Dear Editor,
DNA methylation is a key epigenetic regulatory approach for many biological processes, such as genomic imprinting, epigenetic memory maintenance, aging, and neural development. In addition to 5-methylcytosine, DNA methylation at N6-deoxyadenosine (N6-mA) is the most prevalent DNA modification in prokaryotes [1]. With the advances in various deep sequencing techniques, recent studies have started to find N6-mA in eukaryotes, including humans and rodents [2–7]. In several studies [5, 7, 8], ALKBH1 has been identified as the demethylase that erases DNA methylation at N6-mA. Importantly, in the latest co-crystal structure study [9], mammalian ALKBH1 was revealed to serve as an N6-mA demethylase for unpairing DNA. In contrast to demethylase, the identification of N6-mA methyltransferases in mammalian cells remains inconclusive. Either METTL4 or N6AMT1 has been suggested to be the putative DNA N6-mA methyltransferase [8, 10].
Functionally, in mouse embryonic stem cells, N6-mA regulated by ALKBH1 is enriched at the LINE-1 transposon and function to silence gene transcription [2]. In human mesenchymal stem cells (MSCs), a decreased level of DNA N6-mA regulated by ALKBH1 is necessary for osteogenic differentiation via promoting ATF4 transcription [7]. The level of N6-mA is drastically higher in human glioblastoma stem cells and primary human tumor samples [5], suggesting a functional role of DNA N6-mA in tumorigenesis. Interestingly, N6-mA DIP (DNA immunoprecipitation) sequencing has revealed that many peaks with N6-mA modification are enriched in genes regulating neurodevelopment, suggesting that N6-mA functions to repress neurodevelopmental genes. Indeed, in the mouse brain, the overall level of DNA N6-mA in the prefrontal cortex is significantly elevated under stress [3]. However, detailed genome-wide analysis of N6-mA enrichment found that there is also region-specific loss of N6-mA, which is associated with up-regulated neurodevelopmental genes.
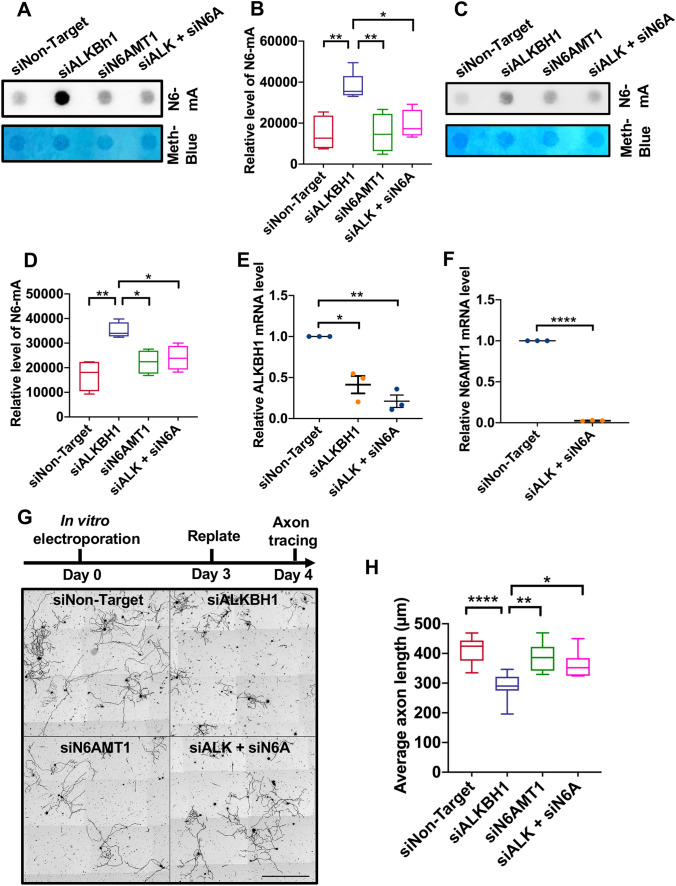
Axonal injury and regeneration are well known to involve neuronal stress responses and induce changes in gene transcription [11]. Therefore, we first measured the levels of DNA N6-mA modification using a N6-mA-specific antibody validated in several previously published studies [3, 5, 7]. To determine if ALKBH1 acts as an N6-mA demethylase in neurons, we first used siRNAs against ALKBH1 in differentiated neuronal CAD cells and then measured the levels of N6-mA. CAD cells are Cath.-a-differentiated; they are a variant of a CNS (central nervous system) catecholaminergic cell line established from a brain tumor. In serum-free conditions, CAD cells stop dividing, differentiate into post-mitotic neurons and form neuronal processes. The dot blot results showed that knocking down ALKBH1 in CAD cells (siALKBH1) resulted in a significantly higher level of N6-mA. Importantly, double knock-down of both ALKBH1 and the DNA N6-mA methyltransferase N6AMT1 (siALKBH1 + siN6AMT1) rescued the level of N6-mA back to the control condition (Fig. 1A, B). These results provide evidence that ALKBH1 and N6AMT1 function as the N6-mA demethylase and methyltransferase, respectively, in CAD cells. Interestingly, knocking down N6AMT1 alone did not significantly decrease the level of N6-mA (Fig. 1A, B), suggesting that the endogenous level of DNA N6-mA in neuronal cells might already be low in the control condition. To confirm that the N6-mA dot signals were indeed from genomic DNAs rather than interference with the N6-methylated RNA, we found that DNase A almost completely eliminated the methylene-blue and dot blot signals detected with the specific N6-mA antibody (not shown).
Fig. 1.
ALKBH1 acts as a DNA N6-deoxyadenosine (N6-mA) demethylase in mouse sensory neurons to regulate axon regeneration in vitro. A Representative dot blot images showing how knocking down ALKBH1 and N6AMT1, either alone or together, affects N6-mA levels in CAD cells. B Quantification of data as in A (n = 4–5 independent experiments; *P <0.05, **P <0.01, one-way ANOVA followed by Tukey’s multiple comparisons test). C Representative dot blot images showing how knocking down ALKBH1 and N6AMT1, either alone or together, affects N6-mA levels in adult mouse sensory neurons. D Quantification of data as in C (n = 4 independent experiments; *P <0.05, **P < 001, one-way ANOVA followed by Tukey’s multiple comparisons test). E Real-time PCR analysis showing significantly reduced mRNA levels of ALKBH1 3 days after knocking down ALKBH1 (siALKBH1) or together with N6AMT1 (siALK + siN6A) (n = 3 independent experiments; P = 0.0315 for siALKBH1 and 0.0092 for siALK + siN6A, one-sample t-test). F Real-time PCR analysis showing significantly reduced mRNA level of N6AMT1 3 days after knocking down ALKBH1 and N6AMT1 (n = 3 independent experiments; ****P <0.0001, one-sample t-test). G Upper, timeline of the culture and replate experiments; lower, representative images of cultured sensory neurons 24 h post-replating after knocking down ALKBH1, N6AMT1, or together with siRNAs (siALKBH1, siN6AMT1, or siALK + siN6A). Scale bar, 500 μm. H Quantification of average lengths of the longest neurites (n = 7–10 independent experiments; *P <0.05, **P <0.01, ****P <0.0001, one-way ANOVA followed by Tukey’s multiple comparisons test).
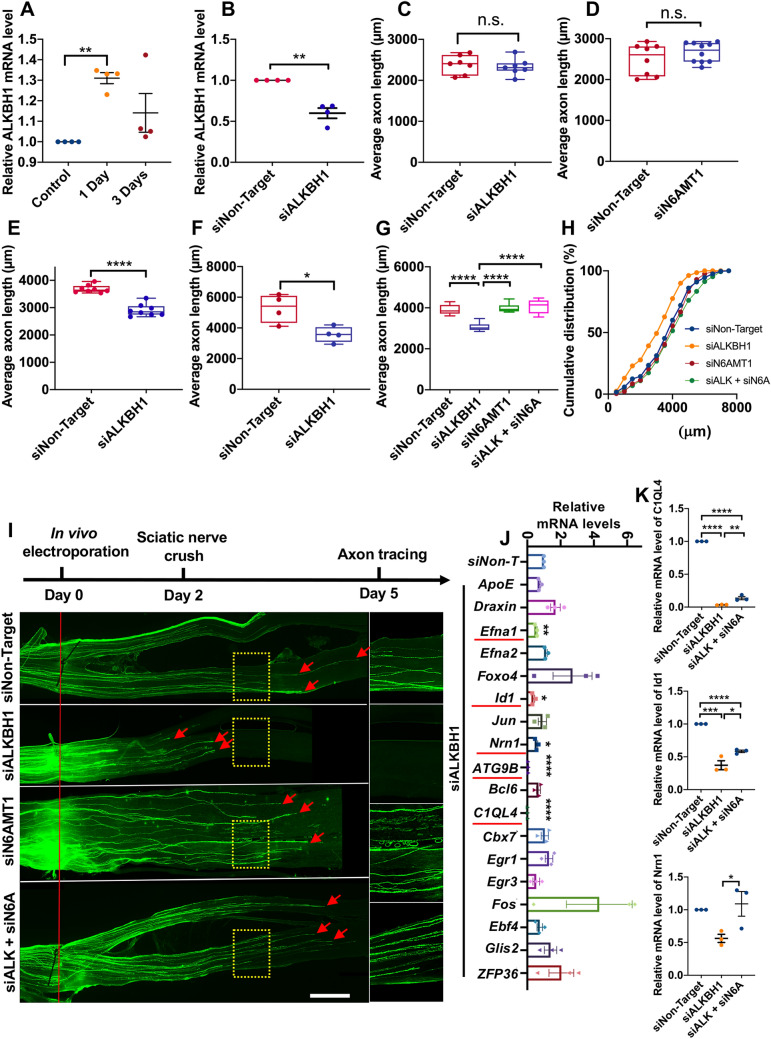
We next performed the same sets of experiments using cultured adult mouse primary sensory neurons from the dorsal root ganglion (DRG). The results showed that knocking down ALKBH1 also led to a significantly increased level of N6-mA, which was significantly reduced by co-deleting N6AMT1 (Fig. 1C, D), clearly showing that ALKBH1 is the DNA N6-mA demethylase in adult DRG neurons. To investigate whether ALKBH1 functions in sensory neurons to regulate regenerative axon growth, we used our previously established culture-replate model [12], which allowed us to evaluate regenerative axon regrowth from the neuronal soma after the genes of interests had been knocked down. Specifically, adult mouse sensory neurons were first transfected with either siNon-Target, siALKBH1, siN6AMT1, or siALKBH1 + N6AMT1 via electroporation and cultured for 3 days. The real-time PCR results showed that siALKBH1 or siN6AMT1 transfection resulted in a significantly reduced level of ALKBH1 or N6AMT1 (Fig. 1E, F). The neurons were then re-suspended and replated for another 24 h before fixation (Fig. 1G). Quantification of the average lengths of axons 24 h after replating demonstrated that knocking down ALKBH1 significantly impaired regenerative axon growth (Fig. 1G, H). Although knocking down N6AMT1 alone had little effect, it significantly rescued the axon growth defect induced by deleting ALKBH1 (Fig. 1G, H). Together, these results demonstrated that ALKBH1 and its regulation of DNA N6-mA demethylation are necessary for sensory axon regeneration in vitro.
To determine the role of ALKBH1 in sensory axon regeneration in vivo, we first found that the mRNA level of ALKBH1 was significantly elevated 1 day after peripheral nerve injury and returned to the control level after 3 days (Fig. 2A). We then performed in vivo electroporation [13] with either siNon-Target or siALKBH1 into adult sensory neurons and performed sciatic nerve crush injury 2 or 3 days after the electroporation. Real-time PCR data showed that 3 days after in vivo electroporation of siALKBH1, its mRNA level in DRGs was significantly reduced (Fig. 2B) compared with that in control DRGs (siNon-Target). Our recent time-course analysis of sensory axon regeneration [14] showed that the rate of axon regeneration is slow in the first 2 days and switches to a faster rate ~3 days after nerve injury. Therefore, we examined sensory axon regeneration at 2, 3, and 4 days after the nerve crush. The results showed that 2 days after the nerve crush when the axon regeneration rate was still low, knocking down ALKBH1 did not have a significant effect (Fig. 2C). Similarly, knocking down N6AMT1 also had no effect on sensory axon regeneration 2 days after injury (Fig. 2D). In contrast, knocking down ALKBH1 led to significantly reduced spontaneous sensory axon regeneration 3 and 4 days after the nerve crush (Fig. 2E, F), indicating that ALKBH1 is necessary for fast sensory axon regeneration in vivo. Similar to the in vitro results, knocking down N6AMT1 alone had little effect on sensory axon regeneration 3 days after the nerve crush (Fig. 2G–I), whereas it significantly rescued the regeneration impaired by ALKBH1 deletion (Fig. 2G–I). In summary, these results demonstrated that ALKBH1 and lower DNA N6-mA levels are necessary for fast sensory axon regeneration in vivo.
Fig. 2.
ALKBH1 and N6AMT1 coordinate to regulate sensory axon regeneration in vivo. A Real-time PCR analysis showing significantly elevated mRNA level of ALKBH1 in sensory neurons post-injury (n = 4 independent experiments; P = 0.0014, one-sample t-test). B Real-time PCR analysis showing significantly reduced mRNA level of ALKBH1 in vivo 3 days after electroporation (n = 4 independent experiments; P = 0.0076, one-sample t-test). C Quantification of average length of regenerating sensory axons showing no significant difference between the control and the ALKBH1 knockdown group 2 days after nerve injury (n = 7 mice in each condition; P = 0.6387, n.s., not significant, unpaired Student’s t-test). D Quantification of average length of regenerating sensory axons showing no significant difference between the control and the N6AMT1 knockdown group 2 days after nerve injury (n = 7 mice in the control and 10 in the siN6AMT1 group; P = 0.2715, n.s., not significant, unpaired Student’s t-test). E, F Quantification of average length of regenerating axons showing that knocking down ALKBH1 impairs sensory axon regeneration 3 (E) and 4 days (F) after nerve injury (n = 8 mice in each condition for 3 days, ****P <0.0001; n = 4 mice in each condition for 4 days, P = 0.018, unpaired Student’s t-test). G Quantification of average length of regenerating axons 3 days after nerve injury under conditions shown in I (n = 7–9 mice in each condition; ****P <0.0001, one-way ANOVA followed by Tukey’s multiple comparisons test. H Cumulative distribution curves showing that knocking down N6AMT1 rescues the axon regeneration impaired by ALKBH1 knockdown. I Upper, timeline of the experiment; lower, representative images showing that knocking down N6AMT1 reverses the sensory axon regeneration impaired by down-regulation of ALKBH1 3 days after nerve injury (right column, enlarged images of areas in the dashed yellow boxes; red line, crush sites; red arrows, regenerating axon tips; scale bars, 1 mm for the left panel and 0.5 mm for the right panel). J Real-time PCR analyses of changed transcription of 18 genes in sensory neurons after knocking down ALKBH1. The 5 significantly down-regulated genes are underlined in red. n = 3 independent experiments; P = 0.0096 for Efna1, 0.0118 for Id1, and 0.0202 for Nrn1, P <0.0001 for ATG9B or C1QL4, one-sample t-test. K Real-time PCR analysis of mRNA levels of C1QL4 (P <0.0001, n = 3), Id1 (P = 0.0001, n = 3), and Nrn1 (P = 0.0368, n = 3) after knocking down ALKBH1 or double-knockdown of ALKBH1 and N6AMT1. One-way ANOVA followed by Tukey’s multiple comparisons test, independent experiments. Data are represented as the mean ± SEM. *P <0.05, **P <0.01, ***P <0.001, ****P <0.0001, compared to control if not designated.
Previous studies [3, 5] in neurons or glial cells have shown that ALKBH1 and N6-mA function to repress the expression of many neurodevelopmental genes. Based on these studies, we first performed gene ontology (GO) analysis on a public RNA-seq dataset (GSE117632) acquired from patient-derived human glioblastoma cells with or without Alkbh1-knockdown [5]. When running GO analysis on the differentially-expressed genes (>1 reads per kilobase of transcript, per million mapped reads) with 2-fold or greater down-regulation upon ALKBH1-knockdown, the biological process termed regulation of neuron differentiation (GO: 0045664) was enriched. Among these 65 genes, we selected 18 genes-of-interest for real-time qPCR analysis in mouse DRG neurons. The results showed that among the 18 genes 5 were significantly down-regulated by ALKBH1 knockdown: Efna1, Id1, Nrn1, ATG9B, and C1QL4 (Fig. 2J). None of the genes was significantly up-regulated upon ALKBH1 knockdown, consistent with the idea that DNA N6-mA mainly functions to condense chromatin and repress gene expression. Moreover, simultaneously knocking down the methyltransferase N6AMT1 significantly rescued the mRNA levels of Id1, Nrn1, and C1QL4 (Fig. 2K). Importantly, some of these genes have been shown to regulate axon growth or regeneration, such as Nrn1 that encodes the protein neuritin [15], suggesting that ALKBH1 might support sensory axon regeneration by reducing the neuronal DNA N6-mA level and the subsequent up-regulation of these neurodevelopmental genes.
There is emerging evidence that the intrinsic ability of mammalian axon regeneration is epigenetically regulated [16]. Here, we demonstrated that sensory axon regeneration was regulated by the N6-mA demethylase ALKBH1, providing an important physiological function for ALKBH1 and DNA N6-mA modification. Importantly, we provided both in vitro and in vivo data that sensory axon regeneration was supported by ALKBH1, likely mediated by its role as the DNA N6-mA demethylase. Moreover, co-deletion of the methyltransferase N6AMT1 completely rescued the axon regeneration impaired by ALKBH1 down-regulation both in vitro and in vivo, further confirming the important function of DNA N6-mA in the regulation of sensory axon regeneration. We also found that knocking down ALKBH1 resulted in down-regulation of neurodevelopmental genes, whereas peripheral nerve crush resulted in up-regulation of ALKBH1 and neurodevelopmental genes. Therefore, our results provide a potential mechanism underlying such rejuvenation of gene expression during axon regeneration. Thus, it will be of great importance in the future to determine if the level of DNA N6-mA in mature CNS neurons is high, which might underlie their low intrinsic regeneration ability, and if overexpression of ALKBH1 can enhance CNS axon regeneration.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The work was supported by grants from the National Institutes of Health (R01NS085176, R01GM111514, R01EY027347, R01EY030883, and R01EY031779), the Craig H. Neilsen Foundation (259450), and the BrightFocus Foundation (G2017037).
Conflict of interest
The authors declare no competing interests.
References
- 1.Alderman MH, 3rd, Xiao AZ. N(6)-Methyladenine in eukaryotes. Cell Mol Life Sci. 2019;76:2957–2966. doi: 10.1007/s00018-019-03146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao B, Cheng Y, Wang Z, Li Y, Chen L, Huang L, et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun. 2017;8:1122. doi: 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Zhao Q, Wei W, Lin Q, Magnan C, Emami MR, et al. The DNA modification N6-methyl-2'-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction. Nat Neurosci. 2019;22:534–544. doi: 10.1038/s41593-019-0339-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q, et al. N6-methyladenine DNA modification in glioblastoma. Cell. 2018;175(1228–1243):e1220. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Zhao S, Nelakanti RV, Lin K, Wu TP, Alderman MH, 3rd, et al. N6-methyladenine in DNA antagonizes SATB1 in early development. Nature. 2020;583:625–630. doi: 10.1038/s41586-020-2500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C, Liu Y, Li X, Zou J, Zou S. DNA N6-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res. 2016;4:16033. doi: 10.1038/boneres.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao CL, Zhu S, He M, Chen, Zhang Q, Chen Y, et al. N6-methyladenine DNA modification in the human genome. Mol Cell 2018, 71: 306–318 e307. [DOI] [PubMed]
- 9.Zhang M, Yang S, Nelakanti R, Zhao W, Liu G, Li Z, et al. Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA. Cell Res. 2020;30:197–210. doi: 10.1038/s41422-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kweon SM, Chen Y, Moon E, Kvederaviciute K, Klimasauskas S, Feldman DE. An adversarial DNA N6-methyladenine-sensor network preserves polycomb silencing. Mol Cell. 2019;74(1138–1147):e1136. doi: 10.1016/j.molcel.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li WY, Zhang WT, Cheng YX, Liu YC, Zhai FG, Sun P, et al. Inhibition of KLF7-targeting microRNA 146b promotes sciatic nerve regeneration. Neurosci Bull. 2018;34:419–437. doi: 10.1007/s12264-018-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saijilafu, Hur EM, Liu CM, Jiao Z, Xu WL, Zhou FQ. Signalling regulates mammalian axon regeneration by inducing the expression of Smad. Nat Commun 2013, 4: 2690. [DOI] [PMC free article] [PubMed]
- 13.Saijilafu, Hur EM, Zhou FQ. Genetic dissection of axon regeneration via in vivo electroporation of adult mouse sensory neurons. Nat Commun 2011, 2: 543. [DOI] [PMC free article] [PubMed]
- 14.Gao Y, Hu YW, Duan RS, Yang SG, Zhou FQ, Wang RY. Time course analysis of sensory axon regeneration in vivo by directly tracing regenerating axons. Neural Regen Res. 2020;15:1160–1165. doi: 10.4103/1673-5374.270315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang T, Li H, Zhang S, Liu F, Wang D, Xu J. Nrn1 overexpression attenuates retinal ganglion cell apoptosis, promotes axonal regeneration, and improves visual function following optic nerve crush in rats. J Mol Neurosci. 2021;71:66–79. doi: 10.1007/s12031-020-01627-3. [DOI] [PubMed] [Google Scholar]
- 16.Qian C, Zhou FQ. Updates and challenges of axon regeneration in the mammalian central nervous system. J Mol Cell Biol. 2021;12:798–806. doi: 10.1093/jmcb/mjaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.