Abstract
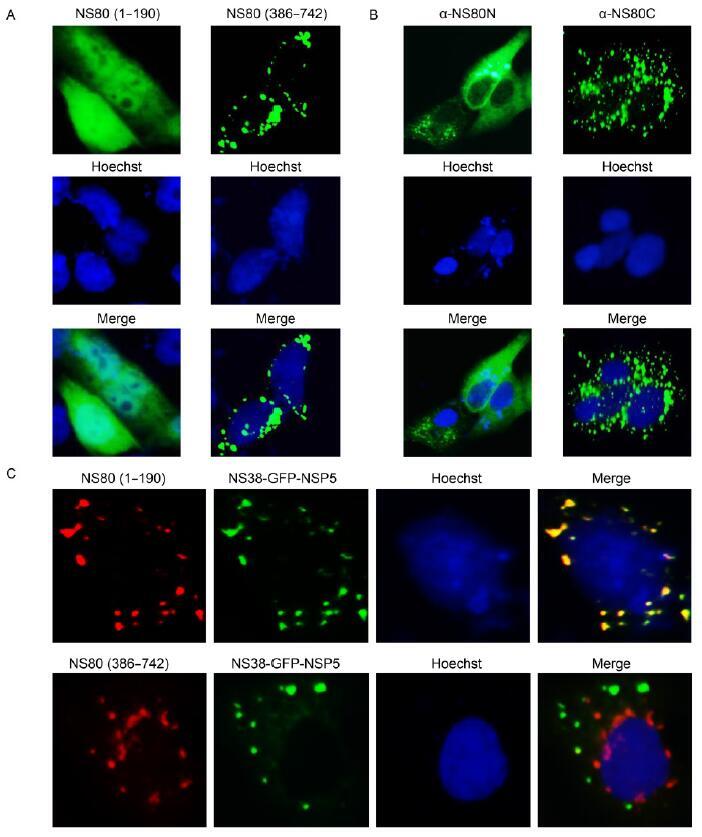
Aquareovirus species vary with respect to pathogenicity, and the nonstructural protein NS80 of aquareoviruses has been implicated in the regulation of viral replication and assembly, which can form viral inclusion bodies (VIBs) and recruit viral proteins to its VIBs in infected cells. NS80 consists of 742 amino acids with a molecular weight of approximately 80 kDa. Interestingly, a short specific fragment of NS80 has also been detected in infected cells. In this study, an approximately 58-kDa product of NS80 was confirmed in various infected and transfected cells by immunoblotting analyses using α-NS80C. Mutational analysis and time course expression assays indicated that the accumulation of the 58-kDa fragment was related to time and infection dose, suggesting that the fragment is not a transient intermediate of protein degradation. Moreover, another smaller fragment with a molecular mass of approximately 22 kDa was observed in transfected and infected cells by immunoblotting with a specific anti-FLAG monoclonal antibody or α-NS80N, indicating that the 58- kDa polypeptide is derived from a specific cleavage site near the amino terminus of NS80. Additionally, different subcellular localization patterns were observed for the 22-kDa and 58-kDa fragments in an immunofluorescence analysis, implying that the two cleavage fragments of NS80 function differently in the viral life cycle. These results provide a basis for additional studies of the role of NS80 played in replication and particle assembly of the Aquareovirus.
Keywords: Aquareovirus, nonstructural protein NS80, cleavage fragments, subcellular localization, functional analysis
Footnotes
ORCID: 0000-0003-4681-0060
References
- Arnold MM, Murray KE, Nibert ML. Formation of the factory matrix is an important, though not a sufficient function of nonstructural protein mu NS during reovirus infection. Virology. 2008;375:412–423. doi: 10.1016/j.virol.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attoui H, Fang Q, Mohd Jaafar F, Cantaloube JF, Biagini P, de Micco P, de Lamballerie X. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae) J Gen Virol. 2002;83:1941–1951. doi: 10.1099/0022-1317-83-8-1941. [DOI] [PubMed] [Google Scholar]
- Becker MM, Peters TR, Dermody TS. Reovirus sigma NS and mu NS proteins form cytoplasmic inclusion structures in the absence of viral infection. J Virol. 2003;77:5948–5963. doi: 10.1128/JVI.77.10.5948-5963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nunez A, Menaya-Vargas R, Benavente J M-, Costas J. Avian reovirus microNS protein forms homooligomeric inclusions in a microtubule-independent fashion, which involves specific regions of its C-terminal domain. J Virol. 2010;84:4289–4301. doi: 10.1128/JVI.02534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering TJ, Arnold MM, Miller CL, Hurt JA, Joyce PL, Nibert ML. Carboxyl-proximal regions of reovirus nonstructural protein muNS necessary and sufficient for forming factory-like inclusions. J Virol. 2005;79:6194–6206. doi: 10.1128/JVI.79.10.6194-6206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering TJ, Kim J, Miller CL, Piggott CD, Dinoso JB, Nibert ML, Parker JS. Reovirus nonstructural protein mu NS recruits viral core surface proteins and entering core particles to factory-like inclusions. J Virol. 2004;78:1882–1892. doi: 10.1128/JVI.78.4.1882-1892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes SM, Hyatt AD, Eaton BT. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J Gen Virol. 1993;74:525–530. doi: 10.1099/0022-1317-74-3-525. [DOI] [PubMed] [Google Scholar]
- Busch LK, Rodriguez-Grille J, Casal JI, Martinez-Costas J, Benavente J. Avian and mammalian reoviruses use different molecular mechanisms to synthesize their {micro}NS isoforms. J Gen Virol. 2011;92:2566–2574. doi: 10.1099/vir.0.036459-0. [DOI] [PubMed] [Google Scholar]
- Cai L, Sun X, Shao L, Fang Q. Functional investigation of grass carp reovirus nonstructural protein NS80. Virol J. 2011;8:168. doi: 10.1186/1743-422X-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH. Subnanometer- resolution structures of the grass carp reovirus core and virion. J Mol Biol. 2008;382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhu J, Hui WH, Zhang X, Honig B, Fang Q, Zhou ZH. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Fang Q. Functional Analyses of Mammalian Reovirus Nonstructural Protein muNS and Expression. Virol Sin. 2009;24:1–8. doi: 10.1007/s12250-009-3016-5. [DOI] [Google Scholar]
- Fan C, Shao L, Fang Q. Characterization of the nonstructural protein NS80 of grass carp reovirus. Arch Virol. 2010;155:1755–1763. doi: 10.1007/s00705-010-0753-6. [DOI] [PubMed] [Google Scholar]
- Fan C, Zhang L, Lei C, Fang Q. Expression and Identification of Inclusion Forming-related Domain of NS80 Nonstructural Protein of Grass Carp Reovirus. Virol Sin. 2009;24:194–201. doi: 10.1007/s12250-009-3028-1. [DOI] [Google Scholar]
- Fang Q, Ke LH, Cai YQ. Growth characterization and high titre culture of GCHV. Virol Sin. 1989;3:314–319. [Google Scholar]
- Fang Q, Shah S, Liang Y, Zhou ZH. 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China C Life Sci. 2005;48:593–600. doi: 10.1360/062004-105. [DOI] [PubMed] [Google Scholar]
- Fields BN, Raine CS, Baum SG. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology. 1971;43:569–578. doi: 10.1016/0042-6822(71)90282-0. [DOI] [PubMed] [Google Scholar]
- Guo H, Sun X, Yan L, Shao L, Fang Q. The NS16 protein of aquareovirus-C is a fusion-associated small transmembrane (FAST) protein, and its activity can be enhanced by the nonstructural protein NS26. Virus Res. 2013;171:129–137. doi: 10.1016/j.virusres.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Ivanovic T, Boulant S, Ehrlich M, Demidenko AA, Arnold MM, Kirchhausen T, Nibert ML. Recruitment of cellular clathrin to viral factories and disruption of clathrin-dependent trafficking. Traffic. 2011;12:1179–1195. doi: 10.1111/j.1600-0854.2011.01233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WT, Lin FL, Wang YC, Shih WL, Lee LH, Liu HJ. Intracellular cleavage of sigmaA protein of avian reovirus. Virus Res. 2010;149:71–77. doi: 10.1016/j.virusres.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Chappell JD, Danthi P, Dermody TS. Genespecific inhibition of reovirus replication by RNA interference. J Virol. 2006;80:9053–9063. doi: 10.1128/JVI.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ooms LS, Chappell JD, Dermody TS. Identification of functional domains in reovirus replication proteins muNS and mu2. J Virol. 2009;83:2892–2906. doi: 10.1128/JVI.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PW, Hayes EC, Joklik WK. Characterization of antireovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology. 1981;108:134–146. doi: 10.1016/0042-6822(81)90533-X. [DOI] [PubMed] [Google Scholar]
- Mertens P, Attoui H, Duncan R, Dermody T, et al. The Double Stranded RNA viruses. In: Andrew M K, Adams Michael J, Carstens Eric B, Lefkowitz Elliot J, et al., editors. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Oxford: Elsevier; 2011. pp. 497–650. [Google Scholar]
- Miller CL, Broering TJ, Parker JS, Arnold MM, Nibert ML. Reovirus sigma NS protein localizes to inclusions through an association requiring the mu NS amino terminus. J Virol. 2003;77:4566–4576. doi: 10.1128/JVI.77.8.4566-4576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Arnold MM, Broering TJ, Hastings CE, Nibert ML. Localization of mammalian orthoreovirus proteins to cytoplasmic factory-like structures via nonoverlapping regions of microNS. J Virol. 2010;84:867–882. doi: 10.1128/JVI.01571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Broering TJ, Kim J, Higgins DE, Nibert ML. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol. 2002;76:4483–4496. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel AA, Rockemann DD, Hetrick FM, Samal SK. Identification of grass carp haemorrhage virus as a new genogroup of aquareovirus. J Gen Virol. 1999;80:2399–2402. doi: 10.1099/0022-1317-80-9-2399. [DOI] [PubMed] [Google Scholar]
- Shao L, Fan C, Maj E, Fang Q. Molecular characterization of nonstructural protein NS38 of grass carp reovirus. Virol Sin. 2010;25:123–129. doi: 10.1007/s12250-010-3115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Guo H, Yan LM, Liu H, Fang Q. Aquareovirus NS80 recruits viral proteins to its inclusions, and its C-terminal domain is the primary driving force for viral inclusion formation. PLoS One. 2013;8:55334. doi: 10.1371/journal.pone.0055334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AH, Chen LB, Fields B N. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV- 1 cells. Virology. 1982;120:399–411. doi: 10.1016/0042-6822(82)90040-X. [DOI] [PubMed] [Google Scholar]
- Shaw AL, Samal SK, Subramanian K, Prasad BV. The structure of aquareovirus shows how the different geometries of the two layers of the capsid are reconciled to provide symmetrical interactions and stabilization. Structure. 1996;4:957–967. doi: 10.1016/S0969-2126(96)00102-5. [DOI] [PubMed] [Google Scholar]
- Silverstein SC, Schur PH. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology. 1970;41:564–566. doi: 10.1016/0042-6822(70)90178-9. [DOI] [PubMed] [Google Scholar]
- Smith RE, Furuichi Y. Gene mapping of cytoplasmic polyhedrosis virus of silkworm by the full-length mRNA prepared under optimized conditions of transcription in vitro. Virology. 1980;103:279–290. doi: 10.1016/0042-6822(80)90187-7. [DOI] [PubMed] [Google Scholar]
- Touris-Otero F, Martinez-Costas J, Vakharia VN, Benavente J. Avian reovirus nonstructural protein microNS forms viroplasm- like inclusions and recruits protein sigmaNS to these structures. Virology. 2004;319:94–106. doi: 10.1016/j.virol.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Varela R, Martinez-Costas J, Mallo M, Benavente J. Intracellular posttranslational modifications of S1133 avian reovirus proteins. J Virol. 1996;70:2974–2981. doi: 10.1128/jvi.70.5.2974-2981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener JR, Bartlett JA, Joklik WK. The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein mu 2 and the major nonstructural protein mu NS, respectively. Virology. 1989;169:293–304. doi: 10.1016/0042-6822(89)90154-2. [DOI] [PubMed] [Google Scholar]
- Yan L, Liu H, Li X, Fang Q. The VP2 protein of grass carp reovirus (GCRV) expressed in a baculovirus exhibits RNA polymerase activity. Virol Sin. 2014;29:86–93. doi: 10.1007/s12250-014-3366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Zhang J, Guo H, Yan S, Chen Q, Zhang F, Fang Q. Aquareovirus NS80 initiates efficient viral replication by retaining core proteins within replication-associated viral inclusion bodies. PLoS One. 2015;10:0126127. doi: 10.1371/journal.pone.0126127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ji G, Liu H, Zhang K, Liu G, Sun F, Zhu P, Cheng L. Cryo-EM structure of a transcribing cypovirus. Proc Natl Acad Sci U S A. 2012;109:6118–6123. doi: 10.1073/pnas.1200206109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Guo H, Chen Q, Zhang F, Fang Q. The N-Terminal of aquareovirus NS80 is required for interacting with viral proteins and viral replication. PLoS One. 2016;11:0148550. doi: 10.1371/journal.pone.0148550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]


