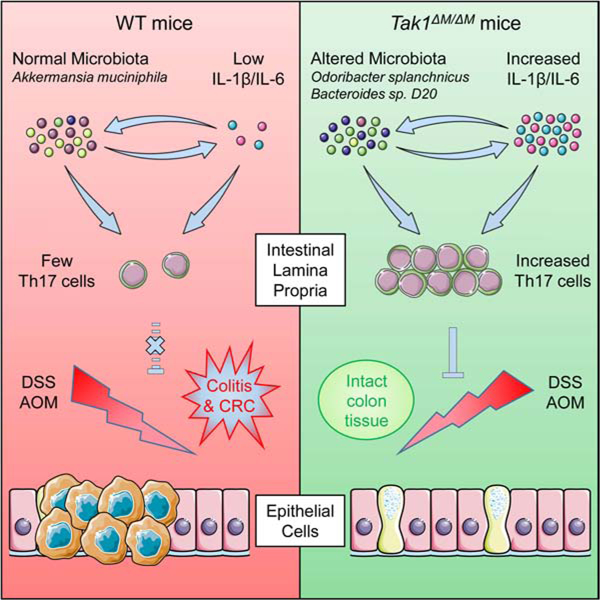
SUMMARY
Microbiota plays critical roles in regulating colitis and colorectal cancer (CRC). However, it is unclear how the microbiota generates protective immunity against these disease states. Here, we find that loss of the innate and adaptive immune signaling molecule TAK1 in myeloid cells (Tak1∆M/∆M) yields complete resistance to chemical-induced colitis and CRC through microbiome alterations that drive protective immunity. Tak1∆M/∆M mice exhibit an altered microbiota that is critical for resistance, with antibiotic-mediated disruption ablating protection and Tak1∆M/∆M microbiota transfer conferring protection against colitis or CRC. The altered microbiota of Tak1∆M/∆M mice promotes IL-1β and IL-6 signaling pathways, which are required for induction of protective intestinal Th17 cells and resistance. Specifically, Odoribacter splanchnicus is abundant in Tak1∆M/∆M mice and sufficient to induce intestinal Th17 cell development, and confer resistance against colitis and CRC in wild-type mice. These findings identify specific microbiota strains and immune mechanisms that protect against colitis and CRC.
Keywords: TAK1 signaling, acute colitis, colon cancer, microbiota, innate immunity, Th17 cells, Odoribacter splanchnicus, Bacteroides sp. D20
Graphical Abstract
eTOC blurb
Xing et al. report that myeloid-specific Tak1-deficient mice are completely resistant to colitis and colon cancer, and show that the microbiota of these mice has the ability to induce intestinal Th17 cells and confer protective immunity. Additionally, the work identifies Odoribacter splanchnicus as a single protective strain.
INTRODUCTION
Colorectal cancer (CRC) is the third leading cause of cancer-related death in the United States (Markowitz and Bertagnolli, 2009; Tenesa and Dunlop, 2009), and colonic inflammation is a major risk factor for CRC development. Individuals diagnosed with ulcerative colitis (UC), one of the most common types of inflammatory bowel diseases (IBDs), have increased risk of developing CRC (Jess et al., 2012; Xavier and Podolsky, 2007). Although innate immune cells, cytokines, and microbiota components contribute to colitis and CRC in a context-dependent manner (Neurath, 2014; West et al., 2015), the precise mechanisms remain unclear.
Microbiota have been identified as an important regulator of immune cell activation, inflammation, and cancer development through nuclear factor kappa B (NF-κB), type I interferon, and inflammasome activation (Alexander et al., 2014; Rooks and Garrett, 2016). Local immune system interacts with gut microbiota to control the immune responses, tissue damage, and cancer development (Geva-Zatorsky et al., 2017; Rosshart et al., 2017; Surana and Kasper, 2017; Wlodarska et al., 2017). Metagenomic profiling has identified multiple prominent CRC-associated bacteria enriched in patients (Ternes et al., 2020). As most studies link microbiota composition (a mixed population) to the disease progression, some species are reported to promote colitis and CRC (Geva-Zatorsky et al., 2017; Rosshart et al., 2017; Surana and Kasper, 2017; Wlodarska et al., 2017); however, only a few bacterial strains were identified as immune modulators that induce potent protective immunity.
Proinflammatory cytokines, such as interleukin (IL)-6 and IL-1β, can contribute to inflammation-associated cancer (Elinav et al., 2013; Hunter and Jones, 2015; Lopetuso et al., 2013; Neurath, 2014; West et al., 2015). IL-6 promotes colitis-associated CRC by activating the STAT3 pathway in colonic epithelial cells (Bollrath et al., 2009; Grivennikov et al., 2009; Wang et al., 2014b), and IL-1β facilitates intestinal inflammatory pathology and gastric cancer (Coccia et al., 2012; Tu et al., 2008). In addition to the key functions in normal and neoplastic epithelial cells, IL-1β and IL-6, in combination with TGF-β and IL-23, are required for the development of T helper (Th) 17 cells (Artis and Spits, 2015; Dong, 2008; Korn et al., 2009; Littman and Rudensky, 2010; McGeachy and Cua, 2008; Ouyang et al., 2008; Weaver and Hatton, 2009). Th17 cells are constitutively present in the intestinal lamina propria (LP) due to the activation by microbial flora, especially segmented filamentous bacteria (SFB) (Atarashi et al., 2015; Ivanov et al., 2009; Sano et al., 2015), and play a critical role in the autoimmune diseases and host defense against bacteria and fungi (Dong, 2008; Korn et al., 2009). A recent study suggested that colonic Th17 cells may have immune-suppressive function (Esplugues et al., 2011).
Transforming growth factor-β-activated kinase-1 (TAK1), also known as mitogen-activated protein kinase kinase kinase 7 (encoded by MAP3K7), is an essential component of innate and adaptive immune signaling with cell-type-dependent function (Ajibade et al., 2013). Although TAK1 is a positive regulator in T cells (Wan et al., 2006), myeloid-specific Tak1-deficient (Tak1∆M/∆M) mice produce large amounts of IL-1β, IL-6, and TNF-α, and rapidly die during lipopolysaccharide (LPS)-induced septic shock, suggesting a negative regulator function (Ajibade et al., 2012). However, the role of TAK1 deficiency in microbiota, inflammation, and cancer development remains obscure.
In this study, we report that Tak1∆M/∆M mice are completely resistant to dextran sulfate sodium (DSS)-induced colitis and azoxymethane (AOM)/DSS-induced CRC. The resistance of Tak1∆M/∆M mice relies on altered composition of gut microbiota and innate immune signaling (IL-1β and IL-6), due to their roles in activating the expansion of protective Th17 cells. With metagenomic sequencing to dissect the key microbial candidates, we identify Odoribacter splanchnicus as a single bacterial strain that induces Th17 cells and protects mice against colitis and CRC. Overall, our findings provide important insights into mechanisms by which microbiota interact with innate immune signaling to control protective immunity against cancer.
RESULTS
Tak1 Deficiency in Myeloid Lineage Renders Mice Resistance to DSS-induced Colitis and Colon Cancer
To determine whether key innate immune pathways are involved in inflammation and associated cancer, we generated mice with myeloid-specific deletion of Tak1, Mekk3, Myd88, or P38 (by Lyz2-Cre) and challenged them with 5% DSS to induce acute colitis. Tak1flox/flox;Lyz2-Cre+/+ (Tak1∆M/∆M) mice were completely resistant to DSS treatment without great loss of body weight (Figures 1A and 1B). In contrast, WT, Mekk3∆M/∆M, Myd88∆M/∆M, and P38∆M/∆M mice remained sensitive to DSS treatment, died within 9 days, and manifested 30% loss of body weight (Figure S1A). Importantly, like WT mice, the littermate controls (Tak1Flox/Flox;Lyz2-Cre−, or Tak1F/F) were also completely sensitive to DSS treatment, compared to Tak1∆M/∆M mice (Figure S1B). Consistently, DSS-treated Tak1∆M/∆M mice did not manifest diarrhea or hematochezia, and had longer colons, compared to WT mice with high clinical scores (Figure 1C). H&E staining further revealed lack of damage or injury to colonic epithelial tissues (including proximal, middle, and distal regions) in Tak1∆M/∆M mice, with significantly lower pathology scores than WT mice (Figures 1D, S1C, and S1D). Similar alteration on histopathology was observed between littermate controls (Tak1F/F) and Tak1∆M/∆M mice (Figures S1E and S1F). For epithelial barrier integrity, we examined mucin-producing goblet cells and tight junction proteins [Claudin-3 (CLDN3), Zonulin, and Occludin] in colonic tissues, and found them unchanged in DSS-treated Tak1∆M/∆M mice, but markedly lost in WT mice (Figures 1D and S1G). These data suggest that TAK1 deficiency in myeloid cells confers the resistance to colitis with maintenance of the integrity of epithelial tissues and tight junction barrier.
Figure 1. Tak1 Deficiency in Myeloid Lineage Renders Mice Resistance to DSS-induced Colitis and Colon Cancer.
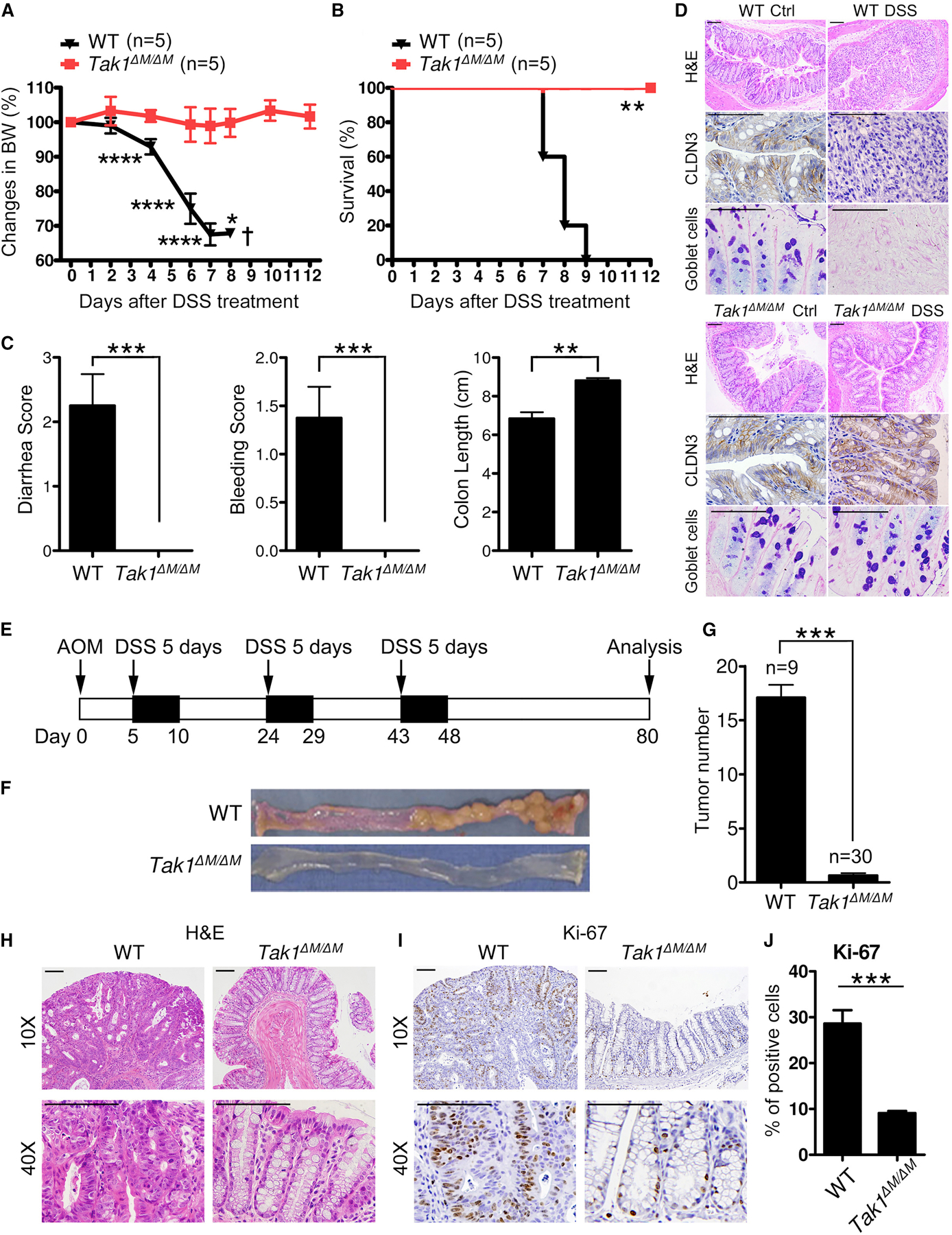
(A-C) Body weight changes, survival, and clinical scores (diarrhea, bleeding, colon length) in WT and Tak1∆M/∆M mice after 5% DSS treatment.
(D) H&E staining (10X), IHC staining of CLDN3 (40X), and AB/PAS staining (40X) on colon sections (day 5 after DSS treatment, scale bar: 100 µm).
(E) Strategy for AOM/DSS-induced colon cancer model.
(F-G) Colon tumors and tumor number analysis in WT and Tak1∆M/∆M mice on day 80 after AOM/DSS treatment.
(H-J) H&E and IHC staining of Ki-67 on colon sections (day 80 after AOM/DSS treatment, scale bar: 100 µm), and the analysis of Ki-67 positive cells.
Statistical analyses: Student’s unpaired t test (A, C, G, and J) and Mantel-Cox log-rank test (B). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
See also Figure S1.
Next, we tested Tak1∆M/∆M mice in AOM/DSS-induced CRC (Figure 1E). As expected, numerous tumors were developed in the middle and distal regions of colon in WT mice; in contrast, Tak1∆M/∆M mice manifested no or a few (1–2 only) colonic tumors (Figures 1F and 1G). Similar results were obtained between littermate controls (Tak1F/F) and Tak1∆M/∆M mice (Figures S1H and S1I). Colonic crypts in WT mice are entirely filled with tumor cells, which formed large tumor bulks; whereas Tak1∆M/∆M mice manifested little or no disruption of the colonic epithelial structures, with a significantly lower number of proliferating colonic epithelial cells (Figures 1H–1J). These results indicate that Tak1∆M/∆M mice are resistant to colitis-associated CRC.
Microbiota in Tak1∆M/∆M Mice Are Altered and Critically Required for Resistance to Colonic Inflammation
Commensal microbiota have been implicated to modulate colitis, CRC, and checkpoint blockade immunotherapy (Sears and Pardoll, 2018; Sivan et al., 2015; Spranger et al., 2017; Vetizou et al., 2015; Wang and Wang, 2017). In DSS colitis model, depletion of microbiota exacerbated tissue damage and shortened survival (Rakoff-Nahoum et al., 2004). To determine whether microbiota contributes to the colitis-resistance in Tak1∆M/∆M mice, a cocktail of antibiotics (Abx) was used to remove gut microbiota, and successful elimination of commensal bacteria was determined by the loss of colonies and enlarged cecum (Figures S2A and S2B). Abx-treated Tak1∆M/∆M mice lost their resistance to DSS-induced colitis, experienced considerable loss of body weight, and died within 10 days, consistent with damaged epithelial structure of colonic tissues, compared to control Tak1∆M/∆M mice (Figures 2A and 2B).
Figure 2. Microbiota in Tak1∆M/∆M Mice Are Altered and Critically Required for Resistance to Colonic Inflammation.
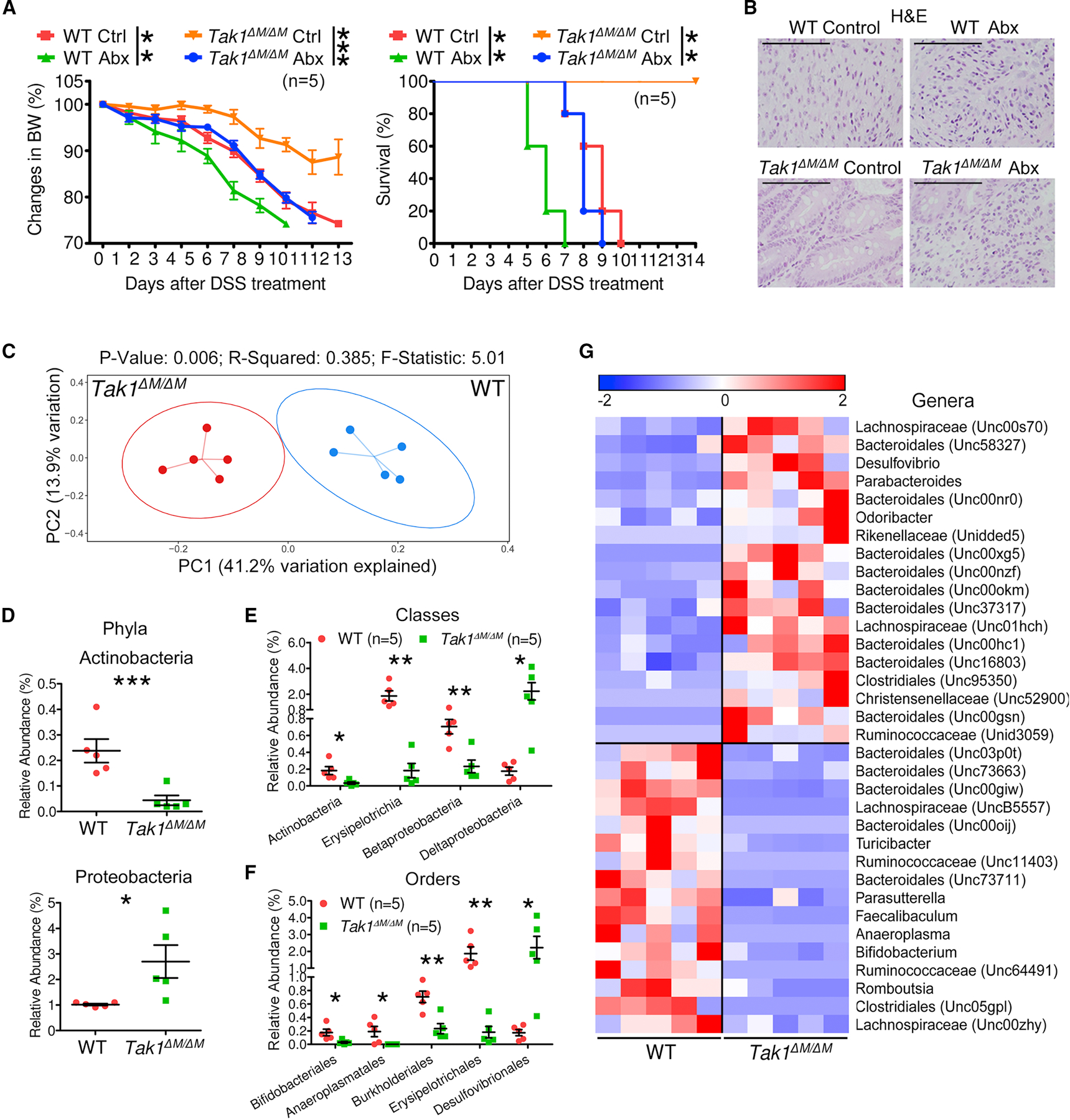
(A-B) Body weight changes, survival, and tissue damage in WT and Tak1∆M/∆M mice with combined Abx pretreatment for 4 weeks and 5% DSS for 5 days.
(C-G) The 16S rRNA gene profiling data for fecal microbiome from 8 weeks old WT and Tak1∆M/∆M mice. (C) PCoA plot of bacterial beta-diversity. (D-G) Relative abundance of significantly altered taxa at the rank of phylum, class, order, and genus (including unspecified taxa).
Statistical analyses: Student’s unpaired t test (A-left, D-F), Monte Carlo permutation test (C), and Mantel-Cox log-rank test (A-right). *p<0.05; **p<0.01; ***p<0.001.
To evaluate the microbiota composition in Tak1∆M/∆M mice, fecal samples were collected for purification of the bacterial genomic DNA (gDNA) and 16S rRNA sequencing of the amplified V4 region. The principal coordinate analysis (PCoA) plot of bacterial beta-diversity showed a significant shift of microbiota composition between WT and Tak1∆M/∆M mice (Figure 2C). Comparative analyses were performed to identify the taxa with significantly altered relative abundance at various ranks. Although the major phyla (such as Bacteroidetes and Firmicutes) did not change significantly, a decrease of Actinobacteria and an increase of Proteobacteria were detected (Figure 2D). Additional subpopulations were identified at the class and order levels (Figures 2E and 2F). Specifically, we observed 18 increased and 16 decreased genera in Tak1∆M/∆M mice, as shown in the heatmap plot (Figure 2G). Similar changes were found between littermate controls (Tak1F/F) and Tak1∆M/∆M mice. In particular, some of the altered genera in littermates were the same as observed in WT mice (Figures S2C–S2E). Despite some alterations in microbiota in Tak1∆M/∆M mice after DSS treatment, Odoribacter remained at a high level (Figures S2F–S2H). These results suggest that altered microbiota composition in Tak1∆M/∆M mice is required for resistance to DSS-induced colitis.
Specific Gut Microbiota in Tak1∆M/∆M Mice Can Generate Protective Immunity in WT Mice against Colitis and Colon Cancer
To determine the functional importance of specific microbiota composition in Tak1∆M/∆M mice, we cohoused WT and Tak1∆M/∆M mice and measured their microbiota by 16S rRNA sequencing. PCoA plot showed that the cohoused WT and Tak1∆M/∆M mice had a unified pattern of microbiota composition, which was intermediate but significantly differed from those of single-housed controls (Figure 3A). Comparative analyses detected intermediate microbial distribution of taxa at the phylum, order, and genus levels in the cohoused mice, whereas significantly varied relative abundances were observed in single-housed WT and Tak1∆M/∆M mice (Figures 3B, S3A and S3B). After AOM/DSS treatment, cohoused WT and Tak1∆M/∆M mice developed intermediate number of colonic tumors, compared to single-housed controls (Figures 3C and 3D). Consistently, we observed intermediate levels of tumor cell infiltration in colonic crypts and epithelial cell proliferation in cohoused WT and Tak1∆M/∆M mice, compared with single-housed controls (Figures S3C and S3D). After 5% DSS treatment, cohoused WT mice had slightly ameliorated survival compared with control WT mice; in contrast, cohoused Tak1∆M/∆M mice became partially sensitive to acute colitis, with significantly decreased survival compared to control Tak1∆M/∆M mice (Figure 3E). Immunostaining of CLDN3, Zonulin, and Occludin further showed intermediate levels of colonic tissue damage and barrier integrity in cohoused WT and Tak1∆M/∆M mice (Figures S3E and S3F), suggesting that specific microbiota composition is critical for the prevention of colitis and CRC in Tak1∆M/∆M mice.
Figure 3. Specific Gut Microbiota in Tak1∆M/∆M Mice Can Generate Protective Immunity in WT Mice against Colitis and Colon Cancer.
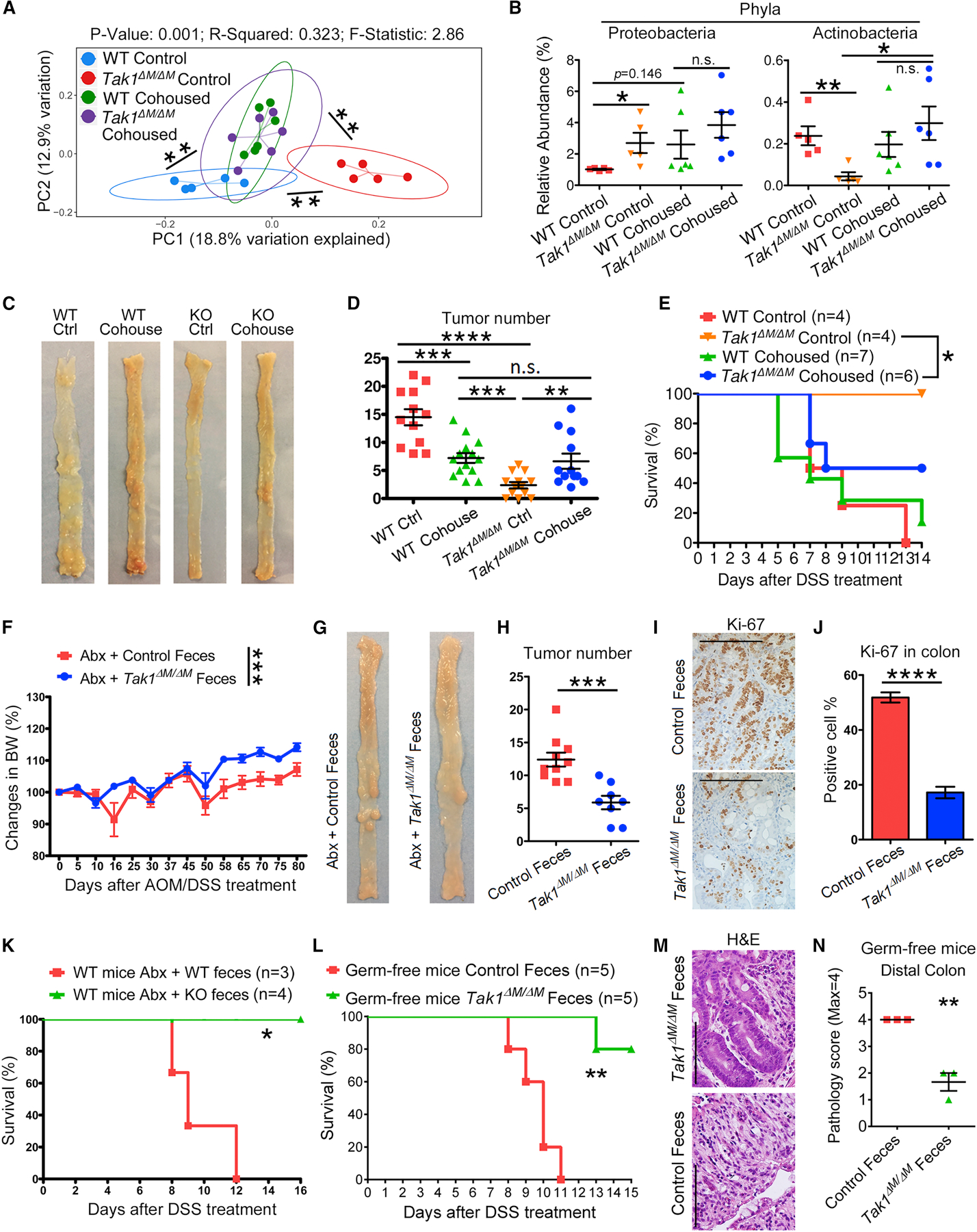
(A-B) The 16S rRNA gene profiling data for fecal microbiome from control and cohoused WT and Tak1∆M/∆M mice. (A) PCoA plot of bacterial beta-diversity. (B) Relative abundance of significantly altered taxa at phylum level.
(C-D) Colon tumors and tumor number analysis in control and cohoused WT and Tak1∆M/∆M mice on day 80 after AOM/DSS treatment.
(E) Survival of control and cohoused WT and Tak1∆M/∆M mice after 5% DSS treatment.
(F-J) Recipient WT mice with Abx-pretreatment and fecal material transfer were treated with AOM/DSS. (F-H) Body weight changes, colon tumors on day 80, and tumor number analysis.
(I-J) IHC staining of Ki-67 on colon sections (scale bar: 100 µm) and analysis of positive cells.
(K) Survival of Abx-treated WT mice with fecal material transfer and 5% DSS treatment.
(L) Survival of germ-free WT mice with fecal material transfer and 4% DSS treatment.
(M-N) H&E staining (40X, scale bar: 100 µm) and histopathology scores on colon sections from fecal microbiota transferred germ-free WT mice after DSS treatment.
Statistical analyses: Student’s unpaired t test (B, D, F, H, J, and N), Monte Carlo permutation test (A), and Mantel-Cox log-rank test (E, K, and L). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
We then investigated whether the transfer of gut microbiota from Tak1∆M/∆M mice can be exploited as a therapeutic approach to ameliorate the cancer development in WT mice. After Abx-pretreatment and fecal microbiota transfer, AOM/DSS was administered to induce colon cancer. The WT mice treated with Tak1∆M/∆M microbiota manifested significant amelioration of body weight loss, and had decreased number of tumors than control mice treated with WT fecal components (Figures 3F–3H). Ki-67 staining showed decreased proliferation of colonic cells in mice treated with Tak1∆M/∆M fecal microbiota, compared with the high level in controls (Figures 3I and 3J). The microbiota recipient mice were also treated with 5% DSS, and mice treated with Tak1∆M/∆M fecal material were completely resistant to acute colitis, unlike controls with WT microbiota that remained sensitive (Figure 3K). Importantly, similar results were obtained in fecal transferred germ-free WT mice, further demonstrating the protective function of Tak1∆M/∆M microbiota (Figures 3L and S3G). Pathological analysis on H&E sections indicated significantly ameliorated colon epithelial histopathology in germ-free mice transferred with Tak1∆M/∆M microbiota (Figures 3M and 3N). Overall, these results indicate the specific microbiota in Tak1∆M/∆M mice can be transferred to WT mice to enhance resistance to colitis and CRC.
Innate Immune Signaling Pathways Are Required for Resistance against Colitis and Colon Cancer in Tak1∆M/∆M Mice
Innate immune system is the first line of host defense against pathogens that provides microorganism recognition and plays a critical role in mediating inflammation and cancer development (Alexander et al., 2014). Therefore, we measured the innate immune cytokines and found both serum and intestinal levels of IL-1β and IL-6 were elevated in Tak1∆M/∆M mice (Figures 4A, 4B, and S4A), in agreement with our previous study (Ajibade et al., 2012). Abx treatment in Tak1∆M/∆M mice resulted in a considerable reduction of IL-6 (Figure 4A), but not IL-1β (Figure 4B), suggesting that microbiota specific to Tak1∆M/∆M mice regulate IL-6 production.
Figure 4. Innate Immune Signaling Pathways Are Required for Resistance against Colitis and Colon Cancer in Tak1∆M/∆M Mice.
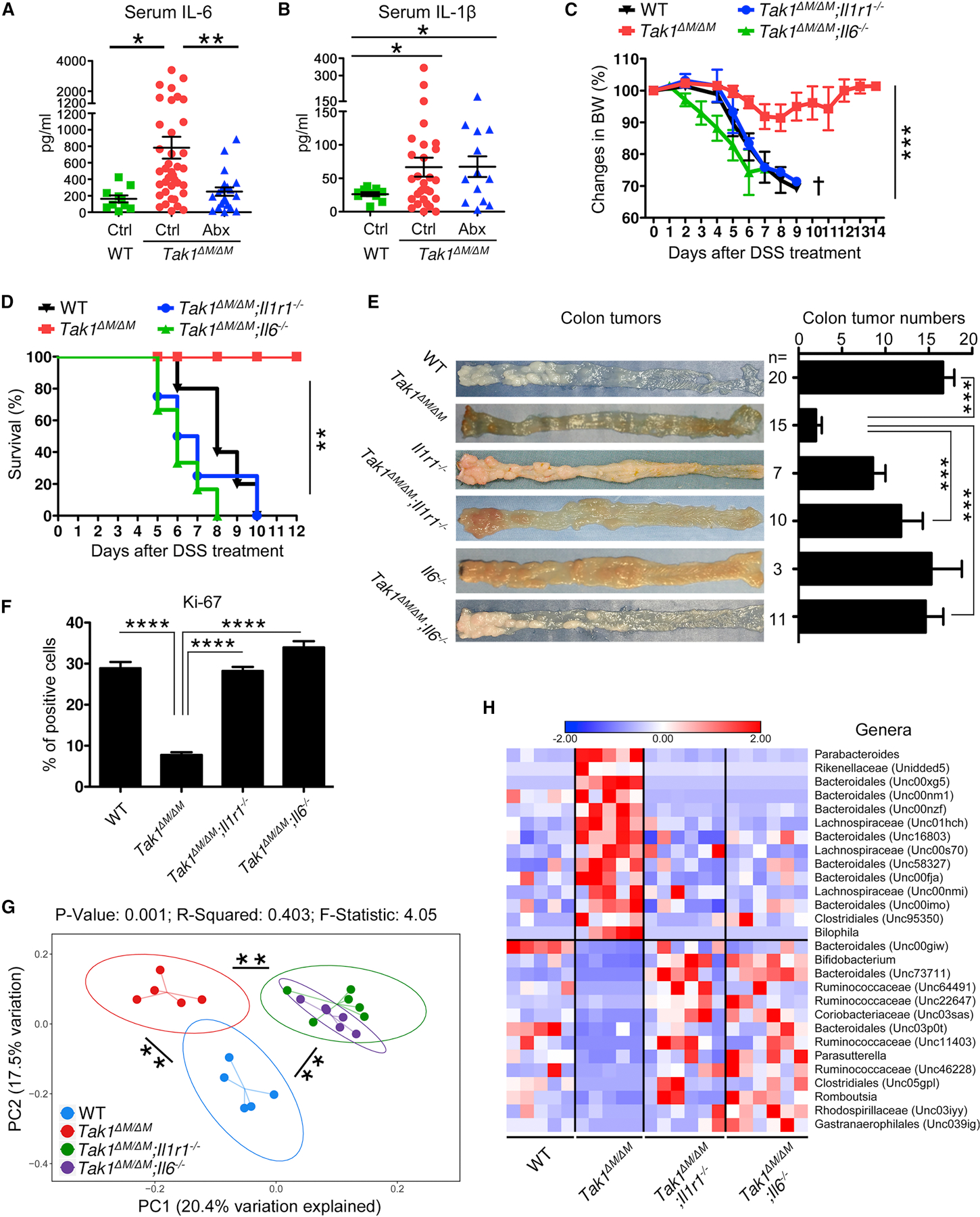
(A-B) Serum IL-6 and IL-1β in WT, Tak1∆M/∆M, and Abx-treated Tak1∆M/∆M mice (for 4 weeks).
(C-D) Body weight changes and survival of different strains after 5% DSS treatment.
(E-F) Colon tumors in different strains on day 80 after AOM/DSS treatment, tumor number analysis, and Ki-67 positive cells on colon sections.
(G-H) The 16S rRNA gene profiling data for fecal microbiome from WT, Tak1∆M/∆M, and double knockout mice. (G) PCoA plot of bacterial beta-diversity. (H) Relative abundance of significantly altered taxa at genus level, including unspecified taxa.
Statistical analyses: Student’s unpaired t test (A-C, E, and F), Monte Carlo permutation test (G), and Mantel-Cox log-rank test (D). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
Next, to investigate the roles of IL-1β and IL-6 signaling pathways, we generated Tak1∆M/∆M;Il1r1−/− and Tak1∆M/∆M;Il6−/− double knockout (DKO) mice. Ablation of IL-1R1 or IL-6 in Tak1∆M/∆M mice completely converted the resistance to sensitive phenotype upon 5% DSS treatment (Figures 4C and 4D), consistent with severe tissue damage and loss of epithelial barrier function (Figure S4C). WT and single knockout (Il1r1−/− and Il6−/−) control mice were sensitive to DSS-induced colitis (Figure S4B), as reported previously (Gonzalez-Navajas et al., 2010; Grivennikov et al., 2009). With AOM/DSS treatment, we demonstrated that both DKO strains manifested an increase in the colonic tumor incidents compared with Tak1∆M/∆M mice (Figure 4E). Extent of colon dysplasia and the proliferation of colonic epithelial cells were significantly increased in DKO mice compared with those in Tak1∆M/∆M mice (Figures S4D and 4F). Subsequent experiments using bone marrow chimeric mice demonstrated that adoptive transfer of Tak1∆M/∆M bone marrow, but not Tak1∆M/∆M;Il1r1−/− bone marrow, could inhibited tumor development (Figures S4E and S4F), suggesting the important effects of IL-1β on CRC-resistance are mainly mediated by bone-marrow-derived immune cells, but not epithelial cells. Overall, our studies suggest that IL-1R1- and IL-6-mediated signaling pathways are required for the resistance of Tak1∆M/∆M mice to colitis and CRC.
To further investigate potential interactions of commensal microbiota with key innate immune signaling in Tak1∆M/∆M mice, fecal samples from WT, Tak1∆M/∆M, and DKO mice were collected and subjected to 16S rRNA sequencing. PCoA plot revealed that deletion of IL-1R1 or IL-6 results in significant changes in gut microbiota composition compared with that in Tak1∆M/∆M mice (Figure 4G). The microbiota of the two DKO strains were similar to each other and differed from the microbiota of WT controls. Subsequent analyses demonstrated that multiple significant changes of various taxa in Tak1∆M/∆M mice were dramatically reversed in the DKO strains (Figures S4G and S4H). Similarly, on the genus level, significantly expanded taxa in Tak1∆M/∆M mice recovered in the DKO strains to WT levels, while the reduced taxa in Tak1∆M/∆M mice were elevated in DKO mice, in some cases even exceeding the levels in WT controls (Figure 4H). Therefore, these data indicate that the altered microbiota composition in Tak1∆M/∆M mice is directly linked to the IL-1R1 and IL-6 signaling pathways.
Th17 Cells in Tak1∆M/∆M Mice Are Controlled by Innate Immune Signaling Pathways and Microbiota and Are Required for Resistance to Colitis and CRC
Because bone marrow derived immune cells and active IL-1R1 and IL-6 signaling pathways are required for the resistance to colitis and CRC in Tak1∆M/∆M mice, we purified immune cells from various organs of WT and Tak1∆M/∆M mice for flow cytometry analysis. A substantial unexpected increase of Th17 population (CD4+/IFN-γ−/IL-17A+) was detected in mesenteric lymph nodes (mLN), Peyer’s patches (PP), and LP in small intestine and colonic tissues of Tak1∆M/∆M mice (Figures 5A and 5B), but not in spleen or peripheral lymph nodes (Figure S5A), compared with those in WT mice. Although total levels of CD4+ cells were consistently increased, substantial changes in other immune populations (such as Th1, Th2, Treg cells, neutrophils, and macrophages) between WT and Tak1∆M/∆M mice were not detected (Figures S5B, S5E, and S5F). CD8+ cells were also slightly increased; however, their overall abundance remained very low in colonic tissue of Tak1∆M/∆M mice. Consistent with the increase in Th17 cells, we observed elevated levels of IL-17A in Tak1∆M/∆M mice (Figure S6B). After DSS treatment, Tak1∆M/∆M mice still maintained high percentage of Th17 cells in colon and IL-17A level in serum, compared with WT mice (Figures S6A and S6B).
Figure 5. Th17 Cells in Tak1∆M/∆M Mice Are Controlled by Innate Immune Signaling Pathways and Microbiota and Are Required for Resistance to Colitis and CRC.
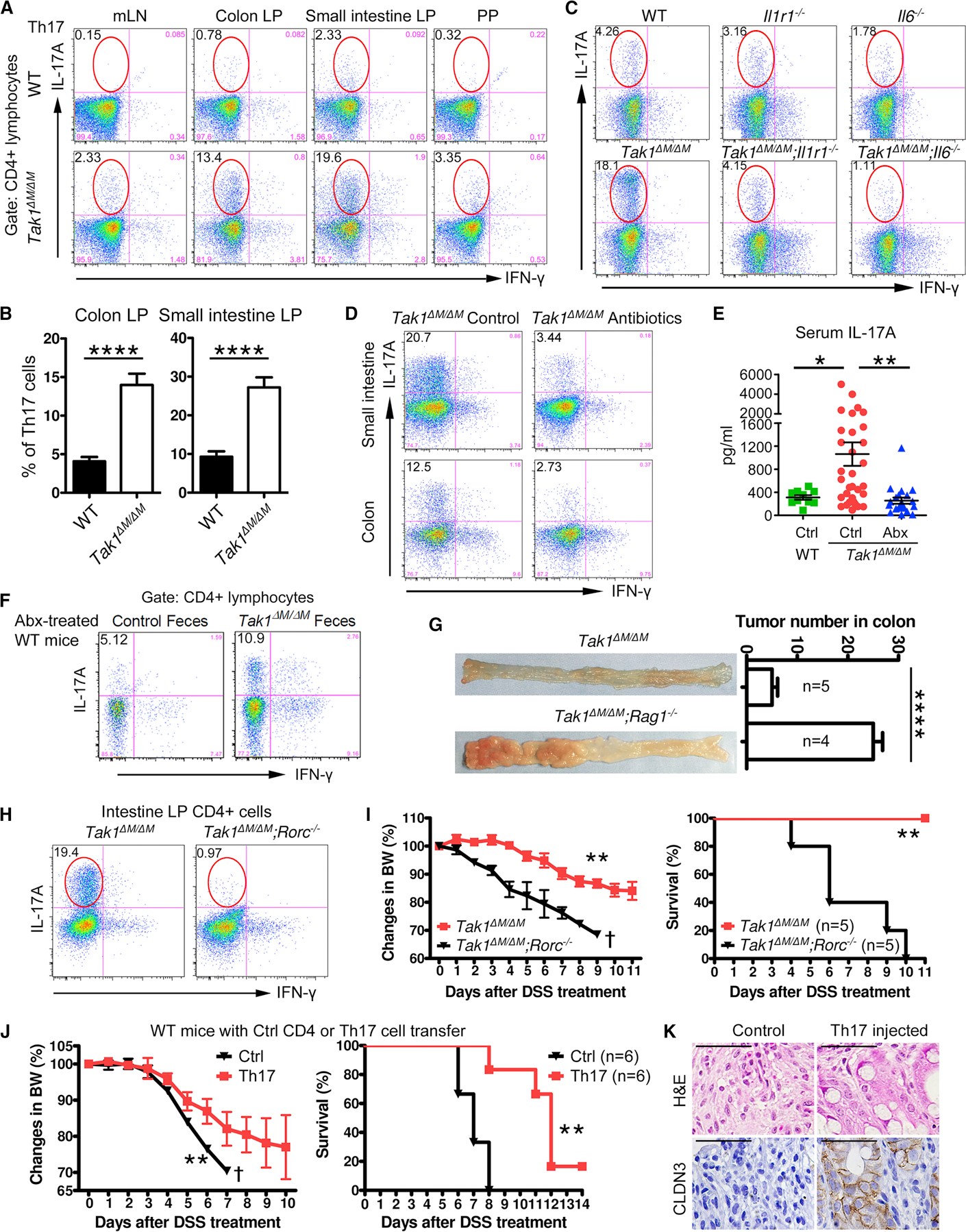
(A) Flow cytometry of Th17 cells (IL-17A+/IFN-γ−) in gated CD4+ lymphocytes from mLN, colon and small intestine LP, and PP.
(B) Th17 cell percentage in colon and small intestine from WT and Tak1∆M/∆M mice.
(C) Flow cytometry of colon LP Th17 cells (IL-17A+/IFN-γ−) in gated CD4+ lymphocytes from different mouse strains.
(D) Flow cytometry of intestinal LP Th17 cells (IL-17A+/IFN-γ−) in gated CD4+ lymphocytes from control and Abx-treated Tak1∆M/∆M mice (for 4 weeks).
(E) Serum IL-17A in WT, Tak1∆M/∆M, and Abx-treated Tak1∆M/∆M mice (for 4 weeks).
(F) Flow cytometry of intestinal LP Th17 cells (IL-17A+/IFN-γ−) in gated CD4+ lymphocytes from WT mice with Abx-pretreatment, fecal material transfer, and AOM/DSS treatment (on day 80).
(G) Colon tumors and tumor number analysis in Tak1∆M/∆M and Tak1∆M/∆M;Rag1−/− mice with AOM/DSS treatment (day 80).
(H-I) Intestinal Th17 cells (IL-17A+/IFN-γ−) in Tak1∆M/∆M and Tak1∆M/∆M;Rorc−/− mice, and their body weight changes and survival after 5% DSS treatment.
(J-K) Intestinal LP lymphocytes were purified from Tak1∆M/∆M;Il17-GFP+ mice. Control CD4+ (CD4+/GFP−) and Th17 cells (CD4+/GFP+) were sorted and injected intraperitoneally (i.p.) to WT recipients. (J) Body weights and survival after 5% DSS treatment. (K) H&E and IHC staining of CLDN3 on colon sections.
Statistical analyses: Student’s unpaired t test (B, E, G, I-left, and J-left) and Mantel-Cox log-rank test (I-right and J-right). *p<0.05; **p<0.01; ****p<0.0001.
See also Figures S5 and S6.
We then aimed to determine whether IL-1R1 and IL-6 signaling pathways are required for Th17 cell development. The results indicated that the ablation of IL-1R1 or IL-6 in Tak1∆M/∆M mice led to a substantial decrease in Th17 population to the WT control level (Figures 5C and S5C), suggesting the elevated IL-1β and IL-6 in Tak1∆M/∆M mice are required for the increase of Th17 cells.
Then, we investigated whether the specific commensal microbiota are required for the increase of Th17 cells in Tak1∆M/∆M mice, and observed significantly decreased Th17 population in Abx-treated Tak1∆M/∆M mice (Figures 5D and S5D). The elevated serum level of IL-17A in Tak1∆M/∆M mice was also considerably reduced after Abx treatment (Figure 5E). Microbiota transfer experiments further showed that intestinal Th17 cells were substantially increased in both Abx-pretreated and germ-free WT mice transferred with microbiota from Tak1∆M/∆M mice, consistent with increased IL-17A productions in serum and colon tissues (Figures 5F and S5G–S5K), indicating the critical role of Tak1∆M/∆M-specific microbiota in promoting Th17 cell development.
Although IL-17-producing γδ T cells are recently reported to control the gut permeability in DSS-colitis model (Lee et al., 2015), we did not observe any appreciable change in TCR-γδ+/IL-17A+ T cells. Instead, a considerable increase in TCR-β+/IL-17A+ T cells (CD4+ Th17) was observed in Tak1∆M/∆M mice and germ-free mice transferred with Tak1∆M/∆M microbiota (Figures S5L and S5M). Collectively, these results suggest that specific components of microbiota and IL-6/IL-1β signaling in Tak1∆M/∆M mice are required for the development of intestinal Th17 cells.
Intestinal Th17 Cells Protect Tak1∆M/∆M Mice from DSS-induced Colitis and Colon Cancer
To determine whether microbiota-induced Th17 cells play a key role in mediating the resistance of Tak1∆M/∆M mice against colitis and CRC, we generated Tak1∆M/∆M;Rag1−/− mice, which lack B and T cells (including Th17 cells) (Mombaerts et al., 1992), and observed a significant loss of body weights and increased mortality rate after 5% DSS treatment, compared with Tak1∆M/∆M mice (Figure S6C). Consistently, we found that Tak1∆M/∆M;Rag1−/− mice developed significantly more tumors than Tak1∆M/∆M mice, with enhanced cell proliferation (Ki-67+ cells) in colonic epithelia (Figures 5G and S6D). Because RORγt, encoded by Rorc gene, is the master transcription factor required for Th17 cell differentiation (Artis and Spits, 2015; Bluestone et al., 2009), we next generated Tak1∆M/∆M;Rorc−/− mice to genetically deplete Th17 cells. As expected, intestinal Th17 cells were specifically deleted in Tak1∆M/∆M;Rorc−/− mice (Figure 5H). Upon 5% DSS treatment, Tak1∆M/∆M;Rorc−/− mice became completely sensitive, with significant loss of body weights (Figure 5I).
To further determine whether intestinal Th17 cells in Tak1∆M/∆M mice are sufficient to mediate direct protection, we generated Tak1∆M/∆M;Il17-GFP+ mice, which expresses transgenic GFP in IL-17A-producing cells. Control (CD4+/GFP−) and Th17 (CD4+/GFP+) cells were sorted from purified intestinal LP immune cells, and directly injected into WT recipient mice. After 5% DSS treatment, Th17 cell transferred mice had significantly ameliorated body weights and survival than controls, consistent with reduced colon tissue damage and epithelial barrier impairment (Figures 5J and 5K). These data suggest that intestinal Th17 cells in Tak1∆M/∆M mice are necessary and sufficient to confer the in vivo protective immunity against colitis.
Identification of the Dominant Microbiota Species/Strains in Tak1∆M/∆M Mice
To identify the dominant microbiota species or strains that are essential for the resistance to colitis and CRC, we performed whole shotgun metagenomics analysis in WT and Tak1∆M/∆M mice and identified 48 increased and 31 reduced species in the gut microbiota of Tak1∆M/∆M mice (Figures 6A and 6B). Significantly changed taxa on the genus level are shown in the heatmap (Figure S6E). SFB and B. fragilis have been reported to induce Th17 cell differentiation (Ivanov et al., 2009; Wu et al., 2009); however, their levels were very low in Tak1∆M/∆M mice.
Figure 6. Identification of the Dominant Microbiota Species/Strains in Tak1∆M/∆M Mice.
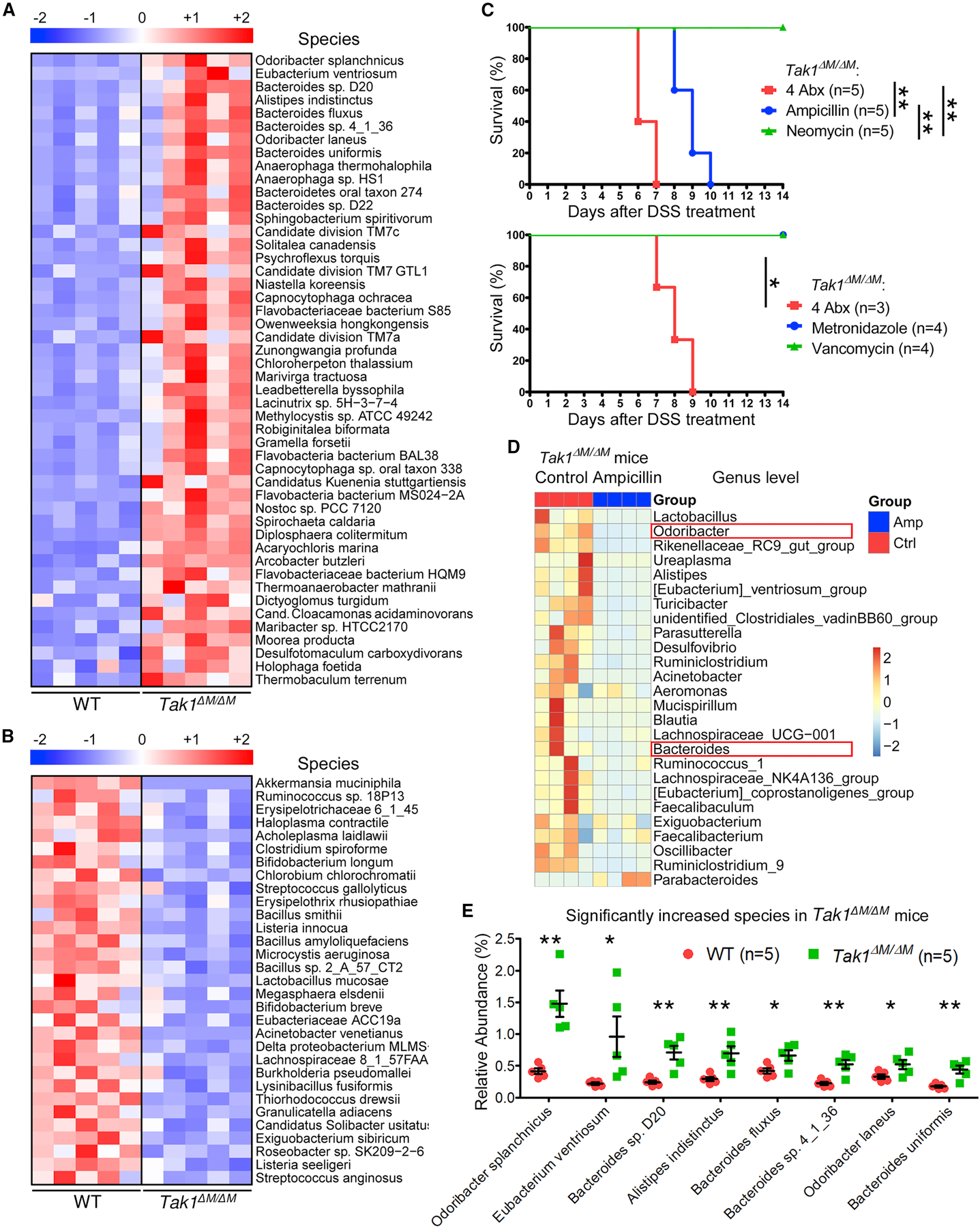
(A-B) Whole shotgun metagenomic sequencing for fecal microbiome from WT and Tak1∆M/∆M mice. Heatmap for relative abundance of significantly increased (A) and decreased (B) species.
(C) Survival of Tak1∆M/∆M mice with combined or single Abx pretreatment and 5% DSS.
(D) The 16S rRNA profiling for fecal microbiome in control and ampicillin-treated Tak1∆M/∆M mice (for 4 weeks). Relative abundance of taxa at genus level with most dramatic decrease.
(E) Significantly increased species in Tak1∆M/∆M mice with the highest abundance from metagenomic sequencing.
Statistical analyses: Student’s unpaired t test (E) and Mantel-Cox log-rank test (C). *p<0.05; **p<0.01.
To further screen the potentially important microbiota components responsible for the resistance to colitis, we treated Tak1∆M/∆M mice with mixed and single Abx in each group; and found that treatment with ampicillin, but not other Abx, abolished the ability of Tak1∆M/∆M mice to resist DSS-induced colitis (Figure 6C). The 16S sequencing of fecal materials from control and ampicillin-treated Tak1∆M/∆M mice identified protective bacterial candidates (boxed in red) at the genus level (Figure 6D), which showed the most pronounced decrease after ampicillin treatment and were also listed as top increased species in Tak1∆M/∆M mice (Figure 6E). Based on these data, 4 candidate bacterial strains were selected for further functional studies, including Odoribacter splanchnicus (OS), Bacteroides sp. 4_1_36 (B4), Bacteroides sp. D20 (BD), and Bacteroides uniformis (BU).
Identification of Protective Microbiota Strains for Colitis and Colon Cancer
To identify protective single microbiota strains, Abx-treated WT mice were transferred with OS, B4, BD, or BU. Successful bacteria colonization and similar levels of abundance in these 4 groups were validated (Figures S6F and S6G). After DSS treatment, all 4 strains partially and significantly extended the mouse survival; OS and BD induced stronger protection with 50% survival at the endpoint compared with B4 and BU that induced moderate protection with 30% survival (Figure 7A). Nontoxigenic Bacteroides fragilis (NTBF), as a positive control, provided partial and significant protection as reported (Chan et al., 2019). We further found the combination of 4 strains lead to better survival than single bacteria (Figure S7A), suggesting that multiple key microbiota strains act in concert to generate protective immunity against colitis.
Figure 7. Identification of Protective Microbiota Strains for Colitis and Colon Cancer.
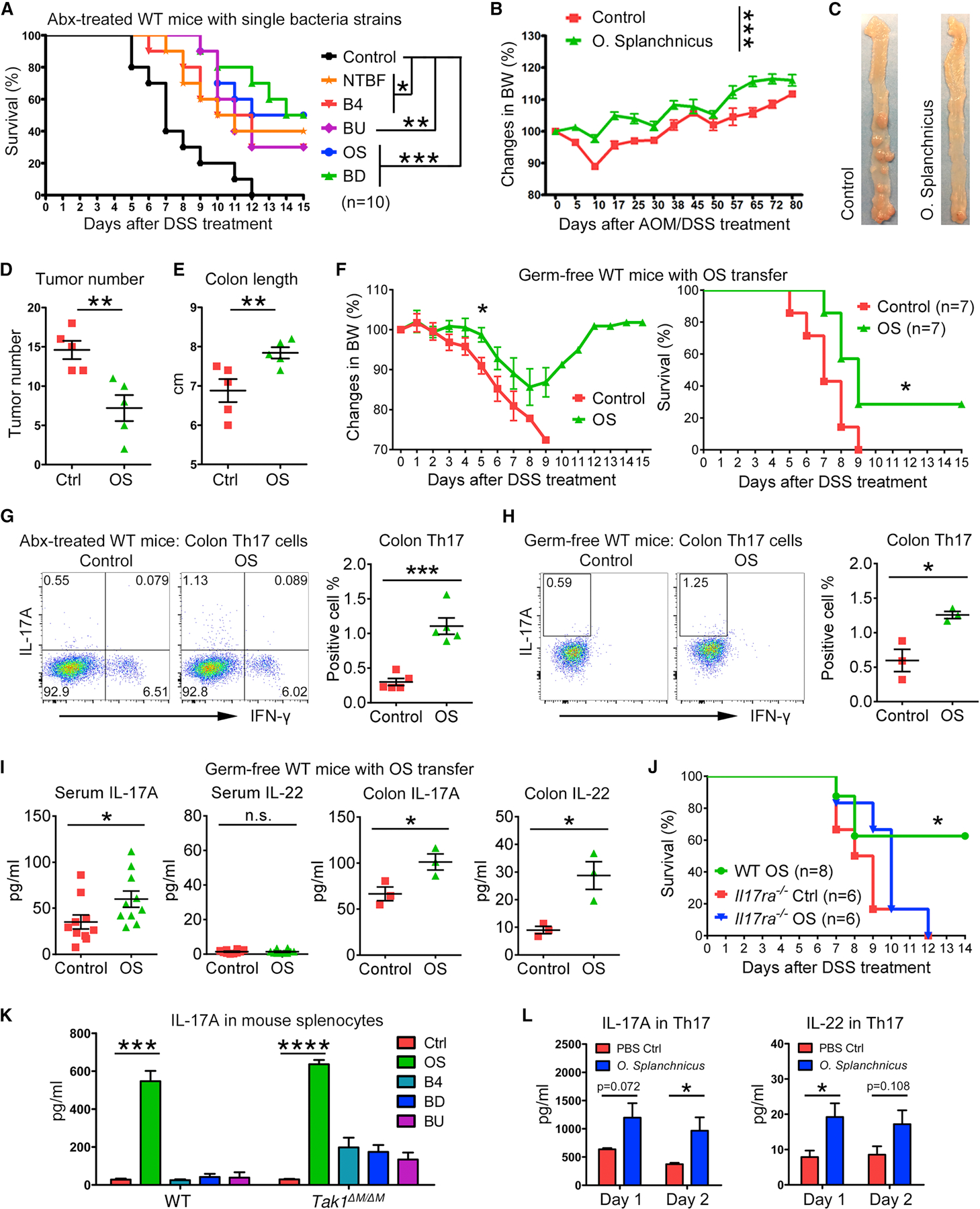
(A) Survival of Abx-treated WT mice with single bacteria strain transfer and 5% DSS treatment.
(B-E) Body weight changes, colon tumors (on day 80), tumor number analysis, and colon length in Abx-treated WT mice with O. splanchnicus (OS) transfer and AOM/DSS treatment.
(F) Body weight changes and survival of germ-free WT mice with OS transfer and 3% DSS treatment.
(G-H) Flow cytometry of colon LP Th17 cells (IL-17A+/IFN-γ−) in gated CD4+ lymphocytes from Abx-treated and germ-free WT mice with OS transfer.
(I) Cytokines in serum and colon tissues from control and OS-transferred germ-free WT mice.
(J) Survival of Abx-treated WT and Il17ra−/− mice with OS transfer and 5% DSS treatment.
(K) IL-17A production in splenocytes from WT and Tak1∆M/∆M mice, after 1 day treatment of PBS control or autoclaved single bacteria strains.
(L) Intestinal LP Th17 cells were sorted from Tak1∆M/∆M;Il17-GFP+ mice for cytokine productions after autoclaved OS treatment.
Statistical analyses: Student’s unpaired t test (B, D-E, F-left, G-I, K-L) and Mantel-Cox log-rank test (A, F-right, and J). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
See also Figures S6 and S7.
Furthermore, OS, as the top increased species in Tak1∆M/∆M mice compared with both WT and littermate controls (Figure S6H), was selected to investigate the induction of protective immunity against CRC. In Abx-treated WT mice, OS inoculation led to significantly ameliorated body weights after AOM/DSS treatment and decreased tumor numbers compared with control mice (Figures 7B–7D). The increase of colon length in OS-treated mice was indicative of a healthy status with potentially lower inflammation (Figure 7E). Importantly, germ-free mice were monocolonized with OS single strain (Figure S7B). After DSS treatment, the control mice were completely sensitive, while OS-transferred germ-free mice had significantly extended survival, with limited loss and complete recovery of body weights (Figure 7F). These data suggest that OS alone can induce significant protective immunity against colitis and CRC.
We next investigated the immune responses to OS transfer, and found significant increases of colon Th17 cells in both Abx-pretreated and germ-free WT mice, consistent with elevated levels of IL-17A and IL-22 in the colon and/or serum after OS colonization (Figures 7G–7I and S7C). Notably, without Abx pretreatment, OS transfer in WT mice could lead to a higher percentage of colon Th17 cells (Figures S7D and S7E) than those observed in germ-free or Abx-treated mice, indicating other microbiota strains may synergistically enhance the Th17 cell development induced by OS. To demonstrate the requirement of Th17 cells in mediating OS-induced protective immunity, we treated Il17ra−/− mice with Abx and OS, and found that they were similarly sensitive to DSS-induced colitis with or without OS administration (Figures 7J and S7F), suggesting that OS protects host against colitis by inducing intestinal Th17 cells and cytokines.
To determine the potential mechanisms of how single bacterial strains induce protective immunity against colitis and CRC, we tested the effects of OS, B4, BD, and BU on stimulating cytokine productions by immune cells. In neutrophils and macrophages, IL-1β could only be induced by OS, whereas IL-6 was dramatically stimulated by all 4 strains (Figure S7G). Importantly, OS induced higher levels of IL-17A in total splenocytes than that detected in the case of B4, BD, or BU (Figure 7K). As certain Th17 cytokines that may have protective functions against colitis (Lee et al., 2015; Sugimoto et al., 2008), IL-17A and IL-22 were induced by OS stimulation in the cells from spleen and/or mLN (Figure S7H). Furthermore, intestinal Th17 cells were obtained from Tak1∆M/∆M;Il17-GFP+ mice, and their production of IL-17A and IL-22 after OS treatment were observed (Figure 7L). Overall, our data indicate that a single bacterial strain identified from Tak1∆M/∆M mouse microbiota can induce innate immune cytokine production and stimulate Th17 cells to confer protective immunity against colitis and CRC.
DISCUSSION
In this study, we demonstrate that Tak1∆M/∆M mice are completely resistant to chemically induced colitis and CRC, different from most of the reported models that are more sensitive to colitis than WT mice (Terzic et al., 2010; Wirtz et al., 2007), thus providing a unique opportunity to determine the mechanisms and identify single bacterial strains responsible for the protective immunity. Several CRC-associated bacteria have been identified in cancer patients and in animal models (Belkaid and Hand, 2014), however, only very few bacteria were found to contribute to protective immunity against colitis and CRC. Using this unique resistant mouse model, we identified numerous bacterial taxa that have been implicated in human Crohn’s disease and cancer development: (1) Lachnospiraceae is dramatically increased in resistant Tak1∆M/∆M mice and decreased in Crohn’s disease patients (Gevers et al., 2014), and negatively correlates with CRC in mouse recipients of human fecal transplant (Baxter et al., 2014). Administration of Clostridium immunis (in Lachnospiraceae family) protects mice from DSS-induced colitis (Surana and Kasper, 2017). (2) Alistipes indistinctus was also considerably increased in the gut flora of Tak1∆M/∆M mice, and the antitumor efficacy of this species in cancer immunotherapy was demonstrated (Routy et al., 2018). (3) Odoribacter splanchnicus and Odoribacter laneus are two major species with significantly increased abundance in Tak1∆M/∆M mice. Consistently, Odoribacter ranks among the top increased genera in resistant laboratory animals treated with microflora of wild mice (Rosshart et al., 2017). (4) Akkermansia muciniphila is significantly reduced, and this species demonstrated the highest significant decrease in Tak1∆M/∆M mice. Indeed, Akkermansia muciniphila is positively associated with the induction of CRC in mice recipients of human fecal transplant (Baxter et al., 2014). Collectively, these data suggest that specifically altered (increased or decreased) microbiota species in Tak1∆M/∆M mice are relevant to their ability to inhibit or promote colitis and cancer development.
To illustrate the functions of specific microbiota in Tak1∆M/∆M mice, we demonstrate that Abx-mediated microbiota depletion converts the resistant phenotype to sensitive phenotype. Cohousing and fecal transfer approaches suggest that the microbiota of Tak1∆M/∆M mice can be transferred to WT mice to confer the resistance to colitis and CRC. Moreover, we provide compelling evidence to demonstrate that microbiota are critically required to activate innate immune signaling with subsequent cytokine production and to promote Th17 cell development. Although IL-1β or IL-6 alone can promote colonic inflammation and colitis-associated CRC (Bollrath et al., 2009; Coccia et al., 2012; Grivennikov et al., 2009; Tu et al., 2008; Wang et al., 2014b), Tak1∆M/∆M mice produce high levels of IL-1β and IL-6 while still maintaining a resistant phenotype. Depletion of the microbiota considerably reduces IL-6 production; Importantly, ablation of IL-1R1 or IL-6 signaling pathways alters the composition of microbiota, reduces the number of Th17 cells in the intestinal LP, and sensitizes the animals against colitis and CRC, suggesting that microbiota modulate protective immunity through IL-1R1 and IL-6 signaling pathways.
Th17 cells and IL-17A show context-dependent roles in mediating colitis and CRC, probably due to the following mechanisms: (1) Different tumor models (Apc mutation model versus chemically induced model). IL-17A promotes the tumor development in genetically induced (Apcmin/+) CRC models (Chae et al., 2010; Grivennikov et al., 2012; Wang et al., 2014a; Wu et al., 2009), but displays a protective effect in most of the colitis-associated CRC models (O’Connor et al., 2009; Ogawa et al., 2004; Yang et al., 2008), via regulation of epithelial permeability, barrier function, and tissue repair (Lee et al., 2015; Maxwell et al., 2015; Song et al., 2015). In existing tumor cells and intact colon epithelial cells, IL-17A may act through altered signaling pathways and show distinct roles. (2) Different animal models (DSS-sensitive versus resistant models). Most previous studies using knockout mouse models show a decreased Th17 population or loss of key Th17 cytokines, resulting in a more sensitive phenotype than WT mice in response to DSS-induced colitis. By contrast, our unique animal model (Tak1∆M/∆M) significantly increased Th17 cell population and were completely resistant to DSS-induced colitis. A recent report shows that Fam64a−/− mice decreased Th17 cells and ameliorated colitis and CRC (Xu et al., 2019), but Th17 cells in Fam64a−/− mice were not tested for their proinflammatory or suppressive role. (3) Different cytokine environment. Besides the number of Th17 cells, their function could be influenced by several key cytokines (IL-6, IL-1β and IL-23). With the presence of IL-23 or serum amyloid A proteins, Th17 cells could acquire a pathogenic pro-inflammatory phenotype, compared with non-pathogenic Th17 cells induced by IL-6 and TGF-β (Ghoreschi et al., 2010; Lee et al., 2020). Th17 cells generated in different environments may further secrete distinct cytokines and show pro-inflammatory or immune-suppressive functions (Esplugues et al., 2011).
Although gut microbiota have been linked to cancer development and immunotherapy (Honda and Littman, 2016; Hooper et al., 2012; Rooks and Garrett, 2016; Thaiss et al., 2016; Zitvogel et al., 2016), little is known about specific bacterial strains that induce and modulate antitumor immunity. Recently, a combination of 11 bacterial strains improves the antitumor efficacy of checkpoint inhibitors (Tanoue et al., 2019); however, a similar effect of a single strain was not demonstrated. In this work, we identified a single bacterial strain (Odoribacter splanchnicus) that confers significant resistance to colitis and CRC, due to induced Th17 cells and enhanced cytokine productions. Notably, Th17 cell differentiation is mainly directed by specific microbiota strains (Honda and Littman, 2016), and some bacteria species have been identified (Chan et al., 2019; Ivanov et al., 2009; Wu et al., 2009; Yang et al., 2014). By contrast, other CD4 subsets were not affected by specific Tak1∆M/∆M microbiota in our model.
Based on our findings, we propose a multiple-step working model: 1) altered microbiota interplay with innate immune signaling to produce cytokines (IL-1β and IL-6) in Tak1∆M/∆M mice; 2) microbiota and key cytokines stimulate dramatic expansion of Th17 cells; and 3) Th17 cells and IL-17A mediate the resistance to colitis and CRC. In summary, our study reported a unique colitis-resistant mouse model, demonstrated the interaction of microbiota and innate immune signaling in regulating Th17 cells, tissue integrity, and inflammation, and identified a single bacterial strain that can trigger protective immunity against cancer.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Rong-Fu Wang (rongfuwa@usc.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The 16S and shotgun metagenomics raw sequence data reported in this paper were deposited in NCBI Sequence Read Archive (SRA) with the accession number of PRJNA495072.
Data analyses for microbiota sequencing are performed following published articles (described in detail in “QUANTIFICATION AND STATISTICAL ANALYSIS” section), in which the sources of code for computational data analysis pipeline are available.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and In Vivo Procedures
As described previously (Ajibade et al., 2012), Tak1flox/flox mice (gift from Dr. Michael Schneider, Baylor College of Medicine) were crossed with Lyz2-Cre mice (Jackson Laboratory) to generate the Tak1∆M/∆M mouse strain. C57BL/6 (WT), Il1r1−/−, Il6−/−, Rag1−/−, and Rorc−/− mice were purchased from Jackson Laboratory. Il17-GFP+ strain is a gift from Dr. Scott Durum (National Institutes of Health). Tak1∆M/∆M mice were then crossed with these mice to generate Tak1∆M/∆M;Il1r1−/−, Tak1∆M/∆M;Il6−/−, Tak1∆M/∆M;Rag1−/−, Tak1∆M/∆M;Rorc−/−, and Tak1∆M/∆M;Il17-GFP+ strains. In all experiments, 7 to 9 weeks old female mice were used unless specifically described; and the mice in different groups were single housed unless stated otherwise. The littermate control mice (Tak1flox/flox;Lyz2-Cre−) were weaned at 3 weeks old and housed separately from the Tak1∆M/∆M mice after the genotyping. All the mice were housed under a 12:12 light:dark cycle, and maintained in specific pathogen-free facilities that are accredited by Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Houston Methodist Research Institute and University of Southern California.
In acute colitis model, mice were administered 5% DSS (molecular weight 36–50 kDa; MP Biomedicals) in drinking water for 5 days followed by regular water for 2 weeks. In colon cancer model, mice were i.p. injected AOM (Sigma-Aldrich, 10 mg/kg body weight) on day 0, and provided with 2% DSS through drinking water on days 5–10, 24–29, and 43–48, then sacrificed on day 80 for colon tumor analysis.
To generate bone marrow chimeric mice, recipient mice were γ-irradiated at 950 rad, and intravenously injected with 10 million fresh bone marrow cells from donor mice after 24 hours. Recipient mice were treated with antibiotic water (0.5 mg/ml Baytril, Bayer) for two weeks starting from 2 days before irradiation, followed by 4 weeks of regular water. Six weeks after irradiation, chimeric mice were treated for future experiments.
For antibiotic treatment, freshly weaned (3–4 weeks old) WT B6 and Tak1∆M/∆M mice were treated with a combination of various antibiotics (ampicillin, 1 mg/ml; neomycin, 1 mg/ml; metronidazole, 1 mg/ml; vancomycin, 0.5 mg/ml) through drinking water. After 4 weeks, cecum was dissected from control and antibiotic-treated mice for the validation of successful microbiota ablation, as indicated by the enlargement of cecum. Fecal samples were also collected, resuspended in sterile PBS, filtered with a 70 µm strainer, and applied on antibiotic-free agar plates prepared with brain heart infusion (BHI) medium. Both anaerobic (37°C for 3 days) and aerobic cultures (37°C for 2 days) were performed for the confirmation of microbiota depletion. After the validation, mice were then treated with 5% DSS for the induction of acute colitis.
For animal cohouse, age- and sex-matched WT and Tak1∆M/∆M mice were freshly weaned (3–4 weeks old) from their parents, and the littermates were randomly separated into two groups. In cohouse group, WT and Tak1∆M/∆M mice were raised in the same cages at 1:1 ratio (2 WT and 2 knockout mice in each cage) after weaning, while in control groups, the mice were raised in separate cages to maintain their differences on microbiota composition. After 4 weeks, fecal materials were collected from control single-housed mice and cohoused mice for 16S sequencing to validate the similarity of microbiota in cohoused ones. Then the mice were treated with 5% DSS to induce acute colitis and AOM/DSS for colon cancer.
For fecal materials or cultured bacteria transfer, freshly weaned (3–4 weeks old) WT B6 mice were pretreated with a combination of various antibiotics (ampicillin, 1 mg/ml; neomycin, 1 mg/ml; metronidazole, 1 mg/ml; vancomycin, 0.5 mg/ml) for 4 weeks, and were randomly separated into different groups for the oral gavage of fecal materials or single bacteria strains. The viably preserved fecal microbial communities were transferred by oral gavage every week for 4 weeks into the mice. Fecal pellets were freshly collected from WT B6 mice and Tak1∆M/∆M mice, and stored in −80°C freezer. Cryopreserved fecal stocks were thawed and resuspended in sterile PBS (1 ml per fecal pellet), and filtered with a 70 µm strainer. Fecal stocks from the same genotype were mixed so that each recipient mouse received the same 0.2 mL suspension every week. For the gavage of single bacteria strains, the number of anaerobically cultured bacteria were evaluated based on reading of the spectrophotometer at Optical Density of 600 nm. Each week, 5×108 bacteria/strain were transferred to each mouse with 0.2 ml PBS. Mice that received the same fecal stocks or bacteria species were housed in the same cage. The mice were treated with AOM/DSS or 5% DSS after 4 weeks from the initial transfer.
Germ-free mice were purchased from Taconic Biosciences at 3–4 weeks old and housed in autoclaved sterile microisolators with positively pressured HEPA-filtered sterile air, irradiated food, and autoclaved water. Mouse handling and weekly cage changes were performed by investigators wearing sterile gowns, masks, and gloves in a sterile biosafety hood. Fecal microbiota or OS single strain were transferred to the germ-free mice once a week for 4 weeks, similar as in Abx-treated mice. The sterility in control mice (treated with sterile PBS) was examined every two weeks by the fecal material culture on antibiotic-free BHI agar plates. Successful colonization in bacteria-transferred mice were validated after 4 weeks by fecal material culture and PCR.
METHOD DETAILS
Histology, Immunohistochemistry, and Microscopy
Fresh colon pieces were fixed with 3.7% formalin for 24 hours, and then sent to the histology core at Baylor Breast Care Center for further processing and H&E staining. Alcian blue and periodic acid schiff (AB/PAS) staining was performed as per manufacturer’s instruction (Newcomer Supply) to show Goblet cells. For IHC staining, unstained tissue sections were deparaffinized in xylene, rehydrated in graded ethanol solutions, and washed in tap water. Antigen retrieval was achieved by boiling the slides in a pressure cooker for 3 min in a citrated buffer (10 mM trisodium citrate, pH 6.0). After 10 min treatment with 3% H2O2, tissue sections were blocked with 5% normal goat serum in TBST for 1 hour at room temperature, incubated with primary antibodies at 4°C overnight, and with EnVision Polymer-HRP secondary antibodies (Dako) at room temperature for 30 min. After the application of DAB chromogen (Vector), tissue sections were stained with hematoxylin, dehydrated, and mounted. The slides were analyzed by a pathologist and the investigators. Images were acquired using the Olympus BX61 microscope along with DP71 digital camera (Olympus).
After the IHC staining of Ki-67, slides were read under microscope and fields were randomly selected for analysis. Pictures were taken at 40X magnification. The positive and negative cell numbers were counted in each field. For each mouse, 4–5 fields were used for quantification, with the animal number of 3–5 for each group.
Histopathology score in colitis model
Formalin-fixed and paraffin-embedded colons were processed and stained with H&E according to standard procedures. Tissues were scored on a 0–4 system, according to published criteria (Erben et al., 2014; Maxwell et al., 2015): 0, normal; 1, mild inflammation with less than 10% loss of epithelial structure (crypts), focal enterocyte hyperplasia; 2, moderate inflammation with 10%–30% of crypt loss, multifocal enterocyte hyperplasia, goblet cell loss; 3, marked inflammation with 30%–50% of crypt loss, diffuse enterocyte hyperplasia, few goblet cells; 4, severe inflammation with over 50% of crypt loss, diffuse enterocyte hyperplasia, mucosal ulceration. Each data point represents an individual mouse.
16S rRNA gene sequencing
As previous described (Rosshart et al., 2017), 16S rRNA gene sequencing methods were adapted from the methods developed for the Earth Microbiome Project and Human Microbiome Project. Bacterial genomic DNA was extracted using QIAamp Fast DNA Stool Mini Kit (QIAGEN). For the microbiota comparison in WT, Tak1∆M/∆M mice, cohoused mice, and DKO mice, samples were submitted to Alkek Center for Metagenomics and Microbiome Research in Baylor College of Medicine for library generation and sequencing. The 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2×250 bp paired-end protocol yielding pair-end reads. The primers used for amplification (515F-806R) contain adapters for MiSeq sequencing and single-end barcodes allowing pooling and direct sequencing of PCR products on the Illumina MiSeq platform. For the microbiota comparison in littermate controls, Tak1∆M/∆M mice, and DSS-treated Tak1∆M/∆M mice, samples were submitted to Novogene for library generation, sequencing, and data analyses.
Whole shotgun metagenomic sequencing
Fresh fecal samples were collected from WT and Tak1∆M/∆M mice, immediately frozen on dry ice, and submitted to Novogene for further procedures. Whole metagenomic DNA was extracted using QIAamp DNA Microbiome Kit (QIAGEN). DNA obtained from each sample was used for library construction with NEBNext Ultra II-E7645 (New England Biolabs), which were then shotgun sequenced in the Illumina HiSeq Platform using Hiseq 4000 SBS kit, PE150 (Illumina).
Bacteria colonization PCR
Fecal bacterial genomic DNA was extracted from bacteria-transferred mice using QIAamp Fast DNA Stool Mini Kit (QIAGEN). PCR was performed with GenDEPOT amfiSure PCR Master Mix, followed by the electrophoresis on a 2% agarose gel. Real-time QPCR was performed using an Applied Biosystems 7500 Fast Real-Time PCR system with SYBR Green master mix (Applied Biosystems). Relative abundances of specific taxa were normalized with the universal bacteria primers.
Isolation of Intestinal LP Lymphocytes
Briefly, the fat, connective tissue, Peyer’s patches, and feces were removed from colon. The colons were then cut into ~0.5-cm pieces and incubated in 2 mM EDTA/PBS/FBS for 30 min at 37°C with stirring. After EDTA treatment, the small intestine and colon pieces were washed in PBS/FBS solution and incubated in 0.5 mg/ml Collagenase-IV (Sigma)/RPMI-1640/FBS for 90 min at 37°C with stirring. After collagenase digestion, the supernatant which contains LP cells, were centrifuged, and the cell pellets were re-suspended in 44% Percoll (GE)/RPMI-1640/FBS. The 44% Percoll/cell suspension was slowly added onto the surface of 67% Percoll/RPMI-1640/FBS. After centrifugation, the LP cells were at the 44% to 67% interface.
Flow Cytometry and Sorting
For surface marker staining, cells were directly stained in a cocktail of fluorochrome-conjugated antibodies (eBioscience or BD Biosciences) for 30 min on ice, washed twice in PBS/FBS, and resuspended in PBS/FBS for flow cytometry. For intracellular staining, cells were activated with PMA, Ionomycin and protein transport inhibitor for 4 hours at 37°C. Then the cells were resuspended in Fix/Perm buffer (BD) and treated for 20 min on ice, washed with Perm/Wash buffer (BD), and stained with the antibody cocktail for 30 min on ice. After washing twice, cells were re-suspended in PBS/FBS for flow cytometry. BD LSR II analyzer was used to distinguish different subsets. For the sorting of Th17 cells, intestinal LP lymphocytes were isolated from Tak1∆M/∆M;Il17-GFP+ mice, enriched with Untouched Mouse CD4 Cells Kit following manufacturer’s instruction, and then stained with PE-CD4 antibody for 10 min at room temperature. After washing twice with RPMI-1640/FBS, cells were filtered with 40 µm strainer, and resuspended in RPMI-1640/FBS for sorting. BD FACSAria II sorter were used to acquire the control CD4+ (PE+/GFP−) and Th17 population (PE+/GFP+).
Enzyme-Linked Immunosorbent Assay (ELISA)
Briefly, the plates were coated with capture antibodies (eBioscience or MBL) at 4°C overnight, and blocked with PBS/BSA for an hour at room temperature. Samples and standards were incubated in the plates for 2 hours at room temperature. The plates were treated with biotinylated detection antibodies (eBioscience or MBL) for 1 hour, and HRP chromogen (Thermo) for 30 min at room temperature. Tetramethylbenzidine (TMB; Sigma) was added to the plates for 5–10 min and the reactions were stopped with an equal volume of 2 M H2SO4. The absorbance of each well was read at 450 nm on BioTek Synergy 2 Microplate Reader.
In Vitro Immune Cell Stimulation by Microbiota Strains
Peritoneal macrophages and neutrophils were obtained by intraperitoneal injection of 1.5 ml 3% (v/v) thioglycollate (BD), and peritoneal cavities were flushed after 4 hours (for neutrophils) or 3 days (for macrophages) with RPMI-1640 media with 2% FBS. Total immune cells from spleens and mLNs were collected by mashing the cells through the 40-µm strainer into the petri dish containing RPMI-1640 media with 2% FBS. Red blood cells were lysed by ACK buffer for 5 min at room temperature. Bacteria antigens for the candidate strains were prepared by resuspending 6 × 10 9 bacteria in 20 ml PBS, and autoclaving for 20 minutes. For in vitro stimulation, cells were cultured in 96-well U-bottom plates (0.1 million/well), with or without 20 ng/ml mouse IL-2 for enhancing T cell activity, and treated with autoclaved bacteria suspension (1:10 dilution to final concentration). After 4 hours (for macrophages), 20 hours (for neutrophils), or 48 hours (for total splenocytes and mLN cells), the culture supernatants were collected to examine the cytokine production by ELISA.
Th17 cells were sorted from intestinal LP in Tak1∆M/∆M;Il17-GFP+ mice, and seeded (5 × 104 cells/well) in 96-well U-bottom plates. Total splenocytes from the same mice were irradiated as antigen-presenting cells (APCs). After the overnight pre-incubation with autoclaved bacterial lysates (1:10 dilution), the APCs (2 × 105 cells/well) were added into the Th17 cells in 96-well plates or empty wells for baseline controls. Mouse IL-2 (20 ng/ml) was added for enhancing Th17 cell activity. After 24 and 48 hours, the culture supernatants were collected to examine the cytokine production by ELISA.
QUANTIFICATION AND STATISTICAL ANALYSIS
General Statistical analysis:
Descriptive statistics, including means, standard deviations, medians, and ranges, were computed for each group and analyzed with Student’s t-test or for multiple comparisons, with ANOVA. Data is presented as mean ± standard error of the mean (SEM). Differences in mice survival were evaluated with Mantel-Cox log-rank test. The sample size for each experiment is included in the results section and the associated figure legend. All analyses were performed with GraphPad Prism 5 (GraphPad Software, La Jolla, CA). P-values < 0.05 were considered significant.
16S rRNA gene compositional analysis
Data analysis for 16S rRNA sequencing was performed as previous described (Rosshart et al., 2017). Briefly, 16S rRNA gene sequences were clustered into Operational Taxonomic Units (OTUs). Abundances were recovered by mapping the demultiplexed reads to the OTUs. The rarefied OTU tables were generated for downstream analyses using a visualization toolkit developed at the CMMR named ATIMA (Agile Toolkit for Incisive Microbial Analyses). Significance of categorical variables are determined using the non-parametric Mann-Whitney test for two category comparisons or the Kruskal-Wallis test when comparing three or more categories. Correlation between two continuous variables is determined with R’s base “lm” function for linear regression models, where p values indicate the probability that the slope of the regression line is zero. PCoA plots employ the Monte Carlo permutation test to estimate p values, which are further adjusted for multiple comparisons with the FDR algorithm.
Pairwise PERMANOVA tests with false discovery rate adjustments for multiple comparisons were applied using the adonis function in the vegan package to the unweighted and weighted UniFrac metric to determine differences in microbial community composition. Differences in taxonomic abundance and differences between pairwise unweighted UniFrac distances were assessed with the Kruskal-Wallis test with a false discovery rate adjustment for multiple comparisons in R version 3.2.1. Heatmaps were generated under R package ‘pheatmap’ (The R Foundation, https://cran.r-project.org/). Additionally, multiple other helper functions and graphing tools were utilized in the R environment.
Indicator species analysis, as implemented by the indicspecies package, was used to identify OTUs that were most indicative of WT or Tak1∆M/∆M group based upon both probability of occurrence and abundance in those groups. Those OTUs that were found to be significant indicators through permutation tests and subsequent False-Discovery-Rate adjustment were further screened to ensure that we identified highly indicative OTUs. Therefore, we removed those indicator OTUs that did not occur in at least 75% of the mice for which they were indicative, and we further removed indicator OTUs that were not observed at an average relative abundance of greater than 0.1% for the samples shown.
After the analysis of 16S sequencing data, detailed information are listed in Supplement Tables S1 and S2, with key data used for figure generation.
Whole shotgun metagenomic sequencing analysis
Raw reads were filtered to trim nucleotides from the 3’ end using a quality threshold of 30 and remove adaptor contamination and low-quality. As a consequence, an average of 92.6% high quality reads was obtained from all samples.
The high-quality reads of the HiSeq platform were then aligned to the Mus musculus complete genome by SOAP2 using the criterion of identity ≥ 90%. Sequence-based gene abundance profiling was performed as previous described (Li et al., 2014). The relative abundances of phyla, genera, species and KOs were calculated by the sum of the relative abundance of their annotated genes. At species level, the fold change of relative abundance over 1.5 was considered as increased taxa, while the fold change less than 0.5 considered as reduced taxa. The alpha diversity (within-sample diversity) was quantified by the Shannon index using the relative abundance profile at gene, genus and KO levels as described (Li et al., 2014). The beta diversity (between-sample diversity) was calculated using Bray-Curtis distance (R 3.2.5, vegan package 2.4–4).
After the analysis of metagenomic sequencing data, detailed information are listed in Supplement Table S3, with key data used for figure generation.
Supplementary Material
Supplement Table S1. Altered Gut Microbiota Composition in Tak1∆M/∆M Mice Are Normalized by Cohousing with Control WT Mice (related to Figures 2 and 3)
Spreadsheet 1. 16S microbial sequencing data: Reads per sample
Spreadsheet 2. 16S microbial sequencing data: Raw OTU counts
Spreadsheet 3. 16S microbial sequencing data: Rarefied OTU counts
Spreadsheet 4. 16S microbial sequencing data: Alpha Diversity
Spreadsheet 5. 16S microbial sequencing data: Taxonomy
Spreadsheet 6. 16S microbial sequencing data at kingdom level
Spreadsheet 7. 16S microbial sequencing data at phylum level
Spreadsheet 8. 16S microbial sequencing data at class level
Spreadsheet 9. 16S microbial sequencing data at order level
Spreadsheet 10. 16S microbial sequencing data at family level
Spreadsheet 11. 16S microbial sequencing data at genus level
Spreadsheet 12. 16S microbial sequencing data: U.UniFrac DM
Spreadsheet 13. 16S microbial sequencing data: W.UniFrac DM
Spreadsheet 14. 16S microbial sequencing data: U.UniFrac PCoA
Spreadsheet 15. 16S microbial sequencing data: W.UniFrac PCoA
Spreadsheet 16. 16S microbial sequencing data: U.UniFrac tSNE
Spreadsheet 17. 16S microbial sequencing data: W.UniFrac tSNE
Supplement Table S2. IL-1R1 and IL-6 Signaling Pathways are Required for Shaping Microbiota in Tak1∆M/∆M Mice (related to Figure 4)
Spreadsheet 1. 16S microbial sequencing data: Reads per sample
Spreadsheet 2. 16S microbial sequencing data: Raw OTU counts
Spreadsheet 3. 16S microbial sequencing data: Rarefied OTU counts
Spreadsheet 4. 16S microbial sequencing data: Alpha Diversity
Spreadsheet 5. 16S microbial sequencing data: Taxonomy
Spreadsheet 6. 16S microbial sequencing data at kingdom level
Spreadsheet 7. 16S microbial sequencing data at phylum level
Spreadsheet 8. 16S microbial sequencing data at class level
Spreadsheet 9. 16S microbial sequencing data at order level
Spreadsheet 10. 16S microbial sequencing data at family level
Spreadsheet 11. 16S microbial sequencing data at genus level
Spreadsheet 12. 16S microbial sequencing data: U.UniFrac DM
Spreadsheet 13. 16S microbial sequencing data: W.UniFrac DM
Spreadsheet 14. 16S microbial sequencing data: U.UniFrac PCoA
Spreadsheet 15. 16S microbial sequencing data: W.UniFrac PCoA
Spreadsheet 16. 16S microbial sequencing data: U.UniFrac tSNE
Spreadsheet 17. 16S microbial sequencing data: W.UniFrac tSNE
Supplement Table S3. Altered Gut Microbiota Composition in Tak1∆M/∆M Mice by Metagenomic Sequencing (related to Figure 6)
Spreadsheet 1. Metagenomic sequencing data at superkingdom level
Spreadsheet 2. Metagenomic sequencing data at phylum level
Spreadsheet 3. Metagenomic sequencing data at class level
Spreadsheet 4. Metagenomic sequencing data at order level
Spreadsheet 5. Metagenomic sequencing data at family level
Spreadsheet 6. Metagenomic sequencing data at genus level
Spreadsheet 7. Metagenomic sequencing data at species level
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FC: Anti-mouse CD3 | Thermo Fisher | Clone: 17A2 |
| FC: Anti-mouse CD4 | Thermo Fisher | Clone: RM4-5 |
| FC: Anti-mouse CD8 | Thermo Fisher | Clone: 53-6.7 |
| FC: Anti-mouse IL-17A | Thermo Fisher | Clone: eBio17B7 |
| FC: Anti-mouse IL-4 | Thermo Fisher | Clone: BVD6-24G2 |
| FC: Anti-mouse Foxp3 | Thermo Fisher | Clone: FJK-16s |
| FC: Anti-mouse RORγt | Thermo Fisher | Clone: B2D |
| FC: Anti-mouse CD11b | Thermo Fisher | Clone: M1/70 |
| FC: Anti-mouse Ly-6G | Thermo Fisher | Clone: RB6-8C5 |
| FC: Anti-mouse F4/80 | Thermo Fisher | Clone: BM8 |
| FC: Anti-mouse CD45 | Thermo Fisher | Clone: 30-F11 |
| FC: Anti-mouse IFN-γ | BD Biosciences | Clone: XMG1.2 |
| FC: Anti-mouse TCRβ | Thermo Fisher | Clone: H57-597 |
| FC: Anti-mouse TCRγδ | Thermo Fisher | Clone: eBioGL3 |
| ELISA: Anti-mouse IL-1β purified | Thermo Fisher | Cat#: 14-7012-85, RRID: AB_468397 |
| ELISA: Anti-mouse IL-1β biotin | Thermo Fisher | Cat#: 13-7112-85, RRID: AB_466925 |
| ELISA: Anti-mouse IL-6 purified | Thermo Fisher | Cat#: 14-7061-85, RRID: AB_468423 |
| ELISA: Anti-mouse IL-6 biotin | Thermo Fisher | Cat#: 13-7062-85, RRID: AB_466911 |
| ELISA: Anti-mouse IL-17A purified | Thermo Fisher | Cat#: 14-7175-85, RRID: AB_763569 |
| ELISA: Anti-mouse IL-17A biotin | Thermo Fisher | Cat#: 13-7177-85, RRID: AB_763571 |
| IHC: Anti-mouse Claudin 3 | Thermo Fisher | Cat#: 34-1700, RRID: AB_2533158 |
| IHC: Anti-mouse Zonulin (Haptoglobin) | Bioss | Cat#: BS-1808R, RRID: AB_10856942 |
| IHC: Anti-mouse Occludin | Bioss | Cat#: BS-1495R, RRID: AB_10855189 |
| IHC: Anti-mouse Ki-67 | Cell Signaling | Cat#: 12202, RRID: AB_2620142 |
| IHC: HRP-linked rabbit 2nd antibody | DAKO | Cat#: K4003, RRID: AB_2630375 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dextran Sulfate Sodium Salt (DSS) | MP Biomedicals | Cat#: 02160110 |
| Azoxymethane (AOM) | Sigma-Aldrich | Cat#: A5486 |
| Ampicillin | Thermo Fisher | Cat#: BP1760-25 |
| Neomycin | Thermo Fisher | Cat#: BP266925 |
| Metronidazole | Thermo Fisher | Cat#: ICN15571025 |
| Vancomycin | Thermo Fisher | Cat#: BP29581 |
| Percoll centrifugation media | GE Healthcare | Cat#: 17089101 |
| Collagenase IV | Sigma-Aldrich | Cat#: C5138 |
| Poly-HRP Streptavidin | Thermo Fisher | Cat#: N200 |
| TMB (3,3’,5,5’-Tetramethylbenzidine) | Sigma-Aldrich | Cat#: T5525 |
| Critical Commercial Assays | ||
| Alcian Blue/PAS Stain Kit | Newcomer Supply | Cat#: 91022A |
| Dynabeads Untouched Mouse CD4 Cells Kit | Thermo Fisher | Cat#: 11415D |
| QIAamp Fast DNA Stool Mini Kit | QIAGEN | Cat#: 51604 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#: 554715 |
| DAB Peroxidase (HRP) Substrate Kit | Vector Laboratories | Cat#: SK-4100, RRID: AB_2336382 |
| Mouse IL-22 Uncoated ELISA Kit | Thermo Fisher | Cat#: 88-7422-88, RRID: AB_2575121 |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT: C57BL/6J | The Jackson Laboratory | Cat# JAX:000664, RRID: IMSR_JAX:000664 |
| Mouse: Il1r1−/−: B6.129S7-Il1r1tm1Imx/J | The Jackson Laboratory | Cat# JAX:003245, RRID: IMSR_JAX:003245 |
| Mouse: Il6−/−: B6;129S2-Il6tm1Kopf/J | The Jackson Laboratory | Cat# JAX: 002254, RRID: IMSR_JAX: 002254 |
| Mouse: Lyz2-Cre: B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Cat# JAX: 004781, RRID: IMSR_JAX: 004781 |
| Mouse: Il17-GFP+ | Laboratory of Scott Durum | N/A |
| Mouse: Tak1flox/flox | Laboratory of Michael Schneider | N/A |
| Mouse: Rorc−/−: B6.129P2(Cg)-Rorctm2Litt/J | The Jackson Laboratory | Cat# JAX: 007572, RRID: IMSR_JAX:007572 |
| Mouse: Rag1−/−: B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | Cat# JAX: 002216, RRID: IMSR_JAX:002216 |
| Mouse: Germ-free WT C57BL/6 | Taconic Biosciences | Cat# Taconic: GF-B6 |
| Mouse: Il17ra−/−: B6.Cg-Il17ratm2.2Koll/J | The Jackson Laboratory | Cat# JAX: 033431, RRID: IMSR_JAX: 033431 |
| Bacterial Strains | ||
| Odoribacter splanchnicus | ATCC | 29572 |
| Bacteroides sp. 4_1_36 | BEI Resources | HM-258 |
| Bacteroides sp. D20 | BEI Resources | HM-189 |
| Bacteroides uniformis | ATCC | 8492 |
| Nontoxigenic Bacteroides fragilis (NTBF) | Laboratory of Cynthia Sears | N/A |
| Oligonucleotides | ||
| Genotyping (Tak1flox/flox) Common: 5’-GCACAGAAAATGCACAGTGCTC-3’ | (Liu et al., 2006) | N/A |
| Genotyping (Tak1flox/flox) WT: 5’-GCTTGGGACAGGCTGGTAAAG-3’ | (Liu et al., 2006) | N/A |
| Genotyping (Tak1flox/flox) Mutant: 5’-CTTACAAGCCGAATTCCAGCA-3’ | (Liu et al., 2006) | N/A |
| Genotyping (Tak1flox/flox) Excised: 5’-CTCCTCCACTCCGCCCCTAC-3’ | (Liu et al., 2006) | N/A |
| Genotyping (Il1r1) F-WT: 5’-GGTGCAACTTCATAGAGAGATGA-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il1r1) F-Mutant: 5’-CTCGTGCTTTACGGTATCGC-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il1r1) R-Common: 5’-TTCTGTGCATGCTGGAAAAC-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il6) F-Common: 5’-TTCCATCCAGTTGCCTTCTTGG-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il6) R-WT: 5’-TTCTCATTTCCACGATTTCCCAG-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il6) R-Mutant: 5’-CCGGAGAACCTGCGTGCAATCC-3’ | The Jackson Laboratory | N/A |
| Genotyping (Lyz2-Cre) Mutant: 5’-CCCAGAAATGCCAGATTACG-3’ | The Jackson Laboratory | N/A |
| Genotyping (Lyz2-Cre) Common: 5’-CTTGGGCTGCCAGAATTTCTC-3’ | The Jackson Laboratory | N/A |
| Genotyping (Lyz2-Cre) WT: 5’-TTACAGTCGGCCAGGCTGAC-3’ | The Jackson Laboratory | N/A |
| Genotyping (Rag1) F: 5’-TCTGGACTTGCCTCCTCTGT-3’ | The Jackson Laboratory | N/A |
| Genotyping (Rag1) R: 5’-CATTCCATCGCAAGACTCCT-3’ | The Jackson Laboratory | N/A |
| Genotyping (Rorc) Common F: 5’-CCCCCTGCCCAGAAACACT-3’ | The Jackson Laboratory | N/A |
| Genotyping (Rorc) WT R: 5’-GGATGCCCCCATTCACTTACTTCT-3’ | The Jackson Laboratory | N/A |
| Genotyping (Rorc) Mutant R: 5’-CGGACACGCTGAACTTGTGG-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il17-GFP) F: 5’-GACTTCAAGGAGGACGGCAACAT-3’ | Laboratory of Scott Durum | N/A |
| Genotyping (Il17-GFP) R: 5’-GAGAGACCATGATGGTCACTGGA-3’ | Laboratory of Scott Durum | N/A |
| Genotyping (Il17ra) WT F: 5’-GCATCCCAGACCAGCATTAG-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il17ra) WT R: 5’-CCAGCTGTCATCCAGACAAG-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il17ra) Mutant F: 5’-GCCTCCAAGTCTAGCTTTGCT-3’ | The Jackson Laboratory | N/A |
| Genotyping (Il17ra) Mutant R: 5’-CCTAAGGAGGTCCGGAATGT-3’ | The Jackson Laboratory | N/A |
| QPCR (OS) F: 5’-ATGTAATGATGAGCACTCTAACGG-3’ | (Tong et al., 2011) | N/A |
| QPCR (OS) R: 5’-GGCTTTTGAGATTGGCATCC-3’ | (Tong et al., 2011) | N/A |
| QPCR (Universal bacteria) F: 5’-ACTCCTACGGGAGGCAGCAGT-3’ | (Ayres et al., 2012) | N/A |
| QPCR (Universal bacteria) R: 5’-ATTACCGCGGCTGCTGGC-3’ | (Ayres et al., 2012) | N/A |
| QPCR (Bacteroidetes) F: 5’-CATGTGGTTTAATTCGATGAT-3’ | (Queipo-Ortuno et al., 2012) | N/A |
| QPCR (Bacteroidetes) R: 5’-AGCTGACGACAACCATGCAG-3’ | (Queipo-Ortuno et al., 2012) | N/A |
| QPCR (Bacteroides) F: 5’-GGTTCTGAGAGGAGGTCCC-3’ | (Ayres et al., 2012) | N/A |
| QPCR (Bacteroides) R: 5’-GCTGCCTCCCGTAGGAGT-3’ | (Ayres et al., 2012) | N/A |
| Deposited Data | ||
| 16S and shotgun metagenomics raw sequence data | This paper | NCBI SRA: PRJNA495072 |
| Software and Algorithms | ||
| GraphPad Prism 5 | GraphPad Software | N/A |
| ATIMA (Agile Toolkit for Incisive Microbial Analyses) | Alkek Center for Metagenomics and Microbiome Research, Baylor College of Medicine | http://atima.jplab.net |
| R software v3.2.1 | The R Foundation for Statistical Computing | https://cran.r-project.org/ |
HIGHLIGHTS.
Myeloid-specific Tak1-deficient mice are completely resistant to colitis and CRC
Specific microbiota in Tak1∆M/∆M mice plays a key role in protective immunity
Innate immune pathways and gut Th17 cells are required for protective immunity
O. splanchnicus alone induces Th17 cells and confers resistance to colitis and CRC
ACKNOWLEDGMENTS
We thank Michael Schneider (Baylor College of Medicine) for providing Tak1flox/flox mice, Scott Durum (National Institutes of Health) for Il17-GFP+ mice; Cynthia Sears (Johns Hopkins University) for providing NTBF; Wei Wang (MD Anderson Cancer Center) for histopathological scoring; Julian Hurdle (Texas A&M University) for anaerobic bacteria culture; Chen Qian, Xin Liu, Linfeng Li, Bo Ning, Qingtian Li, and Xilai Ding (Houston Methodist Research Institute) for technical advices; Changfu Yao (Cedars-Sinai Medical Center) and Pengfei Liu (Baylor College of Medicine) for scientific advices; Chongming Jiang, Huahui Ren, and Kaifu Chen (Houston Methodist Research Institute) for support on data analysis; Kalpana Mujoo and John Gilbert for scientific editing. This work was in part supported by grants from the NCI, NIH (R01CA101795 and 1U54CA210181-01), and Department of Defense (DoD) CDMRP BCRP (BC151081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ajibade AA, Wang HY, and Wang RF (2013). Cell type-specific function of TAK1 in innate immune signaling. Trends in immunology 34, 307–316. [DOI] [PubMed] [Google Scholar]
- Ajibade AA, Wang Q, Cui J, Zou J, Xia X, Wang M, Tong Y, Hui W, Liu D, Su B, et al. (2012). TAK1 negatively regulates NF-kappaB and p38 MAP kinase activation in Gr-1+CD11b+ neutrophils. Immunity 36, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander KL, Targan SR, and Elson CO 3rd (2014). Microbiota activation and regulation of innate and adaptive immunity. Immunol Rev 260, 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D, and Spits H (2015). The biology of innate lymphoid cells. Nature 517, 293–301. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Trinidad NJ, and Vance RE (2012). Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nature medicine 18, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter NT, Zackular JP, Chen GY, and Schloss PD (2014). Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluestone JA, Mackay CR, O’Shea JJ, and Stockinger B (2009). The functional plasticity of T cell subsets. Nature reviews Immunology 9, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, et al. (2009). gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer cell 15, 91–102. [DOI] [PubMed] [Google Scholar]
- Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, and Bothwell AL (2010). Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 107, 5540–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JL, Wu S, Geis AL, Chan GV, Gomes TAM, Beck SE, Wu X, Fan H, Tam AJ, Chung L, et al. (2019). Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal immunology 12, 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, and Maloy KJ (2012). IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. The Journal of experimental medicine 209, 1595–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C (2008). TH17 cells in development: an updated view of their molecular identity and genetic programming. Nature reviews Immunology 8, 337–348. [DOI] [PubMed] [Google Scholar]
- Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, and Flavell RA (2013). Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nature reviews Cancer 13, 759–771. [DOI] [PubMed] [Google Scholar]
- Erben U, Loddenkemper C, Doerfel K, Spieckermann S, Haller D, Heimesaat MM, Zeitz M, Siegmund B, and Kuhl AA (2014). A guide to histomorphological evaluation of intestinal inflammation in mouse models. International journal of clinical and experimental pathology 7, 4557–4576. [PMC free article] [PubMed] [Google Scholar]
- Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W Jr., Rongvaux A, Van Rooijen N, Haberman AM, et al. (2011). Control of TH17 cells occurs in the small intestine. Nature 475, 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. (2017). Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943 e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. (2010). Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Navajas JM, Law J, Nguyen KP, Bhargava M, Corr MP, Varki N, Eckmann L, Hoffman HM, Lee J, and Raz E (2010). Interleukin 1 receptor signaling regulates DUBA expression and facilitates Toll-like receptor 9-driven antiinflammatory cytokine production. The Journal of experimental medicine 207, 2799–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. (2009). IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, and Jones SA (2015). IL-6 as a keystone cytokine in health and disease. Nature immunology 16, 448–457. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jess T, Rungoe C, and Peyrin-Biroulet L (2012). Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 10, 639–645. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, and Kuchroo VK (2009). IL-17 and Th17 Cells. Annual review of immunology 27, 485–517. [DOI] [PubMed] [Google Scholar]
- Lee JS, Tato CM, Joyce-Shaikh B, Gulan F, Cayatte C, Chen Y, Blumenschein WM, Judo M, Ayanoglu G, McClanahan TK, et al. (2015). Interleukin-23-Independent IL-17 Production Regulates Intestinal Epithelial Permeability. Immunity 43, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, et al. (2020). Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 180, 79–91 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32, 834–841. [DOI] [PubMed] [Google Scholar]
- Littman DR, and Rudensky AY (2010). Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858. [DOI] [PubMed] [Google Scholar]
- Liu HH, Xie M, Schneider MD, and Chen ZJ (2006). Essential role of TAK1 in thymocyte development and activation. Proceedings of the National Academy of Sciences of the United States of America 103, 11677–11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopetuso LR, Chowdhry S, and Pizarro TT (2013). Opposing Functions of Classic and Novel IL-1 Family Members in Gut Health and Disease. Frontiers in immunology 4, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz SD, and Bertagnolli MM (2009). Molecular origins of cancer: Molecular basis of colorectal cancer. The New England journal of medicine 361, 2449–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, Stevens E, Bigler J, Davis JA, Rottman JB, et al. (2015). Differential Roles for Interleukin-23 and Interleukin-17 in Intestinal Immunoregulation. Immunity 43, 739–750. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, and Cua DJ (2008). Th17 cell differentiation: the long and winding road. Immunity 28, 445–453. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, and Papaioannou VE (1992). RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877. [DOI] [PubMed] [Google Scholar]
- Neurath MF (2014). Cytokines in inflammatory bowel disease. Nature reviews Immunology 14, 329–342. [DOI] [PubMed] [Google Scholar]
- O’Connor W Jr., Kamanaka M, Booth CJ, Town T, Nakae S, Iwakura Y, Kolls JK, and Flavell RA (2009). A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nature immunology 10, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A, Andoh A, Araki Y, Bamba T, and Fujiyama Y (2004). Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin Immunol 110, 55–62. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Kolls JK, and Zheng Y (2008). The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28, 454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queipo-Ortuno MI, Boto-Ordonez M, Murri M, Gomez-Zumaquero JM, Clemente-Postigo M, Estruch R, Cardona Diaz F, Andres-Lacueva C, and Tinahones FJ (2012). Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am J Clin Nutr 95, 1323–1334. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, and Medzhitov R (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. [DOI] [PubMed] [Google Scholar]
- Rooks MG, and Garrett WS (2016). Gut microbiota, metabolites and host immunity. Nature reviews Immunology 16, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, et al. (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028 e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, et al. (2015). An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 163, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, and Pardoll DM (2018). The intestinal microbiome influences checkpoint blockade. Nat Med 24, 254–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Dai D, He X, Zhu S, Yao Y, Gao H, Wang J, Qu F, Qiu J, Wang H, et al. (2015). Growth Factor FGF2 Cooperates with Interleukin-17 to Repair Intestinal Epithelial Damage. Immunity 43, 488–501. [DOI] [PubMed] [Google Scholar]
- Spranger S, Dai D, Horton B, and Gajewski TF (2017). Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer cell 31, 711–723 e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, and Mizoguchi A (2008). IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. The Journal of clinical investigation 118, 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana NK, and Kasper DL (2017). Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et al. (2019). A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605. [DOI] [PubMed] [Google Scholar]
- Tenesa A, and Dunlop MG (2009). New insights into the aetiology of colorectal cancer from genome-wide association studies. Nature reviews Genetics 10, 353–358. [DOI] [PubMed] [Google Scholar]
- Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, and Letellier E (2020). Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism? Trends in microbiology 28, 401–423. [DOI] [PubMed] [Google Scholar]
- Terzic J, Grivennikov S, Karin E, and Karin M (2010). Inflammation and colon cancer. Gastroenterology 138, 2101–2114 e2105. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Zmora N, Levy M, and Elinav E (2016). The microbiome and innate immunity. Nature 535, 65–74. [DOI] [PubMed] [Google Scholar]
- Tong J, Liu C, Summanen P, Xu H, and Finegold SM (2011). Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe 17, 64–68. [DOI] [PubMed] [Google Scholar]
- Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B, Betz KS, Penz-Oesterreicher M, Bjorkdahl O, Fox JG, et al. (2008). Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer cell 14, 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YY, Chi H, Xie M, Schneider MD, and Flavell RA (2006). The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nature immunology 7, 851–858. [DOI] [PubMed] [Google Scholar]
- Wang K, Kim MK, Di Caro G, Wong J, Shalapour S, Wan J, Zhang W, Zhong Z, Sanchez-Lopez E, Wu LW, et al. (2014a). Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 41, 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, and Wang HY (2017). Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell research 27, 11–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang K, Han GC, Wang RX, Xiao H, Hou CM, Guo RF, Dou Y, Shen BF, Li Y, et al. (2014b). Neutrophil infiltration favors colitis-associated tumorigenesis by activating the interleukin-1 (IL-1)/IL-6 axis. Mucosal immunology 7, 1106–1115. [DOI] [PubMed] [Google Scholar]
- Weaver CT, and Hatton RD (2009). Interplay between the Th17 and Treg cell lineages: a (co-)evolutionary perspective. Nature reviews Immunology 9, 883–889. [DOI] [PubMed] [Google Scholar]
- West NR, McCuaig S, Franchini F, and Powrie F (2015). Emerging cytokine networks in colorectal cancer. Nature reviews Immunology 15, 615–629. [DOI] [PubMed] [Google Scholar]
- Wirtz S, Neufert C, Weigmann B, and Neurath MF (2007). Chemically induced mouse models of intestinal inflammation. Nature protocols 2, 541–546. [DOI] [PubMed] [Google Scholar]
- Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, et al. (2017). Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 22, 25–37 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15, 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, and Podolsky DK (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434. [DOI] [PubMed] [Google Scholar]
- Xu ZS, Zhang HX, Li WW, Ran Y, Liu TT, Xiong MG, Li QL, Wang SY, Wu M, Shu HB, et al. (2019). FAM64A positively regulates STAT3 activity to promote Th17 differentiation and colitis-associated carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America 116, 10447–10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z, et al. (2008). Regulation of inflammatory responses by IL-17F. The Journal of experimental medicine 205, 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, Ayyoub M, Routy B, and Kroemer G (2016). Microbiome and Anticancer Immunosurveillance. Cell 165, 276–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table S1. Altered Gut Microbiota Composition in Tak1∆M/∆M Mice Are Normalized by Cohousing with Control WT Mice (related to Figures 2 and 3)
Spreadsheet 1. 16S microbial sequencing data: Reads per sample
Spreadsheet 2. 16S microbial sequencing data: Raw OTU counts
Spreadsheet 3. 16S microbial sequencing data: Rarefied OTU counts
Spreadsheet 4. 16S microbial sequencing data: Alpha Diversity
Spreadsheet 5. 16S microbial sequencing data: Taxonomy
Spreadsheet 6. 16S microbial sequencing data at kingdom level
Spreadsheet 7. 16S microbial sequencing data at phylum level
Spreadsheet 8. 16S microbial sequencing data at class level
Spreadsheet 9. 16S microbial sequencing data at order level
Spreadsheet 10. 16S microbial sequencing data at family level
Spreadsheet 11. 16S microbial sequencing data at genus level
Spreadsheet 12. 16S microbial sequencing data: U.UniFrac DM
Spreadsheet 13. 16S microbial sequencing data: W.UniFrac DM
Spreadsheet 14. 16S microbial sequencing data: U.UniFrac PCoA
Spreadsheet 15. 16S microbial sequencing data: W.UniFrac PCoA
Spreadsheet 16. 16S microbial sequencing data: U.UniFrac tSNE
Spreadsheet 17. 16S microbial sequencing data: W.UniFrac tSNE
Supplement Table S2. IL-1R1 and IL-6 Signaling Pathways are Required for Shaping Microbiota in Tak1∆M/∆M Mice (related to Figure 4)
Spreadsheet 1. 16S microbial sequencing data: Reads per sample
Spreadsheet 2. 16S microbial sequencing data: Raw OTU counts
Spreadsheet 3. 16S microbial sequencing data: Rarefied OTU counts
Spreadsheet 4. 16S microbial sequencing data: Alpha Diversity
Spreadsheet 5. 16S microbial sequencing data: Taxonomy
Spreadsheet 6. 16S microbial sequencing data at kingdom level
Spreadsheet 7. 16S microbial sequencing data at phylum level
Spreadsheet 8. 16S microbial sequencing data at class level
Spreadsheet 9. 16S microbial sequencing data at order level
Spreadsheet 10. 16S microbial sequencing data at family level
Spreadsheet 11. 16S microbial sequencing data at genus level
Spreadsheet 12. 16S microbial sequencing data: U.UniFrac DM
Spreadsheet 13. 16S microbial sequencing data: W.UniFrac DM
Spreadsheet 14. 16S microbial sequencing data: U.UniFrac PCoA
Spreadsheet 15. 16S microbial sequencing data: W.UniFrac PCoA
Spreadsheet 16. 16S microbial sequencing data: U.UniFrac tSNE
Spreadsheet 17. 16S microbial sequencing data: W.UniFrac tSNE
Supplement Table S3. Altered Gut Microbiota Composition in Tak1∆M/∆M Mice by Metagenomic Sequencing (related to Figure 6)
Spreadsheet 1. Metagenomic sequencing data at superkingdom level
Spreadsheet 2. Metagenomic sequencing data at phylum level
Spreadsheet 3. Metagenomic sequencing data at class level
Spreadsheet 4. Metagenomic sequencing data at order level
Spreadsheet 5. Metagenomic sequencing data at family level
Spreadsheet 6. Metagenomic sequencing data at genus level
Spreadsheet 7. Metagenomic sequencing data at species level
Data Availability Statement
The 16S and shotgun metagenomics raw sequence data reported in this paper were deposited in NCBI Sequence Read Archive (SRA) with the accession number of PRJNA495072.
Data analyses for microbiota sequencing are performed following published articles (described in detail in “QUANTIFICATION AND STATISTICAL ANALYSIS” section), in which the sources of code for computational data analysis pipeline are available.


