Abstract
Metabolic flux analysis (MFA) is widely used to estimate intracellular fluxes. Conventional MFA, however, is limited to continuous cultures and the mid-exponential growth phase of batch cultures. Dynamic MFA (DMFA) has emerged to characterize time-resolved metabolic fluxes for the entire culture period. Here, the linear DMFA approach was extended using B-spline fitting (B-DMFA) to estimate mass balanced fluxes. Smoother fits were achieved using reduced number of knots and parameters. Additionally, computation time was greatly reduced using a new heuristic algorithm for knot placement. B-DMFA revealed that Chinese hamster ovary cells shifted from 37 °C to 32 °C maintained a constant IgG volume-specific productivity, whereas the productivity for the controls peaked during mid-exponential growth phase and declined afterward. The observed 42% increase in product titer at 32 °C was explained by a prolonged cell growth with high cell viability, a larger cell volume and a more stable volume-specific productivity.
Abbreviations: B-DMFA, B-spline fitting dynamic metabolic flux analysis; CHO, Chinese hamster ovary; DMFA, dynamic metabolic flux analysis; L-DMFA, linear dynamic metabolic flux analysis; MFA, metabolic flux analysis; SSR, variance-weighted sum of squared residuals
Keywords: Dynamic, Metabolism, Flux analysis, CHO cells, Temperature shift, B-spline curve fitting
Highlights
-
•
New dynamic MFA framework using B-spline (B-DMFA) generates smooth fit.
-
•
B-DMFA performs better than linear DMFA when fitting fast dynamic changes.
-
•
Heuristic algorithm for knot placement dramatically reduced computation time.
-
•
Temperature shifted cultures maintain a constant IgG volume specific productivity.
-
•
CHO cells shifted to 32 °C have a 42% higher IgG titer due to larger cell volume.
1. Introduction
For over 20 years, metabolic flux analysis (MFA) has proven a powerful tool to quantitatively characterize cell metabolic phenotypes (Bonarius et al., 1996, Nielsen, 2003, Vallino and Stephanopoulos, 1993). MFA estimates intracellular fluxes using experimental measurements and mass balances. The quantification of metabolic fluxes has facilitated a better understanding of biological systems (Boghigian et al., 2010), including the identification of bottlenecks in product formation (Burleigh et al., 2011, Nyberg et al., 1999) and metabolic regulation (Fendt and Sauer, 2010). MFA is routinely used in media design (Altamirano et al., 2006, Martínez et al., 2010, Xing et al., 2011) and metabolic engineering (Becker et al., 2005) in order to improve metabolic phenotypes.
Conventional MFA assumes constant intracellular fluxes, a condition only satisfied by continuous cultures at steady state and during the mid-exponential phase of batch cultures. Most industrial cultures display dynamic metabolic changes (Matasci et al., 2008, Meadows et al., 2010), such as a gradual decline in cell-specific growth rate during batch and fed batch (Antoniewicz et al., 2007, Niklas et al., 2011) or diauxic growth (Mahadevan et al., 2002). Frequently, the most relevant metabolic behaviour occurs during transient conditions, where conventional MFA is no longer applicable.
Monoclonal antibody production by CHO cells illustrates the point. Conventional MFA has been used to study the Warburg effect observed during the initial growth phase, yet all cultures display this phenotype during the initial exponential phase. What distinguishes successful cultures is the transition from exponential to stationary growth phase, whether the transition is invoked naturally (Ahn and Antoniewicz, 2011, Martínez et al., 2013), caused by medium design (Altamirano et al., 2006) or deliberate manipulation of culture conditions, such as a temperature shift (Bollati-Fogolin et al., 2005). A metabolic shift from lactate production to lactate consumption during this transition aids longevity and productivity of the culture (Altamirano et al., 2006, Ma et al., 2009, Martínez et al., 2013, Niklas et al., 2011, Young, 2013).
Several approaches have been used to estimate dynamic flux distributions (Antoniewicz, 2013). While all approaches assume pseudo-steady state for intracellular metabolism, i.e. the changes in internal metabolite concentrations are trivial compared to the fluxes (Varma and Palsson, 1994), they differ in terms of how dynamics in the data are captured. The simplest approach is to manually divide the time series data into distinct metabolic phases and estimate the flux profile of each phase by MFA (Ahn and Antoniewicz, 2011, Altamirano et al., 2006, Martínez et al., 2013, Niklas et al., 2011). This approach produces an average flux estimate for each phase but fails to capture the temporal evolution of metabolic fluxes. It is also difficult to maintain a homogenous precision among determined rates, which are dependent on the consistency and frequency of data points contained in each phase. Newer approaches overcome these limitations by first generating smooth functions to fit the measured metabolite concentrations and subsequently estimating instantaneous rates based on the functions’ slope (Lequeux et al., 2010, Llaneras and Pico, 2007, Niklas et al., 2011). Then MFA is applied to each time point to generate a continuous flux distribution profile. The independent smoothing of individual concentration profiles followed by differentiation ignores mass conservation, potentially introducing significant bias into the flux estimates. Dynamic MFA (DMFA) overcomes this issue by using free fluxes as basis for the fitting functions and a least-square approach to fit the full dataset simultaneously (Leighty and Antoniewicz, 2011).
The original DMFA implementation uses piecewise linear functions to describe the time profile of free fluxes (Leighty and Antoniewicz, 2011). While simple in form, the linear assumption creates rates with unnatural, non-smooth breakpoints and struggle to fit higher-order dynamics, which do commonly occur since the time profile of absolute rates tend to trace a sigmoid curve (e.g., Monod model). In this study, we have extended the linear method by using B-spline functions when performing DMFA (B-DMFA), bringing in capabilities for higher order fit and the use of knot multiplicity. Knots are equivalent to the so-called DMFA time points (Leighty and Antoniewicz, 2011).
B-splines, or basis splines, are often used in computer-aided geometrical design and software packages to approximate data due to their rich mathematical structure and the robust numerical algorithms available (Prautzsch et al., 2002). B-splines and their integrals can be formulated such that they are linear with respect to the model parameters, and therefore all statistical and computational advantages shown in the linear DMFA (L-DMFA) implementation are retained (Leighty and Antoniewicz, 2011). Moreover, by exploiting the local support property of B-splines, a fast heuristic algorithm was developed for knot placement, which is by far the most time consuming task when performing DMFA. The performance of the B-DMFA algorithm was compared to the current L-DMFA approach (Leighty and Antoniewicz, 2011) using a simulated diauxic growth dataset of yeast fermentation (Sonnleitner and Kappeli, 1986). Demonstrating application to real data, B-DMFA was used to determine flux profiles of CHO cultures in order to investigate the effects of temperature shift on CHO cells metabolism.
2. Materials and methods
2.1. Dynamic metabolic flux analysis (DMFA) using B-Spline fitting
Metabolic fluxes (v(t)) are calculated by performing mass balance around each metabolite in a metabolic network, which is represented by a stoichiometric matrix (S) for the internal metabolites (cint) and a matrix R for the external metabolites (cext)
| (1) |
Eq. (1) can be divided in balanced (internal) and not balanced (external) metabolites. It is assumed that internal metabolites are at pseudo steady-state, i.e., the fluxes to and away from any internal metabolite are far greater than the net accumulation of the metabolite itself (Varma and Palsson, 1994)
| (2) |
| (3) |
In conventional MFA, metabolic fluxes are assumed to be constant, and the internal metabolite balances are mathematically formulated by Eq. (3) with constant . In DMFA, metabolic fluxes are allowed to change over time to reflect the dynamics of the measured concentrations and rates.
The vector of fluxes (v(t)) can be expressed as a function of the null space of the balanced stoichiometric matrix S and a set of free fluxes (u(t)):
| (4) |
Leighty and Antoniewicz (2011) calculated internal fluxes using a time-dependent formulated as a linear spline function. A smoother fit, however, can be achieved by formulating using higher order B-splines as shown in Eq. (5) (Curry and Schoenberg, 1947, Curry and Schoenberg, 1966).
| (5) |
Under B-spline terminology, m is the number of time points (called knots), k is the order of the polynomial segments of the B-spline (order k means that the curve is made of polynomial segments of degree k−1), is the ith control point, and is the ith normalized B-spline blending function of order k. The control points and blending functions define the shape of the B-spline (Prautzsch et al., 2002). The normalized blending functions are estimated by the Cox–de Boor recurrence relation (Cox, 1972, De Boor, 1972):
| (6) |
| (7) |
In this study, a quadratic B-spline (k=3) was used to fit the free flux vector, thus the metabolic fluxes can be expressed as:
| (8) |
where and
The time profiles of metabolite concentrations are described using the integral of Eq. (2), thus the integral of Eq. (8) is required. The indefinite integral of B-spline function of 3rd order is:
| (9) |
where
The B-spline functions for metabolite concentrations, after the rearrangement of , can subsequently be expressed in matrix form as:
| (10) |
where and
For a pre-determined knot series, the control points (matrix and the initial metabolite concentrations (vector are linear with respect to the model. The complete time series of metabolic fluxes can be estimated by solving a single optimization problem (Antoniewicz, 2013). The optimization estimates free fluxes and initial metabolite concentrations by minimizing the variance-weighted sum of squared residuals (SSR) between estimated and measured external metabolite concentrations ( and ) and external rates (if available (Leighty and Antoniewicz, 2011):
| (11) |
where Wi and Wrj are diagonal matrices containing the inverse of measurement variances of metabolite i and external rates j for every time point, respectively.
The optimization is linear with respect to P and , thus the solution to the problem can be solved explicitly using the equation (Leighty and Antoniewicz, 2011)
| (12) |
where H and J are the Hessian and Jacobian matrixes, respectively. From this point onwards, the formulation is identical to the L-DMFA approach (Leighty and Antoniewicz, 2011), namely the method used to estimate standard deviations for the estimated metabolic fluxes.
The B-DMFA algorithm was scripted in MATLAB, and is available from the corresponding author upon request.
2.2. Heuristic algorithm to determine the placement and number of knots
Finding the least-square solution of B-splines is a simple linear problem once the knots sequence has been specified, otherwise the problem is non-linear and difficult to solve. There are various parameterization approaches to specify the knot sequence, such as uniformly spaced or chord length (Prautzsch et al., 2002), however, these approaches are mainly designed to fit one curve at a time. Since we are simultaneously fitting a large number of curves using a single knot sequence, we developed a heuristic algorithm that uses the SSR within knot spans to guide the placement of knots (see Supplementary materials for details). The algorithm was conceived based on the fact that the shape of B-splines can be controlled locally, i.e., adjusting positions of proximal knots have limited distal effects. Generally there are more time series data points than there are piecewise polynomials required to fit the experimental data. As such, the algorithm starts from no internal knots and progressively adds knots until the SSR is reduced to an acceptable threshold. A region of data with greater dynamic behaviour will correspondingly have higher density of knots. This avoids issues related to over-fitting and oscillation.
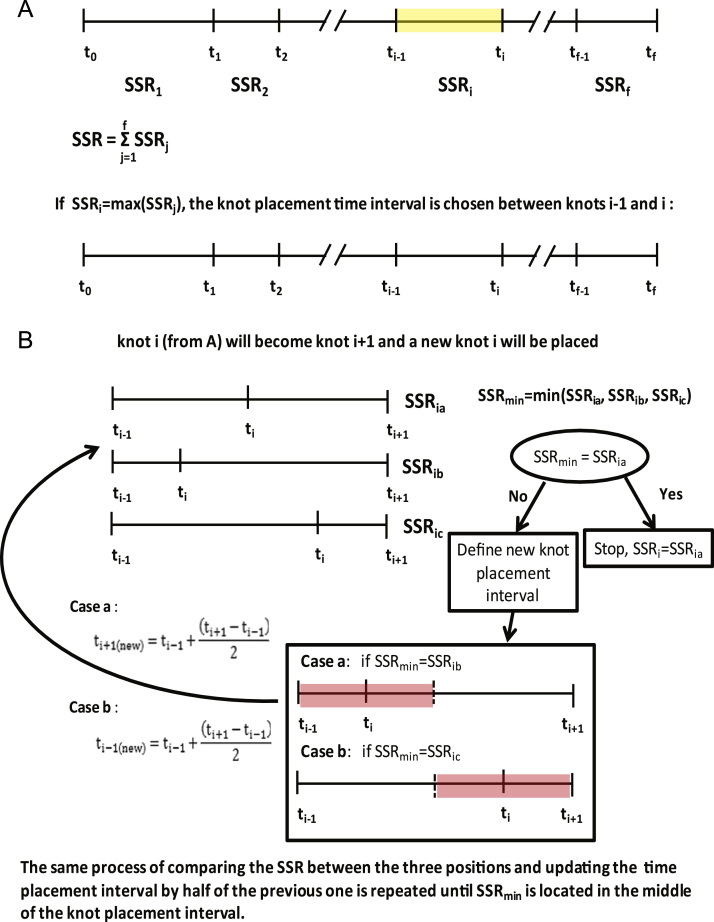
Briefly, the algorithm places a knot into the time span that has the largest contribution to the overall SSR (Fig. 1A). The position of each added knot is determined by a greedy trial-and-error approach, guided by SSR trial calculations. The SSR of placing a knot in either the middle of the time span or the middle of the half right or left of the defined time interval is compared (Fig. 1B). If the smallest SSR is given by placing the knot in the middle position, then it is kept as part of the final knot sequence. Otherwise, the focus is shifted to the right or left half of the current time span, chosen based on the knot position that gives the smallest SSR. The previous smallest SSR is compared again with the options of placing the new knot in the middle of the right or left half of the new time span (half of the previous time interval), and the knot position that generates the smallest SSR is selected. The process is repeated, with the time span segment becoming narrower, until the best knot position is found to be placed at the middle of the last defined time span. The algorithm starts again for the next knot. After the placement of the second and subsequent knots, the position of all knots are adjusted again to minimize SSR in order to account for the effects of other knots, whereby a better position for the ith knot is checked between the time interval defined by i-1th knot and i+1th knot. The whole process terminates when the minimum SSR is below the chi-square threshold.
Fig. 1.
(A) Selecting knot placement time interval. The time interval that has the greatest contribution to the total SSR is selected for placing a knot. (B) Placing knot i within the selected time interval. The position of the knot i is determined by an iterative process. First, the SSRs of placing the knot in the middle of the knot placement time interval, or the middle of the left or right half of the knot placement time interval are compared. The process is stopped if placing the knot in the middle of the knot placement time interval gives the smallest SSR. Otherwise the knot placement time interval is updated to the right or left half of the initial time interval, depending on which knot position gives the smallest SSR. Then the process is repeated on the new reduced knot placement time interval.
2.3. Metabolic model
The metabolic model was derived from the Mus musculus GeM (Quek and Nielsen, 2008) as previously described (Quek et al., 2010). The metabolic model contains the TCA cycle, pentose phosphate pathway (PPP) and pathways for the synthesis of essential biomass precursors (e.g., fatty acids, steroids, glycogen, and nucleotides). Under the current DMFA implementation, a fully determined model is required. The original model is undetermined by one degree of freedom, due to the possible localization of malic enzymes in both mitochondria and cytoplasm. Here, we assumed the mitochondrial enzyme to be inactive. Further modifications were made to the model to reflect differences in substrate consumption and by-product formation: (1) the uptake of hypoxanthine, choline, myo-inositol and ethanolamine were removed; (2) proline degradation pathway was replaced by its biosynthetic pathway; and (3) an IgG1 antibody production reaction was added (IgG1 composition shown in Table S1). L-alanyl-L-glutamine dipeptide was treated as free glutamine and alanine, and the decomposition of glutamine was assumed to be negligible since its extracellular accumulation is low. A biomass equation was used, with the precursor compositions obtained from a previous CHO cell culture study (Martínez et al., 2013). The average dry weight for the control cells at exponential growth phase was measured to be 350 [pg/cell]. The model in SBML format is provided in Supplementary materials.
2.4. Cell culture
The CHO cell line XL99-Ab2, producing an IgG1 antibody (Ab2), is a derivative of CHO-K1 (ATCC 61-CCL) adapted from adherent growth in serum containing medium to suspension growth in EX-CELL302 serum free medium at the University of New South Wales. The XL99-Ab2 cell line was further adapted to EX-CELL CD CHO Fusion (Sigma-Aldrich, Castle Hill, Australia) medium supplemented with 8 mM GlutaMAX (L-alanyl-L-glutamine; Life Technologies Australia, Mulgrave, Australia), 400 μg/ml Geneticin (Life Technologies Australia, Mulgrave, Australia) and 0.2% v/v Anti-Clumping Agent (Life Technologies Australia, Mulgrave, Australia). XL99-Ab2 cells were cultivated in four 1L shake flasks with a working volume of 200 ml in humidified Infors shaking incubators set at 37°C, 7.5% CO2 and 130 rpm. After 72.5 h, two shake flasks were transferred to a humidified incubator set at 32 °C with otherwise identical settings. Samples for viable cell density and cell diameter measurements as well as antibody and metabolite concentration analysis were taken twice a day. Cell number, viability and cell diameter were determined using a CedeX cell counter (Innovatis, Bielefeld, Germany). Ammonia concentrations were measured using a Nova Bioprofiler FLEX (Nova Biomedical, Waltham, USA). For extracellular metabolites and antibody analysis, 500 μl of cell suspension was removed from the cell cultures, centrifuged at 200g for 5 min and supernatants were frozen on dry ice and stored at −80 °C until further processing.
2.5. Extracellular metabolites and antibody concentration analysis
Glucose, lactate, L-alanyl-L-glutamine and amino acid concentrations were measured using HPLC as described previously (Dietmair et al., 2010). Antibody concentrations were determined using surface plasmon resonance with a Biacore T-100 system (GE Healthcare, Mansfield, Australia) and the human antibody capture kit (GE Healthcare, Mansfield, Australia) was used for the immobilization of an Ab2 binding α-Hu IgG FC-specific antibody onto the CM5 sensor chips. Samples were diluted 5–10 times in a 96 well plate and protein-A purified Ab2 was used to obtain a standard curve for quantification.
3. Results and discussion
3.1. Comparing B-DMFA against L-DMFA
The implementation of L-DMFA is formally identical to B-DMFA with order (k) 2, except that L-DMFA uses an optimization algorithm to specify knot sequence (Leighty and Antoniewicz, 2011). A diauxic growth model of yeast on glucose (Nielsen and Villadsen, 2003, Sonnleitner and Kappeli, 1986) was used to compare their performances (Fig. 2). In the simulated batch culture, S. cerevisiae initially metabolize glucose by an oxidoreductive metabolism. While the uptake rate of glucose is described by Monod kinetics, the oxidation of glucose is additionally limited by the respiratory capacity. Excess pyruvate therefore overflows to ethanol. As glucose is consumed, its consumption rate decreases to a point where spare respiratory capacity allows the reconsumption of ethanol. The uptake rate of ethanol is described by Monod kinetics that includes glucose inhibition. The complex dynamics of the oxygen consumption rate is shown in Fig. 2.
Fig. 2.
Simulated data of yeast diauxic growth in batch culture. Initially, cells metabolize glucose and produce ethanol and CO2. Ethanol is consumed when glucose is depleted. Transients in the oxygen consumption rate and CO2 production rate around the depletion of glucose and ethanol are shown.
The goodness-of-fit by L-DMFA (Leighty and Antoniewicz, 2011) and by B-DMFA were compared using the described simulated dataset. The dataset consisted of concentrations of glucose, ethanol, biomass, and rates of oxygen and carbon dioxide at 30 min intervals from 0 to 15 h. Initial glucose and biomass concentrations were set to 2 g/L and 0.1 g/L. Measurement errors were assumed to be 5% (assuming a minimum error of 0.01) (Fig. 3).
Fig. 3.
Fitting of simulated dataset of yeast diauxic growth by L-DMFA and B-DMFA, and simulations computation time. In (A)–(E) the simulated data points are represented by ◊, and the fitting of L-DMFA and B-DFMA are represented by black and red lines, respectively. In (F) the computation time (in an Intel®Core™ i5 CPU with 8 GB RAM) by L-DMFA and B-DMFA with increasing number of sampling points is shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The two approaches were comparable in terms of the ability to fit concentration datasets (Fig. 3 A–C). B-DMFA, however, gave a smoother fit for the oxygen and CO2 rates, particularly at time points when glucose and ethanol approached depletion (Fig. 3 D and E). Unlike L-DMFA, B-DMFA was able to trace the spike in the rates by allowing insertion of multiple knots at the same time point. Therefore, B-DMFA generates a better fitting for dynamic experimental rates that contain drastic changes in a short period of time.
A quantitative comparison of the fitting between both approaches was carried out (Table 1). Particularly, we examined how sampling frequency would affect performance. B-DMFA and L-DMFA were performed on ten datasets for each sampling frequency (15, 30 and 60 min sampling intervals). Each dataset was generated by corrupting the simulated concentrations and rates with normally distributed random noise (5% standard deviation). Since the upper limit of SSR in DMFA vary depending on the number of knots, a proxy for goodness-of-fit (SSRo) was used, which measures the discrepancy between the fitted and the noise-free models. This allows us to compare the goodness-of-fit for the various fitted models using the simulated noise-free dataset as a common basis. SSRo was found to be similar for both B-DMFA and L-DMFA. However, B-DMFA achieved this with a smaller number of internal knots (time points) and parameters than L-DMFA (Table 1), demonstrating the higher fitting power achieved using B-spline fitting.
Table 1.
Comparison of B-DMFA, L-DMFA and L-DMFA using the heuristic knot placement algorithm. For each sampling frequency ten datasets with noise (5% standard deviation) were simulated, for sampling every 15, 30 and 60 min. The datasets were analyzed with B-DMFA, L-DMFA and L-DMFA using the heuristic algorithm for knots placement. The SSR with respect to a dataset without noise (SSRo), the computation time (in an Intel®Core™ i5 CPU with 8 GB RAM), the number of internal knots required for fitting and the number of parameters were determined. The goodness-of-fit was similar between the three approaches, however, B-DMFA required less parameters. B-DMFA is also a much faster approach than L-DMFA, mainly due to the knots placement heuristic algorithm.
| B-DMFA | L-DMFA | L-DMFA+heuristic algorithm | ||
|---|---|---|---|---|
| 15 min | 94.8±24.2 | 125.9±32.8 | 97.1±22.5 | |
| SSRo | 30 min | 65.1±14.3 | 75.7±18.0 | 65.5±15.9 |
| 1 h | 49.0±12.6a | 51.1±11.5a | 50.2±12.2b | |
| 15 min | 23.2±13.2 | 3,543±3,119 | 36.3±12.3 | |
| Time (s) | 30 min | 9.5±2.9 | 826.2±413.4 | 22.5±13.5 |
| 1 h | 5.6±3.0a | 140.5±40.1a | 10.1±8.0b | |
| 15 min | 7±2 | 15±15 | 9±1 | |
| N° Internal knots | 30 min | 6±1 | 11±7 | 9±3 |
| 1 h | 6±1a | 8±4a | 9±4b | |
| 15 min | 40±6 | 69±61 | 45±6 | |
| N° Parameters | 30 min | 37±4 | 51±27 | 46±12 |
| 1 h | 36±5a | 40±14a | 46±17b |
One out of ten datasets did not converge with a 95% CI, thus the dataset was omitted from the analysis.
Two out of ten datasets did not converge with a 95% CI, thus the datasets were omitted from the analysis.
B-DMFA was found to be significantly faster than L-DMFA (Table 1). We attributed this to the heuristic algorithm for knots placement. L-DMFA uses an optimization approach, which begins with a maximum potential DMFA time points (e.g., 29 in the 15 min case) and removes one by one until the fit is still within the 95% confidence intervals (CI) (Leighty and Antoniewicz, 2011), while the B-DMFA heuristic algorithm adds knots starting from zero. This was verified when the knot placement heuristic algorithm was implemented in the original L-DMFA approach, and as a consequence, the computation times were significantly improved. Interestingly the B-DMFA approach remains the fastest (Table 1). The SSRo indicates that the resulting fits were similar to the original L-DFMA implementation. Nonetheless, B-DMFA remained the faster approach compared to the original L-DMFA with or without the heuristic algorithm, especially for datasets presenting higher sampling frequency (Table 1).
Fig. 3F shows that the B-DMFA approach is significantly less sensitive to the number of sampling time points than L-DMFA. At 16 time points, B-DMFA algorithm took 7 s to generate a statistically acceptable flux distribution, while L-DMFA took 115 s; at 91 sampling points, B-DMFA was two orders of magnitude faster (Fig. 3F). The longest computation time using L-DMFA was approximately 1 h. This computation time can be considered reasonable, however, when dealing with large models that describes the full cell metabolism (more than 100 reactions), and more experimental measurement (the full set of amino acids concentrations for a ten days cell culture) the computation time could be in the order of days.
The heuristic algorithm for knot placement was validated to be sound by a Monte Carlo approach. By randomly sampling 10,000 sets of knot sequences with a fixed number of knots, the best SSR was still 37% greater despite the Monte Carlo approach requiring 8.5 min to complete compared to the 12 s required by the heuristic algorithm. When sample size was increased to 100,000, the best SSR was 25% less than the heuristic algorithm, but only four instances returned SSR better than the algorithm (Figure S1). The sets of statistically acceptable knot sequences randomly generated were not very different compared to the knot sequence determined by the algorithm. The largest difference was 6% of the full time span when compared to the best set. However, the Monte Carlo approach took 1.4 h to completion.
3.2. Effects of temperature shift on CHO cells size and productivity
Growth profile, antibody concentration, mean cell diameter and cell specific productivity for the two control cultures (37 °C) and two temperature-shifted cultures (32 °C) are shown in Fig. 4. The log plot of cell number (Fig. 4A) showed that cell-specific growth rates for all four cultures were constant at about 0.025 to 0.029 h −1 during early exponential phase, but began to decline after 96 h. This is expected for shake flask cultures, as pH is not controlled. At 100 h, the cell-specific growth rates for the temperature-shifted and control cultures were between 0.012–0.016h−1 and 0.020–0.023 h−1, respectively, suggesting a reduced growth rate and slower metabolism for the temperature-shifted cultures. A lower peak cell density was also observed for the temperature-shifted cultures. Both observations are consistent with the literature (Kumar et al., 2007, Moore et al., 1997). Cultures reached peak cell density at 168 h and 192 h for the control and temperature-shifted cultures, respectively. Cell viabilities for all four cultures were above 90% for the remaining culture duration up to 240 h. Cell viability was slightly higher for the temperature-shifted cultures than the control cultures. The reduction of culture temperature from 37 °C to 32 °C appears to have reduced the cell growth rate and overall metabolism.
Fig. 4.
Viable cell density, antibody concentration, cell size and specific productivity. (A) XL99-Ab2 cells were cultivated in four shake flasks, two cultures were temperature-shifted at 72.5 h (32 °C-1 and 32 °C-2) and the other two were maintained at 37 °C throughout the whole experiment (37 °C-1 and 37 °C-2). Approximately 24 h after the temperature shift the growth rate decreased and peak viable cell density was lower in temperature-shifted cells compared to cells at 37 °C. (B) Final antibody titer was higher in temperature-shifted cells by about 42%. (C) An increase in cell diameter was observed for cells at 32 °C on day 5, approximately two days after the temperature shift. (D) Cell specific productivity (Qp) based on cell number increased for cells cultivated at 32 °C while cell volume normalized Qp did not show an increased productivity for temperature-shifted cells. Qp after 72.5 h was estimated until cells entered stationary phase.
The final antibody titer was on average 42% higher in the temperature-shifted cultures compared to the controls. As a consequence of a smaller integral viable cell density and a higher antibody titer, the average cell-specific productivity (Qp) appeared to be larger for the temperature-shifted cultures (Fig. 4D). Cells in the temperature-shifted cultures, however, showed a significant increase in cell diameter compared to the controls despite all cultures showing similar cell sizes before the temperature shift (Fig. 4C). Metabolic fluxes and antibody yields were therefore normalized to cell volume instead of cell number due to the observed differences in cell sizes. Since cell volume is proportional to cell mass (Frame and Hu, 1990), it is more accurate to express metabolic fluxes and antibody yields in terms of cell volume instead of cell number when cell size varies (Nielsen et al., 1997). The difference in average Qp was eliminated once Qp was normalized against cell volume (Fig. 4D).
Several studies of CHO cell cultures have reported an enhanced Qp when temperature was reduced from 37 °C to mild hypothermia (Al-Fageeh et al., 2006, Bollati-Fogolin et al., 2005, Fox et al., 2004, Furukawa and Ohsuye, 1998, Kumar et al., 2007, Yoon et al., 2003), but none of these studies accounted for the role of cell size. Temperature-shifted CHO cells typically accumulate in the G1 phase (Kumar et al., 2007, Moore et al., 1997), and might therefore be expected to be smaller in size than cells in S or G2 phases. However, an increase in size has been observed for cells arrested in G1 phase by means other than temperature shift (Al-Rubeai et al., 1992, Bi et al., 2004, Carvalhal et al., 2003, Lloyd et al., 2000). It has previously been found that cell-specific productivity correlates with cell size (Bi et al., 2004, Lloyd et al., 2000) and our data similarly showed that volumetric productivity was largely constant even with an almost 40% increase in observed cell volume.
3.3. B-DMFA captures dynamic metabolic changes caused by temperature shift
Dynamic flux analysis of CHO cell cultures was performed using the B-DMFA framework. The experimentally measured metabolite concentrations (amino acids, glucose, lactate and ammonia), antibody concentration and cell volume were used as input. Cell number was not used due to differences in cell diameter observed between the control and the temperature-shifted cultures (Fig. 4C). The second control culture (37 °C‐2) ) had a smaller inoculum concentration than the first (37 °C‐2), ), thus the 37 °C-2dataset was shifted back by 8.5 h. A total of 408 measurements (24 concentrations×17 time points) per culture were fitted (Fig. 5). Since oxygen uptake was not measured, the measurement set has only one degree of redundancy with respect to the model inputs and outputs (Quek et al., 2010). The calculated residual error therefore reflects the consistency of nitrogen balance among the measurements.
Fig. 5.
Fitted concentrations from CHO cell cultures by B-DMFA. The experimental data are presented by points and the B-spline fit by lines. Data for cell cultures at constant 37 °C are shown in pink (37°C‐1) and red (37°C‐2), and for temperature-shifted cultures in cyan (32°C‐1) and blue (32°C‐2). Measurements before 24 h were omitted from the analysis due to their high uncertainty. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The B-splines were generally able to accurately trace all of the measured concentrations for both control and temperature-shifted cultures (Fig. 5). The number of internal knots used and parameters fitted ranged from 0 to 2 and 93 to 141, respectively, for the four cultures; computation times were less than seven seconds. The fitting (Figure S2), and the number of knots and parameters used by L-DMFA were comparable with B-DMFA. However, the computation time increased by 3 orders of magnitude and the estimated fluxes are C0 continuous (Figure S3). The knot selection by B-DMFA was validated by a Monte Carlo approach using 100,000 random samples of knot sequence; SSR was improved by only 3% at most.
Compared to the temperature-shifted cultures, the control cultures displayed faster depletion of glucose, glutamine and serine, and accumulation of lactate, ammonia and glycine (Fig. 5). The time-resolved absolute fluxes estimated by B-DMFA were consistent with these general observations (Fig. 6). The biomass precursor (glycogen, steroids, fatty acids and nucleotides) reactions presented the biggest difference in flux between control and temperature-shifted cultures, particularly between 80 and 180 h a higher flux was observed for the cells at 37 °C. The fluxes for serine aldolase (glycine production from serine) and glutaminase (glutamate production from glutamine) were also higher in a similar fashion to the biomass precursor reactions, but glycolytic fluxes were only marginally higher. Lactate dehydrogenase flux was higher for the control cultures; in agreement with the lactate concentration profile (Fig. 5, Fig. 7). TCA cycle flux was similar under both culture conditions. The absolute fluxes clearly showed the transient changes in metabolism as the cultures progress (Fig. 6). Particularly in the control cultures, the glutamine consumption flux was elevated when cell-specific growth rate was beginning to decline at 100 h, which coincided with the switch from lactate production to consumption (Fig. 7).
Fig. 6.
Absolute intracellular fluxes over time for control and temperature-shifted cultures. Fluxes in [mM/h], time scale in hours. For reversible fluxes positive flux is taken from left to right or downward. The figure shows the estimated dynamic fluxes from 40 to 200 h (for full culture duration see Figure S4). Fluxes for the control cultures are shown in pink (37°C‐1) and red (37°C‐2), and for the temperature-shifted cultures in cyan (32°C‐1) and blue (32°C‐2). Fluxes calculated by B-DMFA are shown with solid lines and the 95% CIs are shown with dashed lines. Clear flux differences were observed between the control and temperature-shifted cultures. Specially for the biomass precursor reactions, control cultures showed higher fluxes than temperature-shifted cultures between 80 and 180 h. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Concentration, absolute fluxes and volume specific fluxes for glucose, lactate and glutamine. Concentration in [mM], absolute fluxes in [mM/h], volume specific fluxes in [mmol/gDW/cell] and time scale in hours. The data for the control cultures are shown in pink (37°C‐1) and red (37°C‐2), and the temperature-shifted cultures in cyan (32°C‐1) and blue (32°C‐2).The glucose and glutamine consumption fluxes were higher for the control cultures, whereas they were more stable over time for the temperature-shifted cultures. Similarly, the lactate flux was higher for the control cultures until 168 h. At stationary phase cells began to consume lactate. Differences in fluxes between the control and temperature-shifted cultures were reduced when fluxes were expressed as specific fluxes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The differences in absolute fluxes between the two conditions can partially be attributed to differences in cell density, as cultures at 37 °C grew faster than temperature-shifted cells and concomitantly accumulated more biomass. Volume-specific fluxes were therefore estimated to check if normalized fluxes were still different. While fluxes still showed transient behaviors, flux differences between the two culture conditions were diminished in general (Figure S5). Glutaminase and lactate dehydrogenase fluxes for the control cultures were still significantly higher compared to the temperature-shifted cultures (Fig. 7), but glycolytic fluxes were only marginally higher. The disparity between glycolytic and lactate fluxes suggests that, for the control cultures, a larger fraction of pyruvate was derived from other carbon sources in addition to glucose compared to temperature-shifted cultures. Biomass precursor reactions were kept at higher fluxes between 95 and 168 h for the control cultures, which is proportional to the higher cell-specific growth rate of the control cultures during that period of time. Cells entered stationary phase at 168 h, showing a reduction in their biomass precursor requirements and concomitantly most intracellular fluxes. An interesting exception are the TCA cycle fluxes, which appeared to be similar and constant throughout the whole culture for both conditions. Yoon et al. (2003) showed no change in cell-specific glucose, glutamine and lactate fluxes when temperature of CHO cell culture was reduced from 37 °C to 33 °C, but a significant increase in all three fluxes when temperature was reduced to 30 °C (Yoon et al., 2003).
Although the average volumetric Qp was the same for the two conditions (Fig. 4D) during exponential phase, the time-resolved volumetric Qp profile revealed by B-DMFA was very different (Fig. 8). The volumetric Qp for the temperature-shifted cultures was largely constant at about 6–7 pg cell−1day−1 for the entire culture duration, despite the decline in cell-specific growth rate. In contrast, volumetric Qp for the control cultures peaked between 8–10 pg cell−1day−1 at 90 h, before gradually declining to 0. The higher titer achieved in the temperature-shifted cultures could be attributed to a more stable volumetric Qp (Fig. 8).
Fig. 8.
Volume specific productivity (Qp) over time for control and temperature-shifted cultures. The figure shows the estimated volume specific Qp from 40 to 200 h. The data for the control cultures are shown in pink (37 °C-1) and red (37 °C-2), and the temperature-shifted cultures in cyan (32 °C-1) and blue (32 °C-2). The cell number was normalized by cell volume. The temperature-shifted cultures have an almost constant volumetric Qp over the full culture. On the other hand, cells at constant temperature have a peak in volumetric Qp around 90 h and then a decline. However, on average volumetric Qp is comparable in both conditions from 72.5 h to 168 h. Note that the 95% CIs of the Qp estimated by B-DMFA were large before the temperature shift due to the low cell number. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Overall, B-DFMA was able to quantify changes in cell metabolism for the whole cell culture duration. While we expect the temperature shift to induce transient metabolic behaviors in the cell cultures, B-DFMA was able to account for the gradual decline in cell-specific growth rate and dynamic consumption and production profiles. The changes observed in the latter were not expected for the control cultures. None of the metabolic fluxes were constant over time for any of the culture conditions (Fig. 6, Fig. 7, S4 and S5). The gradual decline in cell-specific growth rate may be pH-related, and possibly due to the accumulation of lactate and ammonia. While B-DMFA was designed for resolving transient metabolic behaviors, it was crucial in showing that volume-specific productivity of IgG1 was significantly more stable in the temperature-shifted cultures compared to the controls. These observations would have been missed if average fluxes were used, highlighting the benefit of B-DMFA compared to conventional MFA.
As other MFA techniques, the B-DMFA approach is unable to resolve fluxes of parallel and cyclic pathways. This challenge can be solved with isotopic tracer experiments (Ahn and Antoniewicz, 2011, Quek et al., 2010). In the analysis of CHO cell culture we were unable to resolve cytosolic versus mitochondrial malic enzyme flux and had to assume that all flux was cytosolic. Moreover, the volume specific fatty acids production was estimated to decrease during the cell culture (Figure S3) in agreement with the reduction of volume specific cell growth rate. However, fatty acids production significantly in excess of growth requirements was recently reported for CHO cells in stationary phase based on a 13C isotopic tracer study (Ahn and Antoniewicz, 2013). The fate of the lipids–lipid accumulation or futile turnover through beta-oxidation—has not yet been resolved and this phenomenon was therefore not considered in the current model. B-DMFA framework can be enhanced with isotopic tracer experiments to resolve parallel and cyclic pathways, and to estimate production fluxes of metabolites that are not uniquely part of a biomass equation. The full resolution of mammalian cells metabolic fluxes over-time still remains a challenge.
Since the current paper was submitted, another paper on dynamic metabolic flux analysis (Vercammen et al., 2014) was published describing the use of B-spline functions to fit the time profile of free fluxes. The approach of Vercammen et al. differs significantly from B-DMFA including the optimization approach and the approach to knot placement. Vercammen et al. directly estimate specific fluxes through a non-linear optimization approach, which includes irreversibility constraints, while B-DMFA (like the original L-DMFA approach) estimates absolute fluxes by an optimization problem that is linear with respect to the parameters and can be solved analytically. Whereas, B-DMFA employs a heuristic method to determine number and position on B-splines knots; Vercammen et al.’s approach uses Akaike’s model discrimination criterion for knots addition, adding one knot in one B-spline at a time. Therefore, the resulting B-spline functions to fit the free fluxes can have different number of internal knots. The final location of the knots is estimated by the non-linear optimization. Vercammen et al’s approach additionally includes in the non-linear optimisation the choice of optimal basis for the null space. Since Vercammen et al optimise all fitting parameters, it is expected that the fit is more efficient, i.e., a fit within the observed measurement errors is achieved with fewer fitted parameters than with B-DMFA. The price is the requirement to solve a non-trivial, non-linear optimization problem, which is computationally demanding even for the moderate size problem presented in their paper and may not be viable for dynamic flux analysis for large-scale networks. In contrast, B-DMFA with a heuristic knot-placement algorithm was shown to be capable of resolving dynamic fluxes in a large CHO model including 408 experimental measured data in less than seven seconds.
4. Conclusions
We have demonstrated the usefulness of applying conventional B-spline notation and formulation to calculate dynamic fluxes without being restricted to any specific order of fitting. B-DMFA is capable of generating smooth fit for complete time series experimental data, as well as incorporating statistical estimation of dynamic metabolic fluxes in one step using redundant datasets. Using a higher-order fit, B-DMFA was shown to perform better in fitting dynamic external rates compared to L-DMFA. The heuristic algorithm developed to determine the knots sequence was validated to be accurate and was demonstrated to be much faster than the optimization method used in the L-DMFA approach (Leighty and Antoniewicz, 2011). By resolving the dynamic changes in metabolic fluxes in temperature-shifted XL99-Ab2 culture, we showed that the reduction of growth and overall metabolic rates by inducing mild hypothermia may have benefited the CHO cell culture in terms of enhancing the stability of recombinant protein production, leading to an observed increase in final antibody titer by 42%. This leaves room to speculate whether the slower cell metabolism and the reduced accumulation of lactate and ammonia have a positive effect on the stability of recombinant protein productivity.
Acknowledgements
The authors would like to thank Mr Michael Wang for the metabolomic analysis, which was performed within the Queensland Node of Metabolomics Australia.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2015.06.001.
Appendix A. Supplementary material
Supplementary material
References
- Ahn W.S., Antoniewicz M.R. Metabolic flux analysis of CHO cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab. Eng. 2011;13:598–609. doi: 10.1016/j.ymben.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Ahn W.S., Antoniewicz M.R. Parallel labeling experiments with [1,2-(13)C]glucose and [U-(13)C]glutamine provide new insights into CHO cell metabolism. Metab. Eng. 2013;15:34–47. doi: 10.1016/j.ymben.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Al-Fageeh M.B., Marchant R.J., Carden M.J., Smales C.M. The cold-shock response in cultured mammalian cells: harnessing the response for the improvement of recombinant protein production. Biotechnol. Bioeng. 2006;93:829–835. doi: 10.1002/bit.20789. [DOI] [PubMed] [Google Scholar]
- Al-Rubeai M., Emery A.N., Chalder S., Jan D.C. Specific monoclonal-antibody productivity and the cell cycle-comparisons of batch, continuous and perfusion cultures. Cytotechnology. 1992;9:85–97. doi: 10.1007/BF02521735. [DOI] [PubMed] [Google Scholar]
- Altamirano C., Illanes A., Becerra S., Cairo J.J., Godia F. Considerations on the lactate consumption by CHO cells in the presence of galactose. J. Biotechnol. 2006;125:547–556. doi: 10.1016/j.jbiotec.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Antoniewicz M.R. Dynamic metabolic flux analysis-tools for probing transient states of metabolic networks. Curr. Opin. Biotechnol. 2013 doi: 10.1016/j.copbio.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Antoniewicz M.R., Kraynie D.F., Laffend L.A., Gonzalez-Lergier J., Kelleher J.K., Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab. Eng. 2007;9:277–292. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Klopprogge C., Zelder O., Heinzle E., Wittmann C. Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl. Environ. Microb. 2005;71:8587–8596. doi: 10.1128/AEM.71.12.8587-8596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J.X., Shuttleworth J., Ai-Rubeai M. Uncoupling of cell growth and proliferation results in enhancement of productivity in p21(C1P1)-arrested CHO cells. Biotechnol. Bioeng. 2004;85:741–749. doi: 10.1002/bit.20025. [DOI] [PubMed] [Google Scholar]
- Boghigian B.A., Seth G., Kiss R., Pfeifer B.A. Metabolic flux analysis and pharmaceutical production. Metab. Eng. 2010;12:81–95. doi: 10.1016/j.ymben.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Bollati-Fogolin M., Forno G., Nimtz M., Conradt H.S., Etcheverrigaray M., Kratje R. Temperature reduction in cultures of hGM-CSF-expressing CHO cells: effect on productivity and product quality. Biotechnol. Prog. 2005;21:17–21. doi: 10.1021/bp049825t. [DOI] [PubMed] [Google Scholar]
- Bonarius H.P., Hatzimanikatis V., Meesters K.P., de Gooijer C.D., Schmid G., Tramper J. Metabolic flux analysis of hybridoma cells in different culture media using mass balances. Biotechnol. Bioeng. 1996;50:299–318. doi: 10.1002/(SICI)1097-0290(19960505)50:3<299::AID-BIT9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Burleigh S.C., van de Laar T., Stroop C.J., van Grunsven W.M., O'Donoghue N., Rudd P.M., Davey G.P. Synergizing metabolic flux analysis and nucleotide sugar metabolism to understand the control of glycosylation of recombinant protein in CHO cells. BMC Biotechnology. 2011;11:95. doi: 10.1186/1472-6750-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhal A.V., Marcelino I., Carrondo M.J.T. Metabolic changes during cell growth inhibition by p27 overexpression. Appl. Microbiol. Biotechnol. 2003;63:164–173. doi: 10.1007/s00253-003-1385-5. [DOI] [PubMed] [Google Scholar]
- Cox M.G. The numerical evaluation of B-splines. IMA J. Appl. Math. 1972;10:134–149. [Google Scholar]
- Curry H.B., Schoenberg I.J. On Spline distributions and their limits—the Polya distribution functions. Bull. Am. Math. Soc. 1947;53 1114-1114. [Google Scholar]
- Curry H.B., Schoenberg I.J. On Pólya frequency functions IV: the fundamental spline functions and their limits. J. d'anal. Math. 1966;17:71–107. [Google Scholar]
- De Boor C. On calculating with B-splines. J. Approx. Theory. 1972;6:50–62. [Google Scholar]
- Dietmair S., Timmins N.E., Gray P.P., Nielsen L.K., Krömer J.O. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal. Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Fendt S.M., Sauer U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst. Biol. 2010:4. doi: 10.1186/1752-0509-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox S.R., Patel U.A., Yap M.G.S., Wang D.I.C. Maximizing interferon-gamma production by Chinese hamster ovary cells through temperature shift optimization: Experimental and modeling. Biotechnol. Bioeng. 2004;85:177–184. doi: 10.1002/bit.10861. [DOI] [PubMed] [Google Scholar]
- Frame K.K., Hu W.S. Cell-volume measurement as an estimation of mammalian-cell biomass. Biotechnol. Bioeng. 1990;36:191–197. doi: 10.1002/bit.260360211. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Ohsuye K. Effect of culture temperature on a recombinant CHO cell line producing a C-terminal alpha-amidating enzyme. Cytotechnology. 1998;26:153–164. doi: 10.1023/A:1007934216507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Gammell P., Clynes M. Proliferation control strategies to improve productivity and survival during CHO based production culture—a summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology. 2007;53:33–46. doi: 10.1007/s10616-007-9047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighty R.W., Antoniewicz M.R. Dynamic metabolic flux analysis (DMFA): a framework for determining fluxes at metabolic non-steady state. Metab. Eng. 2011;13:745–755. doi: 10.1016/j.ymben.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Lequeux G., Beauprez J., Maertens J., van Horen E., Soetaert W., Vandamme E., Vanrolleghem P.A. Dynamic metabolic flux analysis demonstrated on cultures where the limiting substrate is changed from carbon to nitrogen and vice versa. J. Biomed. Biotechnol. 2010 doi: 10.1155/2010/621645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaneras F., Pico J. A procedure for the estimation over time of metabolic fluxes in scenarios where measurements are uncertain and/or insufficient. BMC Bioinformatics. 2007:8. doi: 10.1186/1471-2105-8-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D.R., Holmes P., Jackson L.P., Emery A.N., Al-Rubeai M. Relationship between cell size, cell cycle and specific recombinant protein productivity. Cytotechnology. 2000;34:59–70. doi: 10.1023/A:1008103730027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N.N., Ellet J., Okediadi C., Hermes P., McCormick E., Casnocha S. A single nutrient feed supports both chemically defined NS0 and CHO fed-batch processes: improved productivity and lactate metabolism. Biotechnol. Prog. 2009;25:1353–1363. doi: 10.1002/btpr.238. [DOI] [PubMed] [Google Scholar]
- Mahadevan R., Edwards J.S., Doyle F.J. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys. J. 2002;83:1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez V., Gerdtzen Z.P., Andrews B.A., Asenjo J.A. Viral vectors for the treatment of alcoholism: Use of metabolic flux analysis for cell cultivation and vector production. Metab. Eng. 2010;12:129–137. doi: 10.1016/j.ymben.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Martínez V.S., Dietmair S., Quek L.E., Hodson M.P., Gray P., Nielsen L.K. Flux balance analysis of CHO cells before and after a metabolic switch from lactate production to consumption. Biotechnol. Bioeng. 2013;110:660–666. doi: 10.1002/bit.24728. [DOI] [PubMed] [Google Scholar]
- Matasci M., Hacker D.L., Baldi L., Wurm F.M. Recombinant therapeutic protein production in cultivated mammalian cells: current status and future prospects. Drug Discov. Today: Technol. 2008;5:e37–e42. doi: 10.1016/j.ddtec.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Meadows A.L., Karnik R., Lam H., Forestell S., Snedecor B. Application of dynamic flux balance analysis to an industrial Escherichia coli fermentation. Metab. Eng. 2010;12:150–160. doi: 10.1016/j.ymben.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Moore A., Mercer J., Dutina G., Donahue C.J., Bauer K.D., Mather J.P., Etcheverry T., Ryll T. Effects of temperature shift on cell cycle, apoptosis and nucleotide pools in CHO cell batch cultures. Cytotechnology. 1997;23:47–54. doi: 10.1023/A:1007919921991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. It is all about metabolic fluxes. J. Bacteriol. 2003;185:7031–7035. doi: 10.1128/JB.185.24.7031-7035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J., Villadsen J. Kluwer Academic/Plenum Publishers; New York; London: 2003. Bioreaction Engineering Principles. [Google Scholar]
- Nielsen L.K., Reid S., Greenfield P.F. Cell cycle model to describe animal cell size variation and lag between cell number and biomass dynamics. Biotechnol. Bioeng. 1997;56:372–379. doi: 10.1002/(SICI)1097-0290(19971120)56:4<372::AID-BIT3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Niklas J., Schrader E., Sandig V., Noll T., Heinzle E. Quantitative characterization of metabolism and metabolic shifts during growth of the new human cell line AGE1.HN using time resolved metabolic flux analysis. Bioprocess Biosyst. Eng. 2011;34:533–545. doi: 10.1007/s00449-010-0502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg G.B., Balcarcel R.R., Follstad B.D., Stephanopoulos G., Wang D.I. Metabolic effects on recombinant interferon-gamma glycosylation in continuous culture of Chinese hamster ovary cells. Biotechnol. Bioeng. 1999;62:336–347. doi: 10.1002/(sici)1097-0290(19990205)62:3<336::aid-bit10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Prautzsch H., Boehm W., Paluszny M. Bezier and B-Spline Techniques. Springer; Berlin: 2002. [Google Scholar]
- Quek L.E., Dietmair S., Kromer J.O., Nielsen L.K. Metabolic flux analysis in mammalian cell culture. Metabol. Eng. 2010;12:161–171. doi: 10.1016/j.ymben.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Quek L.E., Nielsen L.K. On the reconstruction of the Mus musculus genome-scale metabolic network model. Genome Inform. 2008;21:89–100. [PubMed] [Google Scholar]
- Sonnleitner B., Kappeli O. Growth of saccharomyces-cerevisiae is controlled by its limited respiratory capacity—formulation and verification of a hypothesis. Biotechnol. Bioeng. 1986;28:927–937. doi: 10.1002/bit.260280620. [DOI] [PubMed] [Google Scholar]
- Vallino J.J., Stephanopoulos G. Metabolic flux distributions in corynebacterium-glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 1993;41:633–646. doi: 10.1002/bit.260410606. [DOI] [PubMed] [Google Scholar]
- Varma A., Palsson B.O. Metabolic flux balancing—basic concepts, scientific and practical use. Bio-Technol. 1994;12:994–998. [Google Scholar]
- Vercammen D., Logist F., Impe J. Dynamic estimation of specific fluxes in metabolic networks using non-linear dynamic optimization. BMC Syst. Biol. 2014;8:132. doi: 10.1186/s12918-014-0132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z.Z., Kenty B., Koyrakh I., Borys M., Pan S.H., Li Z.J. Optimizing amino acid composition of CHO cell culture media for a fusion protein production. Process. Biochem. 2011;46:1423–1429. [Google Scholar]
- Yoon S.K., Song J.Y., Lee G.M. Effect of low culture temperature on specific productivity, transcription level, and heterogeneity of erythropoietin in chinese hamster ovary cells. Biotechnol. Bioeng. 2003;82:289–298. doi: 10.1002/bit.10566. [DOI] [PubMed] [Google Scholar]
- Young J.D. Metabolic flux rewiring in mammalian cell cultures. Curr. Opin. Biotechnol. 2013 doi: 10.1016/j.copbio.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material