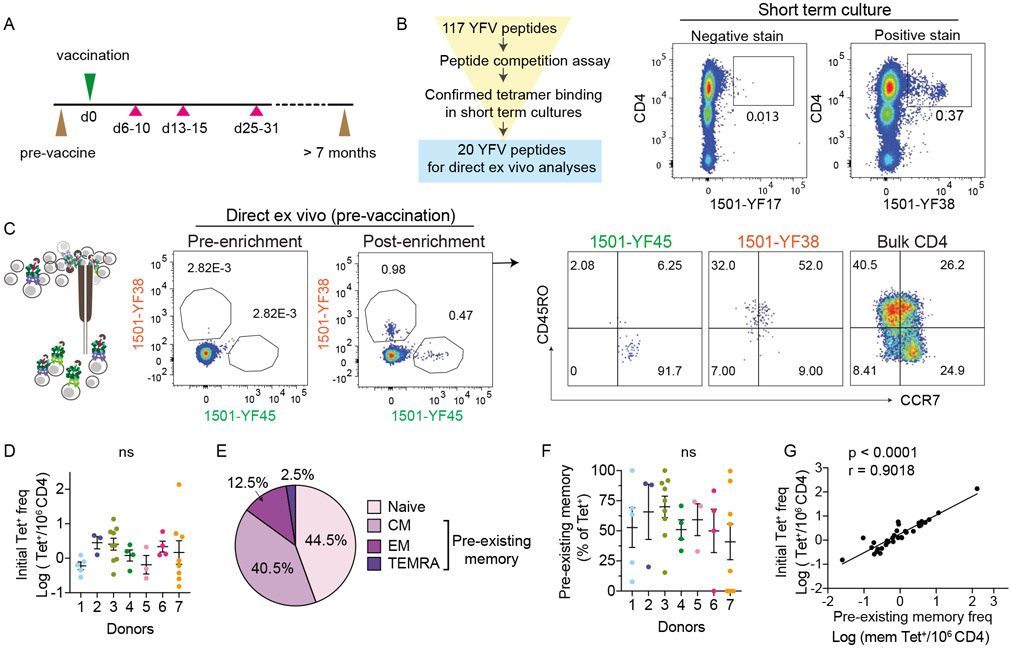
We immunized seven YFV naive healthy subjects with the YFV vaccine. These individuals had no history of YFV exposure and were confirmed to be YFV seronegative (Table S1). All volunteers received one vaccine dose and had blood taken before vaccination, 6-10 days, 13-15 days, 25-31 days, and 7 to 34 months after vaccination (Fig. 1A). To enable longitudinal analyses of virus-specific T cells using pMHC tetramers, we recombinantly expressed five HLA-DR monomers matched to donor class II HLA-DR alleles, DRA/B1*0301, 0401, 0407, 0701, 1501. An initial set of 117 peptides was selected based on prior studies and in silico prediction (de Melo et al., 2013; James et al., 2013). Peptide binding to the five HLA-DR alleles of interest was confirmed by competition assay. To focus on relevant YFV epitopes recognized by vaccine-induced T cells, peripheral blood mononuclear cells (PBMC) obtained after vaccination from each individual were stimulated with the selected peptide pool and stained with tetramers (Fig 1B). Tetramers that positively stained cultured cells were then selected for direct ex vivo analyses (Table S2 and Fig. S1A). Tetramer staining was coupled with magnetic column-based enrichment to enable enumeration of rare tetramer-labeled T cells in the unprimed repertoire (Fig. 1C and S1B). Cells were co-stained with anti-CD45RO and anti-CCR7 to delineate the baseline differentiation state of virus-specific T cells. In total, we analyzed 36 YFV-specific CD4+ populations using 20 unique epitopes for 5 HLA-DRs in 7 individuals. This revealed CD4+ precursor frequencies that ranged from 0.15 to 137 cells per million CD4+ T cells (Fig. 1D). In agreement with prior studies on pre-existing memory T cells (Su et al., 2013), memory markers were abundantly expressed by YFV-specific T cells before vaccination (Fig. 1E). Collectively, 40.5% of tetramer+ precursors expressed a central memory phenotype (CM, CD45RO+CCR7+), 12.5% expressed an effector memory phenotype (EM, CD45RO+CCR7−), 2.5% expressed a TEMRA phenotype (CD45RO−CCR7−), and the remaining 44.5% were comprised of naive phenotype T cells (CD45RO−CCR7+). The abundance of pre-existing memory varied between T cells specific for different epitopes within the same individual and correlated with the size of precursor population (Fig. 1F-G). We looked for shared phenotypes between T cell specificities but did not find a consistent pattern. Cells that recognized the same epitope expressed distinct frequencies and differentiation states in different donors (Fig. S1C). These data indicated that humans can call upon highly variable and heterogeneous T cell repertoires in response to a novel viral infection, including a substantial number of T cells that have already acquired memory differentiation and are abundant before antigen exposure.
Figure 1: YFV-specific CD4+ T cells in YFV unexposed individuals.
(A) Overview of the study protocol. At indicated time points, phlebotomy (pink triangles) or leukopheresis (brown triangles) were performed to obtain serial blood samples before and after vaccination. (B) Outline shows the selection process for peptides used in direct ex vivo analyses. Donor 3 PBMCs obtained on post-vaccine day 14 was stimulated with YFV peptide pool for 3 weeks. Example plots show representative staining for a peptide that was not selected (negative stain) or selected (positive stain) for further analyses. (C) Direct ex vivo staining, pre- and post-magnetic enrichment of representative YFV tetramer+ cells using blood obtained before vaccination. Anti-CD45RO and CCR7 staining identified tetramer+ cells that primarily expressed a naïve (1501-YF45) or memory (1501-YF38) phenotype before vaccination. Bulk CD4+ T cells are shown for comparison (right). (D) The frequency of YFV tetramer+ CD4+ T cell across 7 healthy subjects was quantified by direct ex vivo staining. Each dot represents data from a distinct YFV-specific population, repeated an average of 3.3 times (± 2.3). Donors 1 (n = 5), 2 (n = 3), 3 (n = 9), 4 (n = 4), 5 (n = 3), 6 (n = 4), 7 (n = 8). (E) Differentiation phenotype of tetramer+ cells before vaccination (n = 36): naïve (CD45R−CCR7+), central memory (CM, CD45RO+CCR7+), effector memory (EM, CD45RO+CCR7−), and TEMRA (CD45R−CCR7−). (F) Abundance of pre-existing memory T cells as a percentage of tetramer+ cells shown in D. (G) Correlation between the frequency of tetramer+ T cell and pre-existing memory T cells within each population (n = 36). For D and F, data are shown as mean ± SEM. No statistical difference between donors were identified using Welch’s ANOVA. For G, association was measured by Spearman correlation. Also see Fig. S1.
Post-vaccine dynamics of primary CD4+ T cell response
We next sought to measure the immune dynamics of YFV-specific populations. We used tetramers to monitor vaccine-induced T cell responses during the first month and beyond 7 months after vaccination. Multiple virus-specific populations were tracked simultaneously and identified across time using the same tetramers. This showed that the overall peak of response converged on days 13-15, followed by a typical contraction phase and the establishment of a new baseline at a memory time point (Fig. 2A). To mark activated T cells, tetramer+ cells were stained for ICOS, a CD28-related costimulatory molecule induced by TCR signaling (Hutloff et al., 1999). ICOS staining peaked around 2 weeks after vaccination and waned thereafter (Fig. 2B). The incomplete downregulation of ICOS expression a month later suggested that some virus-specific T cells remained activated many weeks post vaccination, possibly in response to antigen persistence after viral clearance (Akondy et al., 2017; Akondy et al., 2009). Quantification of activated T cells using CD38 expression demonstrated a similar pattern (Fig. 2C, S2A). Notably, the summary kinetics do not fully describe the variation in antigen-specific T cell responses. Distinct YFV-specific populations from the same individual generated different magnitude of responses that varied in the rate of expansion and contraction (Fig. 2D-E). The measured peak frequency differed by over 270-fold and ranged between 1.7 to 462 cells per million (Fig. S2B). The maximal effector response generally emerged 2 weeks after vaccination, but a few populations peaked earlier or later and the timing was independent of the initial phenotype (Fig.S2C-D).
Figure 2: The kinetics of primary CD4+ T cell response after vaccination.
(A-C) Frequency of tetramer+ cells (A) and their activation status as measured by MFI of ICOS (B) or CD38 (C) at the indicated time points (n = 36). (D) Example of two tetramer+ populations that showed distinct response kinetics. (E) Plots show changes in tetramer staining for the same populations across time as in D (top) and a time-dependent increase in CD45RO and ICOS expression (bottom). Experiments were performed with the consideration of expected T cell frequency and sample availability (pre-vaccine: 40 million CD4; d7, 14, 28: 10 million PBMCs; d657: 10 million CD4). Plots are representative of 3 experiments. For A-C, mixed-effects analyses were performed. For D, multiple t-test was performed and corrected using Holm-Sidak method. Data are represented as mean ± SEM. * p < 0.05, *** p < 0.001, **** p < 0.0001. Also see Fig. S2.
Vaccination boosts rare virus-specific populations
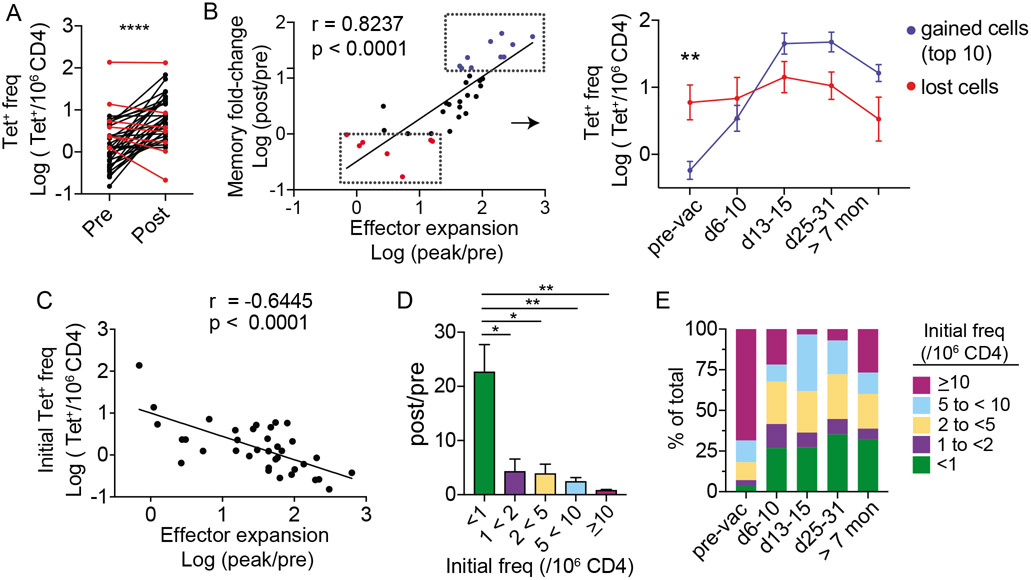
We compared the frequency of each antigen-specific population at least 7 months after vaccination with its baseline frequency and found that YFV-specific T cells did not uniformly become more numerous after immunization. While the vaccine induced an overall increase in YFV-specific T cells, 7 out of 36 post-vaccine populations became less abundant after vaccination (19.4%) (Fig. 3A). Next, we investigated if populations that failed to gain memory cells also generated a weaker effector response. The magnitude of effector expansion was measured as a fold change between the peak frequency and the pre-vaccine baseline (peak/pre). The efficiency by which each population established a memory pool was quantified by dividing the frequency at a memory time point with its starting frequency (post/pre). This comparison showed a tight correlation between the magnitude of change from baseline during early and late post-vaccine time points (Fig. 3B, left). To better understand this relationship, we followed T cell dynamics over time for the top 10 boosted populations and the bottom 7 that lost cells after vaccination. This revealed a clear vaccine-induced repositioning of immunodominance hierarchy. Cells that started off at higher baseline continued to be the dominant populations within the first week after vaccination, but they soon became surpassed by cells that generated a larger effector response in the subsequent time points (Fig. 3B, right). Consistent with this, precursor size negatively correlated with effector expansion and the rarest precursor subset acquired the largest gain in memory cells after vaccination (Fig. 3C-D). As a group, the relative abundance of T cells that started with an initial frequency of under 1 cell per million represented 3.5% of all tetramer+ cells before vaccination and increased to 32.4% after vaccination (Fig. 3E). Thus, previously rare antigen-specific populations were preferentially recruited as part of an effective response to a first-time infectious challenge.
Figure 3: Vaccination preferentially recruits rare tetramer+ precursors.
(A) Plot shows the frequency of tetramer+ T cells. Line connects the same population before (pre) and over 7 months after vaccination (post) (n = 36). Populations that became less abundant > 7 months after vaccination are marked in red. (B) Correlation between fold-change in effector response and fold-change at a memory time point. Red in B identifies the same red populations in A. The top ten most expanded populations are indicated in blue. Each dot represents a distinct tetramer+ population. Right panel shows longitudinal changes in T cell frequency for the blue and red subsets. (C) Correlation between effector expansion and pre-vaccine frequency. (D) Tetramer+ cells are grouped by pre-vaccination frequencies (<1 (n = 15),1 to < 2 (n = 6), 2 to < 5 (n = 8), 5 to < 10 (n = 5), ≥10 (n = 2)). Bar-graph shows fold-change in frequency at a post-vaccine memory time point for cells in each bin. (E) The relative abundance of cells in each bin is plotted as a percentage of all tetramer+ cells at the indicated time points. For A, paired t-test was performed. For B, association was measured by Pearson correlation. For C, Spearman correlation was used. Differences between blue and red subsets in B were tested by multiple t-test and corrected by Holm-Sidak method. For D, Welch’s ANOVA was used. Data are represented as mean ± SEM. * p < 0.05, ** p < 0.01, **** p < 0.0001.
Pre-existing memory T cells generate heterologous responses
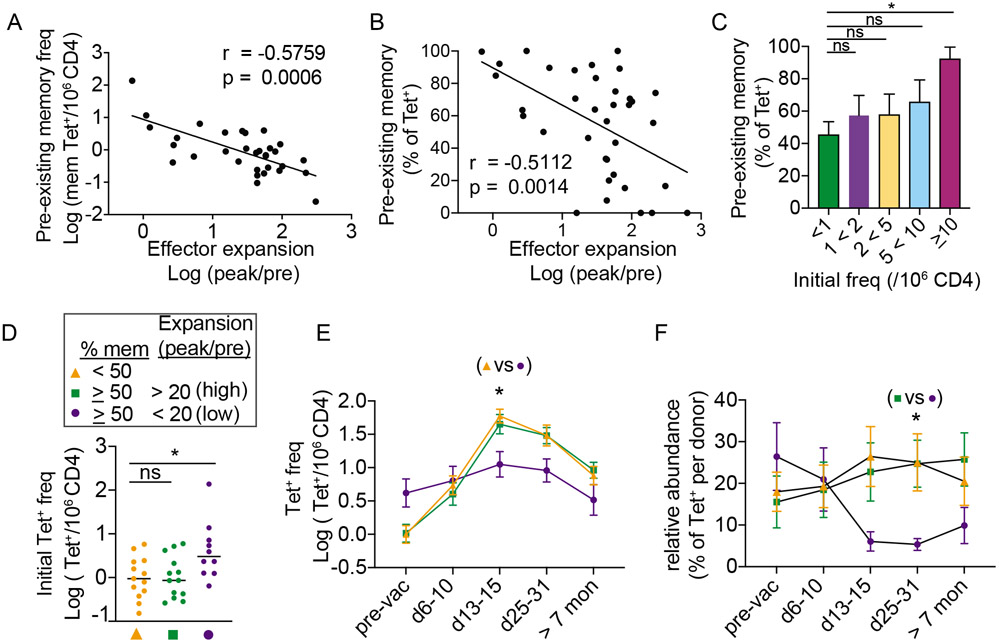
The small magnitude of expansion generated by abundant precursors was unexpected. Our data contrasted with studies in mice, where precursor frequency positively correlated with effector responses (Moon et al., 2007; Nelson et al., 2015). One explanation for this may be the presence of pre-existing memory T cells in the human pre-immune repertoire, which is largely absent among CD4+ T cells from adult mice. In support of this idea, the frequency of pre-existing memory T cells inversely correlated with effector response (Fig. 4A). Pre-existing memory T cell abundance, as a percentage of tetramer+ population, was also negatively associated with vaccine-induced expansion (Fig. 4B). While pre-existing memory phenotype was most highly expressed by the largest and the least responsive precursor subset, we noted that the distribution of memory phenotype was broad and included low frequency cells that expanded well (Fig. 4C). Using a simple midway cutoff to separate pre-vaccine populations into those that contained under 50% or at least 50% of pre-existing memory cells, we showed that memory-dominant populations generated a broad range of effector responses after vaccination (Fig. S3A-B). Next, we investigated features associated with differential capacity for expansion. Because a key goal of vaccination is to form vaccine-specific memory cells, we defined weak response using the fold-expansion generated by the 7 populations that failed to gain cells after vaccination (20, mean peak/pre value plus 2 standard deviation). We then partitioned memory-dominant populations into high (> 20) or low (<20) response groups. This showed that low-response memory populations were more abundant compared to the other groups before vaccination (Fig. 4D). Following vaccination, the low-response group was initially more abundant but became surpassed by other-YFV-specific T cells. By contrast, both naïve-dominant and high-response memory groups were subdominant during early post-vaccine period but subsequently reached a higher frequency in the later phases of the response (Fig. 4E-F). Taken together, these data identified altered responsiveness in a subset of memory precursors and highlighted the connection between the pre-immune repertoire and post-immune response.
Figure 4: Pre-existing memory T cells generate heterologous responses.
(A-B) Correlation between effector response and the frequency (A) or the relative abundance (B) of tetramer+ cells that expressed a memory phenotype prior to vaccination. (C) Bar-graph shows the average memory phenotype in each bin as a percentage of tetramer+ cells. (D) Tetramer+ T cells were divided into 3 subsets according to the frequency of memory phenotype cells before vaccination and the magnitude of maximal change from the baseline. Scatter plot shows the distribution of pre-vaccination frequency for each subset. (E) Average frequency of tetramer+ cells in each group at the indicated time points. (F) Relative abundance of tetramer+ cells in each group, calculated as a percentage of tetramer+ cells from the same individual. For A, association was measured by Spearman correlation. For B, Pearson correlation was used. For C and D, Welch’s ANOVA was used. For E-F, mixed effects analysis and Tukey correction was performed. Data are represented as mean ± SEM. * p < 0.05. Also see Fig. S3.
Proliferative and functional potentials are intact in pre-existing memory T cells
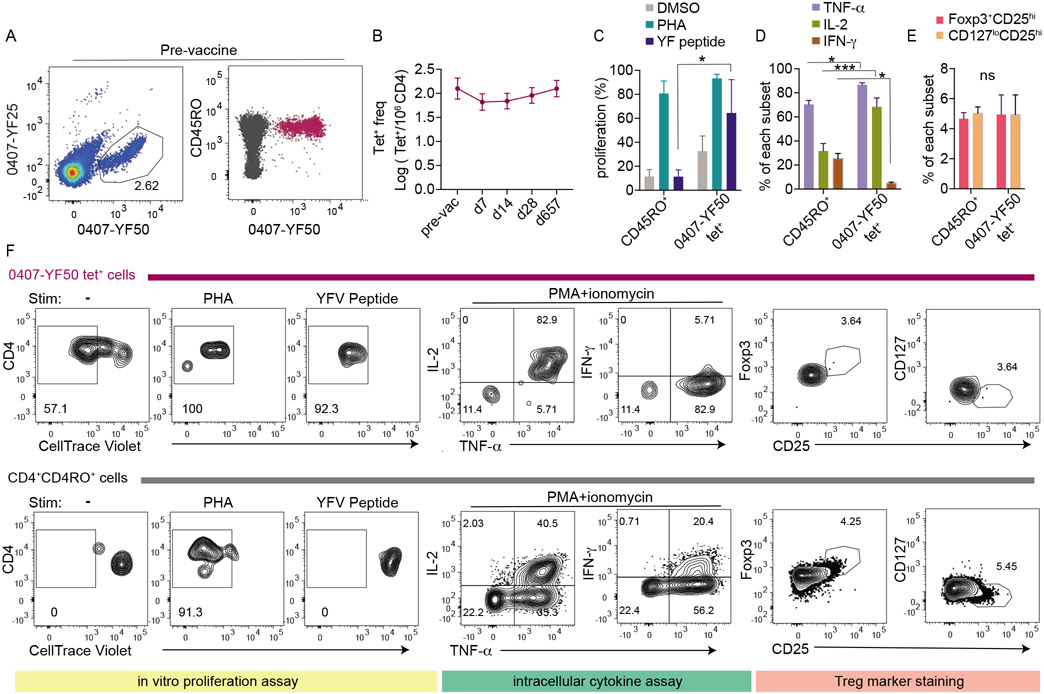
We sought to understand why some memory precursors expanded well whereas others did not. We first analyzed ICOS staining to look for a difference in T cell activation. The activation kinetics appeared diminished for the low-response memory group, although the differences from better responding cells were not statistically significant (Fig. S3C). Next, we examined the distribution of memory subsets by CD45RO and CCR7 expression, which was similar between groups with distinct response kinetics (Fig. S3D). To further investigate T cell responsiveness, we focused our analyses on one precursor population that predominately expressed a memory phenotype before vaccination, did not expand after vaccination, and was sufficiently abundant in the baseline blood sample to be isolated for in vitro studies (Fig. 5A-B). In vitro proliferation assay was performed by sorting equal numbers of CellTrace Violet-labeled 0407-YF50 tetramer+ cells or CD4+CD45RO+ memory cells into wells containing monocyte-derived dendritic cells. Cultures were treated with vehicle control (DMSO), Phytohaemagglutinin (PHA), or YFV peptide and analyzed after 5 days. We found that YFV tetramer+ cells proliferated robustly to PHA and cognate peptide, despite of an apparent lack of response to vaccine in vivo (Fig. 5C, 5F). These data indicated that these tetramer+ cells did not have an intrinsic defect to engage antigens or to undergo proliferation. Consistent with this, stimulated 0407-YF50 tetramer+ cells produced higher levels of TNF-α and IL-2 but less IFN-γ compared to background CD4+ T cells, suggesting a competent but skewed functional phenotype (Fig. 5D, 5F). Because the presence of regulatory T cells (Tregs) within an antigen-specific population could impact the robustness of effector response (Su et al., 2016), we investigated whether 0407-YF50 tetramer+ cells contained a higher proportion of Tregs. We used Foxp3+CD25hi or CD127loCD25hi phenotypes to identify Tregs and showed that the abundance of Tregs was not increased within 0407-YF50 tetramer+ cells (Fig. 5E-F). Collectively, these data argued against impaired responsiveness as a general explanation for why some pre-existing memory T cells expanded weakly after vaccination.
Figure 5: Low-responding T cells proliferate and produce cytokines in vitro.
(A) Tetramer staining and CD45RO expression of a representative low-response population in the pre-vaccination sample (0407-YF50 from donor 7). (B) Response kinetics of 0407-YF50 tetramer+ cells across time. Each time point is representative of 3-5 experiments. (C) Five hundred CD45RO+CD4+ T cells and 0407-YF50 tetramer+ T cells were cultured for 5-day with monocyte-derived dendritic cells and treated with DMSO, PHA, or YF peptide as indicated. Bar-graph summaries the percentage of proliferating cells with low CellTrace Violet (CTV) staining. (D) The frequency of CD45RO+CD4+ T cells and tetramer+ cells that positively stained for TNF-α, IL-2, or IFN-γ. (E) The frequency of CD45RO+CD4+ T cells and tetramer+bcells that were Foxp3+CD25hi or CD127loCD25hi (F) Plots show representative staining for CTV, cytokines, CD25, Foxp3, or CD127. C-E summarized data from 2-3 experiments using cells from pre-vaccine samples. Significance was determined by two-way ANOVA with Sidak’s multiple comparison test. Data are represented as mean ± SEM. * p < 0.05, *** p < 0.001. Also see Fig. S3.
Vaccination reshapes the baseline virus-specific repertoire to improve repertoire fitness
T cells do not act in isolation but instead interact with other T cells to generate a collective response. Early adoptive transfer experiments have clearly demonstrated a dependence of T cell response on cell number, whereby transferring a higher numbers of transgenic T cells limited the ability these cells expand in recipient host (Badovinac et al., 2007; Hataye et al., 2006). This observation has been linked to competition for limiting antigens (Kedl et al., 2000; Smith et al., 2000), which also leads to selective expansion of high affinity T cells in a polyclonal repertoire (Busch and Pamer, 1999; Malherbe et al., 2004). Competition between endogenous T cells may explain why some YFV-specific populations generated weak responses to vaccination. To investigate this idea, we used median staining intensity (MFI) of tetramer staining as a way to approximate the strength of TCR-ligand interaction and examined tetramer MFI at baseline and following vaccination.
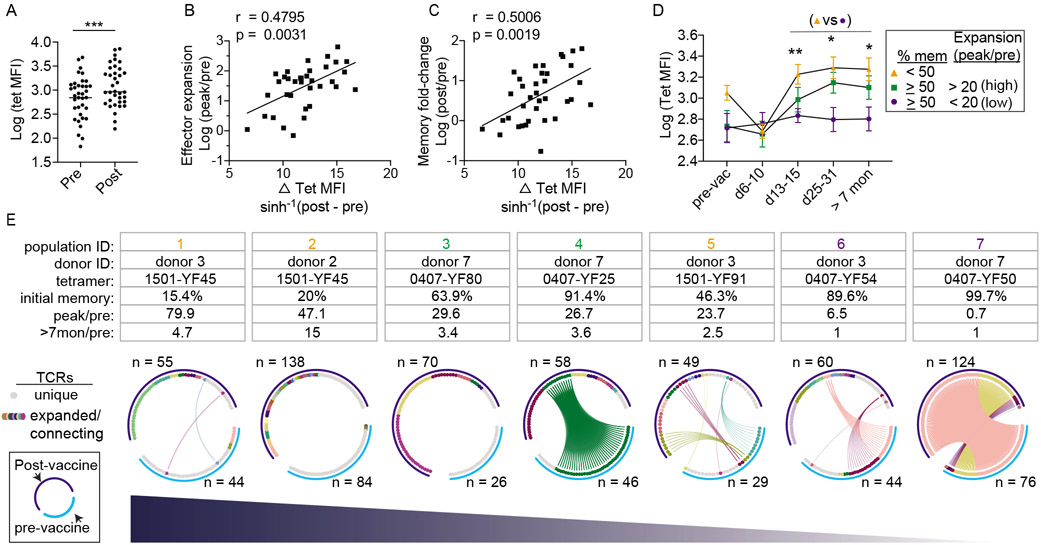
Tetramer+ populations were divided by pre-vaccine memory phenotype (< 50% or ≥ 50% memory). In agreement with the possibility of cross-reactivity, populations containing a higher percentage of memory cells stained more weakly for tetramers before vaccination (Fig. S4A). However, differences in tetramer binding at baseline was not predictive of a better or worse response. Limiting the analyses to populations that stained as well as those in the naive-dominant group did not change the overall patterns of expansion (Fig. S4B-D). In contrast to the lack of an association between effector expansion and pre-vaccine tetramer MFI, a significant correlation emerged after vaccination (Fig. S4E-F). We found tetramer+ cells stained more strongly for tetramers in the post-vaccine samples (Fig. 6A). Consistent with selective recruitment for higher avidity T cells, the gain in post-vaccine tetramer MFI correlated with the magnitude of effector response and the boost in memory T cells (Fig. 6B-C). Serial analyses of tetramer+ cells showed that the increase in post-vaccine tetramer MFI was limited to populations that had expanded, providing further support for coupling of effective response with selective T cell recruitment (Fig. 6D). Collectively, these data suggested that vaccine improved the binding of virus-specific repertoire to YFV after vaccination
Figure 6: Effective response is coupled with selective T cell recruitment and reorganization of clonal hierarchy.
(A) Plot summarizes tetramer MFI before and at least 7 months after vaccination (n = 36). (B-C) Correlation between the gain in tetramer MFI (MFI > 7 months after vaccination – baseline MFI) and fold-change in effector (B) or memory response (C). (D) Changes in tetramer MFI over time for three groups of YFV-specific T cells as indicated. (E) Circos plots summarize TCR sequences from 7 YFV-specific populations obtained from 3 donors. Each plot represents a single tetramer+ population from one donor and includes data from cells isolated before vaccination (light blue arc) or over 7 months after vaccination (dark blue arc). Gray marks unique TCRs. Other colors indicate expanded TCRs and TCRs found in both time points. TCRs of the same amino acid sequence share the same color, are ordered by frequency, and are connected by lines if found in both visits. For A, paired t-test was performed. For B and C, association was measured by Pearson correlation. For D, mixed effects analysis and Tukey correction was performed. *p < 0.05, **p < 0.01. Also see Fig. S4-6.
Our findings are consistent with a model in which intra-clonal competition between cells of the same specificity brings out the stronger-binding constituents of a population that has an edge over their less competitive neighbors. To dissect the selection process at a single cell level, we performed TCR sequencing on 903 individually sorted cells encompassing 7 YFV-specific populations from 3 donors using blood drawn before and at least 7 months after vaccination (Fig. 6E, Fig. S5). Each circos plot represented TCRs from the same tetramer+ population, shown in separate arcs for the pre-vaccine and post-vaccine time points. TCRs were ordered by frequency and connected by lines if the same TCRβ sequences were shared between blood obtained from separate visits (Table S3). Before vaccination, primarily naïve populations contained mostly unique clonotypes. Expanded TCRs were found in memory-dominant populations, consistent with prior antigen-driven proliferation in the in vivo setting. The degree of clonal expansion was particularly prominent for the most memory-skewed population (population 7, same as 0407-YF50 in Fig. 5), which contained 2 highly expanded clonotypes that occupied 92% of repertoire space. After vaccination, three distinct patterns were observed. For populations that started with mostly unique TCR sequences, vaccination focused the immune repertoire onto a diverse set of T cell clones that were minimally shared with TCR sequences from the baseline sample (populations 1, 2, 3). For the most clonal population that generated minimal effector response, the post-immune TCR composition remained remarkably similar to the pre-vaccination repertoire (population 7). For the remaining populations, vaccination established a new clonal hierarchy that replaced the initial repertoire. None of highest ranking clonotypes in the pre-vaccine repertoire remained the most abundant after antigen exposure, although some TCRs detected in the pre-vaccine samples persisted in the post-vaccine blood (populations 4, 5, 6). A clear example of this was population 4, which started with a single dominant expanded clonotype that accounted for 83% of T cells sequenced but became less abundant after vaccination (21%). This clonotype was displaced by a new TCR that was not identified in the baseline sample, suggesting that the T cell(s) expressing the dominant post-vaccine TCR was likely rare before vaccination. The recruitment of previously rare T cells into the repertoire reduced the space occupied by pre-expanded TCRs and contributed to a more balanced and diverse clonotype distribution after vaccination (Fig. S6A-B). Notably, vaccine-specific memory repertoire can change in the absence of a numerical gain for memory cells after exposure (population 6).
Having an inventory of diverse T cells may be necessary to evolve the repertoire, as recruitment of rare precursors failed to occur when the majority of TCRs before vaccination were already expanded (population 7). To test this idea, we quantified the frequency of singly occurring TCRs as a percentage of total unique clonotypes in the pre-vaccine sample (diversity reserve). Four tetramer-labeled populations were independently sampled to test diversity reserve’s sensitivity to cell number differences. We found diversity reserve to be a highly repeatable measurement and can be reliably quantified within the range of 8 to 84 well-defined tetramer+ cells (Fig. S6C). Next, we investigated how baseline diversity relates to T cell response after vaccination and found an association between diversity reserve and the magnitude of early expansion (Fig. S6D). Higher diversity reserve further correlated with a larger gain in memory cells (Fig. S6E), suggesting that having a reservoir of rare T cell clones contributed to the expansion and recruitment of virus-specific T cells. Collectively, these data provided clear evidence that vaccination reshaped the clonal composition of virus-specific T cells. For pre-existing memory repertoires that already contained expanded sequences, the availability of alternative TCRs provided the opportunity for recruiting T cells that may be initially rare but are more responsive to viral antigens.
Discussion
We tracked YFV-specific T cells over time to elucidate characteristics that enabled successful effector response and the recruitment of memory cells. The data described here represent a comprehensive longitudinal study of CD4+ T cell response to YFV and include responses to multiple YFV antigens in the same individual. The tetramer-based enrichment approach provided the sensitivity necessary to detect rare precursor T cells and enabled characterization of antigen-specific T cells directly ex vivo in a manner that was not complicated by potential changes from in vitro cultures. Our analyses of the precursor repertoire in YFV naive individuals showed high level of heterogeneity on population and single cell levels. Tetramer-labeled populations differed by over 900-fold in precursor frequency, contained variable abundance of pre-existing memory T cells, and displayed a broad range of TCR diversity. Even though the donors in this study had undergone vigorous screening for YFV exposure, YFV-specific T cells expressing a memory phenotype were identified by tetramers in all individuals before vaccination. These findings are consistent with past studies on other pathogen-specific memory CD4+ T cells in unexposed healthy adults (Kwok et al., 2012; Su et al., 2013), as well as more recent analyses on pre-existing memory T cells to SARS-CoV-2 (Bacher et al., 2020; Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Nelde et al., 2020; Peng et al., 2020; Weiskopf et al., 2020). Together, these datasets provide strong evidence that pre-existing memory T cells are prevalent in the adult human repertoire.
A key unresolved question is how pre-existing memory T cells impact human response to novel vaccines and pathogens. Memory cells have generally been viewed a superior source of protective immunity. The advantage of memory cells has been ascribed to their larger starting number and faster division rate (Kathryn et al., 2013; Pihlgren et al., 1996; Rogers et al., 2000; Veiga-Fernandes et al., 2000). However, naïve cells have also been shown to share similar proliferation rate as memory T cells (Stock et al., 2006; Zimmermann et al., 1999), or generate a greater magnitude of expansion compared to memory T cells (Martin et al., 2012b; Mehlhop-Williams and Bevan, 2014). Modes of stimulation and the choice of transgenic T cells likely contributed to differences in these observations. Here, we provide a detailed view of human T cell kinetics of naive and memory population in the context of primary immunization. Our data did not find a universally better response by memory cells but instead highlighted the heterogeneity within the pre-existing memory compartment. Pre-existing memory cells included those that were highly abundant during the immediate post-vaccine period but were numerically disadvantaged for the remainder of the response. Our data indicate that at least some of these populations were capable of making cytokines. Their functional activity, in combination with a higher initial abundance, may position them as key early responders to infection. As T cell response evolve, low frequency precursors expanded preferentially and dominated the later phases of the response. We suggest that the convergence of these distinct T cell kinetics may provide superior immune coverage from pathogens. Skewing of one subset of pre-existing memory T cells in favor of the other could off-set this balance and promote suboptimal T cell responses to infections.
In addition to reorganizing the immunodominance hierarchy on a population level, vaccination also alters the clonal composition of virus-specific T cells. A widely held view is that infectious challenges narrow the TCR repertoire (Busch and Pamer, 1999). Vaccination did indeed selectively expand certain T cell clones from a diverse naïve precursor repertoire. However, pre-existing memory repertoire already contained oligoclonal T cells, which likely have been selected by prior exposures to cross-reactive antigens and may not respond optimally to the current pathogen. In expanded populations that maintained a reserve of diverse clones, vaccination was capable of overriding the pre-established clonal hierarchy to select for rare T cells that were more responsive to vaccine antigens. This resulted in an overall increase in the diversity of the expanded clonotypes after vaccination. Having a diverse TCR repertoire has been directly linked to protective T cell responses and host survival in mice (Messaoudi et al., 2002). For fast evolving pathogens, the diversity in T cell composition may additionally limit escape variants as mutations emerge, and thus an important factor to consider in a vaccine response. The mechanism of selection likely involves competition for limited resources such as cytokines and/or antigens. Ligand sensitivity may provide additional advantages when access to antigens is limited by large numbers of cells recognizing and competing for the same pMHC complex. Although our data do not directly address how T cells become selected, the finding that tetramer MFI increased after vaccination suggests preferential recruitment of high avidity T cells. Thus, vaccination restructures T cell hierarchy and leads to improved repertoire fitness. Peripheral education of virus-specific repertoire to arm the immune system with the most responsive T cells may be one key mechanism for why vaccines are protective.
In conclusion, we have linked precursor repertoire to the primary human CD4+ T cell response in the setting of YFV vaccine. These data are broadly relevant for T cell responses to other novel pathogens, including SARS-CoV-2. The identification of memory T cells that can recognize SARS-CoV-2 before exposure has led to the speculation of pre-existing protective immunity (Bacher et al., 2020; Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Nelde et al., 2020; Peng et al., 2020; Weiskopf et al., 2020). However, whether pre-existing memory cells facilitate a faster and more robust response to SARS-CoV-2 to limit the severity of infection remains an open question. Our finding that over half of YFV-specific precursors expressed a memory phenotype suggest that this type of memory cells is not a rare finding in unexposed healthy adults. The observations that memory-dominant precursors generate distinct types of post-immune dynamics further highlight the complexity of human pre-existing memory responses. Given varied influences of past immune interactions on the baseline human T cell composition, a critical function of vaccination may be to re-prioritize the repertoire to ensure that the most relevant T cells are recruited for the response to later infection. The impact of advanced age and underlying health conditions on this process may be critical parameters to consider in future designs of effective vaccines against novel pathogens.
Limitation of the Study
Our data were generated from serial blood samples and therefore unable to detect cells that have entered peripheral tissues and other vaccine-induced responses outside of the circulatory compartment. We also do not know how pre-existing memory cells were acquired. Given the extent of interaction between humans and the microbial environment, we suspect that the microbiome and its many antigens likely play a key role in shaping the human precursor repertoire. In addition, flexible engagement of TCRs to different pMHC complexes could enabled some T cells elicited by past infections, dietary antigens, and other environmental exposures to be detected as memory cells to a new pathogen. T cells could also undergo homeostatic turnover and acquire a memory phenotype in the absence of foreign antigens (Haluszczak et al., 2009; Kawabe et al., 2017). The combined influences from past exposures and basal signals likely contribute to the observed differences in precursor frequency and memory phenotype for the same viral epitope in different donors. A key unresolved question is how pre-existing memory cells differ from classic pathogen-induced memory in response to antigen stimulation. Our data demonstrated that pre-existing memory T cells have the capacity to respond, but more studies are needed to evaluate their antigen sensitivity and ability to compete for antigens. Finally, this study focuses on T cell dynamics from a numerical perspective. Future studies should explore functional aspects of the pre-existing memory repertoire to fully understand how pre-existing T cell states contribute to protection or pathology for human diseases.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Laura F. Su (laurasu@upenn.edu)
Material Availability
This study did not generate new unique reagents.
Data and Code Availability
Data were analyzed uses existing computational packages. The datasets generated during this study are available at Mendeley Data (Bartolo, Laurent; Pan, Yi-Gen (2021), “YF TCR data”, doi: 10.17632/75xhmfrpm6.1)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Samples
Healthy adults were screened for travel history to YFV endemic regions (tropical areas of Africa and Central and South America) and prior YFV vaccination by questionnaire. YFV neutralization assay were performed to evaluate antibody response to YFV. All donors were HLA typed (Histogenetics, NY). Seven participants confirmed to have no prior YFV exposure by serologic test and carry the appropriate HLA allele (HLA-DRB1*0301, 0401, 0407, 0701, 1501) were enrolled for the study. Participants received a single dose of 17D live-attenuated yellow fever vaccine strain subcutaneously (YF-VAX®, Sanofi Pasteur). Leukopheresis was performed prior to and 210-1011 days after vaccination. Phlebotomy for whole blood was obtained 6-10 days, 13-15 days, 25-31 days after vaccination. All samples were de-identified and obtained with IRB regulatory approval from the University of Pennsylvania. Subject characteristics are shown in Table S1.
Primary cells
Human samples preparation
PBMCs were isolated from blood samples through density gradient centrifugation as manufacture instruction (Ficoll-Paque, GE Healthcare). T cells and monocytes were enriched from leukopheresis products (RosetteSep, STEMCELL Technologies). Cells were cryopreserved in fetal bovine serum (FBS) with 10% DMSO for later analysis.
Dendritic cells preparation
Dendritic cells were generated according to McCurley et al. (McCurley and Mellman, 2010). In brief, 10 million monocytes were maintained in culture medium (RPMI1640 containing 10% FBS and 25 mM HEPES) supplemented with 100ng/mL GM-CSF + 6.5ng/mL IL-4 (BioLegend). Cells were re-supplied with fresh monocyte medium containing cytokines on day 3. After 5 or 6 days, cells were harvested and treated with 100ng/ml LPS (Sigma) overnight and cultured with sorted CD4+ cells.
Cell lines
Hi5 cells (ThermoFisher) were maintained by insect cell culture medium (ESF921, Expression Systems) supplemented with 0.02% gentamicin at 28°C. Vero cells (ATCC) were maintained by DMEM (Corning) supplemented with 10% FBS at 37°C.
METHOD DETAILS
YFV-17D focus reduction neutralization tests (FRNT)
Approximately 300 foci of YFV-17D were added to two-fold serial dilutions of heat-inactivated serum. Virus-serum mixtures were then incubated for 1 h at 37°C and then added to 2.5E4 Vero cells/well in 96-well flat-bottom plates. Virus-serum mixtures were incubated with cells at 37°C for 1hour, washed, and then overlayed with a 1.5% Methyl Cellulose:1xPa (DMEM) medium. After 40 hours of incubation at 37°C, plates were fixed with 4% paraformaldehyde (Electron Microscopy Sciences), permeabilized, blocked, and stained by sequential incubation with a biotin-conjugated 4G2 monoclonal antibody (ATCC), streptavidin-HRP (MP Biomedicals), and TrueBlue peroxidase substrate (KPL). Images were collected and foci were counted on an ImmunoSpot (Cellular Technology Ltd). FRNT50 titers were reported as the highest reciprocal dilution giving a focus count ≤ the 50% neutralization cutoff, and the geometric mean was computed for technical duplicates.
Epitope selection
YFV peptide candidates included sequences identified in previous studies (de Melo et al., 2013; James et al., 2013) and additional peptides predicted to bind HLA-DRB1*0301, 0401, 0407, 0701, 1501 with a consensus percentile under 20 using IEDB analyses resource (Paul et al., 2015). To validate HLA binding, a low stringency peptide competition assay was performed to identify peptides that can compete off biotinylated control peptides by at least 30%. Biotinylated sequences were HA306 (GGPKYVKQNTLKLAT) for DRB1*0401 and 0407, CLIP103 (CGGGPVSKMRMATPLLMQA) for DRB1*0701 and 1501, and TT511 (CGGKIIVDYNLQSK) for DRB1*0301. Biotinylated control peptides were exchanged with HLA-DR at 1:2 molar ratio in the presence of 5 times excess of a test peptide and detected using horseradish peroxidase (HRP)-conjugated streptavidin (BD Biosciences). PBMCs obtained after vaccination from each donor were stimulated with DR-binding peptides at 0.4ug/ml per peptide for 3 weeks and stained with tetramers. For production of tetramers, HIS-tagged DR protein monomers were produced by Hi5 insect cells and purified using Ni-NTA (Qiagen) and size exclusion columns (AKTA, GE Healthcare). Protein biotinylation, peptide exchange, and tetramerization were performed using standard protocols as previously described. (Day et al., 2003; Su et al., 2013). See Table S2 for peptide sequences used for generating the tetramer for direct ex vivo analyses.
Direct ex vivo T cell analyses and cell sorting
Precursor analyses were performed using 30 to 100 million CD3+ or CD4+ enriched T cells. For post-vaccination analyses, 10 million PBMCs (up to 1 month after vaccination) or 10 to 30 million CD3+ or CD4+ enriched T cells (> 7 months) were used. Tetramer staining was carried out as previously described (Su et al., 2013). In brief, cells were stained at room temperature for 1 hour using 5ug of each tetramer in 50ul reaction. Up to 4 tetramers were combined in the same reaction. Tetramer tagged cells were enriched by adding anti-PE and/or anti-APC magnetic beads and passing the mixture through a magnetized column (Miltenyi). The tetramer-enriched samples were further stained with live/dead dyes, exclusion markers (anti-CD19 and anti-CD11b, BioLegend), and surface markers (anti-CD3, anti-CD4, anti-CD45RO and anti-CCR7, BioLegend) for 30 minutes at 4°C. Additional surface antibodies were used to mark T cell activation (anti-CD38 and anti-ICOS, BioLegend) or Treg phenotype (anti-CD25 and anti-CD127, BioLegend) depending on the experiment. For Foxp3 staining, cells were fixed, permeabilized, and stained with anti-Foxp3 antibody (ThermoFisher) using eBioscience Foxp3/Transcription Factor Staining Buffer Set according to manufacture instruction (ThermoFisher). Samples were acquired by flow cytometry using LSRII (BD) or sorted on FACS Aria (BD). Frequency calculation was obtained by mixing 1/10th of sample with 200,000 fluorescent beads (Spherotech) for normalization (Su et al., 2013). Data analyses were performed using FlowJo (BD).
T cell stimulation and proliferation assays
Cell proliferation was performed by labeling CD4+ T cells with 2uM CellTrace Violet (CTV, ThermoFisher) for 5 minutes at room temperature. After CTV labeling, cells were stained with tetramers, surface markers, enriched and sorted as above for 0407-YF50 tetramer+ cells and CD45RO+CD4+ T cells (400-500 cells/well). T cells were cultured with DMSO, PHA-M (1:100, ThermoFisher), or YF50 peptide (50ug/ml) together with 100,000 peripheral monocyte derived dendritic cells per well. CTV staining was analyzed after 5 days in culture. For cytokine analyses, tetramer enriched cells were collected in a 96-well plate and stimulated with phorbol myristate acetate (PMA, 5ng/mL, Sigma) and ionomycin (500ng/mL, Sigma) in the presence monensin (2uM, Sigma) and Brefeldin A (5ug/mL, Sigma) for 5-hours as previously described (Del Alcazar et al., 2019; Wendel et al., 2018). After stimulation, cells were restained with tetramers, followed by intracellular cytokine staining with anti-TNF-a, anti-IL-2, and anti-IFN-γ antibodies (BioLegend) using BD Cytofix/Cytoperm Fixation/Permeabilization Kit according to manufacturer protocol (BD).
Single-cell TCR sequencing and analyses
Single cell TCR Sequencing by nested PCRs was performed using the primer sets and the protocol as previously described in Han et al. (Han et al., 2014). In brief, reverse transcription was performed with CellsDirect One-Step qRT-PCR kit according to the manufacturer’s instructions (CellsDirect, Invitrogen) using a pool of 5’ TRVB-region specific primers and 3’ C-region primers. The cDNA library was amplified using a second set of multiple internally nested V-region and C-region primers with HotStarTaq DNA polymerase kit (Qiagen). The final PCR reaction was performed on an aliquot of the second reaction using a primer containing common base sequence and a third internally nested Cβ primer. PCR products were gel purified (Qiagen) and sequenced on NextSeq 500/550 (Illumina) by 300 cycle pair-end reaction. Output data (.bcl files) were converted to fastq format using bcl2fastq software. Reads 1 and read 2 were converted into one paired end read using pandaseq (Masella et al., 2012). Data were demultiplexed by the unique combinations of plate, row, and column nucleotide barcodes. Consensus TCRβ sequences were identified using the V(D)J alignment software MiXCR (Bolotin et al., 2015). Successfully aligned sequences were first thresholded by a read count of 200 reads per sequence. In wells containing multiple TCRβ chains above this cutoff, dual chains (two productive chains in a single cell) were called when both chains were separated by a read count ratio of 0.5 or more. In cases for which more than 2 chains passed this criterion or when a TCR sequence was not unique to one tetramer, data were discarded as ambiguous. For downstream analyses, data wrangling was perform using the tidyverse package. TCRs common to pre- and post-vaccine visits were identified using the most abundant TCRβ sequence if more than one sequence was found at a given position. Circos plots were made using the circlize package of R software (Gu et al., 2014).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data transformation was performed using logarithmic function or hyperbolic sine transformation () when data contained zero values. Assessment of normality was performed using D’Agostino-Pearson test. Spearman was used if either of the two variables being correlated was non-normal. Otherwise, Pearson was used to measure the degree of association. The best-fitting line was calculated using least squares fit regression. Statistical comparisons were performed using two-tailed Student’s t-test or Wilcoxon signed-rank test, using a p-value of <0.05 as the significance level. Multiple-way comparisons were performed using ANOVA or mixed effect model and corrected for multiple comparisons. Welch’s correction was applied for unequal variance. Statistical analyses were performed using GraphPad Prism. Lines and bars represent mean and variability is represented by standard error of the mean (SEM). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Supplementary Material
YFV-specific TCR sequences (relates to Figure 6).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD45RO | Biolegend | Cat# 304238; RRID:AB_2562153 or Cat# 304233; RRID:AB_11219394 |
| Mouse anti-human CD197 (CCR7) | Biolegend | Cat# 353234; RRID:AB_2563867 or Cat# 353208; RRID:AB_11203894 |
| Mouse anti-human CD4 | Biolegend | Cat# 344634; RRID:AB_2566017 or Cat# 317408; RRID:AB_571951 |
| Mouse anti-human CD3 | Biolegend | Cat# 300472; RRID:AB_2687178 |
| Armenian hamster anti-human/mouse/rat CD278 (ICOS) | Biolegend | Cat# 313528; RRID:AB_2566126 |
| Mouse anti-human CD38 | Biolegend | Cat# 303508; RRID:AB_314360 |
| Mouse anti-human CD11b | Biolegend | Cat# 301342; RRID:AB_2563395 or Cat# 301308; RRID:AB_314160 |
| Mouse anti-human CD19 | Biolegend | Cat# 302210; RRID:AB_314240 or Cat# 302218; RRID:AB_314248 |
| Mouse anti-human CD25 antibody | Biolegend | Cat# 302630; RRID:AB_11126749 |
| Mouse anti-human CD127 antibody | Biolegend | Cat# 351322; RRID:AB_10897104 |
| Mouse anti-human IFN-γ antibody | Biolegend | Cat# 502506; RRID:AB_315231 |
| Mouse anti-human TNF-α antibody | Biolegend | Cat# 502928; RRID:AB_2561315 |
| Rat anti-hunab IL-2 antibody | Biolegend | Cat# 500338; RRID:AB_2562632 |
| Rat anti-human Foxp3 antibody | ThermoFisher | Cat# 56-4776-41; RRID:AB_1582210 |
| Biotin-conjugated 4G2 monoclonal antibody | ATCC | Cat# HB-112; RRID:CVCL_J890 |
| Bacterial and Virus Strains | ||
| YFV-17D | BEI Resources | NR-115 |
| Biological Samples | ||
| Healthy Adult Human Peripheral Blood Sample | Hospital of the University of Pennsylvania | https://www.pennmedicine.org/ |
| Pre-Immune Yellow Fever Virus Serum | BEI Resources | Cat# NR-42565 |
| Early-Immune Yellow Fever Virus Antiserum (Polyclonal Antiserum) | BEI Resources | Cat# NR-29337 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LIVE/DEAD™ Fixable Dead Cell Stain Kits | Invitrogen | Cat# L34966 or Cat# L34976 |
| CellTrace™ Violet Cell Proliferation Kit | ThermoFisher | Cat# C34557 |
| Recombinant Human IL-2 | Peprotech | Cat# 200-02 |
| Recombinant Human IL-7 | Peprotech | Cat# 200-07 |
| Recombinant Human IL-15 | Peprotech | Cat# 200-15 |
| Recombinant Human GM-CSF (carrier-free) | Biolegend | Cat# 572904 |
| Recombinant Human IL-4 (carrier-free) | Biolegend | Cat# 574006 |
| YF23 (NS3 313 - 327); Seq: ESATILMTATPPGTS | This paper | N/A |
| YF25 (NS3 448 - 462); Seq: KGPLRISASSAAQRR | This paper | N/A |
| YF31 (NS4b 37 - 51); Seq: GAAWTVYVGIVTMLS | This paper | N/A |
| YF38 (NS5 398 - 412); Seq: EEFIAKVRSHAAIGA | This paper | N/A |
| YF42 (NS5 756 - 770); Seq: ACLSKAYANMWSLMY | This paper | N/A |
| YF44 (NS5 867 - 881); Seq: IHLVIHRIRTLIGQE | This paper | N/A |
| YF45 (NS3 147 - 161); Seq: GEVIGLYGNGILVGD | This paper | N/A |
| YF48 (EE 43 - 59); Seq: ISLETVAIDRPAEVRKV | This paper | N/A |
| YF50 (EE 241 - 257); Seq: TIRVLALGNQEGSLKTA | This paper | N/A |
| YF51 (EE 301 - 317); Seq: TDKMFFVKNPTDTGHGT | This paper | N/A |
| YF53 (NS3 151 - 167); Seq: GLYGNGILVGDNSFVSA | This paper | N/A |
| YF54 (NS3 367 - 383); Seq: LPSIRAANVMAASLRKA | This paper | N/A |
| YF57 (NS5 7 - 23); Seq: TLGEVWKRELNLLDKRQ | This paper | N/A |
| YF69 (NS3 355 - 371); Seq: WILADKRPTAWFLPSIR | This paper | N/A |
| YF80 (EE 313 - 329); Seq: TGHGTVVMQVKVSKGAP | This paper | N/A |
| YF81 (NS3 283 - 299); Seq: WEVIIMDEAHFLDPASI | This paper | N/A |
| YF91 (NS5 367 - 383); Seq: PPAGTRKIMKVVNRWLF | This paper | N/A |
| YF93 (EE 457 - 473); Seq: MGAVLIWVGINTRNMTM | This paper | N/A |
| YF108 (NS5 325 - 341); Seq: VIKILTYPWDRIEEVTR | This paper | N/A |
| YF111 (NS5 619 - 635); Seq: VQLIRMAEAEMVIHHQH | This paper | N/A |
| HA306; Seq: GGPKYVKQNTLKLAT | This paper | N/A |
| CLIP103; Seq: CGGGPVSKMRMATPLLMQA | This paper | N/A |
| TT511; Seq: CGGKIIVDYNLQSK | This paper | N/A |
| Ni-NTA Agarose | QIAGEN | Cat# 30230 |
| Methyl Cellulose | Sigma-Aldrich | Cat# M0512-250G |
| Amphotericin B | Gibco | Cat# 15290-018 |
| TrueBlue peroxidase substrate | KPL | Cat# 5510-0030 |
| AccuCount Fluorescent Particles | Spherotech | Cat# ACFP-50-5 |
| Horseradish peroxidase (HRP)-conjugated streptavidin | BD Biosciences | Cat# 554066 |
| HRP | MP Biomedicals | Cat# ICN55563 |
| BSA | Sigma-Aldrich | Cat# A8022-500G |
| PFA | Electron Microscopy Sciences | Cat# 15714-S |
| Triton-X | Roche | Cat# 10789704001 |
| TBS 10X | Thermo Scientific Pierce | Cat# 37520 |
| Tween | Fisher Bioreagent | Cat# BP337100 |
| HEPES | Corning | Cat# 25-060-Cl |
| L-glut | Mediatech | Cat# MT25-0050CI |
| NaHCO3 | Sigma-Aldrich | Cat# S4019-500G |
| Phorbol myristate acetate (PMA) | Sigma-Aldrich | Cat# P8139-5MG |
| Ionomycin calcium salt from Streptomyces conglobatus | Sigma-Aldrich | Cat# I0634-5MG |
| Brefeldin A | Sigma-Aldrich | Cat# B7651-5MG |
| Monensin | Biolegend | Cat# 420701 |
| Lipopolysaccharides | Sigma-Aldrich | Cat# L4391 |
| ESF 921 Insect Cell Culture Medium | Expression System | Cat# 96-001-01 |
| Critical Commercial Assays | ||
| RosetteSep™ Human Monocyte Enrichment Cocktail | STEMCELL | Cat# 15068 |
| RosetteSep™ Human CD4+ T Cell Enrichment Cocktail | STEMCELL | Cat# 15062 |
| RosetteSep™ Human T Cell Enrichment Cocktail | STEMCELL | Cat# 15061 |
| HotStarTaq DNA polymerase kit | Qiagen | Cat# 203205 |
| NextSeq 500/550 Mid Output Kit v2.5 (300 Cycles) | Illumina | Cat# 20024905 |
| CellsDirect™ One-Step qRT-PCR Kit | Invitrogen™ | Cat# 11753-500 |
| MinElute Gel Extraction Kit | Qiagen | Cat# 28604 |
| BD Cytofix/Cytoperm™ Fixation/Permeablization Kit | BDBioscience | Cat# 554714 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | ThermoFisher | Cat# 00-5523-00 |
| Deposited Data | ||
| Yellow fever virus-specific TCR sequencing data | This paper | Mendeley Data, V1, doi:10.17632/75xhmfrpm6.1 |
| Experimental Models: Cell Lines | ||
| Hi5 insect cells | ThermoFisher | Cat# B85502 |
| Vero cells | ATCC | CCL-81 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| FlowJo (Version: 10.6.0) | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism 6 | GraphPad | https://www.graphpad.com/ |
| R Version 3.6.3 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| RStudio Version 1.3.959 | R studio | https://www.rstudio.com/ |
| Bcl2fastq | Illumina | https://support.illumina.com/ |
| Pandaseq | Masella, A., et al. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC bioinformatics. DOI: 10.1186/1471-2105-13-31 | https://github.com/neufeld/pandaseq |
| MiXCR | Bolotin, D., et al. (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nature methods. DOI: 10.1186/1471-2105-13-31 | https://github.com/milaboratory/mixcr |
| Circlize Version:0.4.11 | Gu, Z. (2014) circlize implements and enhances circular visualization in R. Bioinformatics. DOI:10.1093/bioinformatics/btu393 | https://CRAN.R-project.org/package=circlize |
| Other | ||
| LS Column | Miltenyi Biotec | Cat# 130-042-401 |
| Superdex® 200 Increase 10/300 GL | Sigma | Cat# GE28-9909-44 |
| Graphical abstract | This paper | BioRender |
Highlights.
Class II tetramer enrichment identifies YFV-reactive T cells in unexposed adults
CD4+ T cell precursor states predict post-vaccine dynamics
Single-cell TCR sequencing reveals reorganization of clonal hierarchy by vaccination
Repertoire diversity facilitates robust peripheral T cell selection
Acknowledgments:
We thank Lea Williams, Tammy Ruozhang Xu, and Ning Jiang for helpful discussions. We thank our study subjects for their participation.
Funding:
NIH R01AI134879 (L.F.S), VA Merit Award IMMA-020-15F (L.F.S), National Cancer Institute P30-CA14051 (M.E.B), and Schmidt Futures, the V Foundation (M.E.B).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: None
References
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, et al. (2009). The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol 183, 7919–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P, Rosati E, Esser D, Martini GR, Saggau C, Schiminsky E, Dargvainiene J, Schroder I, Wieters I, Khodamoradi Y, et al. (2020). Low-Avidity CD4(+) T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans with Severe COVID-19. Immunity 53, 1258–1271 e1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Haring JS, and Harty JT (2007). Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity 26, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 12, 380–381. [DOI] [PubMed] [Google Scholar]
- Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, et al. (2020). SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587, 270–274. [DOI] [PubMed] [Google Scholar]
- Busch DH, and Pamer EG (1999). T cell affinity maturation by selective expansion during infection. J Exp Med 189, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, et al. (2003). Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest 112, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo AB, Nascimento EJ, Braga-Neto U, Dhalia R, Silva AM, Oelke M, Schneck JP, Sidney J, Sette A, Montenegro SM, and Marques ET (2013). T-cell memory responses elicited by yellow fever vaccine are targeted to overlapping epitopes containing multiple HLA-I and -II binding motifs. PLoS neglected tropical diseases 7, e1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Alcazar D, Wang Y, He C, Wendel BS, Del Rio-Estrada PM, Lin J, Ablanedo-Terrazas Y, Malone MJ, Hernandez SM, Frank I, et al. (2019). Mapping the Lineage Relationship between CXCR5(+) and CXCR5(−) CD4(+) T Cells in HIV-Infected Human Lymph Nodes. Cell reports 28, 3047–3060 e3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, et al. (2020). Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501 e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, and Brors B (2014). circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. [DOI] [PubMed] [Google Scholar]
- Haluszczak C, Akue AD, Hamilton SE, Johnson LD, Pujanauski L, Teodorovic L, Jameson SC, and Kedl RM (2009). The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med 206, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Glanville J, Hansmann L, and Davis MM (2014). Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol 32, 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hataye J, Moon JJ, Khoruts A, Reilly C, and Jenkins MK (2006). Naive and memory CD4+ T cell survival controlled by clonal abundance. Science 312, 114–116. [DOI] [PubMed] [Google Scholar]
- Hutloff A, Dittrich AM, Beier KC, Eljaschewitsch B, Kraft R, Anagnostopoulos I, and Kroczek RA (1999). ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature 397, 263–266. [DOI] [PubMed] [Google Scholar]
- James EA, LaFond RE, Gates TJ, Mai DT, Malhotra U, and Kwok WW (2013). Yellow fever vaccination elicits broad functional CD4+ T cell responses that recognize structural and nonstructural proteins. J Virol 87, 12794–12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathryn AF, Jason MS, Stephen CJ, Vaiva V, and David M (2013). Preexisting high frequencies of memory CD8+ T cells favor rapid memory differentiation and preservation of proliferative potential upon boosting. Immunity 39, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe T, Jankovic D, Kawabe S, Huang Y, Lee PH, Yamane H, Zhu J, Sher A, Germain RN, and Paul WE (2017). Memory-phenotype CD4(+) T cells spontaneously generated under steady-state conditions exert innate TH1-like effector function. Sci Immunol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, and Marrack P (2000). T cells compete for access to antigen-bearing antigen-presenting cells. J Exp Med 192, 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok WW, Tan V, Gillette L, Littell CT, Soltis MA, LaFond RB, Yang J, James EA, and DeLong JH (2012). Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J Immunol 188, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, et al. (2020). SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. [DOI] [PubMed] [Google Scholar]
- Malherbe L, Hausl C, Teyton L, and McHeyzer-Williams MG (2004). Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity 21, 669–679. [DOI] [PubMed] [Google Scholar]
- Martin JR, Jeffrey CN, and John TH (2012a). Pathogen-Specific Inflammatory Milieux Tune the Antigen Sensitivity of CD8(+) T Cells by Enhancing T Cell Receptor Signaling. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MD, Condotta SA, Harty JT, and Badovinac VP (2012b). Population dynamics of naive and memory CD8 T cell responses after antigen stimulations in vivo. J Immunol 188 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, and Neufeld JD (2012). PANDAseq: paired-end assembler for illumina sequences. BMC bioinformatics 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, et al. (2020). Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley N, and Mellman I (2010). Monocyte-derived dendritic cells exhibit increased levels of lysosomal proteolysis as compared to other human dendritic cell populations. PLoS One 5, e11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhop-Williams ER, and Bevan MJ (2014). Memory CD8+ T cells exhibit increased antigen threshold requirements for recall proliferation. J Exp Med 211, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, and Nikolich-Zugich J (2002). Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science 298, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, and Jenkins MK (2007). Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lubke M, Bauer J, Rieth J, Wacker M, et al. (2020). SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. [DOI] [PubMed] [Google Scholar]
- Nelson RW, Beisang D, Tubo NJ, Dileepan T, Wiesner DL, Nielsen K, Wuthrich M, Klein BS, Kotov DI, Spanier JA, et al. (2015). T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 42, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar JJ, Khanna KM, and Lefrancois L (2008). Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28, 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Lindestam Arlehamn CS, Scriba TJ, Dillon MB, Oseroff C, Hinz D, McKinney DM, Carrasco Pro S, Sidney J, Peters B, and Sette A (2015). Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods 422, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, et al. (2020). Broad and strong memory CD4 (+) and CD8 (+) T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlgren M, Dubois PM, Tomkowiak M, Sjogren T, and Marvel J (1996). Resting memory CD8+ T cells are hyperreactive to antigenic challenge in vitro. J Exp Med 184, 2141–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PR, Dubey C, and Swain SL (2000). Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J Immunol 164, 2338–2346. [DOI] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, et al. (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028 e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Wikstrom ME, and Fazekas de St Groth B (2000). Visualizing T cell competition for peptide/MHC complexes: a specific mechanism to minimize the effect of precursor frequency. Immunity 13, 783–794. [DOI] [PubMed] [Google Scholar]
- Stock AT, Jones CM, Heath WR, and Carbone FR (2006). Cutting edge: central memory T cells do not show accelerated proliferation or tissue infiltration in response to localized herpes simplex virus-1 infection. J Immunol 177, 1411–1415. [DOI] [PubMed] [Google Scholar]
- Su LF, Del Alcazar D, Stelekati E, Wherry EJ, and Davis MM (2016). Antigen exposure shapes the ratio between antigen-specific Tregs and conventional T cells in human peripheral blood. Proc Natl Acad Sci U S A 113, E6192–E6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, and Davis MM (2013). Virus-Specific CD4(+) Memory-Phenotype T Cells Are Abundant in Unexposed Adults. Immunity 38, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Fernandes H, Walter U, Bourgeois C, McLean A, and Rocha B (2000). Response of naive and memory CD8+ T cells to antigen stimulation in vivo. Nat Immunol 1, 47–53. [DOI] [PubMed] [Google Scholar]
- Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, et al. (2020). Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel BS, Del Alcazar D, He C, Del Rio-Estrada PM, Aiamkitsumrit B, Ablanedo-Terrazas Y, Hernandez SM, Ma KY, Betts MR, Pulido L, et al. (2018). The receptor repertoire and functional profile of follicular T cells in HIV-infected lymph nodes. Sci Immunol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MA, Ravkov EV, and Bevan MJ (2008). Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity 28, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Lee SY, and Bevan MJ (2009). Complete but curtailed T-cell response to very low-affinity antigen. Nature 458, 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C, Prevost-Blondel A, Blaser C, and Pircher H (1999). Kinetics of the response of naive and memory CD8 T cells to antigen: similarities and differences. Eur J Immunol 29, 284–290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
YFV-specific TCR sequences (relates to Figure 6).
Data Availability Statement
Data were analyzed uses existing computational packages. The datasets generated during this study are available at Mendeley Data (Bartolo, Laurent; Pan, Yi-Gen (2021), “YF TCR data”, doi: 10.17632/75xhmfrpm6.1)