Abstract
Rationale and Objectives
Computer-aided methods have been widely applied to diagnose lesions on breast MRI. The first step was to identify abnormal areas. A deep learning Mask R-CNN was implemented to search the entire set of images and detect suspicious lesions.
Materials and Methods
Two DCE-MRI datasets were used, 241 patients acquired using non-fat-sat sequence for training, and 98 patients acquired using fat-sat sequence for testing. All patients have confirmed unilateral mass cancers. The tumor was segmented using FCM clustering algorithm to serve as the ground truth. Mask R-CNN was implemented with ResNet-101 as the backbone. The neural network output the bounding boxes and the segmented tumor for evaluation using the Dice Similarity Coefficient (DSC). The detection performance, and the trade-off between sensitivity and specificity, was analyzed using free response ROC.
Results
When the pre-contrast and subtraction image of both breasts were used as input, the false positive from the heart and normal parenchymal enhancements could be minimized. The training set had 1,469 positive slices (containing lesion) and 9,135 negative slices. In 10-fold cross-validation, the mean accuracy=0.86 and DSC=0.82. The testing dataset had 1,568 positive and 7,264 negative slices, with accuracy=0.75 and DSC=0.79. When the obtained per-slice results were combined, 240/241 (99.5%) lesions in the training and 98/98 (100%) lesions in the testing datasets were identified.
Conclusions
Deep learning using Mask R-CNN provided a feasible method to search breast MRI, localize, and segment lesions. This may be integrated with other AI algorithms to develop a fully-automatic breast MRI diagnostic system.
Keywords: Breast MRI, Fully-Automatic Detection, Deep Learning, Mask R-CNN
INTRODUCTION
Breast cancer is one of the most common cancers worldwide and a leading cause of cancer death in women. Detection and treatment in its early stage can increase patient survival (1). Breast MRI is a well-established clinical imaging modality for diagnosis of breast cancer, and also for screening of high-risk women (2–5).
In the clinical setting, the evaluation is done by radiologists’ visual interpretation. The suspicious abnormality should be identified first, and then further characterized for diagnosis. Since many sequences with thin slices are acquired to cover the entire breast with hundreds of images, radiologists need to spend time to carefully evaluate the entire dataset. Therefore, the reading is usually done with the assistance of computer-aided diagnosis (CAD) software, which is used to generate subtraction images, maximum intensity projection (MIP), color-coded DCE wash-out maps, and DCE time course, etc., and displays them together on the workstation for evaluation. The morphological and DCE temporal information is then interpreted by a radiologist and combined to determine the level of malignancy based on BI-RADS descriptors (6).
The diagnostic sensitivity and specificity of breast MRI can be affected by several factors, e.g. radiologists’ experience (7, 8), magnetic field strength (9), and DCE-MRI protocol (10–12). The current CAD can improve workflow efficiency and diagnostic accuracy, especially for patients with multiple or satellite lesions (13). Many CAD studies have further attempted to characterize abnormal lesions and give a final diagnosis (14–17). Most of them apply computer algorithms to extract features and build a diagnostic model, but not very successful due to the limited information provided by pre-defined features (18). For developing a fully-automatic CAD system, the first required task is to detect abnormal areas, which is rarely reported.
In recent years, artificial intelligence (AI) algorithms, particularly deep learning, have demonstrated remarkable progress in medical image analysis for performing many clinical tasks. Convolutional Neural Network (CNN) is a common deep learning method that can be applied to give a probability of malignancy for identified lesions (19,20). It can be further applied to perform a search in the entire MRI dataset to detect abnormal lesions (18,21,22). Patch-based CNN is used to discriminate whether each patch (a small portion of images) belongs to a lesion or not (21,23–25). Another approach uses CNN (26,27) or Mask Regional-Convolutional Neural Network (R-CNN) (28–30) to search the whole image or feature map to detect and localize the lesion.
The purpose of this study is to implement Mask R-CNN to search and detect suspicious lesions in breast MR images (29,30). The architecture provides a flexible and efficient framework for parallel evaluation of region proposal (attention), object detection, and segmentation (30–32). After the location of the lesion is detected, the tumor is further segmented, and the result is compared to the ground truth.
MATERIALS AND METHODS
Patients and datasets
The Institutional Review Board approved this retrospective study, and the requirement of informed consent was waived. The inclusion criteria were consecutive patients receiving breast MRI for diagnosis or staging of suspicious lesions, and confirmed to have histologically-proven cancer. Only patients with unilateral mass lesions in one breast were selected, to ensure that the contralateral breast was normal and could serve as a good reference based on symmetry. The exclusion criteria were patients who received prior procedure or treatment (e.g. neoadjuvant chemotherapy or hormonal therapy) before MRI. Two well-curated datasets were used to perform this study. A dataset obtained from one hospital with 241 patients (mean age 49 y/o, range 30–80 y/o) was used for training. Another dataset from a different hospital with 98 patients (mean age 49 y/o, range 22–67 y/o) was used for testing. Since only mass lesions were considered, the invasive ductal carcinoma (IDC) was the dominating histological subtype. The patient and tumor information, and the composition of positive and negative images in both datasets, are listed in Table 1.
Table 1:
Summary of lesions and image compositions in the training and testing datasets
| Training (N=241) | Testing (N=98) | |
|---|---|---|
| Age Range, years old | 30 – 80 (mean 49) | 22 – 67 (mean 49) |
| 1-D Tumor Size Range | 0.6 – 6 cm | 0.5 – 5 cm |
| Invasive Ductal Cancer (IDC) % | 82.0 % | 77.6 % |
| MR System (DCE sequence) | Siemens 3T (non-fat-sat) | Siemens 1.5 T (fat-sat) |
| Total Image Number | 10,604 | 8,832 |
| Positive Image Number (with lesion) | 1,469 | 1,568 |
| Negative Image Number (without lesion) | 9,135 | 7,264 |
| 2-D Tumor Area Range on Positive Slices | 0.35 – 13.1 (median 1.94) cm2 | 0.26 – 11.0 (median 2.42) cm2 |
MR protocols
For the training dataset, breast MRI was performed on a 3T scanner (Siemens Trio-Tim, Erlangen, Germany). The DCE-MRI consisted of 7 frames, including one pre-contrast and six post-contrast acquisitions using non-fat-sat sequence, with TR/TE=280/2.6 msec, flip angle=65°, matrix size=512×343, field of view=34cm, and slice thickness=3mm. The testing dataset was acquired using a 1.5T scanner (Siemens Magneton Skyra, Erlangen, Germany). DCE-MRI was acquired using a fat-suppressed three-dimensional fast low angle shot (3D-FLASH) sequence with one pre-contrast and four post-contrast frames, with TR/TE=4.50/1.82 msec, flip angle=12°, matrix size=512×512, field of view=32cm, and slice thickness=1.5mm.
Tumor Segmentation
The tumor was segmented on the subtraction image generated by subtracting the pre-contrast image from the 2nd DCE post-contrast image, using the fuzzy c-means (FCM) clustering algorithm (33). This DCE frame was selected based on the high tissue contrast between the boundary of the mass lesion (often showing strong peripheral enhancement) and the adjacent normal tissue. A square ROI was placed on maximum intensity projection to indicate the location. The tumor within the selected ROI was enhanced using an un-sharp filter with a 5×5 kernel constructed using the inverse of the two-dimensional Laplacian filter. FCM was applied to obtain the membership map of all voxels indicating the likelihood of each voxel belonging to the tumor or the non-tumor cluster. Then, the 3-dimensional connected-component labeling and whole filling was applied to finalize the ROI. The result was verified by an experienced radiologist and corrected if necessary. Based on the segmented tumor mask, the smallest bounding box covering the lesion was computed for evaluating the deep learning detection result.
Deep learning network
The deep learning detection algorithm was implemented using a custom architecture derived from the Mask R-CNN (29,30), shown in Figure 1. This method has been successfully implemented for the detection of hemorrhage on non-contrast brain CT, and detailed methods were described there (30). Firstly, various pre-defined shapes and distribution of bounding boxes were placed to identify a potential abnormality in the entire image. Then the bounding boxes were ranked based on the likelihood. Several bounding boxes on each slice with the highest probabilities were extracted to generate region proposals to locate specific regions. These composite region proposals were pruned using non-maximum suppression and used as input into a classifier to determine whether these regions belonged to lesion or non-lesion. For the detected lesion, a segmentation network was added to determine the tumor boundary with binary masks. In this study, we used ResNet101 as the feature pyramid network (FPN) to work as the backbone (34). The number of input channel was 3. The inputs from the FPN bottom-up pathway were added to the feature maps of the top-down pathway using a projection operation to match matrix dimensions, as shown in Figure 1. The final loss function was focal loss including a term for L2 regularization of the network parameters (35). All models were trained with Adam optimization. The learning rate was set to 0.0001 with momentum term 0.5 to stabilize training (36). This study was implemented in Python 3.6 using the open-source TensorFlow 1.4 library (Apache 2.0 license) (37). Experiments were performed on a GPU-optimized workstation with four NVIDIA GeForce GTX Titan X cards (12GB, Maxwell architecture).
Figure 1.

Mask R-CNN architecture. Hybrid 3D-contracting (middle block) and 2D-expanding (right block) fully convolutional feature-pyramid network architecture used for the mask R-CNN backbone. The architecture incorporates both traditional 3 ×3 filters as well as bottleneck 1×1–3×3–1×1 modules (left block). The contracting arm is composed of 3D operations and convolutional kernels. The number of input channel is 3.
Training and evaluation
In the training dataset, 10-fold cross-validation was used to evaluate the performance. The final trained network was applied to the independent dataset for testing. Since it was not reliable to detect a very small lesion < 3 mm, if a lesion was detected only on a single slice without involving any of the neighboring slices, it was dismissed.
Evaluation of tumor localization and segmentation
After a lesion was identified, the Intersection over Union (IoU), defined as the ratio between the predicted tumor bounding box and the ground truth box, was calculated. On the positive slice which contained lesion, the prediction was true positive (TP) if IoU was ≥ 0.5. The case with IoU < 0.5 was false negative (FN). On the image slice which did not contain lesion, if no bounding box was detected, the prediction was true negative (TN); if any lesion was detected, it was false positive (FP). After the per-slice results were obtained, they were combined to give per-lesion detection. If any positive slice in a lesion was identified, the lesion was determined as positive.
The detection performance was evaluated using the free response ROC analysis (38). First, the FROC was plotted using the TP rate (correctly localized lesions in all lesions) vs. the number of FP per image by varying the threshold, which could be used to assess the detection sensitivity at different levels of false-positives, and thus, to evaluate the trade-off. Then, the Alternative FROC (AFROC) was plotted using the TP rate vs. the probability of FP images in all negative images, and the AUC was extracted for evaluating the overall performance. To investigate the performance in lesions of different sizes, they were divided into two groups, above and below the median tumor area, and performed separate analysis. For each true positive lesion, the segmented tumor was compared to ground truth using the Dice Similarity Coefficient (DSC).
RESULTS
Determination of three inputs into network
The Mask R-CNN architecture allows 3 input channels. Initially, the pre-contrast image, post-contrast image, and the subtraction image of the diseased breast were used as inputs. Figure 2 shows an example acquired using the fat-sat sequence in the testing dataset, in which the strong parenchymal enhancements in bilateral breasts are identified as possible lesions. When the post-contrast image was replaced by the subtraction image from the contralateral normal breast, the symmetry could be used to eliminate the false detection of bilateral parenchymal enhancements. After exploration using different images as inputs and comparing the results, finally the three inputs were determined as pre-contrast image (used to identify chest region), and the subtraction images from the diseased breast and the contralateral normal breast.
Figure 2.

One case example from a 62-year-old patient with a small mass lesion in the testing dataset, who also shows strong parenchymal enhancements in both breasts. (a) Pre-contrast image acquired using fat-sat sequence; (b) The post-contrast image; (c) The subtraction image; (d) Tumor detection result searched by the algorithm when using pre-contrast, post-contrast and subtraction images as inputs. Two large blue boxes are the detection output from Mask R-CNN. The box in the left breast (right side of image) correctly encloses the cancer, but it also contains the surrounding parenchymal enhancements much larger than the size of the cancer. Another blue box in the right breast (left side of image) wrongly detects the parenchymal enhancements, thus a false positive result. (e-g) When the subtraction image, contralateral subtraction image, and pre-contrast images are used as inputs, the small cancer is correctly diagnosed with probability=0.87.
Performance in the training dataset
For the training set, the performance was evaluated using 10-fold cross-validation, 9-fold for training and 1-fold for validation. The final results from all cases were combined. Based on the IoU, there were 1,245 TP, 7,834 TN, 1,301 FP, and 224 FN cases. The sensitivity of tumor detection was 0.85, the specificity was 0.86, and the overall accuracy was 0.86. In the 1,245 true positives, the tumor was segmented and compared to the ground truth to calculate the DSC. The mean value was 0.82, ranging from 0.64 to 0.97 in 10-fold cross-validation. All performance results are summarized in Table 2. Figures 3–5 show three case examples in the training dataset acquired using the non-fat-sat sequence, illustrating different detection results.
Table 2:
Summary of lesion detection and segmentation performance using Mask R-CNN in the training and testing datasets, using per-slice based analysis
| Training (N=241) | Testing (N=98) | |
|---|---|---|
| Per-Lesion Detection Rate | 240/241 (99.5%) | 98/98(100%) |
| Per-Slice Detection Sensitivity (%) | 1,245 / 1,469 (85%) | 1,254 / 1,568 (80%) |
| Per-Slice Detection Specificity (%) | 7,834 / 9,135 (86%) | 5,396 / 7,264 (74%) |
| Per-Slice Detection Accuracy (%) | 9,079 / 10,604 (86%) | 6,650 / 8,832 (75%) |
| Lesion Segmentation Dice Score Range | 0.64 – 0.97 (mean 0.82) | 0.31 – 0.97 (mean 0.79) |
| AFROC AUC (all lesions) | 0.82 | 0.71 |
| AFROC AUC (large lesions above median) | 0.93 | 0.85 |
| AFROC AUC (small lesions below median) | 0.67 | 0.54 |
| FROC Sensitivity at 2 FP/Image (all lesions) | 0.83 | 0.81 |
| FROC Sensitivity at 2 FP/Image (large lesions) | 0.94 | 0.88 |
| FROC Sensitivity at 2 FP/Image (small lesions) | 0.69 | 0.63 |
Figure 3.

True positive case example from a 41-year-old patient with a strongly enhanced mass lesion in the training dataset. (a) Post-contrast image acquired using non-fat-sat sequence; (b) The subtraction image; (c) Tumor detection result searched by the algorithm. The segmented tumor is highlighted by green color, and used as the ground truth. The red box is the output from Mask R-CNN, which correctly detects the location of the cancer with probability=0.99, a true positive result. (d) The subtraction image of the contralateral normal breast. (e) The pre-contrast image, used as one input to identify the breast region, so the enhancements from the heart can be excluded. (c-e) are used as the 3 inputs into the Mask R-CNN.
Figure 5.
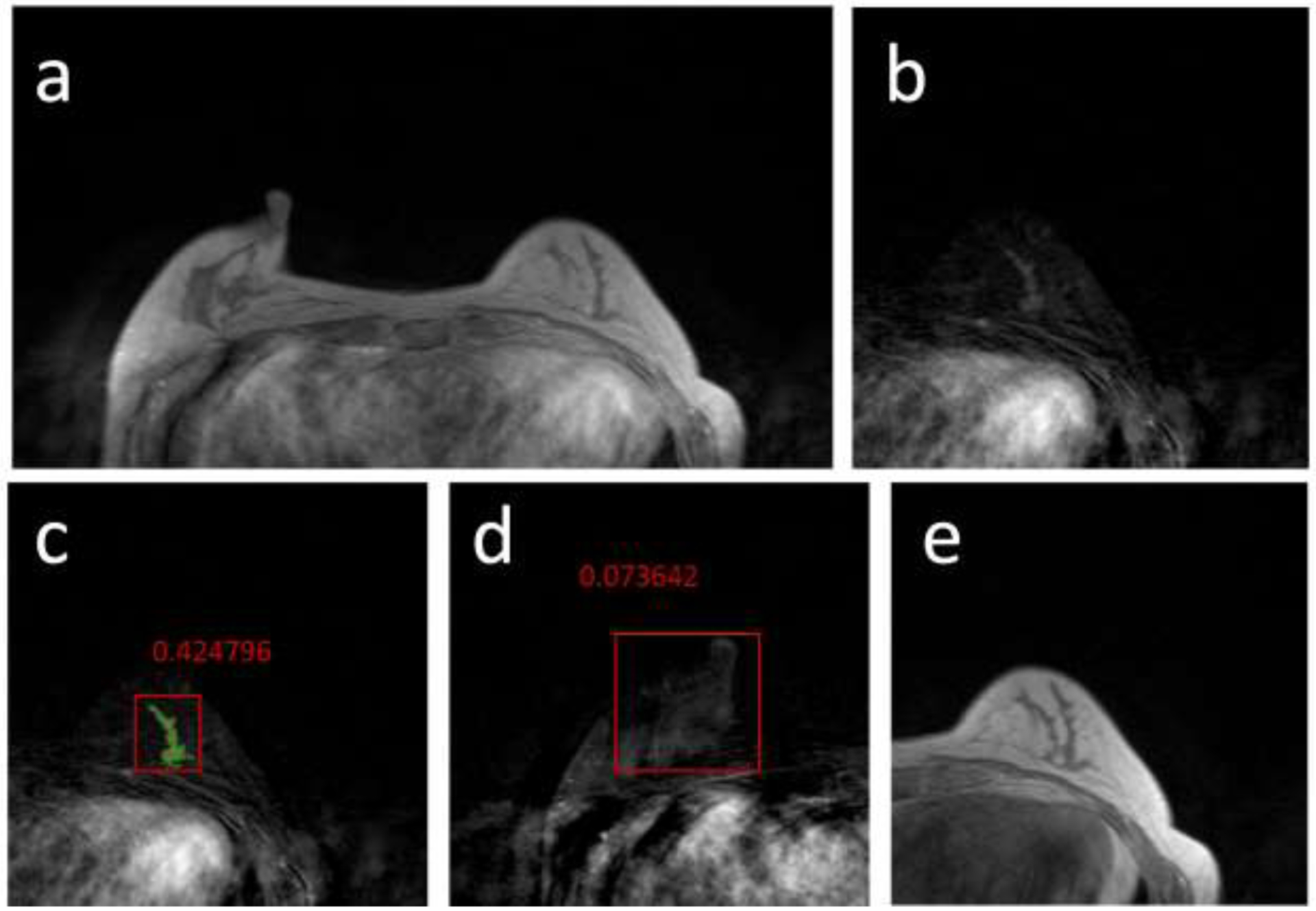
False Negative case example from a 57-year-old patient with a mildly enhanced, but pathologically confirmed cancer in the training dataset. (a) Post-contrast image acquired using non-fat-sat sequence; (b) The subtraction image of the breast which contains the tumor; (c) Tumor detection result searched by the algorithm. The segmented tumor is highlighted by green color, and used as the ground truth. The red box is the result of Mask R-CNN, with the malignant probability=0.42, a false negative result. However, other slices of this lesion show probability >0.5, so the FN detection on this slice does not affect the final per-lesion accuracy. (d) The subtraction image of the contralateral normal breast. A true negative result, with probability=0.08, is marked as an example. (e) The pre-contrast image, used as one input to identify the breast region.
Performance in the testing dataset
The testing dataset was acquired using the fat-sat sequence, and the difference can be compared between Figure 2 and Figures 3–5. Although the tissue contrast in the breast on images acquired using fat-sat and non-fat-sat sequences is different, the contrast in the junction between the chest wall muscle and the lung is similar. The other two inputs are subtraction images, which do not vary much between fat-sat and non-fat-sat sequences. Therefore, the model developed from the training set was directly applied to the testing set. The results showed 1,254 TP, 5,396 TN, 1,895 FP, and 314 FN slices. The sensitivity was 0.80, the specificity was 0.74, and the overall accuracy was 0.75. In true positive cases, the range of DSC for segmented tumors was 0.31–0.97, with the mean of 0.79.
Factors associated with false detection
To understand the possible factors leading to false predictions, we further analyzed tumor size, tumor enhancement, parenchymal enhancement, and tumor locations in the different diagnostic groups. The small tumor was difficult to be detected, which was the main reason for FN prediction. The mean tumor area calculated from all slices was significantly larger in TP compared to FN groups (area 3.55 cm2 vs. 0.42 cm2, p<0.01). Although we used subtraction images of both breasts as inputs, and indeed it helped to eliminate the parenchymal enhancements that showed clear symmetry (as shown in Figure 2), yet not all parenchymal enhancements could be eliminated in this way, especially for the enhanced tissues in small areas, as illustrated in the false positive example shown in Figure 4d. For the tumor segmentation, the difference between the predicted tumor and the ground truth was mainly coming from the parenchymal enhancement, especially for cases with severe field inhomogeneity from a strong bias field.
Figure 4.
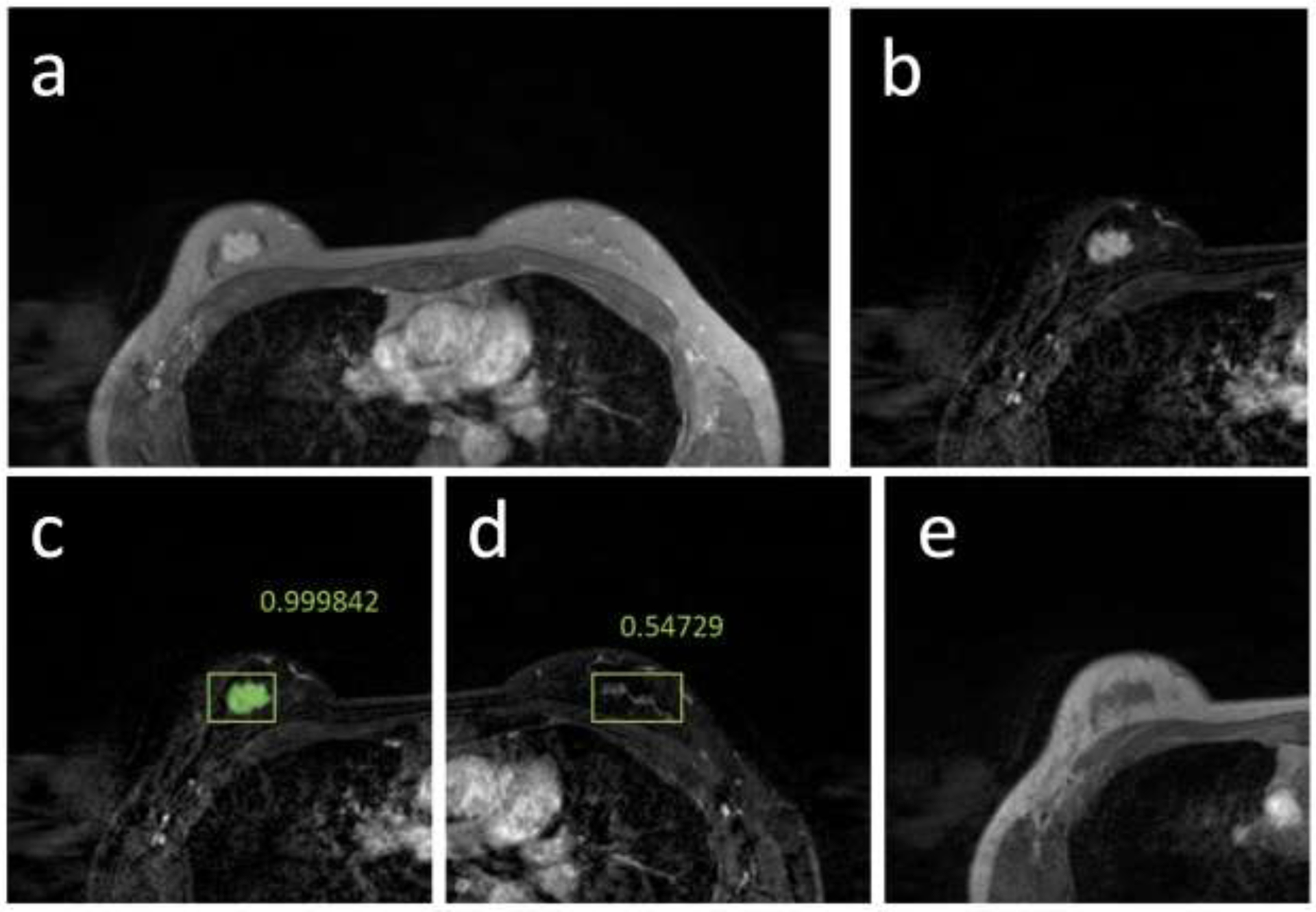
True positive and false positive case example from a 39-year-old patient with a strongly enhanced mass lesion in the training dataset. (a) Post-contrast image acquired using non-fat-sat sequence; (b) The subtraction image; (c) Tumor detection result searched by the algorithm. The red box is the output from Mask R-CNN, which correctly detects the location of the cancer with probability=0.99. (d) The subtraction image of the contralateral normal breast. In this breast, an area with probability=0.54 is detected, a false positive result. (e) The pre-contrast image, used as one input to identify the breast region.
Free Response ROC analysis
The FROC and Alternative FROC (AFROC) results are shown in Figure 6. In FROC, when using the false positive of 2 lesions per image as the reference, the detection sensitivity was 0.83 in the training set and 0.81 in the testing set. The AUC in AFROC was 0.82 in the training and 0.71 in the testing sets, respectively. We also performed analyses in large and small lesion groups, separated using the median tumor area, 1.94 cm2 in the training and 2.42 cm2 in the testing datasets, and the results are listed in Table 2. As expected, the performance was better in the large lesion group.
Figure 6.
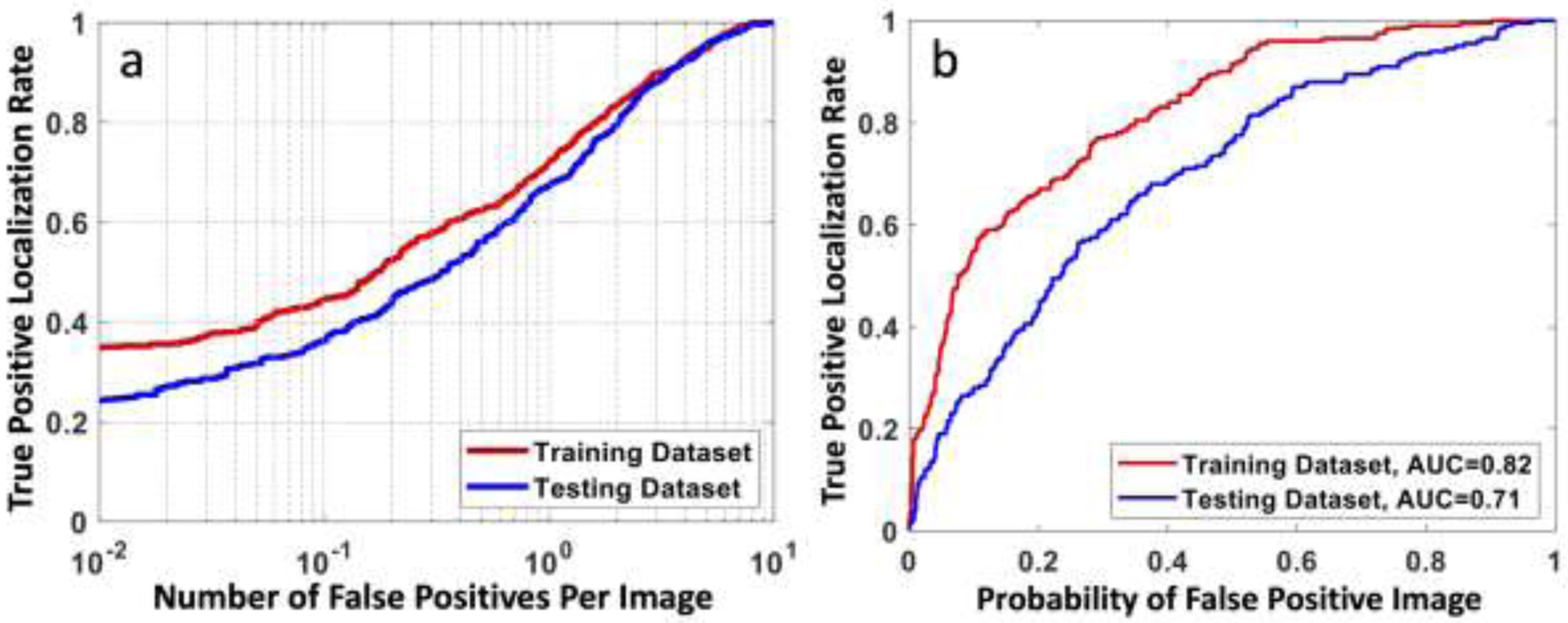
(a) The free response ROC curve calculated using all lesions in the training and testing datasets, by plotting the true positive rate (correctly localized lesions in all lesions) vs. the number of false-positives per image. When using the false positive of 2 lesions per image as the reference, the detection sensitivity is 0.83 in the training set and 0.81 in the testing set. (b) The Alternative FROC (AFROC) curve by plotting the true positive rate vs. the probability of false-positive images in all negative images.
Per-Patient diagnosis
All deep learning analysis was done using per-slice basis. The detected lesions from all slices were combined to give per-patient diagnosis. This was usually done based on the most aggressive finding. If there was a positive detection with IoU ≥ 0.5 found on any slice in a lesion, the lesion was determined as a cancer. In the training set, 240/241 cancers were identified, and the detection sensitivity was 99.5%. The only missing case as one mildly enhanced 7 mm DCIS. In the testing set, all 98/98 cancers were identified, with sensitivity of 100%. Therefore, although the sensitivity analyzed using per-slice basis in the training and testing datasets was 0.80–0.85, when the results were combined to give per-lesion diagnosis, the sensitivity was approaching 100%.
DISCUSSION
In this study, we implemented a fully automatic deep learning method using Mask R-CNN for detection of breast cancer by searching the entire set of MR images. Many studies have investigated the value of machine learning, including radiomics and deep learning, for differentiation of benign and malignant lesions. These studies mainly focused on characterization of already identified abnormal lesions. The detection was a much more challenging task, especially in MRI where many images were acquired to cover the entire breast. Our results showed that Mask R-CNN was a feasible method. In per-slice analysis, the mean accuracy was 0.86 and 0.75 in the training and testing datasets, respectively. When the per-slice results were combined to give per-lesion results, the cancer detection sensitivity was 99.5% in the training dataset and 100% in the testing dataset. In detected lesions, the segmented tumor was also in good agreement with the ground truth, with DSC of 0.82 in the training dataset and 0.79 in the testing dataset.
The chest region includes the enhancement from the heart. While it is very easy for a human reader to dismiss this, the task is difficult for the computer. One commonly used strategy is to segment the breast first and only perform the search within the breast (21,27), but this requires one more pre-processing step and not easy to achieve a clean breast segmentation. Deep learning offers a fully-automatic strategy. We demonstrated that by including the pre-contrast image as one input, which demarcated the background and the chest region well, it provided anatomic information and helped to dismiss enhancements from the heart in the chest region. The results also demonstrated that by including the contralateral subtraction image as one input, it helped to eliminate false positives coming from parenchymal enhancements, as shown in Figure 2. Using bilateral breast symmetry is very important in radiologists’ visual interpretation, and it can be implemented in deep learning as well, by including the contralateral breast as one input.
Deep learning is an emerging method that has been shown capable of searching and detecting abnormalities in pathology images. For example, Bejnordi et al. (39) used multiple AI methods to detect breast cancer lymph node metastases on pathology whole-slide images. For radiology, the earliest application is for detecting pneumonia on chest X-Ray (40). For breast lesions, most studies were for 2D mammography and then extended to DBT. Kooi et al. applied a patch-based method to mammography (24), which divided the whole image into many small portions for local recognition. Samala et al. applied the method to DBT by using a pre-trained model from mammography (25). Besides the patch-based method, another feasible method is weakly supervised learning. Kim et al. utilized this method to detect and localize lesions from the 4-view digital mammograms (26), similar to the reading of radiologists in clinics. A residual neural network was implemented with input of 4 views. The feature maps before the global pooling layers were extracted to give the probability maps, indicating the detected lesion location and the level of suspicion. After the lesion was identified, it could be further segmented and characterized to make a diagnosis as benign or malignant. This streamlined procedure has been implemented as a commercial product. Ribli et al. implemented the faster R-CNN algorithm using VGG16 as a backbone network to detect lesions on digital mammograms (28) and reached a sensitivity of 0.9.
For lesion detection on breast MRI, because many images were acquired with different pulse sequences, it was much more challenging compared to the detection on mammography and DBT. Wang et al. (41) designed a Siamese Network to detect metastatic lesions in the spine using the patch-based method. Dalmış et al. applied the patch-based method to localize breast lesions on DCE-MRI (21). The candidate areas for patch extraction were first identified by U-net, and then a Siamese neural network was applied for detection, using the 3D patch and the symmetrical patch from the contralateral breast as inputs. This method obtained a sensitivity of 0.83 for mass tumors. Although this work was for breast MRI, the main goal was to characterize the identified candidate lesions and make correct diagnosis as cancers vs. benign/normal tissues, thus the major effort was to solve a diagnosis problem not a detection problem. Interestingly, they also included the symmetric area in the contralateral normal breast as the reference for comparison to improve diagnostic accuracy. Another strategy was to implement weakly supervised learning to predict the presence of cancer in DCE-MRI, by Zhou et al. (27). A dense net was applied within the segmented breast areas, and the suspicious lesion locations were calculated from the feature maps. Based on the detection results, a conditional random field was employed to estimate the tumor boundary, but the DSC was only 0.51. This approach was aiming to predict the presence of suspicious areas, not using the labeled lesions for supervised training. The Mask R-CNN used in our study was different from these studies, thus the results could not be directly compared. The region proposal network was applied to search suspicious regions within the entire image, which has been shown as a sensitive approach, as in (28); and also, including the contralateral normal breast as one input could utilize the symmetry to improve the specificity, as in (21). The Mask R-CNN used in the present study has been applied to search, detect, and diagnose brain hemorrhage on head CT, and achieved a very high accuracy of 0.97 (30).
For most object detection algorithms, a high sensitivity is associated with high false positives. The Mask R-CNN is not a single shot algorithm, and thus can increase specificity (29). In all of the selected regions, they were ranked to extract those with high probabilities. Then the bounding boxes were regressed to generate lesion masks. Furthermore, if a lesion was only detected on a single slice without involving neighboring slices, it was smaller than 3 mm and unlikely to be a true lesion. These additional processing steps could improve specificity while maintaining a reasonable sensitivity. The drawback was that, compared to other architectures, the training became much more complicated, and might take longer and need more training cases.
The major limitation was the small case number and the unbalanced data. For each patient, the number of positive imaging slices containing the lesion was much smaller than the negative slices, which was an inherent problem for lesion detection on MRI. The unbalanced input might lead to unstable training. Second, the training dataset was non-fat-sat images, and the testing dataset was fat-sat images. Although this represented a realistic clinical scenario, not an optimal setting for evaluating the performance of the developed model. However, since both fat-sat and non-fat-sat images showed a good contrast between chest wall and lung, and the subtraction images were of similar quality, the developed model using non-fat-sat images could be applied to fat-sat images and achieved reasonable accuracy. For different datasets, such as images acquired using different protocols or different MR systems, transfer learning could be applied, i.e. to use part of the testing dataset to re-tune the developed model (42). Third, only histologically confirmed cancer cases were analyzed in our study, which was the very first step to prove that Mask R-CNN detection is feasible. The method will need to be tested in more realistic breast MRI datasets that include benign findings and completely normal examinations, e.g. from a screening population. After an abnormal lesion is identified, it can be further characterized to rule out malignancy, and deep learning can also be applied to differentiate cancers from benign lesions and normal tissues (43–44). Lastly, we only included mass lesions in this study, so the major histological type was IDC. DCIS and ILC were more likely to present as non-mass-like enhancements and excluded, thus their case number was too small to perform a separate subtype analysis. For non-mass-like enhancements, since the exact pathological extent of the lesion is unclear, it is difficult to be accurately segmented by computer algorithms to provide ground truth for calculating DSC, thus they are excluded in this work as an initial proof-of-principle study. Nonetheless, we anticipate that the developed model can also be used to detect non-mass lesions when the tissue enhancement contrast is high. If they can be detected, even if the tumor ROI cannot be precisely segmented, the information may be sufficient for radiologists and oncologists to decide an optimal treatment plan.
CONCLUSION
In summary, we implemented a deep learning method using Mask R-CNN algorithm to search the entire set of breast MRI to detect abnormal lesions. The algorithm allowed the search on the whole image without prior breast segmentation, and reached the accuracy of 0.86 in per-slice basis analysis in the training dataset. The inclusion of the pre-contrast image and the contralateral subtraction image as inputs could help to eliminate false positives coming from the heart and the normal parenchymal enhancements, and achieved a high specificity of 0.86. In the detected lesions, the DSC of the segmented tumor compared to ground truth was 0.82. The model developed from non-fat-sat images could be applied to fat-sat images acquired using a different MR system. When the obtained per-slice results from one lesion were combined to give per-lesion result, 240/241 lesions in the training, and 98/98 lesions in the testing datasets were correctly identified. The results suggest that Mask R-CNN has the potential to be implemented for the detection of candidate lesions in breast MRI, which can be further integrated with lesion characterization algorithms to develop a fully-automatic, deep learning-based, breast MRI diagnosis system.
ACKNOWLEDGEMENT
This study was supported in part by NIH/NCI R01 CA127927 and R21 CA208938.
List of Abbreviations:
- 3D-FLASH
three-dimensional fast low angle shot
- AFROC
Alternative Free-response ROC
- AI
Artificial Intelligence
- BI-RADS
Breast Imaging Reporting and Data System
- CAD
Computer-Aided Diagnosis
- CNN
Convolutional Neural Network
- DCE
Dynamic Contrast-Enhanced
- DSC
Dice Similarity Coefficient
- FCM
Fuzzy C-Means
- FPN
Feature Pyramid Network
- FROC
Free-response ROC
- IoU
Intersection over Union
- MIP
Maximum Intensity Projection
- TE
Echo Time
- TR
Repetition Time
- R-CNN
Regional Convolutional Neural Network
- ROC
Receiver Operating Characteristic
- ROI
Region Of Interest
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sites A (2014) SEER cancer statistics review 1975–2011. Bethesda, MD: National Cancer Institute [Google Scholar]
- 2.Kuhl C (2007) The current status of breast MR imaging part I. Choice of technique, image interpretation, diagnostic accuracy, and transfer to clinical practice. Radiology 244:356–378 [DOI] [PubMed] [Google Scholar]
- 3.Kuhl CK (2007) Current status of breast MR imaging part 2. Clinical applications. Radiology 244:672–691 [DOI] [PubMed] [Google Scholar]
- 4.Montemurro F, Martincich L, Sarotto I, et al. (2007) Relationship between DCE-MRI morphological and functional features and histopathological characteristics of breast cancer. European radiology 17:1490–1497 [DOI] [PubMed] [Google Scholar]
- 5.Raikhlin A, Curpen B, Warner E, Betel C, Wright B, Jong R (2015) Breast MRI as an adjunct to mammography for breast cancer screening in high-risk patients: retrospective review. AJR Am J Roentgenol. 204:889–897 [DOI] [PubMed] [Google Scholar]
- 6.Ikeda DM, Hylton NM, Kinkel K, et al. (2001) Development, standardization, and testing of a lexicon for reporting contrast-enhanced breast magnetic resonance imaging studies. J Magn Reson Imaging. 13:889–895 [DOI] [PubMed] [Google Scholar]
- 7.Renz D, Baltzer P, Kullnig P, et al. (2008) Clinical value of computer-assisted analysis in MR mammography. A comparison between two systems and three observers with different levels of experience. RoFo: Fortschritte auf dem Gebiete der Rontgenstrahlen und der Nuklearmedizin 180:968–976 [DOI] [PubMed] [Google Scholar]
- 8.Lehman CD, Blume JD, DeMartini WB, Hylton NM, Herman B, Schnall MD (2013) Accuracy and interpretation time of computer-aided detection among novice and experienced breast MRI readers. American Journal of Roentgenology 200:W683–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djilas-Ivanovic D, Prvulovic N, Bogdanovic-Stojanovic D, et al. (2012) Breast MRI: intraindividual comparative study at 1.5 and 3.0 T; initial experience. Journal of BU ON: official journal of the Balkan Union of Oncology 17:65–72 [PubMed] [Google Scholar]
- 10.Pediconi F, Catalano C, Occhiato R, et al. (2005) Breast lesion detection and characterization at contrast-enhanced MR mammography: gadobenate dimeglumine versus gadopentetate dimeglumine. Radiology 237:45–56 [DOI] [PubMed] [Google Scholar]
- 11.Pediconi F, Catalano C, Padula S, et al. (2008) Contrast-enhanced MR mammography: improved lesion detection and differentiation with gadobenate dimeglumine. American Journal of Roentgenology 191:1339–1346 [DOI] [PubMed] [Google Scholar]
- 12.Martincich L, Faivre-Pierret M, Zechmann CM, et al. (2011) Multicenter, double-blind, randomized, intraindividual crossover comparison of gadobenate dimeglumine and gadopentetate dimeglumine for breast MR imaging (DETECT Trial). Radiology 258:396–408 [DOI] [PubMed] [Google Scholar]
- 13.Gubern-Mérida A, Vreemann S, Martí R, et al. (2016) Automated detection of breast cancer in false-negative screening MRI studies from women at increased risk. European journal of radiology 85:472–479 [DOI] [PubMed] [Google Scholar]
- 14.Chang Y-C, Huang Y-H, Huang C-S, Chen J-H, Chang R-F (2014) Computerized breast lesions detection using kinetic and morphologic analysis for dynamic contrast-enhanced MRI. Magnetic resonance imaging 32:514–522 [DOI] [PubMed] [Google Scholar]
- 15.Dorrius MD, Jansen-van der Weide MC, van Ooijen PM, Pijnappel RM, Oudkerk M (2011) Computer-aided detection in breast MRI: a systematic review and meta-analysis. European radiology 21:1600–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renz DM, Böttcher J, Diekmann F, et al. (2012) Detection and classification of contrast-enhancing masses by a fully automatic computer-assisted diagnosis system for breast MRI. Journal of Magnetic Resonance Imaging 35:1077–1088 [DOI] [PubMed] [Google Scholar]
- 17.Vignati A, Giannini V, De Luca M, et al. (2011) Performance of a fully automatic lesion detection system for breast DCE-MRI. Journal of Magnetic Resonance Imaging 34:1341–1351 [DOI] [PubMed] [Google Scholar]
- 18.Codari M, Schiaffino S, Sardanelli F, Trimboli RM (2019) Artificial Intelligence for Breast MRI in 2008–2018: A Systematic Mapping Review. American Journal of Roentgenology 212:280–292 [DOI] [PubMed] [Google Scholar]
- 19.Lee J-G, Jun S, Cho Y-W, Lee H, Kim GB, Seo JB, Kim N (2017) Deep Learning in Medical Imaging: General Overview. Korean Journal of Radiology 18:570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-masni MA, Al-antari MA, Park J-M, et al. (2018) Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Computer methods and programs in biomedicine 157:85–94 [DOI] [PubMed] [Google Scholar]
- 21.Dalmış MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern-Mérida A (2018) Fully automated detection of breast cancer in screening MRI using convolutional neural networks. Journal of Medical Imaging 5:014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheth D, Giger ML (2020) Artificial intelligence in the interpretation of breast cancer on MRI. J Magn Reson Imaging 51:1310–1324 [DOI] [PubMed] [Google Scholar]
- 23.Yap MH, Pons G, Martí J, et al. (2017) Automated breast ultrasound lesions detection using convolutional neural networks. IEEE journal of biomedical and health informatics 22:1218–1226 [DOI] [PubMed] [Google Scholar]
- 24.Kooi T, Litjens G, Van Ginneken B, et al. (2017) Large scale deep learning for computer aided detection of mammographic lesions. Medical image analysis 35:303–312 [DOI] [PubMed] [Google Scholar]
- 25.Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K (2016) Mass detection in digital breast tomosynthesis: Deep convolutional neural network with transfer learning from mammography. Medical physics 43:6654–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E-K, Kim H-E, Han K, Kang BJ, Sohn Y-M, Woo OH, Lee CW (2018) Applying data-driven imaging biomarker in mammography for breast cancer screening: preliminary study. Scientific reports 8:2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Luo LY, Dou Q, et al. (2019) Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging 50:1144–1151 [DOI] [PubMed] [Google Scholar]
- 28.Ribli D, Horváth A, Unger Z, Pollner P, Csabai I (2018) Detecting and classifying lesions in mammograms with deep learning. Scientific reports 8:4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He K, Gkioxari G, Dollár P, Girshick R (2017) Mask r-cnn. In: Computer Vision (ICCV), 2017 IEEE International Conference on. IEEE. pp. 2980–2988 [Google Scholar]
- 30.Chang P, Kuoy E, Grinband J, et al. (2018) Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT. American Journal of Neuroradiology 39:1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohit Malhotra K, Davoudi A, Siegel S, Bihorac A, Rashidi P (2018) Autonomous detection of disruptions in the intensive care unit using deep mask R-CNN. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. pp. 1863–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couteaux V, Si-Mohamed S, Nempont O, et al. (2019) Automatic knee meniscus tear detection and orientation classification with Mask-RCNN. Diagnostic and interventional imaging 100:235–242 [DOI] [PubMed] [Google Scholar]
- 33.Nie K, Chen J-H, Hon JY, Chu Y, Nalcioglu O, Su M-Y (2008) Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Academic radiology 15:1513–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 770–778 [Google Scholar]
- 35.Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2020) Focal loss for dense object detection. IEEE transactions on pattern analysis and machine intelligence 42:318–327 [DOI] [PubMed] [Google Scholar]
- 36.Kingma D, Ba J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980 [Google Scholar]
- 37.Abadi M, Barham P, Chen J, et al. (2016) TensorFlow: A System for Large-Scale Machine Learning. In: OSDI. pp. 265–283 [Google Scholar]
- 38.Chakraborty DP, Winter LHL (1990) Free-response methodology: alternate analysis and a new observer performance experiment. Radiology 174:873–881 [DOI] [PubMed] [Google Scholar]
- 39.Bejnordi BE, Veta M, van Diest PJ, et al. (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318:2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajpurkar P, Irvin J, Zhu K, et al. (2017) CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv preprint arXiv:171105225 [Google Scholar]
- 41.Wang J, Fang Z, Lang N, Yuan H, Su M-Y, Baldi P (2017) A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Computers in Biology and Medicine 84:137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Chen JH, Lin Y, et al. (2020) Prediction of Breast Cancer Molecular Subtypes on DCE-MRI Using Convolutional Neural Network with Transfer Learning between Two Centers. European Radiology (accepted manuscript ID#: EURA-D-20–01789R2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Zhang Y, Chang KT, et al. (2020) Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning With Consideration of Peritumor Tissue. J Magn Reson Imaging 51(3):798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurttakal AH, Erbay H, İkizceli T, Karaçavuş S (2020) Detection of breast cancer via deep convolution neural networks using MRI images. Multimed Tools Appl 79:15555–15573 [Google Scholar]


