Abstract
Avian infectious bronchitis virus (IBV) is causing considerable economic losses in the world poultry industry. The main difficulty of prevention and control of IB disease is the numerous genotypes and serotypes. The genetic analysis of IBV was mainly based on the S1 gene which played an important role in infectivity. In the study, One hundred and thirty-nine strains of avian infectious bronchitis virus were isolated from chickens showing signs of disease in southern China during the period from April 2019 to March 2020. The nucleotide and amino acid sequences from the isolated field strains were compared to 22 published references. Nucleotide homologies ranged from 64.5% to 100% and amino acid homologies ranging from 70% to 99.8%. Six genotype IBV strains were co-circulating in southern China. QX-type was still the most dominant genotype. Alignment of nucleotide and amino acid sequences of S1 gene revealed that the substitutions, insertions and deletions are widely among isolated strains. Recombination analysis showed that there is a large number of recombinant strains amongst these isolates, forming new sub branches, subtypes and variants. Therefore, long-term continuing surveillance is necessary for IBV prevention and control.
Key words: IBV, epidemiology, S1 gene, southern China
INTRODUCTION
Avian infectious bronchitis (IB) is an acute and highly contagious disease characterized by upper respiratory symptoms, air sacculitis, proventriculitis, nephritis, enteritis and egg production drop, resulting in considerable economic losses in the poultry industry, globally (Yu et al., 2001; Cook et al., 2012; Sjaak de Wit et al., 2011). The causative agent of the diseases avian infectious bronchitis virus (IBV), which belongs to the genus gamma Coronavirus of the family Coronaviridae of the order Nidovirales (Cavanagh, 2005). Similar to other coronaviruses, avian infectious bronchitis virus genome consists of a linear, non-segmented, positive-sense, and single-stranded RNA approximately 27 kilobases (kb) in length and encodes four structural proteins: nucleocapsid (N) protein, envelope (E) protein, membrane (M) glycoprotein, and spike (S) glycoprotein (Sutou et al., 1988; Liu et al., 2009).
The glycoprotein S is a precursor protein, and subsequently cleaved into two non-covalently bound polypeptides (S1 and S2) by proteases (Stern and Sefton, 1982). S1 subunit forming the tip of the viral spike, and S2 subunit anchoring the S1 into viral membrane. The S1 glycoprotein is the main infectious and pathogenic protein of IBV, and plays a critical role in viral attachment and neutralizing antibodies induction (Cavanagh, 1983; Cavanagh et al., 1986; Shang et al., 2018). Interestingly, the differences in the S1 subunit contribute to significant divergences in the viral serotype as well as the poor cross-protection (Cavanagh et al., 1986; Koch et al., 1990), leading to serotype-specific host immunity against IBV. Currently, the rapid variation of S1 gene and the poor cross-protection between the different serotypes pose significant challenges to control this disease.
Considering the antigenic variability of IBV and the poor cross-protection of commercial vaccine against different IBV strains, the characterization of IBV strains based on genotype or serotype is beneficial to prevent this disease (Jones et al., 2004). However, the lack of standard positive serum makes the identification of serotype impossible. The variability in the S1 protein is considered as the basis for genotype and serotype classification, definition of antigen characteristics, induction of poor cross-protection of commercial vaccines, and emergence of IBV variants. Thus, genotype classification based on the S1 gene becomes the primary method for classifying IBV strains because of the rapid evolution of S1 gene and the importance of S1 in virus-host interactions and immune responses (Mo et al., 2013; Valastro et al., 2016). Till now, at least 32 genotypes have been identified based on the S1 gene worldwide, including QX-type, 4/91-type, LDT3-type, HN08-type, TW-type, TC07-2-type and Mass-type (Yu et al., 2001; Ji et al., 2011).
Long-term continuing surveillance is significantly important to control this disease. With the aim of understanding the epidemiology and the characteristics of IBV strains circulating in southern China, a long-term surveillance program was performed in southern China. This study covered the monitoring data from April 2019 to March 2020. Here, we described in detail the results of this study, including the distribution, characterization, and recombination events of IBV isolates in southern China. The present work provides information on the molecular epidemiology of IBV, which will be beneficial for the control of IBV in Chinese poultry industry.
MATERIALS AND METHODS
Ethical Approval
All animal experiments were approved by the Committee of the Ethics on Animal Care and Experiments at South China Agricultural University (approval ID: SYXK-2019-0136). Permission was issued for the field studies, and all of the owners of the farms gave informed consent to conduct the study on this site.
Samples Collection
From April 2019 to March 2020, an epidemiological survey was performed on yellow-feathered broiler chicken farms in southern China, including Guangdong, Guangxi, Fujian, Yunnan, Hunan, Hubei, Sichuan, Jiangsu, Anhui and Zhejiang provinces. All the farms were visited monthly to collect clinical samples (including kidney and trachea) from sick birds displaying respiratory signs, nephritis and egg production drop.
Isolation, Purification, and Identification of IBV
A total of 420 swab samples were collected for IBV detection. Virus isolation was performed using SPF chicken embryonated eggs according to the previous description (Han et al., 2011). Briefly, clinical samples were frozen and thawed three times, homogenized in PBS containing penicillin (100 U/mL) and streptomycin (100 μg/mL), clarified by centrifugation, and sterilized by filtration using a 0.22-μm filter. The supernatant was inoculated into the 9-day-old SPF chicken embryonated eggs by the allantoic sac route, and blind passage till the occurrence of embryonic lesions. Virus purification was performed using the limiting dilution method (Hebberecht et al., 2019). Briefly, the allantoic fluid was filtered by the filter, and added 0.1 mL into 0.9 mL PBS for 10 times to obtain 10−1~10−8 viral dilutions. The 9-day-old SPF chicken embryonated eggs was inoculated with each dilution, and harvested after 72 hours. According to the pathological characteristics and the reverse transcription-polymerase chain reaction (RT-PCR) assay results, the highest dilution of allantoic fluid confirmed IBV positive was used to inoculating chicken embryonated eggs three times.
The presence of IBV was identified and verified by RT-PCR as previously described (Liu et al., 2006; Feng et al., 2018). Briefly, specific primer pairs (sense primer: 5’-CCTAAGAACGGTTGGAAT-3’ and anti-sense primer: 5’-TACTCTCTACACACACAC-3’) were designated based on the M gene sequence, RNA was extracted from the allantoic fluids using a Viral DNA/RNA Miniprep Kit (Axygen, USA) following the manufacturer's instructions, and subjected to RT-PCR for IBV detection.
Cloning and Sequencing of S1 Gene
Specific primer sets for amplifying the entire S1 gene (sense primer: 5’-AACTGAACAAAAGACCGACT-3’ and anti-sense primer: 5’-CAAAACCTGCCATAACTAACA-3’) were designated based on the nucleotide sequence of LX4 strain (Accession No. AY338732). RT-PCR was carried out to amply the entire S1 gene using a high-fidelity pfu DNA polymerase (Q5™ High-Fidelity DNA polymerase, NEB) under the following conditions: 98°C for 10 s, 45 cycles of 98°C for 10 s, 56°C for 30 s, and 72°C for 30 s, and 72°C for 10 min.The amplicons were cloned into a pEASY-Blunt plasmid, and sequenced commercially (Sangon Biotech, Shanghai). Sequence analyses were carried out using the DNAStar software (Madison, WI, USA).
Sequence Alignment and Phylogenetic Analysis
Sequence alignments were performed with nucleotide sequences of IBV isolates and other IBV strains retrieved from GenBank using DNASTAR software (Madison, WI, USA). Phylogenetic analysis was performed based on the nucleotide sequences of S1 gene by neighbor-joining method by using MEGA version 7.0. The bootstrap values were determined from 1,000 replicates of the original data. All the reference IBV strains used in this study are provided in Table 1.
Table 1.
IBV reference strains used in this study.
| Strain | Origin | Year | Genotype/serotype | Motif a | Accession No. |
|---|---|---|---|---|---|
| YX10 | China | 2010 | QX-type | HRRRR | JX840411 |
| D90 | China | Vaccine strain | QX-type | HRRRR | MF508703 |
| DY05 | China | 2005 | QX-type | HRRRR | GQ265928 |
| A2 | China | 1996 | QX-type | HRRRR | AY043312 |
| LX4 | China | 1999 | QX-type | HRRRR | AY338732 |
| QXIBV | China | 1997 | QX-type | HRRRR | AF193423 |
| 4/91 | UK | 1991 | 4/91 serotype | RRSRR | AF093794 |
| TA03 | China | 2003 | 4/91 serotype | RRSRR | AY837465 |
| 7/93 | UK | 1993 | 4/91 serotype | RRSRR | Z83979 |
| HN08 | China | 2008 | HN08-type | RRFRR | GQ265940 |
| SAIBK | China | 2009 | HN08-type | RRFRR | DQ288927 |
| ArkDPI | USA | 1981 | Arkansas serotype | HRSRR | EU418976 |
| Ark99 | USA | 1973 | Arkansas serotype | HRSRR | M99482 |
| H52 | Netherlands | Vaccine strain | Mass serotype | RRFRR | AF352315 |
| H120 | Netherlands | Vaccine strain | Mass serotype | RRFRR | EU822341 |
| Ma5 | USA | Vaccine strain | Mass serotype | RRFRR | AY561713 |
| TW3468/07 | China | 2007 | TW-type | RRSRR | EU822336 |
| TW2575/98 | China | 1998 | TW-type | RRFRR | DQ646405 |
| TC07-2 | China | 2007 | TC07-2-type | HRRKR | GQ265948 |
| CK/CH/GD/KP10 | China | 2010 | TC07-2-type | HRHKR | HQ018919 |
| CK/CH/GD/NC10 | China | 2010 | TC07-2-type | HRRKR | HQ018903 |
| LDT3 | China | Vaccine strain | LDT3-type | RRFRR | KR608272 |
Recombination Analysis
Putative recombination analysis was performed based on the S1 gene of IBV isolates using the Recombination Detection Program 4 (RDP v.4.97) software. A total of 9 methods (including RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, PhyLPro, LARD and 3Seq) were employed to analyze the putative recombination events. Recombination events only supported by five of these methods with a P-value adjusted to 0.05 which were regarded as positive. The potential recombination events were further verified by SimPlot software (version 3.5.1).
RESULTS
Clinical Characteristics of IBV Isolates in Southern China
From April 2019 to March 2020, a total of 420 clinical samples were collected from sick chickens in southern China. The onset age of sick chickens mainly ranged between 5 and 60 days. All the IBV strains were isolated from chickens showing typical respiratory clinical signs such as difficulty in breathing, nasal discharge, sneezing, tracheal rales, watery eyes, lethargy, and mucosal thickening with serous or catarrhal exudates in the nasal passages, sinuses, and trachea, or showing typical nephritis symptoms such as enlarged and pale kidneys, frequently with urate deposits in the tubules, severe dehydration and weight loss. All the 420 field strains were confirmed IBV positive by observation of curled and dwarfed embryos, RT-PCR and sequencing, and 139 IBV strains were identified.
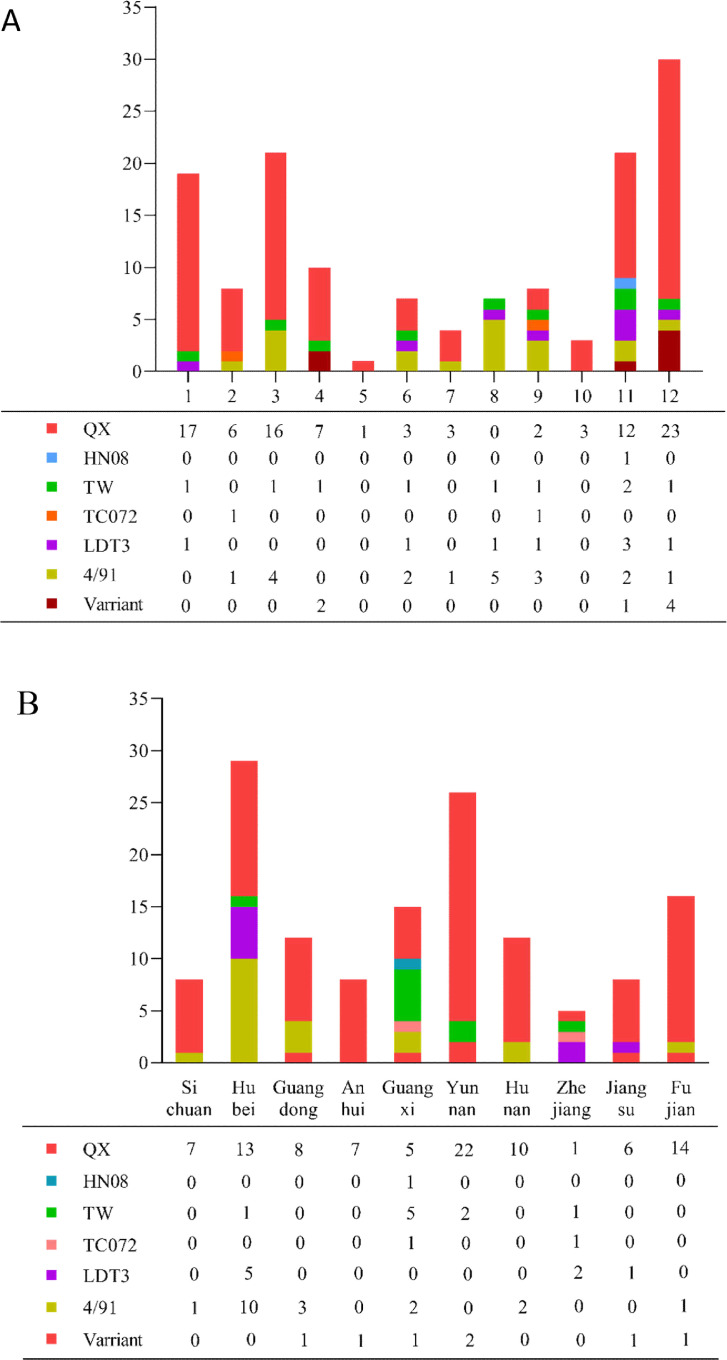
The information of IBV isolation and identification was statistically analyzed: 139 field IBV strains circulating in poultry industry were isolated from dead or diseased chickens in 10 provinces in southern China, including Yunnan (n = 26), Hubei (n = 29), Hunan (n = 12), Guangxi (n = 15), Jiangsu (n = 8), Fujian (n = 16 ), Guangdong (n = 12), Zhejiang (n = 5), Anhui (n = 8) and Sichuan (n = 8). Yunnan, Hubei, Fujian, and Guangxi were the four regions with the largest number of IBV isolates (Figure 1B). Moreover, 109 strains (78.4%) were collected in winter and spring seasons (Figure 1A).
Figure 1.
(A) The number of IBV strains isolated from each month is shown by genotype from April 2019 to March 2020. (B) The number of IBV strains isolated from each province is shown by genotype from April 2019 to March 2020.
Diversity Analysis of the S1 Gene of IBV Isolates
Due to the importance of S1 protein in serotyping and cross-protection, S1 gene of IBV isolates were sequenced for diversity analysis. As a result, a total of 420 nucleotide sequences of S1 gene were obtained, but only 139 sequences were identified because many IBV isolates from the same chicken flock had identical sequences. Within the S1 genes obtained in this study, the nucleotide sequences were 1614~1640 nt in length, and the corresponding deduced amino acid (aa) numbers of S1 protein were 538 aa (2.2%, 3/139), 539 aa (5.8%, 8/139), 540 aa (39.6%, 55/139), 541 aa (5%, 7/139), 542 aa (7.9%, 11/139), 543 aa (18%, 25/139), 544 aa (7.2%, 10/139), 545 aa (9.4%, 13/139) and 546 aa (5%, 7/139) (Supplementary material Table 2). Interestingly, the diverse S1 genes contained mutations, insertions and/or deletions, resulting in the differences of nucleotides and different numbers of nucleotides and deduced amino acids. All the sequences of S1 gene of IBV isolates have been submitted to GenBank, and the accession numbers and information of IBV strains were listed (Supplementary material Table 1).
Because the protease cleavage site motif is essential for cleaving during viral maturation period (Jackwood et al., 2001), we analyzed the motif sequence of the cleavage site in this study. As a result, a total of 8 cleavage site motifs were identified. Compared to the cleavage site motifs of the reference strains, the motif HRRRR (49.6%, 69/139), RRF(S)RR (27.3%, 38/139), HRHRR (15.1%, 21/139) and HRRKR (5%, 7/139) were frequently observed in IBV isolates. Sometimes, the cleavage recognition motif tends to be associated with genotypes. A degree of preference can be observed, with HRRRR presenting mostly in QX-type strains, and RRSRR in 4/91 type IBV strain. Interestingly, three cleavage site motifs were first identified, including RRFSR (0.7%, 1/139), RRLRR (0.7%, 1/139) and RRCRR (1.4%, 2/139), indicating the continuously evolution of IBV in south China. The cleavage site motifs of IBV have become complex and diverse in many regions with the spread of IBV in worldwide. Multiple sequence alignment based on the nucleotide sequence of S1 gene of the 139 IBV studied strains from this study and other strains retrieved from GenBank was performed using the Clustal X program. The nucleotide homology of S1 gene between isolates was 64.5% to 100%, while that between isolates and reference strains was 70% to 99.8%. These results indicated that the S1 gene of the field isolates was highly variable.
Furthermore, several insertions, deletions and mutations were observed in S1 gene of 139 isolated field strains (Supplementary material Table 2). Consistent with previous reports, three hypervariable regions (HVRs) were found according to the sequence analysis of S1 gene. Most variation in the deduced amino acid sequences of 139 field strains occurred in the regions 3-79, 61-136 and 154-432 (comparison with the S1 sequence of H120 strain). Generally, most of the isolates had deletions of N at site 25, G/S at site 130, G at site 96, and S at site 123.
Phylogenetic Analysis of IBV Isolates
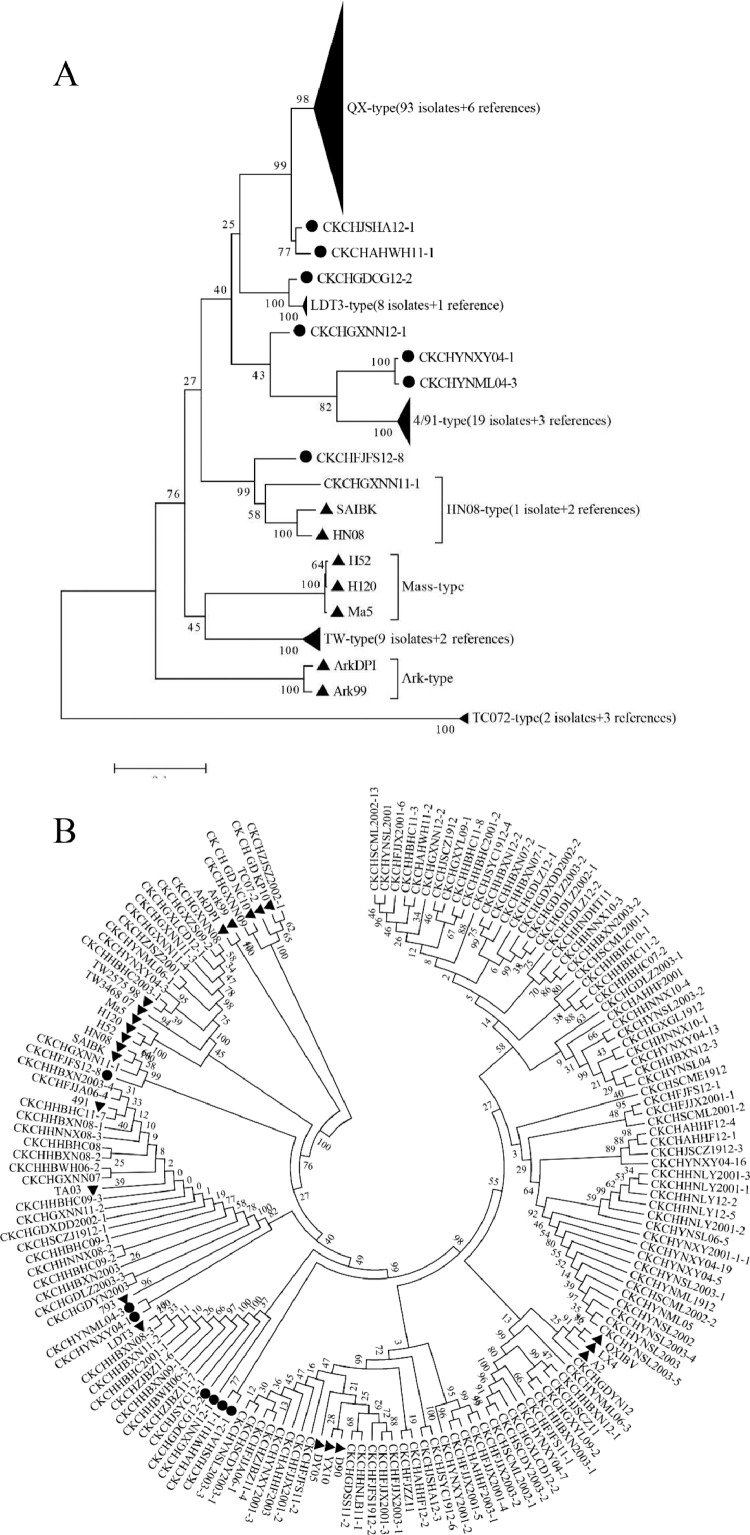
To determine the genetic relationships among the IBV strains, phylogenetic analysis was performed using the nucleotide sequences of the S1 gene of the 139 isolates and 22 reference strains. As a result, 132 IBV isolates and most reference strains were clustered into six groups: QX-type (66.9%, 93/139), 4/91-type (13.7%, 19/139), TW-type (6.5%, 9/139), LDT3-type (5.8%, 8/139), HN08-type (0.7%, 1/139), TC07-2-type (1.4%, 2/139). Additionally, seven field isolates CK/CH/YNML04-3, CK/CH/GXNN12-1, CK/CH/GDCG12-2, CK//CH/JSHA12-1, CK/CH/AHWH11-1, CK/CH/YNXY04-1 and CK/CH/FJFS12-8 were clustered into individual genotypes without reference strains based on their S1 genes and termed variants (Figure 2).
Figure 2.
Phylogenetic tree of 139 isolates and 22 reference strains for S1 genes of IBV (starting at the AUG translation initiation codon and ending at the cleavage recognition motifs). A) Phylogenetic tree of S1 gene of isolates from April 2019 to March 2020 (Traditional). B) Phylogenetic tree of S1 gene of isolates in from April 2019 to March 2020 (Circle). The phylogenetic tree was constructed using the MEGA version 5.0 by the neighbor joining method with No. of differences model and setting bootstrap 1,000 replicates. The reference strains are marked with ▲. Seven variant strains that were marked with ●. Unmarked strains are field strains isolated in this study.
QX-type strains were the most prevalent strain among the surveyed provinces (Figure 1B). The proportion and geographical distribution indicated that QX-type strain was the predominant genotype of IBV circulating in southern China. All the QX-type isolates shared a high degree of nucleotide (92%) and amino acid (92.6%) homology with QX-type vaccine strains, but shared low homology at nucleotide (86%) and amino acid (86.2%) levels with other vaccine strains (H120, H52, Ma5, 4/91, and LDT3-A).
The 4/91-type isolates were the second prevalent strain in southern China. A total of 19 strains (13.7%) were isolated from Guangxi, Hunan, Hubei, Fujian, Guangdong and Sichuan. The homology of nucleotide and amino acid among the 4/91-type isolates were 93.5% and 93%, respectively. The homology of nucleotide and amino acid sequences between the 4/91-type isolates and vaccine strains were 95.5% and 93.7%, respectively. In contrast, the 4/91-type isolates shared a lower degree of nucleotide (80%) and amino acid (78.8%) similarity with the vaccine strains (H120, H52, Ma5, QX and LDT3-A).
The TW-type isolates were the third most prevalent strain in southern China. A total of 9 strains were isolated from Guangxi, Hubei, Yunnan and Zhejiang. The nucleotides and amino acid homologies between TW-type isolates and TW2575/98 reference strain were 92.8% ~ 99.4% and 94.6% ~ 97.8%, but their nucleotide and amino acid homology with other Chinese mainland strains and vaccine strains (H120, H52, 4/91, QX and LDT3) were lower than 84.9% and 83.3% respectively. These observations may suggest different serotypes and low protection between vaccine strains and TW-type isolated strains.
The other 11 isolates belonged to LDT3-type (n = 8), TC07-2-type (n = 2), and HN08-type (n = 1). The LDT3-type isolates shared high identity with reference strains LDT3 at nucleotide and amino acid levels, and TC072-type isolates shared highly identity with TC07-2 strains. In contrast, the homology of nucleotide and amino acid sequences between the HN08-type isolates and vaccine strain HN08 was 88.7% and 87.1%, respectively.
Recombination Analysis of IBV Isolates
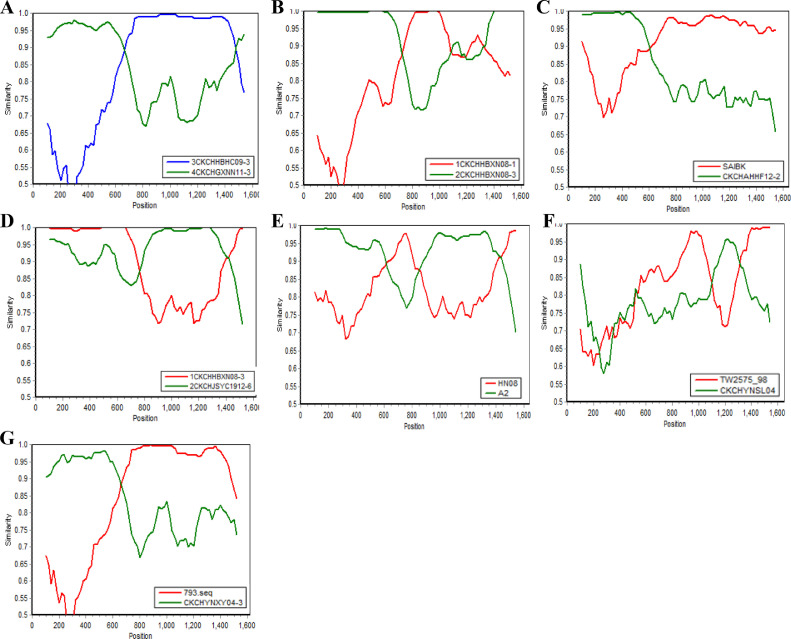
Strains were considered as recombinants if any crossover event appeared between two putative parental strains. It has been reported that the recombination events can occur in S1 gene of IBV (Wang et al., 1993), so we have identified putative recombination events in this study. As a result, evidence of recombination was found in 7 IBV strains using RDP package (Figure 3). As shown in Table 2, the variant CK/CH/GDCG12-2 was derived from recombination between SAIBK (major parent) and CK/CH/AHHF12-2 (minor parent), with recombination breakpoints mapping to positions 106 (beginning breakpoint) and 629 (ending breakpoint); The variant CK/CH/YNML04-3 was derived from recombination between CK/CH/GXNN11-3 (major parent) and CK/CH/HBHC09-3 (minor parent), the recombination region located in the nucleotide site 646–1468, with 99.1% similar to CK/CH/HBHC09-3 (4/91-type); The isolate CK/CH/GXNN12-1 appears to be a recombinant between strain CK/CH/HBXN08-3 as the major parent and strain CK/CH/HBXN08-1 as the minor parent; the recombination occurring between nucleotides 723–1091, which was 99.7% similar to CK/CH/HBXN08-1 (4/91-type), while the rest of the S1 gene was 97.8% similar to CK/CH/HBXN08-3 (LDT3-type. The variant CK/CH/JSHA12-1 was derived from recombination between CK/CH/JSYC1912-6 (major parent) and CK/CH/HBXN08-3 (minor parent), with recombination breakpoints mapping to positions 1395 (beginning breakpoint) and 1615 (ending breakpoint). The variant CK/CH/AHWH11-1 was a recombinant between A2 (major parent) and HN08 (minor parent), the recombination occurred between nucleotides 762-845; The isolate CK/CH/HBYNXY04-1 appears to be a recombinant with strain CK/CH/YNXY04-3(TW-type) as the major parent and 793 (4/91-type) as the minor parent, with recombination breakpoints mapping to positions 646 (beginning breakpoint) and 1621 (ending breakpoint). The isolate CK/CH/FJFS12-8 appears to be a recombinant with strain TW2575-98 as the major parent and CK/CH/YNSL04 (QX-type) as the minor parent, with recombination breakpoints mapping to positions 1101 (beginning breakpoint) and 1270 (ending breakpoint) (Table 2).
Figure 3.
SimPlot analysis of variant strains CK/CH/YNML04-3, CK/CH/GXNN12-1, CK/CH/GDCG12-2, CK/CH/JSHA12-1, CK/CH/AHWH11-1, CK/CH/YNXY04-1 and CK/CH/FJFS12-8. (A)The S1 gene of CK/CH/YNML04-3 was from a recombination of the reference strain CK/CH/GXNN11-3 (TW-type) and reference strain CK/CH/HBHC09-3 (4/91-type). (B) The S1 gene of CK/CH/GXNN12-1 was from a recombination of the reference strain CK/CH/HBXN08-3 (LDT3-type) and reference strain CK/CH/HBXN08-1 (4/91-type). (C) The S1 gene of CK/CH/GDCG12-2 was from a recombination of the reference strain SAIBK (HN08-type) and reference strain CK/CH/AHHF12-2 (4/91-type). (D) The S1 gene of CK//CH/JSHA12-1 was from a recombination of the reference strain CK/CH/JSYC1912-6 (QX-type) and reference strain CK/CH/HBXN08-3 (LDT3-type). (E) The S1 gene of CK/CH/AHWH11-1 was from a recombination of the reference strain A2 (QX-type) and reference strain HN08 (HN08-type). (F) The S1 gene of CK/CH/HBYNXY04-1 was from a recombination of the reference strain 793 (491-type) and reference strain CK/CH//YNXY04-3 TW-type). (G) The S1 gene of CK/CH/FJFS12-8 was from a recombination of the reference strain TW2575-98 (TW-type) and reference strain CK/CH/YNSL04 (QX-type). The y-axis gives the percent identity within a sliding window 200 bp wide centered on the position plotted, with a step size between plots of 20 bp.
Table 2.
S1 gene recombination in variant IBV strains.
| Breakpoint positions |
Major parenta |
Minor parentb |
Detection methods | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Begin | End | Strain | Genotype | Similarity | Strain | Genotype | Similarity | P value* | R G B M C S T |
| CK/CH/YNML04-3 | 646 | 1468 | CK/CH/GXNN11-3 | TW-type | 96% | CK/CH/HBHC09-3 | 491-type | 99.1% | 5.02 × 10−44 | + + + + + + + |
| CK/CH/GXNN12-1 | 723 | 1091 | CK/CH/HBXN08-3 | LDT3-type | 97.8% | CK/CH/HBXN08-1 | 491-type | 99.7% | 4.61 × 10−33 | + - + + + + + |
| CK/CH/GDCG12-2 | 106 | 629 | SAIBK | HN08-type | 97.1% | CK/CH/AHHF12-2 | 491-type | 98.5% | 7.71 × 10−17 | + + + + + + + |
| CK//CH/JSHA12-1 | 1395 | 1615 | CK/CH/JSYC1912-6 | QX-type | 94.9% | CK/CH/HBXN08-3 | LDT3-type | 99.5% | 1.05 × 10−19 | + + - + + + + |
| CK/CH/AHWH11-1 | 762 | 845 | A2 | QX-type | 96.4% | HN08 | HN08-type | 92.3% | 1.77 × 10−11 | + + + + - + + |
| CK/CH/HBYNXY04-1 | 646 | 1621 | CK/CH/YNXY04-3 | TW-type | 94.2% | 793 | 4/91-type | 96% | 4.43 × 10−31 | + + + + + + + |
| CK/CH/FJFS12-8 | 1101 | 1270 | TW2575-98 | TW-type | 78.6% | CK/CH/YNSL04 | QX-type | 97.1% | 3.58 × 10−13 | + + + + + + - |
Major parent = Sequence closely related to the transferred fragment in the recombinant.
Minor parent = Sequence most closely related to the sequence surrounding the transferred fragment.
P value of RDP method.
R: RDP G: GENECONV B: Bootscan M: Maxchi C: Chimaera S: SiScan T: Tree Topo.
DISCUSSION
There are two major factors accounting for the genetic diversity of IBV. Firstly, mutations, insertions and deletions frequently occurred in the IBV genomes due to the inaccuracy of the coronavirus RNA-dependent RNA polymerase (RDRP) (Mahmood et al., 2011; Feng et al., 2017). Secondly, unique random template switching during RNA replication may result in a high frequency of homologous RNA recombination (Cavanagh et al., 2005). Thereby, the variant strains and new genotypes of IBV are continuously emerging, worldwide.
The commercial attenuated vaccines against IBV (including QX, H120, H52, Ma5, and 4/91), have been widely used in Chinese poultry industry. However, IB disease still frequently break out in commercial poultry farms in China due to the poor cross-protection (Jackwood et al., 2001; Lim et al., 2011a). The genotypes or serotypes of IBV strains circulating in Chinese poultry industry are different from the currently used vaccine strains (Yu et al., 2001; Xu et al., 2007). Currently, the commercial vaccines against IBV used in southern China are attenuated strains, and always used with multi-genotypes combination, such as VG / GA + H120 + 491 + LDT3, LaSota + H120 + 491 and Clone30 + H120 + D90 + 491(Xu et al., 2021). However, the attenuated strains in commercial vaccines still need to be updated due to the evolutionary adaptation of the virus strain. Therefore, the long-term molecular epidemiological surveillance is of great significance for the prevention and control of IB disease.
This study is a part of our ongoing surveillance program, which focuses on investigation of IBV molecular epidemiology and identification of circulating IBV strains in commercial poultry farms in south China. In this study, a total of 139 IBV field strains were isolated from diseased or dead chickens in southern China during April 2019 to March 2020. Most diseased chickens were found at the age of 10-25 days, and few chickens were infected with IBV before 10 days. We speculated that the maternal antibody might provide a degree of protection against the circulating strains. Most of IBV infected broilers showed typical respiratory and nephritis symptoms, and few IBV infected broiler showed air sacculitis and peritonitis symptoms, indicating that the respiratory and nephron pathogenic IBV strains remained to be prevalent in south China (Li et al., 2010; Jackwood, 2012).
S protein was cleaved into S1 and S2 subunits when it was in viral maturation period, and the protease cleavage site motif was essential for cleaving (Jackwood et al., 2001). In the study, a total of 8 cleavage site motifs were identified in 139 IBV isolates, including HRRRR, RRFRR, HRHRR, HRRKR, RRLRR, RRFSR, and RRCRR. Previous studies showed that HRRRR was the main existing cleavage site motif of isolated IBV strains in China (Bing et al., 2007; Li et al., 2010). Consistent with these reports, HRRRR (69/139), RRF(S) RR (38/139), HRHRR (21/139) and HRRKR (7/139) were the predominant cleavage site motifs. Interestingly, three cleavage site motifs (RRLRR, RRFSR and RRCRR) were first identified in the present study, indicating the continuously evolution of IBV in south China.
S1 glycoprotein containing the neutralizing epitopes could induce the neutralizing antibodies and involve the serotype evolution, phenotype change and genetic diversity. In this study, we found that S1 genes of the IBV isolates possess different nucleotide in length, with series of mutations, insertions and deletions. It has been reported that the serotype differences of IBVs generally correlated with variations in HVR Hypervariable region of S1 protein, and the division of genotype was partly according to the HVR (Cavanagh and Davis, 1988; Moore et al., 1997; Cavanagh, 2007). Considering the variations in these IBV isolates, the genetic mutation of IBV HVRs may provide the explanation about the evolution of IBV in south China.
There are 6 different groups of IBV strains circulating in southern China, including QX-type, HN08-type, TW-type and LDT3-type, 4/91-type and Mass-type strains. Besides China, QX IBV has spread all over the world in the past 20 years, causing serious losses to laying hens and broilers on commercial farms (Franzo et al., 2017). In 1996, QX-type strain was first isolated in Shandong province, China (Kiss et al., 2015). Our long-term surveillance indicated QX-type strain was mainly associated with typical nephritis and pseudostratified clinical symptoms during 2011 to 2015 (Feng et al., 2017). In this study, we found that QX is the most predominant genotype in southern China, accounting for 66.9% of the isolates from April 2019 to March 2020. All these data provide evidence that QX-type strain is the dominant genotype in China. Currently, vaccines against QX-type strain are widely used to prevent and control IBV (Ji et al., 2020; Ren et al., 2020). However, the recombination and mutation frequently occurred under immunization pressure and natural selection, resulting in the continuous emergence of IBV variants; The 4/91-type and LDT3-type strains accounted for 13.7% and 5.8% of the total isolates, respectively. Both genotypes have their corresponding commercial vaccine, which are approved for use in Chinese poultry industry, since vaccination reduces the incidence of IB disease, but also increases the risk of recombination with field strains. The common circulation of multiple IBV genotypes and the increase in the proportion of IBV variants accelerate the possibility of virus recombination (Ali et al., 2018; Yan et al., 2019; Zhang et al., 2020), which poses a great challenge to IBV vaccination control. Therefore, it is necessary to continue monitoring the prevalence and evolution of the 4/91-type and LDT3-type IBV strains and other co-circulating viral strains in the field. TW-type IBV strains accounted for 6.5% of the field isolates in this study. Consistent with previous studies, the proportion of TW-type strains increased year by year (Yan et al., 2016). Therefore, it is urgent to develop new TW type IB vaccine to prevent the outbreak of TW-IB.
Recombination is an important mechanism for IBV evolution (Lee and Jackwood, 2001; Su et al., 2016). The unique discontinuous transcription system and the viral polymerase “jumping” contribute to the high RNA recombination frequency in the genome of IBV (Zou et al., 2010). The recombination always occurs when the viral polymerase switches from one template to another during genomic synthesis in a host (Lee and Jackwood, 2001). In this study, seven variants with different parental genotypes were detected by recombination analysis. The recombination events took place between the 4/91 and TW, LDT3 or HN08 genotype, and between the QX and TW, LDT3 or HN08 genotype, producing new variant strains and subgroups. These results suggested that recombination between different genotypes may contribute to the emergence of novel IBV variants. Additionally, our data indicated that 4/91-type strains involved the recombination events, providing clues that 4/91 has become an important gene donor in the genetic evolution of IBV. More effort will be required to monitor the recombination of IBV in the future.
Currently, the attenuated vaccine strains against IBV are widely used to control IB disease globally, controlling the occurrence of IB disease to a certain extent. However, the immune selective pressure is considered as an important reason for the occurrence of novel genotypes or serotypes of IBV (Lee and Jackwood, 2001). IBV possesses highly genomic mutation and recombination characteristics under immune selective pressure, causing the development of new genotypic groups under high-intensity vaccination (Pohuang et al., 2011). Actually, the probability of gene recombination between vaccine strains and epidemic strains is increasing in recent years (Lim et al, 2011b). More effort will be required to focus on the occurrence of IBV novel strains in the future.
Acknowledgments
This work was supported by the Guangdong Provincial Promotion Project on Preservation and Utilization of Local Breed of Livestock and Poultry (4000-F18260), National Natural Science Foundation of China (31802206, 32072842), and the Construction Project of Modern Agricultural Science and Technology Innovation Alliance in Guangdong Province (2020KJ128).
Ethical Approval and Consent to Participate: Our animal research was conducted under the guidance of the SCAU's Institutional Animal Care and Use Committee. The chicken sampling procedures were approved by the Animal Care and Use Committee of Guangdong Province, China.
Availability of Data and Materials: The nucleotide sequences of 139 IBV isolated strains have been deposited in GenBank under the accession no. MT766911 to MT769003 and MW042820- MW042867. The authors declare that all data supporting the findings of this study are available within the article.
Disclosures
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101169.
Appendix. Supplementary materials
References
- Ali A., Kilany W.H., Zain El-Abideen M.A., El Sayed M., Elkady M. Safety and efficacy of attenuated classic and variant 2 infectious bronchitis virus candidate vaccines. Poult. Sci. 2018;97:4238–4244. doi: 10.3382/ps/pey312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing G.X., Liu X., Pu J., Liu Q.F., Wu Q.M., Liu J.H. Different genotypes of nephropathogenic infectious bronchitis viruses co-circulating in chicken population in China. Virus Genes. 2007;35:333–337. doi: 10.1007/s11262-007-0100-5. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J. Gen. Virol. 1983;64(Pt 12):2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Evolution of avian coronavirus IBV: sequence of the matrix glycoprotein gene and intergenic region of several serotypes. J. Gen. Virol. 1988;69(Pt 3):621–629. doi: 10.1099/0022-1317-69-3-621. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67(Pt 7):1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Picault J.P., Gough R., Hess M., Mawditt K., Britton P. Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol.. 2005;34:20–25. doi: 10.1080/03079450400025414. [DOI] [PubMed] [Google Scholar]
- Cook J.K.A., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- Feng K.Y., Chen T., Zhang X., Shao G.M., Cao Y., Chen D.K., Lin W.C., Chen F., Xie Q.M. Molecular characteristic and pathogenicity analysis of a virulent recombinant avain infectious bronchitis virus isolated in China. Poult. Sci. 2018;97:3519–3531. doi: 10.3382/ps/pey237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K., Wang F., Xue Y., Zhou Q., Chen F., Bi Y., Xie Q. Epidemiology and characterization of avian infectious bronchitis virus strains circulating in southern China during the period from 2013-2015. Sci. Rep. 2017;7:6576. doi: 10.1038/s41598-017-06987-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo G, Massi P, Tucciarone CM, Barbieri I, Tosi G, Fiorentini L, Ciccozzi M, Lavazza A, Cecchinato M, Moreno A. Think globally, act locally: phylodynamic reconstruction of infectious bronchitis virus (IBV) QX genotype (GI-19 lineage) reveals different population dynamics and spreading patterns when evaluated on different epidemiological scales. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Sun C., Yan B., Zhang X., Wang Y., Li C., Zhang Q., Ma Y., Shao Y., Liu Q., Kong X., Liu S. A 15-year analysis of molecular epidemiology of avian infectious bronchitis coronavirus in China. Infect. Genet. Evol. 2011;11:190–200. doi: 10.1016/j.meegid.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebberecht L., Vancoillie L., Schauvliege M., Staelens D., Demecheleer E., Hardy J., Mortier V., Verhofstede C. Single genome sequencing of near full-length HIV-1 RNA using a limiting dilution approach. J. Virol. Methods. 2019 doi: 10.1016/j.jviromet.2019.113737. [DOI] [PubMed] [Google Scholar]
- Jackwood M.W. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56:634–641. doi: 10.1637/10227-043012-Review.1. [DOI] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Callison S.A., Lee C.W., Plaza H., Wade E. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 2001;45:366–372. [PubMed] [Google Scholar]
- Ji J., Gao Y., Chen Q., Wu Q., Xu X., Kan Y., Yao L., Bi Y., Xie Q. Epidemiological investigation of avian infectious bronchitis and locally determined genotype diversity in central China: a 2016–2018 study. Poult. Sci. 2020;99:3001–3008. doi: 10.1016/j.psj.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Xie J., Chen F., Shu D., Zuo K., Xue C., Qin J., Li H., Bi Y., Ma J., Xie Q. Phylogenetic distribution and predominant genotype of the avian infectious bronchitis virus in China during 2008-2009. Virol. J. 2011;8:184. doi: 10.1186/1743-422X-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.C., Savage C.E., Naylor C.J., Cook J.K.A., El-Houadfi M.A. Possible North African progenitor of the major European infectious bronchitis variant (793B, 4/91, CR88). Proceedings of the IV International Symposium on Avian Coronaand Pneumovirus Infections; Rauischholzhausen, Germany; 2004. [Google Scholar]
- Kiss I., Mató T., Homonnay Z.G., Kojer J., Farsang A., Bálint Á., Palya V. Survey indicates circulation of 4/91 and QX-type infectious bronchitis viruses in Hungary in 2014 - Short communication. Acta Vet. Hung. 2015;63:382–388. doi: 10.1556/004.2015.036. [DOI] [PubMed] [Google Scholar]
- Koch G., Hartog L., Kant A., Van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71(Pt 9):1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Li L., Xue C., Chen F., Qin J., Xie Q., Bi Y., Cao Y. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet. Microbiol. 2010;143:145–154. doi: 10.1016/j.vetmic.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T.H., Lee H.J., Lee D.H., Lee Y.N., Park J.K., Youn H.N., Kim M.S., Lee J.B., Park S.Y., Choi I.S., Song C.S. An emerging recombinant cluster of nephropathogenic strains of avian infectious bronchitis virus in Korea. Infect. Genet. Evol. 2011;11:678–685. doi: 10.1016/j.meegid.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim TH, Kim MS, Jang JH, Lee DH, Park JK, Youn HN, Lee JB, Park SY, Choi IS, Song CS. Live attenuated nephropathogenic infectious bronchitis virus vaccine provides broad cross protection against New Variant Strains. Poult. Sci. 2011;91:89–94. doi: 10.3382/ps.2011-01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Zhang Q.X., Chen J.D., Han Z.X., Liu X., Feng L., Shao Y.H., Rong J.G., Kong X.G., Tong G.Z. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch. Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang X., Wang Y., Li C., Han Z., Shao Y., Li H., Kong X. Molecular characterization and pathogenicity of infectious bronchitis coronaviruses: Complicated evolution and epidemiology in China caused by cocirculation of multiple types of infectious bronchitis coronaviruses. Intervirology. 2009;52:223–234. doi: 10.1159/000227134. [DOI] [PubMed] [Google Scholar]
- Mahmood Z.H., Sleman R.R., Uthman A.U. Isolation and molecular characterization of Sul/01/09 avian infectious bronchitis virus, indicates the emergence of a new genotype in the Middle East. Vet. Microbiol. 2011;150:21–27. doi: 10.1016/j.vetmic.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M.L., Li M., Huang B.C., Fan W.S., Wei P., Wei T.C., Cheng Q.Y., Wei Z.J., Lang Y.H. Molecular characterization of major structural protein genes of avian coronavirus infectious bronchitis virus isolates in Southern China. Viruses. 2013;5:3007–3020. doi: 10.3390/v5123007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.M., Jackwood M.W., Hilt D.A. Identification of amino acids involved in a serotype and neutralization specific epitope within the s1 subunit of avian infectious bronchitis virus. Arch. Virol. 1997;142:2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohuang T., Chansiripornchai N., Tawatsin A., Sasipreeyajan J. Sequence analysis of S1 genes of infectious bronchitis virus Isolated in Thailand during 2008–2009: Identification of natural recombination in the field isolates. Virus Genes. 2011;43:254–260. doi: 10.1007/s11262-011-0635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G., Liu F., Huang M., Li L., Shang H., Liang M., Luo Q., Chen R. Pathogenicity of a QX-like avian infectious bronchitis virus isolated in China. Poult. Sci. 2020;99:111–118. doi: 10.3382/ps/pez568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Luo C., Zhang W., Li F. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaak de Wit J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.F., Sefton B.M. Coronavirus proteins: structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J. Virol. 1982;44:804–812. doi: 10.1128/jvi.44.3.804-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutou S., Sato S., Okabe T., Nakai M., Sasaki N. Cloning and sequencing of genes encoding structural proteins of avian infectious bronchitis virus. Virology. 1988;165:589–595. doi: 10.1016/0042-6822(88)90603-4. [DOI] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Junker D., Collisson E.W. Evidence of natural recombination within the S1 gens of infectious bronchitis virus. Virology. 1993;192:710–716. doi: 10.1006/viro.1993.1093. [DOI] [PubMed] [Google Scholar]
- Xu C., Zhao J., Hu X., Zhang G. Isolation and identification of four infectious bronchitis virus strains in China and analyses of their S1 glycoprotein gene. Vet. Microbiol. 2007;122:61–71. doi: 10.1016/j.vetmic.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Xu G., Ma S., Cheng J., Zhao Y., Zhang G. An attenuated TW-like infectious bronchitis virus strain has potential to become a candidate vaccine and S gene is responsible for its attenuation. Vet. Microbiol. 2021;254:109014. doi: 10.1016/j.vetmic.2021.109014. [DOI] [PubMed] [Google Scholar]
- Yan S.H., Chen Y., Zhao J., Xu G., Zhao Y., Zhang G.Z. Pathogenicity of a TW-like strain of infectious bronchitis virus and evaluation of the protection induced against it by a QX-like strain. Front. Microbiol. 2016;7:1653. doi: 10.3389/fmicb.2016.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Sun Y., Huang X., Jia W., Xie D., Zhang G. Molecular characteristics and pathogenicity analysis of QX-like avian infectious bronchitis virus isolated in China in 2017 and 2018. Poult. Sci. 2019;98:5336–5341. doi: 10.3382/ps/pez351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Jiang Y., Low S., Wang Z., Nam S.J., Liu W., Kwang J. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 2001;45:416–424. [PubMed] [Google Scholar]
- Zhang X., Deng T., Lu J., Zhao P., Chen L., Qian M., Guo Y., Qiao H., Xu Y., Wang Y., Li X., Zhang G., Wang Z., Bian C. Molecular characterization of variant infectious bronchitis virus in China, 2019: Implications for control programmes. Transbound. Emerg. Dis. 2020;67:1349–1355. doi: 10.1111/tbed.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N.L., Zhao F.F., Wang Y.P., Liu P., Cao S.J., Wen X.T., Huang Y. Genetic analysis revealed LX4 genotype strains of avian infectious bronchitis virus became predominant in recent years in Sichuan area, China. Virus Genes. 2010;41:202–209. doi: 10.1007/s11262-010-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.