Abstract
The glycoprotein α-dystroglycan helps to link the intracellular cytoskeleton to the extracellular matrix. A unique glycan structure attached to this protein is required for its interaction with extracellular matrix proteins such as laminin. Up to now, this is the only mammalian glycan known to contain ribitol phosphate groups. Enzymes in the Golgi apparatus use CDP-ribitol to incorporate ribitol phosphate into the glycan chain of α-dystroglycan. Since CDP-ribitol is synthesized in the cytoplasm, we hypothesized that an unknown transporter must be required for its import into the Golgi apparatus. We discovered that CDP-ribitol transport relies on the CMP-sialic acid transporter SLC35A1 and the transporter SLC35A4 in a redundant manner. These two transporters are closely related, but bulky residues in the predicted binding pocket of SLC35A4 limit its size. We hypothesized that the large binding pocket SLC35A1 might accommodate the bulky CMP-sialic acid and the smaller CDP-ribitol, whereas SLC35A4 might only accept CDP-ribitol. To test this, we expressed SLC35A1 with mutations in its binding pocket in SLC35A1 KO cell lines. When we restricted the binding site of SLC35A1 by introducing the bulky residues present in SLC35A4, the mutant transporter was unable to support sialylation of proteins in cells but still supported ribitol phosphorylation. This demonstrates that the size of the binding pocket determines the substrate specificity of SLC35A1, allowing a variety of cytosine nucleotide conjugates to be transported. The redundancy with SLC35A4 also explains why patients with SLC35A1 mutations do not show symptoms of α-dystroglycan deficiency.
Keywords: dystroglycan, glycosylation, nucleotide transport, SLC35A1, SLC35A4, sialic acid, ribitol
Abbreviations: FKRP, fukutin-related protein; FKTN, fukutin; LBB, laminin-binding buffer; MAL II, Maackia amurensis lectin II; NST, nucleotide sugar transporter; PNA, peanut agglutinin
The dystrophin-associated glycoprotein 1 (DAG1) gene gives rise to a single protein that is subsequently cleaved into two fragments that remain attached to each other. The C-terminal part, β-dystroglycan, is a transmembrane protein anchored to the actin cytoskeleton through interaction with the dystrophin protein (1). The N-terminal extracellular part, α-dystroglycan, binds to extracellular matrix components, like laminin, agrin, perlecan, or neurexin (1). This interaction depends on the assembly of a complex glycan on α-dystroglycan. Defects in several enzymes required for the biogenesis of this glycan lead to a group of congenital syndromes characterized by muscle, brain, and eye symptoms. These syndromes have been coined dystroglycanopathies and present a vast clinical spectrum ranging from mild muscular weakness with late onset to severe congenital muscular dystrophy with brain and eye involvement (muscle–eye–brain disease, Walker–Warburg syndrome, and Fukuyama congenital muscle dystrophy) (2, 3, 4, 5). The investigation of genes mutated in affected patients has led to the discovery of many steps in this process and a better understanding of α-dystroglycan glycosylation as well as more generally of protein O-mannosylation.
The α-dystroglycan protein contains three domains: the C-terminal domain, which binds to the extracellular domain of β-dystroglycan, the mucin domain characterized by several mucin-type-O-glycosylation sites as well as a key O-mannosyl glycan important for ligand binding (referred to later as core M3), and the N-terminal domain that is required for complete glycosylation and is subsequently shed by proteolytic cleavage (5).
The ligand-binding epitope of α-dystroglycan is assembled on the core M3 glycan, which has been extensively studied and consists of an O-linked phosphoryl(C6)-mannose extended with a GlcNAc and a GalNAc. It is formed by sequential action of protein O-mannosyltransferases 1 and 2 (5), protein O-linked mannose β-1,2-N-acetylglucosaminyltransferase (6), β-1,3-N-acetylgalactosaminyltransferase 2 (7), and protein-O-mannose kinase (8). In the Golgi apparatus, this glycan is further extended by fukutin (FKTN) and fukutin-related protein (FKRP) with a tandem repeat of ribitol phosphate (9, 10, 11). This structure then acts as a scaffold to which the laminin-binding moiety is attached, with multiple repetitions of a disaccharide consisting of xylose and glucuronic acid that are added by the enzymes transmembrane protein 5 (12) and β-1,4-glucuronyltransferase 1 (13, 14) for the first repetition and subsequently by the proteins LARGE1 or LARGE2 (LARGE xylosyl- and glucuronyltransferase 1 or 2) (15).
FKTN and FKRP in the Golgi apparatus use CDP-ribitol, which is synthesized by the enzyme isoprenoid synthase domain–containing protein in the cytoplasm (9, 10, 11). Yet, the transporter required for the entry of CDP-ribitol into the Golgi remained elusive and is the subject of the presented work. Nucleotide sugars are transported across membranes by specialized proteins known as nucleotide sugar transporters (NSTs) belonging to the SLC35 gene family. These proteins transport nucleotide sugars into the endoplasmic reticulum or the Golgi apparatus while simultaneously exporting the corresponding nucleoside monophosphate (16). Recent structural studies have revealed the concerted conformational changes that are required for this transport (17). However, the substrate specificity of several transporters is still unknown (18). Likewise, we are only starting to understand how these transporters distinguish between related substrates (17, 19).
Interestingly, two studies indicated that the Golgi apparatus CMP-sialic acid transporter SLC35A1 could be implicated in CDP-ribitol transport. The first line of evidence comes from a study aiming to identify factors required for the entry of Lassa virus, which is dependent on the interaction with fully glycosylated α-dystroglycan. Using the haploid cell line HAP1, the authors revealed that inactivation of SLC35A1, the CMP-sialic acid transporter, prevented functional glycosylation of α-dystroglycan and Lassa virus entry (20). Second, SLC35A1 harboring mutations observed in patients were unable to rescue the α-dystroglycan glycosylation deficiency in SLC35A1 KO HAP1 cell lines (21). Of note, when sialylation was inhibited by a fluorinated sialic acid analog, α-dystroglycan still bound to its ligand laminin, suggesting that sialylation is not required for functional α-dystroglycan glycosylation. This indicated that the deficiency of α-dystroglycan glycosylation in HAP1 SLC35A1 KO cells was not because of a depletion of CMP-sialic acid in the lumen of the Golgi apparatus (21).
Here, we demonstrate that SLC35A1 and its paralog SLC35A4 play a redundant role in the glycosylation of α-dystroglycan. Using a series of mutants, we reveal that the large binding site of SLC35A1 allows the promiscuous transport of both CMP-sialic acid and CDP-ribitol. Our findings explain why patients with SLC35A1 deficiency do not show clinical symptoms resembling a dystroglycanopathy.
Results
SLC35A1 is required for dystroglycan glycosylation in HAP1 but not in human embryonic kidney 293 cells
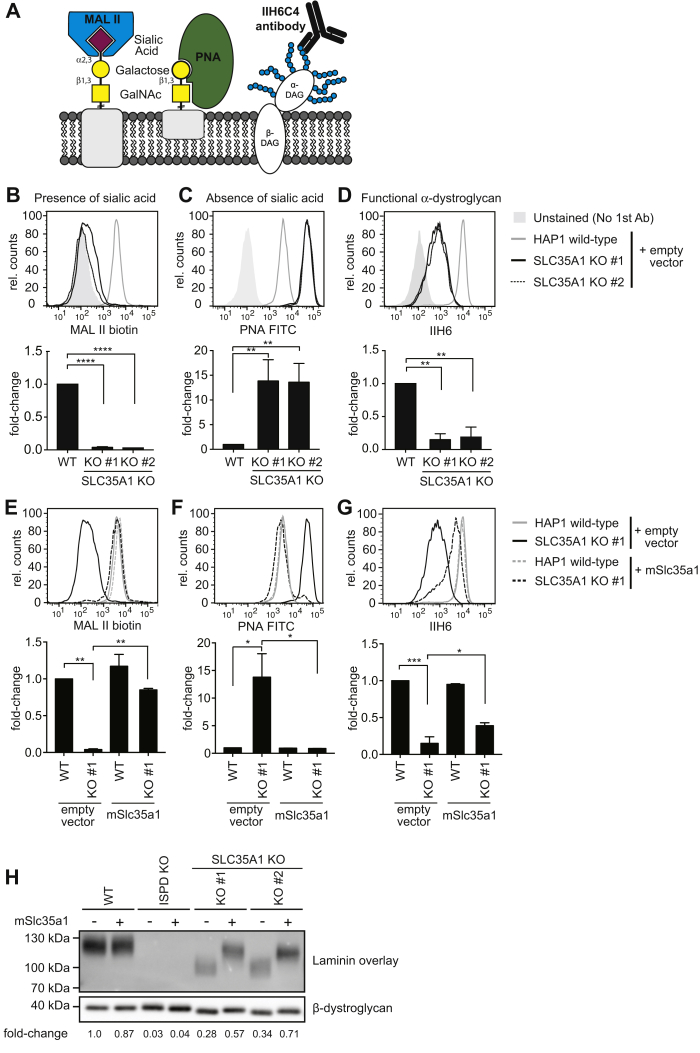
To confirm the results obtained by Jae et al. (20), we first knocked out SLC35A1 in HAP1 cells using CRISPR/Cas9 and assessed α-dystroglycan glycosylation by flow cytometry using the antibody IIH6 recognizing the functional α-dystroglycan glycan as well as protein sialylation by Arachis hypogaea lectin (peanut agglutinin [PNA]) recognizing nonsialylated Galβ1–3GalNAc and Maackia amurensis lectin II (MAL II), binding to α-2,3-linked sialic acid residues (Fig. 1A).
Figure 1.
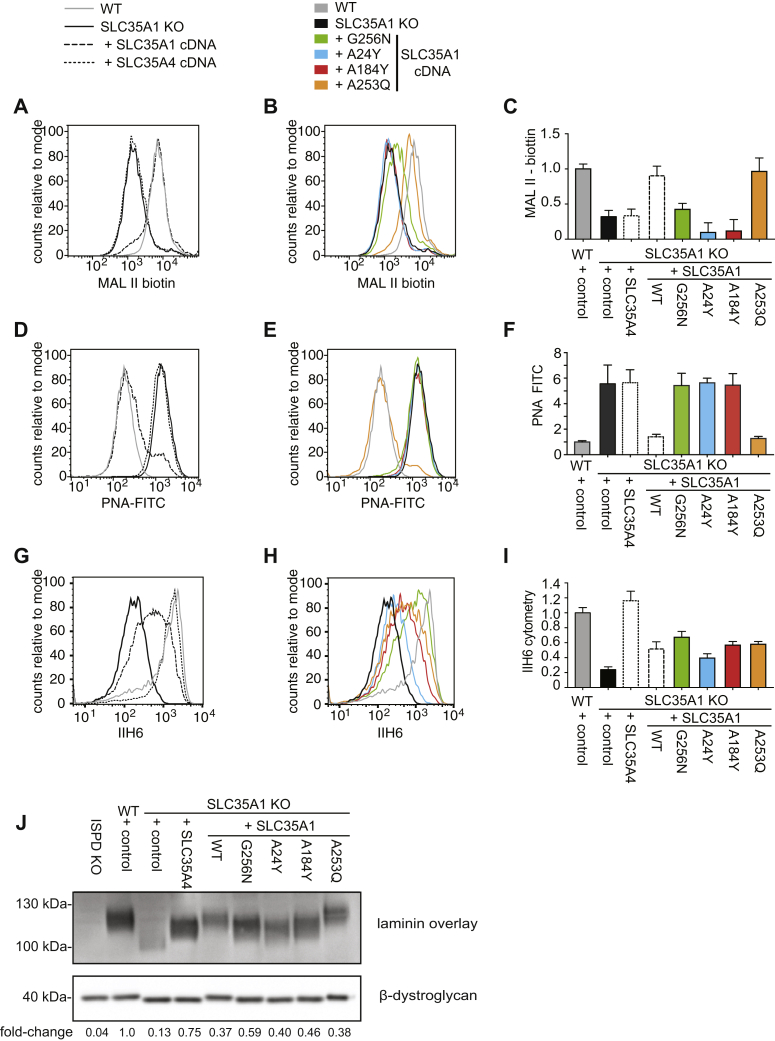
Inactivation of SLC35A1 compromises sialylation and assembly of the laminin-binding glycan of α-dystroglycan.A, schematic representation of tools used to assess sialylation and the assembly of the laminin-binding glycan. B–G, flow cytometry with MAL II lectin, peanut agglutinin (PNA), and the antibody IIH6 was performed on wildtype or SLC35A1 KO HAP1 cells that were transduced with a lentivirus driving expression of murine Slc35a1 complementary DNA or an empty vector control. Two independent clones (#1 and #2) were analyzed. Bar graphs underneath flow cytometry histograms represent means ± SEM of the mean fluorescence intensity (MFI) observed in three independent experiments. Note that the presented experiment was performed in parallel to the experiment presented for Figure 3, explaining why the empty vector controls are identical. H, samples from the indicated HAP1 cell lines were subjected to laminin overlay or β-dystroglycan Western blot. MAL II, Maackia amurensis lectin II.
As expected, inactivation of SLC35A1 led to a simultaneous decrease in MAL II staining and increase in PNA staining indicating a general decrease in protein sialylation, which was restored upon re-expression of mSlc35a1 (Fig. 1, B, C, E, and F). SLC35A1 inactivation also led to a decrease in α-dystroglycan glycosylation as measured by flow cytometry using the IIH6 antibody, which was also rescued by re-expression of mSlc35a1 (Fig. 1, D and G). To assess the laminin-binding capacity of α-dystroglycan in SLC35A1 KO cells, we performed a laminin overlay assay (Fig. 1H). The signal intensity (as a surrogate marker for laminin binding) was decreased in SLC35A1 KO cells. In addition, the apparent molecular weight of α-dystroglycan was reduced, likely because of a loss of sialylation and the loss of the laminin-binding glycan. We also observed that the apparent molecular weight of the β-dystroglycan band was reduced, consistent with the known sialylation of this protein. Re-expression of SLC35A1 in KO cell completely rescued the changes in lectin binding (Fig. 1, E and F) and normalized the migration pattern of β-dystroglycan (Fig. 1H, lower panel), consistent with a complete recovery of sialylation because of the known role of SLC35A1 in CMP-sialic acid transport. In contrast, the signal intensity and apparent molecular weight in laminin overlay, as well as the signal in flow cytometry with the antibody IIH6 (Fig. 1, G and H), were increased but did not reach levels observed in wildtype cells. This indicated that the formation of the ribitol phosphate–containing glycan was only partially rescued. Thus, the lower levels of SLC35A1 achieved upon re-expression completely rescued sialylation, whereas higher levels seemed to be required to ensure the formation of the laminin-binding glycan.
Our observations could be explained if sialylation was required for the function of a key enzyme involved in the biogenesis of the laminin-binding glycan. However, it has been shown previously that inhibition of sialylation by a synthetic fluorinated sialic acid analog does not affect the biogenesis of the functional glycan (21). Thus, a more likely explanation is that SLC35A1 not only transports CMP-sialic acid but also CDP-ribitol, albeit potentially with a lower efficiency. Of note, SLC35A1 KO cells still retained some signal in the laminin overlay assay (Fig. 1H) and in flow cytometry with the IIH6 antibody, which recognizes the intact α-dystroglycan glycan (Fig. 1, D and G). This indicates that while SLC35A1 might be the major transporter for CDP-ribitol in HAP1 cells, other transporters might contribute.
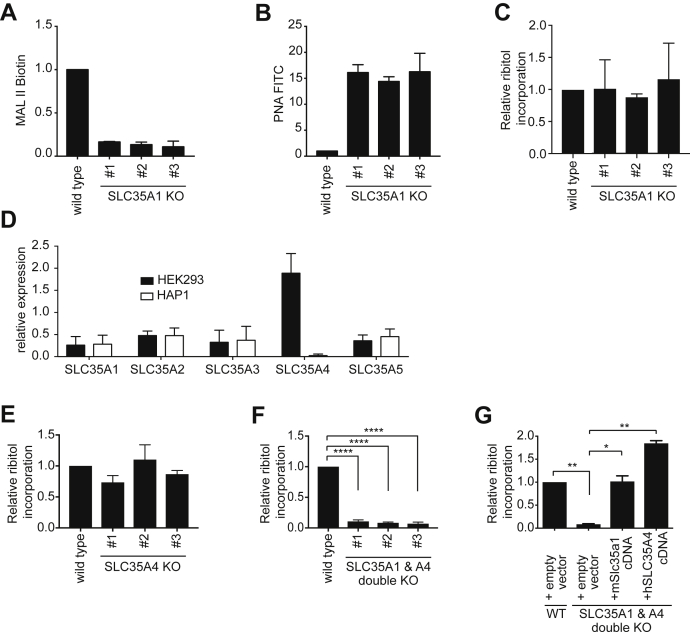
To quantitatively assess the incorporation of ribitol into α-dystroglycan, we chose human embryonic kidney 293 (HEK293) cells, which we can engineer to produce large amounts of α-dystroglycan secreted in the culture medium. When we knocked out SLC35A1 using CRISPR/Cas9 in these cells, we again observed strong changes in MAL II and PNA lectin flow cytometry, indicating a global deficiency in sialylation (Fig. 2, A and B). We then purified α-dystroglycan from the culture medium and assessed ribitol incorporation by GC/MS as described previously (9). To our surprise, incorporation of ribitol into α-dystroglycan was completely unaffected when SLC35A1 was inactivated in HEK293 cells (Fig. 2C).
Figure 2.
SLC35A1 and SLC35A4 play redundant role in CDP-ribitol transport in HEK293 cells.A and B, wildtype HEK293 or three different SLC35A1 KO HEK293 cell lines were analyzed by flow cytometry with MAL II (A) and PNA lectin (B). Means ± SD of the mode of fluorescence intensity of two independent experiments are shown. C, ribitol incorporation into matriglycan was assessed by GC/MS on α-dystroglycan (aa 1–485) affinity purified from wildtype and three different SLC35A1 KO HEK293 clones. Means ± SD of three independent experiments are shown. D, RT-quantitative PCR of the indicated transcripts in HEK293 and HAP1 cells. Values were normalized to the mean of b2-microglobulin (B2M) and TATA-binding protein (TBP) and are presented as means ± SD of three samples. E–G, ribitol incorporation was determined as described for C, but in SLC35A4 KO HEK293 clones (E), SLC35A1 and SLC35A4 double KO HEK293 clones (F), as well as double KO HEK293 clones where expression of either mSlc35a1 or SLC35A4 was restored by lentiviral transduction (G). Empty vector indicates transduction with the empty lentiviral backbone. Values are means ± SD of three independent experiments. HEK293, human embryonic kidney 293 cells; MAL II, Maackia amurensis lectin II; PNA, peanut agglutinin.
SLC35A1 and SLC35A4 perform a partially redundant function in α-dystroglycan glycosylation
SLC35A1 was required for the formation of the laminin-binding glycan of α-dystroglycan in HAP1 cells but not required for the incorporation of ribitol into α-dystroglycan in HEK293 cells. This led us to hypothesize that two transporters might play a redundant role in CDP-ribitol transport in HEK293 cells. Assuming that these transporters may belong to the same family, we performed an RT-quantitative PCR analysis of all members of the SLC35A family in HEK293 and HAP1 cells. This revealed that SLC35A4, a family member with largely unknown function (22), was expressed at much higher mRNA levels in HEK293 cells than in HAP1 cells (Fig. 2D). We also reasoned that a candidate CDP-ribitol transporter should be expressed in the brain and muscle, where α-dystroglycan has important functions. Based on RNA-Seq gene expression data obtained from the Genotype-Tissue Expression portal (Fig. S1) (23), among SLC35A family members, SLC35A1, and especially SLC35A4 show highest expression levels in skeletal muscle and different brain regions. While SLC35A5 reaches comparable levels in the brain, SLC35A2 and SLC35A3 expression levels were consistently lower.
We therefore hypothesized that SLC35A1 and SLC35A4 might play a redundant role in CDP-ribitol transport in HEK293 cells. To test this hypothesis, we proceeded to knock out SLC35A4 in parental HEK293 cells as well as in the previously engineered SLC35A1 KO HEK293 cells. We then purified a secreted α-dystroglycan fragment (aa 1–485) from the supernatant and analyzed ribitol incorporation. This revealed that inactivation of SLC35A4 alone does not significantly affect ribitol incorporation (Fig. 2E). In contrast, inactivation of both SLC35A1 and SLC35A4 simultaneously almost completely abolished ribitol incorporation in the glycan of α-dystroglycan (Fig. 2F). This effect is rescued upon re-expression of either protein in the double KO cells (Fig. 2G), indicating that both proteins independently perform a redundant function in ribitol incorporation into α-dystroglycan.
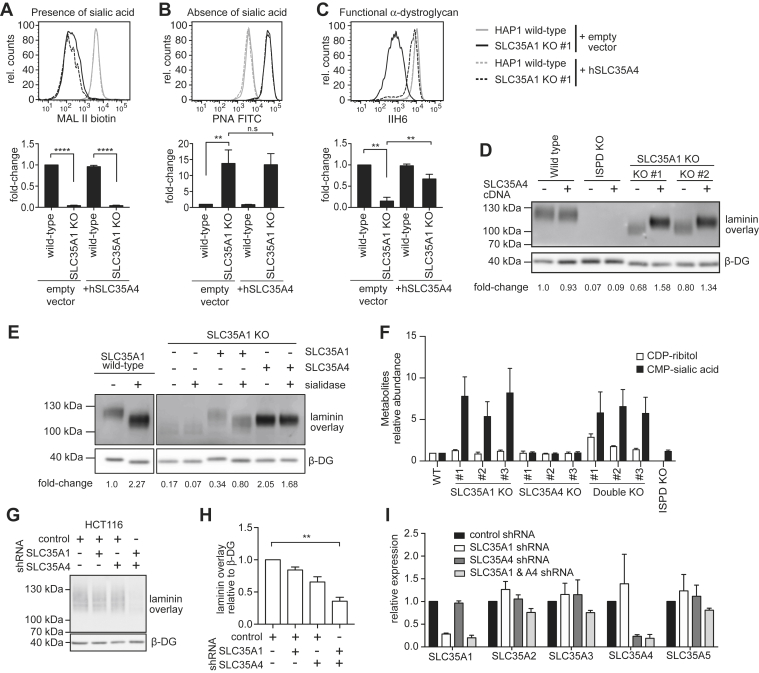
To further investigate the degree of functional redundancy of SLC35A1 and SLC35A4, we went back to HAP1 cells, where we can easily assess endogenous fully glycosylated α-dystroglycan and overall sialylation. We infected SLC35A1 KO HAP1 cells with recombinant lentiviruses driving expression of SLC35A4. Expression of SLC35A4 did not affect general protein sialylation as measured by PNA and MAL II staining using flow cytometry (Fig. 3, A and B). In contrast, we observed that α-dystroglycan glycosylation, assessed by flow cytometry using the IIH6 antibody (Fig. 3C) and laminin overlay (Fig. 3D), was rescued upon SLC35A4 expression. Similar to Figure 1H, we observed again that inactivation of SLC35A1 decreased both binding to laminin and the apparent molecular weight of α-dystroglycan (Figs. 1H and 3D). Interestingly, while SLC35A4 completely rescued the signal in laminin-binding assay to levels exceeding the ones observed in wildtype cells, it only partially rescued the decrease in apparent molecular weight (Fig. 3D, lanes 6 and 8; Fig. 3E) caused by SLC35A1 inactivation. Expression of SLC35A4 did not normalize neither the migration pattern of β-dystroglycan (Fig. 3D) nor lectin binding (Fig. 3, A and B) in SLC35A1 KO cells, suggesting that SLC35A4 does not contribute to CMP-sialic acid transport. This conclusion is also supported by the observation that treatment with sialidase led to a lower apparent molecular weight α- and β-dystroglycan in wildtype and SLC35A1-rescued KO cell lysates, whereas no difference was observed when SLC35A4-expressing SLC35A1 KO cell lysates were treated (Fig. 3E).
Figure 3.
SLC35A1 and SLC35A4 perform redundant functions during the formation of the laminin-binding glycan, but SLC35A1 is crucial for sialylation.A–C, flow cytometry with MAL II lectin, peanut agglutinin (PNA), and the antibody IIH6 was performed on wildtype or SLC35A1 KO HAP1 cells that were transduced with a lentivirus driving expression of murine Slc35a4 complementary DNA or an empty vector control. Bar graphs underneath flow cytometry histograms represent means ± SEM of the mean fluorescence intensity (MFI) observed in three independent experiments. Note that the presented experiment was performed in parallel to the experiment presented for Figure 1, explaining why the empty vector controls are identical. D, laminin overlay performed on wildtype, SLC35A1 KO and ISPD KO HAP1 cells upon expression of SLC35A4 (or an empty vector control). E, samples from the indicated HAP1 cell lines were subjected to laminin overlay or β-dystroglycan Western blot. Where indicated, sialidase digest was performed before laminin overlay. The presented data are from one gel, where two lanes in the middle were removed. F, quantification of CDP-ribitol and CMP-sialic acid (i.e., CMP-N-acetylneuraminic acid) in the indicated cell lines. Relative intracellular CDP-ribitol and CMP-sialic acid levels in HEK293 cells knocked out for SLC35A1 and/or SLC35A4 as measured by LC/MS. The ISPD KO cells were used as a negative control for CDP-ribitol measurements. Means ± SEM of three independent experiments are shown, and values were normalized within each experiment to levels in wildtype (control) cells. G and H, laminin overlay and β-dystroglycan Western blot (G) as well as quantitative evaluation of three independent experiments (H) obtained in HCT116 cells that express SLC35A1, SLC35A4, or nonsilencing control shRNAs. I, RT-quantitative PCR analysis for the indicated mRNAs in cell lines described for G and H. HEK293, human embryonic kidney 293 cells; ISPD, isoprenoid synthase domain–containing protein; MAL II, Maackia amurensis lectin II.
Thus, we conclude that both transporters are redundant in their contribution to the formation of the laminin-binding glycan, but that only SLC35A1 can transport CMP-sialic acid allowing normal sialylation.
Next, we quantified CDP-ribitol and CMP-sialic acid (i.e., CMP-N-acetylneuraminic acid) levels in the SLC35A1 and SLC35A4 single and double KO cells by LC/MS. Inactivation of SLC35A1 led to significant increases in total cellular CMP-sialic acid levels, consistent with the notion that this nucleotide is not efficiently used because of the absence of its transporter (Fig. 3F). Cellular CDP-ribitol levels were unaffected or increased when SLC35A1 and/or SLC35A4 was knocked out. This is consistent with our hypothesis that deficient α-dystroglycan glycosylation in these cell lines is not because of a problem in CDP-ribitol synthesis but rather further downstream.
HAP1 cells mainly expressed SLC35A1, whereas HEK293 cells relied on both a redundant function of SLC35A1 and SLC35A4 to assemble the laminin-binding glycan. To test whether a similar redundancy might exist in other settings, we also tested the effect of the depletion of these two proteins in a third cell line. For that purpose, we knocked down both proteins using shRNAs and assessed laminin-binding capacity of α-dystroglycan in HCT116 via laminin overlay (Fig. 3, G and H). Knockdown resulted in a decrease of more than 80% on the mRNA level (Fig. 3I). Single knockdown of either SLC35A1 or SLC35A4 showed no significant effect on the laminin-binding capacity of α-dystroglycan (lanes 2 and 3). However, knocking down both SLC35A1 and SLC35A4 led to a strong decrease of laminin-binding capacity (lane 4).
Taken together, our data demonstrate that SLC35A4 and SLC35A1 act in a redundant manner in the assembly of the laminin-binding glycan of α-dystroglycan. Yet, SLC35A4 is not able to overcome the loss of CMP-sialic acid transport in SLC35A1 KO cells, indicating that SLC35A4 cannot transport CMP-sialic acid.
The size of the binding pocket determines specificity for CDP-ribitol and CMP-sialic acid
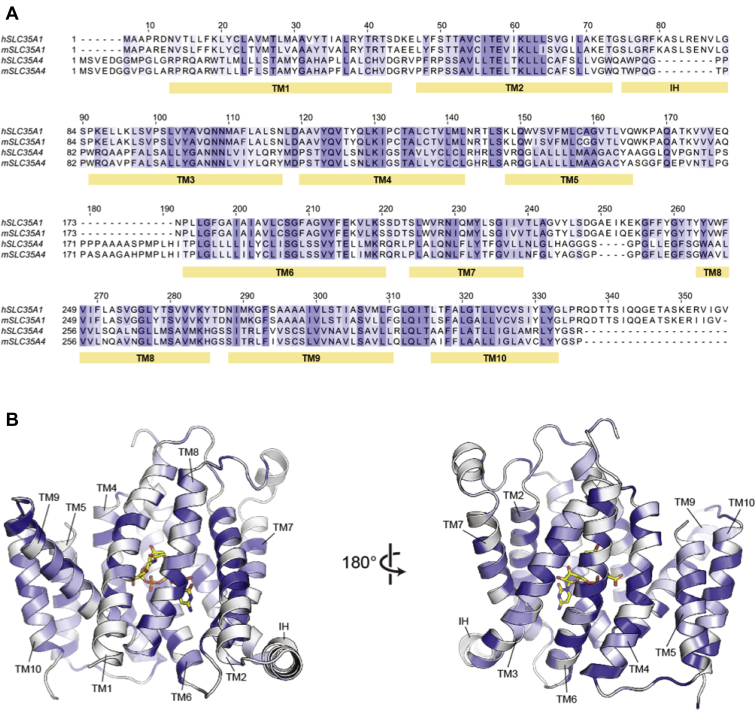
One of us recently determined the X-ray crystal structure of the CMP-sialic acid transporter SLC35A1 (17), which provides a framework for starting to understand the molecular mechanisms of substrate recognition in SLC35A4. There is moderate overall sequence conservation between SLC35A4 and SLC35A1 (24% identity) (Fig. 4); however, nearly all the residues in SLC35A1 that interact with the cytidine group of CMP are conserved in SLC35A4, including Tyr214 and Ser261 (Fig. 4, Fig. S2, and Fig. 5, A and B). We previously proposed that these two residues are critical determinants in conferring nucleotide selectivity since they are Gly and Ala residues, respectively, in the protein products of the SLC35A2 and SLC35A3 genes—the UDP–Gal/GalNAc transporter and the UDP–GlcNAc transporter (NGT), respectively (17). The fact that these cytidine-interacting residues are conserved between CMP-sialic acid transporter and SLC35A4 suggests that SLC35A4 may be able to selectively transport CMP and other cytidine phosphate–coupled molecules (e.g., CDP-ribitol, CDP-ethanolamine).
Figure 4.
Sequence comparison between SLC35A1 and SLC35A4.A, sequence alignments between human and mouse SLC35A1 (CST) as well as human and mouse SLC35A4. The blue shading indicates the degree of residue conservation. The yellow bars under the sequences indicate the location of the indicated transmembrane (TM) domain. The interfacial alpha helix (IH) connecting TMs 2 and 3 is also indicated. The alignments were done using Jalview using a threshold of 40% to indicate the degree of residue conservation. B, the structure of mCST in complex with CMP-sialic acid is shown as a cartoon representation with the CMP-sialic acid shown as yellow sticks. The blue coloring indicates the degree of residue conservation in the same manner as in the sequence alignment in panel A. The top is the Golgi-facing side of the protein, whereas the bottom faces the cytoplasm. mCST, mouse CMP-sialic acid transporter.
Figure 5.
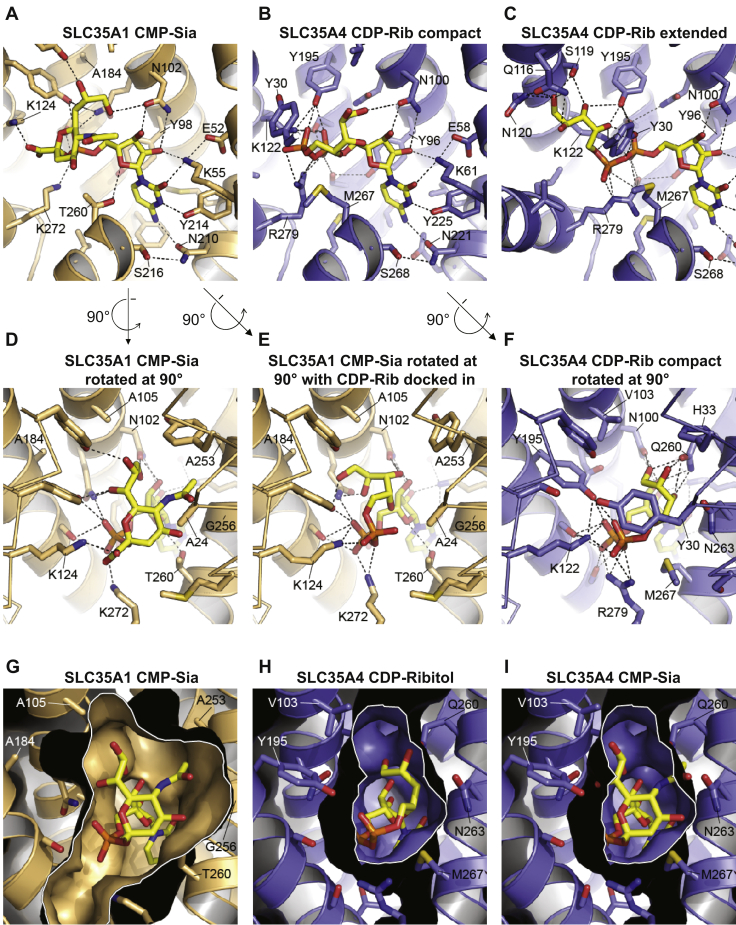
Comparison of substrate-binding sites in SLC35A1 and SLC35A4.A, the structure of CST in complex with CMP-Sia (yellow molecule) is shown. B and C, the structural homology model of SLC35A4 is shown with CDP-ribitol (yellow molecule) computationally docked in either the “compact” (B) or “extended” (C) conformation. In panels A–C, portions of transmembranes (TMs) 1 and 8, which are in front of the substrate, are hidden for clarity. D, a rotated view of the structure of CST in complex with CMP-Sia shown in panel A. E, the same view of the CST structure as in panel D, but with CDP-ribitol computationally docked in. F, a rotated view of what is shown in panel B: the model of SLC35A4 with CDP-ribitol in the compact conformation. In panels D–F, Cα traces are shown for TMs 1 and 7 (instead of cartoon ribbons) for clarity. In panels A–F, key residues are indicated, and select polar interactions are indicated by dashed lines as a visual aid. G, in a view that is similar to that shown in panel D, a surface representation is shown for the CST structure in complex with CMP-Sia. H and I, in a view that is similar to that shown in panel F, a surface representation is shown for the model of SLC35A4 with CDP-ribitol in the compact conformation (H) or with CMP-Sia manually placed into the cavity to show the clashes that would occur between the Sia moiety and several cavity-lining side chains. In panels G–I, a white border outlines the sliced view through the substrate-binding cavity. In all panels, only side chains that are within 5 Å of the substrate are shown for clarity. CMP-Sia, CMP-sialic acid; CST, CMP-sialic acid transporter.
There is also high sequence conservation between SLC35A1 and SLC35A4 in the region where SLC35A1 interacts with the negatively charged groups of CMP and CMP-sialic acid (Fig. 4, Fig. S2, and Fig. 5, A–F). SLC35A1 makes extensive contacts with both the phosphate and the C-1 carboxyl of the sialic acid moiety of CMP-sialic acid through several residues including Lys124 and Lys272 (Fig. 5, A and D). These positively charged residues are conserved in all members of the SLC35A subfamily and are also likely important for interacting with the alpha and beta phosphates of the UDP-coupled sugar substrates of UDP–Gal/GalNAc transporter and UDP–GlcNAc transporter (17). Conservation of these residues in SLC35A4 supports the concept that this protein may also be able to bind and transport negatively charged phosphate-containing molecules.
On the other hand, SLC35A4 does not share any significant sequence conservation with SLC35A1 in the cavity surrounding the binding site for the sialic acid moiety of CMP-sialic acid (Fig. 4, Fig. S2, and Fig. 5, D–I). This cavity in SLC35A1 is very large, which is necessary to accommodate the bulky sialic acid (Fig. 5, D and G). We previously postulated that most of the substrate specificity in CMP-sialic acid comes from interactions between the protein and the nucleotide moiety of the substrate. Indeed, there are not many interactions between the protein and the sialic acid moiety of CMP-sialic acid and also CMP alone can bind with high affinity and inhibit transport, whereas sialic acid alone cannot (17). Of note, this lack of specificity to a certain extent might be desirable since sialic acids not only comprise the most frequent form, N-acetylneuraminic acid, but also rarer variants (24). It also suggests that SLC35A1 should be able to transport other cytidine phosphate–coupled molecules, especially when the cytidine phosphate–coupled molecule is smaller than a sialic acid (i.e., CDP-ribitol, CDP-ethanolamine). Indeed, docking CDP-ribitol into SLC35A1 shows that there is ample room to accommodate a phosphoribitol in the canonical sugar-binding site (Fig. 5E).
Our structural homology model of SLC35A4 suggests that its substrate-binding cavity is much smaller than the equivalent cavity in SLC35A1 (Fig. 5, G–I). This is on account of the change of residues with small side chains to those with larger side chains. For example, Ala24, Ala105, Ala184, Ala253, Gly256, and Thr260 in SLC35A1 are instead Tyr30, Val103, Tyr195, Gln260, Asn263, and Met267, respectively, in SLC35A4 (Fig. S2 and Fig. 5, D–I). Docking CDP-ribitol into the structural homology model of SLC35A4 indicates that CDP-ribitol may bind in two possible modes, which we term “compact” and “extended” (Fig. 5, B and C). These binding modes seem to be dictated by the rotamer conformation that Tyr30 adopts. In the compact mode, the ribitol moiety is oriented such that it is adjacent to the ribose moiety of the cytidine where it can interact with His33, Gln260, and Asn263 (Fig. 5, B and F). In the extended mode, the ribitol instead reaches over to the other side of the cavity where it can interact with Gln116, Ser119, and Asn120 (Fig. 5C). A smaller substrate-binding cavity may be important for selectively coordinating the ribitol moiety of CDP-ribitol, which is much smaller than a sialic acid. In fact, comparison of the model of SLC35A4 with the CMP-sialic acid–bound SLC35A1 structure suggests that Tyr195, Asn263, Met267, and perhaps Tyr30 would clash with the sialic acid moiety (Fig. 5I), suggesting that SLC35A4 would not be capable of transporting CMP-sialic acid.
To test the hypothesis that the size of the catalytic pocket of SLC35A1 largely explains its capacity to transport both CMP-sialic acid and CDP-ribitol, we generated a series of point mutants in SLC35A1 that limit the size of its catalytic pocket (Fig. 6). Subsequently, we expressed these mutants, wildtype SLC35A1 or SLC35A4 in SLC35A1 KO HAP1 cells. To identify mutants that had lost the capacity to transport CMP-sialic acid, we first assessed which mutants were unable to normalize elevated CMP-sialic acid levels in SLC35A1 KO cells (Fig. S3). In parallel, we performed a laminin overlay assay to determine which transporters still allowed CDP-ribitol transport. This revealed that all SLC35A1 mutants still allowed the formation of the laminin-binding glycan to an extent that was comparable to the situation when wildtype SLC35A1 was re-expressed (Fig. S3A), suggesting that all these mutants still permit CDP-ribitol transport. In contrast, three mutants were unable to normalize elevated CMP-sialic acid in SLC35A1 KO cells (Fig. S3B), suggesting that they had lost the capacity to transport CMP-sialic acid. For a more detailed analysis, we then focused on these mutations (G256N, A24Y, and A183Y), as well as one mutation that behaved like the wildtype protein (A253Q). First, we assessed sialylation using staining with MAL II or PNA (Fig. 6, A–F). Similar to SLC35A4, the mutants G256N, A24Y, and A184Y were unable to rescue the reduced MAL II staining or the increased PNA staining in SLC35A1 KO cells, whereas the mutant A253Q and wildtype SLC35A1 were able to do so. This indicated that mutations G256N, A24Y, and A184Y prevent transport of CMP-sialic acid possibly by restricting the size of the binding pocket. However, an alternative hypothesis would be that these mutations completely inactivate transport function. To test this possibility, we assessed glycosylation of α-dystroglycan using the glycan-specific antibody IIH6 and a laminin overlay assay (Fig. 6, F–J). Given that α-dystroglycan glycosylation depends on the transport of CDP-ribitol into the Golgi apparatus, we assessed whether mutants rescued the phenotype of SLC35A1 KO cells. In these experiments, we observed that expression of all four mutants rescued IIH6 staining and laminin overlay signal intensity to a similar extent as wildtype SLC35A1. This indicated that these mutants were still able to transport CDP-ribitol although they were unable to transport CMP-sialic acid. Interestingly, the three mutants that had failed to rescue the sialylation deficiency (G256N, A24Y, and A184Y; Fig. 5, A–F) only partially rescued the reduction of the apparent size of α-dystroglycan in SLC35A1 KO cells (Fig. 6J). Given that part of the apparent size reduction in SLC35A1 KO cell lines is likely attributed to the lack of sialylation (Fig. 3E), our data corroborate the conclusion that mutations G256N, A24Y, and A184Y restrict the binding site and thereby limit the substrate spectrum of SLC35A1 to CDP-ribitol.
Figure 6.
Restricting the catalytic site of SLC35A1 abolishes CMP-sialic acid but still allows CDP-ribitol transport.A–I, HAP1 wildtype or SLC35A1 KO cells engineered to express SLC35A1 or the indicated SLC35A1 mutants were analyzed by flow cytometry with MAL II lectin, PNA, and IIH6 antibody (A and B, D and E, and G and H). Flow cytometry data were divided into two panels for the sake of clarity. Quantification of three independent experiments is shown in panels C, F, and I. Means ± SEM or mean fluorescence intensities are shown. J, laminin overlay and β-dystroglycan Western blot were performed in the same cell lines as described previously in addition to ISPD KO cells. Quantification of the relative signal obtained in laminin overlay was normalized to the β-dystroglycan Western blot signal. ISPD, isoprenoid synthase domain–containing protein; MAL II, Maackia amurensis lectin II; PNA, peanut agglutinin.
Discussion
The sequential incorporation of two ribitol phosphate residues is required to assemble the functional glycan on α-dystroglycan. In the cytoplasm, the enzyme isoprenoid synthase domain–containing protein synthesizes CDP-ribitol, which is then used by FKTN and FKRP in the Golgi apparatus. However, it was still unknown how CDP-ribitol was transported into the Golgi apparatus. In this article, we provide evidence that SLC35A1, the Golgi CMP-sialic acid transporter, and SLC35A4, a transporter of unknown function, are involved in the transport of CDP-ribitol into the Golgi apparatus.
Previous publications (20, 21) had revealed that genetic inactivation of the CMP-sialic acid transporter SLC35A1 in HAP1 cells led to a strongly reduced α-dystroglycan glycosylation independently of its role in sialylation (21). Given that CDP-ribitol to a certain extent resembles CMP-sialic acid, it was tempting to speculate that SLC35A1 can also act as a CDP-ribitol transporter. Mutations in SLC35A1 in humans lead to intellectual disability and bleeding diathesis (25, 26, 27). Platelet-specific inactivation of SLC35A1 in mice led to reduced platelet counts (28). Cells from affected patients show a deficiency in sialylation and CMP-sialic acid uptake into the Golgi apparatus (26). Interestingly, there was no molecular or clinical evidence that these patients suffered from a deficiency in α-dystroglycan glycosylation, even though re-expression of wildtype SLC35A1 corrected the deficiency of α-dystroglycan glycosylation in SLC35A1 KO HAP1 cells. All these observations indicated that SLC35A1 might have a role in CDP-ribitol transport, but they highlighted the possibility that in many cell types, this transport can be done by other transporters. In the present study, we resolve this conundrum by revealing that SLC35A1 and SLC35A4 play a redundant role in allowing ribitol phosphate incorporation into the glycan of α-dystroglycan. In fact, our data are even compatible with the contribution of additional transporters to CDP-ribitol transport, which might allow for the formation of the residual laminin-binding glycan in SLC35A1 KO HAP1 cells (Figs. 1H and 3, D and E). In any case, our findings provide a plausible explanation why so far no dystroglycanopathy patients with mutations in SLC35A4 have been described.
Up to now, very little is known about the function of SLC35A4. Based on its homology with other members of the SLC35A family, it has been suggested to be a potential UDP-sugar transporter (18). However, genetic inactivation of SLC35A4 did not lead to any significant defect in protein glycosylation (22, 29). In an effort to further delineate the function of SLC35A4, Sosicka et al. (22) attempted to reveal molecular properties and made a series of observations. First, using epitope-tagged proteins they provided some evidence that SLC35A4 is not efficiently forming homodimers nor heterodimers with other SLC35A family members (with the exception of SLC35A5). Second, using selective permeabilization of intracellular membranes, they suggested that the last helix of SLC35A4 potentially might show a different topology than shown in the SLC35A1 crystal structure and predicted other structures. These data could be interpreted as evidence that SLC35A4 may not be a functional NST at all. Thus, these data might seem in stark contrast to our data where we show that SLC35A4 clearly rescued the deficient ribitol incorporation into α-dystroglycan observed in SLC35A1 KO cells. However, even though these experiments have been performed very carefully, the data from Sosicka et al. relied largely on epitope-tagged proteins. Often, the addition of an epitope tag does not affect protein function, but in some cases, it can alter protein–protein interactions or protein folding. Hence, it is conceivable that endogenous SLC35A4 protein behaves differently than epitope-tagged overexpressed protein. As such, our observations together with the modeling of the SLC35A4 structure based on the recently produced SLC35A1 structure indicate that SLC35A4 indeed directly contributes to CDP-ribitol transport.
Our observation that SLC35A1 and SLC35A4 play a redundant role in ensuring ribitol phosphorylation leads to the question why two different transporters for CDP-ribitol should exist in the Golgi apparatus. Interestingly, the crystal structure of SLC35A1 revealed that substrate recognition mainly occurs by specific interactions with the nucleotide part of the nucleotide sugar (17). Given that NSTs exchange the corresponding nucleoside monophosphate against the nucleotide sugar, this likely allows the recognition of alternating substrates. Sialic acid is much bulkier and more rigid than ribitol, which is extremely flexible. Therefore, it is not surprising that CDP-ribitol can fit in the binding pocket of SLC35A1, even though this transporter might be primarily dedicated to CMP-sialic acid. In reverse, the large pocket of SLC35A1 likely requires some flexibility since it has to accommodate different CMP-sialic acid species. While humans mainly have CMP-N-acetylneuraminic acid, in other species, other sialic acids (such as CMP-N-glycolylneuraminic acid, CMP-deaminoneuraminic acid) might also need to be transported (24). This led us to postulate that the major difference between SLC35A1 and SLC35A4 might be the size of the binding pocket. Using site-directed mutagenesis, we introduced into the SLC35A1-binding sites bulkier residues resembling the ones that are present in SLC35A4. Consistent with our expectations, such mutations were unable to rescue the sialylation defect in SLC35A1 KO cells but still rescued deficient α-dystroglycan glycosylation (i.e., ribitol phosphorylation).
Recognition of the nucleotide part seems to be of key importance for NSTs (17, 30). Yet, the precise structure of the sugar part seems to be less critical, since some transporters transport a wide variety of sugar conjugates (31, 32). Our observations underline that the size of the binding pocket might be a major determinant in some cases. Transport of less bulky compounds might be considered a nonspecific activity, but our observations concerning SLC35A1 indicate that they might very well be biologically relevant.
Experimental procedures
Plasmid construction
We obtained a mouse Slc35a1 complementary DNA clone from GE Dharmacon (accession number: BC012252; clone ID: 4190458) and amplified the open reading frame by PCR using Q5 polymerase (New England Biolabs) adding a C-terminal FLAG tag and flanking NheI–EcoRI sites using the primers mSlc35a1_NheI-Sense and mSlc35a1_EcoRI-Reverse (for primers, see Table S1). The resulting PCR product was digested with the restriction endonucleases NheI and EcoRI and cloned into the XbaI and EcoRI sites of the plasmid pUB82 resulting in the plasmid pUB86. The plasmid pUB82 is a lentiviral expression vector based on the plasmid pLVX-PURO (Clontech), where expression is driven by a truncated SV40 promoter that allows low levels of expression (details available upon request).
The hSLC35A4 open reading frame was amplified by PCR with HEK293 complementary DNA as a template adding a C-terminal FLAG tag and BbsI restriction sites using the primers hSLC35A4-BbsI_Sense and hSLC35A4-BbsI_Reverse (for primers, see Table S1). The resulting PCR product was digested using the restriction endonuclease BbsI and cloned into the XbaI and EcoRI plasmid pUB82 leading to the plasmid pUB90.
Expression constructs for mutant hSLC35A4 and mSlc35a1 were cloned by Gibson assembly (NEBuilder HiFi; New England Biolabs). For this, two overlapping fragments of the open reading frames were amplified by PCR using primers containing the desired mutation (for primers, see Table S1). In order to allow expression in mammalian cells, the assembly was set up with the plasmid pUB82 digested with the enzymes XbaI and EcoRI.
The guide RNAs to knock out genes were cloned into the BbsI site of the pX459 (Addgene; #62988) vector after annealing and phosphorylation of corresponding oligonucleotides (33). The targeted sequence for each gene is listed in Table S2 with the protospacer adjacent motif sequence in underlined lower case.
For knockdown experiments, shRNAs targeting human SLC35A1 and SLC35A4 were inserted into XhoI–EcoRI sites of a lentiviral vector pJG97, which is similar to the miR-E system described by Fellmann et al. (34). Inserts were generated by extension of two overlapping primers (shown in the table later) followed by PCR amplification using the primers miR-E-XhoI_Sense and miR-E-EcoRI_Reverse (for primers, see Table S1) in order to add flanking XhoI–EcoRI sites.
Flow cytometry
Cells were washed once with PBS and subsequently incubated with 500 μl of nonenzymatic cell dissociation solution (Sigma) until they detached (20 min). Cells were collected in 4 ml PBS and washed twice with 1 ml of 0.1% fetal calf serum in PBS centrifuging at 500g for 5 min in between. Cells were then incubated with IIH6C4 antibody (1:200 dilution; Merck Millipore) for 40 min, with biotin-labeled MAL II (10 μg/ml; Vector Laboratories) for 40 min or with FITC–labeled PNA (10 μg/ml; Sigma–Aldrich) for 90 min. Subsequently, cells were washed twice as described previously and incubated for 20 min with 1:200 diluted goat antimouse IgM-CFL (Santa Cruz) for the cells incubated with IIH6C4 antibody or with 5 μg/ml of FITC-labeled streptavidin (Vector Laboratories) for the cells incubated with MAL II. All cells were washed twice as described previously, resuspended in 400 μl PBS, and analyzed using a FACSverse flow cytometer (BD Biosciences).
Cell culture
HAP1 cells obtained from Haplogen were cultured in Iscove's modified Dulbecco's Medium. HEK293, HEK293T, and HCT116 cells were a gift from Eric Fearon (University of Michigan) and were cultured in Dulbecco's modified Eagle's medium. Both media contained 4.5 g/l d-glucose, 10% fetal calf serum, 2 mM ultraglutamine I (Lonza), and 100 U/ml penicillin/streptomycin (Lonza).
To generate KO cells, we transfected cells with CRISPR/Cas9 constructs using Lipofectamine 2000 and 1 μg of plasmid in 12-well plates (350,000 cells/well). After 24 h, cells were plated at several dilutions in 10 cm plates and selected with 2 μg/ml puromycin (Thermo Fisher Scientific) for 48 h. Clonal populations were picked approximately 10 days later and transferred into 96-well plates. Genomic DNA was isolated from one replicate 96-well plate, the region encompassing the predicted CAS9 cleavage site was amplified, and the PCR products were analyzed by Sanger sequencing (Genewiz) (9).
Recombinant lentiviruses for overexpression or knockdown experiments were produced by transiently transfecting HEK293T cells with second-generation packaging plasmids psPAX2 and pMD2.G (kind gifts from Didier Trono; Addgene #12260 and #12259), as well as a lentiviral vector using the calcium phosphate coprecipitation method as described previously (35). After 24 h, target cells were infected in the presence of 8 μg/ml polybrene (Sigma). Infected cells were selected for 2 days with appropriate antibiotic or until noninfected control cells were dead. Concentrations were 2 μg/ml for puromycin (Thermo Fisher Scientific) (used for pUB95-96-97 and pJG109) or 300 μg/ml for hygromycin (Invivogen) (used for pUB110-111-112-117).
GC/MS analysis of monosaccharides and ribitol
The composition of the glycan of dystroglycan was assessed by GC/MS after methanolytic depolymerization and trimethylsilyation (9). Briefly, we collected medium from HEK293 cells that had been engineered to express an N-terminal 485 amino acids of α-dystroglycan carrying an SFB tag consisting of S-tag, FLAG tag, and a streptavidin-binding peptide (9). After affinity purification using high-capacity streptavidin Sepharose beads (GE17-5113-01; GE Healthcare Life Sciences), proteins were eluted with 0.1% formic acid and dried down with help of a SpeedVac Concentrator (Thermo Fisher Scientific). Methanolysis was performed by resuspending in 200 μl of methanol containing 1.7 M HCl and incubation for 16 h at 70 °C under N2 using 1 ml hydrolysis tubes (#29570; Thermo Fisher Scientific). After drying down, the detached monosaccharides and sugar alcohols were derivatized with N-methyl-N-trimethylsilyl-trifluoroacetamide (Macherey–Nagel) for 30 min at 37 °C before analysis (9) described before. One microliter of sample was injected in splitless mode at 270 °C, using helium as the carrier gas at a flow rate of 1 ml/min. The GC oven temperature was held at 100 °C for 3 min and increased to 300 °C at 3.5 °C/min. The MS source and quadrupole detector were held at 230 and 150 °C, respectively. Data were acquired in combined selected-ion monitoring and scanning mode (68–700 m/z). MS spectra were obtained by electron impact ionization (70 eV). Compounds were identified by comparison of elution time and specific fragments with the ones observed for pure standards. The areas under the curve of quantifier ions were integrated using Agilent Masshunter quantitative analysis software (Agilent) and subsequently normalized to the total ion current. Presented values are shown relative to the ribitol levels measured in wildtype cells.
Laminin overlay and Western blot
Laminin overlay assays were performed as described previously (9). Briefly, proteins were harvested in PBS 1× containing 1% Triton X-100 and cOmplete Proteinase Inhibitor Cocktail (Roche). Samples were sonicated and clarified by centrifugation (15 min—27,000g—4 °C). After measuring protein concentration using the Pierce bicinchoninic acid protein assay (Thermo Fisher Scientific), 1400 μg of protein were incubated under gentle rotation for 16 h at 4 °C with wheat germ agglutinin coupled to agarose beads (Vector Laboratories). After washing three times with 1 ml of PBS containing 0.1% Triton X-100, beads were incubated for 10 min at 72 °C in the presence of sample buffer (62.5 mM Tris–HCl at pH 8.5; 0.5% lithium dodecylsulfate; 0.1 mM EDTA; 2.5% glycerol; 0.05 mM phenol red; 0.05 mM Coomassie brilliant blue; and 100 mM DTT) (36) to release proteins before resolution on 3% to 8% Tris–acetate gels (Life Technologies) for 75 min at 130 V. Treatment with Arthrobacter ureafaciens sialidase (Roche) was performed during the last wash before elution at a concentration of 10 mU/ml for 30 min. Proteins were transferred overnight onto polyvinylidene membrane. Membranes were blocked with laminin-binding buffer (LBB) (50 mM Tris–HCl at pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 1 mM CaCl2) containing 3% bovine serum albumin (BSA) (Sigma) for 1 h at RT and incubated overnight with 1.15 μg/ml Laminin-111 (Sigma; L2020) in LBB. Membranes were washed three times for 10 min with LBB and then incubated for 2 h at RT with rabbit antilaminin antibody (Sigma; L9393) diluted 1:1000 in LBB containing 3% BSA. After three additional 10 min washes with LBB, they were incubated for 1 h at RT with horseradish peroxidase–coupled donkey anti-rabbit IgG antibody (GE Healthcare; NA934V) diluted 1:15,000 in LBB containing 3% BSA. Finally, chemiluminescent signals were detected using a homemade chemiluminescent detection system (37) following three more 10 min washes in LBB at RT. In parallel to the laminin overlay assay, the lower part of the membranes (below the 70 kDa molecular weight mark) was incubated with anti-β-dystroglycan antibody (Santa Cruz; clone 7D11, sc-33701, 1:1000 dilution) using a conventional Western blot protocol. Uncropped images of the laminin overlay assay and Western blots can be seen in the Supporting information.
LC/MS analysis of metabolites (CDP-ribitol and CMP-N-acetylneuraminic acid)
LC/MS analysis was performed essentially as described before (38) and is based on a method described by Coulier et al. (39). Briefly, 5 μl of sample was resolved using an Inertsil 3 μm particle ODS-4 column (150 × 2.1 mm; GL Biosciences) at a constant flow rate of 0.2 ml/min with an Agilent 1290 HPLC system. Mobile phase A consisted of 5 mM hexylamine (Sigma–Aldrich) adjusted to pH 6.3 with acetic acid (Biosolve BV) and phase B of 90% methanol (Biosolve BV)/10% 10 mM ammonium acetate (Biosolve BV) adjusted to pH 8.5 with ammonia (Merck). The mobile phase profile consisted of the following steps and linear gradients: 0 to 2 min at 0% B; 2 to 6 min from 0 to 20% B; 6 to 17 min from 20 to 31% B; 17 to 36 min from 31 to 60% B; 36 to 41 min from 60 to 100% B; 41 to 51 min at 100% B; 51 to 53 min from 100 to 0% B; and 53 to 60 min at 0% B.
Analytes were identified with an Agilent 6550 ion funnel mass spectrometer operated in negative mode with an electrospray ionization source and the following settings: electrospray ionization source spray voltage 3500 V, sheath gas 350 °C at 11 l/min, nebulizer pressure 35 psig, and drying gas 200 °C at 14 l/min. Compound identification was based on their exact mass and retention time compared with purified standards (Sigma–Aldrich (9)). Extracted-ion chromatograms of the [M–H] forms were integrated using MassHunter software (Agilent), and areas were normalized to total ion current. LC/MS results were obtained in three independent experiments. Within each experiment, the normalized value for the untreated condition for each cell line was set to 100%. Data are shown as means ± SEM across three experiments in Figure 3F and means of two experiments in Fig. S3.
SLC35A4 homology modeling and CDP-ribitol docking
A structural homology model of human SLC35A4 (hSLC35A4) was generated using the SWISS-MODEL Web server (40), using the mSLC35A1-CMP-sialic acid structure as a template. CDP-ribitol was docked into either the mouse CMP-sialic acid transporter (Slc35a1) structure (17) or the hSLC35A4 structural homology model using AutoDock4 (41) with the flexible side-chain covalent docking method, as previously described (42). To start this, CDP-ribitol was first added to either the mSlc35a1 or the hSLC35A4 models. For positioning CDP-ribitol into mSlc35a1, the CMP portion of CDP-ribitol was positioned exactly how the CMP moiety of CMP-sialic acid is oriented in the mSlc35a1-CMP-sialic acid structure. For positioning CDP-ribitol into hSLC35A4, the hSLC25A4 model was first superimposed onto the mSlc35a1 model. CDP-ribitol was then positioned into the hSLC25A4 model in the same orientation as in the mSlc35a1 model. For each model, the cytidine portion of CDP-ribitol was set to act as part of the rigid component of the protein. This greatly reduced the computation required and was justified by the likely case that the CMP part of CDP-ribitol will bind the same way that the CMP part of CMP-sialic acid binds mSlc35a1 and the residues that surround the CMP-binding site in mSlc35a1 are nearly completely conserved in hSLC35A4. The phosphoribitol was treated as a flexible residue. Potential ribitol-interacting residues were also set to be flexible while the rest of the protein was kept rigid. Initial docking runs indicated that the ribitol preferred to bind hSLC35A4 in two main conformations, which we term “compact” and “extended.” The binding mode was dictated by the rotamer conformation of Tyr30. Therefore, to get the lowest energy pose for each mode of binding, two separate docking runs were performed where the rotamer of Tyr30 was constrained in the conformation that allowed the desired binding mode of the ribitol. The ligand poses shown in Figure 4 are the lowest-energy poses out of 1000 docking runs.
Statistical analysis
Differences between groups were assessed by one-way ANOVA followed by pairwise testing with the Holm–Sidak test (43). Experiments were repeated at least three times and presented values are means ± standard error of the means unless indicated otherwise. The exceptions are the data presented in Fig. S3, which were performed twice.
Data availability
All data are contained in the article and the supporting information file.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Julie Graff for technical assistance and Emile Van Schaftingen for continuous support as well as comments on the article.
Author contributions
G. T. B. and M. R. W. funding and supervision. M. R. W. molecular modeling. B. U. generation of knockout cell lines. B. U., S. P., and F. C. site-directed mutagenesis, lentiviral rescue, laminin overlay, and flow cytometry. B. U., S. P., and F. C. GC–MS analyses and LC–MS. All authors contributed to the redaction of the article and have approved of the final article.
Funding and additional information
The authors acknowledge funding from the Belgian National Fund for Scientific Research (FNRS; Crédit de recherche to G. T. B.; FRIA fellowship to B. U., Chargé de Recherche fellowship to S. P.; and WELBIO to G. T. B.), the E-Rare network EUROGLYCAN, Université Catholique de Louvain (Fonds Special de Recherche), the Fondation Marguerite-Marie Delacroix, the Prix Professeur Christian Coërs (to S. P. and B. U.), the European Research Council within the Horizon 2020 program (consolidator grant no. 771704), and the Fonds Maurange.
Biographies

Benoît Ury is a doctoral student in the Department of Biochemistry at the de Duve Institute, UCLouvain in Brussels, Belgium. After a master's degree in biomedical and pharmaceutical sciences, he joined the laboratory of Guido Bommer to unravel the remaining mysteries of α-dystroglycan glycosylation. During these studies, he has acquired extensive experience in a wide range of analytical techniques that he wants to use in the pharmaceutical industry in the future.

Sven Potelle is a postdoctoral researcher in the Department of Biochemistry at the de Duve Institute, UCLouvain in Brussels, Belgium. He obtained extensive training in glycobiology during his PhD with Francois Foulquier in Lille (France) and is fascinated by the diverse mechanisms that link defects in protein glycosylation to human diseases. Currently, he is trying to understand the mechanisms that lead to several inherited disorders of glycosylation, with the goal to eventually develop innovative therapies.
Edited by Gerald Hart
Contributor Information
Matthew R. Whorton, Email: whorton@ohsu.edu.
Guido T. Bommer, Email: guido.bommer@uclouvain.be.
Supporting information
References
- 1.Ibraghimov-Beskrovnaya O., Ervasti J.M., Leveille C.J., Slaughter C.A., Sernett S.W., Campbell K.P. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 2.Endo T. Glycobiology of alpha-dystroglycan and muscular dystrophy. J. Biochem. 2015;157:1–12. doi: 10.1093/jb/mvu066. [DOI] [PubMed] [Google Scholar]
- 3.Live D., Wells L., Boons G.J. Dissecting the molecular basis of the role of the O-mannosylation pathway in disease: Alpha-dystroglycan and forms of muscular dystrophy. Chembiochem. 2013;14:2392–2402. doi: 10.1002/cbic.201300417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michele D.E., Barresi R., Kanagawa M., Saito F., Cohn R.D., Satz J.S., Dollar J., Nishino I., Kelley R.I., Somer H., Straub V., Mathews K.D., Moore S.A., Campbell K.P. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida-Moriguchi T., Campbell K.P. Matriglycan: A novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology. 2015;25:702–713. doi: 10.1093/glycob/cwv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa M., Nakamura N., Nakayama Y., Kurosaka A., Manya H., Kanagawa M., Endo T., Furukawa K., Okajima T. GTDC2 modifies O-mannosylated alpha-dystroglycan in the endoplasmic reticulum to generate N-acetyl glucosamine epitopes reactive with CTD110.6 antibody. Biochem. Biophys. Res. Commun. 2013;440:88–93. doi: 10.1016/j.bbrc.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Stevens E., Carss K.J., Cirak S., Foley A.R., Torelli S., Willer T., Tambunan D.E., Yau S., Brodd L., Sewry C.A., Feng L., Haliloglu G., Orhan D., Dobyns W.B., Enns G.M. Mutations in B3GALNT2 cause congenital muscular dystrophy and hypoglycosylation of alpha-dystroglycan. Am. J. Hum. Genet. 2013;92:354–365. doi: 10.1016/j.ajhg.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida-Moriguchi T., Willer T., Anderson M.E., Venzke D., Whyte T., Muntoni F., Lee H., Nelson S.F., Yu L., Campbell K.P. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341:896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerin I., Ury B., Breloy I., Bouchet-Seraphin C., Bolsee J., Halbout M., Graff J., Vertommen D., Muccioli G.G., Seta N., Cuisset J.M., Dabaj I., Quijano-Roy S., Grahn A., Van Schaftingen E. ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto alpha-dystroglycan. Nat. Commun. 2016;7:11534. doi: 10.1038/ncomms11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanagawa M., Kobayashi K., Tajiri M., Manya H., Kuga A., Yamaguchi Y., Akasaka-Manya K., Furukawa J.I., Mizuno M., Kawakami H., Shinohara Y., Wada Y., Endo T., Toda T. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 2016;14:2209–2223. doi: 10.1016/j.celrep.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Praissman J.L., Willer T., Sheikh M.O., Toi A., Chitayat D., Lin Y.Y., Lee H., Stalnaker S.H., Wang S., Prabhakar P.K., Nelson S.F., Stemple D.L., Moore S.A., Moremen K.W., Campbell K.P. The functional O-mannose glycan on alpha-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife. 2016;5 doi: 10.7554/eLife.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manya H., Yamaguchi Y., Kanagawa M., Kobayashi K., Tajiri M., Akasaka-Manya K., Kawakami H., Mizuno M., Wada Y., Toda T., Endo T. The muscular dystrophy gene TMEM5 encodes a ribitol beta1,4-xylosyltransferase required for the functional glycosylation of dystroglycan. J. Biol. Chem. 2016;291:24618–24627. doi: 10.1074/jbc.M116.751917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Praissman J.L., Live D.H., Wang S., Ramiah A., Chinoy Z.S., Boons G.J., Moremen K.W., Wells L. B4GAT1 is the priming enzyme for the LARGE-dependent functional glycosylation of alpha-dystroglycan. Elife. 2014;3 doi: 10.7554/eLife.03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer T., Inamori K., Venzke D., Harvey C., Morgensen G., Hara Y., Beltran Valero de Bernabe D., Yu L., Wright K.M., Campbell K.P. The glucuronyltransferase B4GAT1 is required for initiation of LARGE-mediated alpha-dystroglycan functional glycosylation. Elife. 2014;3 doi: 10.7554/eLife.03941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inamori K., Yoshida-Moriguchi T., Hara Y., Anderson M.E., Yu L., Campbell K.P. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. 2012;335:93–96. doi: 10.1126/science.1214115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadley B., Maggioni A., Ashikov A., Day C.J., Haselhorst T., Tiralongo J. Structure and function of nucleotide sugar transporters: Current progress. Comput. Struct. Biotechnol. J. 2014;10:23–32. doi: 10.1016/j.csbj.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahuja S., Whorton M.R. Structural basis for mammalian nucleotide sugar transport. Elife. 2019;8 doi: 10.7554/eLife.45221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadley B., Litfin T., Day C.J., Haselhorst T., Zhou Y., Tiralongo J. Nucleotide sugar transporter SLC35 family structure and function. Comput. Struct. Biotechnol. J. 2019;17:1123–1134. doi: 10.1016/j.csbj.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker J.L., Newstead S. Structural basis of nucleotide sugar transport across the Golgi membrane. Nature. 2017;551:521–524. doi: 10.1038/nature24464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jae L.T., Raaben M., Riemersma M., van Beusekom E., Blomen V.A., Velds A., Kerkhoven R.M., Carette J.E., Topaloglu H., Meinecke P., Wessels M.W., Lefeber D.J., Whelan S.P., van Bokhoven H., Brummelkamp T.R. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 2013;340:479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riemersma M., Sandrock J., Boltje T.J., Bull C., Heise T., Ashikov A., Adema G.J., van Bokhoven H., Lefeber D.J. Disease mutations in CMP-sialic acid transporter SLC35A1 result in abnormal alpha-dystroglycan O-mannosylation, independent from sialic acid. Hum. Mol. Genet. 2015;24:2241–2246. doi: 10.1093/hmg/ddu742. [DOI] [PubMed] [Google Scholar]
- 22.Sosicka P., Maszczak-Seneczko D., Bazan B., Shauchuk Y., Kaczmarek B., Olczak M. An insight into the orphan nucleotide sugar transporter SLC35A4. Biochim. Biophys. Acta. 2017;1864:825–838. doi: 10.1016/j.bbamcr.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Carithers L.J., Ardlie K., Barcus M., Branton P.A., Britton A., Buia S.A., Compton C.C., DeLuca D.S., Peter-Demchok J., Gelfand E.T., Guan P., Korzeniewski G.E., Lockhart N.C., Rabiner C.A., Rao A.K. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv. Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schauer R., Kamerling J.P. Exploration of the sialic acid world. Adv. Carbohydr. Chem. Biochem. 2018;75:1–213. doi: 10.1016/bs.accb.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kauskot A., Pascreau T., Adam F., Bruneel A., Reperant C., Lourenco-Rodrigues M.D., Rosa J.P., Petermann R., Maurey H., Auditeau C., Lasne D., Denis C.V., Bryckaert M., de Lonlay P., Lavenu-Bombled C. A mutation in the gene coding for the sialic acid transporter SLC35A1 is required for platelet life span but not proplatelet formation. Haematologica. 2018;103:e613–e617. doi: 10.3324/haematol.2018.198028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng B.G., Asteggiano C.G., Kircher M., Buckingham K.J., Raymond K., Nickerson D.A., Shendure J., Bamshad M.J., University of Washington Center for Mendelian Genomics. Ensslen M., Freeze H.H. Encephalopathy caused by novel mutations in the CMP-sialic acid transporter, SLC35A1. Am. J. Med. Genet. A. 2017;173:2906–2911. doi: 10.1002/ajmg.a.38412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamed M., Ashikov A., Guillard M., Robben J.H., Schmidt S., van den Heuvel B., de Brouwer A.P., Gerardy-Schahn R., Deen P.M., Wevers R.A., Lefeber D.J., Morava E. Intellectual disability and bleeding diathesis due to deficient CMP--sialic acid transport. Neurology. 2013;81:681–687. doi: 10.1212/WNL.0b013e3182a08f53. [DOI] [PubMed] [Google Scholar]
- 28.Ma X., Li Y., Kondo Y., Shi H., Han J., Jiang Y., Bai X., Archer-Hartmann S.A., Azadi P., Ruan C., Fu J., Xia L. Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. Haematologica. 2021;106:759–769. doi: 10.3324/haematol.2019.225987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoder-Agha F., Sosicka P., Escriva Conde M., Hassinen A., Glumoff T., Olczak M., Kellokumpu S. N-acetylglucosaminyltransferases and nucleotide sugar transporters form multi-enzyme-multi-transporter assemblies in Golgi membranes in vivo. Cell. Mol. Life Sci. 2019;76:1821–1832. doi: 10.1007/s00018-019-03032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker J.L., Corey R.A., Stansfeld P.J., Newstead S. Structural basis for substrate specificity and regulation of nucleotide sugar transporters in the lipid bilayer. Nat. Commun. 2019;10:4657. doi: 10.1038/s41467-019-12673-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rautengarten C., Ebert B., Moreno I., Temple H., Herter T., Link B., Donas-Cofre D., Moreno A., Saez-Aguayo S., Blanco F., Mortimer J.C., Schultink A., Reiter W.D., Dupree P., Pauly M. The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11563–11568. doi: 10.1073/pnas.1406073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert B., Rautengarten C., Guo X., Xiong G., Stonebloom S., Smith-Moritz A.M., Herter T., Chan L.J., Adams P.D., Petzold C.J., Pauly M., Willats W.G., Heazlewood J.L., Scheller H.V. Identification and characterization of a Golgi-localized UDP-xylose transporter family from Arabidopsis. Plant Cell. 2015;27:1218–1227. doi: 10.1105/tpc.114.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y., Sinha N., Zuber J. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Jordan M., Schallhorn A., Wurm F.M. Transfecting mammalian cells: Optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cubillos-Rojas M., Amair-Pinedo F., Tato I., Bartrons R., Ventura F., Rosa J.L. Tris-acetate polyacrylamide gradient gels for the simultaneous electrophoretic analysis of proteins of very high and low molecular mass. Methods Mol. Biol. 2012;869:205–213. doi: 10.1007/978-1-61779-821-4_17. [DOI] [PubMed] [Google Scholar]
- 37.Haan C., Behrmann I. A cost effective non-commercial ECL-solution for western blot detections yielding strong signals and low background. J. Immunol. Methods. 2007;318:11–19. doi: 10.1016/j.jim.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 38.Veiga-da-Cunha M., Chevalier N., Stephenne X., Defour J.P., Paczia N., Ferster A., Achouri Y., Dewulf J.P., Linster C.L., Bommer G.T., Van Schaftingen E. Failure to eliminate a phosphorylated glucose analog leads to neutropenia in patients with G6PT and G6PC3 deficiency. Proc. Natl. Acad. Sci. U. S. A. 2019;116:1241–1250. doi: 10.1073/pnas.1816143116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coulier L., Bas R., Jespersen S., Verheij E., van der Werf M.J., Hankemeier T. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 2006;78:6573–6582. doi: 10.1021/ac0607616. [DOI] [PubMed] [Google Scholar]
- 40.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianco G., Forli S., Goodsell D.S., Olson A.J. Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. 2016;25:295–301. doi: 10.1002/pro.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the article and the supporting information file.