Abstract
The avian leukosis virus (ALV) strain DL00766 was isolated from a farm in China. The phylogenetic analysis showed that env had the highest homology with the E subgroup reference strain, ranging from 94.5% to 94.9%, whereas gp85 had the highest homology with the B and E subgroups, which were 89.0% to 91.3% and 91.3% to 91.8%. In addition, point mutation analysis of gp85 showed that a 400 bp long fragment in gp85 of DL00766 had the highest homology with subgroup B, ranging from 90.1% to 97.5%, and only 82.7% to 83.1% with E subgroup. These results indicate, DL00766 may be an AVL subgroup E isolate with a subgroup B-like gp85 region. This is also the first finding that the E subgroup is used as a recombinant subject, and the subgroup B provides a recombinant virus of an exogenous gene.
Key words: ALV-E, ALV-B, reorganization, gp85
INTRODUCTION
Recombination is a universal phenomenon in avian leukosis virus (ALV). For example, Lupiani et al. (2000) found recombinant ALV-B with J-like LTR, Wang et al. (2017) discovered recombinant ALV-B with J-like U3 region from the whole-genome sequence. Nine putative recombinant events were detected by Su et al. (2018) in the genomes of the three newly isolated ALV-K strains, which got high statistical support. Wang et al. (2019) discovered a recombinant strain consisting of three subgroups of ALV-J, ALV-E, and ALV-A in Three-Yellow chickens, Commercial Three-Yellow chickens infected with the recombinant strain showed severe immunosuppression and tumors. The more serious situation caused by the recombination phenomenon is the emergence of new subgroups. For example, the most popular ALV-J all over the world today is also thought to be caused by recombination between an unknown exogenous ALV and an ancient endogenous retrovirus family (EAV-HP, or ev / J) (Smith et al., 1999). In addition, the newly discovered K subgroup is also suspected of being a recombinant virus (Zhao et al., 2018). Therefore, monitoring for recombinant ALV will be the focus of the prevention of ALV. One strain of ALV from commercial chickens was isolated in monitoring, which was initially identified as subgroup E. However, further analysis of the virus env indicated that it was a B and E recombinant virus. The molecular characteristics of the genome of the recombinant virus were reported in this study.
MATERIALS AND METHODS
Clinical Samples
In 2009, the spleen, kidney, and liver of a suspected sick HY-LINE VARIETY BROWN were collected from a chicken farm in Hubei province and were processed separately in an anatomy laboratory.
Isolation and Identification of Virus
The total DNA was extracted from the tissue homogenates as follows (Lili et al., 2014). The primer sets H5 (5′-GGATGAGGTGACTAAGAAAG-3′) and H7 (5′-CGAACCAAAGGTAACACACG-3′) were used for specific detection of ALV-J proviral DNA, which produced a 545 bp PCR product. Primer sets H5 and AD1 (5′-GGGAGGTGGCTGACTGTGT-3′) were used to detect ALV subgroups A to E, which produced 295 to 326 bp PCR products (Smith et al., 1998).
Positive samples according PCR results were subjected to virus isolation in the CEF cells. The procedures for isolation and identification of ALV in the cell culture were performed according to previously described studies (Bagust et al., 2004).
The env, which had been amplified using universal primers (forward primer was 5′-GGATGAGGTGACTAAGAAAG-3′, and the reverse primer was 5′-AGTTAAGCCATGCCCCGTTAC-3′.), after the sequencing results were obtained, comparisons were made by BLAST, which showed closer proximity to the E subgroup. Then the full length sequence named DL00766 (accession number: MH454773) was amplified by using referencing 8 pairs of consecutive and overlapping E subgroup primers (Supplementary Table S1) published by Kong et al. (2008).
RESULTS AND DISCUSSION
The PCR assay showed a sequence of about 300 bp, which proved that the virus was a subpopulation of A-E. The infected CEF cells showed positive result by ACELISA on anti-p27 antibody-coated plates, and uninfected CEF cells for the negative control showed negative result by AC-ELISA, It demonstrated the presence of ALV in the samples. The env amplified using universal primers was sequenced and Blast, showing closer proximity to the E subgroup. In summary, the new isolate named DL00766 was E subtype.
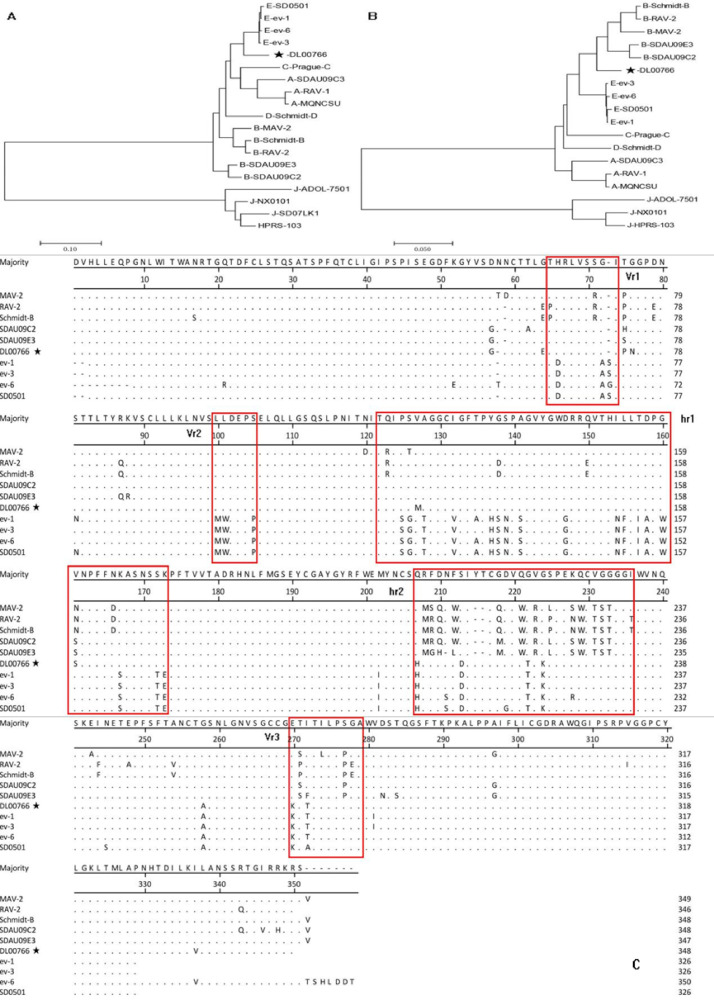
The complete proviral genome sequence of DL00766 was 7521 bp, with the typical arrangement of retroviral genes (i.e., LTR-leader-gag-pol-env-LTR).The major genes of DL00766 was compared with reference strains of each subgroup (Supplementary Table S2) for Clustal W in order to better understand the evolutionary relationship and molecular characteristics of this strain, and it was found that gag and pol genes are well conserved, with identities of 94.0% to 96.7% and 96.1% to 98.4%, respectively. The env gene has the highest homology with the E subgroup reference strain, ranging from 94.5% to 94.9%, whereas it ranged from 55.5% to 93.4% with the other subgroup reference strains. A neighbor-joining tree was drawn by MEGA program (version 7.0.26), and the confidence levels were assessed with 1,000 bootstrap replications. It showed that both DL00766 and the E subgroup reference strains were in the same cluster (Figure 1A). Overall, the results showed that DL00766 belongs to ALV-E. The Blast comparison result showed that in addition to E subgroup, DL00766 also has high homology rate with B subgroup, up to 90% or above. Meanwile, a phylogenetic tree analysis on gp85 of DL00766 was performed and the result showed DL00766 was in a separate cluster between the B and E subgroups (Figure 1B). Further sequence analysis revealed that the gp85 of DL00766 has a region of approximately 430 bp, between 170 and 600 bp, and has the highest homology with the corresponding region of the B subgroup reference strain, which was 90.1% to 97.5%, whereas the homology with the corresponding regions of the E subgroup reference strains was only 82.7% to 83.1%. Bova C A et al. (1986) pointed out that gp85 mainly includes 5 hypervariable regions of Vr1, Vr2, Vr3, hr1 and hr2, and these hypervariable regions determine the species specificity of the subpopulations. Through a point mutation analysis on the amino acid of gp85 of DL00766, 3 hypervariable regions of Vr1, Vr2 and hr1 were found to have similar amino acid compositions to those of the B subgroup reference strain (Vr2 is completely identical), whereas hr2 and Vr3 were found to have similar amino acid compositions to those of the E subgroup reference strains (Figure 1C).
Figure 1.
Phylogenetic analysis of DL00766. Black Black star (★) indicates ALV isolates in this study. (A) Phylogenetic tree of env nucleotide sequences and reference strains; (B) phylogenetic tree of gp85 nucleotide sequences and reference strains; (C) comparative analysis of amino acid mutations in variable regions of gp85 between DL00766 and other ALV-B,E strains. Abbreviation: ALV, avian leukosis virus.
In traditional notions, endogenous viruses are often used as providers of exogenous genes. For instance, J subgroup reference strain HPRS-103 SU-coding domain from endogenous sequences rather than from exogenous virus (Bai et al., 1995).In addition, recombinant viruses have been detected in laying hens, and endogenous viruses provide exogenous genes (Liu et al., 2011). However, in this study, DL00766 had higher homology with the E subgroup reference strain, with a little variation, only gp85 was more specific, and phylogenetic tree analysis showed a single cluster. The hypervariable regions Vr1, Vr2, and hr1 of gp85 also showed amino acid compositions are similar to the B subgroup. These results indicated that recombination of subgroup B fragments may be present between the 170 to 600 bp regions of gp85 of DL00766. The DL00766 isolate is the first time report that the endogenous E subgroup is a recombinant subject, and the exogenous subgroup B provides recombination elements, and finally the recombinant strain was produced.
In addition, where the recombination fragment of DL00766 comes from is also an interesting question. In recent research,the ALV from Chinese flocks are found to mutate frequently, Li et al. (2017) found that the ALV-J quasispecies varied among infectedindividuals even within the same flock, and the dominant quasispecies continued to evolve both for their proportion and gene mutation, Meng et al. (2018) found that the env gene was more variable in 6 ALV-J isolates (SDAU1701–SDAU1706), especially the gp85 protein, which shared only 88.2% to 91.9% identity with the reference strains. When the recombinant fragment of DL00766 was compared with the B subgroup reference strain, the homology with the external reference strains MAV-2, RAV-2 and Schmidt-B were 90.1%, 90.4%, and 90.6%. At the same time, the homology with the internal subgroup B reference strains SDU09C2 and SDU09E2 was 97.5% and 96.3%. This indicates that the subgroup B strain involved in the recombination might originate from China. It is speculated that it may be that the flock originally carried the endogenous ALV and later infected with the Chinese popular ALV-B, and then they recombined in the chicken vivo.
ALV-E, as an endogenous virus, is generally less active or noncarcinogenic. The study was derived from an ALV long-term monitoring project. Usually, the staff only collect samples of chickens with suspected symptoms, which make us suspect that DL00766 has a certain pathogenicity. However, It requires further research to reveal the pathogenicity of DL00766 and the relationship between pathogenicity and recombination.
It was found that a 430 bp recombination of ALV-B in gp85 by phylogenetic analysis of DL00766 in this study. Moreover, this study presents the first finding that the endogenous E subgroup is a recombinant subject, and the subgroup B provides an exogenous gene for a recombinant virus and it may cause pathogenesis of the E subgroup. The emergence of this kind of isolate reminds us to pay attention to the phenomenon of recombination and speed up the purification process of the chicken flock and strengthen the monitoring of ALV.
Acknowledgments
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation of Heilongjiang Province of China (LH2019C013) and the Programs of the State Forestry Administration of Wildlife Protection and Nature Reserve Management Division (2019076018) and National Public Welfare Industry Agricultural Research Project (201203055) and a grant from the Modern Agro-industry Technology Research System in China (CARS-42-G07) and National key research and development program (2018YFD0502201-11).
Disclosures
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101137.
Appendix. Supplementary materials
REFERENCES
- Bai. J., Payne L.N., Skinner M.A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J. Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagust T.J., Fenton S.P., Reddy M.R. Detection of subgroup J avian leukosis virus infection in Australian meat-type chickens. Aust. Vet. J. 2004;82:701–706. doi: 10.1111/j.1751-0813.2004.tb12163.x. [DOI] [PubMed] [Google Scholar]
- Kong Y.B., Zhang X.X., Jiang S., Zhao Q., Sun Y.N. Sequence analysis for the complete provial genome of endogenous avian leukosis virus strain SD0501. Chin. J. Virol. 2008;24:53–58. [PubMed] [Google Scholar]
- Lupiani B., Hunt H., Silva R., Fadly A. Identification and characterization of recombinant subgroup J avian leukosis viruses (ALV) expressing subgroup A ALV envelope. Virology. 2000;276:37–43. doi: 10.1006/viro.2000.0539. [DOI] [PubMed] [Google Scholar]
- Liu. C., Zheng S., Wang Y., Jing L., Gao H. Detection and molecular characterization of recombinant avian leukosis viruses in commercial egg-type chickens in China. Avian Pathol. 2011;40:269–275. doi: 10.1080/03079457.2011.560932. [DOI] [PubMed] [Google Scholar]
- Li Y., Fu J., Cui S., Meng F., Cui Z., Fan J., Chang S., Zhao P. Gp85 genetic diversity of avian leukosis virus subgroup J among different individual chickens from a native flock. Poult. Sci. 2017;96:1100–1107. doi: 10.3382/ps/pew407. [DOI] [PubMed] [Google Scholar]
- Meng F., Li Q., Zhang Y., Cui Z., Chang S., Zhao P. Isolation and characterization of subgroup J Avian Leukosis virus associated with hemangioma in commercial Hy-Line chickens. Poult. Sci. 2018;97:2667–2674. doi: 10.3382/ps/pey121. [DOI] [PubMed] [Google Scholar]
- Smith L.M., Brown S.R., Howes K., McLeod S., Arshad S.S., Barron G.S., Venugopal K., McKay J.C., Payne L.N. Development and application of polymerase chain reaction (PCR) tests for the detection of subgroup J avian leukosis virus. Virus Res. 1998;54:87–98. doi: 10.1016/s0168-1702(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Smith L.M., Toye A.A., Howes K., Bumstead N., Payne L.N., Venugopal K. Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J. Gen. Virol. 1999;80(Pt 1):261–268. doi: 10.1099/0022-1317-80-1-261. [DOI] [PubMed] [Google Scholar]
- Su Q., Li Y., Li W., Cui S., Tian S., Cui Z., Zhao P., Chang S. Molecular characteristics of avian leukosis viruses isolated from indigenous chicken breeds in China. Poult. Sci. 2018;97:2917–2925. doi: 10.3382/ps/pex367. [DOI] [PubMed] [Google Scholar]
- Wang P., Yang Y., Lin L., Li H., Wei P. Complete genome sequencing and characterization revealed a recombinant subgroup B isolate of avian leukosis virus with a subgroup J-like U3 region. Virus Genes. 2017;53:927–930. doi: 10.1007/s11262-017-1493-4. [DOI] [PubMed] [Google Scholar]
- Wang P., Shi M., He C., Lin L., Li H., Gu Z., Li M., Gao Y., Huang T., Mo M., Wei T., Wei P. A novel recombinant avian leukosis virus isolated from gamecocks induced pathogenicity in Three-Yellow chickens: a potential infection source of avian leukosis virus to the commercial chickens. Poult. Sci. 2019;98:6497–6504. doi: 10.3382/ps/pez548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Rao M., Liao M., Cao W. Phylogenetic analysis and pathogenicity assessment of the emerging recombinant subgroup K of avian leukosis virus in South China. Viruses. 2018;10:194. doi: 10.3390/v10040194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.