Abstract
In 2019 coronavirus disease (COVID-19), whose main complication is respiratory involvement, different organs may also be affected in severe cases. However, COVID-19 associated cardiovascular manifestations are limited at present. The main purpose of this study was to identify potential candidate genes involved in COVID-19-associated heart damage by bioinformatics analysis. Differently expressed genes (DEGs) were identified using transcriptome profiles (GSE150392 and GSE4172) downloaded from the GEO database. After gene and pathway enrichment analyses, PPI network visualization, module analyses, and hub gene extraction were performed using Cytoscape software. A total of 228 (136 up and 92 downregulated) overlapping DEGs were identified at these two microarray datasets. Finally, the top hub genes (FGF2, JUN, TLR4, and VEGFA) were screened out as the critical genes among the DEGs from the PPI network. Identification of critical genes and mechanisms in any disease can lead us to better diagnosis and targeted therapy. Our findings identified core genes shared by inflammatory cardiomyopathy and SARS-CoV-2. The findings of the current study support the idea that these key genes can be used in understanding and managing the long-term cardiovascular effects of COVID-19.
Abbreviations: COVID-19, the coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; DEGs, differentially expressed genes; GEO, Gene Expression Omnibus; PPI, the protein-protein interaction; STRING, the search tool for the retrieval of interacting genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genome; ACE2, angiotensin-converting enzyme 2
Keywords: COVID-19, Cardiac remodeling, Differential expression, Bioinformatics analysis
1. Introduction
Declared as a global epidemic on March 2020, Coronavirus Disease-2019 (COVID-19) continues to cause hundreds of thousands of morbidity and mortality worldwide (Organization, 2020). The most common clinical symptoms of the disease include cough, shortness of breath, fever, headache, and fatigue. Recent studies have supported the hypothesis that SARS-CoV-2 also leads to the emergence of many complications as it targets multiple organs and tissues such as the kidneys, large intestine, and heart, especially (Bradley et al., 2020; Unudurthi et al., 2020). Ni et al. previously showed that ACE2 (angiotensin-converting enzyme 2), which is used by the virus to enter the cells and trigger complications, is found in many cell types of the body (Ni et al., 2020). It was reported that ACE2 is expressed ubiquitously and its expression highest in the thyroid, heart, testis, kidneys, small intestine, and adipose tissue (Li et al., 2020). Interestingly it was also found that ACE2 showed medium expression levels in the lungs. These findings explain the reason why many other organs especially the heart vulnerable to the virus in addition to the lungs.
Although the principal clinical complication of COVID-19 is respiratory involvement, recent studies have shown that cardiovascular (CV) risk factors and pre-existing cardiovascular diseases (CVDs) increase susceptibility to COVID-19 (Bansal, 2020; Guo et al., 2020). The fact that the patients with COVID-19 have signs of myocardial involvement and cardiac injury (with an average 25%) that contribute to 40% of deaths, supports the view that COVID-19 is primarily a cardiovascular targeted disease (Akhmerov and Marbán, 2020; Siripanthong et al., 2020). However, information about other COVID-19 associated cardiovascular manifestations is restricted at present and remains unclear. Myocarditis, also known as inflammatory cardiomyopathy or inflammation of the heart muscle, is an uncommon cardiovascular disease primarily manifest as sudden death (Cooper Jr, 2009). The viral infection is one of the most important causes of myocarditis, and thus many viruses have been implicated as the most common viruses associated with myocarditis (Kühl et al., 2005; Tschöpe et al., 2020). Developing myocarditis is rare, but because it affects the heart muscle and the heart's electrical system it may cause severe long-term cardiovascular consequences such as dilated cardiomyopathy (DCM) with chronic heart failure, arrhythmia, and sudden cardiac death (Maron et al., 2009; Becker, 2020; Fischer et al., 2020).
The cardiovascular effects of SARS-CoV-2 infection on the myocarditis axis are still debated. Therefore, filling knowledge gaps by understanding fundamental mechanisms underlying SARS-CoV2-associated cardiac abnormalities is essential for the development of new approaches. The main purpose of this study is to help scientists and researchers by revealing the finest therapeutic targets and diagnostic approaches to use in practice. The present study offers a potential basis to understand the cause and elemental molecular events of COVID-19 related cardiac complications by analyzing mRNA expression datasets from the GEO database.
2. Methods
2.1. Microarray datasets
Two independent publicly available datasets were selected for this study. The GEO datasets GSE150392 and GSE4172 were downloaded from the Gene Expression Omnibus (GEO) database (Barrett et al., 2012). GSE150392 consists of 3 SARS-CoV-2 infected human cardiomyocytes (hiPSC-CMs) and 3 non-infected hiPSC-CMs samples (Sharma et al., 2020). GSE4172 consists of 8 cardiac inflammation (DCMi) patients samples and 4 healthy control patients samples (Wittchen et al., 2007). DCMi group members with parvovirus B19 associated cardiac inflammation were on standard heart failure medication and in New York Heart Association functional class II (NYHA, 1994). Control group members had normal cardiac function and morphology and were negative for cardiac inflammation or viral genome. The detailed characteristics of the groups are summarized in Table 1 . The expression profiling arrays were generated using the Illumina NextSeq 500 platform (GPL18573) for GSE150392 and Affymetrix Human Genome U133 Plus 2.0 Array platform (GPL570) for GSE4172.
Table 1.
Characteristics of the study groups.
| Sample | Source name | Age | Gender | EF% | CI | Inflammation/PVB19 |
|---|---|---|---|---|---|---|
| 1 | Healthy control | 36 | Female | 68 | − | Negative |
| 2 | Healthy control | 46 | Female | 61 | − | Negative |
| 3 | Healthy control | 26 | Female | 74 | − | Negative |
| 4 | Healthy control | 36 | Male | 64 | − | Negative |
| 5 | DCMi | 45 | Male | 34 | + | Positive |
| 6 | DCMi | 62 | Male | 51 | + | Positive |
| 7 | DCMi | 31 | Male | 52 | + | Positive |
| 8 | DCMi | 67 | Male | 43 | + | Positive |
| 9 | DCMi | 60 | Male | 34 | + | Positive |
| 10 | DCMi | 69 | Male | 35 | + | Positive |
| 11 | DCMi | 55 | Female | 31 | + | Positive |
| 12 | DCMi | 31 | Female | 56 | + | Positive |
DCMi; inflammatory cardiomyopathy, EF; ejection fraction, CI; cardiac inflammation, PVB19; parvovirus B19.
2.2. Identification of DEGs
DEGs between SARS-CoV-2 infected hiPSC-CMs and non-infected hiPSC-CMs were screened by using Genevestigator software (Hruz et al., 2008) and the GEO2R tool (Barrett et al., 2013). p-Value <0.05 and a |log2FC| ≥ 0.5 were defined as the cut-off criteria. Finally, a Venn diagram summarizing the overlapping DEGs was generated using The Multiple List Comparator (http://www.molbiotools.com/listcompare.html).
2.3. GO and KEGG enrichment of overlapping DEGs
GO (Gene Ontology) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of overlapped DEGs were performed via the DAVID (The Database for Annotation, Visualization, and Integrated Discovery) online bioinformatics resource (Sherman and Lempicki, 2009) and ToppFun application (Chen et al., 2009). p < 0.05 was chosen as the significance threshold for both GO enrichment and KEGG pathway analysis.
2.4. PPI network construction and identification of candidate hub genes
The protein-protein interaction network (PPI) constructed using the STRING (Search Tool for the Retrieval of Interacting Genes) online tool (Jensen et al., 2009) under the default settings. Thereafter, Cytoscape software was used to visualize and analyze the PPI network (Shannon et al., 2003). The relationships among the DEGs were analyzed by the NetworkAnalyzer plug-in of Cytoscape. Moreover, MCODE (Molecular Complex Detection) plug-in was applied to identify significant clusters in the PPI network. Next, Cytohubba plug-in of Cytoscape was used to screen the hub genes based on the five calculation algorithms; Maximal Clique Centrality (MCC), Degree, EcCentricity, Edge Percolated Component (EPC), and Maximum Neighborhood Component (MNC). To narrow down the number of hub gene candidates in the PPI network, the first 10 genes ranked by each centrality index were selected. Finally, a Venn diagram was drawn for intersections of five algorithms to identify significant hub genes by using the webtool available at InteractiVenn (Heberle et al., 2015).
3. Results
3.1. Identification of DEGs
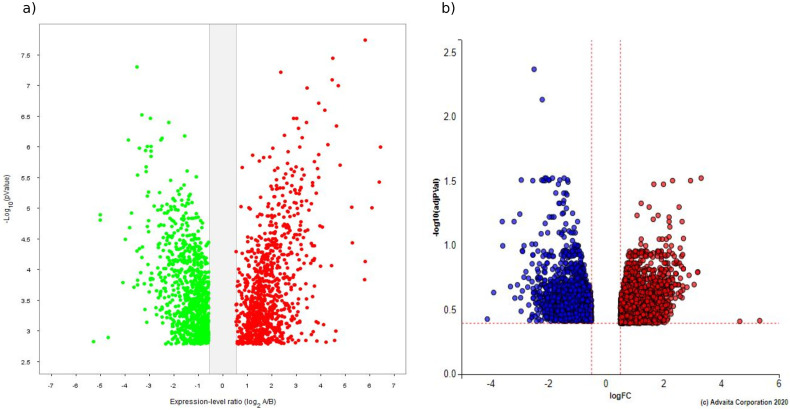
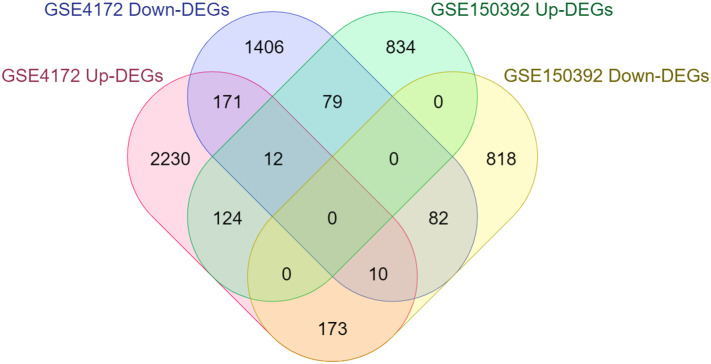
In total, 2157 (1049 upregulated, 1108 downregulated) and 4480 (2720 upregulated, 1760 downregulated) DEGs were extracted from the GSE150392 and GSE4172 datasets, respectively. The volcano plots are shown in (Fig. 1a, b). Furthermore, results showed that a total of 228 DEGs (136 upregulated and 92 downregulated) were shared by two GSE datasets (Fig. 2 ).
Fig. 1.
Identification of differentially expressed genes (DEGs) in the GSE150392 and GSE4172. a) Volcano plot of DEGs in the GSE150392 dataset (red represents upregulated genes with fold changes over 1.4 and green represents downregulated genes with fold changes less than 1.4.), b) volcano plot of DEGs in the GSE4172 dataset (red represents upregulated genes with fold changes over 1.4 and blue represents downregulated genes with fold changes less than 1.4.). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Venn diagram showing the overlapping DEGs between datasets.
3.2. GO term and pathway enrichment analyses
GO and KEGG pathway enrichment analyses were performed using the DAVID website. GO analysis classified the DEGs into three categories: molecular function (MF), biological processes (BP), and cellular components (CC). According to the BP analysis, the upregulated DEGs were mainly associated with the positive regulation of transcription from RNA polymerase II promoter, negative regulation of transcription, DNA-templated, and response to virus, while the downregulated DEGs were significantly involved in cardiac processes such as regulation of cardiac conduction, regulation of cardiac muscle contraction, and ventricular cardiac muscle cell development. For the CC analysis, the upregulated DEGs were mainly enriched in cytosol, membrane, and extracellular space, downregulated DEGs were located in mitochondrion, mitochondrial inner membrane, and cytoplasmic microtubule. In the MF, upregulated DEGs were mainly enriched in transcription factor activity, sequence-specific DNA binding, and RNA polymerase II core promoter proximal region sequence-specific DNA binding, and downregulated DEGs were associated with protein homodimerization activity, transcription factor binding, and chloride channel activity. The detailed results are shown in Fig. 3 . As shown in Table 2 , the upregulated DEGs were significantly enriched in Epstein-Barr virus infection, Protein processing in endoplasmic reticulum, and the downregulated DEGs were significantly enriched in Parkinson's disease, Oxidative phosphorylation, and Cardiac muscle contraction.
Fig. 3.
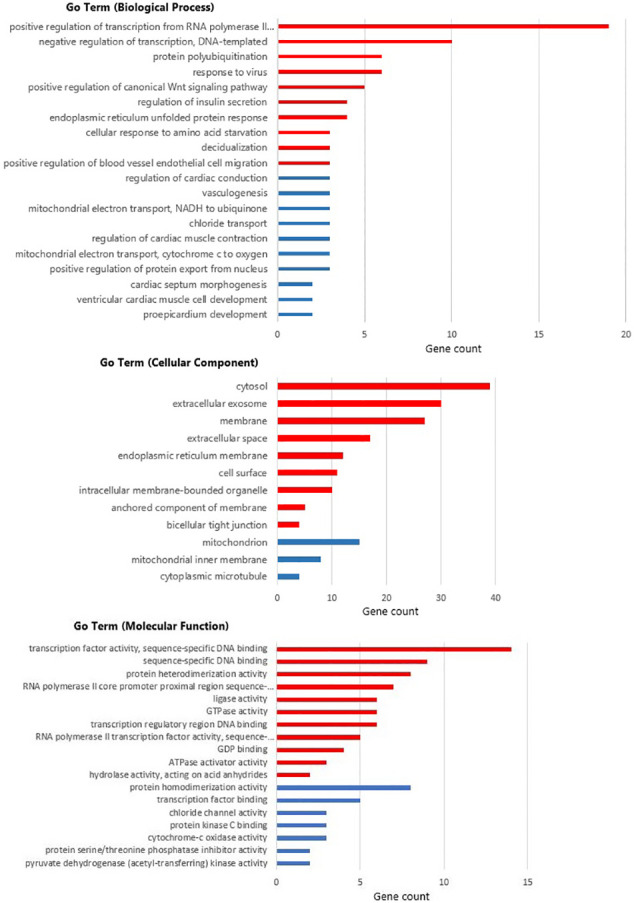
GO analysis of overlapping DEGs. Red bars demonstrate genes of |log2FC| ≥ 0.5, blue bars represent genes of |log2FC| ≤ 0.5. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
The detailed information of the significantly enriched KEGG pathways for the up and downregulated DEGs.
| KEGG pathway | Genes in category | p-Value |
|---|---|---|
| Upregulated DEGs | ||
| Epstein-Barr virus infection | MAP2K3, CDKN1A, PSMD12, JUN, DDX58, EIF2AK2, HLA-E | 5,80E-04 |
| Influenza A | MAP2K3, DNAJB1, JUN, DDX58, HSPA6, EIF2AK2, TLR4 | 3,60E-03 |
| Protein processing in endoplasmic reticulum | DNAJB1, HSPA6, EIF2AK2, CKAP4, UBE2J1, NFE2L2 | 1,50E-02 |
| Pathways in cancer | CDKN1A, JUN, CCND1, CXCR4, FGF2, FOXO1, BIRC3, RUNX1, VEGFA | 1,80E-02 |
| Measles | CCND1, DDX58, HSPA6, EIF2AK2, TLR4 | 2,70E-02 |
| Proteoglycans in cancer | CDKN1A, CCND1, ITPR2, FGF2, TLR4, VEGFA | 2,80E-02 |
| Hepatitis B | CDKN1A, JUN, CCND1, DDX58, TLR4 | 3,50E-02 |
| Rheumatoid arthritis | JUN, TLR4, ATP6V1C1, VEGFA | 3,90E-02 |
| Bladder cancer | CDKN1A, CCND1, VEGFA | 4,80E-02 |
| Downregulated DEGs | ||
| Parkinson's disease | COX7B, NDUFA6, ATP5A1, NDUFB1, PRKACA, SEPT5, COX7C, COX6A2 | 1,90E-06 |
| Oxidative phosphorylation | COX7B, NDUFA6, ATP5A1, NDUFB1, COX7C, COX6A2, LHPP | 1,90E-05 |
| Alzheimer's disease | COX7B, NDUFA6, ATP5A1, NDUFB1, COX7C, COX6A2 | 7,10E-04 |
| Huntington's disease | COX7B, NDUFA6, ATP5A1, NDUFB1, COX7C, COX6A2 | 1,30E-03 |
| Non-alcoholic fatty liver disease (NAFLD) | COX7B, NDUFA6, NDUFB1, COX7C, COX6A2 | 3,90E-03 |
| Cardiac muscle contraction | COX7B, MYL3, COX7C, COX6A2 | 4,10E-03 |
KEGG; Kyoto Encyclopedia of Genes and Genomes.
3.3. Identification and validation of hub genes
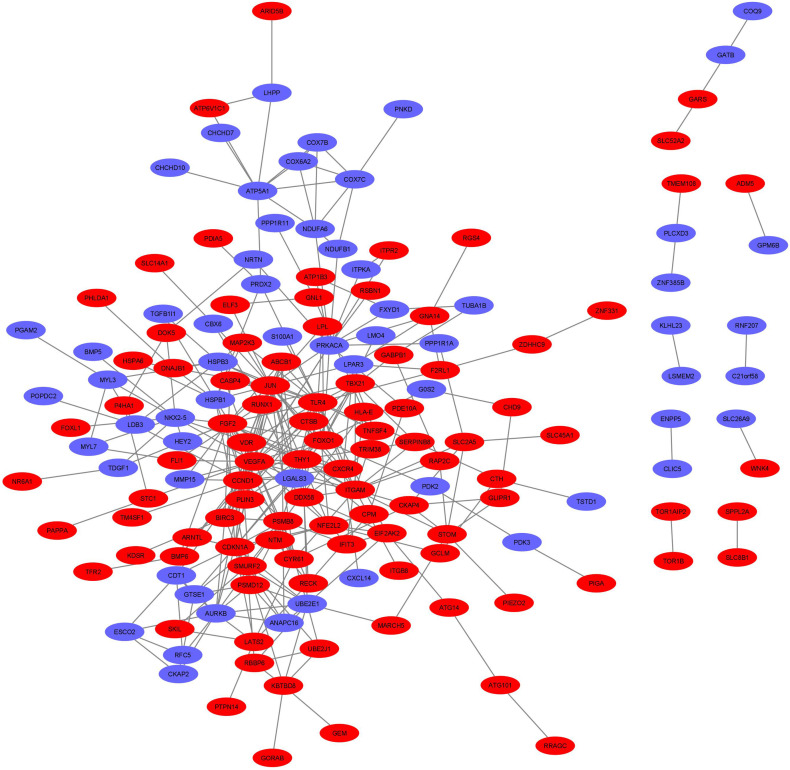
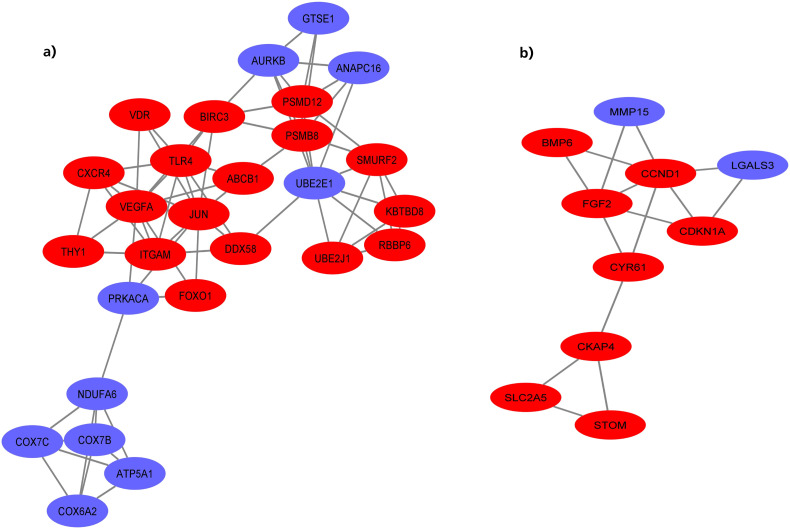
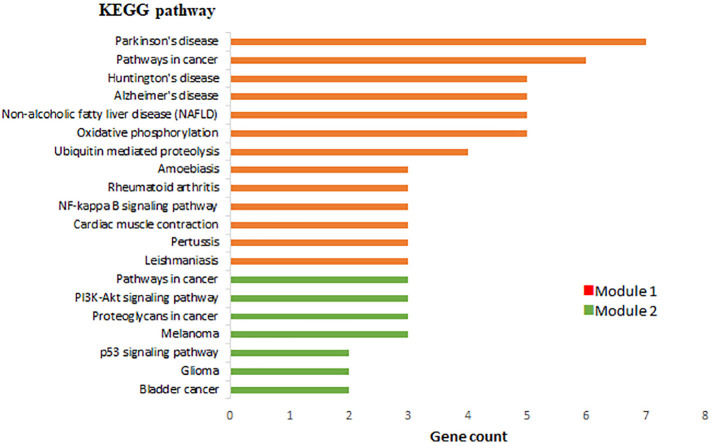
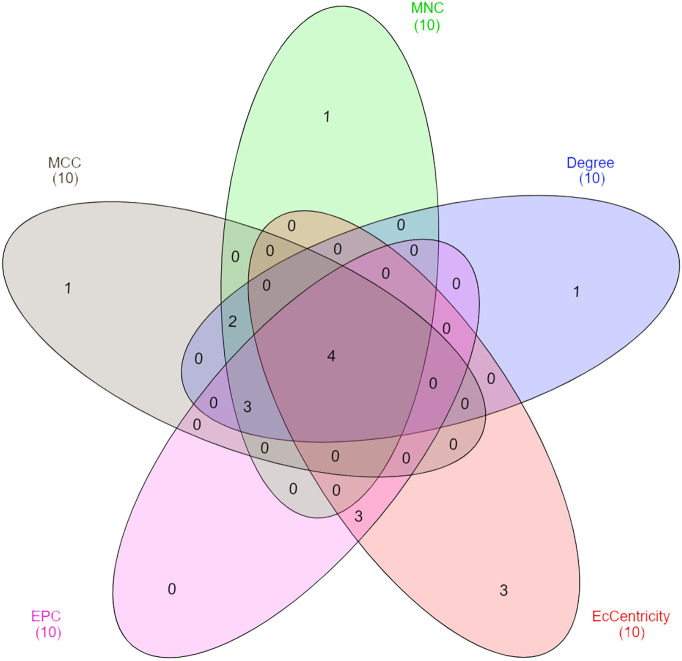
A PPI network of 228 DEGs (160 nodes and 336 edges) was constructed and visualized (Fig. 4 ). Subsequently, this network was imported into Cytoscape for further analysis. Based on the degree of importance, two clusters, which were composed of upregulated and downregulated genes, were then screened from the PPI network using the MCODE plug-in (Fig. 5a, b). Modules with an MCODE score ≥3 and nodes ≥10 were considered for additional analysis. KEGG pathway analysis results of the two most significant modules, performed by DAVID, are shown in Fig. 6 . Next, the top 10 genes evaluated by the five calculation methods were listed (Table 3 ) using the cytoHubba plug-in of Cytoscape. Finally, a total of four genes (FGF2, JUN, TLR4, and VEGFA) shared by five algorithms were identified as hub genes (Fig. 7 ).
Fig. 4.
The whole protein-protein interaction (PPI) network of DEGs (red nodes represent upregulated DEGs and blue nodes represent downregulated DEGs). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Module analysis of PPI network. a) Module 1, b) Module 2 (red nodes represent upregulated DEGs and blue nodes represent downregulated DEGs). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
KEGG pathway analysis of two significant modules in PPI.
Table 3.
Top 10 genes evaluated in the PPI network using five calculation methods and employing CytoHubba in Cytoscape. The overlapping hub genes in the top 10 by five ranked methods respectively in cytoHubba are highlighted in bold.
| Gene | MCC | Gene | MNC | Gene | Degree | Gene | EcCentricity | Gene | EPC |
|---|---|---|---|---|---|---|---|---|---|
| VEGFA | 610.0 | JUN | 25.0 | JUN | 26.0 | JUN | 0.17375 | VEGFA | 28.994 |
| JUN | 609.0 | VEGFA | 24.0 | VEGFA | 26.0 | VEGFA | 0.17375 | JUN | 28.507 |
| FGF2 | 434.0 | CCND1 | 20.0 | TLR4 | 23.0 | TLR4 | 0.17375 | CCND1 | 27.439 |
| CCND1 | 382.0 | TLR4 | 20.0 | CCND1 | 22.0 | FGF2 | 0.17375 | TLR4 | 26.957 |
| CDKN1A | 357.0 | FGF2 | 17.0 | FGF2 | 17.0 | LGALS3 | 0.17375 | FGF2 | 26.208 |
| TLR4 | 285.0 | CDKN1A | 16.0 | CDKN1A | 17.0 | BIRC3 | 0.17375 | CDKN1A | 26.047 |
| ITGAM | 245.0 | ITGAM | 16.0 | ITGAM | 17.0 | CXCR4 | 0.17375 | ITGAM | 24.942 |
| AURKB | 216.0 | AURKB | 13.0 | PRKACA | 15.0 | DDX58 | 0.17375 | LGALS3 | 22.287 |
| PSMB8 | 202.0 | PSMB8 | 10.0 | AURKB | 13.0 | HSPB3 | 0.17375 | CXCR4 | 22.271 |
| PSMD12 | 198.0 | THY1 | 10.0 | PSMB8 | 12.0 | ABCB1 | 0.17375 | BIRC3 | 21.898 |
MCC: Maximal Clique Centrality, MNC: Maximum Neighborhood Component, EPC: Edge Percolated Component.
Fig. 7.
Venn diagram of the intersecting genes derived using five algorithms. MCC: Maximal Clique Centrality, MNC: Maximum Neighborhood Component, EPC: Edge Percolated Component.
4. Discussion
It is well known that non-communicable diseases (NCDs) are the dominant cause of death. According to World Health Organization (WHO) reports, more than two-thirds of all deaths are caused by NCDs, principally cancer, diabetes, cardiovascular diseases (CVDs), and chronic respiratory diseases (Organization, 2010). CVDs are the number 1 cause of death globally (Xing, 2014). Moreover, WHO estimates show that annual CVD mortality is projected to increase by 6 million. Therefore, understanding the underlying molecular mechanisms in the pathogenesis of cardiovascular disease will open the door for further research to recommendations for CVD prevention and treatment, and may spur the development of superb therapeutic strategies. In this study, the potential molecular mechanisms that drive long-term cardiovascular ramifications of COVID-19 infection were evaluated by integrated bioinformatics analysis.
Almost 1.3 million people have died worldwide since COVID-19 emergence at the end of 2019 (Unudurthi et al., 2020). The major hallmark of COVID-19 is respiratory involvement, however accumulating data suggests that additional organs and tissues, such as the large intestine (Villapol, 2020), spleen (Pessoa et al., 2020), kidneys (Benedetti et al., 2020), brain (Iadecola et al., 2020), and heart (Bhatla et al., 2020) are severely affected by the SARS-CoV-2 infection. Recent studies have shown that the mortality rate in COVID-19 patients with CVD is higher compared to other comorbidities (Hessami et al., 2020; Unudurthi et al., 2020). A wide range of cardiovascular complications including acute-onset heart failure, cardiac arrest, arrhythmias, and myocarditis for COVID-19 have been reported (Bhatla et al., 2020; Inciardi et al., 2020). One of the common causes of myocarditis, an inflammatory disease of the heart, is viral infections (Blauwet and Cooper, 2010). Myocarditis is rare, however, it is a life-threatening health problem in almost all age groups (Cooper Jr, 2009). Although coronavirus-induced myocarditis is known, the long-term effect of COVID-19 on myocarditis remains unknown (Alhogbani, 2016). Currently, there is a growing literature examining cardiac involvement in SARS-CoV-2, and several mechanisms have been offered for the potential underlying drivers of COVID-19 related myocarditis (Bavishi et al., 2020). Cardiomyopathies are heart muscle diseases associated with cardiac dysfunction. Myocarditis, when caused by a viral infection, can lead to chronic inflammatory dilated cardiomyopathy (DCMi), which is the major long-term residual effects of myocarditis (Tschöpe et al., 2019). DCMi is characterized by inflammation of the heart muscle and is considered one of the most frequent causes of sudden cardiac death (Basso et al., 2001). Therefore, a clear understanding of the molecular mechanisms involved in cardiac muscle damages and DCMi development is crucial to develop appropriate treatment strategies for CVD patients with COVID-19.
Identification of aberrantly expressed hub genes can help us understand how a process is regulated by a stimulus and will provide a powerful framework for determining accurate therapeutic targets and prevention measures (Ceylan, 2021). Therefore, in the present study, a combined approach of microarray data analysis-bioinformatics tools was used to analyze two different microarray datasets (GSE150392 and GSE4172) to identify potential drivers. A total of 228 mutual DEGs were identified in the two datasets. Finally, according to data analysis results, 4 hub genes (FGF2, JUN, TLR4, and VEGFA) were identified based on our selection criteria.
Recent evidences addressed that hyper-inflammation and cytokine storm damage to multiple organs including heart (Nile et al., 2020; Xu et al., 2020). It has also been reported that cytokines and chemokines levels of patients with laboratory-confirmed COVID-19 infection significantly upregulated (Huang et al., 2020; Rothan and Byrareddy, 2020). Therefore, it is suggested that treatment strategies to reduce hyper-inflammation in COVID-19 patients need to be explored (Ge and He, 2020). Fibroblast growth factors (FGFs) are important mediators that induce cellular migration, cell differentiation, and proliferation (Faul, 2017). Previous studies indicated that FGFs have been associated with pathophysiologic alterations in the heart (Itoh et al., 2016). Among the FGF family members, FGF2 plays important role in the regulation of cardiac differentiation and cardiac remodeling by activating the signaling pathways including PI3K and RAS-MAPK (Ornitz and Itoh, 2015; Yamakawa et al., 2015). Previous studies have reported that exacerbated hypertrophy in cardiac myocytes stimulates by FGF2 (House et al., 2010; Santiago et al., 2014). This function of FGF2 increases the possibility of causing abnormal cardiac reprogramming through cardiac fibrosis (Faul, 2017). In addition, Karatolios et al. (Karatolios et al., 2012) reported that elevated FGF2 levels may also be associated with inflammatory pericardial disorders. Therefore, FGF2 is considered a danger signal molecule in pathological contexts. These previous findings are consistent with our results. In the present study, it was found that FGF2 mRNA expression showed a 3.83 and 2.01 fold increase in SARS-CoV-2 infected and DCMi samples, respectively.
CMs are both producers and targets of VEGFA (vascular endothelial growth factor A), which performs several functions in the heart tissue such as de novo formation of vessels (vasculogenesis) and the formation of new vessels (angiogenesis) (Chen et al., 2006). Hence, the development of a normal cardiovascular system is dependent on normal levels of VEGFA (Haigh, 2008; Braile et al., 2020). It is well known that the coordinated function of multiple cell types in the heart tissue is required for a functional heart. VEGFs are widely expressed in all cell types found in the heart. Therefore, VEGFA expression is tightly managed at a number of levels (Chiu et al., 2013). It has been demonstrated that disruption of VEGFA expression can be deleterious to embryonic heart development, and can also lead to cardiac hypertrophy development (Miquerol et al., 2000; Gogiraju et al., 2019). It was also observed that, in VEGFAhyper mice, an experimental model with elevated VEGFA levels, an increase in VEGFA was found to cause abnormalities in cardiac function (Marneros, 2018). These findings indicate that heart abnormalities are strongly related to increased VEGFA expression that causing progressive cardiac hypertrophy. In addition, it has been demonstrated that ACE2, a critical factor downregulated by SARS-CoV-2 (Verdecchia et al., 2020), antagonizes VEGFA (Yu et al., 2016). Consequently, SARS-CoV-2 leads to upregulation of VEGFA expression by canceling the antagonizing effect of ACE2 on VEGFA. Considering the complexity of the mechanism, further studies are needed to validate the provided associations.
Another factor that can lead to hyper-inflammation when over-stimulated is TLR4 (toll-like receptor 4) (Olejnik et al., 2018). It has been reported that TLRs are involved in a wide variety of functions in various pathological conditions including apoptosis, infectious diseases, neuronal degeneration, and cardiovascular diseases (Gay et al., 2014; Liu et al., 2015). TLR4 is a member of the toll/toll-like receptor family and is activated by several endogenous ligands associated with tissue injury. Previous studies have been shown that TLR4 signaling plays a role in the triggering and progression of many diseases including I/R (ischemia-reperfusion) injury, cancer, and hypertension (Roshan et al., 2016; Biancardi et al., 2017; Patra et al., 2020a). Interestingly, TLR4 level is higher in the heart compared with other TLR members, this can be shown as the reason why TLR4 is mainly involved in different cardiac complications including I/R injury, heart failure, hypertension, and myocarditis (Nishimura and Naito, 2005). Evidence suggests that TLR4 exacerbates the damage to the myocardium through the initiation of the expression of several pro-inflammatory genes and cell surface molecules (Liu et al., 2015). Intriguingly, most recent studies revealed interactions of SARS-CoV-2 and TLRs and also demonstrated that TLR4 may be associated with inflammatory consequences of COVID-19 (Patra et al., 2020b). Increased TLR4 signaling through binding of SARS-CoV-2 spike glycoprotein can be used by the virus to increase ACE2 expression to enter cells (Choudhury and Mukherjee, 2020). Therefore, it is assumed that myocarditis caused by SARS-CoV-2 is due to abnormal TLR4 signaling in COVID-19 patients (Aboudounya and Heads, 2021).
These findings suggest that TLR4 targeted strategies appear to be a promising approach for reducing myocardial inflammation and could create new ideas to treat COVID-19.
Immediate early genes (IEGs) are rapidly and transiently activated genes in response to diverse molecular and cellular events (Bahrami and Drabløs, 2016). Studies have revealed that an increase in IEGs such as c-fos, c-myc, and c-jun is associated with cardiac myocyte hypertrophy (Starksen et al., 1986; Clerk et al., 2002). c-Jun is a critical part of the AP-1(activator protein-1) complex transcription factors responsible for various cellular responses such as apoptosis and survival (Hsu and Hu, 2013). It is known that the CM responds to the pathological hypertrophic stimulus by tuning gene expression (Carreño et al., 2006). Moreover, experimental studies showed that myocardial AP-1 binding actions are considerably altered in cardiac hypertrophy, however, the exact role of c-Jun activity on cardiac hypertrophy is largely unknown since lack of a specific c-Jun inhibitor (Irukayama-Tomobe et al., 2004). Kim-Mitsuyama et al. (Kim-Mitsuyama et al., 2006) have revealed the first time that gene expression of cardiac hypertrophic markers suppressed in experimental models with c-Jun inhibition. It has been also reported that CM hypertrophy was inhibited in the absence of c-Jun's transactivating domain (DNJun) (Brown et al., 1994; Omura et al., 2002). Moreover, Battagello et al. recently reported that transcription factors such as AP-1, ATF-1, and CREB are phosphorylated after virus infection, which can lead to an increase in inflammatory gene expression in the host cell, which can prolongs viral permanence (Battagello et al., 2020). Overall, our results are consistent with those previous studies, and these findings suggest the c-Jun may be used for targeted therapy in pathologic cardiac hypertrophy.
As noted previously, a large number of patients with COVID-19 have been observed to develop new-onset cardiac dysfunctions and sudden cardiac arrest. However, the current knowledge of the COVID-19 and its long-term cardiovascular effects is grossly inadequate. In the present study, several hub genes (FGF2, JUN, TL4, and VEGFA) participating in the progression of inflammatory cardiomyopathy and cardiac remodeling were screened. Although the current study is based on bioinformatics analysis, the results confirm previous findings and contribute additional evidence that suggests SARS-CoV-2 employs a mechanism of upregulation of pathophysiological components to promote cardiac injury and may explain the potential therapeutic targets for late-onset heart disease and other cardiovascular complications among patients with COVID-19.
5. Conclusion
In summary, through GEO data analyses, four hub DEGs were identified as being significantly associated with major remodeling events such as hypertrophy, fibrosis, and metabolic alterations which contribute to new late-onset cardiovascular anomalies in the near future. Given that our analysis is in silico, additional validation experiments are needed to enable a better understanding of cardiovascular events including cardiomyopathy and cardiac remodeling associated with new coronavirus infection.
Data availability
All data used in this study are available in the GEO repository.
CRediT authorship contribution statement
Hamid Ceylan: Investigation, Methodology, Visualization, Writing - Original Draft.
Declaration of competing interest
The author declares that there is no conflict of interest with any financial organization or corporation or individual that can inappropriately influence this work.
References
- Aboudounya M.M., Heads R.J. COVID-19 and toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediat. Inflamm. 2021;2021 doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmerov A., Marbán E. COVID-19 and the heart. Circ. Res. 2020;126:1443–1455. doi: 10.1161/CIRCRESAHA.120.317055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhogbani T. Acute myocarditis associated with novel Middle East respiratory syndrome coronavirus. Ann. Saudi Med. 2016;36:78–80. doi: 10.5144/0256-4947.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami S., Drabløs F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016;62:37–49. doi: 10.1016/j.jbior.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Bansal M. Cardiovascular disease and COVID-19. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:247–250. doi: 10.1016/j.dsx.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso C., Calabrese F., Corrado D., Thiene G. Postmortem diagnosis in sudden cardiac death victims: macroscopic, microscopic and molecular findings. Cardiovasc. Res. 2001;50:290–300. doi: 10.1016/S0008-6363(01)00261-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battagello D.S., Dragunas G., Klein M.O., Ayub A.L.P., Velloso F.J., Correa R.G. Unpuzzling COVID-19: tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission. Clin. Sci. (Lond.) 2020;134:2137–2160. doi: 10.1042/CS20200904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavishi C., Bonow R.O., Trivedi V., Abbott J.D., Messerli F.H., Bhatt D.L. Acute myocardial injury in patients hospitalized with COVID-19 infection: a review. Prog. Cardiovasc. Dis. 2020;63:682–869. doi: 10.1016/j.pcad.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker R.C. Springer; 2020. Anticipating the Long-term Cardiovascular Effects of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C., Waldman M., Zaza G., Riella L.V., Cravedi P. COVID-19 and the kidneys: an update. Front. Med. 2020;7 doi: 10.3389/fmed.2020.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatla A., Mayer M.M., Adusumalli S., Hyman M.C., Oh E., Tierney A., Moss J., Chahal A.A., Anesi G., Denduluri S. COVID-19 and cardiac arrhythmias. Heart Rhythm. 2020;17:1439–1444. doi: 10.1016/j.hrthm.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancardi V.C., Bomfim G.F., Reis W.L., Al-Gassimi S., Nunes K.P. The interplay between angiotensin II, TLR4 and hypertension. Pharmacol. Res. 2017;120:88–96. doi: 10.1016/j.phrs.2017.03.017. [DOI] [PubMed] [Google Scholar]
- Blauwet L.A., Cooper L.T. Myocarditis. Prog. Cardiovasc. Dis. 2010;52:274–288. doi: 10.1016/j.pcad.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braile M., Marcella S., Cristinziano L., Galdiero M.R., Modestino L., Ferrara A.L., Varricchi G., Marone G., Loffredo S. VEGF-A in cardiomyocytes and heart diseases. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.H., Chen T., Birrer M. Mechanism of action of a dominant-negative mutant of c-Jun. Oncogene. 1994;9:791–799. [PubMed] [Google Scholar]
- Carreño J.E., Apablaza F., Ocaranza M.P., Jalil J.E. Cardiac hypertrophy: molecular and cellular events. Rev. Esp. Cardiol. (Engl. Ed.) 2006;59:473–486. [PubMed] [Google Scholar]
- Ceylan H. Identification of hub genes associated with obesity-induced hepatocellular carcinoma risk based on integrated bioinformatics analysis. Med. Oncol. 2021;38:63. doi: 10.1007/s12032-021-01510-0. [DOI] [PubMed] [Google Scholar]
- Chen Y., Amende I., Hampton T.G., Yang Y., Ke Q., Min J.-Y., Xiao Y.-F., Morgan J.P. Vascular endothelial growth factor promotes cardiomyocyte differentiation of embryonic stem cells. Am. J. Phys. Heart Circ. Phys. 2006;291:H1653–H1658. doi: 10.1152/ajpheart.00363.2005. [DOI] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.L., Morgan C.T., Lupton S.J., Lind J.M. Parent of origin influences the cardiac expression of vascular endothelial growth factor (Vegfa) BMC Med. Genet. 2013;14:43. doi: 10.1186/1471-2350-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerk A., Kemp T.J., Harrison J.G., Mullen A.J., Barton P.J., Sugden P.H. Up-regulation of c-jun mRNA in cardiac myocytes requires the extracellular signal-regulated kinase cascade, but c-Jun N-terminal kinases are required for efficient up-regulation of c-Jun protein. Biochem. J. 2002;368:101–110. doi: 10.1042/BJ20021083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L.T., Jr. Myocarditis. N. Engl. J. Med. 2009;360:1526–1538. doi: 10.1056/NEJMra0800028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul C. Cardiac actions of fibroblast growth factor 23. Bone. 2017;100:69–79. doi: 10.1016/j.bone.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Marggraf M., Stark A.W., Kaneko K., Aghayev A., Guensch D.P., Huber A.T., Steigner M., Blankstein R., Reichlin T. Association of ECG parameters with late gadolinium enhancement and outcome in patients with clinical suspicion of acute or subacute myocarditis referred for CMR imaging. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N.J., Symmons M.F., Gangloff M., Bryant C.E. Assembly and localization of toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Ge C., He Y. In Silico prediction of molecular targets of astragaloside IV for alleviation of COVID-19 hyperinflammation by systems network pharmacology and bioinformatic gene expression analysis. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.556984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogiraju R., Bochenek M.L., Schäfer K. Angiogenic endothelial cell signaling in cardiac hypertrophy and heart failure. Front. Cardiovasc. Med. 2019;6:20. doi: 10.3389/fcvm.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J.J. Role of VEGF in organogenesis. Organogenesis. 2008;4:247–256. doi: 10.4161/org.4.4.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:1–7. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessami A., Shamshirian A., Heydari K., Pourali F., Alizadeh-Navaei R., Moosazadeh M., Abrotan S., Shojaei L., Sedighi S., Shamshirian D., Rezaei N. Cardiovascular diseases burden in COVID-19: systematic review and meta-analysis. Am. J. Emerg. Med. 2020 doi: 10.1016/j.ajem.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House S.L., House B.E., Glascock B., Kimball T., Nusayr E., Schultz J.E.J., Doetschman T. Fibroblast growth factor 2 mediates isoproterenol-induced cardiac hypertrophy through activation of the extracellular regulated kinase. Mol. Cell. Pharmacol. 2010;2:143. doi: 10.4255/mcpharmacol.10.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008;2008 doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-C., Hu C.-D. Transcriptional activity of c-Jun is critical for the suppression of AR function. Mol. Cell. Endocrinol. 2013;372:12–22. doi: 10.1016/j.mce.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C., Anrather J., Kamel H. Effects of COVID-19 on the nervous system. Cell. 2020;183:16–27. doi: 10.1016/j.cell.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inciardi R.M., Lupi L., Zaccone G., Italia L., Raffo M., Tomasoni D., Cani D.S., Cerini M., Farina D., Gavazzi E. Cardiac involvement in a patient with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:819–824. doi: 10.1001/jamacardio.2020.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irukayama-Tomobe Y., Miyauchi T., Sakai S., Kasuya Y., Ogata T., Takanashi M., Iemitsu M., Sudo T., Goto K., Yamaguchi I. Endothelin-1–induced cardiac hypertrophy is inhibited by activation of peroxisome proliferator–activated receptor-α partly via blockade of c-jun nh2-terminal kinase pathway. Circulation. 2004;109:904–910. doi: 10.1161/01.CIR.0000112596.06954.00. [DOI] [PubMed] [Google Scholar]
- Itoh N., Ohta H., Nakayama Y., Konishi M. Roles of FGF signals in heart development, health, and disease. Front. Cell Dev. Biol. 2016;4:110. doi: 10.3389/fcell.2016.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatolios K., Moosdorf R., Maisch B., Pankuweit S. Cytokines in pericardial effusion of patients with inflammatory pericardial disease. Mediat. Inflamm. 2012;2012 doi: 10.1155/2012/382082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Mitsuyama S., Izumi Y., Izumiya Y., Namba M., Yoshida K., Wake R., Yoshiyama M., Iwao H. Dominant-negative c-Jun inhibits rat cardiac hypertrophy induced by angiotensin II and hypertension. Gene Ther. 2006;13:348–355. doi: 10.1038/sj.gt.3302670. [DOI] [PubMed] [Google Scholar]
- Kühl U., Pauschinger M., Noutsias M., Seeberg B., Bock T., Lassner D., Poller W., Kandolf R., Schultheiss H.-P. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111:887–893. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- Li M.-Y., Li L., Zhang Y., Wang X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:1–7. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang Y., Cao Z.Y., Wang M.M., Liu X.M., Gao T., Hu Q.K., Yuan W.J., Lin L. Up-regulated TLR 4 in cardiomyocytes exacerbates heart failure after long-term myocardial infarction. J. Cell. Mol. Med. 2015;19:2728–2740. doi: 10.1111/jcmm.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marneros A.G. Effects of chronically increased VEGF-A on the aging heart. FASEB J. 2018;32:1550–1565. doi: 10.1096/fj.201700761RR. [DOI] [PubMed] [Google Scholar]
- Maron B.J., Doerer J.J., Haas T.S., Tierney D.M., Mueller F.O. Sudden deaths in young competitive athletes. Circulation. 2009;119:1085–1092. doi: 10.1161/CIRCULATIONAHA.108.804617. [DOI] [PubMed] [Google Scholar]
- Miquerol L., Langille B.L., Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., Hou C., Wang H., Liu J., Yang D. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24:1–10. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70. doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Naito S. Tissue-specific mRNA expression profiles of human toll-like receptors and related genes. Biol. Pharm. Bull. 2005;28:886–892. doi: 10.1248/bpb.28.886. [DOI] [PubMed] [Google Scholar]
- Olejnik J., Hume A.J., Muhlberger E. Toll-like receptor 4 in acute viral infection: too much of a good thing. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T., Yoshiyama M., Yoshida K., Nakamura Y., Kim S., Iwao H., Takeuchi K., Yoshikawa J. Dominant negative mutant of c-Jun inhibits cardiomyocyte hypertrophy induced by endothelin 1 and phenylephrine. Hypertension. 2002;39:81–86. doi: 10.1161/hy0102.100783. [DOI] [PubMed] [Google Scholar]
- Organization, W.H . 2010. Chapter 1: burden: mortality, morbidity and risk factors (Internet) Global Status Report on Non-communicable Diseases. [Google Scholar]
- Organization, W.H., 2020. WHO Director-General's remarks at the media briefing on 2019-nCoV on 11 February 2020. World Health Organization, Geneva. Available via https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020.
- Ornitz D.M., Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra M.C., Shah M., Choi S. Toll-like receptor-induced cytokines as immunotherapeutic targets in cancers and autoimmune diseases. Semin. Cancer Biol. 2020;64:61–82. doi: 10.1016/j.semcancer.2019.05.002. [DOI] [PubMed] [Google Scholar]
- Patra R., Chandra Das N., Mukherjee S. Targeting human TLRs to combat COVID-19: a solution? J. Med. Virol. 2020;93:615–617. doi: 10.1002/jmv.26387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa M.S.L., Lima C.F.C., Pimentel A.C.F., Costa J.C.G. Multisystemic infarctions in COVID-19: focus on the spleen. Eur. J. Case Rep. Intern. Med. 2020;7 doi: 10.12890/2020_001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshan M.H., Tambo A., Pace N.P. The role of TLR2, TLR4, and TLR9 in the pathogenesis of atherosclerosis. Int. J. Inflamm. 2016;2016 doi: 10.1155/2016/1532832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J.-J., McNaughton L.J., Koleini N., Ma X., Bestvater B., Nickel B.E., Fandrich R.R., Wigle J.T., Freed D.H., Arora R.C. High molecular weight fibroblast growth factor-2 in the human heart is a potential target for prevention of cardiac remodeling. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Garcia G., Jr., Wang Y., Plummer J.T., Morizono K., Arumugaswami V., Svendsen C.N. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep. Med. 2020;1:100052. doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Siripanthong B., Nazarian S., Muser D., Deo R., Santangeli P., Khanji M.Y., Cooper L.T., Jr., Chahal C.A.A. Recognizing COVID-19–related myocarditis: the possible pathophysiology and proposed guideline for diagnosis and management. Heart Rhythm. 2020;17:1463–1471. doi: 10.1016/j.hrthm.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starksen N.F., Simpson P.C., Bishopric N., Coughlin S.R., Lee W., Escobedo J.A., Williams L.T. Cardiac myocyte hypertrophy is associated with c-myc protooncogene expression. Proc. Natl. Acad. Sci. 1986;83:8348–8350. doi: 10.1073/pnas.83.21.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Criteria Committee of the New York Heart Association (NYHA), 1994. Nomenclature and Criteria for Diagnosis of Diseases of the Heart and Great Vessels (9th ed.). Boston: Little, Brown & Co. pp. 253–256.
- Tschöpe C., Cooper L.T., Torre-Amione G., Van Linthout S. Management of myocarditis-related cardiomyopathy in adults. Circ. Res. 2019;124:1568–1583. doi: 10.1161/CIRCRESAHA.118.313578. [DOI] [PubMed] [Google Scholar]
- Tschöpe C., Ammirati E., Bozkurt B., Caforio A.L., Cooper L.T., Felix S.B., Hare J.M., Heidecker B., Heymans S., Hübner N. Myocarditis and inflammatory cardiomyopathy: current evidence and future directions. Nat. Rev. Cardiol. 2020:1–25. doi: 10.1038/s41569-020-00435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unudurthi S.D., Luthra P., Bose R.J., McCarthy J., Kontaridis M.I. Cardiac inflammation in COVID-19: lessons from heart failure. Life Sci. 2020;118482 doi: 10.1016/j.lfs.2020.118482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl. Res. 2020;226:57–69. doi: 10.1016/j.trsl.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittchen F., Suckau L., Witt H., Skurk C., Lassner D., Fechner H., Sipo I., Ungethüm U., Ruiz P., Pauschinger M. Genomic expression profiling of human inflammatory cardiomyopathy (DCMi) suggests novel therapeutic targets. J. Mol. Med. 2007;85:257–271. doi: 10.1007/s00109-006-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, M., 2014. Nanotechnology and nanomaterials for cardiac repair, Cardiac Regeneration and Repair. Elsevier, pp. 3–16.
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H., Muraoka N., Miyamoto K., Sadahiro T., Isomi M., Haginiwa S., Kojima H., Umei T., Akiyama M., Kuishi Y. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Rep. 2015;5:1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Lin Q., Qin X., Ruan Z., Zhou J., Lin Z., Su Y., Zheng J., Liu Z. ACE2 antagonizes VEGFa to reduce vascular permeability during acute lung injury. Cell. Physiol. Biochem. 2016;38:1055–1062. doi: 10.1159/000443056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this study are available in the GEO repository.