Abstract
Avian infectious bronchitis (IB), caused by avian infectious bronchitis virus (IBV), is an acute and highly contagious disease that is extremely harmful to the poultry industry throughout the world. The cross-using of different attenuated live vaccine strains has led to the occurrence of diverse IBV serotypes. In this study, we isolated an IBV strain from a chicken farm in southwest China and designated it CK/CH/SCMY/160315. Construction of a phylogenetic tree based on full S1 gene sequence analysis suggested that CK/CH/SCMY/160315 bears similarity to GI-28, and further comparison of S1 amino acid residues revealed that CK/CH/SCMY/160315 showed mutations and deletions in many key positions between LDT3-A and other GI-28 reference strains. Importantly, CK/CH/SCMY/160315 was identified as a novel recombinant virus derived from live attenuated vaccine strains H120 (GI-1), 4/91 (GI-13), LDT3-A (GI-28), and the field strain LJL/08-1 (GI-19), identifying at least 5 recombination sites in both structural and accessory genes. Pathogenicity analysis indicated that CK/CH/SCMY/160315 caused listlessness, sneezing, huddling, head shaking, and increased antibody levels in the inoculated chickens. To further describe pathogenicity of this novel strain, we assessed viral load in different tissues and conducted hematoxylin and eosin (HE) staining of the trachea, lungs and kidneys. Our results provide evidence for the continuing evolution of IBV field strains via genetic recombination and mutation, leading to outbreaks in the vaccinated chicken populations in China.
Key words: infectious bronchitis virus, recombination, pathogenicity
INTRODUCTION
Infectious bronchitis (IB) is a disease caused by the infectious bronchitis virus (IBV), which is a member of the Gamma-coronavirus genus, the Corona-viridae family, and the Nido-virales order (Walker et al., 2019). IBV poses a major global economic threat, causing a considerable reduction in the quality and quantity of layer chickens (Cavanagh, 2007; Laconi et al., 2020). Infected chickens develop neurological and respiratory symptoms, including disheveled feathers, depression, and respiratory distress (Wu et al., 2016; Xu et al., 2019). Furthermore, chickens infected with IBV also become susceptible to secondary infections with bacteria or other pathogens due to damage to tracheal cilia and therefore display a higher mortality rate (Zhou et al., 2017).
IBV has a single-stranded, positive-sense RNA genome of approximately 27.6 kb in length and thus is one of the largest known RNA viruses. Similar to the genomic RNA (gRNA) of other coronaviruses, two thirds of the IBV genome encode non-structural proteins, such as RNA-dependent RNA polymerase and other accessory and regulatory proteins, while the remaining third encodes 4 structural proteins, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) protein (Franzo et al., 2019). The S protein is critical for antigenic neutralization, hemagglutination, and determination of cell tropism. However, in the Golgi apparatus, spike (S)S protein is cleaved into S1 and S2 subunits by a cellular proteases upon viral invasion, which remains non-covalently linked (de Haan et al., 2004; Yamada and Liu, 2009). Nearly half of the amino acid of the S gene is occupied by the S1 domain, which contains the receptor-binding domain (RBD) and is essential for viral adsorption to the cellular receptor and induction of neutralizing antibodies (Kant et al., 1992; Casais et al., 2003; Promkuntod et al., 2014). In addition, hyper-variable regions (HVRs) of the S1 gene influence the antigenic relatedness and receptor binding with host cells, therefore most of studies have utilized the S1 subunit for IBV genotyping and classification (Valastro et al., 2016; Jiang et al., 2018; Shan et al., 2018; Parsons et al., 2019). The other half of S gene contains transmembrane and C-terminal cytoplasmic tail domains, which is occupied by the S2 domain (Luo and Weiss, 1998). The ecto-domain region of the S2 subunit contains a fusion peptide and 2 heptad repeat regions involved in oligomerization of the S protein, which is required for entry into susceptible cells (Degroot et al., 1987; Tripet et al., 2004).
The most effective way to prevent epidemic IBV is to vaccinate, typically using live attenuated vaccines derived from virulent strains serially passaged in embryonated chicken eggs (Laconi et al., 2020). Commercial attenuated vaccines such as H120, 4/91 and LDT3-A are already frequently used on farms. However, due to the incomplete proofreading mechanisms of coronavirus RNA polymerases and genetic recombination during viral replication, evolution generates extensive genotypic, antigenic, and pathogenic variability in progeny viruses (Cavanagh, 1997; Li et al., 2012; Xu et al., 2019). This frequent variation produces a number of IBV genotypes and serotypes with only limited cross-protective immunity between different serotypes, therefore failures in vaccination immunization are often reported. The QX-like type IBV is a member of the GI-19 group and represents one of the most important IBV genotypes globally, also representing the predominant type of IBV in China since 1999 (Liu et al., 2009). Many reports have pointed out that the QX strain has participated in recombination with vaccine strains and has resulted in changes of the antigenic characteristics of the vaccine strain (Abdel-Moneim et al., 2012; Fellahi et al., 2015). Previous studies have focused on the S1 gene due to its unique antigenicity (Madu et al., 2007; Zhu et al., 2016), but research in recent years has found that non-structural and helper viral proteins of IBV and other coronaviruses may play a regulatory role in viral replication, pathogenicity and immune escape (Armesto et al., 2009; Han et al., 2017). These findings suggest investigation of the whole-genome characteristics of coronaviruses will be of great significance for understanding the relationship between these virus and their antigenicity, tissue tropism, and pathogenicity (van Beurden et al., 2018).
To provide a comprehensive understanding of the genomic characteristics of the novel IBV strain CK/CH/SCMY/160315, we sequenced and compared whole-genome sequences of this novel strain with other reference strains. Furthermore, we also examined the pathogenicity, tissue tropism, and antigenicity in order to elucidate the pathogenic potential of this lineage. The research contributes to our knowledge of IBV evolution.
METHODS AND MATERIALS
Virus Isolation and Passage
The CK/CH/SCMY/160315 strain was isolated from clinical samples (trachea, lung, and kidney) of dead or diseased chickens displaying obvious respiratory symptoms and depression on an avian farm in Southwestern China. Animals had been vaccinated with H120 and LDT3-A. Viral isolation was carried out by inoculating 0.2 mL of 10% tissue homogenates into the allantoic cavity of 9-day-old specific-pathogen-free (SPF) chicken embryos, followed by incubation for 48 h and cooling at 4°C overnight. Allantoic fluid was harvested from the inoculated eggs and centrifuged at low speed (1000 × g) for 15 min. After removing the precipitate, the upper aqueous phase was stored at -80°C until future processing (Liu et al., 2009). The viral titer was determined by inoculating 9-day-old SPF embryonated eggs via their allantoic cavities with 10-fold serial dilutions of the viral stocks in phosphate-buffered saline, and the embryo 50% infectious doses (EID50) were determined with the method of Reed and Muench (Reed and Muench, 1938).
RNA Extraction, Viral cDNA Cloning, and Sequencing
Total RNA was extracted from virus-infected allantoic fluid using TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. cDNA was generated using the PrimeScript RT reagent Kit (Takara Bio Inc., Shiga, Japan) following the supplier's guidelines. The primers and strategies used for cloning the complete and 3′/5′ termini of IBV strains were as previously described (Wu et al., 2016). Where primers did not work due to sequence differences, new primers were designed based on newly determined sequences flanking those genome regions. Each PCR amplicon was cloned into a PMD-19 T vector system (Takara Bio Inc.), and 3 to 5 independent clones were sequenced for each amplicon. Each nucleotide was determined from at least 3 identical results generated from separate PCR products. The nucleotide sequences of the positive clones, determined by Sangon Biological Engineering Technology (Shanghai, China), were then assembled into complete genome sequence using SeqMan II program of DNAstar software package (DNAStar, Madison, WI).
Alignments and Phylogenetic Analysis
The complete S1 genome of IBV reference strains were obtained from GenBank. Sequences were aligned and analyzed using the ClustalW multiple alignment method in the MegAlign program of the DNASTAR software (version 7.1; DNAstar, Madison, WI). Phylogenetic trees were constructed using the Maximum likelihood method in the MEGA software (Tamura, Dudley, Nei, & Kumar, 2007), and bootstrap values were determined from 1,000 replicates of the original data. BLASTN analysis was performed using BLAST website (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and another website of Open Reading Frame Finder (ORF Finder, https://www.ncbi.nlm.nih.gov/orffinder/) was used to find the potential ORFs.
Putative Recombination Analysis
Programs embedded in the Recombination Detection Program 4 (RDP4) software suite were used to identify recombination events in the full-length IBV genome sequence via detection of breaking points using specific algorithms implemented in RDP4. Seven detection methods in RDP (v.4.36), including RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan and 3Seq, were used to confirm recombination events. Only transferred gene fragments where at least 5 detection methods showed a p-value≤1 × 10−14 were accepted. Furthermore, SimPlot (v3.5.1, JHK University, Baltimore, MD) was used to confirm the putative recombination events and precise recombination breakpoints with a 200-bp window sliding along the genome (20-bp step size).
Pathogenicity Testing
One-day-old SPF layer chickens (N = 60) were randomly divided into 2 groups (test and control group, n = 30 each), and housed in different isolators with positive pressure in air-conditioned rooms. All chickens used in this study were obtained from Boehringer Ingelheim Co., ME, China. Chickens aged 1 day in test group were inoculated with CK/CH/SCMY/160315 via oculonasal administration with a dose of 106 EID50/per chick. Chickens in control group were inoculated with sterile PBS (negative controls) using the same routine. All birds were observed twice daily for clinical signs, including tracheal rales, nasal discharge, coughing, eye irritation, and watery diarrhea. Morbidity and mortality were recorded daily. The birds were observed for 21 days post infection (dpi), with food and water provided ad libitum. All sampling and surviving birds were killed humanely using carbon dioxide/oxygen, which was followed by exsanguination.
Blood samples were collected at 4, 8, 12, 16, and 21 dpi from each group. IBV-specific IgG in the sera was assessed using a commercial IBV enzyme-linked immunosorbent assay (ELISA) kit (IDEXX Lab. Inc., Westbrook, ME) following the manufacturer's protocol. At 4, 8, and 12 dpi, 3 chickens from each group were euthanized. Gross lesions were recorded and samples of the trachea, lung, kidney, spleen, liver, cecal tonsil and bursa of fabricius were collected. The samples were processed as follows: after flushing the tissue with 3 mL of PBS and grinding the tissue in liquid nitrogen, the milled powder was transferred to a centrifuge tube where 1 ml TRizol was added already, and the fluids were collected and stored at -80°C until further analysis (Xu et al., 2019). After thawing, RNA was extracted from these samples. Viral loads was quantified by real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR).
A pair of N-gene specific primers (N-F: GCTGCTAAAGGTGCTGATACT; N-R: GCTGTTGATCTTCCACTCCTAC) was designed, and an absolute quantitative method has been used to determine the viral titers in the different tissues using SYBR Green I Real time PCR. Primers were designed by using Primer Premier 5.0 software (Version 5.0, Premier Biosoft, Palo Alto, CA) and synthesized by Sangon Biological Engineering Technology (Shanghai, China). Then, the amplification product was cloned into a pMD19-T vector (Takara) as a positive plasmid to establish the standard curve.
Histopathology
Hematoxylin and eosin (H&E) staining of the lung, tracheas and kidneys tissues was performed for histopathological examination. Briefly, samples collected at necropsy were immediately fixed in 10% neutral buffered formalin, dehydrated in alcohol, embedded in paraffin, and stained with hematoxylin and eosin (HE) for observation using standard light microscopy, as described previously (Ren et al., 2020).
Statistical Analysis
Data are expressed as mean ± standard deviation. Viral titers were analyzed by a Student's t-test using GraphPad Prism for Windows Version 5 (GraphPad Software, La Jolla, CA). For all tests, the following notations are used to indicate significant differences between the groups: *P< 0.05, **P< 0.01, ***P< 0.001.
Animals and Ethics Statement
The viral challenge experiment was approved by the Animal Ethics Committee (AEC) of the College of Life Sciences, Sichuan University (License: SYXK (Chuan) 2013-185). All experimental procedures and animal care strictly followed the guidelines of Animal Management of Sichuan University.
RESULTS
Complete Genome Characterization of CK/CH/SCMY/160315
The complete genome sequence of IBV isolates CK/CH/SCMY/160315 was 27,608 nucleotides in length, excluding the poly-A tail, and we have submitted the full-length sequence to NCBI (accession number: MT505388). The Open Reading Frame Finder (ORF Finder) analysis predicted 10 ORFs and revealed the following genome structure as: 5’UTR-1a-1b-S-3a-3b-E-M-5a-5b-N-3’UTR. In addition, there was also an ORF with the potential to code for a protein of 94 amino acids between gene M and 5a of the strain CK/CH/SCMY/160315, which was identified previously in some IBV strains (Dolz et al., 2012). Our analysis revealed that the genome of CK/CH/SCMY/160315 shares the highest identity with the CK/CH/SCYB/140913 strain (Xia et al., 2016), and has 94.60%,92.25%, 90.82%, and 88.34% identity with the commercial vaccine H120 (GI-1), 4/91 (GI-13), LDT3-A (GI-28), and field strain LJL/08-1 (GI-19), respectively (Zhang et al., 2010; Han et al., 2016). The ORFs, nucleotide similarity, amino-acid identity, and genomic regions present in IBV strains CK/CH/SCMY/160315 and the 4 reference strains used in this study are listed in Table 1.
Table 1.
Sequence length and nucleotide and amino acid sequence identity (%) of different regions of CK/CH/SCMY/160315 compared to 3 representative vaccine strains (H120, 4/91 and LDT3-A) and a field strain (LJL/08-1).
| Length(nt/nn) |
Pairwise % Identity (nt/aa) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Region | ck/CH/SCMY/160315 | 4/91 | H120 | LDT3-A | LJL/08-1 | 4/91 | H120 | LDT3-A | LJL/08-1 |
| complete genome | 27,627 | 27,618 | 27,652 | 27,681 | 27,657 | 92.25(89.02) | 94.60(93.12) | 90.82(76.58) | 88.34(85.77) |
| 5′UTR | 1−528 | 1−528 | 1−528 | 1−529 | 1−526 | 99.81 | 95.27 | 94.18 | 93.16 |
| ORF1a | 529−12,330 (3,933) | 529−12,327 (3,933) | 529−12,300 (3,933) | 530−12,391 (3,963) | 527−12,387 (3,953) | 95.69(96.47) | 99.01(97.79) | 84.45(85.01) | 85.20(86.79) |
| ORF1b | 12,405−20,219 (2,604) | 12,330−20,363 (2,652) | 12,300−20,363 (2,687) | 12,391−20,430 (2,644) | 12,462−20,420 (2,652) | 91.34(95.87) | 94.59(97.34) | 94.09(97.65) | 90.69(96.72) |
| S | 20,313−23,810 (1,165) | 20,314−23,808 (1,165) | 20,314−23,802 (1,162) | 20,381−23,878 (1,165) | 20,371−23,868 (1,165) | 84.06(83.95) | 85.15(85.42) | 98.01(98.54) | 89.01(89.87) |
| 3a | 23,810−23,983 (57) | 23,808−23,981 (57) | 23,802−24,479 (57) | 23,878−24,051 (57) | 23,868−24,041 (57) | 86.78(85.96) | 82.76(82.46) | 99.43(98.25) | 88.54(75.44) |
| 3b | 23,983−24,174 (63) | 23,981−24,175 (64) | 23,975−24,169 (64) | 24,051−24,239 (62) | 24,041−24,232 (63) | 74.36(65.62) | 80(73.44) | 95.76(95.24) | 95.34(93.65) |
| E | 24,155−24,436 (93) | 24,156−24,476 (106) | 24,150−24,479 (109) | 24,223−24,549 (108) | 24,213−24,494 (93) | 85.28(89.25) | 86.67(82.20) | 90.74(87.91) | 98.94(97.85) |
| M | 24,433−25,101 (222) | 24,457−25,125 (222) | 24,451−25,128 (225) | 24,521−25,198 (225) | 24,491−25,162 (223) | 91.89(94.12) | 91.87(93.64) | 89.31(92.27) | 96.28(96.41) |
| 5a | 25,469−25,666 (65) | 25,485−25,682 (65) | 25,488−25,930 (65) | 25,552−25,749 (65) | 25,526−25,723 (65) | 86.87 (86.15) | 88.83 (83.08) | 84.34 (81.54) | 89.88(86.15) |
| 5b | 25,663−25,911 (82) | 25,679−25,927 (82) | 25,682−25,930 (82) | 25,746−26,018 (82) | 25,961−25,812 (49) | 93.17 (91.46) | 91.97 (90.24) | 94.78 (93.90) | 95.33 (93.90) |
| N | 25,854−27,083 (409) | 25,870−27,099 (409) | 25,873−27,102 (409) | 25,937−27,166 (409) | 25,912−27,141 (409) | 94.15 (94.87) | 93.25 (95.60) | 89.13 (92.18) | 89.71 (92.91) |
| 3′UTR | 27,084−27,627 | 27,100−27,618 | 27,102−27,652 | 27,166−27,681 | 27,142−27,657 | 98.69 | 99.24 | 92.66 | 97.82 |
The highest similarity after the genetic comparison is highlighted in bold.
As shown in Table 1, the 4 structural proteins (S, E, M, N) of the CK/CH/SCMY/160315 strain have varying similarities with the reference strains. The spike protein shares 84.06% to 98.01 % nucleotide homology and amino-acid identity (83.95%–98.54%) with reference strains, the region of which was most similar to the LDT3-A strain. However, E protein and M protein exhibited higher nucleotide homology (98.94%, 96.28%) and amino acid identity (97.85%, 96.41%) with LJL/08-1, respectively. A high N gene nucleotide similarity (94.15%) and amino acid similarity (94.87%) was found with strain 4/91. Notably, 2 other regions of CK/CH/SCMY/160315 displayed high nucleotide homology (99.43%, 95.76%) and amino-acid identity (98.25%, 95.24%) with the LDT3-A strain, including accessory protein 3a and 3b, while accessory proteins 5a and 5b exhibited relatively higher nucleotide homology (89.88%, 95.33%) and amino-acid identity (86.15%, 93.90%) with the LJL/08-1 strain. In addition, the 1a gene had a 99.01% nucleic acid and 97.79% amino-acid similarity with the H120 strain. These findings indicate that the CK/CH/SCMY/160315 genome is closely related to the genomes of both vaccine strains (LDT3-A, 4/91 vaccine, and H120) and field strains (LJL/08-1).
Phylogenetic Analysis of the CK/CH/SCMY/160315 Based on the S1 Gene
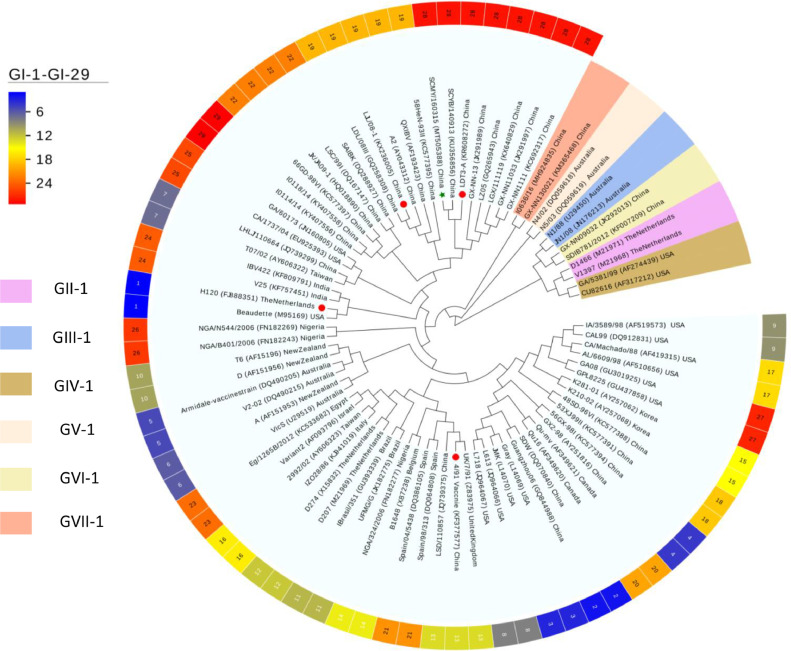
Phylogenetic analysis was performed using complete nucleotide sequences of the S1 gene of CK/CH/SCMY/160315 and 82 reference IBVs strains, which were representative of 36 well-established lineages and 7 genotypes (Valastro et al., 2016; Chen et al., 2017; Ma et al., 2019). The results showed that CK/CH/SCMY/160315 formed a homologous branch with LDT3-A, an attenuated vaccine commonly used in China (Han et al., 2016), but clearly clustered into 2 distinct subgroups within the GI-28 serotype (Figure 1). Another branch of GI-28 contained 5 strains that were endemic in southern China between 2005 and 2011 (Chen et al., 2017).
Figure 1.
Maximum-likelihood phylogeny of the CK/CH/SCMY/160315 strain (green star) and 82 IBV reference strains based on the complete nucleotide sequence of the S1 gene. Four other strains are highlighted with red circles for comparison. The trees were constructed using 1,000 bootstrap replicates with the MEGA 7.0 software. The 82 reference strains include 78 reference strains classified as GI-GVII genotypes and 4 strains (highlighted using a red circle) which were used for whole genome sequence comparison, phylogenetic analysis and the recombination analysis.
Alignment of the S1 gene and inferred amino acid sequences of CK/CH/SCMY/160315 and the GI-28 reference strains showed that for all, the length was 1620 nucleotides, corresponding to a 540-amino acid S1 protein. As illustrated in Figure 2, CK/CH/SCMY/160315 was missing 1 amino acid at positions 287 and 24, respectively, compared to GI-28. Meanwhile, of the 42 identified substitutions between CK/CH/SCMY/160315 and GI-28, 25 were located in the HVR (3, 4 and 18 are located in HVR1, HVR2, and HVR3, respectively), and the remaining 17 mutations were located in the far C-terminus of the S1 subunit of the S protein. However, these two subgroups shared 11 amino acid substitutions in the RBD region, including 1 amino acid deletion in GI-28 references strains. Comparing CK/CH/SCMY/160315 with LDT3-A strains, only 3 mutations were observed, all of which were located in HVR2. Previous research has shown that the amino acid residue at position 69 of the S1 subunit is important for the RBD, and mutations will affect the ability to bind to tracheal cilia (Bouwman et al., 2020). We found that amino acid residue position 69 of the strain CK/CH/SCMY/160315 was changed from I to V, suggesting a difference in antigenicity between the two strains. Moreover, we found CK/CH/SCMY/160315 two specific differences in the S1 amino acid sequence at positions 155H and 121Q which were not found in any other IBV strains.
Figure 2.
Alignment of the deduced amino acid sequences of the S1 subunit of the S1 protein of IBV strains CK/CH/SCMY/160315, LDT3-A, 140913, GX-NN-13, LZ05, GX-NN11033, 111119 and NN1111. Amino acid sequences encoding the 3 hypervariable regions are shown in bold. The yellow box represents differential sites compared with other GI-28 types, while missing sites are demarcated as a red box. Green indicates unique mutation sites, and * represents differential site compared with the LDT3-A strain.
Recombination Analysis
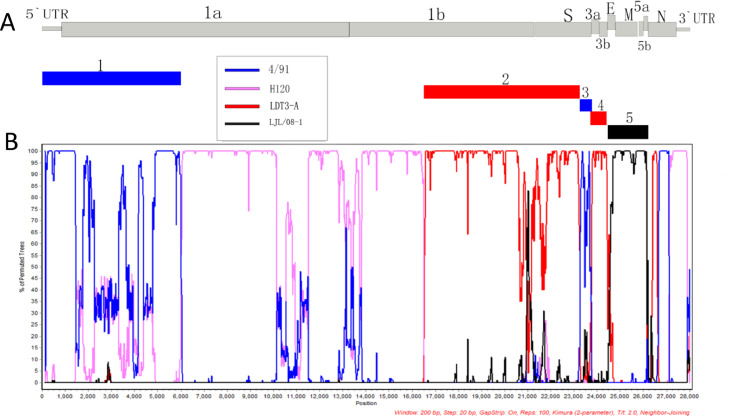
To identify potential recombination events, we used the RDP4 and SimPlot softwares. Five recombination breakpoints within the CK/CH/SCMY/160315 genome were identified from similarity plots. Recombination breakpoints were not preferentially located in any regions, occurring in all genes except the N gene (Figure 3). The results of 7 different methods for recombination detection in the RPD4 software (Table 2) clearly supported the SimPlot results, with CK/CH/SCMY/160315 exhibiting a greater similarity to strain 4/91 in two fragments (from 5′UTR–nt5870; and nt22,885–23,810), including the region of which includes the entire 5′UTR, part of the ORF1a area and S2 section of structure protein S. Although the S2 protein has been considered to be relatively conserved in previous studies (Adzhar et al., 1997), we found that the S2 protein still has the possibility of recombination, which is consistent with results by Abro et al. (Abro et al., 2012) and others. The genomic position from nt16,351-22,884 and nt23,811-24,175 were most similar to LDT3-A. This region comprised most of the ORF1b, part of the S2 gene as well as the entire S1 gene and two accessory proteins 3a, 3b. Interestingly, we found that the two structural proteins E and M, as well as two accessory proteins (5a and 5b), located at nt24,155-25,911, were most similar to the LJL/08-1 strain. This strain belongs to the GI-19 group, a kidney-type pathogenic strain currently circulating in China.
Figure 3.
Genetic characteristics of the CK/CH/SCMY/160315 (A) and Bootscan analysis recombination detection (B). Analysis was carried out using a sliding 200-bp window using 20-bp steps. The y-axis indicates the percentage similarity between the query sequence and the reference sequences, while the x-axis represents gene locus. Reference strains H120 (pink), 4/91 (blue), LDT3-A (red), and LJL/08-1 (black) were used as putative parental strains. In addition, we used 5 rectangles with corresponding colors to associate the genetic characteristics of the IBV strain with the recombination site.
Table 2.
Recombination events of CK/CH/SCMY/160315 as assessed using 7 different methods within the RPD4 software.
| Breakpoints |
Major | Minor |
P value of the detection methods |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Recombinant event | Beginning | Ending | (Similarity) | (Similarity) | RDP | GENECONV | BootScan | MaxChi | Chimaera | SiScan | 3Seq |
| 1 | 9 | 5,870 | H120(97.8%) | 4/91(99.8%) | 5.993 × 10−120 | 1.443 × 10−116 | 5.492 × 10−58 | 2.087 × 10−39 | 1.114 × 10−38 | 1.968 × 10−26 | 1.866 × 10−66 |
| 2 | 16,351 | 22,884 | 4/91(94.2%) | LDT3-A(99.7%) | NS | 2.428 × 10−306 | NS | 2.876 × 10−21 | 3.130 × 10−76 | 3.733 × 10−99 | 2.470 × 10−322 |
| 3 | 22,885 | 23,810 | LDT3-A(86.5%) | 4/91(99.8%) | 1.003 × 10−57 | 2.501 × 10−65 | 3.398 × 10−65 | 2.427 × 10−16 | 2.384 × 10−16 | 8.575 × 10−18 | NS |
| 4 | 23,811 | 24,175 | H120(89.9%) | LDT3-A(97.3%) | 1.228 × 10−24 | 8.905 × 10−24 | 9.431 × 10−24 | 1.843 × 10−18 | 5.718 × 10−17 | 3.337 × 10−25 | 2.220 × 10−15 |
| 5 | 24,155 | 25,911 | 4/91(92.4%) | LJL/08-1(97%) | 6.599 × 10−85 | 9.441 × 10−70 | 2.080 × 10−87 | 1.860 × 10−26 | 1.279 × 10−03 | 2.913 × 10−32 | 2.828 × 10−48 |
| 6 | 26,594 | 27,083 | LDT3-A(90.1%) | 4/91(99.6%) | NS | 9.947 × 10−15 | 3.247 × 10−17 | 5.360 × 10−08 | 1.336 × 10−04 | 8.825 × 10−14 | 3.515 × 10−02 |
Clinical Signs
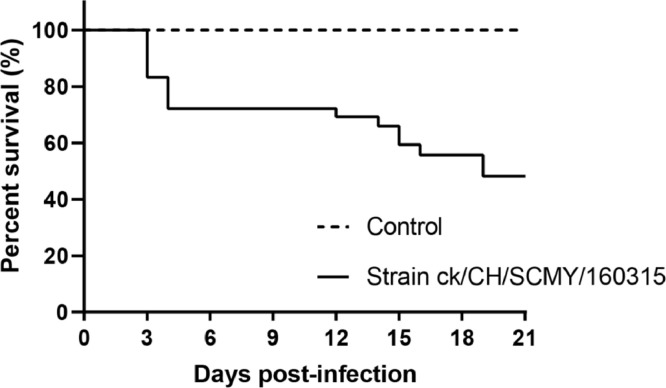
Clinical symptoms such as listlessness, sneezing, huddling and head shaking occured in 4 birds in the test group by 2 dpi, followed by development of symptoms in nearly all birds as well as an 80% morbidity. Six birds died on day 3 after challenge with the strain CK/CH/SCMY/160315, and the peak death rate occurred at 3 to 4 dpi (Figure 4). The death rate was 56.7%, with 17 chickens died during the experimental period. There were no clinical symptoms or deaths observed in the control group birds.
Figure 4.
Survival rates of chickens infected with CK/CH/SCMY/160315.
CK/CH/SCMY/160315 Activates Antibodies and Causes Tissue Damage
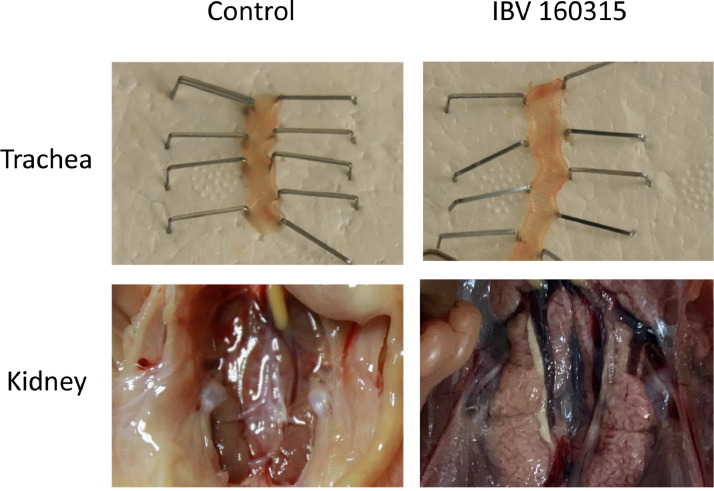
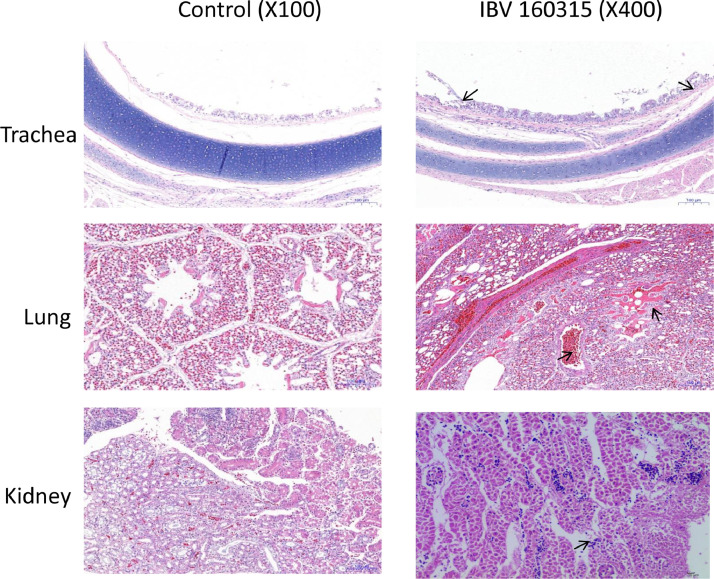
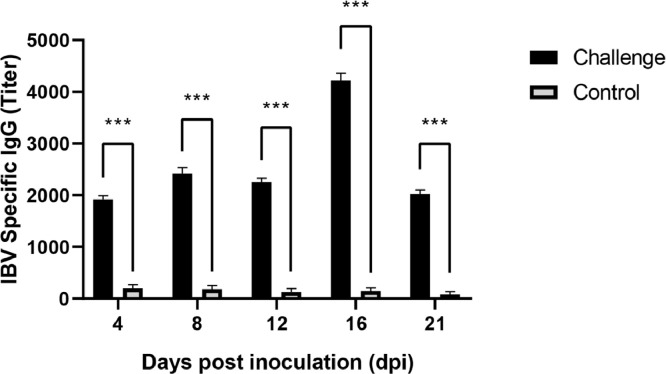
At necropsy, no gross lesions were observed in the control group. In the test group, several lesions were observed, including tracheal catarrhal exudate, mucus and hemorrhage in the trachea and throat, a small amount of mucosal epithelium being shed, and acinar structures in the lamina propria of the mucosa. Lung lesions included hemorrhage and congestion, with numerous red blood cells in the lumen. Typical lesions were pale and swollen kidney, with urate deposition in renal tubule and ureters, and most characteristic histological lesions in the kidney were multifocal necrosis of the renal tubules, and infiltration of inflammatory cells (Figures 5 and 6). Obvious lesions (nephritis) were found in the kidneys of the chicken that died at 12 dpi and a total of 6 chickens dead at 14, 15, 16, and 19 dpi. However, we observed no obvious lesions in the spleen, suggesting that spleen tissues had a lower susceptibility to the IBV strain CK/CH/SCMY/160315. Antibody responses in birds that survived infection were measured at 5 different times post-infection (4, 8, 12, 16, and 21 dpi). Samples collected at all time periods were positive, and the virus antibody titer peaked at 16 dpi (Figure 7).
Figure 5.
Gross lesions in the trachea and kidney of chickens infected with the viruses at 4 days post infection.
Figure 6.
Histopathologic lesions highlight damage to the trachea, lungs, and kidneys. Lesions are indicated by a black arrow.
Figure 7.
Antibody response of chickens after infection the CK/CH/SCMY/160315. The error bars indicate mean ± standard error of mean. Asterisk (*) represents a significant difference from the control group, with significance indicators corresponding to: (∗) significant, P< 0.05; (∗∗) highly significant, P< 0.01; (∗∗∗) very highly significant, P< 0.001.
Content and Changes in Viral Load in Different Tissues During the Experiment
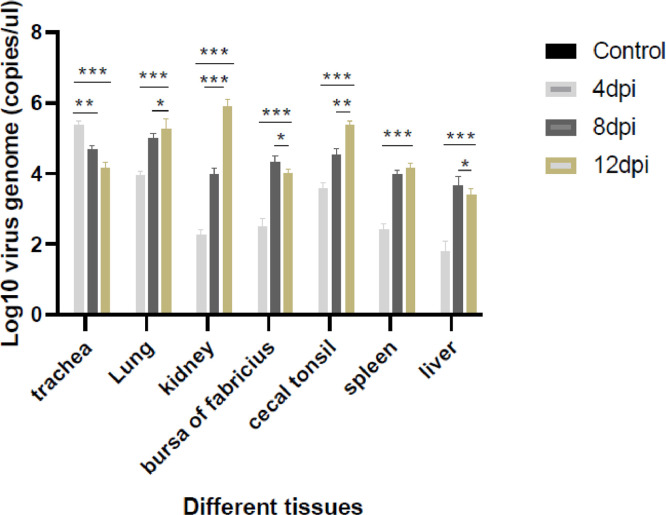
We determined the viral RNA titers in several tissues (trachea, kidney, lung, bursa of fabricius, cecal tonsil, spleen and liver) using a SYBR Green I RT–qPCR assay (Figure 8). The viral DNA levels in the tracheae of the test group peaked at 4 dpi and decreased subsequently. The maximum amount of viral DNA was detected at 8 dpi in the liver and bursa of fabricius of the test group. At 12 dpi, the tissue of lung, kidney, cecal tonsil, and spleen had considerably higher viral loads than other tissues. The viral load was most variable in the kidney, and liver was lower than in other tissues during the entire experimental period. No virus was detected in the control group samples at similar time points.
Figure 8.
Absolute quantitative RT-qPCR was used to determine viral RNA load level of the trachea, kidney, lung, bursa of Fabricius, cecal tonsil, spleen, and liver. Levels between the challenge group and the negative group were statistically significant at 4, 8, and 12. Asterisk (*) denotes the significant difference from the control group, with significance indicators corresponding to: (∗) significant, P< 0.05; (∗∗) highly significant, P< 0.01; (∗∗∗) very highly significant, P< 0.001.
DISCUSSION
The threat of IBV infection is global, as this respiratory virus can cause acute and contagious respiratory infections in poultry, resulting in serious economic losses (Smith et al., 2015). In the present study, we present a novel IBV variant, designated CK/CH/SCMY/160315, which we isolated on a farm in southwestern China in 2016. Flocks had suffered from clinical symptoms of listlessness, sneezing, dyspnea, and respiratory distress. We assessed the whole genome characteristics of this novel strain. Phylogenetic analyses of the S1 gene revealed that CK/CH/SCMY/160315 grouped along the GI-28, a pathogenic kidney IBV (Chen et al., 2017) which is prevalent in southwest China. Meanwhile, located in the same group was the LDT3-A strain, a commonly used vaccine strain in China. In addition, we compared CK/CH/SCMY/160315 and LDT3-A and found that their S1 sequence similarity was as high as 99.8% (data not shown), while the full-length sequence similarity was only 90.82% (Table 1). Comparing S1 amino acid residues, we found that CK/CH/SCMY/160315 had two unique mutations in positions 155H and 121Q. Compared with other GI-28 reference strains, nearly half of the differences of CK/CH/SCMY/160315 were located in the HVR3 region, all of which were shown on HVR2 compared with the case of the most similar strain LDT3-A. It implied that HVR3 on the S1 gene may play an important role in the genetic evolution of different IBVs.
Similarly, to other RNA viruses, IBV continues undergo genetic changes (Kjaerup et al., 2014). The poor replication fidelity of viral polymerase, resulting in frequent point mutation, recombination, and immune pressure selection, is considered the underlying molecular mechanisms for generation of viral variability and diversity which promotes the evolution of IBV (Jackwood et al., 2012). Importantly, recombination events are thought to play a key role for IBV evolution. Recombination events between different strains of IBV could result in the emergence of several novel strains, which have further increased the genetic diversity and complexity of IBVs (Ma et al., 2019; Ren et al., 2019). Recently, various live-attenuated and inactivated vaccines have been widely and extensively used to prevent IB disease on chicken farms in China, and the detection of some novel strains such as 624I, N1/88, I2217–2/16, and JX-17, amongst others, has caused considerable concern, (Quinteros et al., 2016; Xu et al., 2018; Laconi et al., 2019; Huang et al., 2020). These IBV strains were shown to have originated from recombination events between field and vaccine strains, different types of vaccine strains, or different types of field strains. In the present research, we conducted a recombination analysis on the full-length sequence of the strain CK/CH/SCMY/160315 in order to reveal its genetic characteristics. Analysis results show that the CK/CH/SCMY/160315 isolate is a multiple recombinant strain originating from H120, 4/91, LDT3-A and LJL/08-1 strains, all of which are prevalent in the region. The ancestral strains contain 3 vaccine strains, which are attenuated vaccine strains and commonly used in Southwest China (Ma et al., 2019), and the other strain involved in the recombination has a higher mortality rate and is widespread in China (Wang et al., 2020). However, CK/CH/SCMY/160315 displayed a different recombination pattern compared with other reported LDT3-like strains isolated from 2012 to 2019 in China, and recombination events extended to structural genes such as E and M, indicating the complexity of the emergence of new IBV recombination variant in China. The incidence of recombination events between field and IBV vaccine strains warrants attention because the new recombinant virus may lead to changes in IBV virulence in flocks.
A potentially serious consequence of the recombination between vaccine and field strain is that the recombination event may lead to changes in virulence, which could produce naturally weakened vaccine candidate strains or highly pathogenic recombinant strains (Promkuntod et al., 2014). Previous studies have shown that a key factor in the pathogenesis of IBV is its replicase gene, which could cause reverse inheritance. Other studies have shown that this phenomenon may also be related to accessory proteins (Armesto et al., 2009; Laconi et al., 2018). Therefore, we explored the pathogenicity of the novel CK/CH/SCMY/160315 strain. Our results suggest a stronger pathogenicity than for other GI-28 strains (Chen et al., 2017), which can cause a strong immune response in the body. CK/CH/SCMY/160315 induced moderate mortality and exhibited a broader tissue tropism than strains described in previous reports (Han et al., 2016). At necropsy, obvious respiratory and nephritis signs could be seen in the infected flock. Furthermore, we were able to detect the virus in multiple tissues by RT-qPCR, and the amount of viral replication in different tissues varies with the infection time. The trachea, kidneys and lungs are the main sites for IBV replication. As a vaccine strain, LDT3-A is less pathogenic to chickens, while we found that CK/CH/SCMY/160315 is more virulent to chickens in this study. It is speculated that the reason is that the recombination of other genes has changed the pathogenicity of the strain. The results of these pathogenicity experiments show that it is insufficient to evaluate the virulence of the virus strain based on the S1 gene alone. More in-depth research is needed to assess the role of virulence genes, IBV pathogenicity, and gene coordination.
In conclusion, our research shows that the recombination between IBV vaccine strains and field strains is complex and diverse, and the pathogenicity of recombinant strains is no longer determined by S1 alone. In addition, the use of multiple vaccine strains increases the risk of recombination between IBVs, and the recombined strains may exhibit higher pathogenicity. For newly emerging IBV strains, more comprehensive sequence determination and studies of biological characteristics are therefore required. Lastly, continuous monitoring is vital for understanding of the evolutionary characteristics of IBV.
ACKNOWLEDGMENTS
The authors thank Xin Yang for providing the guidance. This work was supported by the Sichuan University, Sichuan Animal Disease Control Center, Southwest University of Science and Technology and Sichuan Sundaily Village Ecological Food Co., Ltd.
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2021.101210.
Appendix. Supplementary materials
REFERENCES
- Abdel-Moneim A.S., Afifi M.A., El-Kady M.F. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch. Virol. 2012;157:2453–2457. doi: 10.1007/s00705-012-1445-1. [DOI] [PubMed] [Google Scholar]
- Abro S.H., Renstrom L.H., Ullman K., Isaksson M., Zohari S., Jansson D.S., Belak S., Baule C. Emergence of novel strains of avian infectious bronchitis virus in Sweden. Vet. Microbiol. 2012;155:237–246. doi: 10.1016/j.vetmic.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhar A., Gough R.E., Haydon D., Shaw K., Britton P., Cavanagh D. Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian. Pathol. 1997;26:625–640. doi: 10.1080/03079459708419239. [DOI] [PubMed] [Google Scholar]
- Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. Plos One. 2009;4:10. doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman, K.M., Parsons, L.M., Berends, A.J., de Vries, R.P., Cipollo, J.F., Verheije, M.H., 2020. Three amino acid changes in avian coronavirus spike protein allow binding to kidney tissue. 94:e01363-01319. [DOI] [PMC free article] [PubMed]
- Casais R., Dove B., Cavanagh D., Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising coronaviridae and arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chen Y.Q., Jiang L., Zhao W.J., Liu L.L., Zhao Y., Shao Y.H., Li H.X., Han Z.X., Liu S.W. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017;198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Stadler K., Godeke G.J., Bosch B.J., Rottier P.J.M. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 2004;78:6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degroot R.J., Luytjes W., Horzinek M.C., Vanderzeijst B.A.M., Spaan W.J.M., Lenstra J.A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolz R., Vergara-Alert J., Perez M., Pujols J., Majo N. New insights on infectious bronchitis virus pathogenesis: characterization of Italy 02 serotype in chicks and adult hens. Vet. Microbiol. 2012;156:256–264. doi: 10.1016/j.vetmic.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellahi S., El Harrak M., Ducatez M., Loutfi C., Koraichi S.I., Kuhn J.H., Khayi S., El Houadfi M., Ennaji M.M. Phylogenetic analysis of avian infectious bronchitis virus S1 glycoprotein regions reveals emergence of a new genotype in Moroccan broiler chicken flocks. Virol. J. 2015;12:116. doi: 10.1186/s12985-015-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo G., Legnardi M., Tucciarone C.M., Drigo M., Martini M., Cecchinato M. Evolution of infectious bronchitis virus in the field after homologous vaccination introduction. Vet. Res. 2019;50:92. doi: 10.1186/s13567-019-0713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.X., Zhang T.T., Xu Q.Q., Gao M.Y., Chen Y.Q., Wang Q.L., Zhao Y., Shao Y.H., Li H.X., Kong X.G., Liu S.W. Altered pathogenicity of a tl/CH/LDT3/03 genotype infectious bronchitis coronavirus due to natural recombination in the 5 '-17 kb region of the genome. Virus Res. 2016;213:140–148. doi: 10.1016/j.virusres.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.X., Zhao W.J., Chen Y.Q., Xu Q.Q., Sun J.F., Zhang T.T., Zhao Y., Liang S.L., Gao M.Y., Wang Q.L., Kong X.G., Liu S.W. Genetic, antigenic, and pathogenic characteristics of avian infectious bronchitis viruses genotypically related to 793/B in China. Vet. Microbiol. 2017;203:125–135. doi: 10.1016/j.vetmic.2017.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.J., Zou C.C., Liu Y., Han Z.L., Xue C.Y., Cao Y.C. A novel low virulent respiratory infectious bronchitis virus originating from the recombination of QX, TW and 4/91 genotype strains in China. Vet. Microbiol. 2020;242:108579. doi: 10.1016/j.vetmic.2020.108579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hall D., Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect Genet. Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Han Z.X., Chen Y.Q., Zhao W.J., Sun J.F., Zhao Y., Liu S.W. Characterization of the complete genome, antigenicity, pathogenicity, tissue tropism, and shedding of a recombinant avian infectious bronchitis virus with a ck/CH/LJL/140901-like backbone and an S2 fragment from a 4/91-like virus. Virus Res. 2018;244:99–109. doi: 10.1016/j.virusres.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant A., Koch G., Vanroozelaar D.J., Kusters J.G., Poelwijk F.A.J., Vanderzeijst B.A.M. Location of antigenic sites defined by neutralizing monoclonal-antibodies on the S1 avian infectious-bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73:591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- Kjaerup R.M., Dalgaard T.S., Norup L.R., Hamzic E., Sorensen P., Juul-Madsen H.R. Characterization of cellular and humoral immune responses after IBV infection in chicken lines differing in MBL serum concentration. Viral. Immunol. 2014;27:529–542. doi: 10.1089/vim.2014.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi A., Listorti V., Franzo G., Cecchinato M., Naylor C., Lupini C., Catelli E. Molecular characterization of whole genome sequence of infectious bronchitis virus 624I genotype confirms the close relationship with Q1 genotype. Transbound. Emerg. Dis. 2019;66:207–216. doi: 10.1111/tbed.13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi A., van Beurden S.J., Berends A.J., Kramer-Kuhl A., Jansen C.A., Spekreijse D., Chenard G., Philipp H.C., Mundt E., Rottier P.J.M., Verheije M.H. Deletion of accessory genes 3a, 3b, 5a or 5b from avian coronavirus infectious bronchitis virus induces an attenuated phenotype both in vitro and in vivo. J. Gen. Virol. 2018;99:1381–1390. doi: 10.1099/jgv.0.001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi A., Weerts E.A.W.S., Bloodgood J.C.G., Marrero J.P.D., Berends A.J., Cocciolo G., de Wit J.J., Verheije M.H. Attenuated live infectious bronchitis virus QX vaccine disseminates slowly to target organs distant from the site of inoculation. Vaccine. 2020;38:1486–1493. doi: 10.1016/j.vaccine.2019.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang X.Y., Wei P., Chen Q.Y., Wei Z.J., Mo M.L. Serotype and genotype diversity of infectious bronchitis viruses isolated during 1985-2008 in Guangxi, China. Arch. Virol. 2012;157:467–474. doi: 10.1007/s00705-011-1206-6. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang X., Gong L., Yan B., Li C., Han Z., Shao Y., Li H., Kong X. Altered pathogenicity, immunogenicity, tissue tropism and 3′-7kb region sequence of an avian infectious bronchitis coronavirus strain after serial passage in embryos. Vaccine. 2009;27:4630–4640. doi: 10.1016/j.vaccine.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z.L., Weiss S.R. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology. 1998;244:483–494. doi: 10.1006/viro.1998.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T.X., Xu L.W., Ren M.T., Shen J., Han Z.X., Sun J.F., Zhao Y., Liu S.W. Novel genotype of infectious bronchitis virus isolated in China. Vet. Microbiol. 2019;230:178–186. doi: 10.1016/j.vetmic.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Chu V.C., Lee H., Regan A.D., Bauman B.E., Whittaker G.R. Heparan sulfate is a selective attachment factor for the avian coronavirus infectious bronchitis virus beaudette. Avian Dis. 2007;51:45–51. doi: 10.1637/0005-2086(2007)051[0045:HSIASA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Parsons L.M., Bouwman K.M., Azurmendi H., de Vries R.P., Cipollo J.F., Verheije M.H. Glycosylation of the viral attachment protein of avian coronavirus is essential for host cell and receptor binding. J. Biol. Chem. 2019;294:7797–7809. doi: 10.1074/jbc.RA119.007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promkuntod N., van Eijndhoven R.E.W., de Vrieze G., Grone A., Verheije M.H. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinteros J.A., Lee S.W., Markham P.F., Noormohammadi A.H., Hartley C.A., Legione A.R., Coppo M.J.C., Vaz P.K., Browning G.F. Full genome analysis of Australian infectious bronchitis viruses suggests frequent recombination events between vaccine strains and multiple phylogenetically distant avian coronaviruses of unknown origin. Vet. Microbiol. 2016;197:27–38. doi: 10.1016/j.vetmic.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Ren G.C., Liu F., Huang M.R., Li L., Shang H.Q., Liang M.L., Luo Q., Chen R.A. Pathogenicity of a QX-like avian infectious bronchitis virus isolated in China. Poult. Sci. 2020;99:111–118. doi: 10.3382/ps/pez568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M.T., Sheng J., Ma T.X., Xu L.W., Han Z.X., Li H.X., Zhao Y., Sun J.F., Liu S.W. Molecular and biological characteristics of the infectious bronchitis virus TC07-2/GVI-1 lineage isolated in China. Infect Genet. Evol. 2019;75:10392. doi: 10.1016/j.meegid.2019.103942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D., Fang S.G., Han Z.X., Ai H., Zhao W.J., Chen Y.Q., Jiang L., Liu S.W. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 2018;250:104–113. doi: 10.1016/j.virusres.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Sadeyen J.R., Cavanagh D., Kaiser P., Burt D.W. The early immune response to infection of chickens with infectious bronchitis virus (IBV) in susceptible and resistant birds. Bmc. Vet. Res. 2015;11:256. doi: 10.1186/s12917-015-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beurden S.J., Berends A.J., Kramer-Kuhl A., Spekreijse D., Chenard G., Philipp H.C., Mundt E., Rottier P.J.M., Verheije M.H. Recombinant live attenuated avian coronavirus vaccines with deletions in the accessory genes 3ab and/or 5ab protect against infectious bronchitis in chickens. Vaccine. 2018;36:1085–1092. doi: 10.1016/j.vaccine.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Dempsey D.M., Dutilh B.E., Harrach B., Harrison R.L., Hendrickson R.C., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M., Rubino L., Sabanadzovic S., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Changes to virus taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2019) Arch. Virol. 2019;164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cui X., Chen X., Yang S., Ling Y., Song Q., Zhu S., Sun L., Li C., Li Y., Deng X., Delwart E., Zhang W. A recombinant infectious bronchitis virus from a chicken with a spike gene closely related to that of a turkey coronavirus. Arch. Virol. 2020;165:703–707. doi: 10.1007/s00705-019-04488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang X., Xu P.W., Zhou L., Zhang Z.K., Wang H.N. Genome sequence and origin analyses of the recombinant novel IBV virulent isolate SAIBK2. Virus Genes. 2016;52:509–520. doi: 10.1007/s11262-016-1337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., He X., Yao K.C., Du L.J., Liu P., Yan Q.G., Wen Y.P., Cao S.J., Han X.F., Huang Y. Phylogenetic and antigenic analysis of avian infectious bronchitis virus in southwestern China, 2012-2016. Infect Genet. Evol. 2016;45:11–19. doi: 10.1016/j.meegid.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.W., Han Z.X., Jiang L., Sun J.F., Zhao Y., Liu S.W. Genetic diversity of avian infectious bronchitis virus in China in recent years. Infect Genet. Evol. 2018;66:82–94. doi: 10.1016/j.meegid.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.W., Ren M.T., Sheng J., Ma T.X., Han Z.X., Zhao Y., Sun J.F., Liu S.W. Genetic and biological characteristics of four novel recombinant avian infectious bronchitis viruses isolated in China. Virus Res. 2019;263:87–97. doi: 10.1016/j.virusres.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang H.N., Wang T., Fan W.Q., Zhang A.Y., Wei K., Tian G.B., Yang X. Complete genome sequence and recombination analysis of infectious bronchitis virus attenuated vaccine strain H120. Virus Genes. 2010;41:377–388. doi: 10.1007/s11262-010-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.S., Zhang M.H., Tian X., Shao H.X., Qian K., Ye J.Q., Qin A.J. Identification of a novel recombinant virulent avian infectious bronchitis virus. Vet. Microbiol. 2017;199:120–127. doi: 10.1016/j.vetmic.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.Z., Luci M., Huang Q.H., Huang Y.Y., Yang S.H., Cui Y.S., Liu C., Tan L.G., Kong Z.J., Xu C.T. Interactive mechanism between avian infectious bronchitis S1 protein T cell peptide and avian MHC I molecule. Virus Res. 2016;215:76–83. doi: 10.1016/j.virusres.2016.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.