Abstract
To characterize the formation mechanism and characteristics of two cointegrate plasmids in Salmonella enterica serotype Enteritidis strain S13, plasmids from strain S13 and three corresponding transconjugants were subjected to whole genome sequencing and analyzed using bioinformatics tools. The traits of two fusion plasmids in transconjugants were characterized by stability and conjugation experiments. Sequence analysis indicated that strain S13 contained four plasmids, including mcr-1-bearing pS13-1, blaCTX–M–55-carrying pS13-2, tet(M)-bearing pS13-3, and floR-carrying pS13-4. IncN1-F33:A–:B– plasmid pS13-2, respectively, fused with IncFI:A–:B– plasmid pS13-3 and IncX1 plasmid pS13-4, which generated two cointegrate plasmids, designated pS13D and pS13F, which involved in two intermolecular replicative mechanisms mediated by IS26 and the novel transposon Tn6952 (ΔTnAS3-IS26-ΔISEcp1-ramA-ΔIS26-ΔTnAS1), respectively. This is the first report of the fusion of the IncN1-F33:A–:B– plasmid and IncFI:A–:B– plasmid mediated by IS26, and with IncX1 plasmid mediated by Tn6952. The formation and evolution of cointegrate plasmids could expand the resistance and host spectrum of fusion plasmids.
Keywords: Salmonella enterica, cointegrate plasmid, IS26, Tn6952, IncN1-F33:A–:B– plasmid
Introduction
Salmonella is an important zoonotic intestinal pathogen and a leading cause of microbial food poisoning (Scallan et al., 2011). In recent years, the widespread application of antibiotics in clinical practice is the driving force of the resistance of salmonella to antimicrobial agents (Deng et al., 2012). The acquisition of genetic material, such as integron gene cassettes, transposons, and resistance plasmids, is the main reason for the rapid development of multidrug resistance (MDR) in Salmonella (Hsu et al., 2006; Wright, 2007; Hu and Li, 2009).
The emergence and spread of fusion plasmids in Enterobacteriaceae pose great public concerns. Conjugative plasmids can capture MDR non-conjugative plasmids through replicative transposition of insertion sequences, such as IS1, IS26, and ISkpn19, thereby expanding the host range of the plasmids (Ohtsubo et al., 1981; He et al., 2015; Chen et al., 2017; Xie et al., 2018; Li et al., 2020). In the present study, the conjugative CTX-M-producing IncN1-F33:A–:B– plasmid was captured by two non-conjugative plasmids in Salmonella, resulting in the rapid transmission of the antibiotic resistance genes tet(M) and floR. F33:A–:B– plasmids are epidemic in Escherichia coli and widely disseminated by acquiring other resistance genes, replicon genes, and plasmids (He et al., 2019). Here, two hybrid plasmids mediated by IS26 and Tn6952 were first characterized in the same Salmonella enterica serotype Enteritidis. Moreover, the traits of two fusion plasmids in transconjugants were further investigated by stability and conjugation experiments.
Materials and Methods
Bacterial Strains
In 2016, a clinical tet(M)-positive Salmonella enterica serotype Enteritidis strain S13 was isolated from a pig in a livestock premises during surveillance of the tet(M) gene in the Henan Province, China. The strain was identified by the VITEK 2 automated identification system (bioMérieux, Marcy-l’Étoile, France) and serotyped according to the Kauffmann–White scheme. Multilocus sequence typing (MLST) of strain S13 was identified according to the protocol recommended at http://mlst.warwick.ac.uk. The genes tet(M), mcr-1, blaCTX–M–55, and floR, and the genetic environment of tet(M) in strain S13 were characterized by polymerase chain reaction (PCR) analysis with the use of the primers listed in Supplementary Table 1.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing to 12 antibiotics of Salmonella strain S13 and its transconjugants was performed using the broth microdilution method and interpreted in accordance with the Clinical and Laboratory Standards Institute, (2017) guidelines. For florfenicol, the resistant breakpoint was interpreted in accordance with the guidelines proposed by the European Committee on Antimicrobial Susceptibility Testing1. E. coli strain ATCC 25922 was used for quality control.
Conjugation Experiments, S1 Nuclease-Pulsed Field Gel Electrophoresis (S1-PFGE), and Southern Blot Hybridization
Conjugation experiments were performed utilizing rifampicin-resistant E. coli strain C600 as the recipient in order to assess the transferability of plasmids to S. enterica strain S13. Four different transconjugants were selected on MacConkey agar plates supplemented with rifampin (400 mg/L) and doxycycline (16 mg/L) or florfenicol (16 mg/L), ceftiofur (16 mg/L), and colistin (2 mg/L). The plasmid profiles of the donor and transconjugant strains, and the location of the tet(M) gene were determined by S1-PFGE and southern blot hybridization. The transfer frequency was calculated as the ratio of the number of transconjugants per recipient. All transconjugants were confirmed by PCR analysis as described previously (Sun et al., 2019).
Whole Genome Sequencing (WGS) and Plasmid Analysis
To investigate alterations to the sizes of the plasmids and their genetic contexts in the donor and transconjugant strains, whole plasmid DNA of strain S13 and the corresponding transconjugants S13D, S13F, and S13S were extracted using the Qiagen Plasmid Midi Kit (Qiagen, Hilden, Germany) and sequenced with the NextSeq 500 Sequencing System (Illumina, Inc., San Diego, CA, United States) and the MinION nanopore sequencing device (Oxford Nanopore Technologies, Oxford, United Kingdom). The complete genome and plasmid sequences were assembled with the Unicycler 0.4.4 assembly pipeline (Wick et al., 2017; Li et al., 2018). The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and the RAST tool were used to annotate the completed plasmid sequence (Aziz et al., 2008). The plasmid sequences were compared with BRIG and Easyfig tool (Alikhan et al., 2011; Beatson, 2011).
Cointegration Assay and Plasmid Stability
To assess the self-transferability of two fusion plasmids, pS13D and pS13F, two cointegration assays were performed via conjugation utilizing the C600 transconjugants S13D and S13F as the donor, respectively, and azide-resistant E. coli J53 as the recipient. The conjugation frequency was calculated as the number of transconjugants per recipient. The stability of fusion plasmids pS13D and pS13F were assessed as described previously (Liu Y. Y. et al., 2020).
Nucleotide Sequence Accession Number
The complete sequences of plasmids pS13-1, pS13-2, pS13-3 (pS13S), pS13-4, and chromosome in S13 and the fusion plasmids pS13D and pS13F were submitted to the GenBank under accession numbers CP047090, CP047091, CP047092, CP047093, CP047094, MT657397, and MT742153, respectively.
Results and Disscussion
Characterization of tet(M)-Bearing S. enterica Stain S13
The S. enterica stain S13 carrying blaCTX–M–55, tet(M), floR, and mcr-1 genes was resistant to amoxicillin, ceftiofur, cefquinome, tetracycline, oxytetracycline, florfenicol, colistin, and sulfamethoxazole-trimethoprim (Supplementary Table 2). S1-PFGE showed that S13 harbored four plasmids, designated as pS13-1 (~96 kb), pS13-2 (~89 kb), pS13-3 (~77 kb), and pS13-4 (~30 kb) (Supplementary Figure 1). Conjugation experiments revealed that the tet(M)-bearing transconjugant S13D harboring a single plasmid with ~167 kb in size, pS13D, the floR-bearing transconjugant S13F carrying the ~128-kb plasmid pS13F, and the blaCTX–M–55-bearing transconjugant S13S bearing the ~89-kb plasmid pS13S had transfer frequencies of 5.36×10–5, 2.11×10–6, and 1.06×10–4, respectively, while no mcr-1-carrying transconjugant was obtained despite repeated attempts (Table 1). The tet(M) gene was located on plasmids pS13-3 and pS13D (Supplementary Figure 1).
TABLE 1.
The characterizations of plasmid carried by strain S13 and its transconjugants.
| Strain | Name | No. plasmids (name) | Size (kb) | Source | Conjugation frequencies | Replication type | Resistance genes |
| Parent strain | S13 | 4, (pS13-1, pS13-2, pS13-3, pS13-4) | ~96 ~89 ~77 ~30 | – | – | – IncN1-F33:A–:B– IncFI:A–:B– IncX1 | mcr-1 blaCTX–M–55tet(M), oqxAB, aadA1, aadA2, blaTEM floR |
| Transconjugants | S13D | 1, (pS13D) | ~167 | pS13-2 pS13-3 | 5.36×10–5 | IncN1–F33:A–:B– IncFI:A–:B– | tet(M), oqxAB, aadA1,aaDA2, blaTEM and blaCTX–M–55 |
| S13F | 1, (pS13F) | ~128 | pS13-2 pS13-4 | 2.11×10–6 | IncN1-F33:A–:B– IncX1 | floR, blaCTX–M–55 | |
| S13S | 1, (pS13S) | ~89 | pS13-2 | 1.06×10–4 | IncX1 | blaCTX–M–55 |
The transconjugant S13D was selected on MacConkey agar plates supplemented with rifampin (400 mg/L) and doxycycline (16 mg/L), S13F was selected on MacConkey agar plates supplemented with rifampin (400 mg/L) and florfenicol (16 mg/L), S13S was selected on MacConkey agar plates supplemented with rifampin (400 mg/L) and ceftiofur (16 mg/L).
Sequence Analysis of Plasmids in S. enterica Strain S13
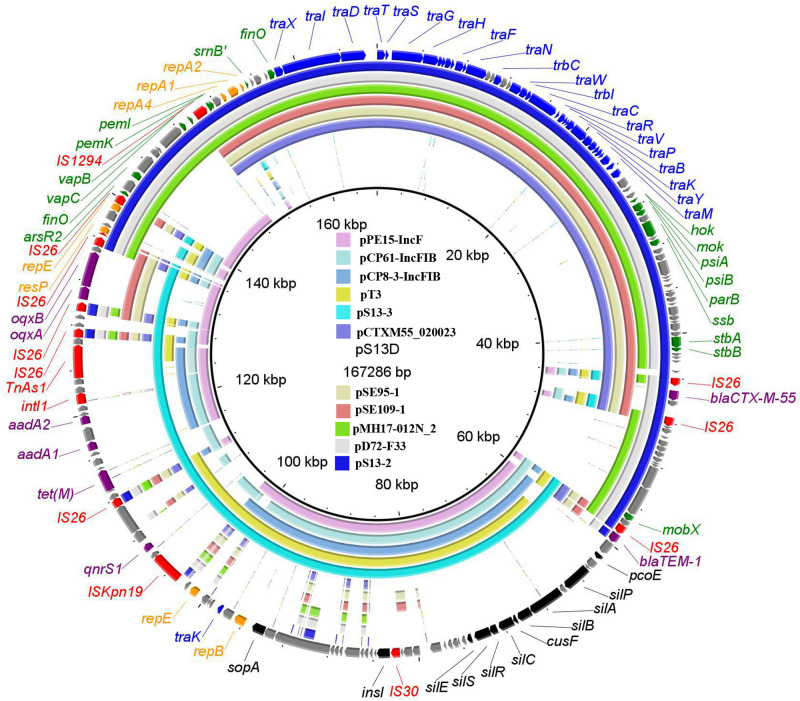
WGS showed that strain S13 has a 4, 744, 425-bp chromosome (G+C content, 50.96%) and four plasmids, pS13-1 (96, 320 bp), pS13-2 (89,179 bp), pS13-3 (77, 279 bp), and pS13-4 (30, 069 bp). The P7 phage-like plasmid pS13-1 carried the mcr-1 resistance gene. BLASTn comparisons showed that pS13-1 was highly similar to the mcr-1-carrying plasmid, as determined by strict structural analysis (Supplementary Figure 2). The pS13-1 plasmid was not self-transmissible and failed to bind to an auxiliary plasmid, although pS13-1-like plasmids are reportedly transferrable via insertion into a conjugative plasmid (He et al., 2019). The plasmid pS13-2 harboring the blaCTX–M–55 gene was a type of IncN1-F33:A–:B– plasmid and possessed the typical backbone of F33:A–:B– plasmids; carried resistance genes and mobile elements in the MDR region, including blaCTX–M–55, ΔblaTEM–1, IS26, and IS1294; and exhibited high homology to other plasmids in E. coli, Citrobacter freundii, and S. enterica (Figure 1).
FIGURE 1.
The whole-plasmid sequence of pS13D and comparisons of pS13-2 and pS13-3, with similar plasmids. The outer ring comprises the CDSs of pS13D. The plasmids in this study included pS13D (MT657397), pS13-2 (CP047092), pS13-3 (CP047093), pCTXM55_020023 (CP025949), pSE95-1 (CP050717), pSE109-1 (CP050710), pMH17-012N_2 (AP018568), pD72-F33 (CP035314), pPE15-IncF (CP041629), pCP61-IncFIB (CP053729), pCP8-3-IncFIB (CP053738), and pT3 (MK656937). Key features of pS13D are highlighted in different colors. Replicon genes are in highlighted in orange, transfer-associated genes in blue, resistance genes in purple, mobile elements in red, virulence related genes in black, and hypothetical proteins in gray.
The tet(M)-bearing plasmid pS13-3 was a novel IncFIA-FIB plasmid possessing the typical structure of IncFIB-type plasmids but lost the tra gene in the transfer region. Online BLASTn analysis revealed that pS13-3 exhibited homology to the E. coli plasmids pPE15-IncF (CP041629), pCP61-IncFIB (CP053729), pCP8-3-IncFIB (CP053738), and pT3 (MK656937) shared 99% identity at 72–84% coverage. As compared with the plasmids mentioned above, the most visible difference of strain S13 was the MDR, which comprised three accessory modules, including the novel tet(M)-bearing transposon Tn6942, the oqxAB resistance module bleO-NimC-IS26-oqxAB-IS26-blaTEM–1, and the segment ISKpn19-TinR-qnrS1 (Figure 1).
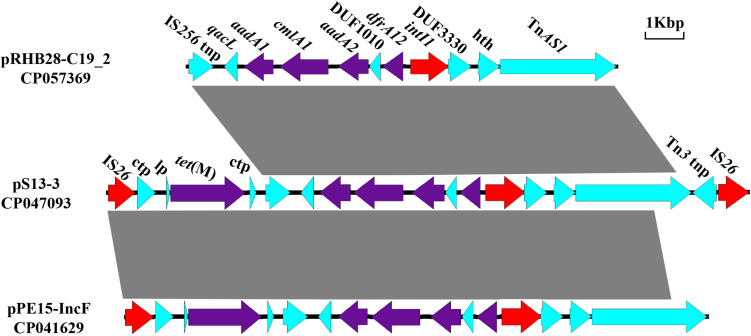
The novel composite transposon Tn6942 (16, 493 kb), harboring the tet(M) resistance gene, and an integron carrying IntI1, aadA1, cmlA1, aadA2, and dfrA12 was flanked by two IS26 elements. Tn6942 was highly similar to that of a fragment in pPE15-IncF recovered from E. coli strain pPE15 in Henan, China. The main difference is that the downstream TnAS1 in Tn6942 was replaced with blaTEM–1 in plasmid pPE15-IncF. Meanwhile, plasmid pRHB28-C19_2 (CP057369) had a similar structure but lacked two parts of segments at both ends, including the tet(M) resistance module [IS26-conjugal transfer protein-LP-tet(M)] and a Tn3 family transposase (Figure 2). Reverse PCR analysis was performed to assess the existence of a circular intermediate of Tn6942 (Supplementary Table 2). There is a circular intermediate IS26-bracked composite transposon carrying tet(M), excising from the S13 strain, revealing that the Tn6942 could stimulate the dissemination of tet(M) gene.
FIGURE 2.
Comparisons of the genetic environment of Tn6942 harboring tet(M) in CP047093, with two other sequences retrieved from the GenBank database. Similar regions are indicated by dotted lines (lp, gene encoding tet(M) leader peptide; tnp, transposage; hth, helix-turn-helix domain-containing protein).
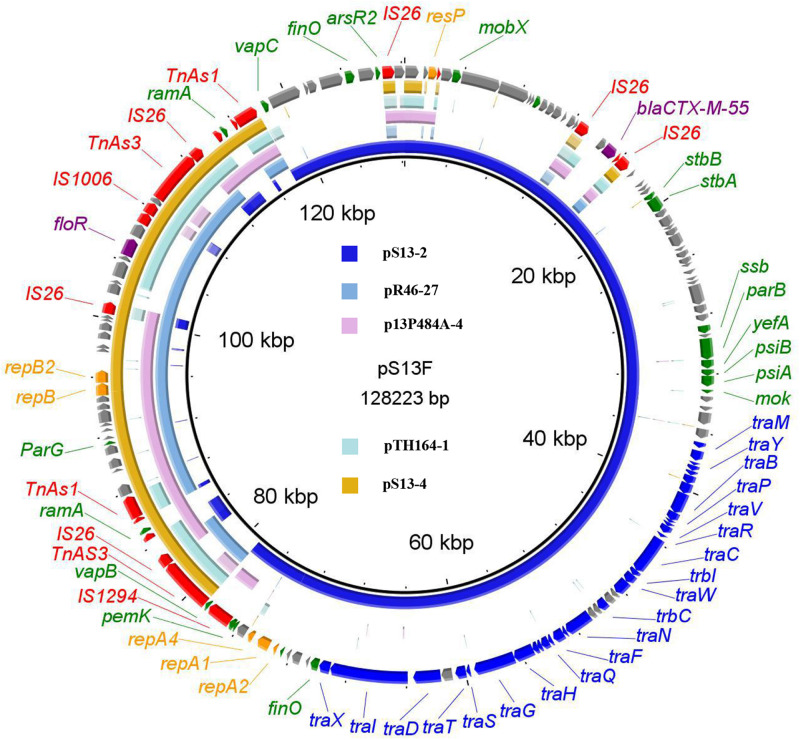
The floR-bearing IncX1 plasmid pS13-4 comprised 42 open reading frames. BLASTn analysis showed that plasmid pS13-4 was most similar to the IncX1 plasmid pR46-27 (CP035774), with 99% identity at 91% coverage (Figure 3). The plasmids pS13-3 and pS13-4 both lost the tra gene region responsible for plasmid conjugation, which may be the reason why these two plasmids were not self-transmissible. However, once inserted into the successfully diffused conjugative plasmid, all resistance genes carried by the plasmids achieved efficient movement. Two cases in point are the formation of conjugative fusion plasmids.
FIGURE 3.
The whole-plasmid sequence of pS13F, comparison of pS13-2 and pS13-4 with similar plasmids. The outer ring comprises the CDSs of pS13F. The plasmids in this study included pS13F (MT742153), pS13-2 (CP047092), pS13-4 (CP047094), pR46-27 (CP035774), p13P484A-4 (CP019284), and pTH164-1 (CP035212). Key features of pS13F are highlighted in the same color as the pS13D.
Proposed Formation Mechanism of Two Fusion Plasmids
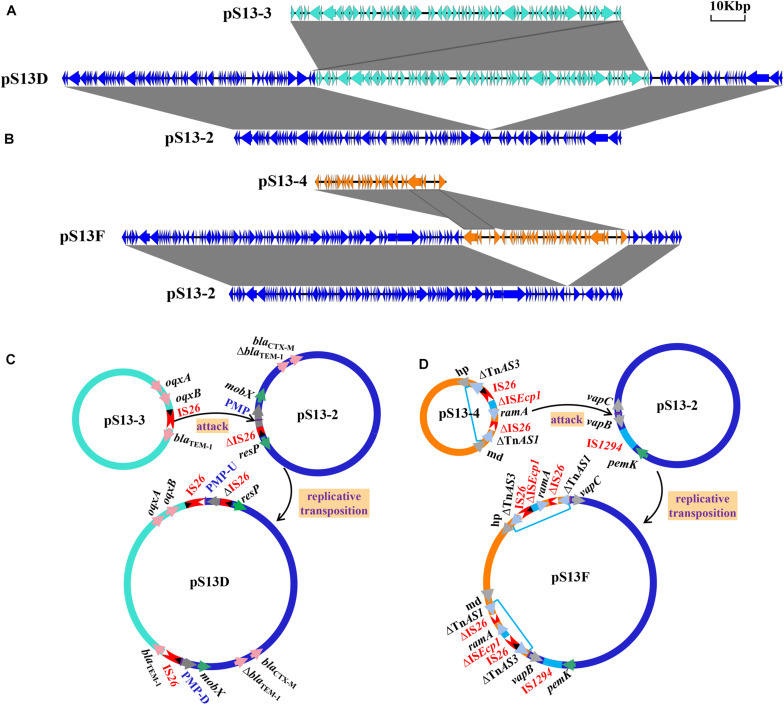
The plasmid pS13D harboring the tet(M) resistance gene was found to be 167,286 kb in size with a G+C content of 51% and contained 173 conserved domains. Further analysis indicated that pS13D was a fusion plasmid with chimeric characteristics consisting of pS13-2, pS13-3, an additional IS26 copy, and an 8-bp sequence target site duplication (TSD) (TTCAAGAT) (Figure 4A). The sequences across the cointegrate junctions were confirmed with the primers Fusion-168-1/2-F/R to link the pS13-3 backbone to the pS13-2 backbone, and the sequences of the PCR amplicons were consistent with that of WGS (Supplementary Table 1). Based on the above sequence analysis, we propose the fusion and resolution model of pS13D shown in Figure 4C. In this model, IS26 between blaTEM–1 and oqxB in pS13-3 captured the conjugative plasmid pS13-2 through attack of a putative mobilization protein (PMP) in the mobA/mobL family, and intermolecular transposition occurred via a replicative transposition mechanism, leading to an 8-bp TSD and an additional copy of IS26 flanking the pS13-3 molecule located upstream of PMP-U. Genetic rearrangement continued as the novel IS26 attacked the TSD (TTCAAGAT) downstream of PMP-D, which subsequently resulted in two TSDs surrounding the insertion fragment and eventually the formation of the hybrid resistance plasmid pS13D.
FIGURE 4.
The mechanism of plasmid fusion, including pS13D and pS13F. The shaded area indicates 100% identity. Colored arrows indicate open reading frames, with pS13-2 in blue, pS13-3 in turquoise, and pS13-4 in orange. (A). Linear sequence comparison of pS13-2, pS13-3, and the fusion plasmid pS13D. (B). Linear sequence comparison of pS13-2, pS13-4, and the fusion plasmid pS13F. (C). A model for the formation of pS13D mediated by IS26. (D) A model for the formation of pS13F mediated by TnAS3-IS26-ΔISEcp1-ramA-ΔIS26-ΔTnAS1. Plasmid names are shown in black. ISs are shown as rectangles (different colors represent different ISs), with triangles representing the terminal inverted repeats (TIRs). Left TIRs, open triangle; right TIRs, closed triangle; pink arrows, resistance genes; gray arrows, proteins. Purple arrows indicate different flanking 8-bp sequences and 5-bp sequences.
IS26, which belongs to the IS6 family of mobile elements, plays a pivotal role in the spread, clustering, and recombination of resistance genes (He et al., 2015; Liu Z. et al., 2020). Examples of the phenomenon of plasmid fusion mediated by IS26 include fusion of IncF33:A–:B– and phage-like plasmids, IncFII and the IncX3 plasmid, IncHI2 and IncFIB plasmids (Chen et al., 2017; Harme and Hall, 2017; Liu Z. et al., 2020). Significantly, the recombination mechanisms of the plasmids described above were all related to the IS26 element located on conjugative plasmids attacking the non-conjugative plasmids. In this study, the non-conjugative IncFI:A–:B–type plasmid captured a IncN1-F33:A–:B– type conjugative helper plasmid, which was mediated by the IS26 element, similar to the fusion of the IncX1 and IncI1 plasmids (Chen et al., 2019).
The hybrid plasmid pS13F harboring the floR gene in the transconjugant S13F was shown to be 128,223 bp in size and belonged to the IncN1-F33:A–:B–/IncX1 plasmid. Sequence alignment revealed that pS13F was obtained through the fusion of pS13-2 (1–78,755 nt; 117,800–128,223 nt) and pS13-4 (78,756–108,824 nt) (Figure 4B). Interestingly, pS13F carried an extra 8970-bp sequence (108,825–117,794 nt) containing a fragment of ΔTnAS3-IS26-ΔISEcp1-ramA-ΔIS26-ΔTnAS1 and an additional 5-bp sequence (TTATA). Based on sequence comparison, the plasmid recombination was mediated by a segment of ΔTnAS3-IS26-ΔISEcp1-ramA-ΔIS26-ΔTnAS1, designated Tn6952, which was located on plasmid pS13-4 (Figure 3 and Supplementary Figure 1). As shown in the model, the 8970-bp segment Tn6952 adjacent to the malate dehydrogenase coding region of pS13-4 attacked the target site (TTATA) of pS13-2 prior to replicative cointegrate formation. Linearized pS13-2 was incorporated into pS13-4 creating the cointegrate pS13F, giving the appearance of a 5-bp TSD (TTATA) and acquired an additional copy of Tn6952 located downstream of the incoming pS13-2 molecule. Subsequently, the TSDs became two direct repeats around the insertion fragment (Figure 4D). To confirm the genetic structures, two primers (Fusion-128-1/2-F/R) were designed, targeting the corresponding fusion regions. Notably, the sequences of the PCR amplicons were consistent with those obtained by WGS, which certified the existence of fusion regions (Supplementary Table 1).
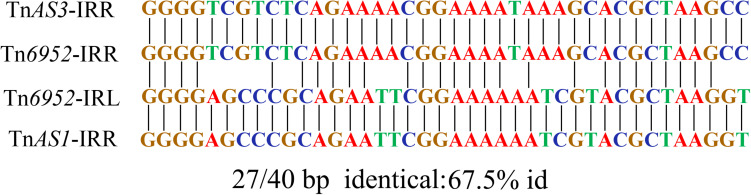
The Tn6952 contained an intact IS26 sequence, two truncated insertion sequences (ΔISEcp1 and ΔIS26), the AraC/XylS family gene ramA, and two Tn3-like element family transposases at both ends. Comparative analysis demonstrated that the segment ΔTnAS3-IS26 existed in Klebsiella pneumoniae plasmid pR46-27 and E. coli plasmid unnamed1 (CP037904). In addition, the fragment ΔISEcp1-ramA-ΔIS26-ΔTnAS1 was observed in E. coli plasmids p13P484A-4 (CP019284) and pEC129_3 (CP038456), and noteworthy, no direct repeat was identified at each extremity of the Tn6952, suggesting that Tn6952 acquired by pS13-4 may have occurred by recombination. To date, there has no report of plasmid fusion mediated by such complex structures, as only the segment of Tn3 carrying mutations in the repressor gene tnpR was able to mediate cointegration of the plasmids, as reported by Ohtsubo et al., who proposed that the repressor could destabilize the cointegrated plasmids through the action of an internal resolution site, while the transposase tnpA and inverted repeat right (IRR) may have taken part in mediating the formation of cointegrated plasmids containing two direct repeats of Tn3 (Ohtsubo et al., 1981). Further analysis showed that both boundaries [IRR and inverted repeat left (IRL)] of Tn6952 shared 100% identity with the IRR of two Tn3-like elements, TnAS3 and TnAS1, respectively (Figure 5). While in transposon Tn6952, the IRR shared 67.5% nucleotide identity with the IRL. The TnAS3 (Kieffer et al., 2019) and ΔTnAS1 (Liu et al., 2017) transposases could mobilize the resistance genes mcr-3 and mcr-5 by IRs, respectively. So, we speculate that the two IRs and transposase of TnAS3 and ΔTnAS1 may be involved in the molecular transposition of plasmid pS13-4 through forming an efficient mobilization unit Tn6952. The ramA gene, which controls expression of the MDR efflux pump genes and influences the virulence of S. enterica could be the facilitator that regulates the stability of Tn6952 (Bailey et al., 2010; Ricci et al., 2014). Therefore, Tn6952 carrying three insertion sequences may accelerate the dissemination of resistance genes and plasmids by cointegration.
FIGURE 5.
Alignment of the IRR and IRL in Tn6952 with the IRs identified on the TnAS3 and TnAS1.
The Biological Features of Two Fusion Plasmids
Stability assays showed that two fusion plasmids, pS13D and pS13F, were appeared to be stable (stability=100%) in E. coli for at least 10 days of passage in an antibiotic-free environment. For the reason that cointegrate pS13D and pS13F could be transferred from the transconjugant S13D and S13F to the recipient C600, the experiments of fitness cost of pS13D and pS13F could not performed accurately. However, to some extent, two fusion plasmids did not exhibit a fitness cost to E. coli C600 (Supplementary Experiments). In addition, pS13D and pS13F were transferred to the E. coli strain J53 at high conjugation frequencies of 8.58×10–3 and 9.2×10–4, respectively, indicating that these two plasmids had relatively high self-transmission rates and stability, and contributed to the dissemination of the resistance genes tet(M), oqxAB, and floR via co-selection, thereby posing a potentially threat to clinical treatment.
Conclusion
A novel IncN1-F33:A–:B– conjugative helper plasmid was identified that could passively fuse with non-conjugative tetracycline and florfenicol resistance-encoding plasmids via two different mechanisms, thereby promoting the conversion into conjugative plasmids transmissible among different species of Enterobacteriaceae. A better understanding of the molecular and triggering mechanisms that contribute to the formation and evolution of cointegration of MDR plasmids is needed to build a foundation to curb further spread of resistance elements among bacterial pathogens via this type of plasmid. New intervention measures are urgently needed to curb formation and dissemination of such elements among Salmonella species and other Enterobacteriaceae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
G-ZH, J-HL, and D-DH conceived and designed the experiments. Y-YL and M-KZ performed the experiments. Y-YL analyzed the data. Y-SP, J-HL, HW, and LY contributed reagents, materials, and analysis tools. Y-YL, D-DH, Y-SP, and G-ZH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was financed by the National Natural Science Foundation of China (32072913) and (32072914).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.676574/full#supplementary-material
(a) S1-PFGE of S. enterica strain S13 and its three transconjugants S13D, S13F, S13S. (b). Southern hybridization of S. enterica strain S13 and the tet(M)-bearing transconjugant S13D with the tet(M) gene as the probe. Marker, Salmonella Braenderup H9812.
Structural comparisons of pS13-4 and the similar plasmids. The outer ring represents the CDSs of the reference sequence pS13-4. The similar plasmids including the E. coli plasmids pD72-mcr1 (CP035313), pZR78 (MF455226), and pGD27-62 (MN232192); S. enterica plasmid pATCC10729_02 (CP019188); and Klebsiella pneumoniae plasmids pMCR_SCKP-LL83 (MF510496) and p160070 -MCR (MG288678).
Primers used for PCR and DNA sequencing in this study.
The MICs of S13 and its transconjugants in this study.
References
- Alikhan N. F., Petty N. K., Zakour N. L. B., Beatson S. A. A. (2011). Blast ring image generator (brig): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., Edwards R. A., et al. (2008). The rast server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. M., Ivens A., Kingsley R., Cottell J. L., Wain J., Piddock L. J. V. (2010). RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192 1607–1616. 10.1128/jb.01517-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics (Oxf.) 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Dong N., Chan W. C., Chen S. (2019). Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microbes Infect. 8 857–865. 10.1080/22221751.2019.1626197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wong H. Y. M., Chan W. C. E. (2017). IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J. Anti. Chemother. 10 2750. 10.1093/jac/dkx238 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Deng X., Ran L., Wu S., Ke B., He D., Yang X., et al. (2012). Laboratory-based surveillance of nontyphoidal Salmonella infections in China. Foodborne Pathog. Dis. 9:305. [DOI] [PubMed] [Google Scholar]
- Harme C. J., Hall R. M. (2017). Targeted conservative formation of cointegrates between two DNA molecules containing IS26 occurs via strand exchange at either IS end. Mol. Microbiol. 106 409–418. 10.1111/mmi.13774 [DOI] [PubMed] [Google Scholar]
- He D., Zhu Y., Li R., Pan Y., Liu J., Yuan L., et al. (2019). Emergence of a hybrid plasmid derived from IncN1-F33:A–:B– and mcr-1-bearing plasmids mediated by IS26. J. Antimicrob. Chemother. 11 3184–3189. 10.1093/jac/dkz327 [DOI] [PubMed] [Google Scholar]
- He S., Hickman A. B., Varani A. M., Siguier P., Dyda F. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. Mbio 6:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. C., Chiu T. H., Pang J. C., Hsuan-Yuan C. H., Chang G. N., Tsen H. Y. (2006). Characterisation of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int. J. Antimicrob. Agents 27 383–391. 10.1016/j.ijantimicag.2005.11.020 [DOI] [PubMed] [Google Scholar]
- Hu X., Li G. (2009). Advances in the relationship between integrons and drug resistance of bacteria. World Notes Antibiot. 30 255–263. [Google Scholar]
- Kieffer N., Nordmann P., Millemann Y., Poirel L. (2019). Functional characterization of a miniature inverted transposable element at the origin of mcr-5 gene acquisition in Escherichia coli. Antimicrob. Agents Chemother. 63:e00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Lu X., Peng K., Liu Y., Xiao X., Wang Z. (2020). Reorganization of mcr-1-bearing large MDR plasmids resolved by nanopore sequencing. J. Antimicrob. Chemother. 75 1645–1647. 10.1093/jac/dkaa046 [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Dong N., Lin D., Yang X., Yin W. M. H., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigaence 3 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Feng Y., Zhang X., McNally A., Zong Z. (2017). New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM–5. Antimicrob. Agents Chemother. 61 e1757–e1717.∗∗e1757-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Liu X. K., Cui X. D., Chen M., Hu G. Z., Pan Y. S., et al. (2020). Characterization of pTS14, an IncF2:A1:B1 Plasmid Carrying tet(M) in a Salmonella enterica Isolate. Front. Microbiol. 11:1523. 10.3389/fmicb.2020.01523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Liu Y., Li R., Wang Z. (2020). Recombination of NDM-5-producing plasmids mediated by IS26 among Escherichia coli. Int. J. Antimicrob. Agents 55:105815. 10.1016/j.ijantimicag.2019.09.019 [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., Mccormick M., Machida Y. (1981). Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb. Symp. Quant. Biol. 45 283–295. [DOI] [PubMed] [Google Scholar]
- Ricci V., Blair J. M., Piddock L. J. (2014). RamA, which controls expression of the MDR efflux pump AcrAB-TolC, is regulated by the Lon protease. J. Antimicrob. Chemother. 69 643–650. 10.1093/jac/dkt432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.p11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. W., Liu Y. Y., Wu H., Wang L. F., Liu J. H., Yuan L., et al. (2019). IS26-flanked composite transposon Tn6539 carrying the tet(M) Gene in IncHI2-Type conjugative plasmids from Escherichia coli isolated from Ducks in China. Front. Microbiol. 9:3168. 10.3389/fmicb.2018.03168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5 175–186. 10.1038/nrmicro1614 [DOI] [PubMed] [Google Scholar]
- Xie M., Li R., Liu Z., Chichan E. W., Chen S. (2018). Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. J. Antimicrob. Chemother. 73 2750–2754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) S1-PFGE of S. enterica strain S13 and its three transconjugants S13D, S13F, S13S. (b). Southern hybridization of S. enterica strain S13 and the tet(M)-bearing transconjugant S13D with the tet(M) gene as the probe. Marker, Salmonella Braenderup H9812.
Structural comparisons of pS13-4 and the similar plasmids. The outer ring represents the CDSs of the reference sequence pS13-4. The similar plasmids including the E. coli plasmids pD72-mcr1 (CP035313), pZR78 (MF455226), and pGD27-62 (MN232192); S. enterica plasmid pATCC10729_02 (CP019188); and Klebsiella pneumoniae plasmids pMCR_SCKP-LL83 (MF510496) and p160070 -MCR (MG288678).
Primers used for PCR and DNA sequencing in this study.
The MICs of S13 and its transconjugants in this study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.