Abstract
Background/Aim: Genetic variations of the CDKN2A and CDK4 gene have been associated to melanoma development. In the present study we investigated the potential associations of CDKN2A and CDK4 gene variants in a colombian population diagnosed with melanoma. Materials and Methods: DNA was extracted from whole blood samples from 85 patients diagnosed with cutaneous melanoma and 166 healthy controls. CDKN2A and CDK4 genes were genotyped using a high-resolution melting assay. Results: A similar distribution of CDKN2A variants 500C>G and 540C>T was found among cases (12% and 31% respectively) and controls (15% and 31% respectively). The CDKN2A variants were present in 36% of acral lentiginous melanoma and 39.47% of lentigo maligna. The haplotype analysis showed an association with susceptibility in the development of melanoma. Conclusion: The presence of haplotype 500G/540C in males is associated with an increased risk of melanoma in a colombian population, especially in subjects with a family history of cancer.
Keywords: Haplotype, melanoma, cancer, variant, susceptibility, germline variant
Approximately 324,635 cases of melanoma are diagnosed annually worldwide with a mortality rate of 22% (1). Melanoma etiology is complex and heterogeneous, involving environmental and genetic factors such as sun exposure, skin color, sex, number, and types of nevi, specific germline variants, and somatic mutations. CDKN2A and CDK4 genes exhibit high penetrance variants associated with melanoma pathogenesis (2,3). Approximately 10% of patients with melanoma who report a positive family history of melanoma have germline pathogenic variants. However, in patients with cutaneous melanoma without positive family history of melanoma, the germline pathogenic variants are usually rare but their presence may increase the risk of melanoma development (4).
CDKN2A gene encodes the protein p16, which inhibits the activity of the CDK4-cyclin complex. This complex phosphorylates the retinoblastoma protein allowing the cell to progress through G1 phase. Besides that, another protein encoded by CDKN2A is p14ARF(ARF), which regulates critical cell-cycle pathways, and it also inhibits MDM2 that leads to ubiquitination of p53 (5,6).
CDK4 gene encodes the catalytic subunit of a heterodimeric Ser/Thr protein kinase, which is involved in the control of the cell cycle (5). Germline variants on the CDK4 gene have been associated with an increased risk of melanoma development. These variants are located in codon 24 in exon 2, affecting the binding site with p16 protein and therefore promoting cell-cycle progression (7). For example, p.R24C and p.R24H variants of CDK4 lead to a 50-fold increased risk of developing melanoma (8,9).
The p16-cyclin D-CDK4/6-retinoblastoma protein (RB1) signaling (CDK4 pathway) is commonly dysregulated in various types of cancers (10). CDKN2A variants such as 500C>G (rs11515) and 540C>T (rs3088440, located in the 3’UTR: untranslated gene region), affect mRNA stability and post-transcriptional regulation of gene expression for both p16 and p14 proteins. Additionally, p.R24C (rs11547328) and p.R24H (rs104894340) variants of CDK4 affect its binding with p16 protein. For that reason, several studies have been conducted to estimate their association with melanoma development (11-13).
In the present study, we evaluated the association between four CDKN2A and CDK4 variants and the development or susceptibility of cutaneous melanoma in a Colombian population and we described the frequencies of these variants in a complex genetic background of our populations.
Materials and Methods
Study participants. The case and control study included 85 melanoma patients and 166 healthy controls, who were recruited between 2018 and 2019 at the Hospital Universitario Centro Dermatológico Federico Lleras Acosta E.S.E in Bogotá D.C, Colombia. Written informed consent was obtained for each patient included in the study. For cases group, the inclusion criteria considered age greater than 18 years and patients diagnosed with cutaneous melanoma. For control group, inclusion criteria included age greater than 18 years and subjects with neither family history of melanoma nor personal history of melanoma. Demographic, morphological, and clinical data was collected during the recruitment period. Variables such as family history of melanoma, history of cancer, nevus count, Fitzpatrick phototype, hair and eyes color were documented.
Institutional Review Board statement. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Research Ethics Committee of the Hospital Universitario Centro Dermatológico Federico Lleras Acosta E.S.E, Bogotá, Colombia. (Grant code 4000.16.6AE).
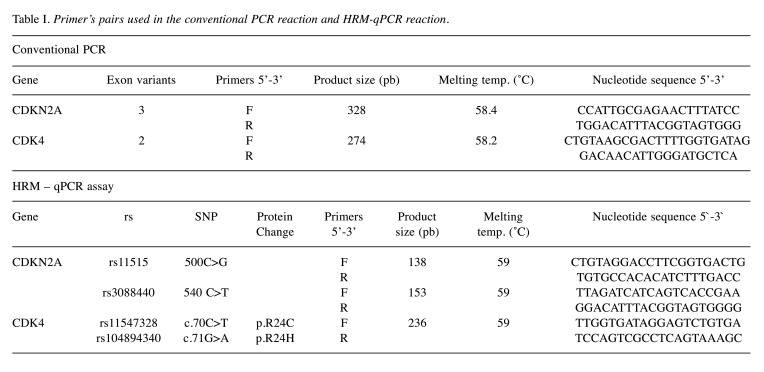
Genomic DNA extraction and genotyping assays. Blood samples were obtained from 251 subjects (85 cases and 166 controls) between years 2018 and 2019. Genomic DNA was extracted from patient’s using a QIAamp® DNA Mini and Blood Mini Handbook kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. After quantitation using UV-Vis spectrometry, DNA samples were stored at –20˚C for future use. PCR conditions were described in the previous study (14). CDKN2A and CDK4 primers are available in Table I.
Table I. Primer’s pairs used in the conventional PCR reaction and HRM-qPCR reaction.
Variant identification of the rs11515, rs3088440, rs11547328 and rs104894340 were carried out by High Resolution Melting analysis (HRM), using CFX96 Touch Tm Real-Time PCR detection system (Bio-Rad, Laboratories, Hercules, CA, USA). The samples were processed with Precision Melt Supermix kit (Bio-Rad, Laboratories). The real-time PCR was conducted in a total volume of 10 ul, containing 20 ng of genomic DNA and 0.5 μM of each primer. The sequence of primers for HRM analysis is listed in Table I. All samples were tested in duplicate. The conditions of qPCR and HRM analysis were: initial denaturation at 95˚C for 2 min; 45 cycles of denaturation at 95˚C for 10 sec, annealing for 30 s (between 53˚C and 65˚C depending on the exon) (Table I), and 72˚C extension cycle for 30 s. To induce heteroduplex formation, the PCR products were subjected to heating at 95˚C for 30 s, at 60˚C for 1 min, and HRM cycle at 65-95 ºC for 10 s/step in 0.2˚C increments.
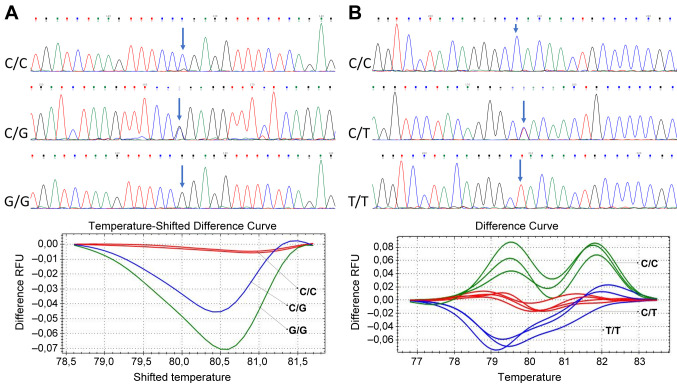
Wild type and mutant DNA, and non-template were used as controls groups in each amplification test. The data was analyzed using the Precision Melt Analysis™ v1.1 Software (Bio-Rad), according to the controls wild type, homozygous or heterozygous. The samples were assigned to different groups. Accuracy of HRM genotypic procedure was confirmed by direct DNA sequencing from several randomly selected samples. The primers of conventional PCR were used to amplify the DNA in the sequencing. The length of the amplicons is listed in Table I. The sequence was performed according to standard methods for Sanger sequencing using a BigDye Terminator V1.1 Cycle Sequencing Kit and ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems®). Variant analysis was performed using the software Chromas (free version). All sequences were confirmed by a second researcher independently (Figure 1).
Figure 1. Identification of 500C>G and 540C>T variants by High-resolution melting (HRM) and sequencing in CDKN2A gene. A). Sequencing and differences curves by HRM, for identification of 500C>G variant. B). Sequencing and differences curve by HRM for identification of 540C>T variant in CDKN2A gene.
Statistical analysis. Stata 16® was used for the Statistical analysis. Hardy Weinberg Equilibrium (HWE) was tested comparing genotype frequencies in both groups using Chi-square test (χ2). Shapiro Wilk test or Wilcoxon rank sum test was used to determinate the normal distribution for each variable. SNPStats program (Institut Catalá d’Oncologia, Barcelona, Spain), Chi-square test and Fisher’s test were applied to determine difference between genotypes, haplotypes, or allele. The genetic association was evaluated calculating Odds Ratio (OR) with a 95% confidence interval, statistical significance was defined as p<0.05. R-Studio was employed to confirm Haplotypes, with the Haplo.Stats package version 1.7.7 (Haplotype Score Tests or Regression Models).
Results
A total of 85 melanoma patients and 166 healthy individuals were included in the study. Demographics and clinical characteristics of cases and control groups was reported in previous studies (14).
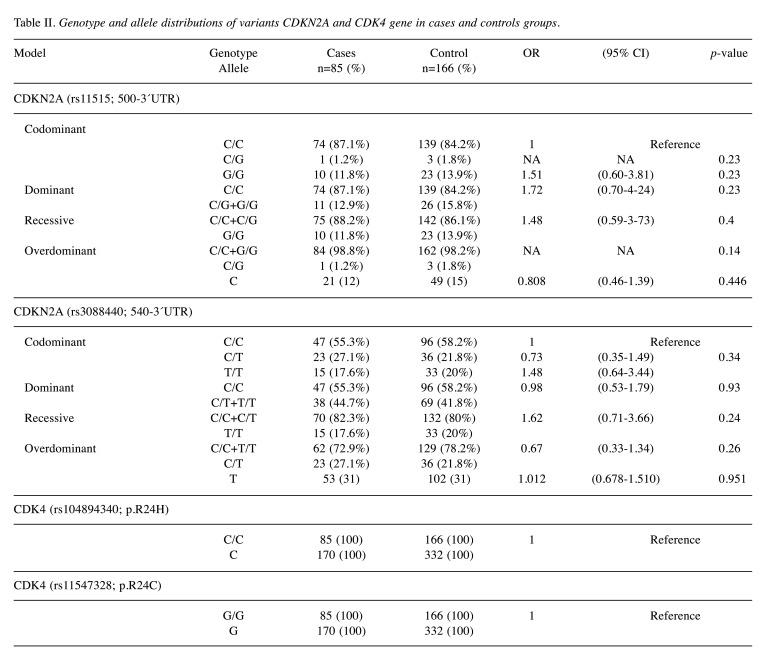
Association between CDKN2A and CDK4 genetic variations and melanoma risk. The genotypic results for CDKN2A 500C>G (rs11515), 540C>T (rs3088440) and CDK4 p.R24C (rs11547328), p.R24H (rs104894340) variants, are shown in Table II. Two genetic variants, rs11515 and rs3088440, were found to be in HWE in the control participants (p<0.05). Our results showed that polymorphisms of all candidate SNPs were not associated with melanoma risk (p>0.05) (Table II).
Table II. Genotype and allele distributions of variants CDKN2A and CDK4 gene in cases and controls groups.
CDKN2A variants 500C>G and 540C>T were present in 12% (11/85) and 31% (38/85) for cases and 15% (26/166) and 31% (69/166) for controls, respectively. The 500C>G variant was homozygous in 12% of the cases and 15% of the controls, meanwhile, the 540C>T variant was homozygous in 18% of the cases and 20% of the controls. On the other hand, CDK4 gene did not present variants in either case and control groups. Four types of genetic models (co-dominant, dominant, recessive and overdominant) were used in the association analysis of CDKN2A variants with susceptibility to develop melanoma (Table II). Our results indicate that p.R24C and p.R24H variants in the CDK4 gene were not present in either group, and 500C>G and 540C>T variants in CDKN2A gene did not display any statistical differences between case and control groups.
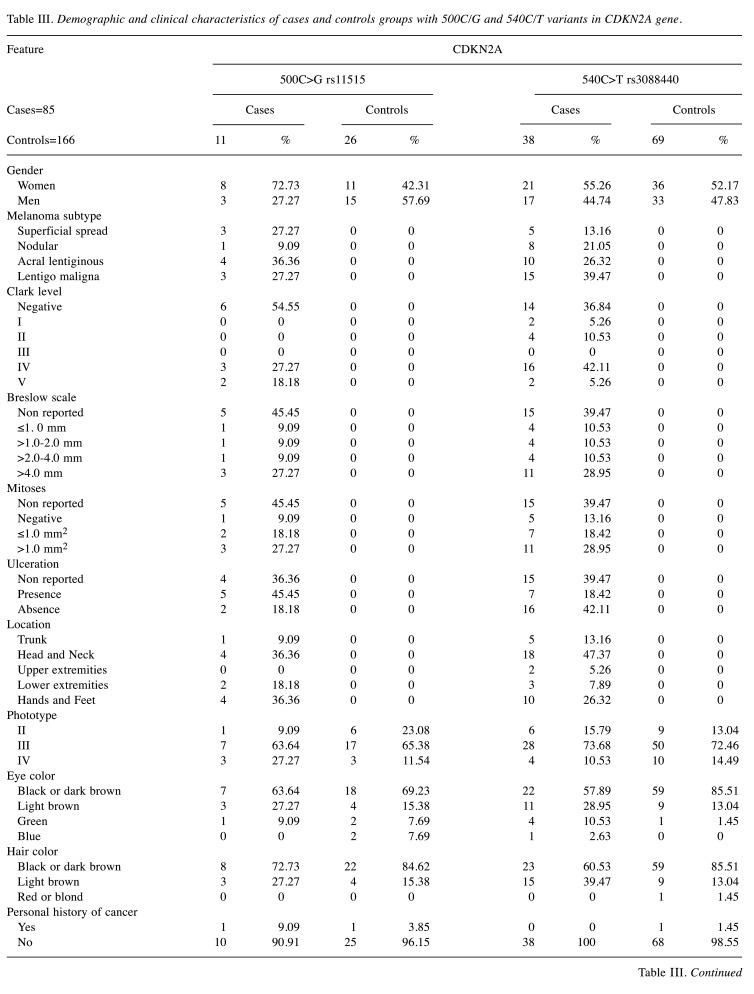
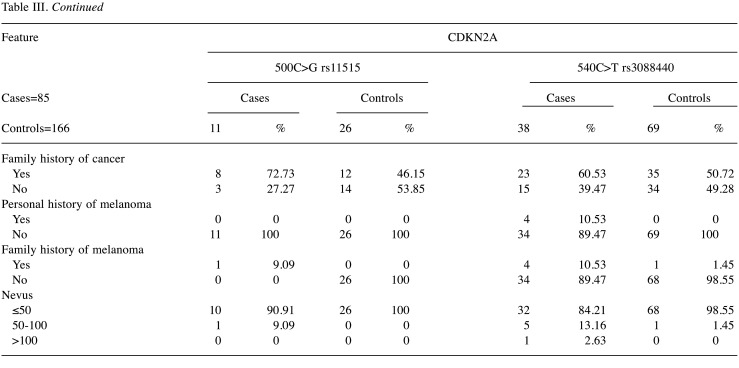
Regarding the 500C>G and 540C>T variants, 72.73% and 55.26% were women, respectively. Besides that, according to melanoma subtype, 36.36% were acral lentiginous melanoma (ALM) and 27.27% were Lentigo Maligna Melanoma (LMM) with 500C>G variant. 540C>T variant was present in 26.32% of ALM and 39.47% of LMM. Five cases and seven controls displayed 2 variants, where 4/5 were women in cases and 3/7 were women in control groups. For the 500C>G variant, 45.45% of cases presented ulceration, conversely 18.42% with 540C>T variant presented ulceration. According to the location of melanoma, the cases that presented the 500C>G variant were in head and neck, and lower extremities (36.36%), and cases with 540C>T variant were located mainly in head and neck (47.37%).
According to the Fitzpatrick phototypes, individuals that have 500C>G variant were phototype II in 63.64% of the cases and 65.38% of control group. Those with the 540C>T variant were phototype II in 73.68% of the cases and 72.46% of control group. Furthermore, 72.73% of the cases with 500C>G variant and 60.53% of the cases with 540C>T variants had a family history of melanoma (Table III).
Table III. Demographic and clinical characteristics of cases and controls groups with 500C/G and 540C/T variants in CDKN2A gene.
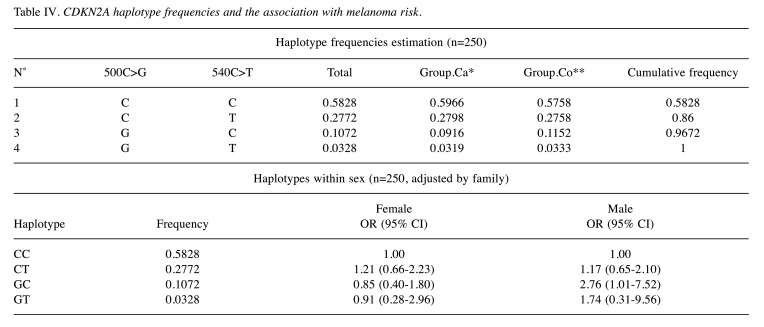
Association between haplotypes and melanoma risk. The most prevalent haplotype combination was 500C-540C in CDKN2A gene, which was present in 59% of cases and 57% of controls, but this analysis did not present any statistical significance between cases and control groups. However, when haplotype interaction analysis with sex and family history cancer was performed, an association was shown in males with haplotype 500G/540C when having a family history of cancer OR=2.76 (95% CI=1.01-7.52) (Table IV).
Table IV. CDKN2A haplotype frequencies and the association with melanoma risk.
Discussion
Due to the low melanoma incidence in the Latin-American region, few studies have been carried out, compared to other populations around the world. An interesting aspect is that reported subtypes of melanoma in the Latin-American region are different to those reported in high-incidence regions. In the Colombian population, several studies have shown that lentigo malignant melanoma is the most frequent with 36.41-52%, followed by acral lentiginous melanoma with 20-24.71% incidence (15). The frequency of melanoma in Latin America varies according to the studied population. For example, in the Argentinian population, the frequencies reported are 33.8% for superficial spreading and 14.5% for nodular melanoma (16) while in Brazil these are 34.45% for superficial spreading and 38.9% for nodular melanoma (17), suggesting that there may be genetic and environmental differences leading to various types of melanomas in this region.
Germline alterations in CDKN2A and CDK4 gene in DNA samples of cases and controls were analyzed to investigate known cancer susceptibility gene variants. The results showed that case and control groups did not display any germline variants in CDK4 gene (p.R24C and p.R24H). A study analyzed an Australian population of 1,109 melanoma patients, but no CDK4 variants were found (18). However, prior studies reported that CDK4 variants frequencies are approximately between 2-15% in Greek (19), Latvian (20), and other populations (5).
Germline variants (500C>G and 540C>T) in the CDKN2A 3’UTR gene have been found in approximately 12-31% of cutaneous melanomas and healthy controls, respectively (21-23), as well as associated with a risk of developing other types of cancer (24). However, it was shown in previous reports, that 14% of patients of 30 Israel melanoma families presented 500C>G germline variants, and other germline variants (25). A total of 285 patients analyzed in a Polish population reported that 21.78% of them presented the 500C>G variant while the 540C>T variant was not found (26), but in an Italian population Pellegrini et al. in 2017, reported frequencies of 38% for the 500C>G variant and 16% for the 540C>T variant (23). Consequently, those variants in CDKN2A 3’UTR were associated with high-risk melanoma in different case-control studies and have been reported the 500C>G and 540C>T variants in CDKN2A gene as a pathogenic gene alteration (21,27). Maccioni et al. in 2013 reported association of 540C>T variants with melanoma risk, OR=1.52 (95%CI=1.14-2.04) (28), and Kumar et al. in 2001 reported an OR=1.71 (95%CI=1.40-3.92) in Sweden population (22).
It was found that 5/11 cases presented both variants (500C>G and 540C>T), where 4/5 were women, and 100% had family history of cancer. The presence of both variants have been reported to be associated with a shorter disease-free survival and poor prognosis (22). The present and other studies confirm that the frequencies of CDKN2A 3’UTR variants depend on different factors, such as selection criteria, melanoma incidence in population analyzed, genetic ancestry and number of patients included in the study. This study reports that the incidence rate of germline variants of females was between 55-72% in cases, and 42-52% in controls groups. Furthermore, it was also found that 60-72% of cases, and 46-50% of controls had a history of other types of neoplasms (22). Multiple studies around the world have evaluated melanoma high-risk genes in patients with family history of melanoma and have reported heterogeneous results for the genetic variants association with the development of melanoma (20,22,23,26,29).
Those variants in CDKN2A 3’UTR had been associated with susceptibility to develop melanoma, in different case and control studies. But they have not been associated with the clinical features of patients or pathological characteristics of melanoma (22). In the present study, the haplotype distribution analysis reported no significant difference in distribution between cases and controls groups. However, it is estimated by haplotype interaction analysis with sex and family history of cancer, that an association exists between the presence of 500G/540C in male and positive family history cancer OR=2.76 (95% CI=1.01-7.52, p<0.05). These data are very important because variants in the 3’UTR region have been correlated with a shorter disease-free survival (30). Variants in CDKN2A 3’UTR are related with changes in p16 mRNA metabolism (transport, stability and translation) and cell-cycle progression, cell activity, and development of melanoma (26,29).
Conclusion
According to our results, frequencies in CDKN2A variants show a similar distribution of CDKN2A 3’UTR variants in cases and control groups. No variants were found in either group for CDK4. Additionally, this study indicates that in the Colombian population the presence of haplotype 500G/540C in males is associated with an increased risk of developing melanoma, especially in subjects with a family history of cancer. These results show that the Colombian population can carry other genetic variants related to the development of melanoma. Future studies should replicate these findings and involve subjects from other geographic regions of Colombia.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Conceptualization: L.D.G.C; methodology: D.T.P and L.D.G.C; software: D.T.P and S.R.G.Q; validation: J.N and L.D.G.C; formal analysis: D.T.P; investigation: D.T.P and L.D.G.C; data curation: S.R.G.Q; Visualization: D.T.P and L.D.G.C; writing–original draft preparation: D.T.P; writing–review and editing: L.D.G.C, J.N and S.R.G.Q; supervision: L.D.G.C; project administration: L.D.G.C; funding acquisition: L.D.G.C. All authors have read, discussed the results, and contributed to the final manuscript.
Acknowledgements
This work was funded by the Hospital Universitario Centro Dermatológico Federico Lleras Acosta E.S.E, Bogotá, Colombia. We thank the staff of the diagnostic support department for the collaboration in obtaining patients and the patients who participate in this study.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein AM, Struewing JP, Chidambaram A, Fraser MC, Tucker MA. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst. 2000;92(12):1006–1010. doi: 10.1093/jnci/92.12.1006. [DOI] [PubMed] [Google Scholar]
- 3.Carr S, Smith C, Wernberg J. Epidemiology and risk factors of melanoma. Surg Clin North Am. 2020;100(1):1–12. doi: 10.1016/j.suc.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Liu T, Tavtigian SV, Boucher K, Kohlmann W, Cannon-Albright L, Grossman D. Targeted germline sequencing of patients with three or more primary melanomas reveals high rate of pathogenic variants. Melanoma Res. 2020;30(3):247–251. doi: 10.1097/CMR.0000000000000645. [DOI] [PubMed] [Google Scholar]
- 5.Puntervoll HE, Yang XR, Vetti HH, Bachmann IM, Avril MF, Benfodda M, Catricalà C, Dalle S, Duval-Modeste AB, Ghiorzo P, Grammatico P, Harland M, Hayward NK, Hu HH, Jouary T, Martin-Denavit T, Ozola A, Palmer JM, Pastorino L, Pjanova D, Soufir N, Steine SJ, Stratigos AJ, Thomas L, Tinat J, Tsao H, Veinalde R, Tucker MA, Bressac-de Paillerets B, Newton-Bishop JA, Goldstein AM, Akslen LA, Molven A. Melanoma prone families with CDK4 germline mutation: Phenotypic profile and associations with MC1R variants. J Med Genet. 2013;50(4):264–270. doi: 10.1136/jmedgenet-2012-101455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casula M, Paliogiannis P, Ayala F, De Giorgi V, Stanganelli I, Mandalà M, Colombino M, Manca A, Sini MC, Caracò C, Ascierto PA, Satta RR, Melanoma Unit of Sassari (MUS) , Lissia A, Cossu A, Palmieri G, Italian Melanoma Intergroup (IMI) Germline and somatic mutations in patients with multiple primary melanomas: A next generation sequencing study. BMC Cancer. 2019;19(1):772. doi: 10.1186/s12885-019-5984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sargen MR, Pfeiffer RM, Yang XR, Tucker MA, Goldstein AM. Variation in cutaneous patterns of melanomagenesis according to germline CDKN2A/CDK4 status in melanoma-prone families. J Invest Dermatol. 2020;140(1):174–181.e3. doi: 10.1016/j.jid.2019.06.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kollmann K, Briand C, Bellutti F, Schicher N, Blunder S, Zojer M, Hoeller C. The interplay of CDK4 and CDK6 in melanoma. Oncotarget. 2019;10(14):1346–1359. doi: 10.18632/oncotarget.26515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheppard KE, McArthur GA. The cell-cycle regulator CDK4: An emerging therapeutic target in melanoma. Clin Cancer Res. 2013;19(19):5320–5328. doi: 10.1158/1078-0432.CCR-13-0259. [DOI] [PubMed] [Google Scholar]
- 10.Guo L, Qi J, Wang H, Jiang X, Liu Y. Getting under the skin: The role of CDK4/6 in melanomas. Eur J Med Chem. 2020;204:112531. doi: 10.1016/j.ejmech.2020.112531. [DOI] [PubMed] [Google Scholar]
- 11.Vargas-Torres SL, Portari EA, Klumb EM, Guillobel HC, de Camargo MJ, Russomano FB, Macedo JM. Association of CDKN2A polymorphisms with the severity of cervical neoplasia in a Brazilian population. Biomarkers. 2014;19(2):121–127. doi: 10.3109/1354750X.2014.881419. [DOI] [PubMed] [Google Scholar]
- 12.Cander S, Karkucak M, Gul OO, Sag SO, Yakut T, Ersoy C, Tuncel E, Erturk E. Association between p16(CDKN2A) C540G polymorphism and tumor behavior in prolactinoma: A single-center study. Biomed Rep. 2014;2(4):589–595. doi: 10.3892/br.2014.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buecher B, Gauthier-Villars M, Desjardins L, Lumbroso-Le Rouic L, Levy C, De Pauw A, Bombled J, Tirapo C, Houdayer C, Bressac-de Paillerets B, Stoppa-Lyonnet D. Contribution of CDKN2A/P16 ( INK4A ), P14 (ARF), CDK4 and BRCA1/2 germline mutations in individuals with suspected genetic predisposition to uveal melanoma. Fam Cancer. 2010;9(4):663–667. doi: 10.1007/s10689-010-9379-9. [DOI] [PubMed] [Google Scholar]
- 14.Tovar-Parra JD, Gutiérrez-Castañeda LD, Gil-Quiñones SR, Nova JA, Pulido L. CDKN2A polymorphism in melanoma patients in colombian population: A case-control study. Biomed Res Int. 2020;2020:7458917. doi: 10.1155/2020/7458917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutiérrez-Castañeda LD, Gamboa M, Nova JA, Pulido L, Tovar-Parra JD. Mutations in the BRAF, NRAS, and C-KIT genes of patients diagnosed with melanoma in Colombia population. Biomed Res Int. 2020;2020:2046947. doi: 10.1155/2020/2046947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loria D, Abriata MG, Santoro F, Latorre C. Cutaneous melanoma in Argentina: An analysis of its characteristics and regional differences. Ecancermedicalscience. 2020;14:1017. doi: 10.3332/ecancer.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vicente ALSA, Crovador CS, Macedo G, Scapulatempo-Neto C, Reis RM, Vazquez VL. Mutational profile of driver genes in Brazilian melanomas. J Glob Oncol. 2019;5:1–14. doi: 10.1200/JGO.19.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoude LG, Gartside M, Johansson P, Palmer JM, Symmons J, Martin NG, Montgomery GW, Hayward NK. Prevalence of Germline BAP1, CDKN2A, and CDK4 mutations in an Australian population-based sample of cutaneous melanoma cases. Twin Res Hum Genet. 2015;18(2):126–133. doi: 10.1017/thg.2015.12. [DOI] [PubMed] [Google Scholar]
- 19.Karagianni F, Njauw CN, Kypreou KP, Stergiopoulou A, Plaka M, Polydorou D, Chasapi V, Pappas L, Stratigos IA, Champsas G, Panagiotou P, Gogas H, Evangelou E, Tsao H, Stratigos AJ, Stefanaki I. CDKN2A/CDK4 status in greek patients with familial melanoma and association with clinico-epidemiological parameters. Acta Derm Venereol. 2018;98(9):862–866. doi: 10.2340/00015555-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veinalde R, Ozola A, Azarjana K, Molven A, Akslen LA, Doniņa S, Proboka G, Cēma I, Baginskis A, Pjanova D. Analysis of Latvian familial melanoma patients shows novel variants in the noncoding regions of CDKN2A and that the CDK4 mutation R24H is a founder mutation. Melanoma Res. 2013;23(3):221–226. doi: 10.1097/CMR.0b013e3283608695. [DOI] [PubMed] [Google Scholar]
- 21.Aitken J, Welch J, Duffy D, Milligan A, Green A, Martin N, Hayward N. CDKN2A variants in a population-based sample of Queensland families with melanoma. J Natl Cancer Inst. 1999;91(5):446–452. doi: 10.1093/jnci/91.5.446. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R, Smeds J, Berggren P, Straume O, Rozell BL, Akslen LA, Hemminki K. A single nucleotide polymorphism in the 3’untranslated region of the CDKN2A gene is common in sporadic primary melanomas but mutations in the CDKN2B, CDKN2C, CDK4 and p53 genes are rare. Int J Cancer. 2001;95(6):388–393. doi: 10.1002/1097-0215(20011120)95:6<388::aid-ijc1069>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini C, Maturo MG, Martorelli C, Suppa M, Antonini A, Kostaki D, Verna L, Landi MT, Peris K, Fargnoli MC. Characterization of melanoma susceptibility genes in high-risk patients from Central Italy. Melanoma Res. 2017;27(3):258–267. doi: 10.1097/CMR.0000000000000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wujcicka W, Zajac A, Szyllo K, Smolarz B, Romanowicz H, Stachowiak G. Association of SNPs in CDKN2A (P14ARF) tumour suppressor gene with endometrial cancer in postmenopausal women. In Vivo. 2020;34(2):943–951. doi: 10.21873/invivo.11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakobson E, Shemesh P, Azizi E, Winkler E, Lassam N, Hogg D, Brookes S, Peters G, Lotem M, Zlotogorski A, Landau M, Safro M, Shafir R, Friedman E, Peretz H. Two p16 (CDKN2A) germline mutations in 30 Israeli melanoma families. Eur J Hum Genet. 2000;8(8):590–596. doi: 10.1038/sj.ejhg.5200505. [DOI] [PubMed] [Google Scholar]
- 26.Przybyła A, Lamperska K, Mackiewicz A. Analysis of sequence variants in the 3’UTR of CDKN2A gene in melanoma patients. Contemp Oncol (Pozn) 2015;19(4):276–279. doi: 10.5114/wo.2015.54227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debniak T, Scott RJ, Huzarski T, Byrski T, Rozmiarek A, Debniak B, Załuga E, Maleszka R, Kładny J, Górski B, Cybulski C, Gronwald J, Kurzawski G, Lubinski J. CDKN2A common variants and their association with melanoma risk: A population-based study. Cancer Res. 2005;65(3):835–839. [PubMed] [Google Scholar]
- 28.Maccioni L, Rachakonda PS, Bermejo JL, Planelles D, Requena C, Hemminki K, Nagore E, Kumar R. Variants at the 9p21 locus and melanoma risk. BMC Cancer. 2013;13:325. doi: 10.1186/1471-2407-13-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamperska KM, Przybyła A, Kycler W, Mackiewicz A. The CDKN2a common variants: 148 Ala/Thr and 500 C/G in 3’ UTR, and their association with clinical course of melanoma. Acta Biochim Pol. 2007;54(1):119–124. [PubMed] [Google Scholar]
- 30.Sauroja I, Smeds J, Vlaykova T, Kumar R, Talve L, Hahka-Kemppinen M, Punnonen K, Jansèn CT, Hemminki K, Pyrhönen S. Analysis of G(1)/S checkpoint regulators in metastatic melanoma. Genes Chromosomes Cancer. 2000;28(4):404–414. doi: 10.1002/1098-2264(200008)28:4<404::aid-gcc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]