Abstract
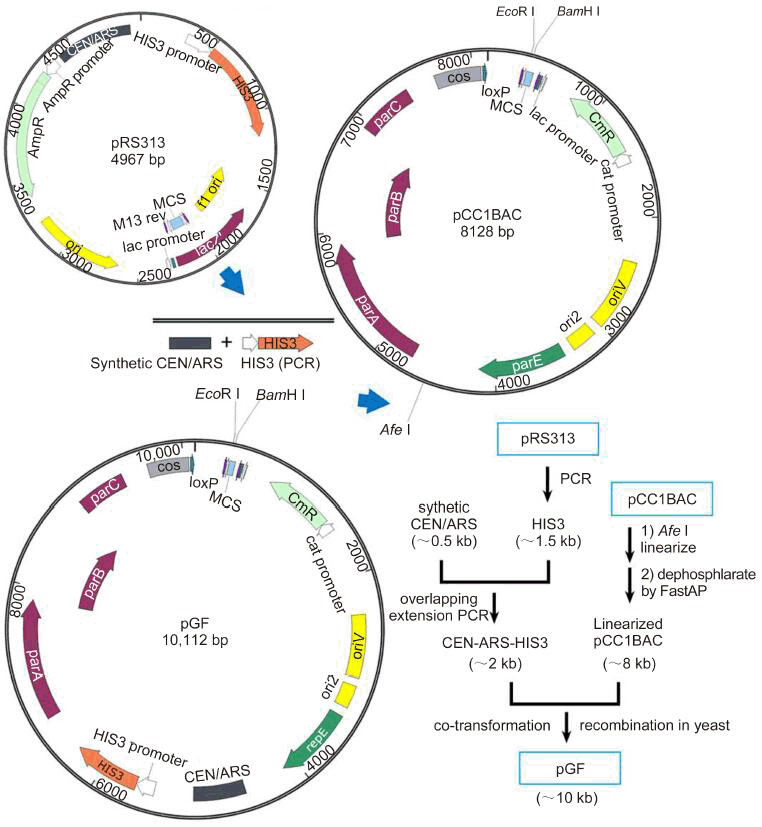
Synthetic biology is a newly developed field of research focused on designing and rebuilding novel biomolecular components, circuits, and networks. Synthetic biology can also help understand biological principles and engineer complex artificial metabolic systems. DNA manipulation on a large genome-wide scale is an inevitable challenge, but a necessary tool for synthetic biology. To improve the methods used for the synthesis of long DNA fragments, here we constructed a novel shuttle vector named pGF (plasmid Genome Fast) for DNA assembly in vivo. The BAC plasmid pCC1BAC, which can accommodate large DNA molecules, was chosen as the backbone. The sequence of the yeast artificial chromosome (YAC) regulatory element CEN6-ARS4 was synthesized and inserted into the plasmid to enable it to replicate in yeast. The selection sequence HIS3, obtained by polymerase chain reaction (PCR) from the plasmid pBS313, was inserted for screening. This new synthetic shuttle vector can mediate the transformation-associated recombination (TAR) assembly of large DNA fragments in yeast, and the assembled products can be transformed into Escherichia coli for further amplification. We also conducted in vivo DNA assembly using pGF and yeast homologous recombination and constructed a 31-kb long DNA sequence from the cyanophage PP genome. Our findings show that this novel shuttle vector would be a useful tool for efficient genome-scale DNA reconstruction.
Electronic Supplementary Material
Supplementary material is available for this article at 10.1007/s12250-016-3730-8 and is accessible for authorized users.
Keywords: synthetic biology, DNA fragment assembly, shuttle vector pGF, transformationassociated recombination (TAR)
Electronic supplementary material
Supplementary material, approximately 142 KB.
Footnotes
These authors contributed equally to this work.
ORCID: 0000-0001-9401-235X
References
- Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, Smith HO. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, Smith HO, Venter JC. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Smith HO, Hutchison CA, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- Hou Z, Xiao G. Total chemical synthesis, assembly of human torque teno virus genome. Virol Sin. 2011;26:181–189. doi: 10.1007/s12250-011-3187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P, Thilly WG. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci U S A. 1989;86:9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci U S A. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kaur J. Primer Based Approach for PCR Amplification of High GC Content Gene: Mycobacterium Gene as a Model. Mol Biol Int. 2014;2014:937308. doi: 10.1155/2014/937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci U S A. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Cho S, Song Y, Kim SC, Cho BK. Emerging tools for synthetic genome design. Mol Cells. 2013;35:359–370. doi: 10.1007/s10059-013-0127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Merryman C, Gibson DG. Methods and applications for assembling large DNA constructs. Metab Eng. 2012;14:196–204. doi: 10.1016/j.ymben.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Noskov VN, Chuang RY, Gibson DG, Leem SH, Larionov V, Kouprina N. Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat Protoc. 2011;6:89–96. doi: 10.1038/nprot.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- Varadaraj K, Skinner DM. Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene. 1994;140:1–5. doi: 10.1016/0378-1119(94)90723-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lin J, Li N, Hu Z, Deng F. Characterization and genomic analysis of a plaque purified strain of cyanophage PP. Virol Sin. 2013;28:272–279. doi: 10.1007/s12250-013-3363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material, approximately 142 KB.


