Abstract
Objective
To quantify the background incidence rates of 15 prespecified adverse events of special interest (AESIs) associated with covid-19 vaccines.
Design
Multinational network cohort study.
Setting
Electronic health records and health claims data from eight countries: Australia, France, Germany, Japan, the Netherlands, Spain, the United Kingdom, and the United States, mapped to a common data model.
Participants
126 661 070 people observed for at least 365 days before 1 January 2017, 2018, or 2019 from 13 databases.
Main outcome measures
Events of interests were 15 prespecified AESIs (non-haemorrhagic and haemorrhagic stroke, acute myocardial infarction, deep vein thrombosis, pulmonary embolism, anaphylaxis, Bell’s palsy, myocarditis or pericarditis, narcolepsy, appendicitis, immune thrombocytopenia, disseminated intravascular coagulation, encephalomyelitis (including acute disseminated encephalomyelitis), Guillain-Barré syndrome, and transverse myelitis). Incidence rates of AESIs were stratified by age, sex, and database. Rates were pooled across databases using random effects meta-analyses and classified according to the frequency categories of the Council for International Organizations of Medical Sciences.
Results
Background rates varied greatly between databases. Deep vein thrombosis ranged from 387 (95% confidence interval 370 to 404) per 100 000 person years in UK CPRD GOLD data to 1443 (1416 to 1470) per 100 000 person years in US IBM MarketScan Multi-State Medicaid data among women aged 65 to 74 years. Some AESIs increased with age. For example, myocardial infarction rates in men increased from 28 (27 to 29) per 100 000 person years among those aged 18-34 years to 1400 (1374 to 1427) per 100 000 person years in those older than 85 years in US Optum electronic health record data. Other AESIs were more common in young people. For example, rates of anaphylaxis among boys and men were 78 (75 to 80) per 100 000 person years in those aged 6-17 years and 8 (6 to 10) per 100 000 person years in those older than 85 years in Optum electronic health record data. Meta-analytic estimates of AESI rates were classified according to age and sex.
Conclusion
This study found large variations in the observed rates of AESIs by age group and sex, showing the need for stratification or standardisation before using background rates for safety surveillance. Considerable population level heterogeneity in AESI rates was found between databases.
Introduction
On 11 March 2020, the World Health Organization declared the outbreak of covid-19, caused by the SARS-CoV-2 virus, a global pandemic. As of March 2021, more than 100 million confirmed cases and 2.7 million deaths have been reported worldwide.1 Vaccines for covid-19 have been developed at unprecedented speed, with phase III clinical efficacy trials reporting results for some vaccines less than a year after WHO declared the pandemic. Since December 2020, several vaccines have been authorised by regulators such as the European Medicines Agency, the US Food and Drug Administration, and the UK Medicines and Healthcare products Regulatory Agency. Large scale immunisation programmes are ongoing worldwide.
Although the speed of vaccine development should be acknowledged, most vaccines have received approval for emergency use only, based on limited trial data. Uncertainty remains about the safety and effectiveness of these vaccines in populations other than those analysed in trials. As with all medicinal products, vaccine safety must continue to be monitored after regulatory authorisation to complement what was learnt during clinical development. Spontaneous adverse event reporting is a foundational component of post-approval pharmacovigilance activities to ensure the safe and appropriate use of medicinal products. Observational healthcare data captured during routine clinical care, such as electronic health records and administrative claims, can augment pharmacovigilance by providing real world information about potential adverse events and the rates of such events in populations of interest. Background rates of adverse events have historically played an important role in monitoring the safety of vaccines by serving as a baseline comparator for observed rates among those vaccinated.2 3 Each new vaccine has potential adverse events of special interest (AESIs) that warrant focused evaluation, based on what is known about previous vaccines and a vaccine’s development.
Regulatory agencies around the world have been preparing safety surveillance strategies for covid-19 vaccines. The FDA Center for Biologics Evaluation and Research published a protocol on background AESI rates to monitor the safety of covid-19 vaccines.4 The vACCine covid-19 monitoring readinESS (ACCESS) project funded by the EMA also included estimation of background AESI rates in its protocol.5 The WHO Council for International Organizations of Medical Sciences recommends using a local population’s own data when defining background rates of AESIs for comparison.6 The Observational Health Data Sciences and Informatics community therefore collaborated to design and execute an international open science study to characterise the background rates of AESIs potentially associated with covid-19 vaccines. We carried out this population based network retrospective cohort study, using observational data from 13 databases in eight countries—Australia, France, Germany, Japan, the Netherlands, Spain, the UK, and the US—to describe, in an epidemiological context, AESIs potentially associated with covid-19 vaccines.
Methods
Data sources
Data were obtained from 13 databases—eight comprising electronic health records and five comprising administrative claims.
The electronic health record databases were: IQVIA Australia Electronic Medical Records (IQVIA_AUSTRALIA); Integrated Primary Care Information (IPCI_NETHERLANDS), a primary care records database from the Netherlands7; IQVIA Longitudinal Patient Data France (IQVIA_FRANCE)8; IQVIA Disease Analyser Germany (IQVIA_GERMANY); Information System for Research in Primary Care (SIDIAP_H_SPAIN),9 a primary care records database that covers more than 80% of the population of Catalonia, Spain; Clinical Practice Research Datalink (CPRD_GOLD_UK), which consists of data collected from UK primary care for all ages10; Columbia University Irving Medical Center (CUMC_US), which covers more than 4.5 million people treated at the New York-Presbyterian Hospital/Columbia University Irving Medical Center in the US; and Optum De-Identified Electronic Health Record Dataset (OPTUM_EHR_US), which covers more than 103 million patients and more than 7000 hospitals and clinics across the US.11
The claims based databases were the Japan Medical Data Center (JMDC_JAPAN)12 and four US administrative claims databases: IBM MarketScan Commercial Claims and Encounters Database (CCAE_US),13 IBM MarketScan Medicare Supplemental and Coordination of Benefits Database (MDCR_US), IBM MarketScan Multi-State Medicaid Database (MDCD_US), and Optum De-Identified Clinformatics Data Mart Database–Socio-Economic Status (OPTUM_SES_US).11
The CPRD-GOLD (UK), IQVIA (France, Germany, and Australia), and IPCI (the Netherlands) databases included primary care data, not information on hospital admissions. Regional electronic health records data such as in the CUMC_US might incompletely capture medical events that are recorded in other healthcare institutions. The claims based sources offered relatively complete data on inpatient, outpatient, and prescriptions and treatment, but lacked measurement data and laboratory results (see table 1 and appendix table 1 for detailed descriptions of the databases).
Table 1.
Characteristics of included populations, stratified by database. Values are numbers (percentages) unless stated otherwise
| CCAE_US | MDCD_US | MDCR_US | OPTUM_EHR_US | OPTUM_SES_US | CUMC_US | CPRD_GOLD_UK | IPCI_NETHERLANDS | SIDIAP_H_SPAIN | IQVIA_FRANCE | IQVIA_GERMANY | IQVIA_AUSTRALIA | JMDC_JAPAN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full name | IBM MarketScan Commercial claims and encounters Database | IBM MarketScan Multi-State Medicaid Database | IBM MarketScan Medicare Supplemental and Coordination of Benefits Database | Optum Dei-Identified Electronic Health Record Data | Optum De-Identified Clinformatics Data Mart Database–Socio-Economic Status | Columbia University Irving Medical Center | Clinical Practice Research Datalink | Integrated Primary Care Information | Information System for Research in Primary Care-Hospitalization Linked Data | IQVIA Longitudinal Patient Data France | IQVIA Disease Analyser Germany | IQVIA Australia Electronic Medical Records | Japan Medical Data Center |
| Data type | Claims | Claims | Claims | EHR | Claims | EHR | EHR | EHR | EHR | EHR | EHR | EHR | Claims |
| Country | US | US | US | US | US | US | UK | Netherlands | Spain | France | Germany | Australia | Japan |
| Total No of patients | 25 315 777 | 12 966 011 | 1 533 709 | 40 955 085 | 18 643 608 | 1 164 196 | 4 532 766 | 1 536 283 | 2 217 536 | 1 746 371 | 9 295 525 | 252 212 | 6 501 991 |
| Person years | 42 889 550 | 23 203 712 | 2 484 782 | 72 328 897 | 32 474 685 | 2 174 312 | 9 638 136 | 3 326 570 | 5 497 613 | 3 008 350 | 16 784 613 | 383 668 | 12 848 482 |
| Age group (years): | |||||||||||||
| 1-5 | 1 256 501 (4.96) | 1 755 796 (13.54) | 0.0 | 1 852 425 (4.52) | 627 032 (3.36) | 40 678 (3.49) | 245 525 (5.42) | 78 848 (5.13) | 99 838 (4.50) | 99 309 (5.69) | 308 728 (3.32) | 13 430 (5.32) | 414 167 (6.37) |
| 6-17 | 4 122 110 (16.28) | 4 188 247 (32.30) | 0.0 | 4 773 000 (11.65) | 1 930 638 (10.36) | 105 520 (9.06) | 635 115 (14.01) | 211 037 (13.74) | 260 102 (11.73) | 268 591 (15.38) | 823 235 (8.86) | 31 780 (12.60) | 1 044 041 (16.06) |
| 18-34 | 6 395 387 (25.26) | 2 885 991 (22.26) | 0.0 | 8 182 549 (19.98) | 3 331 356 (17.87) | 199 020 (17.10) | 946 153 (20.87) | 304 971 (19.85) | 374 994 (16.91) | 328 759 (18.83) | 1 411 620 (15.19) | 50 995 (20.22) | 1 533 866 (23.59) |
| 35-54 | 8 096 864 (31.98) | 2 006 493 (15.48) | 0.0 | 10 737 664 (26.22) | 4 389 220 (23.54) | 300 818 (25.84) | 1 217 618 (26.86) | 394 868 (25.70) | 663 537 (29.92) | 446 804 (25.58) | 2 338 535 (25.16) | 69 872 (27.70) | 2 330 010 (35.84) |
| 55-64 | 4 716 207 (18.63) | 1 004 957 (7.75) | 0.0 | 6 655 199 (16.25) | 2 384 571 (12.79) | 183 612 (15.77) | 594 115 (13.11) | 219 990 (14.32) | 288 494 (13.01) | 229 016 (13.11) | 1 580 565 (17.0) | 36 329 (14.40) | 880 065 (13.54) |
| 65-74 | 728 708 (2.88) | 633 262 (4.88) | 733 157 (47.80) | 4 829 968 (11.79) | 3 106 611 (16.66) | 171 940 (14.77) | 469 682 (10.36) | 180 581 (11.75) | 246 763 (11.13) | 197 816 (11.33) | 1 279 048 (13.76) | 27 272 (10.81) | 279 277 (4.30) |
| 75-84 | 0.0 | 341 267 (2.63) | 536 970 (35.01) | 2 652 453 (6.48) | 1 985 356 (10.65) | 110 883 (9.52) | 290 225 (6.40) | 104 288 (6.79) | 180 903 (8.16) | 117 067 (6.70) | 1 191 402 (12.82) | 15 319 (6.07) | 20 565 (0.32) |
| ≥85 | 0.0 | 149 998 (1.16) | 263 582 (17.19) | 1 271 827 (3.11) | 888 824 (4.77) | 51 725 (4.44) | 134 333 (2.96) | 41 700 (2.71) | 102 905 (4.64) | 59 009 (3.38) | 362 392 (3.90) | 7215 (2.86) | 0.0 |
| Sex: | |||||||||||||
| Female | 13 037 440 (51.50) | 7 322 471 (56.47) | 849 301 (55.38) | 23 220 748 (56.70) | 9 595 675 (51.47) | 693 190 (59.54) | 2 287 698 (50.47) | 783 660 (51.01) | 1 120 373 (50.52) | 926 180 (53.03) | 5 340 273 (57.45) | 137 203 (54.40) | 2 926 702 (45.01) |
| Male | 12 278 337 (48.50) | 5 643 540 (43.53) | 684 408 (44.62) | 17 734 337 (43.30) | 9 047 933 (48.53) | 471 006 (40.46) | 2 245 068 (49.53) | 752 623 (48.99) | 1 097 163 (49.48) | 820 191 (46.97) | 3 955 252 (42.55) | 115 009 (45.60) | 3 575 289 (54.99) |
EHR=electronic health record.
All datasets were previously mapped to the Observational Medical Outcomes Partnership common data model, which is maintained by the Observational Health Data Sciences and Informatics network, an international open science initiative to generate reproducible evidence from observational data.14 This initiative brings together hundreds of researchers from 30 countries, working with health records from around 600 million unique patients in its distributed database. The analysis code was distributed across all centres contributing to Observational Health Data Sciences and Informatics without sharing patient level data.15 16
Study participants
The study period was from 1 January 2017 to 31 December 2019. We defined the target at risk population as people who were observed on 1 January 2017, 1 January 2018, or 1 January 2019 and were observed for at least 365 days before this observation date. The 1 January each year was defined as the index date.
Events of interest
The events of interest in this study were AESIs that might need evaluation after covid-19 vaccination. This list of outcomes was based on the protocol published by the FDA Center for Biologics Evaluation and Research, the prioritised covid-19 vaccine AESI list by the Brighton Collaboration, and previous studies.4 17 We included 15 events: non-haemorrhagic and haemorrhagic stroke, acute myocardial infarction, deep vein thrombosis, pulmonary embolism, anaphylaxis, Bell’s palsy, myocarditis or pericarditis, narcolepsy, appendicitis, immune thrombocytopenia, disseminated intravascular coagulation, encephalomyelitis (including acute disseminated encephalomyelitis), Guillain-Barré syndrome, and transverse myelitis.4
Events were identified by records of the occurrence of conditions based on predefined phenotyping algorithms (eg, diagnosis codes from claims or diagnosis codes and problem lists from electronic health records). Definitions for encephalomyelitis, non-haemorrhagic and haemorrhagic stroke, and acute myocardial infarction also required the record to occur within an inpatient setting in any diagnosis positions, whereas the definition for Guillain-Barré syndrome required the condition to be recorded in an inpatient setting in the primary position. Appendix tables 2 and 3 present the full specifications of all phenotype definitions, including source codes (original codes used in the database) and standard concepts (normative expressions used to represent a unique clinical entity within the Observational Medical Outcomes Partnership common data model, which were mostly SNOMED (Systematized Nomenclature of Medicine) codes in this study).
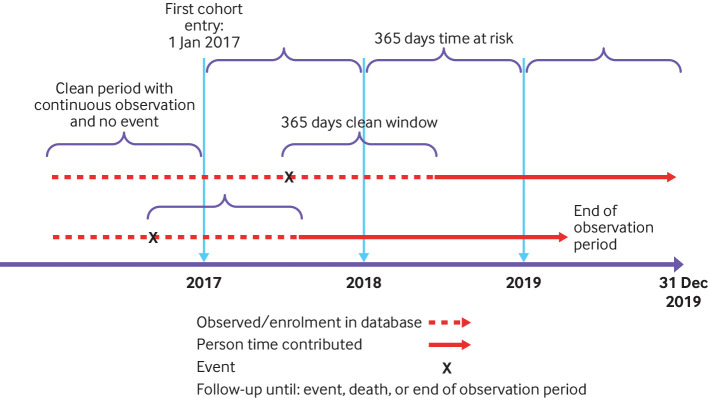
We defined a “clean window” period before each index date, during which qualifying events (AESIs) could not be observed. If an AESI was observed during this period, the participant did not enter the study cohort for that event. If an individual had a qualified event during follow-up, this participant would contribute to the person time of that event cohort after the clean window continually until censored from the cohort.
Figure 1 shows the cohort entry, follow-up, and event definitions. In keeping with the FDA protocol, the clean window was 365 days for all events except anaphylaxis (30 days) and facial nerve palsy and encephalomyelitis (183 days).4
Fig 1.
Study design
As the CPRD-GOLD (UK), IQVIA (France, Germany, and Australia), and IPCI (the Netherlands) databases only included primary care data, we did not use them for events where definition required an inpatient diagnosis.
Statistical analysis
We defined the time at risk as a 365 day period after the index date. Eligible people contributed time at risk from 1 January to 31 December for each qualifying year in 2017 to 2019. Participants were censored if an event occurred during that event’s clean window, at death (if available in the data source), or at the end of their observation period in the database (fig 1). One participant could contribute more than one event. To avoid duplicate counts we used outcome specific prespecified clean windows of 30 to 365 days.
Incidence rates were estimated as the total number of events divided by the person time at risk per 100 000 person years. We calculated the age and sex specific incidence rates in each database and report all rates when the event counts exceeded a minimum cell count of 5. Age was calculated as year of index date minus year of birth and was partitioned into eight mutually exclusive age groups (in years): 1-5, 6-17, 18-34, 35-45, 55-64, 65-74, 75-84, and 85 and older. Age-sex specific rates of each AESI were pooled across all databases using a random effects meta-analysis, with the DerSimonian-Laird method to estimate variance between databases.18 We estimated 95% prediction intervals using the R package “meta”.19 The prediction interval reflects the expected uncertainty if an estimate rate from a new study is included in the meta-analysis.20
Meta-analytic age and sex specific rates were classified using the WHO Council for International Organizations of Medical Sciences thresholds: very common (≥1/10), common (<1/10 to ≥1/100), uncommon (<1/100 to ≥1/1000), rare (<1/1000 to ≥1/10 000), and very rare (<1/10 000).21
All statistical analyses were performed in R software.22 The study protocol and analysis code are available at https://github.com/ohdsi-studies/Covid19VaccineAesiIncidenceCharacterization.
Patient and public involvement
No patients or members of the public were directly involved in the design or analysis of the reported data. Because of covid-19 related restrictions, it has been difficult to interact with relevant patient and public representatives. Some of the contributing databases did, however, involve patients in the evaluation of our data access application.
Results
From the 13 databases, 126 661 070 people contributed 227 043 370 person years of follow-up. The Optum De-Identified Electronic Health Record Dataset (OPTUM_EHR_US) contributed the largest number of patients (n=40 955 085), followed by the IBM MarketScan Commercial Claims and Encounters Database (CCAE_US). Each database captured important population demographics and collectively represented all age and sex subgroups from eight countries (table 1).
Most of the databases included more female than male patients (ranging from 50.5% female patients in SIDIAP_H_SPAIN to 57.5% in IQVIA_GERMANY), except for JMDC_JAPAN (45.0% female patients). The CCAE_US database included patients aged 0-74 years, whereas the MDCR_US database only included patients older than 65 years. The other databases included patients of all ages. Patients aged 35-54 years accounted for the largest proportion of the population in most databases (from 23.5% in OPEUM_SES_US to 35.8% in JMDC_JAPAN). Patients aged 18-34 years, however, accounted for the largest proportion (22.3%) of the IBM MarketScan Multi-State Medicaid Database (MDCD_US) database. The proportion of patients aged 65 years and older ranged from 32.1% in the OPTUM_SES_US database to less than 10% in the CCAE_US, JMDC_JAPAN, and MDCD_US databases. Patients younger than 18 years accounted for 45.8% of the MDCD_US database.
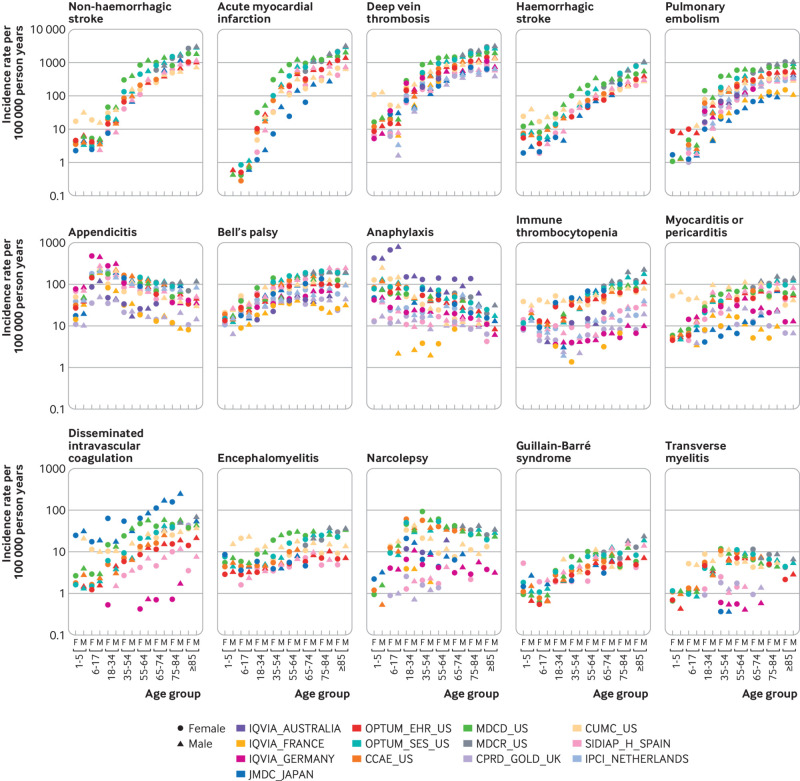
Substantial heterogeneity was observed by age group and sex in the database specific estimates of each event but similar age and sex trends in most databases and the pooled rates (fig 2). For example, the incidence rates of deep vein thrombosis increased with age. In the OPTUM_SES_US data, the incidence rates increased from 20 (95% confidence interval 19 to 22) per 100 000 person years among boys aged 6-17 years to 2030 (2009 to 2051) per 100 000 person years among men aged 75-84 years (see appendix table 4). Similarly, myocardial infarction rates among men increased from 28 (95% confidence interval 27 to 29) per 100 000 person years in those aged 18-34 years to 1400 (1374 to 1427) per 100 000 person years in those older than 85 years in the OPTUM_EHR_US data. The incidence rates for haemorrhagic and non-haemorrhagic stroke, pulmonary embolism, Bell’s palsy, immune thrombocytopenia, Guillain-Barré syndrome, and disseminated intravascular coagulation also increased with age.
Fig 2.
Age and sex stratified incidence rates for 15 adverse events of special interest by database. CCAE_US=IBM MarketScan Commercial Claims and Encounters Database, CPRD_GOLD_UK=Clinical Practice Research Datalink; CUMC_US=Columbia University Irving Medical Center; IPCI_NETHERLANDS=Integrated Primary Care Information; IQVIA_AUSTRALIA=IQVIA Australia Electronic Medical Records; IQVIA_FRANCE=IQVIA Longitudinal Patient Data France; IQVIA_GERMANY=IQVIA Disease Analyser Germany; JMDC_JAPAN=Japan Medical Data Center, MDCD_US=IBM MarketScan Multi-State Medicaid Database, MDCR_US=IBM MarketScan Medicare Supplemental and Coordination of Benefits Database; OPTUM_EHR_US=Optum De-Identified Electronic Health Record Dataset; OPTUM_SES_US=Optum De-Identified Clinformatics Data Mart Database-Socio-Economic Status; SIDIAP_H_SPAIN=Information System for Research in Primary Care-Hospitalization Linked Data
The rates of haemorrhagic stroke were higher in male participants than female participants in most age groups. For example, the incidence rates among those aged 65-74 years in the MDCR_US database were 251 (238 to 265) per 100 000 person years for male participants and 170 (160 to 180) per 100 000 person years for female participants. The incidence rates of acute myocardial infraction, myocarditis or pericarditis, immune thrombocytopenia, and Guillain-Barré syndrome were also higher in male participants than female participants.
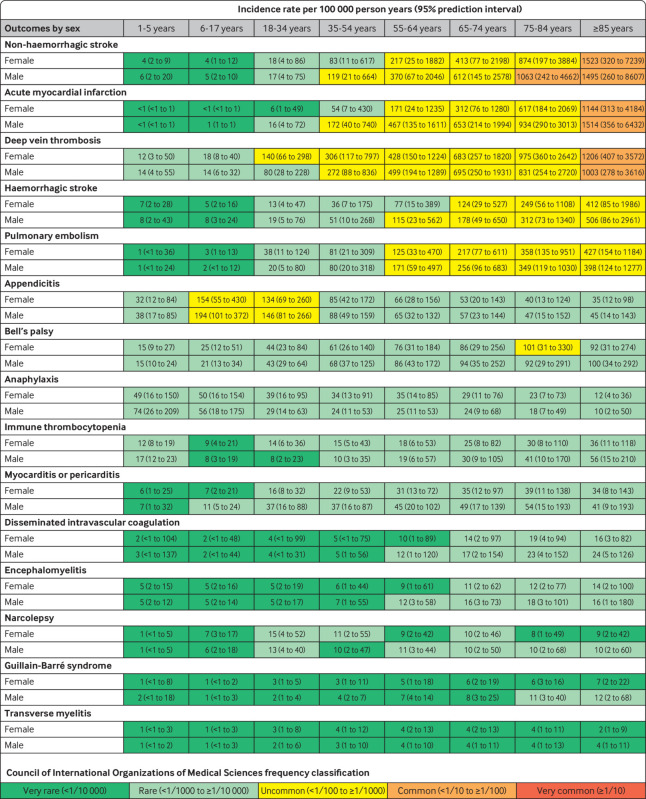
Figure 3 summarises the pooled incidence rates of the 15 AESIs, stratified by age and sex, based on prediction intervals from a meta-analysis of the database estimates. Each age and sex subgroup was classified using the Council for International Organizations of Medical Sciences adverse event frequency system (very common, common, uncommon, rare, or very rare). The AESIs studied spanned the continuum of possible frequencies. The incidence of several events varied substantially by age and was therefore classified differently at different ages. For example, deep vein thrombosis was rare in boy and girl participants younger than 18 years, uncommon in those aged 35-84 years, and common in those 85 years and older. Acute myocardial infarction was very rare (<1/10 000) in women younger than 35 years, rare (<1/1000 to ≥1/10 000) in women aged 35-54 years, uncommon (<1/100 to ≥1/1000) in both men and women aged 55-84 years, and common (<1/10 to ≥1/100) in men and women aged 85 years and older. Anaphylaxis, Bell’s palsy, appendicitis, and immune thrombocytopenia were largely rare in all age groups, although appendicitis was uncommon in those aged 6-34 years. Guillain-Barré syndrome and transverse myelitis were very rare in nearly all subgroups.
Fig 3.
Pooled estimated age and sex stratified incidence rates per 100 000 person years (95% prediction intervals), calculated from meta-analyses
The rates recorded for deep vein thrombosis highlight the population level variation. Thirteen database estimates were obtained for the incidence in women aged 65-74 years, ranging from 387 (370 to 404) per 100 000 person years in CPRD-GOLD to 1443 (1416 to 1470) per 100 000 person years in MDCD_US. The rates in eight databases were less than 650 per 100 000 person years (CPRD-GOLD in the UK; CUIMC in the US; IPCI in the Netherlands; IQVIA in Australia, France, and Germany; JMDC in Japan; and SIDIAP-H in Spain), whereas the rates in three databases were more than twice as high, at more than 1300 per 100 000 person years (MDCD, MDCR, and OPTUM-SES in the US). For women aged 35-54 years, the incidence rates ranged from 159 (151 to 167) per 100 000 person years in Spain (SIDIAP) to 866 (854 to 878) per 100 000 person years in the US (MDCD). Among women aged 75-84 years, the lowest incidence rate was 585 (559 to 612) per 100 000 person years (CPRD-GOLD in the UK) and the highest was 2167 (2126 to 2210) per 100 000 person years (MDCR in the US). No consistent patterns could be found to explain why certain databases yielded higher or lower rates across outcomes. Database specific rates are shown in figure 2, and the raw numbers are available in appendix table 4 and our interactive web app (https://data.ohdsi.org/Covid19VaccineAesiIncidenceCharacterization/).
Discussion
We conducted a multinational network cohort study on the descriptive epidemiology of the AESIs prioritised for post-marketing surveillance of covid-19 vaccines. We report background rates of deep vein thrombosis, pulmonary embolism, stroke, immune thrombocytopenia, and disseminated intravascular coagulation. These events are particularly relevant for covid-19 vaccines as the SARS-COV-2 virus has been observed to affect coagulopathy.23 24 25 26 We assessed the incidence rates of 15 AESIs across 13 databases, eight countries, and four continents. We observed considerable variability with age and sex, emphasising the need for standardisation or stratification of the background rates used for vaccine surveillance. We found substantial heterogeneity between databases, suggesting that, where possible, post-covid-19 vaccine rates should be compared with background, or historical, rates obtained from the same dataset.
Research in context
In vaccine safety surveillance, background incidence rates have been used to estimate the expected number of events in the general population, which is also known as the historical rates comparison method.3 27 Several vaccine safety surveillance guidelines, such as the European Network of Centres of Pharmacoepidemiology and Pharmacovigilance, suggest using this historical rate comparison method, as it increases statistical power to detect rare events and helps to detect signals sooner than other methods. This method has been widely implemented in many countries and by many organisations, including the Vaccine Safety Datalink project in the US28 and the Vaccine Adverse Event Surveillance and Communication project in Europe.3 These background rates are often obtained from the literature or healthcare databases. However, use of historical rate comparisons has some limitations owing to the methods used to obtain the rates, case and population definitions, differences in clinical codes, and geographical and temporal variations.28 We overcame these common limitations by calculating the presented estimates using the same setting and common analysis procedures, phenotyping algorithms, and common data model.
We found substantial population level heterogeneity across data sources for all events, even after standardising outcome definitions and stratifying by age and sex. For example, we observed about a threefold difference between the highest and lowest incidence rates for deep vein thrombosis measured in each database. Previous studies using one or a small number of databases have also observed these variations. US based studies, for example, recorded incidence rates per 100 000 person years for idiopathic thrombocytopenia of around 3 among men and women aged 26-62 years,29 9 among those aged 25-44 years, and 12 among those aged 45-64 years.30 We estimated incidence rates of transverse myelitis ranging from 1 to 4 per 100 000 person years in meta-analyses, depend on age and sex strata. Previous studies have reported overall incidence rates of transverse myelitis ranging from 0.4 to 4.6 per 100 000 person years.27 31
Recorded rates of Bell’s palsy among those older than 65 years have ranged from 4.6 per 100 000 person years in Italian data to 174 per 100 000 person years in US data.30 32 We found similar rates to those previously published using US claims databases and UK general practice data, but higher rates when using Spanish data. A previous study using data from regions in Spain not captured by the SIDIAP database found an incidence rate of 63 per 100 000 person years among those older than 65 years.32 We observed rates of 131 and 182 per 100 000 person years in women and men aged 65-74 years, respectively, using SIDIAP Spanish data (SIDIAP_H_SPAIN). A US study found rates of narcolepsy between 31 and 38 per 100 000 person years among those aged 25-64 years, whereas studies based on European data have found much lower rates of between 0.2 and 2.5 per 100 000 person years for the same age group.3 30 32 We also found higher rates of narcolepsy in US databases than those from other countries. Recently reported data from the ACCESS project also showed heterogeneity in background rates.5 This heterogeneity must be considered when comparing rates across populations.
Most of the studied outcomes also had considerable within source patient level heterogeneity that followed age and sex patterns. We observed that the rates of cardiovascular diseases such as acute myocardial infarction, haemorrhagic and non-haemorrhagic stroke, deep vein thrombosis, and pulmonary embolism increased with age. The incidence of Guillain-Barré syndrome and Bell’s palsy also increased with age. Narcolepsy and appendicitis were more common in younger populations. The patterns observed in our study were generally comparable with those of previous reports.2 3 29 30 32 33 34 Stratification by age and sex and standardisation are likely to be useful analytical strategies to reduce confounding when incidence rates are compared across populations. The observed magnitude of heterogeneity across sources within age and sex subgroups, however, suggests that residual patient level differences will remain, including differences in the distributions of other risk factors, such as comorbidities and medication use.
Comparing published results can be complicated by differing study methods, including the time-at-risk definition, study period, event definitions, population coverage, calendar year, and geographical location.35 Different subgroup definitions also make direct comparison difficult. Previous studies of Guillain-Barré syndrome, for example, have used age strata that do not fully overlap with each other.2 5 29 32 36 37 As we used the same definitions, data model, and analysis with all of our studied databases, the heterogeneity we observed cannot be attributed to variability in analysis. This remaining heterogeneity might have resulted from differences in the underlying populations, healthcare systems, and data capture processes. Although some variability might have been due to systematic error, selection bias, or differential outcome measurement error between databases, some could reflect true population differences, such as socioeconomic status and comorbidities.
As we observed notable differences in incidence rates by age, sex, and database, caution is needed when incidence rates are compared across time or populations. Incidence rates from different sources might be subject to substantial systematic error. Given this heterogeneity, the reported 95% confidence intervals for the database specific rates in our study cannot reflect the systematic errors in the rate estimation. The notably wide prediction intervals for each age and sex subgroup also reflect the substantial population level heterogeneity observed across sources. We observed large variations between electronic health records and claims data sources when using the same analysis and outcome definitions. Variability in rates derived from randomised trials or spontaneous reporting data could be even greater. If observational databases are to be used to inform safety surveillance activities, within database analyses (such as self-controlled case designs or propensity score adjusted comparative cohort designs) may help reduce study bias for any given comparison. Showing consistent effects across databases may further strengthen confidence in results. If observational data are used to derive historical “expected” rates and compared with observed rates of events from another source, then the uncertainty in the background rate must be appropriately integrated to avoid misleading conclusions.
Strengths of this study
The large number of participating databases, geographical coverage, and sizable study population enabled a comprehensive assessment of background incidence rates of AESIs across different healthcare systems and regions worldwide. This study is an example of the collaborative projects possible within the Observational Health Data Sciences and Informatics network. This initiative stores records using a common data model38 and standard vocabularies and develops and uses state-of-the-art methods to draw and validate causal conclusions.39 It has generated impactful evidence in many areas, such as hypertension treatment,40 41 and was able to pivot quickly to generate policy influencing evidence in covid-19 management.42 43 In our study we took advantage of the Observational Medical Outcomes Partnership common data model, which enabled us to use the same study design and analytical code in all databases and to gather results from participating data partners rapidly and without transferring patient level data. All outcome definitions, clinical codes, and phenotype algorithms have been made open source and are available online for review and to maximise reproducibility and reuse. The large scale use of the Observational Medical Outcomes Partnership common data model and open source science strategy has enabled us to generate useful, timely evidence on upcoming covid-19 vaccine safety. We welcome new data partners to run the analyses on their datasets, contribute their results to our web application, and participate in further Observational Health Data Sciences and Informatics studies.
Limitations of this study
The primary limitation of this study is that all outcomes could have been subject to measurement error. As the outcome definitions were based on the presence of specific diagnostic codes and were not validated further, sensitivity or specificity could have been imperfect. Our analysis relied on data from 2017 to 2019 using a target population of all people in each database with more than 365 days of observation indexed on 1 January, 365 days’ time at risk, and outcome specific clean windows to allow for recurrent events. The impact of these design decisions should be explored further.
Some limitations relate to the use of each database. As information on hospital admission was not available in the primary care datasets used (CPRD-GOLD in the UK, IQVIA in France, Germany, and Australia, and IPCI in the Netherlands), events that happened during inpatient visits were not included. The electronic health records data sources were subject to incomplete capture of medical events recorded in other healthcare institutions. The bias of incomplete information was partially mitigated by including only those patients who had at least one year of continuous observation. The five administrative claims data sources offered reliable data capture but lacked data elements such as laboratory test results. The US based claims database did not record death information well. Our within database background rate comparison should have minimised bias related to these database specific limitations, mitigating against such limitations.
Conclusion
Our study assessed the descriptive epidemiology of potential AESIs for covid-19 vaccines. Our study highlights the wide range of adverse effects being monitored, from very rare neurological disorders to more common thromboembolic conditions. We found large variations in the observed rates of AESIs by age group and sex, showing the need for stratification or standardisation before using background rates for safety surveillance. Considerable population level heterogeneity was also found in AESI rates between databases, implying that individual study estimates should be interpreted with caution and that the systematic error associated with database choice should be incorporated into any analysis. We recommend that the same database be used to estimate post-covid-19 vaccine and background rates for comparison in vaccine safety monitoring. The database specific estimates reported here are available in a bespoke interactive web application for regulators and other stakeholders (https://data.ohdsi.org/Covid19VaccineAesiIncidenceCharacterization/). These background rates provide useful real world context to inform public health efforts aimed at ensuring patient safety while promoting the appropriate use of vaccines worldwide.
What is already known on this topic
Background rates of adverse events of special interest (AESIs) have historically played an important role in monitoring vaccine safety
Most studies focused on single or very few events and used different study designs, and none focused on specific AESIs for covid-19 vaccines
No international transcontinental study on background rates of covid-19 vaccine AESIs using the same definitions, data model, and analysis across all databases have been reported
What this study adds
This study found considerable heterogeneity between geographies and databases, suggesting caution when interpreting the differences between observed and expected rates
If possible, the same data source should be used to compare post-covid-19 vaccine (observed) and background (expected) AESI rates for vaccine surveillance
Considerable variability was also found in observed rates of AESIs between age groups and sex, showing the need for standardisation if background rates are used for surveillance purposes
Acknowledgments
This manuscript expresses the opinion of the authors of the paper and may not be understood or quoted as being made on behalf of or reflecting the position of the European Medicines Agency or one of its committees or working parties.
We thank Jennifer A de Beyer (Centre for Statistics in Medicine, University of Oxford) for English language editing.
Web extra.
Extra material supplied by authors
Supplementary information: Appendix tables 1-4 and figure 1
Contributors: XL and AO are joint first authors. DAP, XL, PR, and GH conceived the study. XL, AO, GH, PR, and DAP interpreted the results and wrote the manuscript. RM, AZ, GR, and AS implemented the study. XL, AO, PR, RM, KV, PRR, TDS, and MAS contributed to the study execution (data holders). XL, AO, GH, and PR contributed to the study design. All coauthors contributed to writing the manuscript. All authors approved the final version and had final responsibility for the decision to submit for publication. PRR, MAS, PBR, GH, and DPA are guarantors. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Funding: This work was partially funded by the UK National Institute for Health Research (NIHR), European Medicines Agency, European Health Data and Evidence Network (EHDEN), US Food and Drug Administration CBER BEST initiative (75F40120D00039), and US National Library of Medicine (R01 LM006910). EHDEN has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 806968. The Innovative Medicines Initiative 2 Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. The study funders had no role in the conceptualisation, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Competing interests: All authors have completed the ICMJE disclosure form at http://www.icmje.org/disclosure-of-interest/ and declare the following interests: DPA receives funding from the UK National Institute for Health Research (NIHR) in the form of a senior research fellowship and the Oxford NIHR Biomedical Research Centre. XL receives the Clarendon Fund and Brasenose College Scholarship (University of Oxford) to support her DPhil study. DPA’s research group has received research grants from the European Medicines Agency; the Innovative Medicines Initiative; Amgen, Chiesi, and UCB Biopharma; and consultancy or speaker fees from Astellas, Amgen, and UCB Biopharma. GH and AO receive funding from the US National Institutes of Health (NIH) and the US Food and Drug Administration. KV and PR work for a research group that receives/received unconditional research grants from Yamanouchi, Pfizer-Boehringer Ingelheim, Novartis, GSK, Amgen, and Chiesi, none of which relates to the content of this paper. PBR, RM, AS, GR, and AGS are employees of Janssen Research and Development and shareholders in Johnson and Johnson. MAS receives grants and contracts from the FDA and the US Department of Veterans Affairs within the scope of this research, and grants and contracts from NIH, IQVIA, and Private Health Management outside the scope of this research.
Ethical approval: The protocol for this research was approved by the independent scientific advisory committee for Medicine and Healthcare products Regulatory Agency database research (protocol number 20_000211), the IDIAPJGol (Fundacio Institut Universitari per a la recerca a l’Atencio Primaria de Salut Jordi Gol i Gurina) clinical research ethics committee (project code: 21/007-PCV), the Integrated Primary Care Information governance board (application number 3/2021), and Columbia University institutional review board (AAAO7805).
Data sharing: Patient level data cannot be shared without approval from data custodians owing to local information governance and data protection regulations. Aggregated data, analytical code, and detailed definitions of algorithms for identifying the events are available in a GitHub repository (https://github.com/ohdsi-studies/Covid19VaccineAesiIncidenceCharacterization). The results are available in an interactive web app (https://data.ohdsi.org/Covid19VaccineAesiIncidenceCharacterization).
The lead authors (XL and AO) affirm that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Dissemination to participants and related patient and public communities: We will disseminate a lay summary of our findings through our Twitter and other social media accounts. The involved universities will lead on the dissemination of our results to the lay audience, with help from our communications and outreach teams in the UK, US, and the Netherlands.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. WHO COVID-19 Dashboard. Geneva: World Health Organization 2020; Available at https://covid19.who.int/ (accessed 11 Mar 2021). [Google Scholar]
- 2. Black S, Eskola J, Siegrist C-A, et al. Importance of background rates of disease in assessment of vaccine safety during mass immunisation with pandemic H1N1 influenza vaccines. Lancet 2009;374:2115-22. 10.1016/S0140-6736(09)61877-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wijnans L, Lecomte C, de Vries C, et al. The incidence of narcolepsy in Europe: before, during, and after the influenza A(H1N1)pdm09 pandemic and vaccination campaigns. Vaccine 2013;31:1246-54. 10.1016/j.vaccine.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 4.Center for Biologics Evaluation and Research Office of Biostatistics and Epidemiology. CBER Surveillance Program Background Rates of Adverse Events of Special Interest for COVID-19 Vaccine Safety Monitoring Protocol. https://www.bestinitiative.org/wp-content/uploads/2021/02/C19-Vaccine-Safety-AESI-Background-Rate-Protocol-FINAL-2020.pdf (accessed 11 Mar 2021).
- 5.ACCESS. Background rates of Adverse Events of Special Interest for monitoring COVID-19 vaccines. 2020. http://www.encepp.eu/encepp/viewResource.htm?id=37274 (accessed 11 Mar 2021).
- 6.Report of CIOMS Working Group on Vaccine Safety. CIOMS Guide to Active Vaccine Safety Surveillance. https://cioms.ch/wp-content/uploads/2020/04/240WEB-CIOMS-Guide-AVSS-20170202-protected.pdf (accessed 11 Mar 2021). [DOI] [PubMed]
- 7. Vlug AE, van der Lei J, Mosseveld BM, et al. Postmarketing surveillance based on electronic patient records: the IPCI project. Methods Inf Med 1999;38:339-44. 10.1055/s-0038-1634402 [DOI] [PubMed] [Google Scholar]
- 8. Jouaville SL, Miotti H, Coffin G, et al. Validity and limitations of the longitudinal patient database France for use in pharmacoepidemiological and pharmacoeconomics studies. Value Health 2015;18:A18 10.1016/j.jval.2015.03.115. [DOI] [Google Scholar]
- 9. García-Gil MdelM, Hermosilla E, Prieto-Alhambra D, et al. Construction and validation of a scoring system for the selection of high-quality data in a Spanish population primary care database (SIDIAP). Inform Prim Care 2011;19:135-45. [DOI] [PubMed] [Google Scholar]
- 10. Herrett E, Gallagher AM, Bhaskaran K, et al. Data resource profile: Clinical practice research datalink (CPRD). Int J Epidemiol 2015;44:827-36. 10.1093/ije/dyv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wallace PJ, Shah ND, Dennen T, Bleicher PA, Crown WH. Optum Labs: building a novel node in the learning health care system. Health Aff (Millwood) 2014;33:1187-94. 10.1377/hlthaff.2014.0038 [DOI] [PubMed] [Google Scholar]
- 12. Nagai K, Tanaka T, Kodaira N, Kimura S, Takahashi Y, Nakayama T. Data resource profile: JMDC claims databases sourced from Medical Institutions. J Gen Fam Med 2020;21:211-8. 10.1002/jgf2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen L. The Truven health MarketScan databases for life sciences researchers. 2017. https://www.ibm.com/uk-en/products/marketscan-research-databases
- 14. Hripcsak G, Schuemie MJ, Madigan D, Ryan PB, Suchard MA. Drawing reproducible conclusions from observational clinical data with OHDSI. Yearb Med Inform 2021. 10.1055/s-0041-1726481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voss EA, Makadia R, Matcho A, et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc 2015;22:553-64. 10.1093/jamia/ocu023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform 2015;216:574-8. [PMC free article] [PubMed] [Google Scholar]
- 17.bc-coordinator. Priority list of adverse events of special interest: COVID-19. 2020. https://brightoncollaboration.us/priority-list-aesi-covid/ (accessed 11 Mar 2021).
- 18. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 19. Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health 2019;22:153-60. 10.1136/ebmental-2019-300117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riley RD, Higgins JPT, Deeks JJ. Interpretation of random effects meta-analyses. BMJ 2011;342:d549. 10.1136/bmj.d549 [DOI] [PubMed] [Google Scholar]
- 21.The Council for International Organizations of Medical Sciences (CIOMS). Guidelines for Preparing Core Clinical-Safety Information on Drugs Second Edition – Report of CIOMS Working Groups III and V. Geneva: 1999. [Google Scholar]
- 22.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2021. https://www.R-project.org/.
- 23. Wise J. Covid-19: European countries suspend use of Oxford-AstraZeneca vaccine after reports of blood clots. BMJ 2021;372:n699. 10.1136/bmj.n699 [DOI] [PubMed] [Google Scholar]
- 24.Coronavirus vaccine - weekly summary of Yellow Card reporting. https://www.gov.uk/government/publications/coronavirus-covid-19-vaccine-adverse-reactions/coronavirus-vaccine-summary-of-yellow-card-reporting (accessed 22 Mar 2021).
- 25.CoViD Vaccines and thrombotic events: Possibility of mRNA translation and spike protein synthesis by platelets? Published Online First: 15 March 2021. https://www.bmj.com/content/372/bmj.n699/rr-6 (accessed 22 Mar 2021).
- 26.COVID-19 Vaccine AstraZeneca: benefits still outweigh the risks despite possible link to rare blood clots with low blood platelets. 2021. https://www.ema.europa.eu/en/news/covid-19-vaccine-astrazeneca-benefits-still-outweigh-risks-despite-possible-link-rare-blood-clots (accessed 22 Mar 2021).
- 27. Black S, Eskola J, Siegrist C-A, et al. Importance of background rates of disease in assessment of vaccine safety during mass immunisation with pandemic H1N1 influenza vaccines. Lancet 2009;374:2115-22. 10.1016/S0140-6736(09)61877-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yih WK, Kulldorff M, Fireman BH, et al. Active surveillance for adverse events: the experience of the Vaccine Safety Datalink project. Pediatrics 2011;127(Suppl 1):S54-64. 10.1542/peds.2010-1722I [DOI] [PubMed] [Google Scholar]
- 29. Klein NP, Ray P, Carpenter D, et al. Rates of autoimmune diseases in Kaiser Permanente for use in vaccine adverse event safety studies. Vaccine 2010;28:1062-8. 10.1016/j.vaccine.2009.10.115 [DOI] [PubMed] [Google Scholar]
- 30. Baker MA, Nguyen M, Cole DV, Lee GM, Lieu TA. Post-licensure rapid immunization safety monitoring program (PRISM) data characterization. Vaccine 2013;31(Suppl 10):K98-112. 10.1016/j.vaccine.2013.04.088 [DOI] [PubMed] [Google Scholar]
- 31. Rasmussen TA, Jørgensen MRS, Bjerrum S, et al. Use of population based background rates of disease to assess vaccine safety in childhood and mass immunisation in Denmark: nationwide population based cohort study. BMJ 2012;345:e5823. 10.1136/bmj.e5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Willame C, Dodd C, van der Aa L, et al. Incidence rates of autoimmune diseases in European healthcare databases: A contribution of the ADVANCE project. Drug Saf 2021;44:383-95. 10.1007/s40264-020-01031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hense S, Schink T, Kreisel SH, et al. Estimation of background incidence rates of Guillain-Barré syndrome in Germany - a retrospective cohort study with electronic healthcare data. Neuroepidemiology 2014;43:244-52. 10.1159/000369344 [DOI] [PubMed] [Google Scholar]
- 34. Granieri E, Andreasi NG, De Martin P, et al. Incidence study of Guillain-Barré syndrome in the province of Ferrara, Northern Italy, between 2003 and 2017. A 40-year follow-up. Neurol Sci 2019;40:603-9. 10.1007/s10072-018-3688-4 [DOI] [PubMed] [Google Scholar]
- 35. Black SB, Law B, Chen RT, et al. The critical role of background rates of possible adverse events in the assessment of COVID-19 vaccine safety. Vaccine 2021;39:2712-8. 10.1016/j.vaccine.2021.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levison LS, Thomsen RW, Christensen DH, Mellemkjær T, Sindrup SH, Andersen H. Guillain-Barré syndrome in Denmark: validation of diagnostic codes and a population-based nationwide study of the incidence in a 30-year period. Clin Epidemiol 2019;11:275-83. 10.2147/CLEP.S199839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Maas NAT, Kramer MA, Jacobs BC, et al. Guillain-Barré syndrome: background incidence rates in The Netherlands. J Peripher Nerv Syst 2011;16:243-9. 10.1111/j.1529-8027.2011.00356.x [DOI] [PubMed] [Google Scholar]
- 38. Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012;19:54-60. 10.1136/amiajnl-2011-000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schuemie MJ, Ryan PB, Pratt N, et al. Principles of Large-scale Evidence Generation and Evaluation across a network of databases (LEGEND). J Am Med Inform Assoc 2020;27:1331-7. 10.1093/jamia/ocaa103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suchard MA, Schuemie MJ, Krumholz HM, et al. Comprehensive comparative effectiveness and safety of first-line antihypertensive drug classes: a systematic, multinational, large-scale analysis. Lancet 2019;394:1816-26. 10.1016/S0140-6736(19)32317-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hripcsak G, Suchard MA, Shea S, et al. Comparison of cardiovascular and safety outcomes of chlorthalidone vs hydrochlorothiazide to treat hypertension. JAMA Intern Med 2020;180:542-51. 10.1001/jamainternmed.2019.7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lane JCE, Weaver J, Kostka K, et al. OHDSI-COVID-19 consortium . Risk of hydroxychloroquine alone and in combination with azithromycin in the treatment of rheumatoid arthritis: a multinational, retrospective study. Lancet Rheumatol 2020;2:e698-711. 10.1016/S2665-9913(20)30276-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morales DR, Conover MM, You SC, et al. Renin-angiotensin system blockers and susceptibility to COVID-19: an international, open science, cohort analysis. Lancet Digit Health 2021;3:e98-114. 10.1016/S2589-7500(20)30289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information: Appendix tables 1-4 and figure 1