Abstract
Study Objectives
Sleep and wake are opposing behavioral states controlled by the activity of specific neurons that need to be located and mapped. To better understand how a waking brain falls asleep it is necessary to identify activity of individual phenotype-specific neurons, especially neurons that anticipate sleep onset. In freely behaving mice, we used microendoscopy to monitor calcium (Ca2+) fluorescence in individual hypothalamic neurons expressing the vesicular GABA transporter (vGAT), a validated marker of GABA neurons.
Methods
vGAT-Cre mice (male = 3; female = 2) transfected with rAAV-FLEX-GCaMP6M in the lateral hypothalamus were imaged 30 days later during multiple episodes of waking (W), non-rapid-eye movement sleep (NREMS) or REMS (REMS).
Results
372 vGAT neurons were recorded in the zona incerta. 23.9% of the vGAT neurons showed maximal fluorescence during wake (classified as wake-max), 4% were NREM-max, 56.2% REM-max, 5.9% wake/REM max, while 9.9% were state-indifferent. In the NREM-max group, Ca2+ fluorescence began to increase before onset of NREM sleep, remained high throughout NREM sleep, and declined in REM sleep.
Conclusions
We found that 60.2% of the vGAT GABA neurons in the zona incerta had activity that was biased towards sleep (NREM and REMS). A subset of vGAT neurons (NREM-max) became active in advance of sleep onset and may induce sleep by inhibiting the activity of the arousal neurons. Abnormal activation of the NREM-max neurons may drive sleep attacks and hypersomnia.
Keywords: GABA, vGAT, calcium imaging, GCaMP6, neuronal activity, sleep, zona incerta, hypothalamus, vGAT-Cre mice
Statement of Significance.
Sleep like other distinct behaviors is controlled by the activity of neurons that need to be located and mapped. We use the high-throughput miniscope method to monitor calcium fluorescence in GABA neurons in a hypothalamic region implicated in regulating sleep and where many arousal neurons are also located. We find that fluorescence in 60.2% of the GABA neurons is maximal during sleep states, with the neurons beginning to fluoresce prior to the emergence of an EEG waveform indicative of non-rapid-eye movement sleep. We suggest that activation of sleep-anticipatory neurons may initiate sleep and drive hypersomnolence.
Introduction
A distributed network of neurons is hypothesized to regulate the alternation between waking, non-rapid-eye movement sleep (NREMS) and rapid-eye-movement sleep (REMS). Some neurons, such as those containing histamine, norepinephrine, orexin, or serotonin are most active during waking, decrease their activity in NREMS, and become inactive in REMS (reviewed by Ref. [1]). However, the activity profile of the GABA neurons during sleep–wake states is less understood. GABA is the primary inhibitory neurotransmitter in the CNS, but it is not known whether all GABAergic neurons are sleep-active or whether subsets of GABA neurons show different activity associated with waking, NREMS or REMS. This question needs to be answered because then specific subsets of GABA neurons could be manipulated to predictably regulate sleep or wake states, especially in disorders of sleep such as insomnia (too little sleep) or narcolepsy (excessive sleep). Clusters of GABA neurons in the preoptic [2] and lateral hypothalamic [3] areas that discharge maximally in NREMS and REMS have been found. However, there are also GABA neurons in the posterior hypothalamus that are preferentially wake-active and drive arousal [4, 5]. Unlike norepinephrine, serotonin, histamine, and orexin neurons which are distributed in specific brain areas, the GABA neurons are distributed throughout the brain intermingled with other neuronal phenotypes, which makes it difficult to record their activity. The in vivo electrophysiology method is the primary method that has been used to measure activity of neurons during waking, NREMS and REMS [6–11]. However, new tools are needed to rapidly map the activity of phenotype-specific neurons underlying behaviors such as sleep.
A new method that can image neuronal activity was recently introduced [12]. This method measures changes in intracellular levels of calcium (Ca2+) associated with excitatory signaling in neurons [13]. A genetically encoded Ca2+ fluorescent sensor (GCaMP6) expressed in specific neurons (Cre driven gene expression) provides a reliable readout of the neuron’s activity level [14]. The change in Ca2+-induced fluorescence in GCaMP6 expressing neurons is recorded via a miniature single-photon epifluorescence microscope attached to a miniature gradient refractive index (GRIN) lens [12]. Deep-brain Ca2+ imaging has proven to be very useful in confirming existing in vivo electrophysiology data by imaging neurons in quiet waking, NREMS and REMS [15–18]. We recently used the deep-brain Ca2+ imaging system to monitor activity of neurons expressing melanin-concentrating hormone (MCH) [19]. Here, we use the deep-brain Ca2+ imaging method to monitor activity of neurons that express the gene for vesicular GABA transporter (vGAT), a reliable marker of GABA neurons [20, 21].
In the posterior hypothalamus, GABAergic neurons are intermingled with the orexin and histamine neurons that promote arousal, and are closely mingled with MCH neurons that, in contrast, promote sleep [22–24]. However, the vGAT neurons are separate and distinct from the orexin, histamine or MCH neurons [25], and a few vGAT neurons are selectively active in NREMS [3]. In the current study, we monitored activity of the vGAT neurons located dorsal to the lateral hypothalamus and perifornical area and find that even within the same phenotype of neurons in a localized area, there are subsets showing differing patterns of activity during wake–sleep states. Thus, unlike the arousal neurons which are all active in waking but silent in sleep, converging evidence is beginning to show that subsets of GABA neurons have different activity profiles during wake–sleep states.
Methods
Animals and surgery
The VA Institutional Animal Care and Use Committee approved all animal use procedures (protocols # 638,639). The vGAT-IRES-Cre (Jackson Laboratories, JAX stock #016962) mice were bred in our animal facility and those found to be Cre+ (Transnetyx, Inc.) were later chosen for the imaging study (n = 5; male = 3). After surgery, the animals were housed singly in polycarbonate cages with α-cellulose bedding. Standard rodent laboratory food pellets and reverse osmosis treated drinking water were available to the mice ad libitum. The ambient temperature of the housing room was always controlled (25 ± 2°C) while a 12:12 h light–dark cycle (6 am to 6 pm lights on; 500 lux) was strictly maintained. The mice used for this study were 4 months old and had a mean body weight of 34.7g (±1.71g) at the time of surgery (Table 1).
Table 1.
Sex, Age, Body Mass, and Numbers of Neurons Recorded
| Mouse ID | Sex | Age (month) | Body weight (g) | Numbers of vGAT neurons imaged |
|---|---|---|---|---|
| vG058 | M | 4 | 36.4 | 22 |
| vG239 | M | 4 | 39.4 | 93 |
| vG243 | M | 4 | 28.9 | 77 |
| vG247 | F | 4 | 34.5 | 108 |
| vG251 | F | 4 | 34.3 | 72 |
| Average (± SEM) | 4 | 34.7 (1.71) | 78.4 (11.1) | |
| Total | 60%M | 372 |
The mice were deeply anesthetized (2% isoflurane) and, using a stereotaxic apparatus, the genetically encoded Ca2+sensor, GCaMP6M (rAAV-DJ-EF1a-DIO-GCaMP6M, titer: 4 × 108/mL GC, vol: 2–4 µL: Stanford University Vector Core), was slowly injected over a 10 min period into the lateral hypothalamus (relative to Bregma A = −1.6; lateral = 0.7; ventral from brain surface = 4.8 mm). The viral injection cannula was held in place for 45 min and then slowly retracted. After delivery of the virus, a GRIN lens (0.6 mm diameter; 7.3 mm length; Inscopix stock number 1050–002208) was inserted into the same injection cannula track 0.1 mm above the transfection target. At this time, four miniature screw-type electrodes (Plastic One Inc.) were secured to the skull to record the electrocorticogram (EEG) activity. To measure the electromyogram (EMG) activity, two multi-stranded wire electrodes were inserted into the nuchal muscles. The EEG, EMG electrodes, and the GRIN lens were permanently attached to the skull with dental cement. For pain management, mice were administered carprofen (5 mg/kg SC) for 2 days (d). Approximately 21 days later, the mice were anesthetized (2% isofluorane), placed again in the stereotaxic instrument, and using a micromanipulator (Sutter Instruments), a single photon miniature fluorescence miniscope (Inscopix nVista) attached to a baseplate was carefully positioned atop the GRIN lens. Once the best focal plane was achieved, the baseplate was carefully cemented to the headset. The miniscope was detached and the mice returned to the vivarium for recovery.
Approximately 30 days after the injection of the viral vector, the miniscope and the sleep recording cable were attached, and the mice were adapted to the recording environment for 6 to 8 h during the day cycle. The adaptation took place for three consecutive days. At the end of each adaptation period, the miniscope was detached, a protective cover plate was attached to the baseplate and mice were returned to the vivarium.
Acquisition of the Ca2+ imaging and sleep data
Sleep and Ca2+ imaging were done during the second half of the lights-on period. The first half was reserved for adapting the mice to the recording environment. Ca2+ imaging data were recorded with the nVista® software (Inscopix, Inc.) at a sample rate of 10 frames per second, camera gain of 3.0× and under 10% LED blue radiance power (0.1 mW at the tip of the GRIN lens). Each mouse was always imaged at a single focal plane that had the sharpest images of the fluorescent neurons. EEG/EMG raw analogue signals were bandpass filtered (EEG: 0.5–60 Hz; EMG: 100–1 kHz) and amplified by Grass 7P51 amplifiers, and then digitized with the OmniPlex® acquisition system (Plexon, Inc.). OmniPlex also recorded video of the animal’s behavior (CinePlex Studio®). Start/End signals from the miniscope were input to OmniPlex as well. Thus, OmniPlex displayed the acquired signals from five distinct channels: two EEG, one EMG, one nVista recording synchronization signal and one from the video camera recording the behavior of the mouse. For each mouse, the imaging and sleep data were acquired for a mean of 50.9 min (±1.8 min) when multiple episodes of wake, NREMS and REMS were recorded (Table 2).
Table 2.
Summary of Vigilance States during the Calcium Imaging Sessions
| Vigilance State | Active Wake | Quiet Wake | Non-REM sleep | REM sleep transition | REM sleep | All states |
|---|---|---|---|---|---|---|
| Mouse ID | Number of episodes | |||||
| vG058 | 8 | 24 | 19 | 9 | 6 | 67 |
| vG239 | 16 | 34 | 19 | 7 | 5 | 81 |
| vG243 | 2 | 21 | 19 | 10 | 8 | 60 |
| vG247 | 6 | 18 | 17 | 6 | 3 | 50 |
| vG251 | 5 | 22 | 21 | 9 | 3 | 60 |
| Group average | 7 | 23.8 | 19 | 8.2 | 5 | 64 |
| Group S.E.M. | 2.4 | 2.7 | 0.6 | 0.7 | 0.9 | 5.1 |
| Group SUM | 37 | 119 | 95 | 41 | 25 | 318 |
| Mouse ID | Total time in seconds | |||||
| vG058 | 482 | 378 | 1405 | 122 | 275 | 2662 |
| vG239 | 588 | 848 | 1283 | 157 | 371 | 3247 |
| vG243 | 233 | 498 | 1749 | 260 | 501 | 3241 |
| vG247 | 163 | 390 | 2147 | 89 | 214 | 3003 |
| vG251 | 296 | 472 | 1923 | 158 | 275 | 3124 |
| Group average | 352.4 | 517.2 | 1701.4 | 157.2 | 327.2 | 3055.4 |
| Group S.E.M. | 79.3 | 85.8 | 160.1 | 28.7 | 50.2 | 108.0 |
| Group SUM | 1762 | 2586 | 8507 | 786 | 1636 | 15,277 |
| Mouse ID | Length of episodes in seconds (S.E.M.) | |||||
| vG058 | 53.6 (20) | 15.8 (2.6) | 73.9 (12.8) | 13.6 (1.9) | 45.8 (19.0) | NA |
| vG239 | 36.8 (10) | 24.9 (2.6) | 67.5 (14.5) | 22.4 (1.9) | 74.2 (29.5) | NA |
| vG243 | 116.5 (112) | 23.7 (4.0) | 92.1 (15.2) | 26.0 (2.5) | 62.6 (21.1) | NA |
| vG247 | 27.2 (11) | 21.7(3.5) | 126.3 (16.9) | 14.8 (5.2) | 71.3 (32.1) | NA |
| vG251 | 59.2 (35) | 21.5 (2.6) | 91.6 (12.5) | 17.6 (2.4) | 91.7 (37.7) | NA |
| Group average | 58.6 | 21.5 | 90.3 | 18.9 | 69.1 | |
| Group S.E.M . | 37.6 | 3.1 | 14.4 | 2.8 | 27.9 |
Offline processing of sleep and behavioral data
The sleep–wake states were identified based on standardized criteria for EEG/EMG activity patterns, video recordings of the animal’s behavior (CinePlex Editor®; Plexon.com) and the EEG power spectra (0.5–15 Hz bandwidth). Sleep scoring was done manually and blinded using specialized software (Neuroexplorer.com) by placing a line at the transition point between states. Active waking (AW) was identified when mice were behaviorally active (grooming, walking, rearing, etc.) with desynchronized EEG and high/irregular EMG activity. Quiet waking (QW) was identified by the occurrence of desynchronized EEG, lower EMG tone relative to active waking, and diminished behavioral activity. Non-REMS (NREMS) consisted of high amplitude, slow frequency delta waves (0.5–4 Hz) in the EEG together with a low EMG tone relative to waking and a sleeping posture displayed on the video. REM sleep (REMS) was identified by the presence of regular theta activity (4–8 Hz) in the EEG coupled with low EMG activity relative to NREMS or waking. During REMS, the mouse exhibited a relaxed sleeping posture. The video camera’s resolution and field of view made it difficult to detect the phasic aspects of REMS such as muscle twitches or vibrissae movements.
Criteria for identifying transition time points between states
The transition time point (1 s) represents the beginning and end of specific sleep–wake states, and was determined by visual examination of the EEG, EMG, video recording of the mouse’s ongoing behavior, and the EEG power spectra across the 0.5–15 Hz bandwidth. The EEG spectrogram was generated automatically by Neuroexplorer using a 0.5 s sliding window, multiple tapers and 8192 frequency value resolution. The active wake to quiet wake (AW to QW) transition time point represents 30 s on either side of the transition point. The transition point was placed after a period of active waking followed by QW where there was at least 5 s of little or no gross behavioral movements, diminished EMG activity, and EEG power that was same as AW. The quiet wake to active wake (QW to AW) transition progressed in the opposite direction as that described above. The QW to AW transition analysis window was set at 15 s before the transition point and 30 s after that. The quiet wake to NREMS transition (QW to NREMS) point represents 30 s of QW followed by 120 s of NREMS. The transition to NREMS was marked by the appearance of the first high amplitude slow waves in the EEG followed by sustained high EEG power spectra along the 0.5–4 Hz band (delta activity). The REMS onset point began at the start of a sustained pattern of EEG theta activity (4–8 Hz) ending with the sudden emergence of EMG activity (confirmed by video) and the disappearance of the theta activity in the EEG. We took the additional step of identifying a REMS transition state (Rt) which represented the 20 s period before the onset of the REMS episode. The REMS transition is characterized by a mix of delta and theta activity in the EEG (NREMS to Rt to REMS). It is not unusual to see bouts of REMS transition without entry into REMS. An analysis window for the NREMS to Rt to REMS was set at 65 s, that include 15 s of NREMS followed by 20 s of Rt and then 30 s of REMS. Because in almost all instances REMS episodes terminate in quiet wake, we focused our analysis on the REMS to QW state transition. The REMS to QW window consists of 30 s of REMS followed by 15 s of QW. Sleep scoring and identification of transition points were done by a researcher (PJS) blind to the Ca2+ imaging data. These criteria are the same as in our previously published study [19]. Table 3 summarizes the type, number, and analysis status of all state transition for each mouse.
Table 3.
Summary of State Transitions Measured during the Imaging Sessions
| Mouse ID | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Transition | vG058 | vG239 | vG243 | vG247 | vG251 | Total # of transitions (Tt) | % of Tt | Transition odds | Analysis status |
| QW to AW | 4 | 15 | 1 | 3 | 2 | 25 | 8.0 | 0.21 | Included |
| AW to QW | 8 | 15 | 2 | 5 | 4 | 34 | 10.9 | 1.00 | Included |
| QW to NR | 19 | 18 | 18 | 15 | 20 | 90 | 28.8 | 0.76 | Included |
| NR to QW | 10 | 13 | 11 | 8 | 10 | 52 | 16.6 | 0.56 | Included |
| NR to Rt | 8 | 6 | 7 | 6 | 9 | 39 | 6.7 | 0.39 | Included |
| Rt to R | 6 | 4 | 8 | 3 | 3 | 24 | 7.7 | 0.59 | Included |
| R to QW | 5 | 4 | 7 | 3 | 3 | 22 | 7.0 | 0.85 | Included |
| Total/mouse | 60 | 75 | 54 | 43 | 51 | 283 | 90.4 | 0.62^ | |
| Rt to QW | 2 | 2 | 1 | 2 | 6 | 13 | 4.2 | 0.32 | Excluded |
| R to AW | 2 | 1 | 0 | 0 | 0 | 3 | 1.0 | 0.12 | Excluded |
| Rt to AW | 1 | 0 | 1 | 1 | 0 | 3 | 1.0 | 0.07 | Excluded |
| NR to AW | 1 | 0 | 0 | 2 | 2 | 5 | 1.6 | 0.05 | Excluded |
| R to Rt | 0 | 0 | 1 | 0 | 0 | 1 | 0.3 | 0.04 | Excluded |
| QW to Rt | 0 | 1 | 2 | 0 | 0 | 3 | 1.0 | 0.03 | Excluded |
| Rt to NR | 0 | 1 | 0 | 0 | 0 | 1 | 0.3 | 0.02 | Excluded |
| AW to NR | 0 | 0 | 0 | 1 | 0 | 1 | 0.3 | 0.00 | Excluded |
| Total/mouse | 6 | 5 | 5 | 6 | 8 | 27 | 9.6 | 0.08^ | |
Offline processing of Ca2+ imaging data
The imaging data were processed and analyzed in an independent and blind manner using the Inscopix Data Processing software (Inscopix DPS®, Inc.). First, the raw nVista HDF5 files were temporally down-sampled by a factor of 10 (i.e. 10 Hz down 1 Hz) to reduce its file size and decrease the processing time. The down-sampled movie was then cropped to focus the analysis on the GRIN lens field of view. The cropped movie was then processed by the Inscopix DPS to correct for defective pixilation, row noise and dropped frames. The corrected movie was then processed with the program’s spatial bandpass filter (cutoffs; low: 0.005, high: 0.5 pixels), and two iterations of a motion correction algorithm (down to ±2.11 pixels for the x-axis and less than ±2.4 pixels for the y-axis translation). Subsequently, Inscopix DPS calculated a mean image frame or F0 of the entire movie (calculated by each pixel). The mean image F0 represents the mean Ca2+-induced fluorescence intensity across the recording period independently of the sleep states in which the fluorescence occurred. Subsequently a normalized ΔF-F0/F0 movie was automatically generated using the F0 as the reference frame (i.e. Fi - F0/F0 where i indicates every pixel). Next, Inscopix DPS identified clusters of neighboring pixels representing similar ΔF/F0 values across the entire movie. These clusters represent unbiased regions of interest (ROIs). Inscopix DPS extracted the ROIs using a Principal and Independent Component Analysis (PCA→ICA) algorithm. For each mouse, PCA→ICA analysis was set to find a maximum of 150 principal components and 120 independent components. After the PCA→ICA analysis extracted potential ROIs (neurons), the program’s visualization tools were used to manually verify that a fluorescence trace (temporal dimension) was associated with each ROI. The manual verification process was always done by one researcher (AVO) who checked each ROI to determine the following: (1) the ROI had neuronal soma-like morphology, (2) the ROI was associated with Ca2+ peaks traces that featured rapid rise followed by a slower decay, (3) the source of the strongest fluorescence signals always remained within the ROI boundaries, and (4) the fluorescence signal source was actually emitted by the ROI and not by neighboring neurons as a reflection (false positive). The verification process was done frame by frame for the entirety of the recorded movie. Regions of interest that failed the visual inspections were discarded from further analysis. The fluorescence trace for each verified ROI was exported to a spreadsheet where rows represented time (1 s increments) and columns contained all fluorescence values for each verified neuron (ROI). Once on the spreadsheet and using the visualizing tools of Inscopix DPS, covariance values were converted back to the original fluorescence units of ΔF/F0 (expressed as fractions). Fractions were then transformed as percent. Next, fluorescence values were transformed into Z scores using the population mean and standard deviation. Z scores values were later precisely (<100 ms) time matched to the scored sleep–wake data. For each neuron, each Z score value was binned into its corresponding vigilance state (AW, QW, NREMS, REMS transition, and REMS proper), and subsequently, values from all episodes of each given vigilance state were averaged (i.e., Z score average per state).
Data analysis of calcium traces during sleep–wake states
The EEG and EMG signals that were recorded via OmniPlex® (Plexon.com) were imported into a data visualization and physiological analysis software program (Neuroexplorer.com). A researcher (PJS) blind to the Ca2+ imaging data scored the sleep data into sleep–wake states as described previously in “Criteria for identifying transition time points between states.” In each mouse, the standardized fluorescence values for each neuron (ROI) were aligned with the vigilance state (AW, QW, NREMS, RT, and REMS) in 1 s epochs and an average fluorescence corresponding to each state was obtained. The fluorescence value (Z-score) corresponding to each ROI was binned into one of four groups (REM-max, NREM-max, Wake-max, or W-REM-max) based on a 2× criteria. For example, a REM-max ROI had a value in REM sleep that was twice that of the value in the other states. Similarly, a W-REM-max ROI had a fluorescence value in wake that was similar to REM sleep but in each state, the values were twice that of the values in the other states. Those neurons that did not meet the 2× cut-off were placed in the No-change group. Statistical analysis compared the fluorescence between sleep–wake states (AW, QW, NREM, pre-REM, and REM sleep) in each of the five groups.
The time-course of activity during the transition periods was determined for the Wake-Max, NREMS-Max, and REMS-Max groups. Because the time-course of fluorescence is based on 1-s intervals, the Z-scores were expressed as percent of their maximal fluorescence during the transition period. To determine whether neurons in each mouse displayed synchronized activity, the Z-scores were plotted for only the REMS-max group. We chose a REMS episode of average duration and we plotted the fluorescence of the five mice (209 neurons). We did not plot synchronicity in the NREMS-Max group as there were insufficient number of neurons to draw a meaningful conclusion.
Experimental design and statistical analysis
A generalized linear mixed model (GLMM; SPSS-25) with post hoc Bonferroni paired comparisons (sequential adjusted) was chosen to determine statistical significance between the five sleep–wake states. An unstructured diagonal covariance matrix yielded the lowest Information Criteria (Akaike or Bayesian) and the best model fit. The GLMM model was appropriate based on the correlation of the fluorescence data between the neurons, the skewed distribution of the data (Kolmogorov–Smirnov test for normality), and unbalanced design. Statistical significance was evaluated at the p = 0.05 level (two-tailed). Graphs were plotted using GraphPad Prism, Sigma Plot, or Neuroexplorer.
Post-mortem analysis of GCaMP6m expression
After Ca2+ imaging was completed, mice were euthanized with 5% isofluorane and then perfused transcardially with 0.9% saline (10 mL) followed by 10% buffered formalin in 0.1 M PBS (30 mL). The brains were collected and fixed in 10% buffered formalin for 1 week. The brains were equilibrated in 30% sucrose solution in 0.1 M PBS and cut coronally at 40 microns-thickness using a sliding microtome. The sections were washed, mounted onto gelatin-coated slides and cover slipped using fluorescence mounting medium. The fluorescence images were obtained at 10× and 20× using a Leica TCS-SP8 confocal microscope. GCaMP6m positive neurons were directly identified on the specimens because transfected vGAT-Cre neurons specifically expressed the circularly permuted EGFP [26].
Data and material availability
All raw and extracted data, code and materials used in the study are available by writing to the corresponding author. We encourage mining the imaging data set to extend the analysis.
Results
Location of the GRIN lenses and the vGAT transfected neurons
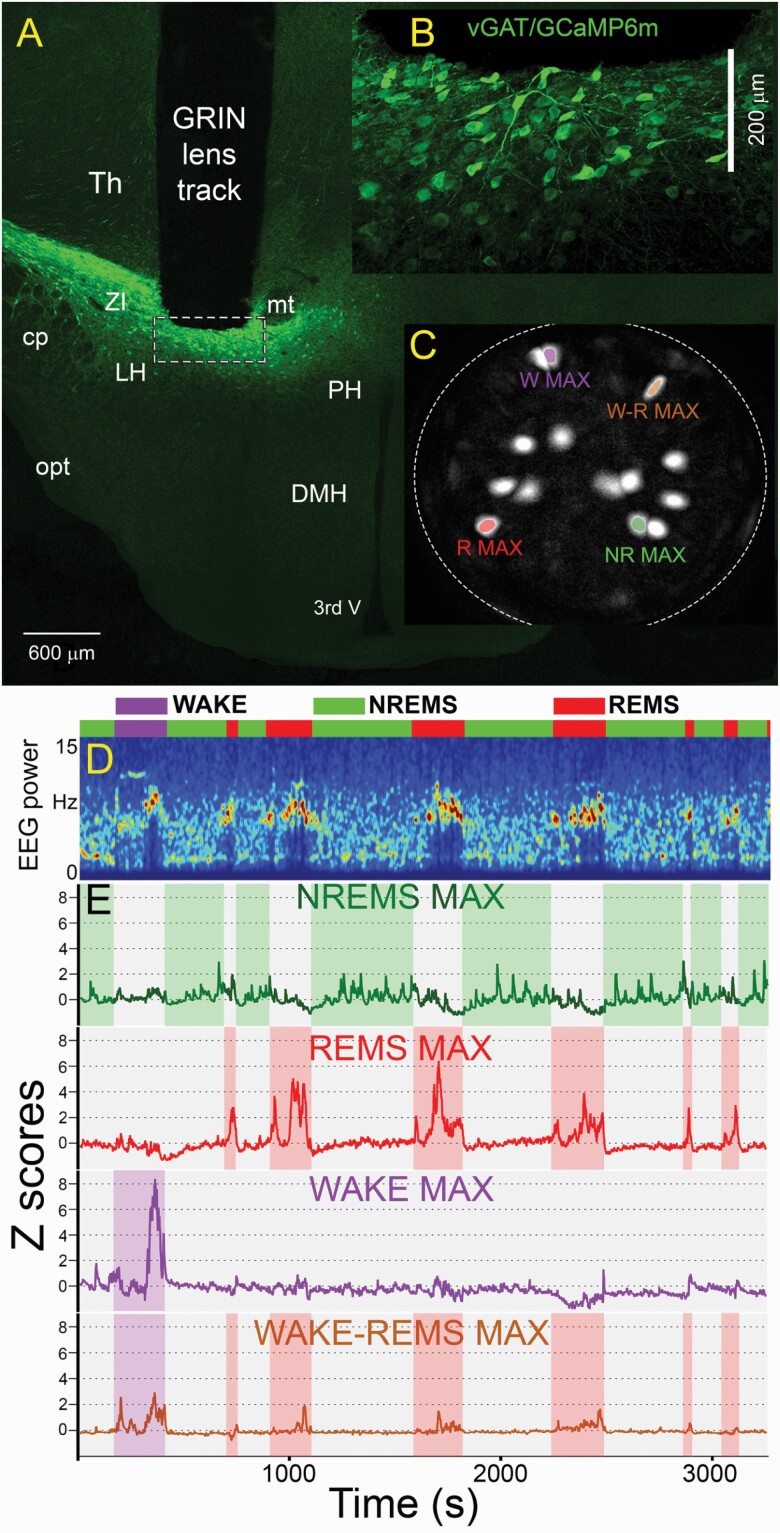
Figure 1 depicts a representative image of the GRIN lens atop a cluster of transfected vGAT neurons that are within the 200 µm working distance of the lens. The underlying neurons were brought into focus by rotating the miniscope (full technical specification at Inscopix.com), and we imaged at a single focal plane that yielded the sharpest and clearest images. In all mice, the transfected vGAT neurons were located most densely within the ventral zona incerta (vZI) with few neurons below the ZI in the dorsal lateral hypothalamus (dLH). We note that this is the same region that contains MCH neurons [19]. The tips of the GRIN lenses ended within the vZI as depicted in Figure 1. The neurons appeared to be very healthy with the GCaMP6M expression mainly found inside the neuron cytoplasm and mostly absent from the neuron’s nucleus (inset B). We noticed that the virus titer of 4 × 108 GC/mL gave us high numbers of transfected vGAT neurons. Because of the dense cluster of vGAT neurons in the vZI area, we were able to image multiple neurons within a single field of view (see panel C in Figure 1). A total of 372 individual neurons were imaged in five vGAT-Cre mice (Table 1).
Figure 1.
Change in fluorescence of the Ca2+ sensor GCaMP6m in vGAT-Cre neurons in the ventral zona incerta and dorsal lateral hypothalamus (vZI-dLH). Panel A depicts in a representative mouse the location of the tip of the GRIN lens atop a dense cluster of vGAT neurons transfected with GCaMP6m. The miniscope can be rotated to focus and we imaged at a focal plane that contained the sharpest images of the neurons. Most transfected neurons were located in the zona incerta and some in the dorsal lateral hypothalamus. Inset B is a higher magnification image of the tip of the GRIN lens (dashed rectangle in panel A). The inset shows numerous vGAT-GCaMP6 containing neurons within the optimal focal plane of the GRIN lens (200 µm). Panel C depicts the field of view of the GRIN lens and shows maximal fluorescence in regions of interest (neurons) after extraction by the PCA-ICA algorithm. Panel D depicts the 1-h sleep–wake session when the fluorescence in the neurons depicted in panel C was measured. The colored horizontal bar identifies wake, NREMS and REMS along with the spectrogram of the EEG. Panel E depicts the change in fluorescence (expressed as ΔF/F0) in the four colored neurons shown in panel C in relation to the wake–sleep states. 3rdV, third ventricle; cp, cerebellar peduncle; DMH, dorsomedial hypothalamus; LH, lateral hypothalamus; mt, mammillothalamic tract; NR, NREMS; opt, optical tract; PH, posterior hypothalamus; R, REMS; Th, Thalamus; W, wake; W-R, wake-REMS; ZI, Zona incerta.
It is useful to note that during the baseplate implant procedure which was done under isofluorane anesthesia we observed fluorescence activity in many vGAT neurons. The fluorescent activity under isofluorane anesthesia was very helpful in bringing into focus the vGAT neurons. We have used the same procedure to image the MCH neurons, but the MCH neurons did not show fluorescence activity under isofluorane anesthesia.
Activity of vGAT neurons in waking and sleep states
Cytoplasmic Ca2+-induced fluorescence in vGAT neurons was recorded during the sleep states as well as during periods of quiet or active waking. Table 2 summarizes sleep parameters such as number of episodes, total sampling time, and mean duration of wake–sleep episodes that occurred during the Ca2+ imaging sessions. Periods of walking, rearing, or grooming were classified as active waking.
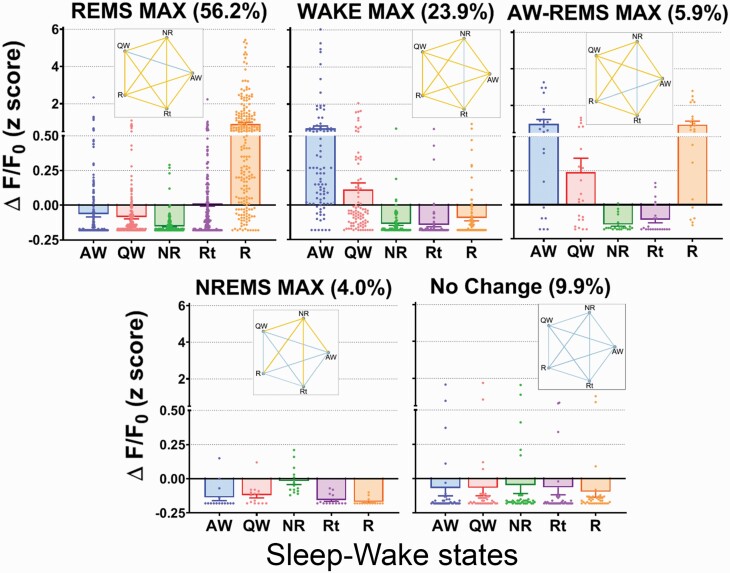
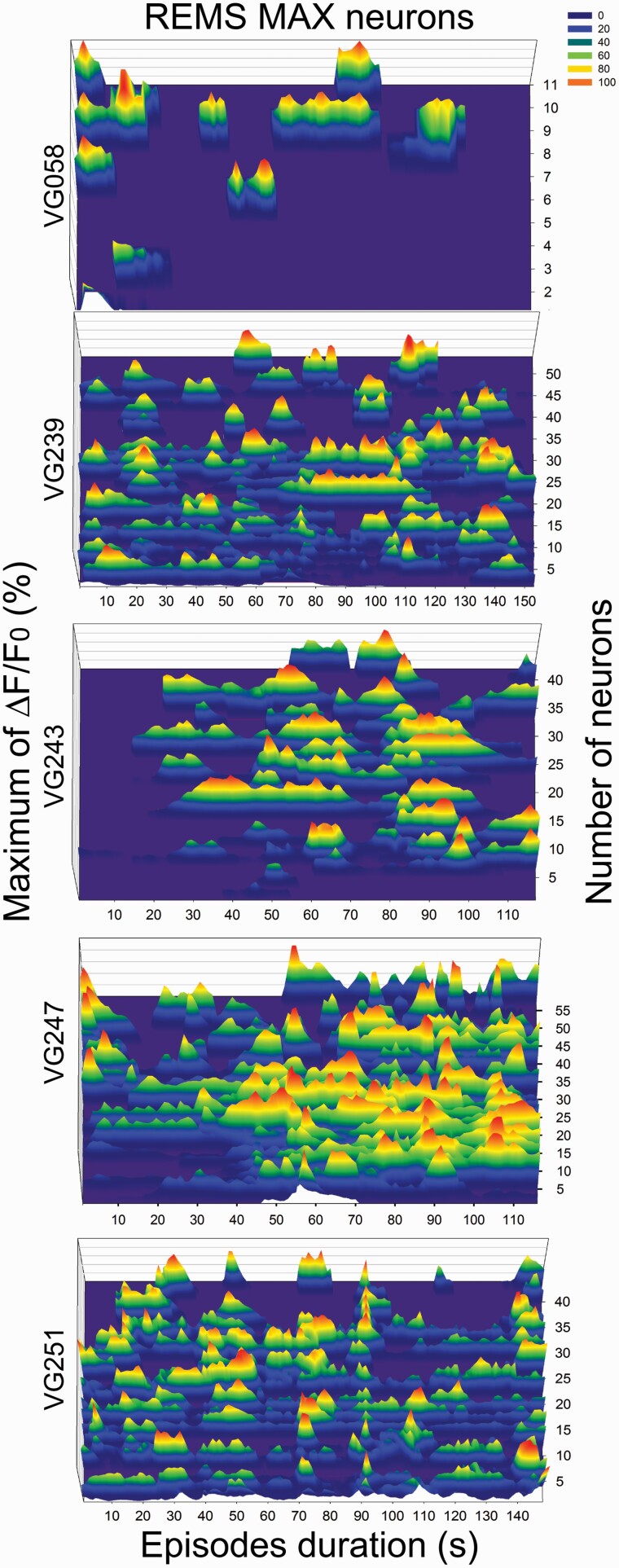
The change in fluorescence (expressed as Z scores) of the 372 vGAT neurons was binned into five groups based on a cutoff equal to or larger than two-times maximal activity in any given vigilance state. Neurons that had maximal fluorescence during REMS (REMS-Max group) represented most of the imaged neurons (56.2%; n = 209). The general linear mixed model found that the average Ca2+ fluorescence during REMS was significantly higher compared to waking, NREMS or REMS transition state (F [1, 4, 040] = 69.007, p < 0.0001). The mean fluorescence in REMS was roughly nine times as high compared to the other vigilance states. REMS-Max vGAT neurons also showed mild fluorescence during the transition into REMS (Figure 2). We examined whether REMS-Max neurons displayed synchronic activity during REMS (Figure 3). Two mice show clear periods of orchestrated activity whereas the others do not. This is in sharp contrast to MCH neurons [19] which showed unambiguous synchronized activity in REMS. We suggest that for vGAT neurons in the ZI, more data are needed.
Figure 2.
Subsets of vGAT GABA neurons in the zona incerta have different profiles of activity during wake–sleep states. 56.2% of the vGAT neurons were most active during REM sleep. A subset of vGAT neurons was most active in NREM sleep. As such, 60.2% of the vGAT neurons were most active in sleep, either NREMS or REM. In contrast, 29.8% were most active in waking or waking+REM sleep. Thus, our study indicates the activity of many of vGAT neurons in the vZI-dLH is biased in favor of sleep, which is not surprising since the most widely prescribed hypnotics promote sleep through a GABA mechanism. Pentagrams indicate statistical difference according to a general linear mixed model. Yellow lines indicate significant pair-wise comparisons (Bonferroni; p < 0.05), whereas the blue connecting lines indicate no significant differences. All values were plotted as scattered individual dots. Each dot represents an individual vGAT neuron. Column height indicates the arithmetic mean while data spread is indicated by error bars (+ or −SEM). AW, active wake; QW, quiet wake; NR, NREMS; R, REMS; Rt, transition into REMS.
Figure 3.
Pattern of fluorescence activity during REM sleep. We have found that 56% (n = 209 neurons) of the vGAT neurons in the zona incerta are most active during REM sleep. To determine whether the fluorescence occurs synchronously, we aligned the calcium peaks relative to the onset of REM sleep in the five mice. Fluorescence values were normalized as percent of maximal activity per mouse. In two mice (VG243 n = 42 and VG251 n = 42 neurons) most of the vGAT neurons fluoresce synchronously while in the other three mice (VG058 n = 11, VG239 n = 54, and VG247 n = 59 neurons) only some show such activity. This contrasts sharply with the synchronous activity shown by neurons containing melanin-concentrating hormone during REM sleep [19]. We suggest that ensemble activity patterns could be used to identify normal versus diseased conditions and gauge the therapeutic efficacy of hypnotics to duplicate normal sleep.
The Wake-Max neurons made up the second most abundant group among all vGAT neurons (23.9%; n = 89). Wake-Max vGAT neurons were significantly more fluorescent when the mice were behaviorally active (grooming, walking, rearing, etc.) as compared to periods of quiet wake, NREMS or REMS (F [4, 435] = 20.204, p < 0.0001). During active behaviors, the Wake-Max neurons had seven-fold more activity than that during quiet wake.
The third group of neurons, in terms of relative abundance, was comprised of vGAT neurons which did not show a relation to any vigilance state (i.e. No Change). The no change vGAT neurons made up 9.9% (n = 37) of all imaged neurons. GLMM identified no significant difference in the average activity across vigilance states (F [4, 180] = 0.149, p = 0.963).
The fourth group was made of neurons that were maximally fluorescent in both active waking and REMS. AW-REMS-Max neurons made up 5.1% (n = 22) of the imaged neurons. In this group, the fluorescence during active waking and REMS was significantly higher compared to the other vigilance states (F [4, 105] = 16.669, p < 0.0001).
Lastly, we found that 4.0% (n = 15) of the imaged neurons showed high fluorescence during NREMS. The NREMS-Max group of vGAT neurons was significantly higher compared to the other vigilance states (F [4, 70] = 11.214, p < 0.0001). On average, the fluorescence in the NREMS-Max neurons was 10 times as compared to the other states. The fluorescence during the other vigilance states was uniformly low and not significantly different from each other (Figure 2).
We could not examine whether the NREMS-Max VGAT neurons show synchronized activity during NREMS because only two mice had more than four neurons (one had 4 and other 6 neurons). Further studies with larger sample sizes will elucidate whether vGAT neurons in the ZI show synchronized activity.
The time-course of activity during transition between vigilance states
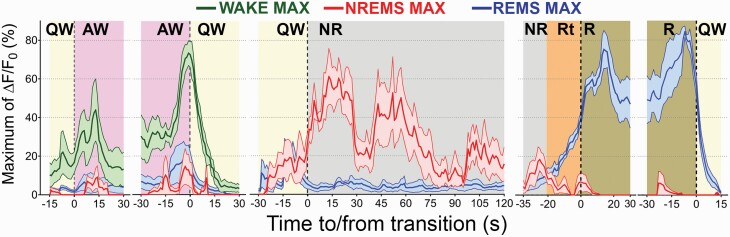
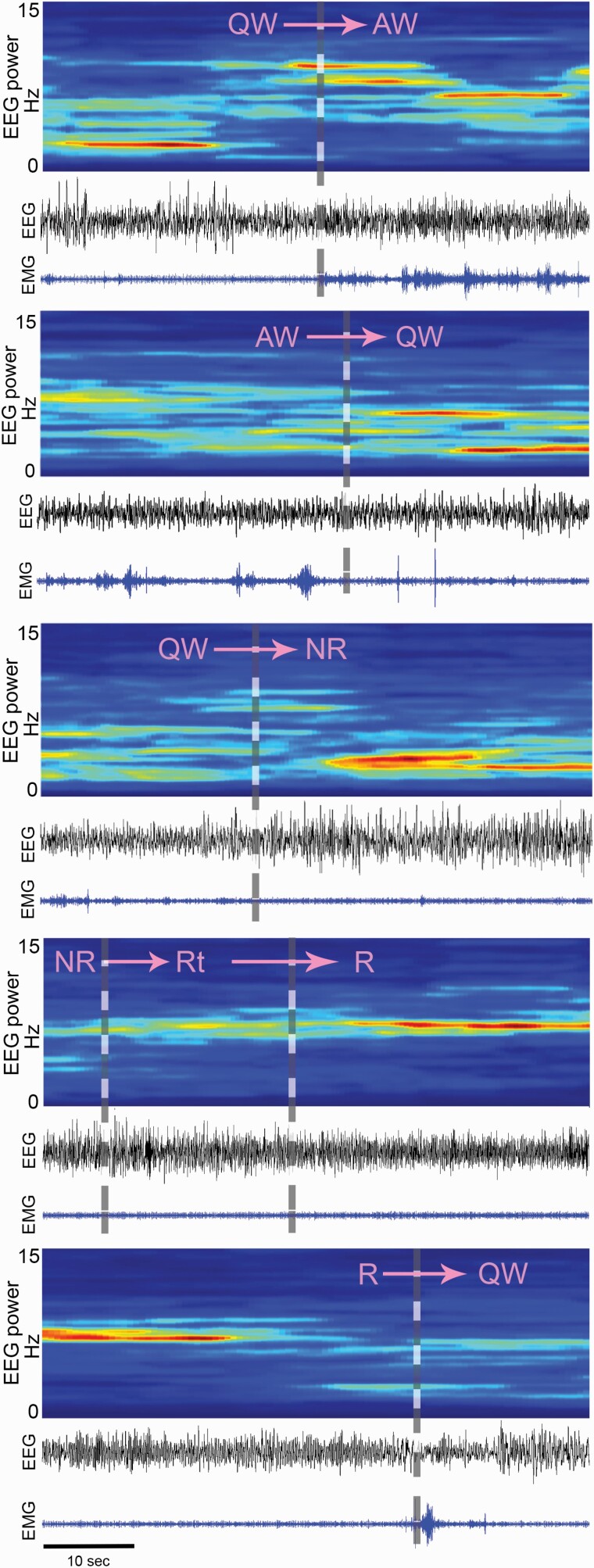
Figure 4 summarizes the time course of the change in the fluorescence signal in the Wake-Max (n = 89, green lines), NREMS-Max (n = 15, red lines) or REMS-Max (n = 209, blue lines) groups. These three groups contained 84.1% of all the neurons imaged in the study. To plot the time-course, the activity of individual neurons was aligned with respect to the onset and offset of each vigilance state and expressed as group mean ±SEM (thick lines ± shadowed areas). We examined the most common state transitions occurring during the time of day of our study (afternoon). We studied the following transitions: QW to AW, AW to QW, QW to NREMS, NR to Rt, Rt to R, and R to QW. Representative examples of these state transitions are illustrated in Figure 5. Regardless of its total duration, all transitions within the categories mentioned above were included in the analysis (Table 3).
Figure 4.
Time course of the change in calcium fluorescence in vGAT neurons in the vZI-dLH during the transitions of the sleep–wake states. The figure shows the time course of fluorescence in three groups: Wake-Max (green; 89 neurons), NREMS-Max (red; 15 neurons), and REMS-Max (blue; 209 neurons). The change in fluorescence, expressed as percent of maximal fluorescence, was aligned to the transition point between vigilance states. Change in fluorescence values were normalized as percent of maximal fluorescence per mouse and then averaged. Solid colored lines represent the averages per group, while shaded bands indicate the ±SEM. Note that in the NREMS-Max and REMS-Max neurons fluorescence increased approximately 30 s before the onset of NREMS. Fluorescence activity in the NREMS-Max group of neurons continued to increase as NREM sleep emerged, and then progressively declined as NREM continues. This suggests that the time course of fluorescence in the NREM-max neurons might be used to gauge sleep homeostasis.
Figure 5.
Classification of transition points between wake–sleep states. The wake–sleep states were partitioned based on EEG, EMG, EEG power spectra and video recording of the animal behavior, criteria that were same as in our previous study [19]. The transition point is indicated by the gray dashed vertical line.
The activity in the Wake-Max neurons increased (~+35%) sharply 10–15 s after the emergence of behavioral activity. As compared to sleep, the activity of Wake-Max neurons remained relatively high as long as the mice were behaviorally active (grooming, walking, rearing, etc.). Mice did not display feeding or drinking during our imaging sessions. The fluorescence quickly diminished once the mice were quietly awake.
The mean fluorescence in the NREMS-Max group began to increase 30 s prior to the onset of NREMS (+35%) and remained high across the NREMS episode. The highest (~75%) level of activity in NREMS-Max neurons occurred 15–30 s after sleep onset and then had a second activity peak spanning from 45 to 60 s after the first peak of activity. The fluorescence in this group decreased during the transition to REMS and in REMS.
The vGAT neurons in the REMS-Max group increased fluorescence during the transition into REMS, reaching a peak (+85%) 20s into the REMS. It is important to note that the end of REMS was always identified by the re-emergence of skeletal muscle tone. However, fluorescence from REMS-Max vGAT neurons did not increase when the EMG activity turned high, that is in active or quiet wake, indicating that these neurons are functionally separate from the Wake-Max neurons.
Discussion
The primary finding of our study was that subsets of vGAT neurons in the vZI-dLH have different profiles of activity during wake–sleep states. We found that 60.2% were most active during sleep (NREMS or REMS) and a separate subset (23.9%) were most active during waking. Moreover, a small (4%) subset of the vGAT neurons increased their activity before the onset of NREM sleep, continued to be active in NREM sleep but then were relatively silent during REM sleep. Our findings linking 60% of the vGAT neurons to sleep states is not entirely surprising because GABA is the primary inhibitory neurotransmitter in the brain. Furthermore, GABA is very important for generation and maintenance of sleep states considering that the most widely prescribed hypnotics enhance GABAergic inhibition across the entire CNS [27]. It remains to be determined whether the sleep active vGAT neurons oppose the activity of the local arousal neurons.
NREMS MAX and REMS MAX neurons
Another research group has already recorded activity of neurons in the same hypothalamic region as our study [3, 8, 28]. They used juxtacellular electrophysiology to record activity of neurons in head-restrained rats (n = 104 neurons in 77 rats) [3]. Post-mortem analysis of a few neurobiotin-filled neurons revealed 10 neurons that were positive for vGAT but negative for melanin-concentrating hormone (MCH). In that study, two of the vGAT neurons were classified as NREMS-Max while eight were designated as REMS-Max [3]. In that study, the two neurons that were vGAT+ NREMS-Max increased their activity during the transition from wake to NREMS [3]. Our results corroborate their results in that we also found NREMS-Max vGAT neurons anticipated sleep onset (QW to NREMS transition) and were maximally active during NREM sleep. The time-course of the increase in action potentials during sleep in their study is also similar to the increase in fluorescence that we observed using the genetically encoded Ca2+ sensor. Like the electrophysiological studies, we also found fewer NREMS-Max neurons compared to REMS-Max neurons. Another electrophysiology study has confirmed that in the ZI there are more neurons that are maximally active during REMS compared to NREMS [11].
Other studies have identified neurons in the preoptic area that anticipate NREMS [29, 30], but the phenotype of those neurons was not determined. In the preoptic area, GABA neurons that project to the arousal histaminergic neurons have been found to be active in NREMS and REMS, but these were not shown to anticipate NREMS [2]. Recently, we [19] and others [31] used the deep-brain Ca2+ imaging method to identify activity of the neurons containing melanin-concentrating hormone (MCH). We did not find any MCH neurons that were active in NREM sleep. Moreover, galanin neurons in the lateral hypothalamus are not NREMS-Max either [32]. Interestingly, we found a gradual downward slope in the time-course of fluorescence of the NREM-Max neurons during NREM sleep (Figure 4). This shows a dissipation of the Ca2+ induced fluorescence during NREM sleep. Moreover, the peak fluorescence appears to cycle with a 30–40 s periodicity (Figure 4) but more data are needed to fully identify link with the EEG. We suggest that the time course of activity could be used to gauge accumulation and dissipation of sleep homeostasis.
WAKE MAX neurons
We found that 23.9% of all imaged vGAT neurons in the vZI/dLH were Wake-Max while 5.9% were Wake-REMS-Max. Altogether nearly, a third of all vGAT neurons show strong activity during wake. Only one Wake-Max neuron was recorded in the electrophysiology study [3] but that is likely because in that study rats were not allowed to freely behave. Juxtacellular electrophysiological recording requires a head-restrained in vivo preparation whereby behavior must be severely constrained. We suggest that allowing animals to move freely is critical to determine the true activity patterns of neurons. We observed that the Wake-Max neurons became most active during grooming, walking, or rearing. We could not observe activity during feeding/drinking as the mice did not engage in these types of behavior during the time of the imaging session. Interestingly Wake-Max vGAT neurons also show anticipatory activity ahead of both QW and NREMS. The pattern of neuronal activation of the Wake-Max neurons suggests that termination of active behaviors demands strong GABAergic inhibition coming from the vZI/dLH. More importantly, in addition to the Wake-Max vGAT neurons, the other two types of sleep active vGAT neurons studied here also anticipate sleep onset. Because altogether these three groups made up 84.1% of all sampled GABA neurons, our study strongly indicates that most of the GABA neurons in the vZI/dLH may play a significant role in making the brain achieve a quiet state (AW to QW transition), especially making it fall asleep (QW to NREMS transition). It can be also realized from our hypothesis that GABA neurons may also be implicated in pathologies associated with high levels of active behaviors like ADHD or hyperarousal states like insomnia of initiation.
Activating vGAT-ZI neurons induces sleep
Other chemical markers such as calbindin, calretinin, galanin, LHx6, neuropeptide Y, parvalbumin, and somatostatin colocalize in different clusters of vGAT hypothalamic neurons [33, 34]. In particular, the chemogenetic activation of the vGAT vZI cluster that co-expresses LHx6, a developmental gene transcriptional repressor, strongly induces both NREMS and REMS [35]. The LHx6+ neurons represent 45% of all vGAT+ neurons within the ventral portion of the ZI area, receive input from many arousal neurons and project to the orexin and GABA neurons residing in the ventral aspect of the LH/PeF area. Indirect readout of the activity using c-FOS suggested the LHx6+ vZI GABAergic neurons were most active during the lights-on phase which is the normal sleeping time in mice [35]. These neurons also showed increased c-FOS in response to increased sleep pressure and sleep rebound. Most importantly, their study found that selective pharmacogenetic activation of the LHx6+ vZI GABAergic neurons significantly enhanced both NREMS and REMS. In contrast, selective inhibition of the LHx6 neurons significantly reduced both types of sleep. Thus, the selective manipulation of the spontaneous activity of the vZI GABAergic neurons supports the role of the GABA neurons within the ventral ZI in inducing both NREMS and REMS. We hypothesize that some of the sleep active neurons we imaged could be LHx6+ GABA neurons. However, further Ca2+ imaging studies in LHx6-Cre mice are needed to test this hypothesis.
While the GABA neurons in the vZI-dLH induce sleep there are also clusters of GABA neurons located more ventrally and medially where pharmacogenetic activation has been found to increase waking, in particular exploratory behaviors such as foraging and digging [4]. In contrast, active behaviors like climbing and grooming were strongly inhibited by stimulation of the wake vGAT hypothalamic neurons. A second research group used optogenetics to stimulate vGAT+ neurons in the ventral part of the LH and found that these neurons drive wake by inhibiting the thalamic reticular GABAergic neurons [5]. At that time, it was unknown whether the activity of single GABA neurons in the ventral part of the LH was biased toward waking [4]. Recently, the same research group used the fiber-photometry method to measure population level Ca2+-induced fluorescence changes from the ventral LH vGAT+ neurons [36]. Similar to our study, they found high levels of activity among GABA LH neurons during wake but also in REMS (WAKE-REMS MAX type). In contrast, they did not observe GABA neurons with maximal activity that occurred ahead of or during NREMS. These authors hypothesized that optogenetic stimulation of these ventral LH GABA neurons drives waking by inhibiting VLPO galanin+ neurons. In contrast to the fiber-photometry study [36], but agreeing with our study, another laboratory used the miniscope method and found a small subset of NREMS-Max vGAT (5%) neurons within the Perifornical/LH area [37]. These researchers also observed Wake-Max, REMS-Max, and state independent GABA neurons as we are reporting here. Like our study, this group used the miniscope to image single neuron calcium transients. The miniscope method underscores the strength of monitoring single neuronal activity as it reveals the existence of subsets of neurons within an apparent homogeneous phenotypic ensemble (i.e. GABAergic hypothalamic neurons).
Altogether, it is understood that the ZI-LH area serves as an integrative node for regulation of wide variety of behaviors [38, 39]. As we are reporting here, there are clusters of GABA neurons that become active prior to the onset of sleep and continue to be active during sleep. The increase in activity of a subset of GABA neurons would inhibit the arousal neurons and make the brain quiet down and eventually fall asleep. Other clusters of GABA neurons in ventral LH would do the opposite driving waking. It is important to also mention that we found that roughly 10% of the vGAT imaged neurons do not show state dependent activity. Similar finding was observed by another miniscope study in the ventral LH [37]. Perhaps the state-independent neurons could be interneurons providing a local inhibitory tone.
A limitation of our Ca2+ imaging study is that even though our data strongly suggest that GABA vZI neurons promote sleep states, our hypothesis relies on correlation data. Correlation does not necessarily imply causation. Further studies where these neurons are specifically manipulated will test our hypothesis. A second limitation of our study is that it is difficult to infer whether the peaks in fluorescence reflect single action potentials or a cluster of action potentials, even though there is a linear relationship between the increase in fluorescence and action potentials [19]. Newer voltage sensors or next generation GCaMP sensors might be able to resolve the temporal pattern of imaged neurons [40]. Even though calcium peaks are not necessarily correlated one-to-one with action potentials, it does not mean that calcium peaks cannot reveal meaningful information about the activity of the neuron. Similarly, chemogenetic or optogenetics also reveal potential functions in neurons even though their rhythm of stimulation does not necessarily mimic the spontaneous cadence of action potentials. Thus, it is not necessary to duplicate the cadence or pattern of action potentials to evoke behavior. Moreover, the optogenetic and pharmacogenetic studies indicate that to induce the behavior not all of the neurons need to be stimulated [41]. Likewise, calcium imaging samples the activity of a subset of neurons but it cannot image the entire population. Nonetheless, the miniscope method is able to image, for the first time, large-scale activity of individual neurons [14] and glia [42, 43]. The change in fluorescence is a measurement of the change in the internal mileu of the neuron, something that action potentials, a binary all-or-none signal, does not measure. Other significant strength of miniscope calcium imaging is that the fluorescence in individual cells can be recorded longitudinally over days to months. This allows for a new understanding of the effect of various treatments on network activity and for comparing normal versus diseased brains.
Acknowledgments
PJS was supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research Development (BLR&D; BX000798; BX004216), and NIH grants NS052287 and NS079940. The contents of this work do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Disclosure statements
Financial Statement. The authors declare no financial conflict of interest.
Non-financial Statement: None.
Author contributions
All authors shared in collecting the data and writing the manuscript. The imaging data were analyzed by CBC and SWL. CBC and SWL share first authorship.
References
- 1. Jones BE. Arousal and sleep circuits. Neuropsychopharmacology. 2020;45(1):6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chung S, et al.. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature. May 25, 2017;545(7655):477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hassani OK, et al. GABAergic neurons intermingled with orexin and MCH neurons in the lateral hypothalamus discharge maximally during sleep. Eur J Neurosci. 2010;32(3):448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Venner A, et al. A Novel Population of Wake-Promoting GABAergic Neurons in the Ventral Lateral Hypothalamus. Curr Biol. 2016;26(16):2137–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Herrera CG, et al. Hypothalamic feedforward inhibition of thalamocortical network controls arousal and consciousness. Nat Neurosci. 2016;19(2):290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aston-Jones G, et al. Activity of norepinephrine-containing locus coeruleus neurons in behaving rats anticipates fluctuations in the sleep-waking cycle. J Neurosci. 1981;1(8):876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. John J, et al. Cataplexy-active neurons in the hypothalamus: implications for the role of histamine in sleep and waking behavior. Neuron. 2004;42(4):619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee MG, et al. Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. J Neurosci. 2005;25(28):6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mileykovskiy BY, et al. Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron. 2005;46(5):787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu MF, et al. Activity of dorsal raphe cells across the sleep-waking cycle and during cataplexy in narcoleptic dogs. J Physiol. 2004;554(Pt 1):202–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koyama Y, et al. State-dependent activity of neurons in the perifornical hypothalamic area during sleep and waking. Neuroscience. 2003;119(4):1209–1219. [DOI] [PubMed] [Google Scholar]
- 12. Ghosh KK, et al. Miniaturized integration of a fluorescence microscope. Nat Methods. 2011;8(10):871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian L, et al. Neural activity imaging with genetically encoded calcium indicators. Prog Brain Res. 2012;196:79–94. [DOI] [PubMed] [Google Scholar]
- 14. Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox J, et al. Calcium imaging of sleep-wake related neuronal activity in the dorsal pons. Nat Commun. 2016;7:10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weber F, et al. Control of REM sleep by ventral medulla GABAergic neurons. Nature. 2015;526(7573):435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weber F, et al. Circuit-based interrogation of sleep control. Nature. 2016;538(7623):51–59. [DOI] [PubMed] [Google Scholar]
- 18. Weber F, et al. Regulation of REM and non-REM sleep by periaqueductal GABAergic neurons. Nat Commun. 2018;9(1):354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blanco-Centurion C, et al. Dynamic network activation of hypothalamic MCH neurons in REM sleep and exploratory behavior. J Neurosci. 2019;39(25):4986–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McIntire SL, et al. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389(6653):870–876. [DOI] [PubMed] [Google Scholar]
- 21. Chaudhry FA, et al. The vesicular GABA transporter, VGAT, localizes to synaptic vesicles in sets of glycinergic as well as GABAergic neurons. J Neurosci. 1998;18(23):9733–9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blanco-Centurion C, et al. Optogenetic activation of melanin-concentrating hormone neurons increases non-rapid eye movement and rapid eye movement sleep during the night in rats. Eur J Neurosci. 2016;44(10):2846–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Konadhode RR, et al. Optogenetic stimulation of MCH neurons increases sleep. J Neurosci. 2013;33(25):10257–10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jego S, et al. Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci. 2013;16(11):1637–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blanco-Centurion C, et al. VGAT and VGLUT2 expression in MCH and orexin neurons in double transgenic reporter mice. IBRO Rep. 2018;4:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagai T, et al. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci U S A. 2001;98(6):3197–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gottesmann C. GABA mechanisms and sleep. Neuroscience. 2002;111(2):231–239. [DOI] [PubMed] [Google Scholar]
- 28. Hassani OK, et al. Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci U S A. 2009;106(7):2418–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szymusiak R, et al. Sleep-related neuronal discharge in the basal forebrain of cats. Brain Res. 1986;370(1):82–92. [DOI] [PubMed] [Google Scholar]
- 30. Szymusiak R, et al. Sleep-waking discharge of basal forebrain projection neurons in cats. Brain Res Bull. 1989;22(2):423–430. [DOI] [PubMed] [Google Scholar]
- 31. Izawa S, et al. REM sleep-active MCH neurons are involved in forgetting hippocampus-dependent memories. Science. 2019;365(6459):1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen KS, et al. A hypothalamic switch for REM and non-REM sleep. Neuron. 2018;97(5):1168–1176.e4. [DOI] [PubMed] [Google Scholar]
- 33. Hernández VM, et al. Parvalbumin+ neurons and Npas1+ neurons are distinct neuron classes in the mouse external globus pallidus. J Neurosci. 2015;35(34):11830–11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qualls-Creekmore E, et al. Galanin-expressing GABA neurons in the lateral hypothalamus modulate food reward and noncompulsive locomotion. J Neurosci. 2017;37(25):6053–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu K, et al. Lhx6-positive GABA-releasing neurons of the zona incerta promote sleep. Nature. August 31, 2017;548(7669):582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Venner A, et al. An inhibitory lateral hypothalamic-preoptic circuit mediates rapid arousals from sleep. Curr Biol. 2019;29(24):4155–4168.e5. [DOI] [PubMed] [Google Scholar]
- 37. Oesch LT, et al. REM sleep stabilizes hypothalamic representation of feeding behavior. Proc Natl Acad Sci U S A. 2020;117(32):19590–19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X, et al. Zona incerta: an integrative node for global behavioral modulation. Trends Neurosci. 2020;43(2):82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Urbain N, et al. Motor cortex gates vibrissal responses in a thalamocortical projection pathway. Neuron. 2007;56(4):714–725. [DOI] [PubMed] [Google Scholar]
- 40. Bayguinov PO, et al. Imaging voltage in genetically defined neuronal subpopulations with a cre recombinase-targeted hybrid voltage sensor. J Neurosci. 2017;37(38):9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Upright NA, et al. Behavioral effect of chemogenetic inhibition is directly related to receptor transduction levels in rhesus monkeys. J Neurosci. 2018;38(37):7969–7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poskanzer KE, et al. Astrocytes regulate cortical state switching in vivo. Proc Natl Acad Sci U S A. 2016;113(19):E2675–E2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ingiosi AMH, et al. A role for astroglial calcium in mammalian sleep and sleep regulation. Curr Biol. 2020;30(22):4373–4383.e7. doi: 10.1061/j.cub.2020.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]