Abstract
Memory B cells seem to be more durable than antibodies and thus crucial for the long-term immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Here we investigate SARS-CoV-2 spike-specific memory B cells and their dependence on CD4+ T cell help in different settings of coronavirus disease 2019 (COVID-19). Compared with severely ill individuals, those who recovered from mild COVID-19 develop fewer but functionally superior spike-specific memory B cells. Generation and affinity maturation of these cells is best associated with IL-21+CD4+ T cells in recovered individuals and CD40L+CD4+ T cells in severely ill individuals. The increased activation and exhaustion of memory B cells observed during COVID-19 correlates with CD4+ T cell functions. Intriguingly, CD4+ T cells recognizing membrane protein show a stronger association with spike-specific memory B cells than those recognizing spike or nucleocapsid proteins. Overall, we identify CD4+ T cell subsets associated with the generation of B cell memory during SARS-CoV-2 infection.
Keywords: SARS-CoV-2, COVID-19, memory B cells, spike, CD4+ T cell, IL-21, CD40L, antibody, recovered, severely ill
Graphical abstract
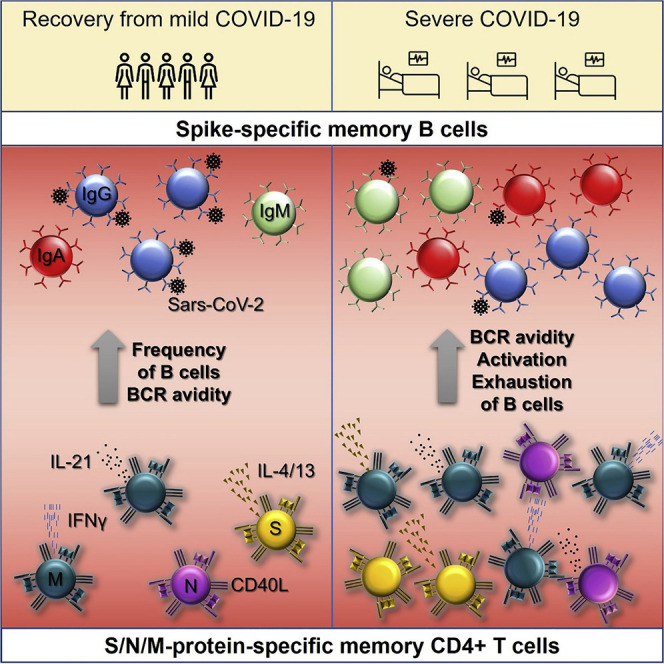
Pušnik et al. report that individuals who recovered from mild COVID-19 develop fewer but functionally superior SARS-CoV-2 spike-specific memory B cells than individuals with severe disease. The development and activation of those cells are associated with distinct CD4+ T cell functions in both clinical outcomes of COVID-19.
Introduction
With as of September 7, 2020 over 103 million infected individuals worldwide and over 2 million deaths, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is an unprecedented public health emergency (World Health Organization, 2020). Despite the immense efforts and engagement of many research institutions worldwide, to date, there is no available vaccine or treatment with proven efficacy against SARS-CoV-2. Furthermore, there is an increasing concern that, similar to other coronavirus infections, infection with SARS-CoV-2 does not lead to formation of long-lived immunological memory (Huang et al., 2020; Kellam and Barclay, 2020). For endemic coronaviruses, it has been demonstrated that antibody levels substantially decrease 6 months after the infection and that reinfections frequently occur 12–36 months following the initial challenge. However, usually, existing partial immunity protects from a more severe course after reinfection (Kissler et al., 2020; Edridge et al., 2020). These data indicate that natural infection but also potentially vaccination can provide only short-period protection against reinfection with SARS-CoV-2. Indeed, recent studies have suggested a rapid decline of virus-specific antibody titers in recovered individuals and a slower decay after the initial contraction phase (Seow et al., 2020; Ibarrondo et al., 2020; Wu et al., 2021). However, although antibody levels may decay, memory B and T cells, as well as plasmablasts, are often maintained following viral infections. Memory B cells specific for the SARS-CoV-2 spike (S) protein were detected in recovered individuals and associated with neutralization of the virus, indicating their protective value (Kreer et al., 2020; Juno et al., 2020; Vaisman-Mentesh et al., 2020). A recent study demonstrated that, unlike antibodies, levels of S-specific memory B cells remain stable during the first 5 months after the infection (Vaisman-Mentesh et al., 2020). If these findings can be extrapolated to a longer period, memory B cells might confer long-lived immunity against SARS-CoV-2. It is therefore of great importance to elucidate the factors underlying the formation of robust memory B cell response against SARS-CoV-2.
It is well established that for the production of high-affinity antibodies capable of binding protein antigens, B cells require helper signals from cognate CD4+ T cells. The presence of virus-specific fully functional CD4+ T cells could therefore be a decisive factor for long-lived immunity. Numerous studies have identified and characterized SARS-CoV-2-specific CD4+ T cells in severely ill individuals and individuals recovered from mild coronavirus disease 2019 (COVID-19) (Grifoni et al., 2020; Juno et al., 2020; Le Bert et al., 2020; Sekine et al., 2020; Weiskopf et al., 2020). The frequency of these cells has been positively associated with the levels of S-specific antibodies in plasma (Grifoni et al., 2020). Moreover, circulating T follicular helper cells specific for the S protein were shown to correlate with the frequency of cognate memory B cells (Juno et al., 2020). Collectively, these findings confirm the dependence of B cell responses on CD4+ T cells during COVID-19. Nevertheless, little is known about the specificity and functions of CD4+ T cells that support the formation of long-lived S-specific B cell memory in SARS-CoV-2-infected individuals. Here we analyzed the properties of S1-specific memory B cells together with S-, N-, and M-specific CD4+ T cell responses in two different disease settings: severe COVID-19 and recovery after mild infection. Furthermore, we correlate CD4+ T cell functions, crucial for the development of B cells, with the quantity and quality of B cell memory.
Results
Subjects recovered from a mild SARS-CoV-2 infection demonstrate lower frequency but better quality of S-specific memory B cells compared with severely ill individuals
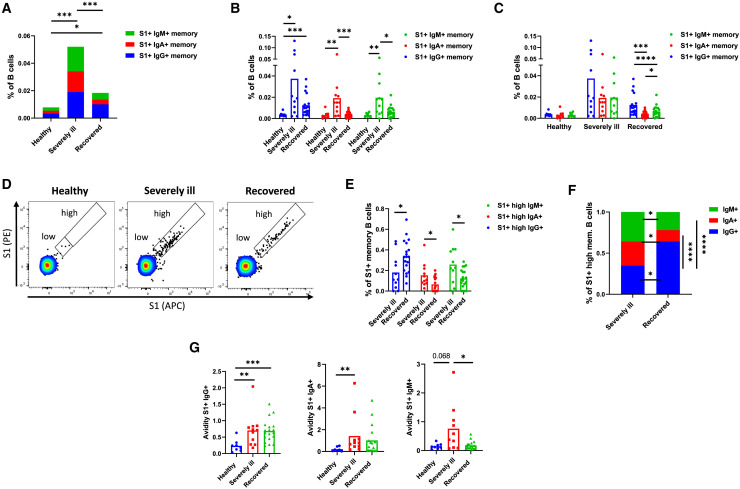
Current data indicate that antibody responses to SARS-CoV-2 infection decay more rapidly in comparison with other viral diseases (Huang et al., 2020; Kellam and Barclay, 2020). In contrast, SARS-CoV-2-specific memory B cells appear to be more persistent and may mediate longer immunity (Vaisman-Mentesh et al., 2020). It is therefore important to investigate memory B cells of SARS-CoV-2-infected individuals and elucidate the factors pivotal for their generation. To address this, we investigated memory B cell responses in peripheral blood of three groups of individuals: those who recovered from mild to moderate symptomatic COVID-19 (referred to as recovered individuals, n = 19), individuals who were hospitalized at the intensive care unit (ICU) as a result of severe COVID-19 (referred to as severely ill individuals, n = 13), and healthy (n = 8) subjects. Demographic data regarding individuals included in our study can be found in Table S1. In particular, we focused on the B cells specific for the S1 subunit of the S protein, which harbors the receptor binding site and might, therefore, be the primary target of neutralizing antibodies. We first compared the frequency of S1-specific memory B cell responses and observed higher levels in individuals with severe COVID-19 (0.076% ± 0.047% of B cells) compared with recovered individuals (0.023% ± 0.014% of B cells), suggesting an augmented B cell response in severely ill patients (p = 0.0005) (Figure 1 A). Considering B cell receptor (BCR) isotype, IgA+ and IgM+ S1-specific memory B cells were more frequent in severely ill individuals. The frequency of IgG+ S1-specific memory B cells was not significantly different between the groups (Figure 1B). Interestingly, IgG was the most frequent isotype among the S1-specific memory B cells of recovered individuals, but not in individuals with severe COVID-19 and healthy individuals (Figure 1C). These findings suggest that individuals who recovered from mild COVID-19 generally develop fewer S1-specific memory B cells that, unlike those of severely ill individuals, predominantly express IgG on their surface.
Figure 1.
S1-specific memory B cells of healthy, severely ill, and recovered individuals
(A) Shown is the mean frequency of memory B cells binding recombinant S1 protein as a percentage of all B cells. Contributions of cells positive for IgM, IgA, and IgG are shown as stacked columns.
(B) Comparison of S1-specific memory B cell levels between the healthy, recovered, and severely ill individuals.
(C) Comparison of BCR-isotype prevalence among the S1-specific memory B cells in three different groups of individuals.
(D) Representative flow cytometry plots demonstrating the capacity of memory B cells to bind the S1 protein.
(E) Comparison of S1-specific memory B cell frequencies binding high amounts of the S1 protein between the recovered and severely ill individuals.
(F) Comparison of BCR-isotype prevalence among the S1-specific memory B cells binding high amounts of the S1 protein.
(G) Avidity of the IgG+, IgA+, or IgM+ on the surface of S1-specific memory B cells. Shown are comparisons for all three groups of individuals. The number of independent experiments represented in the above graphs is n = 8 for healthy, n = 11 for severely ill, and n = 19 for the recovered group. Data are presented as mean ± SD. Differences between the groups were assessed using the Mann-Whitney test or Wilcoxon test for matched data. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Furthermore, we observed that S1-specific memory B cells of healthy individuals, which are most likely cross-reactive memory B cells, predominantly bind low amounts of the S1-probe. Based on the binding capacity of these cells, we defined a cutoff between the B cell populations binding low and high amounts of the S1 protein (Figure 1D). We then compared the frequency of memory B cells binding high amounts of S1 protein between the severely ill and recovered individuals. The data showed that, within the S1+ memory B cell population, recovered individuals have more IgG+ but fewer IgA+ and IgM+ memory B cells binding high amounts of the S1 protein (Figure 1E). This might be because of the differences in the BCR isotype usage, because we observed that, in recovered individuals, memory B cells binding high levels of S1 protein mostly bear IgG BCR. In contrast, all three isotypes were equally represented among the cells of severely ill individuals (Figure 1F). Because IgG is generally considered superior to IgA and IgM, this indicates that recovered individuals might have functionally superior S-specific memory B cells.
We also compared the avidity of BCR present on the surface of S1-specific memory B cells. The avidity was assessed by normalizing the mean fluorescence intensity of bound S1-probe with the level of BCR expression as described previously (Chao et al., 2006; Pape et al., 2018; Smith et al., 2018). Although in the case of IgG and IgA we observed no differences between the severely ill and recovered groups, the avidity of IgM was significantly increased in the severely ill group (Figure 1G).
Collectively, our findings suggest that severely ill SARS-CoV-2-infected individuals generate higher levels of S-specific memory B cells than individuals recovered from mild COVID-19. However, S1-specific memory B cells of recovered individuals have a higher proportion of IgG+ cells with a high binding capacity for the S1 protein.
SARS-CoV-2 membrane (M) protein evokes superior T cell response when compared with S and nucleocapsid (N) proteins
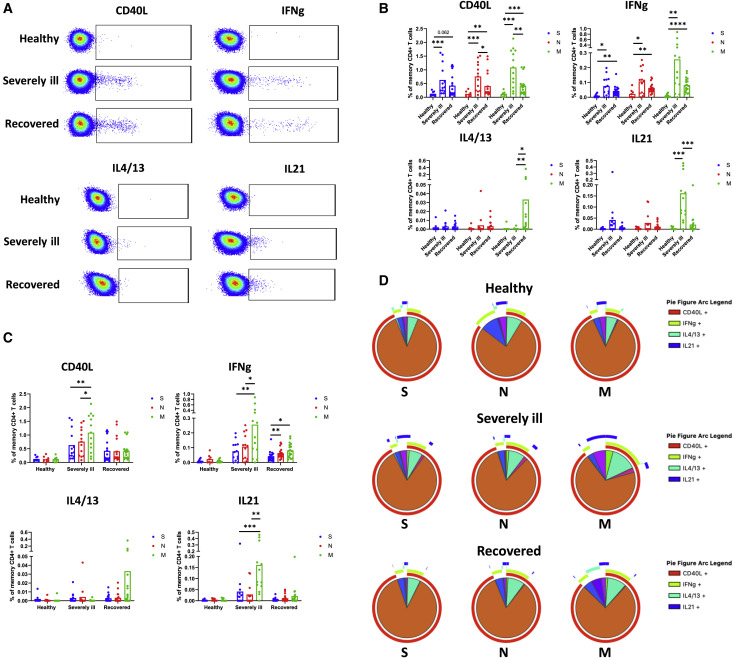
The formation of an efficient antiviral B cell response largely depends on CD4+ T cell help that is delivered in the form of surface signaling molecules and cytokines. The helper capabilities of SARS-CoV-2-specific CD4+ T cells, however, have not been fully described. We, therefore, sought to identify the functions and specificities of CD4+ T cells that are involved in the formation of memory B cell population targeting SARS-CoV-2 S protein. We stimulated peripheral blood mononuclear cells (PBMCs) of SARS-CoV2-infected individuals with pools of overlapping peptides spanning the entire amino acid sequences of the S, M, and N proteins. Four different helper functions were monitored, including the expression of CD40L, IFNγ, IL-4/13, and IL-21 (Figure 2 A). We found that severely ill individuals (0.76% ± 0.5% and 1.09% ± 0.67%, respectively) had a higher frequency of CD40L-expressing CD4+ T cells compared with recovered individuals (0.41% ± 0.43% and 0.42% ± 0.33%, respectively) when stimulated with N or M peptide pools (p = 0.019 and p = 0.0049, respectively). For the S protein, however, the difference between severely ill (0.63% ± 0.54%) and recovered (0.42% ± 0.42%) individuals was not statistically significant (p = 0.11). Similarly, for IFNγ response, severely ill individuals showed the highest mean frequencies of IFNγ-secreting CD4+ T cells specific for S, N, and M proteins (0.076% ± 0.065%, 0.107% ± 0.087%, and 0.256% ± 0.262%, respectively), but the differences were not statistically significant when compared with recovered individuals (0.043% ± 0.036%, 0.063% ± 0.032%, and 0.082% ± 0.048%, respectively) (p = 0.21, p = 0.38, and p = 0.068, respectively). The frequency of IL-4/13-expressing cells was generally low regardless of disease severity and stimulation. However, recovered individuals (0.033% ± 0.09%) showed significantly higher IL-4/13 response than severely ill individuals (0.0004% ± 0.001%) when stimulated with M protein (p = 0.0081). Also, in the case of CD4+ T cells expressing IL-21, no significant differences were observed between the groups for S and N proteins. Conversely, severely ill individuals (0.16% ± 0.15%) showed a good response against the M protein that was significantly higher compared with the recovered individuals (0.021% ± 0.045%) (p = 0.0002) (Figure 2B).
Figure 2.
SARS-CoV-2-specific CD4+ T cell responses of healthy, severely ill, and recovered individuals
(A) Representative flow cytometry plots showing the peptide-induced expression of CD40L, IFNγ, IL-4/13, and IL-21 in memory CD4+ T cells for each group of individuals.
(B) Compared are the frequencies of CD4+ T cells specific for S, N, and M proteins between healthy, severely ill, and recovered individuals. Each of the graphs represents one of the four CD4+ T cell helper functions investigated (expression of CD40L, IFNγ, IL-4/13, and IL-21).
(C) Comparison of CD4+ T cell frequencies responding to S-, N-, and M-based peptide pools.
(D) SPICE analysis of SARS-CoV-2-specific CD4+ T cell responses. Each pie chart depicts the prevalence of polyfunctional subsets within a certain group of individuals for a particular protein stimulus. Arcs surrounding the pie charts show the relative contribution of each of the four investigated CD4+ T cell functions. The number of independent experiments represented in the above graphs is n = 8 for healthy, n = 13 for severely ill, and n = 18 for the recovered group. Data are presented as mean ± SD. Differences between the groups were assessed using the Mann-Whitney test or Wilcoxon test for matched data. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Comparing the specificity of SARS-CoV-2-specific CD4+ T cells differences between the S, N, and M protein mostly became obvious in the case of severely ill individuals. Here we observed significantly increased frequencies of memory CD4+ T cells expressing CD40L, IFNγ, or IL-21 in response to stimulation with the M protein when compared with S (p = 0.0012, p = 0.0068, and p = 0.001, respectively) or N proteins (p = 0.017, p = 0.04, and p = 0.0024, respectively). Furthermore, recovered individuals showed higher frequencies of IFNγ-expressing cells when stimulated with M (p = 0.011) or N (p = 0.004) protein in comparison with S protein, demonstrating relatively low immunogenicity of the latter (Figure 2C).
We next analyzed the polyfunctional profile of SARS-CoV-2-specific CD4+ T cell responses using the SPICE software (Roederer et al., 2011). The majority of SARS-CoV-2-specific CD4+ T cells expressed only CD40L, followed by cells co-expressing CD40L and IFNγ and those expressing exclusively IFNγ. IL-21 or IL-4/13 responses were less frequent. This trend was observed within all subject groups and simulations. Importantly, stimulation of infected individuals with M protein led to a more diverse functional profile with a higher proportion of cells that do not respond solely with CD40L expression (Figure 2D).
Taken together, our findings suggest that, similar to B cell response, the magnitude of SARS-CoV-2-specific CD4+ T cell response increases with the severity of the disease. Moreover, the response to M protein was higher in magnitude and more functionally diverse as compared with S and N proteins.
IL-21-secreting memory CD4+ T cells specific for the M protein are associated with robust S-specific B cell memory in COVID-19 recovered individuals
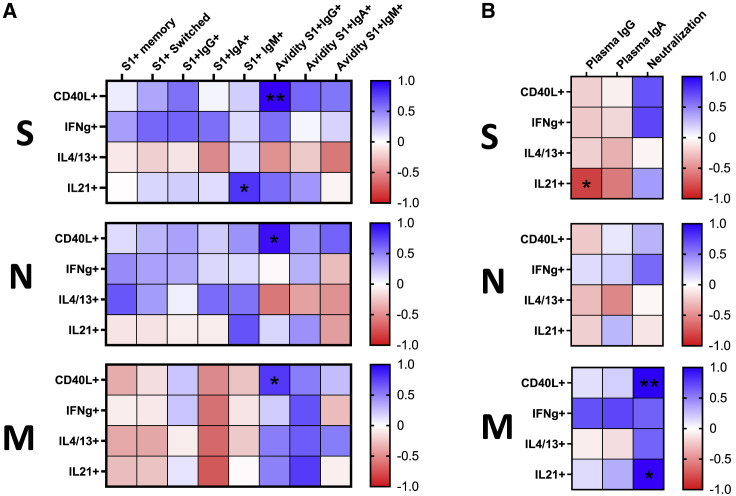
To determine the properties of memory CD4+ T cells governing the quantity and quality of S-specific memory B cells in individuals recovered from mild COVID-19, we compared frequencies of S-, N-, and M-specific memory CD4+ T cells exerting different effector functions with parameters defining quality and quantity of the B cell response, including frequencies of S1-specific bulk, class-switched, IgG+, IgA+, and unswitched memory B cells, along with the BCR avidity of the latter three subsets. Surprisingly, our analysis showed an overall negative relationship between the robustness of S1-specific B cell response and the functions of S-specific CD4+ T cells. The strongest negative association showed cells expressing CD40L and IFNγ that significantly correlated with the frequency of S1+IgA+ memory B cells (r = −0.51, p = 0.031) and avidity of S1-specific IgG-isotype BCR (r = −0.57, p = 0.014), respectively. A similar negative trend was observed for most of the functions of N-specific CD4+ T cells, although none of the correlations showed statistical significance. Conversely, CD4+ T cells recognizing the M protein were positively associated with the quantity and quality of S1-specific memory B cells. In particular, IL-21-secreting cells were significantly correlated with the frequency of bulk (r = 0.56, p = 0.015), as well as IgG (r = 0.52, p = 0.027) and IgA (r = 0.49, p = 0.040) class-switched S1+ memory B cells. Furthermore, this subset was associated with high avidity of IgG-isotype BCR on S1+ memory B cells (r = 0.65, p = 0.0032), suggesting its prominent role in affinity maturation (Figure 3 A). To fully understand the functional profile of memory CD4+ T cells that might be involved in the formation of SARS-CoV-2 S-specific B cell response, we correlated the frequencies of polyfunctional CD4+ T cell subsets expressing different combinations of CD40L, IFNγ, IL-4/13, and IL-21 with the B cell parameters. In concordance with the rest of the data, the strongest association showed memory CD4+ T cells specific for the M protein that either expressed IL-21 alone or in combination with CD40L and IFNγ (Figure S1A).
Figure 3.
Associations between the SARS-CoV-2-specific B cell memory and CD4+ T cell functions in recovered individuals
(A) Heatmaps demonstrate the strength of correlations between the CD4+ T cell functions and parameters determining quantity and quality of S1-specific memory B cell response. Each heatmap shows CD4+ T cell response specific for one of the three proteins. Cells are color-coded concerning Spearman’s correlation coefficient. Significant correlations are marked by asterisks.
(B) Heatmaps depicting correlations between the magnitude of CD4+ T cell helper functions and determinants of humoral response, including levels of S1-specific IgG, IgA, and neutralization capacity. Each heatmap shows CD4+ T cell response specific for one of the three proteins. The number of independent experiments represented in the above graphs is n = 18. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S1.
We next compared the frequency of SARS-CoV-2-specific CD4+ T cells and plasma levels of S1-specific IgA and IgG, along with neutralization capacity. Although we did not observe any significant correlations in recovered individuals, a positive trend was observed between the IFNγ- and IL-4/13-secreting cells specific for the M protein and IgG (r = 0.45, p = 0.061) and IgA (r = 0.45, p = 0.061) levels, respectively (Figure 3B). Further dissecting the CD4+ T cell response into polyfunctional subsets, cells co-expressing CD40L, IFNγ, and IL-21 and cells co-expressing CD40L and IL-21 positively correlated with the plasma IgG levels, while IL4-/13-expressing cells correlated with IgA levels. All subsets were specific for the M protein (Figure S1B).
Taken together, we demonstrated that the frequency of M-specific memory CD4+ T cells expressing IL-21 alone or in combination with IFNγ and CD40L correlates with the frequency and BCR avidity of S1-specific B cell memory.
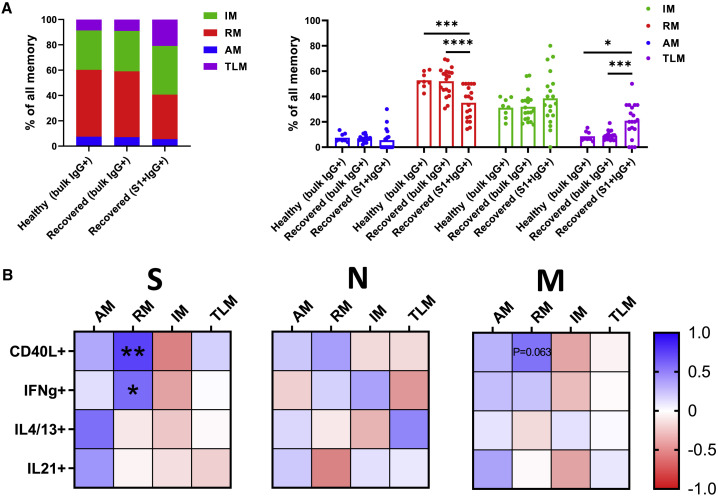
S-specific memory B cells of recovered individuals show an increased proportion of tissue-like memory phenotype together with a reduced frequency of resting memory cells
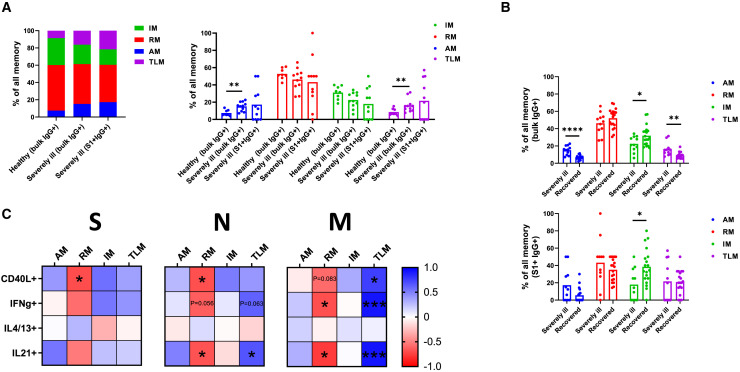
To determine the phenotypic characteristics of memory B cells in SARS-CoV2 infection, we next assessed the phenotypic composition of IgG+ memory B cells in peripheral blood of recovered individuals. The distinctions between activated memory (AM), resting memory (RM), tissue-like memory (TLM), and intermediate memory (IM) were made based on the expression of CD21 and CD27 (AM, CD27+CD21−; RM, CD27+CD21+; TLM, CD27−CD21−; IM, CD27−CD21+). Although bulk IgG+ memory B cells of individuals recovered from mild COVID-19 showed the same memory subset composition as those of healthy individuals, the composition of IgG+ S1-specific cells was significantly altered. In particular, the proportion of RM was decreased among IgG+ S1-specific cells (35% ± 13%) when compared with bulk IgG+ memory B cells of either healthy (53% ± 6%) or recovered individuals (52% ± 11%) (p = 0.0002 and p < 0.0001, respectively). Furthermore, the data revealed a significant increase in the proportion of TLM comparing IgG+ S1-specific cells (21% ± 14%) with bulk IgG+ memory B cells of healthy (9% ± 4%) and recovered individuals (9% ± 4%) (p = 0.019 and p = 0.0003, respectively) (Figure 4 A). These findings suggest an increased proportion of SARS-CoV-2-specific IgG+ memory B cells with activated and exhausted phenotype.
Figure 4.
Memory phenotype of B cells in recovered individuals
(A) Graphs demonstrate proportions of memory subsets within the bulk and S1-specific memory B cell populations as compared with the healthy control. The number of independent experiments is n = 8 for the healthy group and n = 18 for the recovered group. Data are presented as mean ± SD. Differences between the groups were assessed using the Mann-Whitney test or Wilcoxon test for matched data.
(B) Correlative relationships show the impact of CD4+ T cell functions on the memory phenotype of S1+ memory B cells. Each heatmap shows CD4+ T cell response specific for one of the three proteins. Cells are color-coded concerning Spearman’s correlation coefficient. Significant correlations are marked by asterisks. The number of independent experiments is n = 18. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Given the possible role of CD4+ T cell helper functions in sustaining S1-specific memory B cells, we next assessed whether they also have an impact on memory phenotype. Indeed, we observed a significant positive association between the S-specific CD4+ T cells expressing CD40L (r = 0.61, p = 0.0067) or IFNγ (r = 0.47, p = 0.047) and the proportion of RM. A similar trend was observed for the cells expressing CD40L in response to stimulation with the M peptide pool (r = 0.45, p = 0.063) (Figure 4B). Considering the co-expression of molecules providing help to B cells, we observed that numerous subsets specific for S and M proteins are associated with the phenotype of memory B cells. In general, the frequency of IM was negatively correlated, while the frequencies of AM and RM were positively associated with subsets expressing different combinations of CD40L, IFNγ, IL4/13, and IL-21 (Figure S1C).
Collectively, these data suggest a decreased proportion of long-lived highly functional RM phenotype among the IgG+ S1-specific memory B cells of recovered individuals. Although the factors driving skewing of the S1-specific memory B cell compartment remain unknown, S- and M-specific CD4+ T cells expressing different combinations of CD40L, IFNγ, IL4/13, and IL-21 were associated with a low proportion of IM in favor of the AM and RM.
CD40L-expressing CD4+ T cells are associated with affinity maturation of S-specific memory B cells in severely ill individuals
As we elucidated the association between the SARS-CoV-2-specific CD4+ T cell and B cell memory in recovered individuals, we next evaluated this relationship in severely ill individuals. Comparing the same CD4+ T cell functions and B cell parameters, we observed a significant positive association between the frequency of CD40L-expressing CD4+ T cells and the avidity of IgG on the surface of S1+ memory B cells. Interestingly, this was the case for S-specific (r = 0.83, p = 0.0047), N-specific (r = 0.77, p = 0.013), and M-specific CD4+ T cells (r = 0.65, p = 0.049). Moreover, we found a positive correlation between the proportion of IL-21-secreting CD4+ T cells responding to stimulation with M protein and the frequency of S1-specific IgM+ memory B cells (r = 0.66, p = 0.043) (Figure 5 A). Importantly, the latter had a high binding capacity for the S1 protein during severe COVID-19. We also investigated the CD4+ T cell subsets co-expressing different combinations of CD40L, IFNγ, IL-4/13, and IL-21 and their possible role in providing help to S-specific B cells. Notably, memory CD4+ T cells expressing only CD40L correlated with the avidity of IgG on the surface of S1-specific memory B cells regardless of their specificity. Moreover, the subset co-expressing CD40L, IFNγ, and IL-21 and the subsets co-expressing CD40L and IL-21 positively correlated with the avidity of IgG BCR. Both subsets were specific for the S protein (Figure S2A).
Figure 5.
Associations between the SARS-CoV-2-specific B cell memory and CD4+ T cell functions in severely ill ICU patients
(A) Heatmaps demonstrate the strength of correlations between the CD4+ T cell functions and parameters determining quantity and quality of S1-specific memory B cell response. Each heatmap shows CD4+ T cell response specific for one of the three proteins. Cells are color-coded concerning Spearman’s correlation coefficient. Significant correlations are marked by asterisks.
(B) Heatmaps illustrate correlations between the magnitude of CD4+ T cell helper functions and factors describing humoral response (levels of S1-specific IgG, IgA, and neutralization capacity). Each heatmap shows CD4+ T cell response specific for one of the three proteins. The number of independent experiments represented in the above graphs is n = 11. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S2.
Furthermore, we investigated the influence of CD4+ T cell help on plasma IgG and IgA levels and neutralization capacity. Our findings suggest a negative correlation between the frequency of IL-21-secreting CD4+ T cells specific for the S protein and IgG levels (r = −0.71, p = 0.029). Moreover, CD40L- (r = 0.92, p = 0.0036) and IL-21-expressing (r = 0.83, p = 0.017), M-specific CD4+ T cells positively correlated with the level of plasma neutralization (Figure 5B). Among the polyfunctional subsets, CD4+ T cells expressing exclusively IFNγ, in response to any of the three proteins, showed a strong association with the plasma neutralization activity. Two further subsets that correlated with neutralization were M-specific IFNγ- and IL-21-producing cells and CD40L-only-expressing cells (Figure S2B).
Taken together, we demonstrated that CD40L-expressing CD4+ T cells are the best correlate of the high-avidity IgG found on S1-specific memory B cells. Importantly, the plasma neutralization capacity of severely ill individuals was positively associated with CD40L, IFNγ, and IL-21 production in CD4+ T cells.
CD4+ T cells with helper functions are associated with activation and exhaustion of S-specific memory B cells in severely ill individuals
Similarly, as for recovered individuals, we assessed the subset composition of IgG+ memory B cells in severely ill individuals. Strikingly, we found a severely altered phenotype of both bulk IgG+ memory B cells and S1-specific IgG+ memory B cells. Comparing the bulk IgG+ memory B cells, we observed a significant increase of AM (15% ± 5%) in severely ill individuals when compared with healthy (7% ± 3%) individuals (p = 0.0036). Moreover, the proportion of TLM was higher in severely ill (16% ± 8%) than healthy (9% ± 4%) subjects (p = 0.0073), pointing at a severe immune activation ongoing in these patients. In the case of S1-specific IgG+ memory B cells of severely ill individuals, however, the same trend was observed, but the differences were not significant due to the large variability between individuals (Figure 6 A). The high variance can be explained by the low number of detected S1-specific cells in some cases.
Figure 6.
Memory phenotype of B cells in severely ill ICU patients
(A) Graphs demonstrate proportions of memory subsets within bulk and S1-specific memory B cell populations in comparison with the healthy control.
(B) Compared are the frequencies of memory B cell subsets between the severely ill and recovered individuals for bulk IgG+ and S1+ IgG+ memory B cell populations. The number of independent experiments is n = 8 for the healthy group, n = 11 for the severely ill group, and n = 18 for the recovered group. Data are presented as mean ± SD. Differences between the groups were assessed using the Mann-Whitney test or Wilcoxon test for matched data.
(C) Correlation maps show associations between CD4+ T cell functions and memory phenotype of S1+ memory B cells. Each map shows the CD4+ T cell response specific for one of the three proteins. Cells are color-coded concerning Spearman’s correlation coefficient. Significant correlations are marked by asterisks. The number of independent experiments is n = 11. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Comparing the prevalence of memory B cell subsets between the severely ill and recovered individuals, we observed significant differences at the level of bulk IgG+ memory B cells. In particular, the proportion of AM (p < 0.0001) and TLM (p = 0.0013) was significantly higher in severely ill individuals, while the IM (p = 0.029) subset was more frequent in recovered individuals. For the S1-specific IgG+ memory B cells, we did not observe any significant differences between the groups with the exception of IM (p = 0.016) frequency that was higher in recovered individuals (Figure 6B).
We next evaluated the impact of SARS-CoV-2-specific CD4+ T cells on the memory phenotype of IgG+ S1+ memory B cells in severely ill individuals. We found that the proportion of RM phenotype inversely correlated with the frequencies of CD40L-expressing CD4+ T cells specific for S (r = −0.71, p = 0.028) or N (r = −0.69, p = 0.031) protein, IFNγ-secreting CD4+ T cells specific for the M protein (r = −0.69, p = 0.031), and IL-21-secreting CD4+ T cells specific for N (r = −0.68, p = 0.037) or M (r = −0.77, p = 0.013) protein. The same trend was observed in the case of M-specific CD40L-expressing CD4+ T cells (r = −0.58, p = 0.083) and N-specific IFNγ-secreting CD4+ T cells (r = −0.63, p = 0.056). Furthermore, we observed a positive correlation between the proportion of TLM and frequency of CD4+ T cells expressing CD40L in response to stimulation with M protein (r = 0.75, p = 0.017), IFNγ in response to stimulation with the M protein (r = 0.92, p = 0.00056), and IL-21 in response to stimulation with M (r = 0.92, p = 0.00056) or N (r = 0.67, p = 0.047) protein, indicating the involvement of these subsets in B cell activation and exhaustion. A positive trend was also observed between the frequencies of TLM and IFNγ-secreting CD4+ T cells specific for the N protein (r = 0.62, p = 0.063) (Figure 6C). Considering the co-expression of molecules mediating help to B cells, the proportion of TLM was strongly associated with the following memory CD4+ T cells: subset co-expressing CD40L, IFNγ, and IL-21; subset expressing CD40L and IFNγ; subset expressing CD40L and IL-21; and subset expressing only IFNγ. The same polyfunctional CD4+ T cells negatively correlated with the proportion of RM phenotype. These correlations were the most profound in the case of memory CD4+ T cells specific for the M protein, although the same trend was also observed for S and N proteins.
Collectively, these findings suggest skewing of the memory B cell compartment in severely ill SARS-CoV-2-infected individuals toward activated and exhausted phenotypes. This activation can be potentially explained by high frequencies of SARS-CoV-2-specific CD4+ T cells expressing stimulatory proteins, such as CD40L, IFNγ, and IL-21.
Discussion
Seasonal coronavirus infections usually evoke short-lived immunological memory responses, and reinfection can therefore reoccur as soon as 6 months after initial infection (Kissler et al., 2020; Edridge et al., 2020). The same may be true for infection with the novel SARS-CoV-2 because recent studies suggested accelerated waning of antibody responses following COVID-19 (Ibarrondo et al., 2020; Seow et al., 2020). Conversely, SARS-CoV-2-specific memory B cell levels seem to remain constant at least during the first 5 months following the onset of the disease (Vaisman-Mentesh et al., 2020). This makes memory B cells of great interest because they could be the critical component of long-lived immunity against SARS-CoV-2 infection. In our present study, we investigated the possible impact of CD4+ T cell functions on the formation of S-specific B cell memory in individuals recovered from mild SARS-CoV-2 infection and severely ill individuals. We show that in recovered individuals, IL-21 expression is the major function associated with a robust B cell memory. Conversely, in severely ill individuals, CD40L expression was positively correlated with the avidity of IgG but at the same time contributed to activation and exhaustion of the memory B cells.
For long-term protection from SARS-CoV-2 infection, virus-specific memory B cells may be of greater importance than the antibodies whose levels decline rapidly after infection. In particular, memory B cells targeting receptor-binding S protein might be a decisive factor for long-term protection. Several studies have detected S-specific memory B cells in recovered individuals (Kreer et al., 2020; Juno et al., 2020; Vaisman-Mentesh et al., 2020). In line with their findings, we demonstrated the presence of S1+ memory B cells in individuals recovered from mild COVID-19, as well as severely ill ICU patients. Although severely ill individuals developed significantly higher levels of S1-specific memory B cells, recovered individuals showed a higher proportion of memory B cells with IgG BCR that are generally considered superior to IgM or IgA. Moreover, S1-specific memory B cells of recovered individuals harbored a higher proportion of IgG+ cells able to bind high amounts of S1 protein. In the case of severely ill individuals, however, there was a higher proportion of IgA+ and IgM+ memory B cells with increased S1-binding capacity. Overall, these findings suggest that severely ill individuals develop higher levels of S-specific memory B cells than individuals recovered from mild COVID-19 but lack the class-switching to IgG. It is, however, important to consider that differential decay of the specific memory B cells over time could contribute to the observed differences. The extent of this contribution, however, might not be significant because it has been shown that SARS-CoV-2-S-specific memory B cells remain stable at least for the 6 months post-recovery (Vaisman-Mentesh et al., 2020).
One of the main factors driving B cell and humoral responses against viral proteins is CD4+ T cell help. Recent reports confirmed the presence of virus-specific CD4+ T cells in SARS-CoV-2-infected individuals (Grifoni et al., 2020; Juno et al., 2020; Le Bert et al., 2020; Weiskopf et al., 2020; Oja et al., 2020). Although cells reactive against the entire proteome have been identified, the immunodominant regions are those encoding for the S, N, and M proteins (Grifoni et al., 2020; Oja et al., 2020). We next investigated functions of S-, N-, and M-specific CD4+ T cells that are important for B cell help. From the currently available literature, it is not clear whether the magnitude of SARS-CoV-2-specific CD4+ T cell response is higher in severely ill individuals or those recovered from mild disease (Oja et al., 2020; Sekine et al., 2020). Similar to a previous study (Sekine et al., 2020), we have demonstrated that the highest magnitude of SARS-CoV-2-specific CD4+ T cells was observed in severely ill individuals with an exception of IL-4/13-secreting cells that were more frequent in recovered individuals. The frequency of CD4+ T cell response also largely depended on their specificity. In most cases, the highest response was specific for the M protein as compared with S and N. The same trend has been previously observed for the SARS-CoV-2-specific IFNγ response (Sekine et al., 2020). Moreover, the functional composition of M-specific CD4+ T cells showed to be more diverse.
To understand the importance of SARS-CoV-2-specific memory CD4+ T cells in the formation of S1-specific B cell memory, we compared the magnitude of T cell functions and B cell parameters describing the quantity and quality of the response. We demonstrated that in recovered individuals, IL-21-secreting CD4+ T cells specific for the M, but not N or S, protein are strongly associated with the generation of class-switched S1+ memory B cells and increase the affinity of their BCR. In severely ill individuals, however, the affinity maturation of S1+ memory B cells was associated with CD40L-expressing CD4+ T cells specific for any of the three proteins. Furthermore, we investigated the influence of CD4+ T cell functions on the development of S1-specific humoral response. We observed positive correlations between the M-specific polyfunctional CD4+ T cells and plasma IgG and IgA levels. Similarly, previous studies have shown a positive correlation between the magnitudes of CD4+ T cells and plasma IgG and IgA specific for the virus in recovered individuals (Grifoni et al., 2020; Oja et al., 2020). In severely ill patients, the subset associated with high avidity of BCR also positively correlated with the neutralization capacity of respective plasma, suggesting profound importance of CD40L-expressing CD4+ T cells in severe COVID-19. The plasma neutralization capacity of severely ill individuals was also positively associated with IFNγ and IL-21 production in CD4+ T cells.
An important indicator of quality and durability of the memory B cells is their phenotypic traits dividing them into four memory subsets: RM, AM, IM, and TLM. In the peripheral blood of healthy individuals, RM is the prevalent subset; however, during several viral infections its prevalence decreases in favor of atypical subsets like AM and TLM (Oliviero et al., 2015; Burton et al., 2018; Kardava et al., 2014). In particular, the TLM subset showed aberrant properties in settings of HIV infection, like an increased expression of exhaustion and activation markers and a decreased rate of somatic hypermutation (SHM) (Kardava et al., 2014). Similar to the previous study (Oliviero et al., 2020), we found that the individuals who recovered from mild COVID-19 have the same composition of bulk memory B cell compartment as healthy donors. The subset composition of S1-specific memory B cells, however, was significantly altered. We observed a decrease in the frequency of RM and an expansion of the TLM subset. The memory subset composition of the S1-specific B cells was potentially influenced by S- and M-specific CD4+ T cells expressing different combinations of CD40L, IFNγ, and IL-21. In severely ill individuals, the skewing of the B cell compartment was more severe and could be observed for the bulk memory B cell population. In particular, the frequencies of AM and TLM subsets were significantly increased compared with healthy and recovered individuals, while the frequency of IM decreased. These changes were even more profound in the S1-specific memory B cell population but were overall not significant due to the high variability among the individuals. The reason for this variability was the poor condition of some severely ill individuals whose PBMCs consequently had unusually low viability. In these cases, we were not able to detect many viable IgG+ S1-specific B cells causing a high deviation in the memory subset distribution. Of note, the frequency of CD4+ T cells expressing CD40L, IFNγ, and IL-21 positively correlated with S1-specific TLM and inversely with RM, suggesting that high levels of functional CD4+ T cells lead to aberrant activation and exhaustion of memory B cells in severely ill individuals.
Taken together, our data provide a detailed insight into the composition and formation of S1-specific memory B cells in two distinct courses of COVID-19: severe disease and mild cold-like infection. We show that in individuals recovered from mild infection, IL-21 secretion is the primary CD4+ T cell function associated with the quantity and quality of S1-specific memory B cells. In severely ill individuals, however, this role is taken over by the CD40L-expressing CD4+ T cells. Intriguingly, CD4+ T cells specific for the M protein better correlated with the S-specific B cell response compared with those specific for S and N proteins. We believe that our findings provide valuable information for the design of a SARS-CoV-2 vaccine that would provide long-term immune protection. In particular, current vaccine designs could be modified in a way that would engender IL-21-producing CD4+ T cells targeting the M protein to elicit memory B cells with high-avidity BCR. However, further studies are needed that would follow the development of CD4+ T cell responses and S-specific memory B cells over time to assess their longevity.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD3-BV510 (clone UCHT1) | Biolegend | Cat# 300448, RRID: AB_2563468 |
| Mouse anti-human CD19-APC-Cy7 (clone HIB19) | Biolegend | Cat# 302218, RRID: AB_314248 |
| Mouse anti-human CD21-PE-Cy7 (clone Bu32) | Biolegend | Cat# 354912, RRID: AB_2561577 |
| Mouse anti-human CD27-BV605 (clone O323) | Biolegend | Cat# 302830, RRID: AB_2561450 |
| Mouse anti-human IgM-BV785 (clone MHM-88) | Biolegend | Cat# 314544, RRID: AB_2800832 |
| Human anti-human IgA-VioBright 515 (clone REA1014) | Miltenyi Biotec | Cat# 130-116-886, RRID: AB_2727744 |
| Mouse anti-human IgG-BV421 (clone G18-145) | BD | Cat# 562581, RRID: AB_2737665 |
| Mouse anti-human CD3-APC-Cy7 (clone UCHT1) | Biolegend | Cat# 300426, RRID: AB_830755 |
| Mouse anti-human CD4-BV786 (clone SK3) | BD | Cat# 563877, RRID: AB_2738462 |
| Mouse anti-human CD8-BV510 (clone RPA-T8) | Biolegend | Cat# 301048, RRID: AB_2561942 |
| Mouse anti-human CD45RO-BV605 (clone UCHL1) | Biolegend | Cat# 304238, RRID: AB_2562153 |
| Mouse anti-human CD40L-PE-CF594 (clone TRAP1) | BD | Cat# 563589, RRID: AB_2738297 |
| Mouse anti-human CD279-PE-Cy7 (clone EH12.2H7) | Biolegend | Cat# 329918, RRID: AB_2159324 |
| Mouse anti-human CXCR5-FITC (clone J252D4) | Biolegend | Cat# 356912, RRID: AB_2561894 |
| Mouse anti-human IFNγ-PE (clone B27) | Biolegend | Cat# 506507, RRID: AB_315440 |
| Mouse anti-human IL4-BV421 (clone MP4-25D2) | Biolegend | Cat# 500826, RRID: AB_2561679 |
| Mouse anti-human IL13-BV421 (clone JES10-5A2) | Biolegend | Cat# 501916, RRID: AB_2616748 |
| Mouse anti-human AF647 (clone 3A3-N2) | Biolegend | Cat# 513006, RRID: AB_1227661 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (Wuhan strain) | Isolated from patient | N/A |
| Biological samples | ||
| Peripheral blood of Covid-19-recovered individuals | Heinsberg county, NRW, Germany | N/A |
| Peripheral blood of Covid-19-severely ill individuals | University Hospital Bonn, Community Hospital Heinsberg, Germany | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| APC Streptavidin | Biolegend | Cat# 405207 |
| PE Streptavidin | Biolegend | Cat# 405204 |
| Biotinylated SARS-CoV-2 (COVID-19) S1 protein, His, Avitag (MALS verified) | BioCat | Cat# S1N-C82E8-200ug-AC |
| DPBS, no calcium, no magnesium | ThermoFisher Scientific | Cat# 14190094 |
| RPMI 1640 Medium | ThermoFisher Scientific | Cat# 21875034 |
| DMEM, high glucose | ThermoFisher Scientific | Cat# 41965039 |
| MEM Eagle | Pan Biotech | Cat# P03-2710 |
| Sodium azide | Sigma-Aldrich | Cat# S2002-25G, CAS: 26628-22-8 |
| EDTA (0.5 M), pH 8.0, RNase-free | ThermoFisher Scientific | Cat# AM9260G |
| Penicillin-Streptomycin (5,000 U/mL) | ThermoFisher Scientific | Cat# 15070063 |
| L-Glutamine (200 mM) | ThermoFisher Scientific | Cat# 25030081 |
| BD GolgiStop | BD | Cat# 554724 |
| BD GolgiPlug | BD | Cat# 555029 |
| Ionomycin calcium salt | Sigma-Aldrich | Cat# I3909-1ML |
| PMA | Sigma-Aldrich | Cat# P1585-1MG, CAS: 16561-29-8 |
| Dimethyl sulphoxide (DMSO) | Carl Roth | Cat# A994.2 |
| Pancoll human, Density: 1.077 g/ml | Pan Biotech | Cat# P04-60500 |
| Sera Plus, EU approved regions, special processed FBS | Pan Biotech | Cat# P30-3702 |
| Carboxymethylcellulose sodium salt | Sigma-Aldrich | Cat# C5013-1KG |
| OptiPRO SFM | ThermoFisher Scientific | Cat# 12309019 |
| Crystal violet | Carl Roth | Cat# T123.3 |
| Ethanol | Carl Roth | Cat# T171.4 |
| Formaldehyde solution 37% | Carl Roth | Cat# CP10.2 |
| HEPES solution | Sigma-Aldrich | Cat# H0887-20ML |
| Rainbow Fluorescent Particles | Biolegend | Cat# 422905 |
| PepTivator SARS-CoV-2 Prot_S | Miltenyi Biotec | Cat# 130-126-700 |
| PepTivator SARS-CoV-2 Prot_M | Miltenyi Biotec | Cat# 130-126-702 |
| PepTivator SARS-CoV-2 Prot_N | Miltenyi Biotec | Cat# 130-126-698 |
| Critical commercial assays | ||
| Zombie Aqua Fixable Viability Kit | Biolegend | Cat# 423102 |
| FcR Blocking Reagent, human | Miltenyi Biotec | Cat# 130-059-901 |
| CD19 MultiSort Kit, human | Miltenyi Biotec | Cat# 130-055-301 |
| Anti-SARS-CoV-2-ELISA (IgG) | Euroimmun | EI 2606-9601G |
| Anti-SARS-CoV-2-ELISA (IgA) | Euroimmun | EI 2606-9601A |
| Fixation/Permeabilization Solution Kit | BD | Cat# 554714 |
| Experimental models: Cell lines | ||
| VERO C1008 (Vero 76, clone E6, Vero E6) | ATCC | Cat# CRL-1586 |
| Software and algorithms | ||
| FlowJo Software | TreeStar | Version 10.0.7 |
| Prism | GraphPad | Version 9 |
| SPICE | NIH | Version 6.1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hendrik Streeck (hendrik.streeck@ukbonn.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The published article includes all datasets generated during this study. Raw data are available from the lead contact upon request.
Experimental model and subject details
Study participants
Twenty-seven individuals were selected from a cross-sectional study cohort (Streeck et al., 2020). The study was approved by the Ethics Committee of the Medical Faculty of the University of Bonn and informed consent was obtained from all subjects. Nineteen of the twenty-seven enrolled individuals were seropositive for the SARS-CoV-2. These individuals presumably got infected during the super-spreading event while participating in the carnival festivities on 15. February 2020. The samples were taken between the 7. and 9. April 2020 so 52-54 days post-infection. All the individuals were asymptomatic at the time of sampling and reported only mild flu-like symptoms (fever, coughing, loss of taste, loss of smell, or dyspnea) for the time of illness. Eight participants were healthy individuals with no detectable SARS-CoV-2-specific IgA and IgG. Moreover, a cohort of severely ill patients (n = 13) who had been hospitalized with acute respiratory distress syndrome (ARDS) and supported with mechanical ventilation were enrolled. At the time of sampling, all severely ill patients were treated at the intensive care units of the Community Hospital Heinsberg or University Hospital Bonn in Germany. The date of onset and other details about the course of the disease before their hospitalization is not known for these individuals. Further details regarding the study participants are available in Table S1.
Cell lines
Vero E6 cells (VERO C1008, ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), and 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were cultured in a humidified CO2 incubator at 37°C.
Method details
Magnetic isolation of B cells
Cryopreserved PBMC were thawed and rested overnight at 37°C. Next, B cells were isolated, after counting and PBS washing steps, using a magnetic B cell isolation kit following the manufacturer’s protocol (Human CD19 MultiSort Kit, Miltenyi Biotec). In brief, cells were resuspended in MACS buffer (PBS supplemented with 0.5% BSA, and 2nM EDTA) and labeled with anti-CD19 antibody attached to magnetic beads. The cell suspension was then passed through a positive selection column resulting in immobilization of labeled CD19+ cells. The flow-through was collected for the assessment of CD4+ T cell responses. The immobilized fraction was washed out of the column and magnetic beads were enzymatically released from the cells. Isolated bead-free CD19+ cells were used for further experiments.
Assessment of SARS-CoV-2 spike-specific B cells by flow cytometry
Magnetically isolated B cells were resuspended in FACS buffer (PBS supplemented with 2% FCS, 0.05% NaN3, and 2mM EDTA) and incubated with the fluorescently labeled recombinant SARS-CoV-2 S1 protein (Biotinylated SARS-CoV-2 (COVID-19) S1 protein, Acrobiolabs). To minimize the unspecific binding of the probe, S1 protein was conjugated to two different streptavidin-fluorochrome conjugates, streptavidin-PE and streptavidin-APC, in an equimolar ratio. After 15 min at 4°C anti-IgG-BV421 antibody (clone G18-145) was added to the cell suspension and the incubation was continued for another 15 min. Following the binding of S1 probes, cells were washed with PBS and stained for viability (ZombieAqua, Biolegend) 15 min at 4°C. Subsequently, cells were washed with FACS buffer and incubated with a solution of antibodies blocking human Fc receptors (FcR block, Miltenyi Biotec). After 10 min a mixture of fluorescently labeled antibodies binding to surface antigens of B cells was added. The mixture included following fluorescently-labeled antibodies: anti-CD3-BV510 (clone UCHT1), anti-CD27-BV605 (clone O323), anti-IgM-BV785 (clone MHM-88), anti-IgA-VioBright 515 (clone REA1014), anti-CD21-PE-Cy7 (clone Bu32), and anti-CD19-APC-Cy7 (clone HIB19). The staining was performed at 4°C for 15 min. Finally, cells were washed with PBS and acquired on a flow cytometer (BD FACS Celesta). Possible longitudinal fluctuations in laser intensity were monitored every day before the experiment and were compensated using fluorescent beads (Rainbow beads, Biolegend). The avidity of the BCR for S1 protein was assessed based on the MFI of fluorescent S1-probe normalized with the MFI of the corresponding BCR isotype. The data were analyzed with the FlowJo Software version 10.0.7 (TreeStar).
Ex vivo stimulation of CD4+ T cells by peptide pools
Following magnetic isolation of B cells the remaining PBMC were allowed to rest overnight in R10 media (RPMI-1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mM HEPES) at 37°C. The next morning PBMC were plated in 96-wells U bottom plates at a density of 1 × 106 PBMC per well in R10 media. Cells were then stimulated with SARS-CoV-2 PepTivator (Miltenyi Biotec) overlapping peptide pools, spanning the entire sequences of S, N, or M proteins. The final concentration of each peptide was 1 μg/ml for all three pools. Cells were stimulated for 6 hours at 37°C. For negative control cells were incubated with an amount of DMSO equivalent to that present in peptide-stimulated samples. Positive controls were stimulated with PMA (20 ng/ml) and ionomycin (1 μg/ml). One hour after initiation of stimulation Golgi Stop and Golgi Plug (BD Bioscience) were added to inhibit the secretion of cytokines.
Assessment of SARS-CoV-2-specific CD4+ T cell functions by flow cytometry
Stimulated cells were washed with PBS and stained for viability using Zombie Aqua dye (Biolegend) for 20 min at room temperature. After washing with staining buffer (PBS supplemented with 2% FCS), cells were stained extracellularly for 20 min at 4°C with the fluorescently conjugated antibodies: anti-CD8-BV510 (clone RPA-T8; Biolegend) and anti-CD45RO-BV605 (clone UCHL1; Biolegend). Subsequently, cells were washed with staining buffer and incubated in CytoFix/CytoPerm Solution (BD Bioscience) for 20 min at 4°C. Fixed and permeabilized cells were then washed with 1x Perm/Wash Buffer (BD Bioscience) and stained intracellularly with the following fluorescently labeled antibodies: anti-CD3-APC-Cy7 (clone UCHT1; Biolegend), anti-CD4-BV786 (clone SK3; BD Bioscience), anti-IFNγ-PE (clone B27; Biolegend), anti-CD40L-CF594 (clone TRAP1; BD Bioscience), anti-IL4-BV421 (clone MP4-25D2; Biolegend), anti-IL13-BV421 (clone JES10-5A2; Biolegend) and anti-IL21-AF647 (clone 3A3-N2; Biolegend) for 30 min at 4°C. Finally, cells were washed with PBS and acquired on FACS Celesta (BD Bioscience). Possible longitudinal fluctuations in laser intensity were monitored every day before the experiment and were compensated using fluorescent beads (Rainbow beads, Biolegend). The data were analyzed with the FlowJo Software version 10.0.7 (TreeStar). The frequency of peptide-responding cells was calculated by subtracting negative control from the peptide-stimulated sample.
Measurement of SARS-CoV-2 spike-specific plasma IgA, IgG, and neutralization capacity
Cryopreserved plasma samples were thawed and each sample was tested with the Euroimmun IgG and IgA Anti-SARS-CoV-2 ELISA kits as per the manufacturer’s instructions. Briefly, plasma samples were thawed and diluted 1:101 in sample buffer. Subsequently, diluted samples were pipetted into test tubes pre-covered with SARS-CoV-2 spike protein and incubated 60 min at 37°C. Samples were washed three times and subsequently incubated with peroxidase-conjugated anti-IgA or anti-IgG antibody. After the incubation (30 min at 37°C) chromogenic substrate was added and samples were incubated for 30 min at room temperature. Finally, the reaction was stopped by the addition of the stop solution, and absorbance was measured on EUROIMMUN Analyzer I platform. A sample was considered positive if the ratio between the absorbances of the sample and calibrator was above 1.1.
To determine the SARS-CoV-2 neutralization capacity of patient plasma a plaque reduction neutralization test was performed as previously described (Streeck et al., 2020). Briefly, plasma samples were thawed, heat-inactivated for 30 min at 56°C, and centrifuged for 5 min at 5000 rpm. The supernatant was carefully collected and serially two-fold diluted starting with 1: 2 up to 1:16384. Subsequently, 120 μL of each plasma dilution was mixed with 80 plaque-forming units (PFU) of SARS-CoV-2 in 120 μL OptiPRO SFM (GIBCO) cell culture medium. After 1h incubation at 37°C, 200 μL of each dilution mixture were added to wells of a 24 well plate previously seeded with 1.5x105 Vero E6 cells/well. Following 1h incubation at 37°C, the inoculum was removed and cells were overlaid with a 1:1 mixture of 1.5% (w/v) carboxymethylcellulose (Sigma-Aldrich) in 2xMEM (Pan Biotech) supplemented with 4% FBS. After incubation at 37°C and 5% CO2 for three days, the overlay was removed and the 24 well plates were fixed using a 6% formaldehyde solution and stained with 1% crystal violet in 20% ethanol revealing the formation of plaques.
Quantification and statistical analysis
Statistical analysis and graphing of the data were performed using GraphPad Prism and SPICE software. Data are presented as mean ± SD for biological replicates. Differences between the groups were assessed using the Mann-Whitney test or Wilcoxon test for matched data. The strength of correlations was evaluated by Spearman’s test. Correlated parameters are interdependent and were preselected based on our hypothesis and available knowledge on the interplay between B and T cells in viral infections. Therefore, the p values were not adjusted for multiple comparisons. However, we acknowledge that, based on the number of comparisons made, some of these correlations might occur by chance. Statistical significance is indicated by the following annotations: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Statistical tests used to determine the degree of significance and the number of biological replicates shown in each graph can be found in figure legends.
Acknowledgments
We thank volunteers who participated in the Heinsberg study and Heinsberg study team members not directly involved in the current study: Beate M. Kümmerer, Maximilian Baum, Tobias Höller, Christine Fuhrmann, Eva Bartok, Moritz Berger, Lukas Wessendorf, Monika Eschbach-Bludau, Angelika Kellings, Astrid Schwaiger, Martin Coenen, Per Hoffmann, Birgit Stoffel-Wagner, Markus M. Nöthen, Anna-Maria Eis-Hübinger, Martin Exner, Ricarda Maria Schmithausen, and Matthias Schmid. Furthermore, we thank the employees of the Central Diagnostics Laboratory at University Hospital Bonn for the provision of IgG and IgA ELISA measurements.
Author contributions
Conceptualization, J.P. and H.S.; methodology, J.P.; investigation, J.P., E.R., B.S., and R.D.-P.; resources, C.B., and C.P.; writing – original draft, J.P.; writing – review & editing, J.P., H.S., E.R., C.B., C.P., G.H., and G.A.; funding acquisition, H.S. and G.H.; supervision, H.S.
Declaration of interests
The authors declare no competing interests.
Published: June 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109320.
Supplemental information
References
- Burton A.R., Pallett L.J., McCoy L.E., Suveizdyte K., Amin O.E., Swadling L., Alberts E., Davidson B.R., Kennedy P.T., Gill U.S., et al. Circulating and intrahepatic antiviral B cells are defective in hepatitis B. J. Clin. Invest. 2018;128:4588–4603. doi: 10.1172/JCI121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M., Wittrup K.D. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1 doi: 10.1038/nprot.2006.94. 755–68. [DOI] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Kardava L., Moir S., Shah N., Wang W., Wilson R., Buckner C.M., Santich B.H., Kim L.J., Spurlin E.E., Nelson A.K., et al. Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. J. Clin. Invest. 2014;124:3252–3262. doi: 10.1172/JCI74351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020;101:791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Oja A.E., Saris A., Ghandour C.A., Kragten N.A.M., Hogema B.M., Nossent E.J., Heunks L.M.A., Cuvalay S., Slot E., Linty F., et al. Divergent SARS-CoV-2-specific T- and B-cell responses in severe but not mild COVID-19 patients. Eur. J. Immunol. 2020;50:1998–2012. doi: 10.1002/eji.202048908. [DOI] [PubMed] [Google Scholar]
- Oliviero B., Mantovani S., Ludovisi S., Varchetta S., Mele D., Paolucci S., Baldanti F., Mondelli M.U. Skewed B cells in chronic hepatitis C virus infection maintain their ability to respond to virus-induced activation. J. Viral Hepat. 2015;22:391–398. doi: 10.1111/jvh.12336. [DOI] [PubMed] [Google Scholar]
- Oliviero B., Varchetta S., Mele D., Mantovani S., Cerino A., Perotti C.G., Ludovisi S., Mondelli M.U. Expansion of atypical memory B cells is a prominent feature of COVID-19. Cell. Mol. Immunol. 2020;17:1101–1103. doi: 10.1038/s41423-020-00542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape K.A., Maul R.W., Dileepan T., Paustian A.S., Gearhart P.J., Jenkins M.K. Naive B Cells with High-Avidity Germline-Encoded Antigen Receptors Produce Persistent IgM(+) and Transient IgG(+) Memory B Cells. Immunity 48. 2018:1135–1143 e4. doi: 10.1016/j.immuni.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Nozzi J.L., Nason M.C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.J., Hinman R.M., Getahun A., Kim S., Packard T.A., Cambier J.C. Silencing of high-affinity insulin-reactive B lymphocytes by anergy and impact of the NOD genetic background in mice. Diabetologia. 2018;61:2621–2632. doi: 10.1007/s00125-018-4730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck H., Schulte B., Kümmerer B.M., Richter E., Höller T., Fuhrmann C., Bartok E., Dolscheid-Pommerich R., Berger M., Wessendorf L., et al. Infection fatality rate of SARS-CoV2 in a super-spreading event in Germany. Nat. Commun. 2020;11:5829. doi: 10.1038/s41467-020-19509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman-Mentesh A., Dror Y., Tur-Kaspa R., Markovitch D., Kournos T., Dicker D., Wine Y. SARS-CoV-2 specific memory B cells frequency in recovered patient remains stable while antibodies decay over time. medRxiv. 2020 doi: 10.1101/2020.08.23.20179796. [DOI] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2020. WHO coronavirus disease (COVID-19) dashboard.https://covid19.who.int/ [Google Scholar]
- Wu J., Liang B., Chen C., Wang H., Fang Y., Shen S., Yang X., Wang B., Chen L., Chen Q., et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat. Commun. 2021;12:1813. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during this study. Raw data are available from the lead contact upon request.