Graphical abstract
Abbreviations: ABC transporter, ATP-binding cassette transporter; ATP, adenosine-triphosphate; BCRP, breast cancer resistance protein (ABCG2); calcein AM, calcein acetoxymethyl; C@PA, computer-aided pattern analysis; F1–5, pharmacophore features 1–5; IC50, half-maximal inhibition concentration; MDR, multidrug resistance; MRP1, multidrug resistance-associated protein 1 (ABCC1); MOE, molecular operating environment; P-gp, P-glycoprotein (ABCB1); SEM, standard error of the mean; SMILES, simplified molecular input line entry specification; Tc, Tanimotto coefficient
Keywords: Well-studied ABC transporters; ABCB1 (P-gp); ABCC1 (MRP1); ABCG2 (BCRP); Under-studied ABC transporters (e.g., ABCA7); Triple / multitarget / broad-spectrum / promiscuous inhibitor / antagonist; Pan-ABC inhibition / antagonism / blockage (PANABC); Pattern analysis (C@PA); Multitarget fingerprints; Alzheimer's disease (AD); Multidrug resistance (MDR)
Highlights
-
•
Exploratory changes in substructure patterns are well tolerated by C@PA.
-
•
Extended positive substructures support prediction capability.
-
•
Increased biological hit rate of 40% for multitarget pan-ABC transporter inhibition.
-
•
Contribution to major understanding of pattern analysis and multitarget activity.
-
•
Pan-ABC transporter inhibitors as tool for elucidation of multitarget binding site.
Abstract
Computer-aided pattern analysis (C@PA) was recently presented as a powerful tool to predict multitarget ABC transporter inhibitors. The backbone of this computational methodology was the statistical analysis of frequently occurring molecular features amongst a fixed set of reported small-molecules that had been evaluated toward ABCB1, ABCC1, and ABCG2. As a result, negative and positive patterns were elucidated, and secondary positive substructures could be suggested that complemented the multitarget fingerprints. Elevating C@PA to a non-statistical and exploratory level, the concluded secondary positive patterns were extended with potential positive substructures to improve C@PA’s prediction capabilities and to explore its robustness. A small-set compound library of known ABCC1 inhibitors with a known hit rate for triple ABCB1, ABCC1, and ABCG2 inhibition was taken to virtually screen for the extended positive patterns. In total, 846 potential broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors resulted, from which 10 have been purchased and biologically evaluated. Our approach revealed 4 novel multitarget ABCB1, ABCC1, and ABCG2 inhibitors with a biological hit rate of 40%, but with a slightly lower inhibitory power than derived from the original C@PA. This is the very first report about discovering novel broad-spectrum inhibitors against the most prominent ABC transporters by improving C@PA.
1. Introduction
ATP-binding cassette (ABC) transport proteins are ubiquitously present in the human body [1], [2], [3], [4], and hence, promote solute and drug distribution, influencing their pharmacokinetic. However, dysfunction of these efflux pumps contribute also to major human diseases. Amongst these diseases are neurological disorders [2], such as Alzheimer’s disease [2], [5], [6], [7], [8], metabolic diseases and related illnesses [9], such as atherosclerosis [9], but also malignant diseases, such as multidrug-resistant cancer [3], [10], [11], [12], [13], [14]. For example, half of the A and G subclasses of ABC transporters have been identified as contributors in Alzheimer’s disease, correlating their downregulation or defective function with a negative disease development [6], [7]. Another example is multidrug-resistant cancer, where the vast majority of ABC transporters has been associated with the multidrug resistance (MDR) phenotype [12], [13], and many transporters were indeed found to export applied antineoplastic agents out of cancer cells, ultimately protecting these from cell death [11], [14].
Unfortunately, only a small fraction of the 49 existing ABC transporters can be considered as well-studied, in particular ABCB1 [8], [14], [15], [16], [17], [18], [19], ABCB11 [20], [21], [22], ABCC1 [1], [4], [14], [15], [17], [23], [24], and ABCG2 [14], [15], [17], [25]. Less-studied ABC transporters that have found much less attention are ABCC2, ABCC4–5, and ABCC10 [1], [4], [14], [24], [26], as well as – to a lesser extend – ABCA1 [27], [28], [29], [30], ABCB4 [14], ABCC3 [1], [4], [14], [24], as well as ABCC7–9 and ABCC11 [1], [4], [31], [32]. The remaining 34 transporters can be considered as under-studied which cannot be addressed by small-molecule modulators besides very rare exceptions. However, small-molecules would represent a potential tool to monitor, influence, and study these transporters for (i) a general understanding of their mechanism of action, and more importantly, for (ii) their exploration as potential pharmacological targets to develop innovative diagnostics and therapeutics.
As a logical consequence, the number of synthetic approaches to gain novel lead structures and potent modulators of ABC transporters is also very unequally distributed amongst the ABC transport proteins, and very scarce for under-studied ABC transporters. While many hundreds of small-molecule modulators of ABCB1 [14], [15], [16], [17], [18], [19], ABCC1 [1], [4], [14], [15], [17], [23], [24], and ABCG2 [14], [15], [17], [25] exist, synthetic approaches to target other ABC transporters have barely been reported. Rare exceptions are, for example, ABCC4 [33], [34], ABCC8 [35], or ABCC10 [36]. This lack of synthetic approaches is explained by the absence of lead structures as starting point for potential synthesis and lead optimization.
Computational approaches are great tools for lead discovery and subsequent optimization with the support of organic synthesis. They have extensively been used within the past 20 years. The vast majority of reports specified structure-based retrospective computational approaches, in which observed biological effects of compounds were underpinned mostly through molecular docking experiments with cryo-EM structures or homology models. Most pronounced are, again, ABCB1 [37], [38], [39], [40], [41], [42] and ABCG2 [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53]. Less- or under-studied ABC transporters are barely reflected in the literature. In terms of retrospective molecular docking experiments, rare exceptions are ABCB5 [54], ABCB6 [55], or ABCC10 [56], [57]. Ligand-based retrospective approaches are much less present in literature and have been described, for example, for ABCB11 [20], [21], [22], or ABCC2 [58].
Prospective approaches for the discovery of novel lead molecules are generally limited with respect to ABC transporters. Regarding structure-based design through molecular docking approaches, ABCB1 [59], [60], [61] and ABCG2 [43], [62], [63] are most pronounced, but also ABCC4 [64] or ABCC5 [65], [66] have been investigated. Prospective ligand-based design is more preferred, as it does not rely on crystal structures, cryo-EM structures, or homology models of ABC transporters. Similarity search (ABCC1 [67] or ABCC4 [64]), pharmacophore modelling (ABCB1 [61], [68], ABCB11 [69], or ABCC1 [67]), machine learning (ABCB1 [70] or ABCG2 [71]), or other pattern-based approaches [72] were demonstrated as powerful computational tools for lead identification, which eventually led often to virtual screenings and actual hit discovery [64], [67], [69]. However, these approaches always took only one transporter into account, completely leaving out the potential of multitarget inhibition. Multitargeting is a promising approach to explore under-studied ABC transporters by targeting similar or mutually overlapping binding sites [15], [73], [74]. Several pharmacological drugs have already been revealed as (weak) pan-ABC transporter inhibitors (=inhibiting several ABC transporters simultaneously), as for example, benzbromarone (1; ABCB1 [75], ABCB11 [20], ABCC1-6 [23], [24], [58], [76], [77], and ABCG2 [75]), cyclosporine A (2; ABCA1 [27], ABCB1 [16], ABCB4 [78], ABCB11 [20], ABCC1–2 [23], [58], ABCC10 [24], ABCG1–2 [75], [79]), glibenclamide (glyburide, 3; ABCA1 [29], ABCB11 [20], ABCC1 [23], ABCC5 [80], ABCC7–9 [81], [82], [83], ABCG2 [58]), probenecid (4; ABCA8 [84], ABCC1–6 [23], [24], [85], [86], [87], ABCC10 [88]), verapamil (5; ABCA8 [84], ABCB1 [16], ABCB4–5 [54], [78], ABCB11 [89], ABCC1 [23], ABCC4 [90], ABCC10 [88], ABCG2 [58]), or verlukast (MK571, 6; ABCA8 [84], ABCB4 [78], ABCB11 [20], ABCC1–5 [23], [58], [80], [87], [91], ABCC10–11 [24], [92], ABCG2 [58]). Fig. 1 provides the molecular formulae of the most prominent drug-like pan-ABC transporter inhibitors known until today.
Fig. 1.
Drugs and drug-like compounds that were shown in several independent studies to be pan-ABC transporter inhibitors. Cyclosporine A (2) was used as standard ABCB1 inhibitor in the presented study.
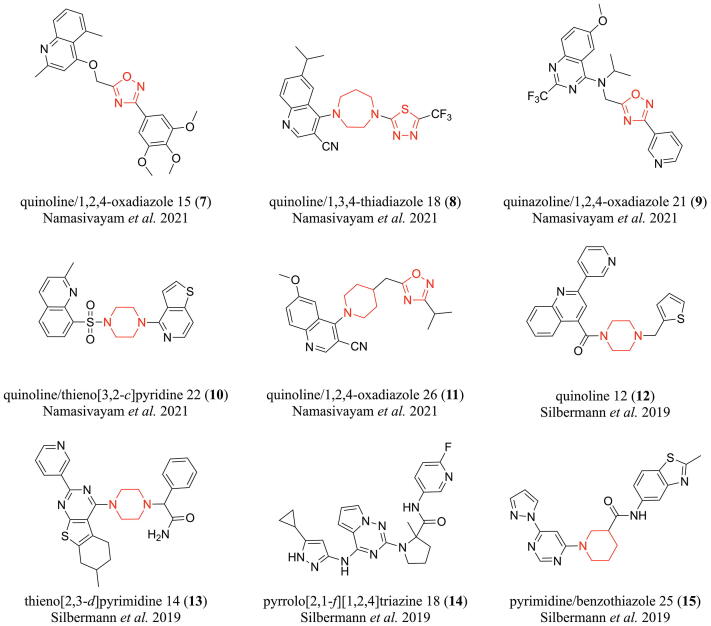
As indicated above, ABCB1, ABCC1, and ABCG2 are the most investigated and understood ABC transporters, and hence, represent model targets for the generation of pan-ABC transporter inhibitors [15]. However, even for these well-studied ABC transporters, only 133 broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors were described to date [15], [43], [45], [62], [67], [75], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126], amongst which only 56 exerted their effects below 10 µM [15], [43], [45], [62], [67], [75], [93], [94], [95], [98], [99], [101], [103], [107], [109], [110], [111], [112], [113], [118], [119], [120], [121], [122], [123], [126], and only 23 showed effects at ≤5 µM [43], [62], [98], [101], [103], [109], [110], [111], [112], [119], [121], [122], [126]. There is generally a lack of highly potent ABCB1, ABCC1, and ABCG2 inhibitors and only a highly limited understanding regarding prediction and discovery of such agents. Recently, we were the first to report on a novel computer-aided pattern analysis (C@PA) approach for the prediction of potent multitarget ABCB1, ABCC1, and ABCG2 inhibitors, discovering compounds 7–11 (Fig. 2) [15]. As the data regarding ABCB1, ABCC1, and ABCG2 is much more advanced than toward other transporters, we continued to improve C@PA’s prediction capabilities, which is reported in the presented study.
Fig. 2.
Broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors obtained from computational approaches. Compounds 7–11 were derived from C@PA as reported by Namasivayam et al. in 2021 [15]. Compounds 12–15 resulted from a combined ligand-based approach using similarity search and pharmacophore modelling as reported by Silbermann et al. in 2019 [67]. The corresponding IC50 values can be found in Table 1. Red mark: suggested secondary positive hits as proposed before [15]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Results and discussion
2.1. Basic scaffold dissection and potential positive hit identification
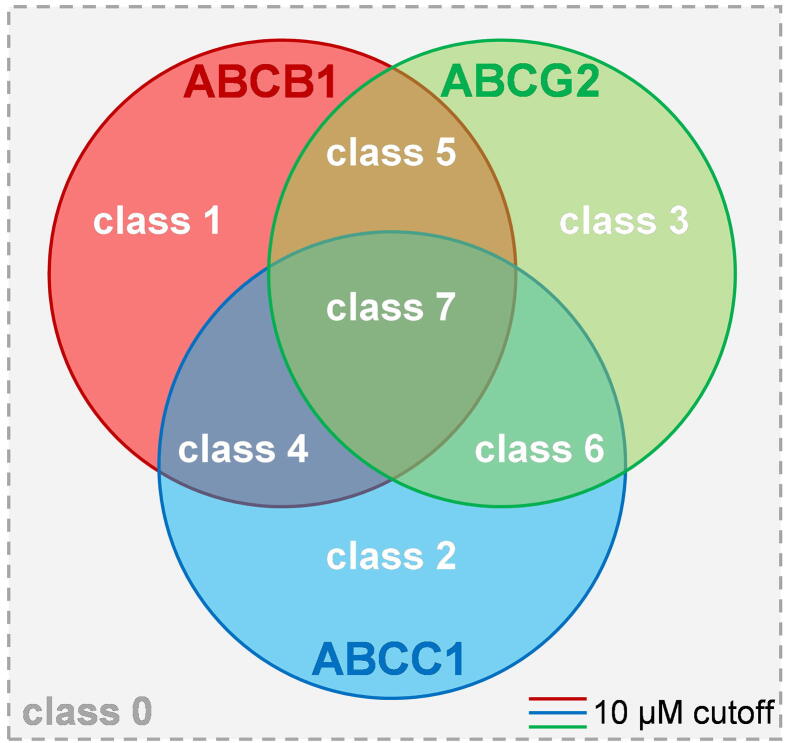
In our latest report about C@PA, we identified so-called ‘multitarget fingerprints’ for the prediction of broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors amongst a manually assembled and curated initial dataset of 1,049 compounds [15]. The model was generated on the basis of (i) the identification of basic scaffolds amongst the most potent known ABCB1, ABCC1, and ABCG2 inhibitors; (ii) the definition of substructures with a positive impact regarding multitarget ABCB1, ABCC1, and ABCG2 inhibition; and (iii) the definition of substructures with a negative impact with respect to multitarget ABCB1, ABCC1, and ABCG2 inhibition. As a result, compounds 8–9 as well as 11 were discovered by a virtual screening as so-called ‘class 7 compounds’ (=IC50 values below 10 µM toward ABCB1, ABCC1, and ABCG2; Fig. 3).
Fig. 3.
Visualization of the classification of modulators of ABCB1, ABCC1, and ABCG2 as proposed earlier [15]: ‘class 7 compounds’ are defined as triple ABCB1, ABCC1, and ABCG2 inhibitors that exert their half-maximal effect against these transporters below 10 µM. This has up to date been reported for 56 compounds [15], [43], [45], [62], [67], [75], [93], [94], [95], [98], [99], [101], [103], [107], [109], [110], [111], [112], [113], [118], [119], [120], [121], [122], [123], [126]. Amongst these molecules are the compounds revealed by C@PA, 8–9 and 11.
In total, 5 multitarget ABCB1, ABCC1, and ABCG2 inhibitors were discovered (7–11) [15], which contained 5 partial structures that were suggested by us as ‘secondary positive hits’: (i) 1,2,4-oxadiazole; (ii) 1,3,4-thiadiazole; (iii) piperazine; (iv) homo-piperazine; and (v) piperidine. In the present study, we extended the positive pattern fingerprints by ‘potential positive hits’ in order to explore their impact on the inhibitory nature of molecules on ABCB1, ABCC1, and ABCG2 function in combination with already known primary positive substructures.
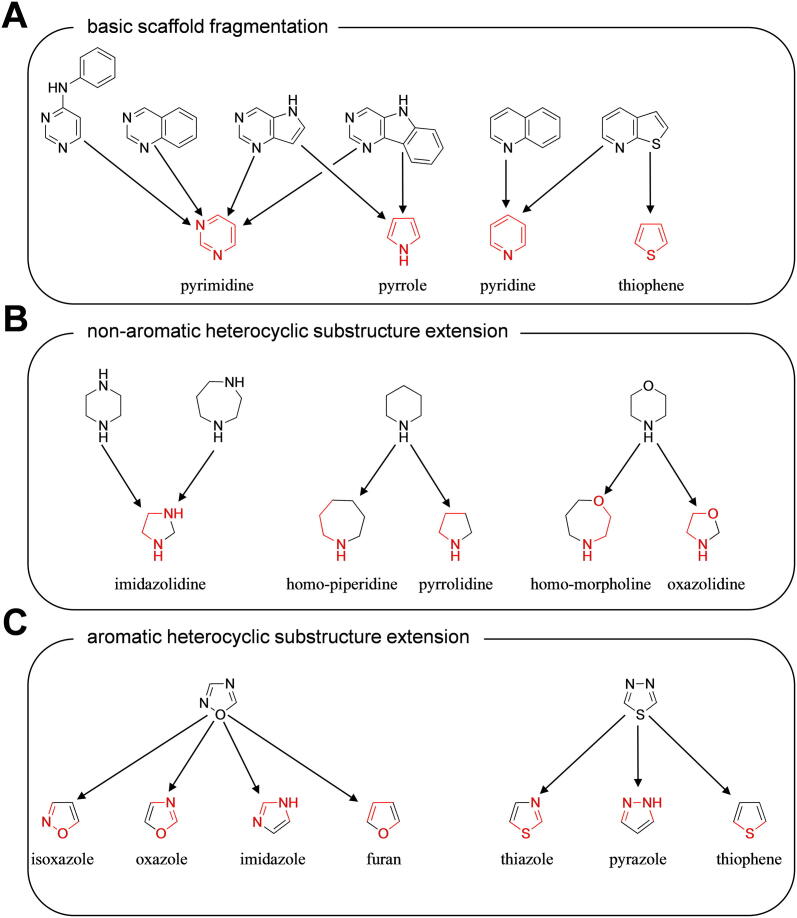
As a first step, we dissected the basic scaffolds (‘Scaffold Fragmentation’; Fig. 4 A) as derived by C@PA [15], which resulted in the first extension of the positive pattern fingerprints with potential positive hits: (i) pyrimidine; (ii) pyrrole; (iii) pyridine; and (iv) thiophene. As a second step, we extended the structural variety of the non-aromatic heterocycles (‘Heterocyclic Substructure Hopping’; Fig. 4 B) as derived and proposed by C@PA: (i) imidazolidine deduced from piperazine and homo-piperazine; (ii) homo-piperidine and pyrrolidine deduced from piperidine; and (iii) homo-morpholine and oxazolidine deduced from morpholine. It must be noted that pyrrolidine was earlier identified by C@PA as a ‘clear negative hit’ [15]. However, for a detailed investigation of piperidine derivatives and their impact on ABCB1, ABCC1, and ABCG2 function, this clear negative hit needed overruling. As a final step, the two found novel aromatic substructures in compounds 7–11, 1,2,4-oxadiazole and 1,3,4-thiadiazole, were extended by five-membered rings that had conserved features of the original substructure (‘Heteroaromatic Substructure Hopping’; Fig. 4 C). Hence, the following aromatic substructures were added to the potential positive hit list: (i) isoxazole; (ii) oxazole; (iii) imidazole; (iv) furan; (v) thiazole; (vi) pyrazole; and (vii) thiophene. Here again, it must be taken note that oxazole was also identified by C@PA as a clear negative hit [15], however, it was now added for a detailed evaluation of 1,2,4-oxadiazole derivatives toward their effect on ABCB1, ABCC1, and ABCG2 function. In total, the 8 'clear positive hits' as derived from C@PA [15] were extended by additional 5 suggested secondary positive hits [15] and 15 deduced potential positive hits. These 20 substructures will in the following be referred to as ‘extended positive hits’ (‘Extended Positive Pattern’).
Fig. 4.
Extension of positive pattern fingerprints: (A) dissection of C@PA-derived basic scaffolds into smaller heteroaromatic units (‘Scaffold Fragmentation’); (B) deduction of non-aromatic heterocycles from C@PA-derived clear positive and secondary positive hits (‘Heterocyclic Substructure Hopping’); and (C) deduction of heteroaromatic five-membered rings from C@PA-derived secondary positive heteroaromatic substructures (‘Heteroaromatic Substructure Hopping’). Red mark: conserved part of the novel substructure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Virtual screening and compound selection
The clear limit of C@PA was the prediction of ABCC1 inhibitors, as the discovered multitarget ABCB1, ABCC1, and ABCG2 inhibitors 7–11 were the only present ABCC1 inhibitors in the biologically evaluated set of 23 compounds [15]. To counteract this effect, we selected a virtual screening data set that favored ABCC1 inhibition as reported by us before [67]. This set of molecules comprised of 1,510 compounds that resulted from a combined virtual screening approach for the prediction of ABCC1 inhibitors. It is known that ~ 23.5% of these compounds comprised of ABCC1 inhibitors. We favored this virtual screening dataset compared to other options because the identified ABCC1 inhibitors (12–15; Fig. 2) were in parallel multitarget ABCB1, ABCC1, and ABCG2 inhibitors, which possibly increased the chance to identify novel lead molecules for broad-spectrum ABCB1, ABCC1, and ABCG2 inhibition.
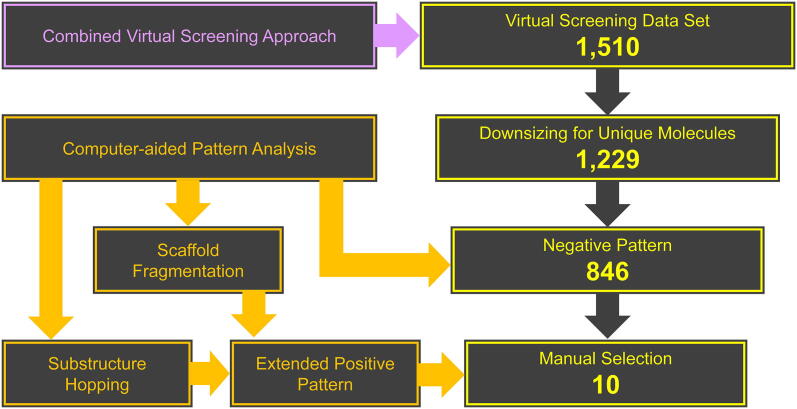
As a first step, the 1,510 compounds were screened for redundant molecules in form of stereoisomers to increase the diversity of the virtual screening data set. In total, 281 were removed, resulting in 1,229 unique compounds. These were in a second step subject to the negative pattern search [15]. While 383 compounds have been eliminated, 846 remained in the virtual screening dataset. Finally, the 846 compounds were screened for the extended positive hits. At least one of these favored substructures was present in these 846 molecules with the following distribution: (i) 1 time: 29 molecules; (ii) 2 times: 277 molecules, (iii) 3 times: 356 molecules; (iv): 4 times: 149 molecules; (v) 5 times: 34 molecules; (vi): 6 times: 1 molecule. From these 846 potential broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors we manually selected and purchased 10 candidates (compounds 16–25; Fig. 5) depending on manner and number of extended positive hits present and general molecular composition, as well as commercial availability and affordability at MolPort® (www.molport.com). Fig. 6 shows the virtual screening flow as exerted in this study.
Fig. 5.
Hit molecules 16–25 derived from the herein presented virtual screening approach as well as the reference ABCC1 and ABCG2 inhibitors, 26 and Ko143 (27), respectively, used in the present study [67], [101]. The corresponding IC50 values of compounds 16–25 can be found in Table 1. Red Mark: extended positive pattern. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Workflow of the herein presented virtual screening approach.
2.3. Biological evaluation
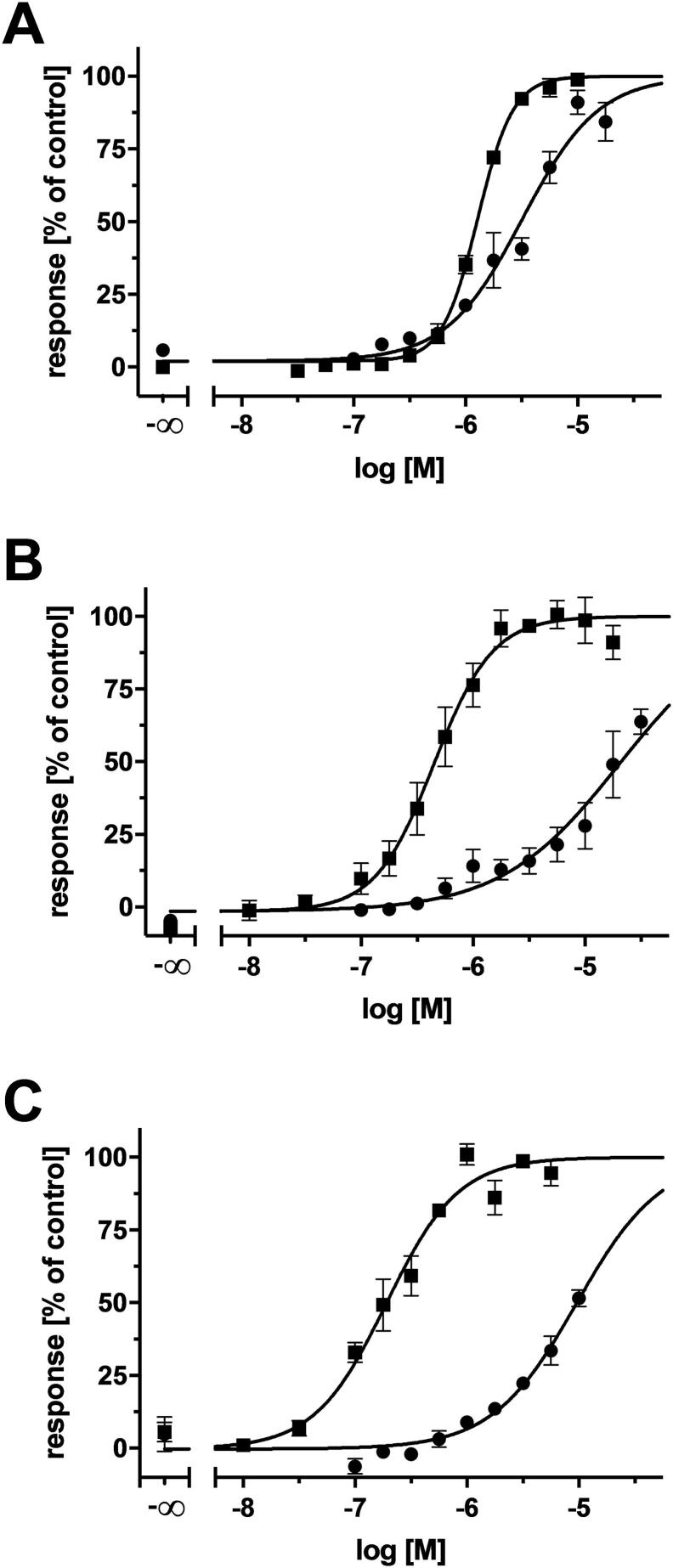
Compounds 16–25 were screened at 10 µM in calcein AM (ABCB1 and ABCC1) as well as pheophorbide A (ABCG2) fluorescence accumulation assays using either ABCB1-overexpressing A2780/ADR, ABCC1-overexpressing H69AR, or ABCG2-overexpressing MDCK II BCRP cells, respectively, as reported earlier [15], [43], [45], [67], [101]. Calcein AM and pheophorbide A are substrates of ABCB1 and ABCC1 as well as ABCG2, respectively, which passively diffuse into the used cells and become extruded by the corresponding ABC transporter. Inhibition of the respective transporter results in the accumulation of these substrates. Calcein AM is subsequently cleaved by intracellular esterases to the fluorescent calcein, while pheophorbide A is already fluorescent. Intracellular fluorescence was determined via microplate reader (calcein AM; ABCB1 and ABCC1) and flow cytometry (pheophorbide A; ABCG2), respectively. Compounds 2 (Fig. 1) and 26–27 (Fig. 5) were used as reference inhibitors against ABCB1, ABCC1, and ABCG2, respectively, as reported before [67], [101]. Fig. 7 provides the screening results for ABCB1 (A), ABCC1 (B), and ABCG2 (C).
Fig. 7.
Preliminary screening of compounds 16–25 against ABCB1 (A), ABCC1 (B), and ABCG2 (C) in calcein AM (A and B) and pheophorbide A (C) assays, respectively, using ABCB1-overexpressing A2780/ADR (A), ABCC1-overexpressing H69AR (B), and ABCG2-overexpressing MDCK II BCRP (C) cells as described earlier [15], [43], [45], [67], [101]. The data were normalized by defining 100% inhibition by the effect value of 10 µM of the reference inhibitors 2 (ABCB1; A), 26 (ABCC1, B), and 27 (ABCG2, C) as reported earlier [67], [101]. Shown is mean ± standard error of the mean (SEM) of at least three independent experiments. Red mark: triple ABCB1, ABCC1, and ABCG2 inhibitors. a no inhibition; b apparent ABCC1 activation (effect at 10 µM: 12.7% ± 2.4%). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As a result, 7 compounds had activities against ABCB1 (16–17, 19, 21–24), while 5 candidates inhibited ABCC1 (16, 18, 22–24), and 8 were active against ABCG2 (16–19, 22–25). Amongst the 10 evaluated compounds, 7 multitarget ABC transporter inhibitors could be identified: 4 triple ABCB1, ABCC1, and ABCG2 inhibitors (16, 22–24), 2 dual ABCB1 and ABCG2 inhibitors (17, 19), and 1 dual ABCC1 and ABCG2 inhibitor (18), which represents a multitarget hit rate of 70%. This even exceeded the very high multitarget hit rate of C@PA of 60.9% as reported earlier [15]. Compounds 21 and 25 were shown to be selective ABCB1 and ABCG2 inhibitors, respectively, while compound 20 did inhibit neither of the evaluated transporters. Table 1 presents the determined IC50 values of the compounds that reached at least 20% [+SEM (standard error of the mean)] compared to the standard ABCB1 (2), ABCC1 (26), and ABCG2 (27) inhibitors.
Table 1.
The determined IC50 values of compounds that resulted in an inhibition level of ≥20% [+ standard error or the mean (SEM)] in the preliminary screening (Fig. 7 A–C) determined in calcein AM (ABCB1 and ABCC1) and pheophorbide A (ABCG2) assays, respectively, applying ABCB1-overexpressing A2780/ADR, ABCC1-overexpressing H69AR, and ABCG2-overexpressing MDCK II BCRP cells, respectively, as described earlier [15], [43], [45], [67], [101]. The reference inhibitors (ABCB1: 2; ABCC1: 26; ABCG2: 27) served as positive controls as already reported earlier [67], [101], defining 100% inhibition. Buffer medium served as a negative control (0%). Shown is mean ± SEM of at least three independent experiments. Light rose mark: IC50 values of the triple ABCB1, ABCC1, and ABCG2 inhibitors 7–15 as reported earlier [15], [67]; dark rose mark: within this work discovered novel multitarget ABCB1, ABCC1, and ABCG2 inhibitors.
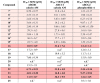 |
The discovery of 4 triple ABCB1, ABCC1, and ABCG2 inhibitors out of 10 candidates represents a biological hit rate of 40%, which was higher than the individual multitarget hit rates as reported in the combined similarity search and pharmacophore modelling approach (23.5%) and C@PA (21.7%) [15], [67]. Amongst these multitarget ABCB1, ABCC1, and ABCG2 inhibitors, compound 23 showed promising inhibitory activities against ABCB1 (4.01 µM), ABCC1 (14.8 µM), and ABCG2 (9.27 µM), almost qualifying it as a class 7 compound (Fig. 3). Besides the above mentioned 56 class 7 compounds [15], [43], [45], [62], [67], [75], [93], [94], [95], [98], [99], [101], [103], [107], [109], [110], [111], [112], [113], [118], [119], [120], [121], [122], [123], [126], further 20 compounds are known which exert their inhibitor effect against ABCB1, ABCC1, and/or ABCG2 up to 15.0 µM [15], [45], [75], [93], [98], [101], [102], [110], [113], [116], [117], [119]. Considering this, compound 23 belongs to the 76 most potent multitarget ABCB1, ABCC1, and ABCG2 inhibitors known until today. Fig. 8 depicts the concentration-effect curves of compound 23 against ABCB1 (A), ABCC1 (B), and ABCG2 (C).
Fig. 8.
Concentration-effect curves of compound 23 (●) against ABCB1 (A), ABCC1 (B), and ABCG2 (C) as obtained in calcein AM (A and B) and pheophorbide A (C) assays, respectively, compared to the reference inhibitors 2 (A;■), 26 (B; ■), and 27 (C; ■) applying ABCB1-overexpressing A2780/ADR (A), ABCC1-overexpressing H69AR (B), and ABCG2-overexpressing MDCK II BCRP (C) cells, respectively, as reported earlier [15], [43], [45], [67], [101]. Shown is mean ± SEM of at least three independent experiments.
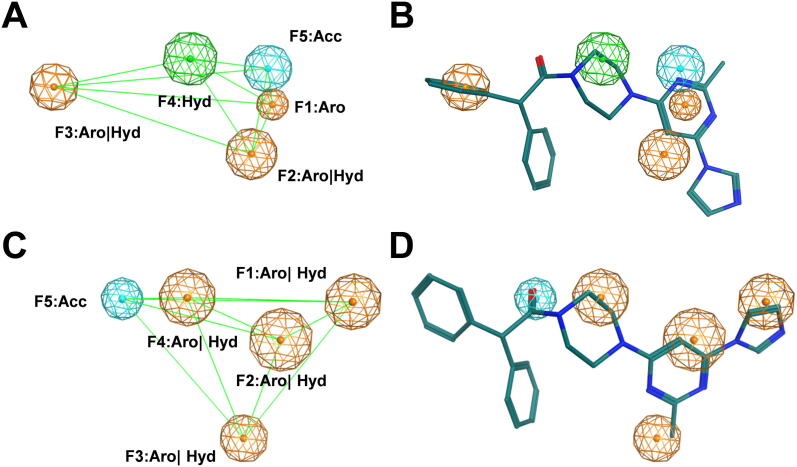
2.4. Pharmacophore modelling
In our previous study, we have explored different ligand-based approaches to validate C@PA [15]. A generated pharmacophore model based on the 6 most potent and diverse class 7 compounds (Supplementary Fig. 1) showed a sensitivity value of 60.4% and a specificity value of 44.5% (C@PA: 62.5% and 90.8%, respectively). Five pharmacophore features were discovered: (i–iv) F1–F4: aromatic/hydrophobic; and (v) F5: acceptor (Fig. 9 A). In the present study, we aimed for an additional investigation of the potential binding properties of compound 23. Hence, we performed a search on the recently presented pharmacophore model [15] for triple ABCB1, ABCC1, and ABCG2 inhibitors [15] by generating conformers of compound 23. As can be seen in Fig. 9 B, compound 23 reflected all five pharmacophore features as derived in the multitarget pharmacophore model [15], which confirms compound 23 as a moderately potent triple ABCB1, ABCC1, and ABCG2 inhibitor. In addition, compound 23 did also reflect all five pharmacophore features as derived from the previously reported similarity search and pharmacophore modelling approach [(i) F1: aromatic; (ii–iii) F2 and F3: aromatic/hydrophobic; (iv) F4: hydrophobic; and (v) F5: acceptor; Fig. 9 C–D] [67]. This suggests that compound 23 represents a good lead molecule for further improvement via synthesis to gain novel potent multitarget ABCB1, ABCC1, and ABCG2 inhibitors focusing ABCC1 inhibition. Furthermore, the findings support the hypothesis of a common multitarget binding site amongst different ABC transporter subfamilies as postulated earlier [15], [74].
Fig. 9.
Pharmacophore model of the most potent triple ABCB1, ABCC1, and ABCG2 inhibitor presented in this work, compound 23. The five multitarget ABCB1, ABCC1, and ABCG2 features [(i–iv) F1–F4: aromatic/hydrophobic; and (v) F5: acceptor] as reported before [15] are depicted (A), to which compound 23 was aligned to (B). In comparison, the five features for ABCC1 inhibition [(i) F1: aromatic; (ii–iii) F2 and F3: aromatic/hydrophobic; (iv) F4: hydrophobic; and (v) F5: acceptor] as reported before [67] are shown (C), and the respective conformer pose of compound 23 (D). The distances between the pharmacophore features are shown as light green lines. While nonpolar hydrogen atoms were omitted, carbon, oxygen, and nitrogen atoms were colored in green, red, and blue, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Conclusions
3.1. Statistical framework of C@PA
The aim of the present study was to extend the knowledge regarding multitarget fingerprints independent from the statistical background as reported previously [15]. This measure was necessary as it is almost impossible to change the statistical distribution of substructures amongst multitarget inhibitors (classes 4–7), specifically class 7 molecules, but also non-multitarget compounds (classes 0–3), unless a compound library of significant size (=hundreds of compounds) compared to the initial dataset of 1,049 compounds of C@PA [15] is synthesized and biologically evaluated on all three transporters. This is unlikely to happen within the next years.
The clear limit of the presented study was to discover class 7 compounds, supporting the threshold values set initially as the selection criteria of C@PA. [15]. These C@PA-derived clear positive hit and clear negative hit substructures are an important framework to obtain potent multitarget ABCB1, ABCC1, and ABCG2 inhibitors [15]. Especially the 32 clear negative hits proved to be of major importance compared to the only 8 found clear positive hits. However, the present work revealed that changes in these substructure compositions are tolerated, indicating an acceptable robustness of C@PA. This can also be visualized when comparing the initial hit rate for multitarget ABCB1, ABCC1, and ABCG2 inhibition of the virtual screening data set (23.5%) [67] with the hit rate of 40% found in the presented work, which indicates that C@PA_1.2 is an even more powerful methodology for the prediction of broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors. Strikingly, the present work demonstrated that the combination of C@PA with other computational approaches, in particular similarity search and pharmacophore modelling, led to a predictive synergism. Hence, the refinement of computer-chemical approaches with improved patterns and data sets may provide even higher biological hit rates in further developed pattern analysis models (e.g., C@PA_1.X).
3.2. Potential of extended positive hits: under-represented substructures
Several defined extended positive hits were reflected in the discovered multitarget ABCB1, ABCC1, and ABCG2 inhibitors 16 and 22–24, namely (i) pyrimidine (24), (ii) pyridine (22–24), (iii) isoxazole (16), (iv) imidazole (23), (v) pyrazole (22 and 24), and pyrrolidine (16). This discovery ultimately showed that the 8 clear positive hits as derived from C@PA [15] may indeed be supported by secondary positive hits, revealing the high potential of substructure extension in C@PA. A detailed analysis of these substructures according to their statistical distribution amongst the 133 known multitarget ABCB1, ABCC1, and ABCG2 inhibitors [15], [43], [45], [62], [67], [75], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126] showed that the substructures isoxazole, imidazole, pyrazole, and pyrrolidine occurred only 1, 3, 1, and 2 times [67], [96], [106], [107], [117], [127], respectively, in these 133 compounds, and were generally only present in 1, 16, 8, and 8 molecules, respectively, of the initial dataset of 1,049 compounds as used in C@PA [15]. Our results indicate that these ‘under-represented substructures’ pose a high exploratory potential for the improvement of C@PA’s prediction capabilities and the discovery of novel pan-ABC transporter inhibitors, as their specific statistical evaluation as exerted in our previous report [15] can easily be changed with a small number of additional compounds.
3.3. Potential of extended positive hits: rejected putative positive substructures
The omnipresent substructures pyrimidine [15], [17] and pyridine [15] must be seen in a different light, as these cannot be regarded on their own as indicators for multitarget ABCB1, ABCC1, and ABCG2 inhibition due to their ubiquitousness. However, our results indicate that these substructures have generally a positive impact on broad-spectrum ABCB1, ABCC1, and ABCG2 inhibition, depending on the composition of and combination with other substructures. Statistically, pyrimidine and pyridine occurred 56 and 28 times, respectively, in the 133 known multitarget ABCB1, ABCC1, and ABCG2 inhibitors [15], [43], [45], [62], [67], [75], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126]. In terms of class 7 compounds, 26 and 14 molecules contained pyrimidine and pyridine, respectively [15]. Indeed, pyrimidine and pyridine could not be considered as clear positive hits in our previous study [15] because many compounds of the other classes 0–6 contained these substructures as well (407 and 209 molecules, respectively). However, these ‘rejected putative positive substructures’ – which, nevertheless, resulted in class 7 molecules in a significant number – must be taken into special consideration for the further improvement of C@PA’s prediction capabilities (e.g., C@PA_1.X). Besides pyrimidine and pyridine, we identified 14 more substructures from the initial data set of 1,049 compounds [15] that should be reconsidered in terms of multitarget ABCB1, ABCC1, and ABCG2 inhibition in particular, and pan-ABC transporter inhibition in general: (i) aniline; (ii) benzoyl; (iii) benzyl; (iv) cyano; (v) 9-deazapurine; (vi) ether; (vii) ethylenediamine; (viii) methoxy; (xiv) methoxyphenyl; (x) phenol; (xi) phenyl; (xii) piperazine; (xiii) pyrrole; and (xiv) resorcin. Cyano, methoxy, and piperazine were already proposed in our previous study as secondary positive hits [15]. Nevertheless, it must clearly be noted that the percentage of occurence of these particular substructures amongst class 7 compounds is rather low. However, they might support other, clearer positive indicators of broad-spectrum ABCB1, ABCC1, and ABCG2 inhibition, enhancing compound potency through their proportionate contribution and combination, which represents a high potential for further developed C@PA-derived models (e.g., C@PA_1.X).
3.4. Outlook: the future of pan-ABC transporter modulators
The present study contributed to a major understanding of pattern analysis and possibilities to extend chemical patterns with the purpose to enhance the prediction rate to obtain biologically active compounds. The statistical distribution of certain substructures that occurred in class 7 or class 4–6 molecules in the initial data set of 1,049 compounds needs revision and re-evaluation, taking the results of the present study into account. We propose a ranking methodology to maximally increase the impact of secondary positive substructures in combination with primary positive hits for the best possible multitarget ABCB1, ABCC1, and ABCG2 inhibition. Deciphering the interconnection between manner, number, as well as the orientational composition of certain substructures and maximal possible impact on ABCB1, ABCC1, and ABCG2 will provide potential candidates for biological screening on other ABC transporters, exploring their nature, function, as well as their suitability as therapeutic or diagnostic drug targets. Furthermore, recent advances in crystallographic methodologies, such as cryo-EM, increasingly provided structural information of ABC transporters of different sub-families. This will allow for the analysis of the ‘multitarget binding site’ [15], [74] with the identified multitarget pan-ABC transporter inhibitors applying a combination of structure-based computational approaches. Using the knowledge derived from C@PA, C@PA_1.2, and potentially C@PA_1.X, new truly multitarget pan-ABC transporter modulators will be derived that could address less- and under-studied ABC transporters to tackle common and rare human diseases.
4. Experimental section
4.1. Computational analysis
4.1.1. Virtual screening dataset
The virtual screening dataset of the 1,510 putative ABCC1 inhibitors was derived by a combined similarity search and pharmacophore modelling approach as described earlier [67]. In short, an initial dataset of 288 known ABCC1 inhibitors with definite IC50 values was collected from ChEMBL [128] and categorized [‘active’ (IC50 < 1 µM); ‘moderate’ (IC50 = 1–10 µM); ‘inactive’ (IC50 > 10 µM)]. Similarity search applying the FTrees algorithm [129], [130] from BioSolveIT GmbH (Sankt Augustin, Germany) was conducted with a Tanimoto coefficient (Tc) of 0.8 by which the database was analyzed according to 4 query molecules (Supplementary Fig. 2) [101], [104], [131], [132]. The flexible alignment tool as well as the MMFF94x force field implemented in MOE (version 2016.08; Chemical Computing Group ULC, Montreal, QC, Canada) were applied for pharmacophore modelling using UNICON [133] to generate the 1000 best (=quality level 3) conformers with a tolerance distance of 1.5 Å and a threshold of 50.0% conservation. Virtual screening was performed with the ZINC12 library [134] consisting of 16,403,865 molecules from which a set of 1,510 molecules as potential ABCC1 inhibitors resulted.
4.1.2. Computer-aided pattern analysis (C@PA)
The computer-aided pattern analysis (C@PA) to predict multitarget ABCB1, ABCC1, and ABCG2 inhibitors was very recently reported [15]. In short, a manually assembled initial dataset of 1,049 compounds that have at least once been assessed for their inhibitory power against ABCB1, ABCC1, and ABCG2 was categorized [‘active’ (IC50 < 10 µM); ‘inactive’ (IC50 ≥ 10 µM)] and classified as class0: inactive compounds; class 1: selective ABCB1 inhibitors; class 2: selective ABCC1 inhibitors; class 3: selective ABCG2 inhibitors; class 4: dual ABCB1 and ABCG2 inhibitors; class 5: dual ABCB1 and ABCG2 inhibitors; class 6: dual ABCC1 and ABCG2 inhibitors; and class 7: triple ABCB1, ABCC1, and ABCG2 inhibitors; Fig. 3). In total, 48 class 7 compounds were identified and analyzed for their basic scaffolds [(i) 4-anilinopyrimidine; (ii) quinazoline; (iii) pyrrolo[3,2-d]pyrimidine; (iv) pyrimido[5,4-b]indole; (v) quinoline; and (vi) thieno[2,3-b]pyrimidine] using the Structure-Activity-Report (SAReport) tool [135] implemented in MOE (version 2019.01). InstantJChem (version 20.15.9) was applied to statistically analyze the initial dataset of 1,049 compounds for 308 commonly occurring chemical substructures [136] and their distribution amongst classes 0–7. ‘Clear positive hits’ [‘Positive Pattern’; (i) isopropyl; (ii) amino; (iii) carboxylic acid ethyl ester; (iv) indole; (v) 3,4,5-trimethoxyphenyl; (vi) morpholine; (vii) thieno[2,3-b]pyrimidine; (viii) sulfone] and ‘clear negative hits’ [‘Negative Pattern’; (i) tert-butyl; (ii) vinyl; (iii) cyclopropyl; (iv) cyclohexyl; (v) anellated cyclopropyl; (vi) anellated cycloheptyl; (vii) dimethylamino; (viii) diethylamino; (ix) nitro; (x) pyrrolidine; (xi) methylene hydroxy; (xii) ethylene hydroxy; (xiii) oxolane; (xiv) carboxylic acid; (xv) carboxylic acid methyl ester; (xvi) biphenyl; (xvii) stilbene; (xviii) 1,2,3-triazole; (xix) 1,2,4-triazole; (xx) tetrazole; (xxi) pyrido[2,3-d]pyrimidine; (xxii) 1,3-dihydroisobenzofuran; (xxiii) chalcone; (xxiv) hydroquinone; (xxv) 2-methoxyphenyl; (xxvi) 3-methoxyphenyl; (xxvii) 2,5-dimethoxyphenyl; (xxviii) 3,5-dimethoxyphenyl; (xxix) unsubstituted thioamide; (xxx) oxazole; (xxxi); urea; (xxxii) thiourea)] were identified. Virtual screening was performed with the ENAMINE REAL drug-like® compound library consisting of 15,547,091 molecules and a set of 1,505 molecule as potential broad-spectrum ABCB1, ABCC1, and ABCG2 inhibitors resulted, from which compounds 7–11 were discovered. Five partial structures were identified amongst these compounds which could be suggested as ‘secondary positive hits’: (i) 1,2,4-oxadiazole; (ii) 1,3,4-thiadiazole; (iii) piperazine; (iv) homo-piperazine; and (v) piperidine.
4.1.3. Scaffold fragmentation, substructure hopping, virtual screening, and compound selection
The C@PA-derived basic scaffolds were dissected using ChemDraw Pro [version 17.1.0.105 (19)] to (i) pyrimidine, (ii) pyrrole, (iii) pyridine, and (iv) thiophene and added to the extended positive hit list. Moreover, the non-aromatic heterocycles piperazine, piperidine, and morpholine were extended to (i) imidazolidine, (ii) homo-piperidine, (iii) pyrrolidine, (iv) homo-morpholine, and (v) oxazolidine. The aromatic substructures 1,2,4-oxadiazole and 1,3,4-thiadiazole were extended to (i) isoxazole, (ii) oxazole, (iii) imidazole, (iv) furan, (v) thiazole, (vi) pyrazole, and (vii) thiophene, and added to the extended positive hit list. In total, 29 extended positive hit substructures including the 8 clear positive hits as defined by C@PA [15] resulted. Subsequently, the 1,510 molecules derived from the combined similarity search and pharmacophore modelling approach [67] were subject to a clear negative hit exclusion (except for pyrrolidine and oxazole), with eventual extended positive hit screening. Depending on price and availability, the 10 candidates 16–25 were manually selected from the residual dataset of 846 potential multitarget ABCB1, ABCC1, and ABCG2 inhibitors and purchased at MolPort® (http://www.molport.com): compound 16 (Name: 2-((2-(3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)pyrrolidin-1-yl)methyl)-5-(5-methyl-3-phenyl-isoxazol-4-yl)-1,3,4-oxadiazole; ZINC ID: 08938070; MolPort® ID: 005–547-575; Link: https://www.molport.com/shop/moleculelink/XFQPDVQEQZTEIY-UHFFFAOYSAN/5547575; Supplier: ENAMINE Ltd.®; Cataloge No.: Z103927872; SMILES: CC1=C(C2=NN=C(O2)CN3CCCC3C4=CC5=C(OCCCO5)C=C4)C(C6=CC=CC=C6)=NO1; purity: ≥90%; compound 17 (Name: 4-(1-(4-fluorophenyl)-1H-imidazole-5-carboxamido)cyclohexyl 1-(4-fluorophenyl)-1H-imidazole-5-carboxylate; ZINC ID: 09672163; MolPort® ID: 004–504-763 Link: https://www.molport.com/shop/moleculelink/PAIVADUCKBGHFQ-UHFFFAOYSA-N/4504763; Supplier: Ukr-OrgSynthesis Ltd.®; Cataloge No.: PB169991190; SMILES: FC1=CC=C(N2C=NC=C2C(NC3CCC(OC(C4=CN=CN4C5=CC=C(F)C=C5)=O)CC3)=O)C=C1; purity: ≥90%; compound 18 (Name: 3-(2-(4-(2-(azepan-1-yl)-2-oxoethyl)piperazin-1-yl)-2-oxoethyl)-5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(3H)-one; ZINC ID: 14239968; MolPort® ID: 005–770-351; Link: https://www.molport.com/shop/moleculelink/LCTNJTHHFHNEAA-UHFFFAOYSA-N/5770351; Supplier: UkrOrgSynthesis Ltd.®; Cataloge No.: PB146323516; SMILES: O=C(N1CCN(CC1)CC(N2CCCCCC2)=O)CN3C=NC4=C(C3=O)C(C5=CC=CS5)=CS4; purity: ≥90%; compound 19 (Name: (4-(benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)(1-(5-(pyrrolidin-1-yl)-1,3,4-thiadiazol-2-yl)-1H-pyrrol-2-yl)methanone; ZINC ID: 15731308; MolPort® ID: 007–821-780; Link: https://www.molport.com/shop/moleculelink/YMCNSQXIHXWXTB-UHFFFAOYSA-N/7821780; Supplier: ChemDiv Inc.®; Cataloge No.: G015-0322; SMILES: O=C(C1=CC=CN1C2=NN=C(N3CCCC3)S2)N4CCN(CC4)CC5=CC=C6OCOC6=C5; purity: ≥90%; compound 20 (Name: 4-(3-(6-(pyridin-2-yl)-4,6,7a,12a-tetrahydro-1H-benzo[4,5]imidazo[1,2-a][1,3,5]triazino[1,2-c][1,3,5]triazin-2(3H)-yl)propyl)morpholine; ZINC ID: 20567796; MolPort® ID 005–912-631; Link: https://www.molport.com/shop/moleculelink/FVOLKPQFOHSUDIUHFFFAOYSAN/5912631; Supplier: Vitas-M Laboratory Ltd.®; Cataloge No.: STK790135; SMILES: N1(CCOCC1)CCCN2CNC3=NC(N4C(N3C2)N=C5C=CC=CC45)C6=CC=CC=N6; purity: ≥90%; compound 21 (Name: N-((1-(5-(4-benzylpiperazin-1-yl)-1,3,4-thiadiazol-2-yl)-1H-pyrrol-2-yl)methyl)-2-(pyrrolidin-1-yl)ethan-1-amine; ZINC ID: 20576334; MolPort® ID: 007–776-896; Link: https://www.molport.com/shop/moleculelink/AMGDZJNOUFPCEC-UHFFFAOYSA-N/7776896; Supplier: ChemDiv Inc.®; Cataloge No.: E985-0683; SMILES: N1(CCCC1)CCNCC2=CC=CN2C3=NN=C(N4CCN(CC4)CC5=CC=CC=C5)S3; purity: ≥90%; compound 22 (Name: 4-(2-(3-(3-(isoquinolin-4-yl)phenyl)-1H-pyrazol-1-yl)ethyl) morpholine; ZINC ID: 23213965; MolPort® ID: 005–039-609; Link: https://www.molport.com/shop/moleculelink/4–3-1–2-morpholin-4-yl-ethyl-1H-pyrazol-3-yl-phenyl-isoquinoline/5039609; Supplier: ChemBridge Corporation®; Cataloge No.: 25121718; SMILES: N1(C=CC(C2=CC=CC(C3=CN=CC4=CC=CC=C34)=C2)=N1)CCN5CCOCC5; purity: ≥90%; compound 23 (Name: 1-(4-(6-(1H-imidazol-1-yl)-2-methylpyrimidin-4-yl)piperazin-1-yl)-2,2-diphenylethan-1-one; ZINC ID: 65362307; MolPort® ID: 016–587-938; Link: https://www.molport.com/shop/moleculelink/DFWDBEHMZWCBIJ-UHFFFAOYSA-N/16587938; Supplier: ChemBridge Corporation®; Cataloge No.: 9210464; SMILES: CC1=NC(N2C=CN=C2)=CC(N3CCN(C(C(C4=CC=CC=C4)C5=CC=CC=C5)=O)CC3)=N1; Purity: ≥90%; compound 24 (Name: 1-(6-(1H-pyrazol-1-yl)pyrimidin-4-yl)-N-benzyl-N-(pyridin-2-yl)piperidine-3-carboxamide; ZINC ID: 70638100; MolPort® ID: 019–920-623; Link: https://www.molport.com/shop/moleculelink/HDSFZCOIEQTPRF-UHFFFAOYSA-N/19920623; Supplier: Life Chemicals Inc.®; Catalog No.: F6175-0779; SMILES: O=C(N(C1=CC=CC=N1)CC2=CC=CC=C2)C3CCCN(C4=CC(N5C=CC=N5)=NC=N4)C3; purity: ≥90%; compound 25 (Name: 3-(2-(4-(6-fluorobenzo[d]isoxazol-3-yl)piperidin-1-yl)-2-oxoethyl)-2-hydroxy-3,4-dihydro-5H-benzo[e][1,4]diazepin-5-one; ZINC ID: 95474733; MolPort® ID: 027–849-694; Link: https://www.molport.com/shop/moleculelink/SBJDVQXJQQGZMH-UHFFFAOYSA-N/27849694; Supplier: Vitas-M Laboratory Ltd.®; Cataloge No.: STL312371; SMILES: OC1=NC2=C(C(NC1CC(N3CCC(C4=NOC5=C4C=CC(F)=C5)CC3)=O)=O)C=CC=C2; purity: ≥90%.
4.2. Biological evaluation
4.2.1. Chemicals
The reference ABCB1 inhibitor 2 as well as the reference ABCG2 inhibitor 27 were purchased from Tocris Bioscience (Bristol, UK). The standard ABCC1 inhibitor 26 was synthesized as described earlier [101]. Calcein AM and pheophorbide A were obtained from Calbiochem [EMD Chemicals (San Diego, USA), supplied by Merck KgaA (Darmstadt, Germany)]. All other chemicals were purchased from Carl Roth (Karlsruhe, Germany), Merck KgaA (Darmstadt, Germany), or Sigma-Aldrich (Taufkirchen, Germany). Compounds 16–26 were stored as 10 mM stock solutions at −20 °C, and dilution series as well as the in-experiment cell culture was performed with Krebs-HEPES buffer [KHB; 118.6 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 4.2 mM NaHCO3, 1.3 mM CaCl2, 1.2 mM MgSO4, 11.7 mM D-glucose monohydrate, and 10.0 mM HEPES (2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid) in doubly distilled water, which was finally adjusted to pH 7.4 with NaOH and sterilized with 0.2 µm membrane filters].
4.2.2. Cell culture
The ABCB1-overexpressing A2780/ADR cells were delivered by European Collection of Animal Cell Culture (ECACC, no. 93112520) and cultivated using RPMI-1640 medium (PAN-Biotech GmbH, Aidenbach, Germany) complemented with fetal bovine serum (FCS; 10%; PAN-Biotech GmbH, Aidenbach, Germany), streptomycin (50 µg/µL; PAN-Biotech GmbH, Aidenbach, Germany), penicillin G (50 U/mL; PAN-Biotech GmbH, Aidenbach, Germany), as well as L-glutamine (2 mM; PAN-Biotech GmbH, Aidenbach, Germany). The ABCC1-overexpressing H69AR cells were provided by American Type Culture Collection (ATCC CRL-11351), which were cultured with RPMI-1640 medium to which FCS (20%), streptomycin (50 µg/µL), penicillin G (50 U/mL), as well as L-glutamine (2 mM) were added. Dr. A. Schinkel (The Netherlands Cancer Institute, Amsterdam, The Netherlands) generously provided the ABCG2-overexpressing MDCK II BCRP cells, which were cultivated in Dulbecco's modified eagle medium (DMEM; Sigma Life Science, Steinheim, Germany) complemented with FCS (10%), streptomycin (50 µg/µL), penicillin G (50 U/mL), as well as L-glutamine (2 mM). The cell lines were stored in a mixture of 90% cell culture medium and 10% DMSO under liquid nitrogen, while cultivation was performed at 37 °C under 5% CO2-humidified atmosphere. After a confluence of 90% was reached, the cells were harvested using a 0.05%/0.02% trypsin-EDTA solution (PAN-Biotech GmbH, Aidenbach, Germany). The processing included centrifugation in a 50 mL falcon (Greiner Bio-One, Frickenhausen, Germany) at 266×g and 4 °C for 4 min (Avanti J-25, Beckmann Coulter, Krefeld, Germany), removal of the supernatant, resuspension in fresh cell culture media, cell counting (CASY TT cell counter with 150 µm capillary, Schärfe System GmbH, Reutlingen, Germany), as well as seeding of cells for either sub-culturing or biological testing.
4.2.3. Calcein AM assay
The inhibitory activity against ABCB1 and ABCC1 was evaluated in a calcein AM assay as reported earlier [15], [67], [101]. Compounds 16–25 were added into a 96-well flat-bottom clear plate (Greiner, Frickenhausen, Germany) at a concentration of 100 µM and 160 µL of cell suspension containing either ABCB1-overexpressing A2780/ADR (30,000 cells/well) or ABCC1-overexpressing H69AR (60,000 cells/well) cells were added. The incubation period at 37 °C under 5% CO2-humidified atmosphere lasted 30 min before 20 µL of a 3.125 µM calcein AM was added to each well, subsequently followed by measurement of fluorescence increase at an excitation wavelength of 485 nm and an emission wave length of 520 nm in 60 sec intervals for 1 h in either POLARstar and FLUOstar Optima microplate readers (BMG Labtech, software versions 2.00R2/2.20 and 4.11-0; Offenburg, Germany). The slope values from the linear fluorescence increase revealed the effect value which has been normalized to the effect value of 10 µM of either compounds 2 (ABCB1) or 26 (ABCC1). As candidates 16–17, 19, and 21–24 as well as 16, 18, 22–24 resulted in significant inhibition (20% + SEM) of ABCB1 and ABCC1, respectively, full-blown concentration-effect curves were generated and IC50 values were calculated applying GraphPad Prism (version 8.4.0, San Diego, CA, USA) using the statistically preferred model (three- or four-parameter logistic equation).
4.2.4. Pheophorbide A assay
The inhibitory activity against ABCG2 was evaluated in a pheophorbide A assay as reported earlier [15], [67]. Each well of a flat-bottom clear 96 well plate was complemented with 20 µL of either of the compounds 16–25 (100 µM), 160 µL of ABCG2-overexpressing MDCK II BCRP cells (45,000 cells/well), as well as 20 µL of a pheophorbide A solution (5 µM), subsequently incubating the plate at 37 °C in a 5% CO2-humidified atmosphere for 120 min. The effect values of compounds 16–25 were measured via flow cytometry [Guava easyCyteTM HT, (Merck Millipore, Billerica, MA, USA; excitation: 488 nm; emission: 695/50 nm)] and compared to the effect value of 10 µM of compound 27. As compounds 16–19 and 22–25 resulted in significant inhibition against ABCG2 (20% + SEM), complete concentration-effect curves were generated as described in 4.2.2.
4.3. Retrospective pharmacophore analysis
In our earlier study, a pharmacophore model was generated to evaluate the performance of C@PA [15]. This model generation has been accomplished on the basis of the 6 most potent and diverse ABCB1, ABCC1, and ABCG2 inhibitors (Supplementary Fig. 1) [43], [99], [101], [103], [110] by aligning these molecules using the flexible alignment tool as described in 4.1.1 applying MOE (version 2019.01) [15]. The best alignment was selected and the pharmacophore model was generated using the consensus method implemented in the Pharmacophore Query Editor with a threshold value of 50.0% and a tolerance distance of 1.2 Å. The conformers of the most potent multitarget ABCB1, ABCC1, and ABCG2 inhibitor in this work, compound 23, were generated using the conformational search tool by selecting the stochastic search method implemented in MOE 2019.01. The default parameters were applied for the conformational search with a maximum limit of 10,000.
CRediT authorship contribution statement
Vigneshwaran Namasivayam: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision. Katja Silbermann: Investigation, Writing - review & editing. Jens Pahnke: Resources, Writing - review & editing, Funding acquisition. Michael Wiese: Resources, Supervision, Writing - review & editing. Sven Marcel Stefan: Conceptualization, Methodology, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
VN thanks ChemAxon for providing an academic research license to their software. JP received funding from Deutsche Forschungsgemeinschaft (DFG; German Research Foundation)/ Germany (DFG 263024513); EFRE und Ministerium für Wirtschaft, Wissenschaft und Digitalisierung achsen-Anhalt/ Germany (ZS/2016/05/78617); Latvian Council of Science/ Latvia (lzp-2018/1-0275); Nasjonalforeningen (16154), HelseSØ/ Norway (2016062, 2019054, 2019055); Barnekreftforeningen (19008); EEA grant/Norway grants Kappa programme (TAČR TARIMAD TO100078); Norges forskningsrådet/ Norway (251290, 260786 PROP-AD, 295910 NAPI, and 327571 PETABC); European Commission (643417). PROP-AD and PETABC are EU Joint Programme - Neurodegenerative Disease Research (JPND) projects. PROP-AD is supported through the following funding organizations under the aegis of JPND - www.jpnd.eu (AKA #301228 – Finland, BMBF #01ED1605 – Germany, CSO-MOH #30000-12631 – Israel, NFR #260786 – Norway, SRC #2015-06795 – Sweden). PETABC is supported through the following funding organisations under the aegis of JPND - www.jpnd.eu (NFR #327571 – Norway, FFG #882717 – Austria, BMBF – Germany, MSMT #8F21002– Czech Republic, VIAA #ES RTD/2020/26 – Latvia, ANR #20-JPW2-0002-04 - France, SRC #2020-02905 – Sweden). The projects receive funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement #643417 (JPco-fuND). SMS receives a Walter Benjamin fellowship of the DFG; 446812474).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.05.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wang J.Q., Yang Y., Cai C.Y., Teng Q.X., Cui Q., Lin J. Multidrug resistance proteins (MRPs): structure, function and the overcoming of cancer multidrug resistance. Drug Resist Updat. 2021;54 doi: 10.1016/j.drup.2021.100743. [DOI] [PubMed] [Google Scholar]
- 2.Gil-Martins E., Barbosa D.J., Silva V., Remião F., Silva R. Dysfunction of ABC transporters at the blood-brain barrier: role in neurological disorders. Pharmacol Ther. 2020;213 doi: 10.1016/j.pharmthera.2020.107554. [DOI] [PubMed] [Google Scholar]
- 3.Pasello M., Giudice A.M., Scotlandi K. The ABC subfamily A transporters: Multifaceted players with incipient potentialities in cancer. Semin Cancer Biol. 2020;60:57–71. doi: 10.1016/j.semcancer.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Sodani K., Patel A., Kathawala R.J., Chen Z.S. Multidrug resistance associated proteins in multidrug resistance. Chin J Cancer. 2012;31:58–72. doi: 10.5732/cjc.011.10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dib S., Pahnke J., Gosselet F. Role of ABCA7 in Human Health and in Alzheimer’s Disease. Int J Mol Sci. 2021;22:4603. doi: 10.3390/ijms22094603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behl T., Kaur I., Sehgal A., Kumar A., Uddin M.S., Bungau S. The interplay of ABC transporters in Aβ translocation and cholesterol metabolism: implicating their roles in Alzheimer's Disease. Mol Neurobiol. 2020;58:1564–1582. doi: 10.1007/s12035-020-02211-x. [DOI] [PubMed] [Google Scholar]
- 7.Pereira C.D., Martins F., Wiltfang J., da Cruz E.S.O.A.B., Rebelo S. ABC transporters are key players in Alzheimer's Disease. J Alzheimers Dis. 2018;61:463–485. doi: 10.3233/JAD-170639. [DOI] [PubMed] [Google Scholar]
- 8.Abuznait A.H., Kaddoumi A. Role of ABC transporters in the pathogenesis of Alzheimer's disease. ACS Chem Neurosci. 2012;3:820–831. doi: 10.1021/cn300077c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Z., Lu Y., Wu T. The impact of ATP-binding cassette transporters on metabolic diseases. Nutr Metab (Lond) 2020;17:61. doi: 10.1186/s12986-020-00478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domenichini A., Adamska A., Falasca M. ABC transporters as cancer drivers: Potential functions in cancer development. Biochim Biophys Acta Gen Subj. 1863;2019:52–60. doi: 10.1016/j.bbagen.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamska A., Falasca M. ATP-binding cassette transporters in progression and clinical outcome of pancreatic cancer: What is the way forward? World J Gastroenterol. 2018;24:3222–3238. doi: 10.3748/wjg.v24.i29.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan S.T., Li Z.L., He Z.X., Qiu J.X., Zhou S.F. Molecular mechanisms for tumour resistance to chemotherapy. Clin Exp Pharmacol Physiol. 2016;43:723–737. doi: 10.1111/1440-1681.12581. [DOI] [PubMed] [Google Scholar]
- 14.Ween M.P., Armstrong M.A., Oehler M.K., Ricciardelli C. The role of ABC transporters in ovarian cancer progression and chemoresistance. Crit Rev Oncol Hematol. 2015;96:220–256. doi: 10.1016/j.critrevonc.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Namasivayam V., Silbermann K., Wiese M., Pahnke J., Stefan S.M. C@PA: Computer-aided pattern analysis to predict multitarget ABC transporter inhibitors. J Med Chem. 2021;64:3350–3366. doi: 10.1021/acs.jmedchem.0c02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Xu H., Ashby C.R., Jr., Assaraf Y.G., Chen Z.S., Liu H.M. Chemical molecular-based approach to overcome multidrug resistance in cancer by targeting P-glycoprotein. P-gpMed Res Rev. 2021;41:525–555. doi: 10.1002/med.21739. [DOI] [PubMed] [Google Scholar]
- 17.He Z.X., Zhao T.Q., Gong Y.P., Zhang X., Ma L.Y., Liu H.M. Pyrimidine: A promising scaffold for optimization to develop the inhibitors of ABC transporters. Eur J Med Chem. 2020;200 doi: 10.1016/j.ejmech.2020.112458. [DOI] [PubMed] [Google Scholar]
- 18.Dong J., Qin Z., Zhang W.D., Cheng G., Yehuda A.G., Ashby C.R., Jr. Medicinal chemistry strategies to discover P-glycoprotein inhibitors: an update. Drug Resist Updat. 2020;49 doi: 10.1016/j.drup.2020.100681. [DOI] [PubMed] [Google Scholar]
- 19.Palmeira A., Sousa E., Vasconcelos M.H., Pinto M.M. Three decades of P-gp inhibitors: skimming through several generations and scaffolds. Curr Med Chem. 2012;19:1946–2025. doi: 10.2174/092986712800167392. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen J.M., Matsson P., Bergström C.A., Hoogstraate J., Norén A., LeCluyse E.L. Early identification of clinically relevant drug interactions with the human bile salt export pump (BSEP/ABCB11) Toxicol Sci. 2013;136:328–343. doi: 10.1093/toxsci/kft197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan R.E., van Staden C.J., Chen Y., Kalyanaraman N., Kalanzi J., Dunn R.T., 2nd A multifactorial approach to hepatobiliary transporter assessment enables improved therapeutic compound development. Toxicol Sci. 2013;136:216–241. doi: 10.1093/toxsci/kft176. [DOI] [PubMed] [Google Scholar]
- 22.Warner D.J., Chen H., Cantin L.D., Kenna J.G., Stahl S., Walker C.L. Mitigating the inhibition of human bile salt export pump by drugs: opportunities provided by physicochemical property modulation, in silico modeling, and structural modification. Drug Metab Dispos. 2012;40:2332–2341. doi: 10.1124/dmd.112.047068. [DOI] [PubMed] [Google Scholar]
- 23.Stefan S.M., Wiese M. Small-molecule inhibitors of multidrug resistance-associated protein 1 and related processes: a historic approach and recent advances. Med Res Rev. 2019;39:176–264. doi: 10.1002/med.21510. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S.F., Wang L.L., Di Y.M., Xue C.C., Duan W., Li C.G. Substrates and inhibitors of human multidrug resistance associated proteins and the implications in drug development. Curr Med Chem. 2008;15:1981–2039. doi: 10.2174/092986708785132870. [DOI] [PubMed] [Google Scholar]
- 25.Peña-Solórzano D., Stark S.A., König B., Sierra C.A., Ochoa-Puentes C. ABCG2/BCRP: Specific and nonspecific modulators. Med Res Rev. 2017;37:987–1050. doi: 10.1002/med.21428. [DOI] [PubMed] [Google Scholar]
- 26.Ivanyuk A., Livio F., Biollaz J., Buclin T. Renal drug transporters and drug interactions. Clin Pharmacokinet. 2017;56:825–892. doi: 10.1007/s40262-017-0506-8. [DOI] [PubMed] [Google Scholar]
- 27.Nagao K., Maeda M., Mañucat N.B., Ueda K. Cyclosporine A and PSC833 inhibit ABCA1 function via direct binding. Biochim Biophys Acta. 1831;2013:398–406. doi: 10.1016/j.bbalip.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Favari E., Zanotti I., Zimetti F., Ronda N., Bernini F., Rothblat G.H. Probucol inhibits ABCA1-mediated cellular lipid efflux. Arterioscler Thromb Vasc Biol. 2004;24:2345–2350. doi: 10.1161/01.ATV.0000148706.15947.8a. [DOI] [PubMed] [Google Scholar]
- 29.Nieland T.J., Chroni A., Fitzgerald M.L., Maliga Z., Zannis V.I., Kirchhausen T. Cross-inhibition of SR-BI- and ABCA1-mediated cholesterol transport by the small molecules BLT-4 and glyburide. J Lipid Res. 2004;45:1256–1265. doi: 10.1194/jlr.M300358-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Becq F., Hamon Y., Bajetto A., Gola M., Verrier B., Chimini G. ABC1, an ATP binding cassette transporter required for phagocytosis of apoptotic cells, generates a regulated anion flux after expression in Xenopus laevis oocytes. J Biol Chem. 1997;272:2695–2699. doi: 10.1074/jbc.272.5.2695. [DOI] [PubMed] [Google Scholar]
- 31.Bardin E., Pastor A., Semeraro M., Golec A., Hayes K., Chevalier B. Modulators of CFTR. Updates on clinical development and future directions. Eur J Med Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113195. [DOI] [PubMed] [Google Scholar]
- 32.Kathawala R.J., Wang Y.J., Ashby C.R., Jr., Chen Z.S. Recent advances regarding the role of ABC subfamily C member 10 (ABCC10) in the efflux of antitumor drugs. Chin J Cancer. 2014;33:223–230. doi: 10.5732/cjc.013.10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M., Urabe T., Okamoto Y., Murakami H., Tatemoto A., Ohno S. Augmentation of murine organ-associated natural immune responses by cepharanthin. Gan To Kagaku Ryoho. 1988;15:127–133. [PubMed] [Google Scholar]
- 34.Tan T.M., Yang F., Fu H., Raghavendra M.S., Lam Y. Traceless solid-phase synthesis and biological evaluation of purine analogs as inhibitors of multidrug resistance protein 4. J Comb Chem. 2007;9:210–218. doi: 10.1021/cc060084t. [DOI] [PubMed] [Google Scholar]
- 35.Tagmose T.M., Schou S.C., Mogensen J.P., Nielsen F.E., Arkhammar P.O., Wahl P. Arylcyanoguanidines as activators of Kir6.2/SUR1K ATP channels and inhibitors of insulin release. J Med Chem. 2004;47:3202–3211. doi: 10.1021/jm031018y. [DOI] [PubMed] [Google Scholar]
- 36.Teijaro C.N., Munagala S., Zhao S., Sirasani G., Kokkonda P., Malofeeva E.V. Synthesis and biological evaluation of pentacyclic strychnos alkaloids as selective modulators of the ABCC10 (MRP7) efflux pump. J Med Chem. 2014;57:10383–10390. doi: 10.1021/jm501189p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao H.L., Gupta P., Cui Q., Ashar Y.V., Wu Z.X., Zeng L. Sapitinib reverses anticancer drug resistance in colon cancer cells overexpressing the ABCB1 transporter. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.574861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo X., Teng Q.X., Dong J.Y., Yang D.H., Wang M., Dessie W. Antimicrobial Peptide reverses ABCB1-mediated chemotherapeutic drug resistance. Front Pharmacol. 2020;11:1208. doi: 10.3389/fphar.2020.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M., Chen X.Y., Dong X.D., Wang J.Q., Feng W., Teng Q.X. NVP-CGM097, an HDM2 inhibitor, antagonizes ATP-binding cassette subfamily B member 1-mediated drug resistance. Front Oncol. 2020;10:1219. doi: 10.3389/fonc.2020.01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Yang D.H., Yang Y., Wang J.Q., Cai C.Y., Lei Z.N. Overexpression of ABCB1 transporter confers resistance to mTOR inhibitor WYE-354 in cancer cells. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21041387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y., Yin D., Ye J., Wei X., Pei Y., Li X. Discovery of Potent Inhibitors against P-Glycoprotein-Mediated Multidrug Resistance Aided by Late-Stage Functionalization of a 2-(4-(Pyridin-2-yl)phenoxy)pyridine Analogue. J Med Chem. 2020;63:5458–5476. doi: 10.1021/acs.jmedchem.0c00337. [DOI] [PubMed] [Google Scholar]
- 42.Liao D., Zhang W., Gupta P., Lei Z.N., Wang J.Q., Cai C.Y. Tetrandrine interaction with ABCB1 reverses multidrug resistance in cancer cells through competition with anti-cancer drugs followed by downregulation of ABCB1 expression. Molecules. 2019;24:4383. doi: 10.3390/molecules24234383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silbermann K., Li J., Namasivayam V., Stefan S.M., Wiese M. Rational drug design of 6-substituted 4-anilino-2-phenylpyrimidines for exploration of novel ABCG2 binding site. Eur J Med Chem. 2021;212 doi: 10.1016/j.ejmech.2020.113045. [DOI] [PubMed] [Google Scholar]
- 44.Wang J.Q., Teng Q.X., Lei Z.N., Ji N., Cui Q., Fu H. Reversal of cancer multidrug resistance (MDR) mediated by ATP-binding cassette transporter G2 (ABCG2) by AZ-628, a RAF kinase inhibitor. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.601400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silbermann K., Li J., Namasivayam V., Baltes F., Bendas G., Stefan S.M. Superior pyrimidine derivatives as selective ABCG2 inhibitors and broad-spectrum ABCB1, ABCC1, and ABCG2 antagonists. J Med Chem. 2020;63:10412–10432. doi: 10.1021/acs.jmedchem.0c00961. [DOI] [PubMed] [Google Scholar]
- 46.Ashar Y.V., Zhou J., Gupta P., Teng Q.X., Lei Z.N., Reznik S.E. BMS-599626, a highly selective Pan-HER kinase inhibitor, antagonizes ABCG2-mediated drug resistance. Cancers (Basel) 2020;12:2502. doi: 10.3390/cancers12092502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z.X., Yang Y., Wang G., Wang J.Q., Teng Q.X., Sun L. Dual TTK/CLK2 inhibitor, CC-671, selectively antagonizes ABCG2-mediated multidrug resistance in lung cancer cells. Cancer Sci. 2020;111:2872–2882. doi: 10.1111/cas.14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Z.X., Peng Z., Yang Y., Wang J.Q., Teng Q.X., Lei Z.N. M3814, a DNA-PK inhibitor, modulates ABCG2-mediated multidrug resistance in lung cancer cells. Front Oncol. 2020;10:674. doi: 10.3389/fonc.2020.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Wang J.Q., Cai C.Y., Cui Q., Yang Y., Wu Z.X. Reversal effect of ALK inhibitor NVP-TAE684 on ABCG2-overexpressing cancer cells. Front Oncol. 2020;10:228. doi: 10.3389/fonc.2020.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J.Q., Li J.Y., Teng Q.X., Lei Z.N., Ji N., Cui Q. Venetoclax, a BCL-2 inhibitor, enhances the efficacy of chemotherapeutic agents in wild-type ABCG2-overexpression-mediated MDR cancer cells. Cancers (Basel) 2020;12:466. doi: 10.3390/cancers12020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kathawala R.J., Espitia C.M., Jones T.M., Islam S., Gupta P., Zhang Y.K. ABCG2 overexpression contributes to pevonedistat resistance. Cancers (Basel) 2020;12:429. doi: 10.3390/cancers12020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Z.X., Yang Y., Teng Q.X., Wang J.Q., Lei Z.N., Wang J.Q. Tivantinib, A c-Met inhibitor in clinical trials, is susceptible to ABCG2-mediated drug resistance. Cancers (Basel) 2020;12:186. doi: 10.3390/cancers12010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krapf M.K., Gallus J., Namasivayam V., Wiese M. 2,4,6-substituted quinazolines with extraordinary inhibitory potency toward ABCG2. J Med Chem. 2018;61:7952–7976. doi: 10.1021/acs.jmedchem.8b01011. [DOI] [PubMed] [Google Scholar]
- 54.Saeed M.E.M., Boulos J.C., Elhaboub G., Rigano D., Saab A., Loizzo M.R. Cytotoxicity of cucurbitacin E from Citrullus colocynthis against multidrug-resistant cancer cells. Phytomedicine. 2019;62 doi: 10.1016/j.phymed.2019.152945. [DOI] [PubMed] [Google Scholar]
- 55.Polireddy K., Khan M.M., Chavan H., Young S., Ma X., Waller A. A novel flow cytometric HTS assay reveals functional modulators of ATP binding cassette transporter ABCB6. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H., Patel A., Wang Y.J., Zhang Y.K., Kathawala R.J., Qiu L.H. The BTK Inhibitor Ibrutinib (PCI-32765) Overcomes Paclitaxel Resistance in ABCB1- and ABCC10-Overexpressing Cells and Tumors. Mol Cancer Ther. 2017;16:1021–1030. doi: 10.1158/1535-7163.MCT-16-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H., Kathawala R.J., Wang Y.J., Zhang Y.K., Patel A., Shukla S. Linsitinib (OSI-906) antagonizes ATP-binding cassette subfamily G member 2 and subfamily C member 10-mediated drug resistance. Int J Biochem Cell Biol. 2014;51:111–119. doi: 10.1016/j.biocel.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsson P., Pedersen J.M., Norinder U., Bergström C.A., Artursson P. Identification of novel specific and general inhibitors of the three major human ATP-binding cassette transporters P-gp, BCRP and MRP2 among registered drugs. Pharm Res. 2009;26:1816–1831. doi: 10.1007/s11095-009-9896-0. [DOI] [PubMed] [Google Scholar]
- 59.Yang Y., Ji N., Cai C.Y., Wang J.Q., Lei Z.N., Teng Q.X. Modulating the function of ABCB1: in vitro and in vivo characterization of sitravatinib, a tyrosine kinase inhibitor. Cancer Commun (Lond) 2020;40:285–300. doi: 10.1002/cac2.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wise J.G., Nanayakkara A.K., Aljowni M., Chen G., De Oliveira M.C., Ammerman L. Optimizing targeted inhibitors of P-glycoprotein using computational and structure-guided approaches. J Med Chem. 2019;62:10645–10663. doi: 10.1021/acs.jmedchem.9b00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmeira A., Sousa E., Vasconcelos M.H., Pinto M., Fernandes M.X. Structure and ligand-based design of P-glycoprotein inhibitors: a historical perspective. Curr Pharm Des. 2012;18:4197–4214. doi: 10.2174/138161212802430530. [DOI] [PubMed] [Google Scholar]
- 62.Antoni F., Wifling D., Bernhardt G. Water-soluble inhibitors of ABCG2 (BCRP) - A fragment-based and computational approach. Eur J Med Chem. 2021;210 doi: 10.1016/j.ejmech.2020.112958. [DOI] [PubMed] [Google Scholar]
- 63.Yang Y., Ji N., Teng Q.X., Cai C.Y., Wang J.Q., Wu Z.X. Sitravatinib, a tyrosine kinase inhibitor, inhibits the transport function of ABCG2 and restores sensitivity to chemotherapy-resistant cancer cells in vitro. Front Oncol. 2020;10:700. doi: 10.3389/fonc.2020.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y., Yuan X., Xiao Z., Jin H., Zhang L., Liu Z. Discovery of novel multidrug resistance protein 4 (MRP4) inhibitors as active agents reducing resistance to anticancer drug 6-Mercaptopurine (6-MP) by structure and ligand-based virtual screening. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0205175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kashgari F.K., Ravna A., Sager G., Lyså R., Enyedy I., Dietrichs E.S. Identification and experimental confirmation of novel cGMP efflux inhibitors by virtual ligand screening of vardenafil-analogues. Biomed Pharmacother. 2020;126 doi: 10.1016/j.biopha.2020.110109. [DOI] [PubMed] [Google Scholar]
- 66.Sager G., Ørvoll E., Lysaa R.A., Kufareva I., Abagyan R., Ravna A.W. Novel cGMP efflux inhibitors identified by virtual ligand screening (VLS) and confirmed by experimental studies. J Med Chem. 2012;55:3049–3057. doi: 10.1021/jm2014666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silbermann K., Stefan S.M., Elshawadfy R., Namasivayam V., Wiese M. Identification of Thienopyrimidine Scaffold as an Inhibitor of the ABC Transport Protein ABCC1 (MRP1) and Related Transporters Using a Combined Virtual Screening Approach. J Med Chem. 2019;62:4383–4400. doi: 10.1021/acs.jmedchem.8b01821. [DOI] [PubMed] [Google Scholar]
- 68.Palmeira A., Rodrigues F., Sousa E., Pinto M., Vasconcelos M.H., Fernandes M.X. New uses for old drugs: pharmacophore-based screening for the discovery of P-glycoprotein inhibitors. Chem Biol Drug Des. 2011;78:57–72. doi: 10.1111/j.1747-0285.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 69.Ritschel T., Hermans S.M., Schreurs M., van den Heuvel J.J., Koenderink J.B., Greupink R. In silico identification and in vitro validation of potential cholestatic compounds through 3D ligand-based pharmacophore modeling of BSEP inhibitors. Chem Res Toxicol. 2014;27:873–881. doi: 10.1021/tx5000393. [DOI] [PubMed] [Google Scholar]
- 70.Klepsch F., Vasanthanathan P., Ecker G.F. Ligand and structure-based classification models for prediction of P-glycoprotein inhibitors. J Chem Inf Model. 2014;54:218–229. doi: 10.1021/ci400289j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang D., Lei T., Wang Z., Shen C., Cao D., Hou T. ADMET evaluation in drug discovery. 20. Prediction of breast cancer resistance protein inhibition through machine learning. J Cheminform. 2020;12:16. doi: 10.1186/s13321-020-00421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Broccatelli F., Carosati E., Neri A., Frosini M., Goracci L., Oprea T.I. A novel approach for predicting P-glycoprotein (ABCB1) inhibition using molecular interaction fields. J Med Chem. 2011;54:1740–1751. doi: 10.1021/jm101421d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stefan S.M. Multi-target ABC transporter modulators: what next and where to go? Future Med Chem. 2019;11:2353–2358. doi: 10.4155/fmc-2019-0185. [DOI] [PubMed] [Google Scholar]
- 74.Stefan K., Leck L.Y.W., Namasivayam V., Bascuñana P., Huang M.-L.-H., Riss P.J. Vesicular ATP-binding cassette transporters in human disease: relevant aspects of their organization for future drug development, Future. Drug Discovery. 2020;2:FDD51. [Google Scholar]
- 75.Ivnitski-Steele I., Larson R.S., Lovato D.M., Khawaja H.M., Winter S.S., Oprea T.I. High-throughput flow cytometry to detect selective inhibitors of ABCB1, ABCC1, and ABCG2 transporters. Assay Drug Dev Technol. 2008;6:263–276. doi: 10.1089/adt.2007.107. [DOI] [PubMed] [Google Scholar]
- 76.Zelcer N., Saeki T., Reid G., Beijnen J.H., Borst P. Characterization of drug transport by the human multidrug resistance protein 3 (ABCC3) J Biol Chem. 2001;276:46400–46407. doi: 10.1074/jbc.M107041200. [DOI] [PubMed] [Google Scholar]
- 77.El-Sheikh A.A., van den Heuvel J.J., Koenderink J.B., Russel F.G. Effect of hypouricaemic and hyperuricaemic drugs on the renal urate efflux transporter, multidrug resistance protein 4. Br J Pharmacol. 2008;155:1066–1075. doi: 10.1038/bjp.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bieczynski F., Burkhardt-Medicke K., Luquet C.M., Scholz S., Luckenbach T. Chemical effects on dye efflux activity in live zebrafish embryos and on zebrafish Abcb4 ATPase activity. FEBS Lett. 2020 doi: 10.1002/1873-3468.14015. in Press. [DOI] [PubMed] [Google Scholar]
- 79.Cserepes J., Szentpétery Z., Seres L., Ozvegy-Laczka C., Langmann T., Schmitz G. Functional expression and characterization of the human ABCG1 and ABCG4 proteins: indications for heterodimerization. Biochem Biophys Res Commun. 2004;320:860–867. doi: 10.1016/j.bbrc.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 80.Borst P., de Wolf C., van de Wetering K. Multidrug resistance-associated proteins 3, 4, and 5. Pflugers Arch. 2007;453:661–673. doi: 10.1007/s00424-006-0054-9. [DOI] [PubMed] [Google Scholar]
- 81.Berghaus A., Jovanovic S. Technique and indications of extended sublabial rhinotomy (“midfacial degloving”) Rhinology. 1991;29:105–110. [PubMed] [Google Scholar]
- 82.Lim J.G., Lee H.Y., Yun J.E., Kim S.P., Park J.W., Suh S.I. Taurine block of cloned ATP-sensitive K+ channels with different sulfonylurea receptor subunits expressed in Xenopus laevis oocytes. Biochem Pharmacol. 2004;68:901–910. doi: 10.1016/j.bcp.2004.05.050. [DOI] [PubMed] [Google Scholar]
- 83.York N.W., Parker H., Xie Z., Tyus D., Waheed M.A., Yan Z. Kir6.1- and SUR2-dependent KATP over-activity disrupts intestinal motility in murine models of Cantu Syndrome, JCI. Insight. 2020;5 doi: 10.1172/jci.insight.141443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsuruoka S., Ishibashi K., Yamamoto H., Wakaumi M., Suzuki M., Schwartz G.J. Functional analysis of ABCA8, a new drug transporter. Biochem Biophys Res Commun. 2002;298:41–45. doi: 10.1016/s0006-291x(02)02389-6. [DOI] [PubMed] [Google Scholar]
- 85.Horikawa M., Kato Y., Sugiyama Y. Reduced gastrointestinal toxicity following inhibition of the biliary excretion of irinotecan and its metabolites by probenecid in rats. Pharm Res. 2002;19:1345–1353. doi: 10.1023/a:1020358910490. [DOI] [PubMed] [Google Scholar]
- 86.Smeets P.H., van Aubel R.A., Wouterse A.C., van den Heuvel J.J., Russel F.G. Contribution of multidrug resistance protein 2 (MRP2/ABCC2) to the renal excretion of p-aminohippurate (PAH) and identification of MRP4 (ABCC4) as a novel PAH transporter. J Am Soc Nephrol. 2004;15:2828–2835. doi: 10.1097/01.ASN.0000143473.64430.AC. [DOI] [PubMed] [Google Scholar]
- 87.Dalpiaz A., Pavan B. Nose-to-Brain Delivery of Antiviral Drugs: A Way to Overcome Their Active Efflux? Pharmaceutics. 2018;10 doi: 10.3390/pharmaceutics10020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou Y., Hopper-Borge E., Shen T., Huang X.C., Shi Z., Kuang Y.H. Cepharanthine is a potent reversal agent for MRP7(ABCC10)-mediated multidrug resistance. Biochem Pharmacol. 2009;77:993–1001. doi: 10.1016/j.bcp.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 89.Horikawa M., Kato Y., Tyson C.A., Sugiyama Y. Potential cholestatic activity of various therapeutic agents assessed by bile canalicular membrane vesicles isolated from rats and humans. Drug Metab Pharmacokinet. 2003;18:16–22. doi: 10.2133/dmpk.18.16. [DOI] [PubMed] [Google Scholar]
- 90.Bai J., Lai L., Yeo H.C., Goh B.C., Tan T.M. Multidrug resistance protein 4 (MRP4/ABCC4) mediates efflux of bimane-glutathione. Int J Biochem Cell Biol. 2004;36:247–257. doi: 10.1016/s1357-2725(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 91.Videmann B., Mazallon M., Prouillac C., Delaforge M., Lecoeur S. ABCC1, ABCC2 and ABCC3 are implicated in the transepithelial transport of the myco-estrogen zearalenone and its major metabolites. Toxicol Lett. 2009;190:215–223. doi: 10.1016/j.toxlet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 92.Tun-Yhong W., Chinpaisal C., Pamonsinlapatham P., Kaewkitichai S. Tenofovir disoproxil fumarate is a new substrate of ATP-binding cassette subfamily C member 11. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01725-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Telbisz Á., Ambrus C., Mózner O., Szabó E., Várady G., Bakos É. Interactions of potential anti-COVID-19 compounds with multispecific ABC and OATP drug transporters. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Antoni F., Bause M., Scholler M., Bauer S., Stark S.A., Jackson S.M. Tariquidar-related triazoles as potent, selective and stable inhibitors of ABCG2 (BCRP) Eur J Med Chem. 2020;191 doi: 10.1016/j.ejmech.2020.112133. [DOI] [PubMed] [Google Scholar]
- 95.Krapf M.K., Gallus J., Spindler A., Wiese M. Synthesis and biological evaluation of quinazoline derivatives – a SAR study of novel inhibitors of ABCG2. Eur J Med Chem. 2019;161:506–525. doi: 10.1016/j.ejmech.2018.10.026. [DOI] [PubMed] [Google Scholar]
- 96.Sorf A., Novotna E., Hofman J., Morell A., Staud F., Wsol V. Cyclin-dependent kinase inhibitors AZD5438 and R547 show potential for enhancing efficacy of daunorubicin-based anticancer therapy: interaction with carbonyl-reducing enzymes and ABC transporters. Biochem Pharmacol. 2019;163:290–298. doi: 10.1016/j.bcp.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 97.Hofman J., Sorf A., Vagiannis D., Sucha S., Kammerer S., Küpper J.H. Brivanib exhibits potential for pharmacokinetic drug-drug interactions and the modulation of multidrug resistance through the inhibition of human ABCG2 drug efflux transporter and CYP450 biotransformation enzymes. Mol Pharm. 2019;16:4436–4450. doi: 10.1021/acs.molpharmaceut.9b00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Teodori E., Contino M., Riganti C., Bartolucci G., Braconi L., Manetti D. Design, synthesis and biological evaluation of stereo- and regioisomers of amino aryl esters as multidrug resistance (MDR) reversers. Eur J Med Chem. 2019;182 doi: 10.1016/j.ejmech.2019.111655. [DOI] [PubMed] [Google Scholar]
- 99.Krapf M.K., Gallus J., Wiese M. Synthesis and biological investigation of 2,4-substituted quinazolines as highly potent inhibitors of breast cancer resistance protein. ABCG2Eur J Med Chem. 2017;139:587–611. doi: 10.1016/j.ejmech.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 100.Schmitt S.M., Stefan K., Wiese M. Pyrrolopyrimidine derivatives and purine analogs as novel activators of Multidrug Resistance-associated Protein 1 (MRP1, ABCC1) Biochim Biophys Acta Biomembr. 1859;2017:69–79. doi: 10.1016/j.bbamem.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 101.Stefan K., Schmitt S.M., Wiese M. 9-Deazapurines as broad-spectrum inhibitors of the ABC transport proteins P-glycoprotein, multidrug resistance-associated protein 1, and breast cancer resistance protein. J Med Chem. 2017;60:8758–8780. doi: 10.1021/acs.jmedchem.7b00788. [DOI] [PubMed] [Google Scholar]
- 102.Schäfer A., Köhler S.C., Lohe M., Wiese M., Hiersemann M. Synthesis of homoverrucosanoid-derived esters and evaluation as MDR modulators. J Org Chem. 2017;82:10504–10522. doi: 10.1021/acs.joc.7b02012. [DOI] [PubMed] [Google Scholar]
- 103.Krapf M.K., Wiese M. Synthesis and biological evaluation of 4-anilino-quinazolines and -quinolines as inhibitors of breast cancer resistance protein (ABCG2) J Med Chem. 2016;59:5449–5461. doi: 10.1021/acs.jmedchem.6b00330. [DOI] [PubMed] [Google Scholar]
- 104.Schmitt S.M., Stefan K., Wiese M. Pyrrolopyrimidine derivatives as novel inhibitors of multidrug resistance-associated protein 1 (MRP1, ABCC1) J Med Chem. 2016;59:3018–3033. doi: 10.1021/acs.jmedchem.5b01644. [DOI] [PubMed] [Google Scholar]
- 105.Obreque-Balboa J.E., Sun Q., Bernhardt G., König B., Buschauer A. Flavonoid derivatives as selective ABCC1 modulators: synthesis and functional characterization. Eur J Med Chem. 2016;109:124–133. doi: 10.1016/j.ejmech.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 106.Lempers V.J., van den Heuvel J.J., Russel F.G., Aarnoutse R.E., Burger D.M., Brüggemann R.J. Inhibitory potential of antifungal drugs on ATP-binding cassette transporters P-glycoprotein, MRP1 to MRP5, BCRP, and BSEP. Antimicrob Agents Chemother. 2016;60:3372–3379. doi: 10.1128/AAC.02931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mathias T.J., Natarajan K., Shukla S., Doshi K.A., Singh Z.N., Ambudkar S.V. The FLT3 and PDGFR inhibitor crenolanib is a substrate of the multidrug resistance protein ABCB1 but does not inhibit transport function at pharmacologically relevant concentrations. Invest New Drugs. 2015;33:300–309. doi: 10.1007/s10637-015-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cihalova D., Staud F., Ceckova M. Interactions of cyclin-dependent kinase inhibitors AT-7519, flavopiridol and SNS-032 with ABCB1, ABCG2 and ABCC1 transporters and their potential to overcome multidrug resistance in vitro. Cancer Chemother Pharmacol. 2015;76:105–116. doi: 10.1007/s00280-015-2772-1. [DOI] [PubMed] [Google Scholar]
- 109.Hu J., Zhang X., Wang F., Wang X., Yang K., Xu M. Effect of ceritinib (LDK378) on enhancement of chemotherapeutic agents in ABCB1 and ABCG2 overexpressing cells in vitro and in vivo. Oncotarget. 2015;6:44643–44659. doi: 10.18632/oncotarget.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krauze A., Grinberga S., Krasnova L., Adlere I., Sokolova E., Domracheva I. Thieno[2,3-b]pyridines–a new class of multidrug resistance (MDR) modulators. Bioorg Med Chem. 2014;22:5860–5870. doi: 10.1016/j.bmc.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 111.Ma S.L., Hu Y.P., Wang F., Huang Z.C., Chen Y.F., Wang X.K. Lapatinib antagonizes multidrug resistance-associated protein 1-mediated multidrug resistance by inhibiting its transport function. Mol Med. 2014;20:390–399. doi: 10.2119/molmed.2014.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Juvale K., Stefan K., Wiese M. Synthesis and biological evaluation of flavones and benzoflavones as inhibitors of BCRP/ABCG2. Eur J Med Chem. 2013;67:115–126. doi: 10.1016/j.ejmech.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 113.Juvale K., Gallus J., Wiese M. Investigation of quinazolines as inhibitors of breast cancer resistance protein (ABCG2) Bioorg Med Chem. 2013;21:7858–7873. doi: 10.1016/j.bmc.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 114.Colabufo N.A., Contino M., Cantore M., Capparelli E., Perrone M.G., Cassano G. Naphthalenyl derivatives for hitting P-gp/MRP1/BCRP transporters. Bioorg Med Chem. 2013;21:1324–1332. doi: 10.1016/j.bmc.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 115.Cihalova D., Hofman J., Ceckova M., Staud F. Purvalanol A, olomoucine II and roscovitine inhibit ABCB1 transporter and synergistically potentiate cytotoxic effects of daunorubicin in vitro. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0083467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Juvale K., Wiese M. 4-Substituted-2-phenylquinazolines as inhibitors of BCRP. Bioorg Med Chem Lett. 2012;22:6766–6769. doi: 10.1016/j.bmcl.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 117.Pick A., Wiese M. Tyrosine kinase inhibitors influence ABCG2 expression in EGFR-positive MDCK BCRP cells via the PI3K/Akt signaling pathway. ChemMedChem. 2012;7:650–662. doi: 10.1002/cmdc.201100543. [DOI] [PubMed] [Google Scholar]
- 118.Mi Y.J., Liang Y.J., Huang H.B., Zhao H.Y., Wu C.P., Wang F. Apatinib (YN968D1) reverses multidrug resistance by inhibiting the efflux function of multiple ATP-binding cassette transporters. Cancer Res. 2010;70:7981–7991. doi: 10.1158/0008-5472.CAN-10-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Colabufo N.A., Berardi F., Perrone M.G., Cantore M., Contino M., Inglese C. Multi-drug-resistance-reverting agents: 2-aryloxazole and 2-arylthiazole derivatives as potent BCRP or MRP1 inhibitors. ChemMedChem. 2009;4:188–195. doi: 10.1002/cmdc.200800329. [DOI] [PubMed] [Google Scholar]
- 120.Pick A., Klinkhammer W., Wiese M. Specific inhibitors of the breast cancer resistance protein (BCRP) ChemMedChem. 2010;5:1498–1505. doi: 10.1002/cmdc.201000216. [DOI] [PubMed] [Google Scholar]
- 121.Colabufo N.A., Pagliarulo V., Berardi F., Contino M., Inglese C., Niso M. Bicalutamide failure in prostate cancer treatment: involvement of Multi Drug Resistance proteins. Eur J Pharmacol. 2008;601:38–42. doi: 10.1016/j.ejphar.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 122.Dai C.L., Tiwari A.K., Wu C.P., Su X.D., Wang S.R., Liu D.G. Lapatinib (Tykerb, GW572016) reverses multidrug resistance in cancer cells by inhibiting the activity of ATP-binding cassette subfamily B member 1 and G member 2. Cancer Res. 2008;68:7905–7914. doi: 10.1158/0008-5472.CAN-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pawarode A., Shukla S., Minderman H., Fricke S.M., Pinder E.M., O'Loughlin K.L. Differential effects of the immunosuppressive agents cyclosporin A, tacrolimus and sirolimus on drug transport by multidrug resistance proteins. Cancer Chemother Pharmacol. 2007;60:179–188. doi: 10.1007/s00280-006-0357-8. [DOI] [PubMed] [Google Scholar]
- 124.Chearwae W., Wu C.P., Chu H.Y., Lee T.R., Ambudkar S.V., Limtrakul P. Curcuminoids purified from turmeric powder modulate the function of human multidrug resistance protein 1 (ABCC1) Cancer Chemother Pharmacol. 2006;57:376–388. doi: 10.1007/s00280-005-0052-1. [DOI] [PubMed] [Google Scholar]
- 125.Chearwae W., Shukla S., Limtrakul P., Ambudkar S.V. Modulation of the function of the multidrug resistance-linked ATP-binding cassette transporter ABCG2 by the cancer chemopreventive agent curcumin. Mol Cancer Ther. 2006;5:1995–2006. doi: 10.1158/1535-7163.MCT-06-0087. [DOI] [PubMed] [Google Scholar]
- 126.Ozvegy-Laczka C., Hegedus T., Várady G., Ujhelly O., Schuetz J.D., Váradi A. High-affinity interaction of tyrosine kinase inhibitors with the ABCG2 multidrug transporter. Mol Pharmacol. 2004;65:1485–1495. doi: 10.1124/mol.65.6.1485. [DOI] [PubMed] [Google Scholar]
- 127.Dohse M., Scharenberg C., Shukla S., Robey R.W., Volkmann T., Deeken J.F. Comparison of ATP-binding cassette transporter interactions with the tyrosine kinase inhibitors imatinib, nilotinib, and dasatinib. Drug Metab Dispos. 2010;38:1371–1380. doi: 10.1124/dmd.109.031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rarey M., Stahl M. Similarity searching in large combinatorial chemistry spaces. J Comput Aided Mol Des. 2001;15:497–520. doi: 10.1023/a:1011144622059. [DOI] [PubMed] [Google Scholar]
- 130.Rarey M., Dixon J.S. Feature trees: a new molecular similarity measure based on tree matching. J Comput Aided Mol Des. 1998;12:471–490. doi: 10.1023/a:1008068904628. [DOI] [PubMed] [Google Scholar]
- 131.Norman B.H., Lander P.A., Gruber J.M., Kroin J.S., Cohen J.D., Jungheim L.N. Cyclohexyl-linked tricyclic isoxazoles are potent and selective modulators of the multidrug resistance protein (MRP1) Bioorg Med Chem Lett. 2005;15:5526–5530. doi: 10.1016/j.bmcl.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 132.Wang S., Wan N.C., Harrison J., Miller W., Chuckowree I., Sohal S. Design and synthesis of new templates derived from pyrrolopyrimidine as selective multidrug-resistance-associated protein inhibitors in multidrug resistance. J Med Chem. 2004;47:1339–1350. doi: 10.1021/jm0310129. [DOI] [PubMed] [Google Scholar]
- 133.Sommer K., Friedrich N.O., Bietz S., Hilbig M., Inhester T., Rarey M. UNICON: A Powerful and Easy-to-Use Compound Library Converter. J Chem Inf Model. 2016;56:1105–1111. doi: 10.1021/acs.jcim.6b00069. [DOI] [PubMed] [Google Scholar]
- 134.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clark A.M., Labute P. Detection and assignment of common scaffolds in project databases of lead molecules. J Med Chem. 2009;52:469–483. doi: 10.1021/jm801098a. [DOI] [PubMed] [Google Scholar]
- 136.Jordan A.M., Roughley S.D. Drug discovery chemistry: a primer for the non-specialist. Drug Discov Today. 2009;14:731–744. doi: 10.1016/j.drudis.2009.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.