Highlights
-
•
First test in which whey and E. coli are used to obtain L-valine.
-
•
Kinetic properties and kinetic models of L-valine production were studied.
-
•
Highest L-valine production was obtained when whey was used as substrate.
-
•
Highest specific growth rate was achieved when using deproteinized whey.
Keywords: Essential amino acids, Contois, Luedeking-Piret, Kinetic models, Dairy byproduct
Abstract
In this study the effect of the carbon source on L-valine production kinetics using genetically modified E. coli was researched. Glucose, lactose, Whey (W) and deproteinized whey (DW) were tested as carbon sources, keeping the carbon/nitrogen (C/N) ratio constant. Biomass generation and substrate consumption were modeled with Contois and Mass Conservation models, respectively, whereas Mass Conservation Balance and Luedeking-Piret models were used for product obtaining. Results showed that L-valine production is partially associated to growth, with values of 0.485 g L-valine/(g dry cell weight.h), and a product loss effect at a specific rate (β) of 0.019 g L-valine/(g dry cell weight.h) with W. The yield of this product increased 36 % using W concerning glucose or lactose as carbon sources. On the other hand, Mass Balance and Luedeking-Piret models adjust properly to experimental data (R2 >0.90). In conclusion whey is a promising substrate for obtaining L-valine using genetically-modified E. coli.
1. Introduction
Whey (W) is a residue rich in carbon and nitrogen, that corresponds to 90 % of the milk used for cheese production, and contains approximately 55 % of the nutritional load of fresh milk [1]. Particularly, it has a high lactose concentration (45 g/L approximately), between 6−10 g/L protein [2], in addition to minerals [3]. Around 170 million tons of W are produced every year worldwide [4], however, a significant number of cheese manufacturers lack appropriate treatments for its use, thus resulting in its waste into rivers or spilled directly onto the soil [5], which represents an important environmental issue due to its high chemical demand and oxygen biochemistry, which, depending on the type of cheese, might exceed the 40,000 and 30,000 mg O2/L, respectively [6].
Due to its high protein content, this byproduct has been used in the industry of food, cosmetic, and pharmaceutical supplements [1,2]. However, these alternatives are not enough to absorb the amount of W produced and thus reduce the contaminating load that its waste causes [7], which makes researching on its applications a necessary course of action. An alternative for exploiting W is to use it as a carbon source for fermentation processes [8], as it is less expensive than other traditional carbon sources [7].
W has been used in fermentation processes to obtain biostimulants for soils and plants [9], organic acids [10], bioalcohols [8,11], and recombining protein production [12]. However, there are few studies that use W for amino acid production (AA) [13,14], more specifically, there are no reports of its use to obtain L-valine.
The importance of L-valine production lies in the fact that it is an essential amino acid in human nutrition as it helps on the synthesis of biological interest peptides [15,16], in addition to being applied on cosmetics [17]. Currently, L-valine production is carried out mostly by fermentation using bacterial species such as Escherichia coli and Corynebacterium glutamicum [13,[18], [19], [20], [21]], a process in which the search of low-cost substrates is a priority.
On the other hand, the development of kinetic models is a useful tool for bioreactor design and upgrade [22], meanwhile metabolic analysis provides important information for bioprocess improvement [23]. It could be possible to establish a complete analysis on the effect of the carbon source on L-valine production kinetics using genetically modified E. coli with these strategies. Given that metabolism for microbial growth and product formation is a complex biochemical process, experimental data are an approximation to get to know what occurs within such metabolic complexity, but their best application is when they are used through kinetic models that allow the integration of all experimental information and a more holistic visualization of its behavior in order to understand it and extend it to other scenarios of growth improvement processes [24]. The goal of this research was to implement mathematical models that represent cell growth (Contois), substrate consumption (glucose, lactose, W and DW) (mass balance), and L-valine production (mass balance and Luedeking-Piret), to assess in conjunction, the effects of the different carbon sources in L-valine production phenotyping and biosynthesis.
2. Materials and methods
2.1. Strain and growth cultures
E. coli strain (CECT 877), which is innocuous, corresponds to the ATCC® 13005™ strain, obtained from the Spanish Type Culture Collection (Colección Española de Cultivos Tipo – CECT), and modified to favor L-valine production. Powder W was purchased from Cimpa S.A.S. (Bogotá, Colombia) food grade, whereas all other reactive agents used were analytical grade.
Strain activation was performed in nutrient broth at 37 °C for 24 h. glycerol at 50 %, mixed with 500 μL bacterial culture in sterile microtubes (known as seed tubes), that were stored at −80 °C [8], were added. In order to ensure seed tube purity, cultures were carried out in agar Eosin Methylene Blue (EMB) Levine [25].
DW preparation was carried out by autoclaving W at 112 °C for 15 min and centrifugation afterwards (5500 rpm, 20 min, 18 °C), to eliminate precipitated protein [12].
Only the carbon source was changed in 4 production mediums (MP) that were prepared, preserving carbon concentrations equivalent to 0.13 mol C/L. The other components for 1 L were magnesium sulfate heptahydrate (MgSO4*7H2O) 1.505 g, and calcium chloride (CaCl2) 0.014 g. Carbon source content in the mediums was: MP1- 4 g Glucose, MP2- 3.8 g Lactose, MP3- 4.3 g W and MP4- 5.4 mL DW, which were sterilized at 121 °C for 15 min. Salt solution (SS) (sterilized at 121 °C for 15 min) for 1 L of water corresponds to: Disodic phosphate (Na2HPO4) 6 g, monobasic potassium phosphate (KH2PO4) 3 g, ammonium sulfate ((NH4)2SO4) 0.5 g, and sodium chloride (NaCl) 0.5 g [26]. The final concentration of W and DW in production mediums was calculated from lactose initial concentration in powder W (88 % w/w) and the prepared DW mother solution (70 % w/v).
2.2. Batch culture
An E. coli colony in EMB agar was inoculated in 400 mL of nutrient broth, being cultured at 37 °C and 250 rpm in a MaxQ 4000 shaker (Thermo Scientific, Marietta, U.S.A) for 15−18 h (exponential phase). Then, it was aseptically added to a 7.5 L bioreactor (New Brunswick Scientific G628-011, Eppendorf, U.S.A) containing 3.2 L of MP1, MP2, MP3, or MP4 (previously sterilized), and 400 mL SS. Volumes were adjusted so the inoculate concentration was 0.2 g dry cells/L at the beginning of the fermentation. Fermentation in the bioreactor was performed at 37 °C with pH control at 6.9, adjusted with NaOH (2 M); dissolved oxygen level was kept approximately at 20 % saturation by agitation speed adjustment between 300 rpm and 600 rpm [27,28] for 24 h. Sampling was carried out every 3 h to measure optical density (OD) and subsequent sugar (glucose and lactose) and L-valine analysis after centrifugation at 10,000 rpm for 9 min at 4 °C.
2.3. Analytical techniques
2.3.1. Whey characterization
A bromatological characterization of W was carried out using the AOAC [29] methods, determining humidity percentage (method 925.10), protein (method 955.04), ethereal extract (method 963.15) and minerals (method 941.12), by triplicate.
2.3.2. Biomass quantification
Biomass quantification (X) was carried out by OD at 600 nm (OD600) with a Genesys 10S UV–vis spectrophotometer (Thermo Scientific, U.S.A) in which, OD600 measured values were correlated with the weight of dry cells (g DCW/L) using a calibration curve [30].
2.3.3. Substrate quantification
Sugar (glucose and lactose) concentration was quantified by spectrophotometric techniques using the DNS method [31] with some modifications. 0.1 mL of sample were mixed with 0.2 mL of DNS, the reaction occurred at 95 °C for 5 min. After stopping the reaction by thermal impact, 1.5 mL of distilled water was added, and it was tempered for 5 min; wavelength was measured afterward at 540 nm in a Genesys 10S UV–vis spectrophotometer (Thermo Scientific, U.S.A) [32]. Glucose and lactose at concentrations of 0.5 at 8 g/L were used for the calibration curve.
2.3.4. L-valine quantification
L-valine was determined by HPLC following the method described by Cigić et al. [33] with some modifications. Samples, before being injected, went through 0.2 μm cellulose nitrate filters. An AA standard curve (0.5, 0.4, 0.3, 0.2 and 0.1 μmol/mL) was used for quantification (external standard). Analyses were carried out in an HPLC Thermo Ultimate 3000 system (Thermo Fisher Scientific, EE. UU.), equipped with a quaternary pump, an automatic injector, a column thermostat set at 40 °C, and a diode arrangement detector with UV detection at 338 nm to quantify primary AA prederivatized with ortho-phthalaldehyde and 3-marcaptopropionic acid (3-MPA/OPA). Separation was performed in a Zorbax Eclipse AAA-C18 column (4.6 × 75 mm, Agilent, U.S.A), particle size 3.5 μm with Zorbax Eclipse AAA pre-column (4.6 × 12.5 mm, Agilent, U.S.A), particle size 5 μm.
2.4. Kinetic parameter determination
Eq. (1) is obtained considering a batch culture and constant volume, which has limited due to the substrate. This equation describes the increase of cellular concentration. Reorganizing in terms of Eq. (1), and applying analytic solution, it is possible to obtain the specific growth rate (μ) (Eq. (2)), with μ equal to the maximum specific growth rate (μmax) in exponential phase. Calculation of the substrate to biomass yield (YX/S), biomass to product yield (YP/X), and substrate to product yield (YP/S) was carried out through linear regression of dry cell weight, glucose and product concentration data, respectively (Eq. (3)). Values were taken from those obtained in the exponential phase until the final time of the process when the highest value of the product was reached [34].
| (1) |
| (2) |
| (3) |
Where X is biomass concentration in dry cell weight per liter (g DCW/L), S is substrate concentration (glucose or lactose (g/L)), μ is specific growth rate (1/h).
2.5. Kinetic models
2.5.1. Cellular growth models
Non-structured, non-segregated kinetic models for biomass growth, substrate consumption and product formation tested were worked on in conjunction since these processes occur simultaneously. Models used for biomass correspond to the one shown in Eq. (1) and modified Monod (Contois model), as described in Eq. (4) [35].
| (4) |
Where Ks is the saturation constant of Monod (g/L), which is achieved at half μmax.
2.5.2. Substrate consumption model
A substrate (glucose, lactose) mass balance was established based on biomass in the substrate (YXS), as shown in Eq. (5), considering that substrate is used only for growth and the amount used for the product is negligible [24,36].
| (5) |
2.5.3. Product formation model
L-valine production was studied with 2 different models, one based on product mass balance (Eq. (6)) according to biomass yield considering the product (YXP) [37], and another based on the kinetic model proposed by Luedekin-Piret, which corresponds to Eq. (7) [24].
| (6) |
| (7) |
Where P is L-valine concentration (g/L), α is a product formation parameter associated to cellular growth (g/g). β is a product formation parameter non-associated to cellular growth (g L-valine/ (g DCW.h), which can change its sign, (+) if the product is forming, or (-) if it is being consumed. If product formation rate depends only on biomass growth rate and not on biomass as such, β = 0 [38,39].
2.6. Model parameter estimation and validation
A non-linear, Least Squares method was used to estimate kinetic parameters of models described in Eqs. (1), (4), (5), (6), (7) through the “fmincon” optimization function of Matlab R2016a. Models were simultaneously solved by ODE45 of Matlab R2016a Runge-Kutta explicit formula with analysis of residue dispersion. Determination coefficient mathematic expressions () (Eq. (8)) and Mean Squared Error (MSE) (Eq. (9)), were used to test deviation of experimental data obtained concerning data from the simulation. Kinetic parameter values obtained from experimental results were analyzed through Duncan multiple comparisons [40] with a confidence level of 95 %, intending to determine whether there were or not any significant differences between the values obtained among the different substrates.
| (8) |
| (9) |
Where fi, yi, , and nt are the model data, experimental data, average of experimental data, and number of experimental data, respectively.
3. Results and discussion
3.1. Whey bromatological characterization
Through bromatological analysis of w, these values were obtained for humidity, ashes, and fat, respectively: 3.12 ± 0.00; 0.07 ± 0.00; and 1.46 ± 0.25 % (w/w). It is important to highlight that the material used showed a high lactose content (88.08 ± 1.93 %) and protein (4.16 ± 0.12 %). Thus, fermentation mediums prepared at an approximate concentration of 0.13 mol C/L, might contain protein concentrations of around 0.18 g protein/L, being this an extra nitrogen source in fermentation, in addition to be enriched with amino acids and minerals, which are essential for optimal growth of E. coli [4].
3.2. Effect of carbon source on E. coli kinetic behavior
Fig. 1 shows the behavior of biomass, substrate and product based on time, during L-valine production by fermentation with E. coli using four different carbon sources. It can be seen that the carbon source has a remarkable effect on the behavior of these variables in L-valine biosynthesis. The maximum biomass concentration was 1.1 g/L at 15 h, without having any significant differences among the substrates. In all cases, a phase lag between 0 and 3 h was observed (Fig. 1a), and despite a diauxic growth and higher biomass concentration in W at 6 h, followed by DW, glucose and lactose are shown, no significant difference is evident at the end of the process (15 h) among carbon sources. Although glucose has been reported as superior when compared with other substrates [41,42], such differences were not quite notorious in this case. The time to reach the stationary phase was similar in all cases. For DW, it was reached at approximately 13 h, followed by glucose (14 h), lactose and W (15 h). Culture was preserved in this stationary phase in order to observe L-valine production on time, considering that it is a secondary metabolite. Process was stopped when the substrate was depleted and there was no evidence of an increase in product biosynthesis.
Fig. 1.
Batch fermentation kinetics with E. coli at 24 h for biomass growth (a), substrate consumption (b), and L-valine formation (c). Glucose (o); Lactose (□); W (Δ); DW (x). Vertical bars represent standard deviation.
Growth kinetics in which W was used, a diauxic behavior was observed. Additionally, lactose as a carbon source was not consumed completely (Fig. 1b), showing a behavior very similar to that of carbon sources W and DW. When relating substrate depletion in the medium concerning L-valine biosynthesis reduction, especially for W and DW, it could be stated that it is an absorption/degradation phenomenon shown by this product [39], or also to the favoring of this medium to obtain other by-products that lead to L-valine consumption. These results shown in Fig. 1 suggest W favorability not only for growth but also for L-valine production.
Calculation of substrate consumption rates for glucose, lactose, and W was performed between 9 and 15 h, which was the period where the highest decrease took place, with behavior without significant differences among the four carbon sources (p > 0.05), thus obtaining values of 0.39, 0.38, 0.36 g/L.h, for glucose, lactose, and W, respectively (Fig. 1b), whereas for DW, the calculation of consumption rate was between 6 and 12 h and showed a value of 0.39 g/L.h, the same as the consumption rate obtained for glucose. In the case of glucose, this was depleted, while lactose, W and DW reached low values without getting depleted. Similar results were found in studies where traditional carbon sources in the industry were not used [24].
In Fig. 1c is presented L-valine biosynthesis, which, when contrasted with biomass production (Fig. 1a), indicates that there is product biosynthesis associated to growth in the four substrates tested, as stated on other reports [43]. Two significantly different patterns are observed (p < 0.05), one for individual carbon sources (glucose and lactose), and other for whey (W and DW). There is an increase in individual sources within the first 18 h, followed by a mild reduction, while in whey, there is a significant increase in the first 12 h, followed by a reduction (12−18 h), and then a drastic drop (18−21 h). Whey reaches productions up to 36 % higher in L-valine concentration concerning individual carbon sources (lactose and glucose) in the period between 15−18 h. However, after 18 h of culture, there is a reduction in product concentration, possibly due to the absorption/degradation process [39] mentioned before, or it can also occur due to formation of by-products such as L-alanine, L-glycine, and α-ketoglutarate [44].
Kinetic parameter determination performed is summarized in Table 1, which shows μmax and yield obtained with Eq. (2) and (3), respectively. When these values are analyzed, it is noticeable that DW was the carbon source with the highest μmax (p < 0.05), and whey showed a higher yield concerning the values obtained for simple substrates. Product maximum concentration values show a value in parenthesis that refers to the time which such maximum was achieved. L-valine maximum yield and concentrations were obtained for whey, thus indicating them as promising materials, suitable for biosynthesis of this amino acid with E. coli, as reported in the biosynthesis of other metabolites of interest by fermentative means [3]. This may occur due to the amount of soluble protein that both W and DW have, which, despite being deproteinized, may contain some residual peptides [45]. Thus, biomass yield in substrate (Yxs) for all carbon sources studied was similar according to other yield values obtained for E. coli [46], looking at these results, it is possible to establish that microbial growth is favored by all these substrates used as carbon source.
Table 1.
Kinetic parameters in E. coli fermentation with different carbon sources.
| Carbon source | μmax (h−1) | Pmax (g/L) | (g L- l-val/L h) | Yxs (g/g) | Ypx (g/g) | Yps (g/g) |
|---|---|---|---|---|---|---|
| Glucose | 0.149b | 0.104b (15 h) | 0.014b | 0.255ab | 0.103b | 0.026b |
| Lactose | 0.103c | 0.117b (18 h) | 0.018b | 0.267a | 0.106b | 0.030b |
| W | 0.070d | 0.197a (15 h) | 0.048a | 0.239b | 0.263a | 0.066a |
| DW | 0.218ª | 0.187a (18 h) | 0.041a | 0.273a | 0.209a | 0.060a |
μmax: Maximum growth rate; Pmax: Maximum concentration of L-valine obtained; YXS: biomass yield concerning substrate (g DCW/g of substrate (carbon source); Ypx: Product yield concerning biomass (g L-valine /g DCW); Yps: Product yield concerning substrate (g L-valine /g substrate); similar lower case letters in the same column indicates there is not significant statistic difference among treatments (p < 0.05).
3.3. L-valine biosynthesis metabolical analysis
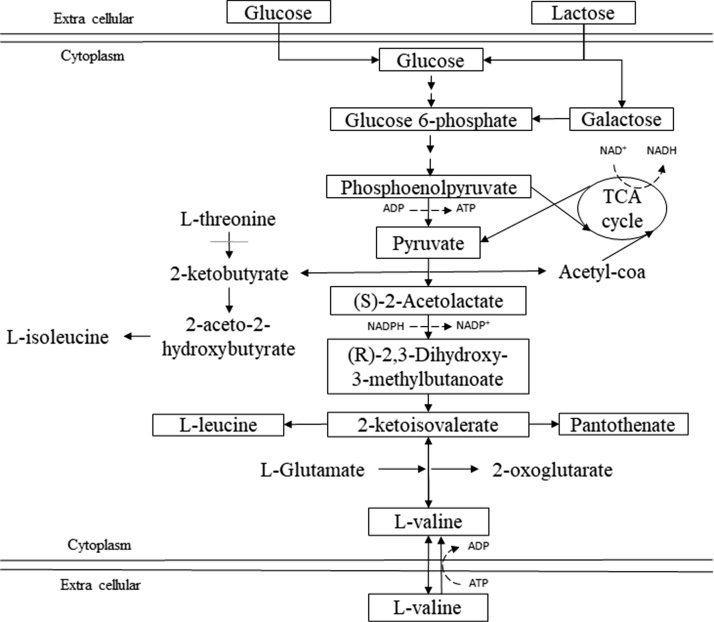
The metabolic route for L-valine biosynthesis is summarized in Fig. 2, in which the type of carbon source used, either glucose or lactose, is considered. Thus, lactose is unfolded into glucose and galactose, which are integrated in the metabolic path as glucose 6-phosphate to continue into glycolysis and its interaction with tricarboxylic (TCA) acid cycle through the pyruvate intermediary metabolite [42]. When analyzing this simplified map of the possible metabolism of the microorganism used on this study (E. coli), L-valine synthesis is identified, carbon flux deviation for biosynthesis of other amino acids such as threonine and isoleucine is notorious, and continues towards leucine biosynthesis, then valine via glutamate and 2-ketoisovalerate consumption, which are the amino acids necessary for L-valine production. Also, there is a clear carbon flux deviation towards isoleucine from pyruvate, which is favored by the presence of threonine, an essential compound for the strain of E. coli used for this work [21]. Being threonine present, the metabolic route is favored to generate 2-acetolactate, which is then addressed to the production of 2-ketoisovalerate, an essential metabolite for the production of other amino acids and metabolic by-products (isobutanol, pantothenate) [47].
Fig. 2.
E. coli biosynthetic paths to L-valine production.
Production of branched chain amino acids such as L-valine, L-isoleucine and L-leucine, consists of several reactions catalyzed by acetohydroxybutanoate synthase enzymes (AHAS). E. coli has these three enzymes AHAS (I, II, and III), which differ in their regulation and biochemical properties. Thus, AHAS I is inhibited by L-valine feedback [18,48,49].
However, given the richness in amino acids that W has [6,50], and the complexity in the final L-valine biosynthesis path from pyruvate, a positive regulatory effect in enzymes could occur [51], which favored valine production (Fig. 1c), situation that happens in complex mediums and not in the minimal ones.
On the other hand, product concentration decrease (W, Fig. 1c) could be explained due to E. coli capacity to catabolize it to generate succinyl-coA, given the glycogenic character of this amino acid [13,49], or also due to the biosynthetic cost of amino acids, which sets a selective restriction to codify amino acids and thus stay alive through time [52].
3.4. Simulation of cellular growth, substrate consumption, and L-valine production under different carbon sources used
L-valine biosynthesis characterization under different carbon sources, cellular growth simulation, substrate consumption, and product formation during the batch process were carried out using MATLAB Software to solve the models proposed (Eq. (1),(4),(5),(6), and (7)) simultaneously. Fig. 3 shows the profiles of experimental and modeled results of substrate consumption, microbial growth and L-valine biosynthesis simultaneously for each substrate analyzed (glucose, lactose, W, and DW). When profiles for all substrates are examined, it is noticeable that data simulated by the models are consistent with experimental data. This way, it can be stated that models tested on this work can predict cell growth behavior, biosynthesis and product absorption/degradation, as well as the substrate consumption profile at the same time.
Fig. 3.
E. coli fermentation kinetics in batch culture, comparison of experimental data Vs model-predicted values. a: Glucose; b: Lactose c: W, and d: DW. Experimental values show the following symbology: (Biomass (x), Substrate (O), Product (+). Model-predicted values show the following symbology: (Biomass (―), Substrate (– • –), Product: Luedeking-Piret (- - -), Mass balance (···).
Table 2 summarizes the values obtained from the kinetic parameters when applying the proposed kinetic models simultaneously. Data obtained from simulation were consistent with the experimental results (μmax and Yxs), except in DW concerning μmax, possibly due to the model arrangement to plot the best simulation curve in all experimental data, which, in the simulation results show a specific maximum growth rate (μmax) of 0.109 h−1 with glucose, 0.103 h−1 with lactose, 0.08 h−1 with W, and 0.119 h-1 with DW.
Table 2.
Parameter estimation and validation of models established for L-valine production in batch fermentation with E. coli using different carbon sources obtained by simulation.
| Estimated kinetic parameters | Glucose | R2 | MSE | Lactose | R2 | MSE | W | R2 | MSE | DW | R2 | MSE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Contois (X); Mass balance (S); Mass balance (P) | ||||||||||||
| μmax (h−1) | 0.106 | 0.919 | 0.002 | 0.103 | 0.953 | 0.001 | 0.083 | 0.894 | 0.002 | 0.119 | 0.928 | 0.002 |
| Ks (g/L) | 0.016 | 0.008 | 0.004 | 0.176 | ||||||||
| Yxs (g/g) | 0.267 | 0.989 | 0.007 | 0.282 | 0.982 | 0.010 | 0.261 | 0.977 | 0.012 | 0.283 | 0.977 | 0.012 |
| Yxp (g/g) | 5.121 | 0.799 | 0.009 | 5.148 | 0.849 | 0.008 | 2.498 | 0.857 | 0.030 | 3.313 | 0.884 | 0.024 |
| β (g Val/g DCW.h) | 0.007 | 0.007 | 0.013 | 0.010 | ||||||||
| Contois (X); Mass balance (S); Luedeking-Piret (P) | ||||||||||||
| μmax (h−1) | 0.106 | 0.918 | 0.002 | 0.103 | 0.953 | 0.001 | 0.083 | 0.894 | 0.002 | 0.119 | 0.928 | 0.002 |
| Ks (g/L) | 0.005 | 0.008 | 0.010 | 0.176 | ||||||||
| Yxs (g/g) | 0.267 | 0.989 | 0.007 | 0.282 | 0.982 | 0.010 | 0.260 | 0.977 | 0.012 | 0.283 | 0.977 | 0.012 |
| α (g/g) | 0.131 | 0.953 | 0.002 | 0.224 | 0.792 | 0.011 | 0.485 | 0.836 | 0.032 | 0.302 | 0.884 | 0.024 |
| β (g Val/g DCW.h) | 0.003 | 0.009 | 0.019 | 0.010 | ||||||||
Val: valine; DCW: dry cell weight; R2: Determination coefficient; MSE: Mean Squared Error.
In Table 2, it is also possible to observe the constant affinity of the microorganism with the substrate or Monod (Ks) saturation constant. This value changes for glucose and W in numbers obtained with Mass balance models and Luedeking-Piret, being the lowest for the whey that showed the highest L-valine concentration value. Additionally, when comparing Ks values for lactose and DW, the value remained constant when making the simulations with the two models described for the product. These values match those reported in literature for this type of microorganism [53,54].
For mass balance model for P, it can be observed that Yxp is higher in pure carbon sources than in complex mediums (Table 2). When observing Fig. 3, L-valine value was higher in the two types of whey used than in glucose and lactose, with a similar biomass behavior in all carbon sources used. On the other hand, β as part of the product formation non-associated to cellular growth in negative values, there are high values in W and DW of β, given that according to L-valine production kinetics (Fig. 1c), the proportion in its reduction after reaching the maximum is considerable compared to fermentations with glucose and lactose, corresponding to the experimental data found.
Results obtained when applying the Luedeking-Piret model for all different carbon sources can also be analyzed in Table 2. Values different from zero were found for the constants related to the rates associated to growth (α) and non-associated to growth (β) for L-valine production, which suggests that product biosynthesis is a process partially associated to growth. These results match others reported for similar models [55]. The rate constant associated to growth for L-valine production (α) for W and DW was 2–3 times higher than lactose and glucose, respectively (Table 2). Also, values in β were found for high W and DW, corresponding to the high adsorption/degradation rate of this amino acid after reaching a maximum, meanwhile there is substrate depletion.
The best model to predict product behavior is Luedeking-Piret, whereas, in the other parameters, they have similar behaviors. Differences among models have also been found and studied [36]. However, on this study, the use of two models (mass balance and Luedeking-Piret) shows that, statistically, there is no significant difference to predict and additionally, for this type of study, that depends more of the substrate type rather than the model type as stated by Xu [36].
3.5. Model validation
Table 2 shows that R2 values are higher than 0.91 for all parameters obtained by Luedeking-Piret for glucose and lactose, and MSE below 2% in every model for all carbon sources used. These results indicate that cellular growth, substrate consumption, L-valine biosynthesis, and degradation simulated data for the carbon sources used showed good adjustment with experimental data concerning time.
Additionally, another quality found in modeling was that the values expected in the models match the error variance, with the MSE not higher than 10 % (Table 2) in every model tested (X, S, and P), which indicates the probability that modeling for predicting and event (cellular growth, substrate consumption, and L-valine production) in time will occur with a high confidence rate [56]. Also, through the analysis of residue dispersion (supplementary material I and II), a linear relation was obtained in all the cases, with a normal distribution of residues and a constant variance in most cases [57].
4. Conclusions
Results indicate that under minimum medium conditions, the highest L-valine production and μmax are obtained with DW. Mass balance and Luedeking-Piret models (R2 >0.90) show the best adjustment to experimental data, besides, to predict the three physiologies in E. coli (cellular growth, substrate consumption and product yield). However, the Luedeking-Piret model is more assertive and suitable for simulation and obtaining results in α and β parameters, proposing product biosynthesis as a process partially associated to growth in all substrates used. Therefore, this kind of work is promising for a green, sustainable and attractive conversion towards the use of this type of by-products (whey) to obtain L-valine as an important additive in food, pharmaceutical and cosmetic industry.
Funding
The authors thank to Gobernación Del Tolima; and the Ministerio de Ciencia Tecnología e Innovación de Colombia (MINCIENCIAS) for the scholarship and financial support [Contract number: FP44842-397].
Declaration of Competing Interest
Authors declare that they do not have any conflict of interest. Funders did not play any part in the design of the study or data collection, analysis, or interpretation; or in writing the manuscript; or in the decision of publishing the results.
Acknowledgements
The authors thank to University of Antioquia, for facilitating laboratories and supporting with researchers; the Gobernación Del Tolima; and the Ministerio de Ciencia Tecnología e Innovación de Colombia (MINCIENCIAS) for the scholarship and financial support [Contract number: FP44842-397].
Contributor Information
Darwin Carranza-Saavedra, Email: darwin.carranza@udea.edu.co.
Claudia Patricia Sánchez Henao, Email: cpatricia.sanchez@udea.edu.co.
José Edgar Zapata Montoya, Email: edgar.zapata@udea.edu.co.
References
- 1.Sharma R. Whey proteins. In: Deeth H.C., Bansal N., editors. Whey Proteins in Functional Foods. Elsevier; 2019. pp. 637–663. [Google Scholar]
- 2.Singh R. Protein byproducts. In: Dhillon G.S., editor. Whey Proteins and Their Value-Added Applications. Elsevier; 2016. pp. 303–313. [Google Scholar]
- 3.Önür Z.Y. Whey every aspect. In: Alapítvány, editor. Innovative Products Produced From Whey. 2020. p. 61. [Google Scholar]
- 4.Yadav J.S.S. Cheese whey: a potential resource to transform into bioprotein, functional/nutritional proteins and bioactive peptides. Biotechnol. Adv. 2015;33(6):756–774. doi: 10.1016/j.biotechadv.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Prazeres A.R., Carvalho F., Rivas J. Cheese whey management: a review. J. of Environ. Manag. 2012;110:48–68. doi: 10.1016/j.jenvman.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Menchik P. Composition of coproduct streams from dairy processing: acid whey and milk permeate. J. Dairy Sci. 2019;102(5):3978–3984. doi: 10.3168/jds.2018-15951. [DOI] [PubMed] [Google Scholar]
- 7.Królczyk J.B. Use of whey and whey preparations in the food industry–a review. Polish J. Food Nutr. Sci. 2016;66(3):157–165. [Google Scholar]
- 8.Pasotti L. Fermentation of lactose to ethanol in cheese whey permeate and concentrated permeate by engineered Escherichia coli. BMC Biotechnol. 2017;17(1):48. doi: 10.1186/s12896-017-0369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caballero P. Obtaining plant and soil biostimulants by waste whey fermentation. Waste Biomass Valor. 2019;11:3281–3292. [Google Scholar]
- 10.Lagoa-Costa B., Kennes C., Veiga M.C. Cheese whey fermentation into volatile fatty acids in an anaerobic sequencing batch reactor. Bioresour. Technol. 2020;308:123226. doi: 10.1016/j.biortech.2020.123226. [DOI] [PubMed] [Google Scholar]
- 11.Sharma D. Microbial cell factories. In: CRC Press, editor. Engineering Microbial Cell Factories for Improved Whey Fermentation to Produce Bioethanol. 2018. pp. 353–370. [Google Scholar]
- 12.Hausjell J. Valorisation of cheese whey as substrate and inducer for recombinant protein production in E. coli HMS174 (DE3) Bioresour. Technol. Rep. 2019;8:100340. [Google Scholar]
- 13.Oldiges M., Eikmanns B.J., Blombach B. Application of metabolic engineering for the biotechnological production of L-valine. Appl. Microbiol. Biotechnol. 2014;98(13):5859–5870. doi: 10.1007/s00253-014-5782-8. [DOI] [PubMed] [Google Scholar]
- 14.D’Este M., Alvarado-Morales M., Angelidaki I. Amino acids production focusing on fermentation technologies–a review. Biotechnol. Adv. 2018;36(1):14–25. doi: 10.1016/j.biotechadv.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi T. Branched‐chain amino acids as pharmacological nutrients in chronic liver disease. Hepatology. 2011;54(3):1063–1070. doi: 10.1002/hep.24412. [DOI] [PubMed] [Google Scholar]
- 16.Janus E. Enhancement of ibuprofen solubility and skin permeation by conjugation with L-valine alkyl esters. RSC Adv. 2020;10(13):7570–7584. doi: 10.1039/d0ra00100g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraf S. Skin targeting approaches in cosmetics. Indian J. Pharm. Educ. Res. Microbiol. 2019;53(4):577–594. [Google Scholar]
- 18.Park J.H. Escherichia coli W as a new platform strain for the enhanced production of L‐Valine by systems metabolic engineering. Biotechnol. Bioeng. 2011;108(5):1140–1147. doi: 10.1002/bit.23044. [DOI] [PubMed] [Google Scholar]
- 19.Park J.H. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. 2007;104(19):7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J.H. Fed‐batch culture of Escherichia coli for L‐valine production based on in silico flux response analysis. Biotechnol. Bioeng. 2010;108(4):934–946. doi: 10.1002/bit.22995. [DOI] [PubMed] [Google Scholar]
- 21.H.T. Huang, (1961). Fermentation process. U.S. Patent No. 2,975,105. Washington, DC: U.S. Patent and Trademark Office.
- 22.Mears L. Mechanistic fermentation models for process design, monitoring, and control. Trends Biotechnol. 2017;35(10):914–924. doi: 10.1016/j.tibtech.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Wendisch V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2019;58:17–34. doi: 10.1016/j.ymben.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q. Kinetic analysis of curdlan production by Alcaligenes faecalis with maltose, sucrose, glucose and fructose as carbon sources. J. Bioresour. Technol. 2018;259:319–324. doi: 10.1016/j.biortech.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P., Melkania U. Impact of furan derivatives and phenolic compounds on hydrogen production from organic fraction of municipal solid waste using co-culture of Enterobacter aerogenes and E. coli. Bioresour. Technol. 2017;239:49–56. doi: 10.1016/j.biortech.2017.04.113. [DOI] [PubMed] [Google Scholar]
- 26.Wu W. Fuel ethanol production using novel carbon sources and fermentation medium optimization with response surface methodology. Int. J. Agric. Biol. Eng. 2013;6(2):42–53. [Google Scholar]
- 27.Wang J., Cheng L.-K., Chen N. High-level production of L-threonine by recombinant Escherichia coli with combined feeding strategies. Biotechnol. Biotechnol. Equip. 2014;28(3):495–501. doi: 10.1080/13102818.2014.927682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu E. Culture conditions for improvement of L-threonine production using a genetically self-cloned L-threonine hyperproducing strain of Escherichia coli K-12. Biosci. Biotechnol. Biochem. 1995;59(6):1095–1098. doi: 10.1271/bbb.59.1095. [DOI] [PubMed] [Google Scholar]
- 29.AOAC . 17ma. ed. Official Methods of Analysis; 2000. Association of Official Analytical Chemists. [Google Scholar]
- 30.Aso Y. Continuous production of d-lactic acid from cellobiose in cell recycle fermentation using β-glucosidase-displaying Escherichia coli. J. Biosci. Bioeng. 2019;127(4):441–446. doi: 10.1016/j.jbiosc.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31(3):426–428. [Google Scholar]
- 32.Yadav J.S.S. Simultaneous single-cell protein production and COD removal with characterization of residual protein and intermediate metabolites during whey fermentation by K. marxianus. Bioprocess Biosyst. Eng. 2013;37(6):1017–1029. doi: 10.1007/s00449-013-1072-6. [DOI] [PubMed] [Google Scholar]
- 33.Cigić I.K. Amino acid quantification in the presence of sugars using HPLC and pre-column derivatization with 3-MPA/OPA and FMOC-Cl. Acta Chim. Slov. 2008;55(3):660–664. [Google Scholar]
- 34.Long C.P. Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab. Eng. 2017;44:100–107. doi: 10.1016/j.ymben.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Öktem Y.A. Recycling and reuse approaches for better sustainability. In: Balkaya N., Guneysu S., editors. Microbial Growth Kinetics of an Anaerobic Acidogenic Bioreactor. 2019. pp. 233–243. [Google Scholar]
- 36.Xu P. Analytical solution for a hybrid Logistic‐Monod cell growth model in batch and continuous stirred tank reactor culture. Biotechnol. Bioeng. 2019;117:873–878. doi: 10.1002/bit.27230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez Henao C.P., Grimaldos Gomez N.A., Quintero Diaz J.C. Producción de ácido clavulánico por fermentación de streptomyces clavuligerus: evaluación de diferentes medios de cultivo y modelado matemático. Dyna. 2012;79(175):158–165. [Google Scholar]
- 38.Aktypis A. Studies on bacteriocin (thermophilin T) production by Streptococcus thermophilus ACA-DC 0040 in batch and fed-batch fermentation modes. Antonie van Leeuwenhoek J. Microbiol. 2007;92(2):207–220. doi: 10.1007/s10482-007-9148-4. [DOI] [PubMed] [Google Scholar]
- 39.Vázquez J.A., Murado M.A. Unstructured mathematical model for biomass, lactic acid and bacteriocin production by lactic acid bacteria in batch fermentation. J. Chem. Technol. 2008;83(1):91–96. [Google Scholar]
- 40.Duncan D.B. Multiple range and multiple F tests. Biometrics. 1955;11(1):1–42. [Google Scholar]
- 41.Bren A. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 2016;6(1):1–10. doi: 10.1038/srep24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aidelberg G. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst. Biol. 2014;8(1):133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanjay K. Kinetics of growth on dual substrates, production of novel glutaminase-free L-asparaginase and substrates utilization by Pectobacterium carotovorum MTCC 1428 in a batch bioreactor. Korean J. Chem. Eng. 2017;34(1):118–126. [Google Scholar]
- 44.Brik Ternbach M. Application of model discriminating experimental design for modeling and development of a fermentative fed‐batch l‐valine production process. Biotechnol. Bioeng. 2005;91(3):356–368. doi: 10.1002/bit.20504. [DOI] [PubMed] [Google Scholar]
- 45.Mulcahy E.M. Characterisation of heat-induced protein aggregation in whey protein isolate and the influence of aggregation on the availability of amino groups as measured by the ortho-phthaldialdehyde (OPA) and trinitrobenzenesulfonic acid (TNBS) methods. Food Chem. 2017;229:66–74. doi: 10.1016/j.foodchem.2017.01.155. [DOI] [PubMed] [Google Scholar]
- 46.Stiefelmaier J. Pink bacteria—production of the pink chromophore phycoerythrobilin with Escherichia coli. J. Biotechnol. 2018;274:47–53. doi: 10.1016/j.jbiotec.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Gu J. Isobutanol and 2-ketoisovalerate production by Klebsiella pneumoniae via a native pathway. Metab. Eng. 2017;43:71–84. doi: 10.1016/j.ymben.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Hao Y. High-yield production of L-valine in engineered Escherichia coli by a novel two-stage fermentation. Metab. Eng. 2020;62:198–206. doi: 10.1016/j.ymben.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Geraskina N.V. Engineering Escherichia coli for autoinducible production of L-valine: an example of an artificial positive feedback loop in amino acid biosynthesis. PLoS One. 2019;14(4):1–16. doi: 10.1371/journal.pone.0215777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dinika I. The roles of Candida tropicalis toward peptide and amino acid changes in cheese whey fermentation. Int. J. Technol. 2019;10(8):1533–1540. [Google Scholar]
- 51.Guo Y. Analysis of acetohydroxyacid synthase variants from branched-chain amino acids-producing strains and their effects on the synthesis of branched-chain amino acids in Corynebacterium glutamicum. Protein Expr. Purif. 2015;109:106–112. doi: 10.1016/j.pep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Zampieri M. Regulatory mechanisms underlying coordination of amino acid and glucose catabolism in Escherichia coli. Nat. Commun. 2019;10(1):1–13. doi: 10.1038/s41467-019-11331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nooshkam M., Varidi M., Bashash M. The Maillard reaction products as food-born antioxidant and antibrowning agents in model and real food systems. Food Chem. 2019;275:644–660. doi: 10.1016/j.foodchem.2018.09.083. [DOI] [PubMed] [Google Scholar]
- 54.Dack R.E., Black G.W., Koutsidis G. The effect of Maillard reaction products and yeast strain on the synthesis of key higher alcohols and esters in beer fermentations. Food Chem. 2017;232:595–601. doi: 10.1016/j.foodchem.2017.04.043. [DOI] [PubMed] [Google Scholar]
- 55.Tan R. Enhancing scleroglucan production by Sclerotium rolfsii WSH-G01 through a pH-shift strategy based on kinetic analysis. Bioresour. Technol. 2019;293:122098. doi: 10.1016/j.biortech.2019.122098. [DOI] [PubMed] [Google Scholar]
- 56.Brandt S. Fourth edition. Springer Sci. & Business; Dordrecht, Switzerland: 2014. Data Analysis - Statistical and Computational Methods for Scientists and Engineers; pp. 1–532. [Google Scholar]
- 57.Wang W., Lu Y. Analysis of the mean absolute error (MAE) and the root mean square error (RMSE) in assessing rounding model. IOP Conf. Ser.: Mater. Sci. Eng. 2018;324 [Google Scholar]