Highlights
-
•
SDH mutations are responsible for the occurrence of head and neck paragangliomas (HNPGLs).
-
•
Deregulation of oxidative phosphorylation (ROS) in association with metabolism may play an important role in HNPGLs.
-
•
NDUFA 2, 4 and 10 showed the most significant deregulaion in HNPGLs, and this deregulation can be corrected by metformin.
-
•
Metformin exerts inhibitory effects on HNPGL cells via reduced level of reactive oxygen species and altered metabolism.
-
•
Proteomic/metabolic profiling of HNPGLs identifies the role of ROS/metabolism as a therapeutic target in clinical settings.
Keywords: Head and neck paragangliomas, Proteomics, Metabolomics, SDH, Metformin
Abstract
Head and neck paragangliomas (HNPGLs) are rare neoplasms that represent difficult treatment paradigms in neurotology. Germline mutations in genes encoding succinate dehydrogenase (SDH) are the cause of nearly all familial HNPGLs. However, the molecular mechanisms underlying tumorigenesis remain unclear. Mutational analysis identified 6 out of 14 HNPGLs harboring clinicopathologic SDH gene mutations. The SDHB gene was most frequently mutated in these patients, and western blot showed loss of SDHB protein in tumors with SDHB mutations. The paraganglioma cell line (PGL-626) was established from a sample that harbored a missense SDHB mutation (c.649C > T). Spectrometric analysis using tandem mass tags identified 151 proteins significantly differentially expressed in HNPGLs compared with normal nerves. Bioinformatics analyses confirmed the high level of enrichment of oxidative phosphorylation and metabolism pathways in HNPGLs. The mitochondrial complex subunits NDUFA2, NDUFA10, and NDUFA4, showed the most significantly increased expression and were localized predominantly in the cytoplasm of PGL-626 cells. The mitochondrial complex I inhibitor metformin exerted dose-dependent inhibitory effects on PGL-626 cells via cooperative down-regulation of NDUFA2, 4, and 10, with a significant decrease in the levels of reactive oxygen species and mitochondrial membrane potential. Further metabolomic analysis of PGL-626 cells showed that metabolites involved in central carbon metabolism in cancer and sphingolipid signaling pathways, pantothenate and CoA biosynthesis, and tryptophan and carbon metabolism were significantly altered after metformin treatment. Thus, this study provides insights into the molecular mechanisms underlying HNPGL tumorigenesis and identifies target correction of metabolic abnormalities as a novel therapeutic approach for this disease.
Introduction
Paragangliomas (PGLs) are highly vascularized neoplasms arising from the neural crest chief cells associated with the parasympathetic (mainly head and neck) or sympathoadrenal (mainly truncal) lineage [1]. The head and neck PGLs (HNPGLs) originate preferentially from the carotid body and the jugular, tympanic, and vagal paraganglia [2]. HNPGLs are generally weakly metastatic, but they infiltrate the adjacent neurovascular structures of the skull base, causing significant morbidities. Germline mutations in the genes encoding succinate dehydrogenase (SDH) subunits or cofactors (SDHA, SDHB, SDHC, SDHD and SDHAF2, here referred to as SDHx) are the cause of nearly all familial HNPGLs [3,4]. Germline mutations in SDHx also occur in up to 30% of HNPGLs considered “sporadic” due to the absence of a family history [5,6], pointing to these HNPGLs being “occult familial” cases [7]. As for SDHD mutations, there is an autosomal dominant transmission with a parent-of-origin effect [8]. A paternal imprinted tumor-suppressor gene mapped to the 11p15 region promoted tumorigenesis of PGLs in synergy with a SDHD mutation [9]. The disease has been reported after maternal transmission of the SDHD mutation in only three cases [[10], [11], [12]]. Even if it is a rare event, it has important clinical implications for SDHD families.
Recently, promoter methylation of SDHC has been described as a novel cause of SDH inactivation in PGLs [13]. Chiara et al. [14] found that the epigenetic modification of SDHC was accompanied by the heterozygous loss of the chromosomal region containing the gene, consistent with the Knudson two-hit model for tumorigenesis. DNA methylation-based epigenetic signatures are becoming valuable cancer biomarkers, and epigenetic therapies have shown the potential for effective treatment of solid cancers [15]. The SDHx mutations in pheochromocytomas (PCCs) and PGLs lead to elevated succinate levels that mediate inhibition of 2-OG-dependent histone and DNA demethylases, thereby causing a CpG island methylator phenotype (CIMP), defined as the methylation of CpG islands of multiple tumor-suppressor genes (TSGs) [16]. The epigenetic inactivation on multiple TSGs may serve as a key mechanism for the progressive behaviors of SDH-mutated HNPGLs as reported in our previous study [2].
SDH plays a central role in energy metabolism as both an enzyme of the tricarboxylic acid (TCA) cycle and as complex II of the mitochondrial respiratory chain, involved in oxidative phosphorylation [17]. Mutation of SDH subunits has been shown to result in accumulation of succinate and inhibition of hypoxia-inducible factor 1 (HIF-1) prolyl hydroxylases, leading to the stabilization of HIF-1, a key factor in the hypoxia response [18,19]. In addition, SDH dysfunction leads to the generation of reactive oxygen species (ROS) via inhibition of SDH in its role as complex II of the respiratory chain. ROS lead directly to the stabilization of HIF-1 [20,21]. These recent mechanistic insights suggest that low oxygen levels have a central role in the genesis of these tumor and clearly suggest a link to high-altitude HNPGLs [4,22,23]. Although experimental data have revealed several pathways involved in hypoxia in HNPGLs, no further investigations have been carried out to profile protein expression in the tumors. At present, surgery is the only effective therapeutic option for HNPGLs. Hence, there is an urgent need to identify potential targets that are active and treatable in this disease. However, there is no commercially available PGL cell lines, making it extremely inconvenient for researchers to perform in vitro drug experiments and in-depth mechanism investigations.
In this study, therefore, we aimed to obtain improved insight into HNPGLs by searching proteomics data for differentially expressed proteins (DEPs) between the tumors and normal tissue samples. We also constructed a protein-protein interaction (PPI) network and performed functional and pathway enrichment analyses of network modules. We showed that the mitochondrial complex I and IV subunits, NDUFA2, NDUFA4, and NDUFA10 had the most significantly deregulated expression in HNPGLs. Interestingly, a PPI “hot spot” was shown in the three proteins with the mitochondrial complex II subunits (SDHA, SDHB, SDHC, and SDHD). Gene-set enrichment analysis demonstrated upregulation of proteins involved in respiratory electron transport and mitochondrial biogenesis and metabolic activities. As metformin behaves as a “weak” mitochondrial poison, due to its Complex I inhibitor activity, this could explain its remarkable clinical ability to prevent many different types of human cancers in diabetic patients, and its effectiveness as an anti-cancer agent in pre-clinical animal models [24]. We further found a significant reduction in the protein expression levels of NDUFA2, NDUFA4 and NDUFA10 together with changes in metabolic capabilities in an immortalized HNPGL cell line in response to metformin treatment.
Materials and methods
Tumor samples
The diagnosis of HNPGLs established on the basis of a clinical otolaryngologic examination, along with computed tomography (CT) scanning, magnetic resonance imaging (MRI), and pathological confirmation after surgery. The tumors were either kept at -80°C until DNA/protein extraction or collected immediately for primary culture after resection. Three cases of great auricular nerves (GANs) obtained from parotidectomy were included as non-tumoral controls. Informed written consent was obtained from all patients donating tissue. This study was approved by the Institutional Review Board of the ethics committee of The Ninth People's Hospital affiliated to School of Medicine, Shanghai Jiao Tong University.
Direct sequencing analysis
Sanger sequencing was conducted to detect point mutations or small deletions/insertions. DNA extraction from the tumor specimens was performed using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China). The whole coding sequence and exon-intron boundaries of the genes were amplified by the polymerase chain reaction (PCR) using standard methods and underwent bidirectional sequencing. The sequence data were analyzed using the Sequencer 4.9 software (Genecode, MI, USA) and compared with the sequences of SDHA (NM_004168.3), SDHB (NM_003000.2), SDHC (NM_003001.3) and SDHD (NM_001276506.1) in GeneBank.
Tandem mass tag (TMT)-based proteomics analysis
Proteomics data collection
Tumor/nerve tissues were lysed, digested, and TMT labeled according to the manufacturer's protocol. In brief, TMT-labeled peptides were separated and analyzed using a nano-UPLC (EASY-nLC1200) coupled with a Q-Exactive mass spectrometry (Thermo Finnigan). Separation was performed with a reversed-phase column (Acclaim PepMap RSLC, Termo Scientifc). Mobile phase A consisted of 0.1% formic acid and 2% acetonitrile, and mobile phase B consisted 80% acetonitrile and 0.1% formic acid. Mass spectrometry (MS) was performed in a data-dependent mode with single full-scan mass spectrum in the orbitrap (m/z 350–1600, 70,000 resolution), and MS/MS scans were carried out in data-dependent mode at 32% collision energy. The resulting MS/MS data were processed using an integrated Andromeda search engine (V.1.5.6.0) integrated into the MaxQuant environment. The protein sequence database (Uniprot_organism_2016_09) was downloaded from UNIPROT, and this database and its reverse decoy were then searched against by MaxQuant software. Trypsin was set as specific enzyme with up to 3 miss cleavage; oxidation [M] and acetyl [protein N-term] were considered as variable modification, and carbamidomethyl [C] was set as fixed modification; false discovery rate (FDR) thresholds for proteins, peptides and modification sites were specified at 1%. Only unmodified unique peptides were used for quantification analysis. The proteins were quantified by the normalized summed peptide intensities. Proteins were considered to be significantly differentially expressed if the TMT ratios were > 1.5 or < 0.67, with a P-value < 0.05. Pheatmap package (version 1.0.8) in R was utilized to perform the hierarchical cluster analysis.
Functional enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database is used to annotate the pathways. The gene ontology (GO) analysis, including biological process (BP), cellular component (CC), and molecular function (MF), helps identify the biological traits for proteomics data. KEGG pathways and GO terms were analyzed with the DAVID database (https://david.ncifcrf.gov/). Statistically significant P-values were set at 0.05 or less.
Protein-protein interaction (PPI)
The STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is a curated knowledge database and used for illustration of predicted interactions of identified proteins and neighbor genes. The proteins expressed significantly differentially in tumors relative to nerve controls were processed in STRING v11.0 (https:// string-db.org/) to obtain high-confidence interaction data (score ≥ 0.7). The PPI network was visualized using the Cytoscape 3.2.1 software (https://cytoscape.org/).
Gene-set enrichment analysis (GSEA)
GSEA allows detection of modest but coordinated changes in expression of protein-encoding genes involved in a common biological function. To determine if the pathways associated with metabolism and oxidative phosphorylation were activated in the tumors, GSEA was performed by the R package clusterProfler (version 3.4.1). The t-statistic mean of the genes was computed in each KEGG pathway using a permutation test with 1000 replications. The up-regulated pathways were defined by a normalized enrichment score (NES) > 0. Gene sets with a P-value less than 0.05 were considered to be significantly enriched or depleted.
Establishment of an immortalized HNPGL cell line (designated PGL-626)
Primary culture
The primary culture of HNPGL was performed according to protocols previously described [23]. Briefly, tumor tissues were cut into 1 mm3 segments, washed with phosphate-buffered saline (PBS), and then incubated in media with enzyme mixture containing 250 U/mL Hyaluronidase Type I-S (#C1639, Sigma-Aldrich) and 160 U/mL Collagenase Type I (#3506, Sigma-Aldrich) in DMEM/F12 (#11330032, Gibco) for 18 h at 37 °C. After the enzymatic incubation of the culture, the cells were re-suspended in DMEM/F12 medium with 10% fetal bovine serum (FBS) (#1414426, Gibco), passed through a 200 mesh filter screen, and finally transferred to 6 cm culture dishes pre-coated with poly-Lornithine and laminin (#354232, BD Biosciences).
Immortalization of primary culture using lentiviral vectors
The HNPGL cultures were immortalized by retroviral-mediated transduction of simian virus 40 (SV40) large-tumor (LT) antigen. The vector pLVX-IRES-puro was purchased from Clontech (CA, USA). SV40-LT expression was evaluated by immunofluorescence staining. The subconfluent culture was infected with lentiviruses for 72 h, and screened with 1.0 μg/mL puromycin (Sigma-Aldrich) for 2 weeks. The selection medium was changed every 2 days, and cell viabilities were assessed under a phase-contrast microscope. Following selection with puromycin, the medium was replaced with DMEM-F12 complete medium containing 5% fetal bovine serum (FBS) (#1414426, Gibco), 5 ng/ml VEGF (#RVEGFI, Gibco), 5ng/ml EGF (#RP-8661,Gibco), 5 ng/ml FGF basic (#RP-8628, Gibco), 15 ng/ml IGF-1 (#RP-10931, Invitrogen), 1ug/ml Hydrocortisone hemisu- cinate (#614157, Sigma-Aldrich), and 50 ug/ml Ascorbic acid (#S3114, Selleck). Cells were maintained in the complete medium and cultured to more than 12 passages.
Immunofluorescence
Cultures were plated on glass slides, fixed with 4 % paraformaldehydees, and permeabilized in 0.3 % Triton X-100. The slides were then blocked with bovine serum albumin before being single- or double-probed with antibodies against S100 (#66616-1-Ig, Proteintech), Chromogranin A (#60135-1-Ig, Proteintech), Synaptophysin (#60191-1-Ig, Proteintech), Vimentin (#60330-1-Ig, Proteintech), SDHA (#ab137040, Abcam), SDHB (#ab1471 4, Abcam), SDHC (#sc-515102, Santa Cruz), SDHD (#SAB3500797, Sigma-Aldrich), NDUFA2 (#ab198196, Abcam), NDUFA4 (#ab129752, Abcam), and NDUFA10 (#ab174829, Abcam). Alexa Fluor® 488 Phalloidin (#A12379, Thermo) was used for the staining of F-actin (filamentous actin) cytoskeleton. The sections were nuclear counterstained with 4,6-Diamidino-2-phenylindole (DAPI, #1985274, Thermo). Images (no z-stack) were acquired using a confocal microscope (LSM 880; Carl Zeiss GmbH, Germany).
Drug treatments in vitro
Cell viability assays
Metformin (#D150959, Sigma-Aldrich) was dissolved in the phosphate-buffered saline (PBS) at a concentration of 100 mM until use. The effect of the drug on cell viability was determined by a Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). Briefly, cells were grown to 50% confluency in 96-well plates before the treatment with PBS control or different concentrations of metformin. At each specified time point, medium was replaced with an equal volume of fresh medium containing CCK-8. Next, cells were incubated for an additional 1 h at 37 °C, and the absorbance of the solution was analyzed at 450 nm using a SpectraMax190 microplate reader (Molecular, USA).
Estimation of reactive oxygen species (ROS) and mitochondrial membrane potential (MMP)
The ROS levels were determined using 2, 7-dichlorodihydrofluorescein diacetate (DCFH-DA, Abcam), a fluorogenic dye that measures intracellular hydroxyl and peroxyl activities. The fluorescent dye Mito-Tracker Red (#C1071S, Beyotime, Shanghai, China) was used to monitor the changes in MMP. Briefly, PGL626 cells (5 × 105 cells/well) were seeded in 24-well plates to achieve about 80% confluence. Subsequently, the medium was replaced with fresh medium with different metformin concentrations of 0, 10 and 20 mM for 48 h. At the end of incubation, the cells were incubated with 10 mM of DCFH-DA or 1 μM Mito-Tracker Red in serum-free medium for 20–30 min in the dark at room temperature. The residual reagent was removed by washing with PBS three times. The fluorescence of DCFH-DA (488 nm excitation/525 nm emission) as well as the Mito-Tracker Red (530 nm excitation/590 nm emission) were captured by a Zeiss confocal laser scanning microscope.
Metabolome analysis
The metabolite extract samples were analyzed by Q Exactive HF-X mass spectrometry (Thermo Scientific, San Jose, USA) on a Vanquish Horizon system (Thermo Scientific). The injection volume of sample was 2 µL and the column temperature was maintained at 40 °C. Mobile phase A consisted of 95 % H2O and 5% acetonitrile containing 0.1% formic acid, and the mobile phase B consisted of 47.5 % acetonitrile, 47.5 % isopropyl alcohol, and 5 % H2O containing 0.1% formic acid. LC gradient was as follows: 100% A at 0 min, 75.5 % A/24.5 % B at 3.5 min, 35 % A/65 % B at 5 min, 0% A/100 % B at 5.5–7.4 min, 48.5 % A/51.5 % B at 7.6 min, 100% A at 7.8–10 min. Electrospray ionization (ESI) source settings were as follows: Aux gas flow rate, 13 arb units; sheath gas flow, 50 arb units; heater temp, 425 °C; capillary temp, 325 °C; spray voltage (+), 3500 V; spray voltage (-), -3500 V. Mass spectral data from 70 to 1050 m/z were collected in the positive ionization mode. Untargeted data analysis was performed using Progenesis QI software (WatersCorporation, Milford, USA). Data were processed in successive treatment steps as peak alignment, peak picking and normalization to obtain data matrix. This matrix was used to perform principal component analysis (PCA). The metabolic features that showed significant changes in the cancer cells with or without Metformin treatment were used to generate heat maps with the ggplot2 package in the R.
Western blotting
Tumor tissues or cells were ultrasonicated in the Lysis Buffer (Beyotime) containing 1mM phenylmethane- sulfonyl fluoride. Tissue/cell extract samples were subjected to Western blotting with antibodies specific for SDHA (#ab137040, Abcam), SDHB (#ab14714, Abcam), HIF-1α (#ab51608, Abcam), NDUFA2 (#ab198196, Abcam), NDUFA4 (#ab129752, Abcam), NDUFA10 (#ab174829, Abcam), cyclinD1 (#C7464, Sigma-Aldrich), cleaved-caspase 3 (# 9664, Cell Signaling), and cytochrome c (#2922, Cell Signaling). The β-actin (# AA128, Beyotime) and GAPDH (# SAB1405848, Sigma-Aldrich) antibodies were used to ensure equal loading of total protein. The blots were detected using a chemiluminescent HRP substrate (Millipore). The intensities of the bands on blots were analyzed using Imager Lab Software (Bio-Rad).
Statistical analysis
Statistical analyses were performed using the SPSS statistics software (SPSS, Chicago, IL). Data were expressed as mean ± standard deviation (SD) from at least three independent experiments. The between- group differences were assessing using one-way analysis of variance (ANOVA) or Student's t-test. P < 0.05 was considered statistically significant.
Results
Mutational analyses revealed significant disease-causing mutations in HNPGLs
Mutational analysis identified 6 out of 14 HNPGLs harboring clinicopathologic SDH gene mutations, including 4 cases with SDHB mutations, one case with a frameshift mutation in SDHD, and one case identified a missense mutation in SDHA. This study included 13 jugular PGLs (JP) and 1 carotid body PGL (CBP). The SDHB gene was most frequently mutated in these patients, and interestingly, 8 out of the 14 samples harbored a silent mutation within the gene (Table 1). Among these, the variation c.18C > A in SDHB occurred more frequently. These synonymous variations were not likely to be disease-causing mutations because they did not cause changes in amino acid sequence. Next, three HNPGL samples with clinicopathologic significant mutations were chosen for further proteomic analysis. The PGL cell line (PGL-626) was constructed from sample T578, which harbored a missense mutation (c.649C > T) in SDHB.
Table 1.
Genetic information of patients with head and neck paragangliomas (HNPGLs).
| Case | Age(yr) | Gender | Diagnosis | Tumor Volume(mm3) | Multiples | Tumor recurrence | Duration of symptoms(yr) | House-Brackmann(HB) grade | Family History | Blood | SDHD | SDHC | SDHB | SDHA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon | Sequence alteration | Codon | Consequence | Exon | Sequence alteration | Codon | Consequence | Exon | Sequence alteration | Codon | Consequence | Exon | Sequence alteration | Codon | Consequence | |||||||||||
| T400 | 38 | M | JP | 26.8 | 2 | 3 | 7 | V | None | None | None | None | None | None | None | None | None | None | E01 | c.18C>A | p.A6A | Synonymous | - | - | - | - |
| T399 | 43 | M | JP | 33.2 | 1 | 2 | 20 | VI | None | None | None | None | None | None | None | None | None | None | E01 | c.18C>A | p.A6A | Synonymous | - | - | - | - |
| T405 | 55 | F | JP | 29.9 | 1 | 0 | 1 | I | None | None | None | None | None | None | None | None | None | None | E01 | c.18C>A | p.A6A | Synonymous | None | None | None | None |
| T408 | 60 | F | JP | 41.2 | 1 | 2 | 3 | VI | None | + | E03 | c.178_182delAAGGC | K60Cfs*7 | Framshift | - | - | - | - | E01 | c.18C>A | p.A6A | Synonymous | - | - | - | - |
| T448 | 37 | F | JP | 38 | 1 | 3 | 6 | VI | None | None | None | None | None | None | None | None | None | None | E01 | c.18C>A | p.A6A | Synonymous | None | None | None | None |
| T679 | 40 | M | JP | 44.2 | 2 | 0 | 5 | VI | None | None | None | None | None | None | - | - | - | - | E01 | c.18C>A | p.A6A | Synonymous | E15 | c.1969G>A | p.V657I | Missense |
| T579 | 46 | F | JP | 37.8 | 3 | 0 | 6 | I | None | None | None | None | None | None | - | - | - | - | None | None | None | None | E15 | c.1911C>T | p.V637V | Synonymous |
| T553 | 55 | M | JP | 35.6 | 1 | 0 | 1 | I | None | None | None | None | None | None | - | - | - | - | E01 | c.18C>A | p.A6A | Synonymous | None | None | None | None |
| T559 | 30 | F | JP | 46.5 | 3 | 0 | 10 | IV | None | + | None | None | None | None | None | None | None | None | E06 | c.541-3C>T | None | splice acceptor site | E07 | c.891T>C | p.P297P | Synonymous |
| T456 | 28 | M | JP | 43.2 | 4 | 0 | 2 | I | None | + | None | None | None | None | None | None | None | None | E02 | c.136C>T | p.R46X | Nonsense | None | None | None | None |
| T478 | 29 | F | JP | 38.1 | 2 | 0 | 6 | VI | None | None | None | None | None | None | None | None | None | None | E01 | c.18C>A | p.A6A | Synonymous | None | None | None | None |
| T578# | 46 | M | JP | 30.2 | 3 | 0 | 7 | VI | None | + | None | None | None | None | None | None | None | None | E07 | c.649C>T | p.R217C | Missense | None | None | None | None |
| T557 | 58 | M | CBP | 29.5 | 3 | 0 | 5 | IV | None | None | - | - | - | - | - | - | - | - | None | None | None | None | None | None | None | None |
| T556 | 39 | M | JP | 40.2 | 3 | 0 | 3 | VI | None | + | - | - | - | - | None | None | None | None | E04 | c.423+1G>A | splice donor site | - | - | - | - | |
indicate the tumor used for the establishment of a cell line.
Proteomics analysis of HNPGLs compared with normal nerve controls
Using proteomics analysis with tandem mass tags, 151 proteins were significantly differentially expressed in HNPGL compared with normal nerves (Supplementary Tables S1 and S2). Among these proteins, 58 and 93 proteins were up- and down-regulated, respectively (Fig. 1A). The top 10 proteins up- or down-regulated in HNPGL are shown in Table 2. Superoxide dismutase 3 (SOD3) was down-regulated in the HNPGL tumor tissue. SOD3 was also down-regulated in a number of malignancies, including breast, lung, pancreas, and colorectal carcinomas, and was related to regulation of HIF-1 expression [[25], [26], [27]]. NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 2 (NDUFA2) showed the most significantly increased expression. NDUFA2, a complex I accessory subunit located in the distal matrix arm, was discovered in patients with neurodegenerative diseases and is also deregulated in some tumors [28]. Isolated complex I deficiency is one of the most common defects of the oxidative phosphorylation system [29]. NDUFA4 encodes a protein that is a subunit of complex IV of the mitochondrial respiratory chain. This protein was significantly upregulated in HNPGLs compared to normal controls. Li et al. [30] demonstrated that up-regulation of NDUFA4 greatly promoted the proliferation, and decreased the apoptosis, of gastric cancer cells through activation of the oxidative phosphorylation pathway. NDUFA10 also belongs to the complex I subunit of the respiratory electron transport chain, and its expression was also increased in the tumors. The major disease-causing mutations in genes coding for subunits of mitochondrial complex II, succinate dehydrogenase (SDHB, SDHC, and SDHD), have been identified in the majority of HNPGLs and lead to dysregulation of oxygen-sensing pathways. For the first time, we demonstrated, using proteomics analysis, that expression of complex I and complex IV subunits of the respiratory electron transport chain was dysregulated in HNPGLs.
Fig. 1.
Proteomic analysis of differentially expressed proteins between HNPGLs and nerve controls (CON). A Hierarchically clustered heat maps of the 151 significantly deregulated proteins in 3 samples from each group. Red represents a high z-score, and blue represents a low z-score; the “row names” HNPGL.1-3 indicate tumors T559, T456 and T578. B–D The GO terms in the biological process (a), cellular component (b) and molecular function (c) categories are depicted. E The bubble chart depicts the top 20 of frequent enrichment pathways corresponding to the deregulated proteins. The color of each dot denotes the P value, and the size of each dot denotes the number of proteins involved in each pathway. con, normal nerve control (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Table 2.
Candidates of differential expressed proteins in HNPGLs compared to nerve controls.
| Gene_symbol | protein_name | FC.PGL–CON |
|---|---|---|
| PMP2 | peripheral myelin protein 2 | 0.05 |
| EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 0.07 |
| MPZ | myelin protein zero | 0.1 |
| FGL2 | fibrinogen-like 2 | 0.11 |
| PRX | periaxin | 0.11 |
| BCAS1 | breast carcinoma amplified sequence 1 | 0.14 |
| SOD3 | superoxide dismutase 3 | 0.14 |
| MBP | myelin basic protein | 0.15 |
| NEFM | neurofilament, medium polypeptide | 0.16 |
| LTF | lactotransferrin | 0.16 |
| NDUFA2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 2 | 5.56 |
| RAB5C | Ras-related protein Rab-5C | 5.27 |
| SLC25A3 | Phosphate carrier protein, mitochondrial | 4.45 |
| FAM21A | family with sequence similarity 21, member A | 4.28 |
| GFAP | glial fibrillary acidic protein | 4.17 |
| NNT | nicotinamide nucleotide transhydrogenase | 3.54 |
| IDH3A | isocitrate dehydrogenase 3 (NAD+) alpha | 3.14 |
| PML | promyelocytic leukemia | 3.09 |
| NDUFA4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 | 2.99 |
| NDUFA10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | 2.70 |
We proceeded with bioinformatic analysis of DEPs in HNPGLs based on proteomics data. We performed gene ontology (GO) enrichment analysis, which provides a common descriptive framework to functionally annotate and classify gene sets. GO categories are organized into three groups: biological process, cellular component, and molecular function. Using a false discovery rate <0.05 threshold, we identified GO terms for biological processes significantly enriched in response to extracellular matrix organization, cell adhesion, response to neuron projection development, neurofilament bundle assembly, and cell development in our study (Fig. 1B). Over-represented cellular components mainly included extracellular exosome, extracellular vesicle, vesicle, membrane-bound organelle, basement membrane, endoplasmic reticulum lumen, cytoplasm, and Golgi lumen (Fig. 1C). For molecular functions, the enriched GO terms were structural molecule activity, protein complex binding, protein binding, superoxide dismutase activity, superoxide dismutase chaperon, and oxidoreductase activity, acting on NAD(P)H (Fig. 1D). We conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEPs in HNPGLs. The results showed that the over-represented DEPs are mainly related to oxidative phosphorylation, thermogenesis, metabolic pathways, pyruvate metabolism, fatty acid metabolism, carbon metabolism, oxytocin signaling pathway, and the citric acid (TCA) cycle signaling pathway (P-value <0.05) (Fig. 1E). Previous studies showed that HNPGL tumorigenesis is the result of mutations in genes encoding the complex II subunit of the mitochondrial respiratory chain, which is involved in the process of oxidative phosphorylation and might also function as a cellular oxygen sensor [31,32]. Our results are basically consistent with those reported in the previous PGL studies.
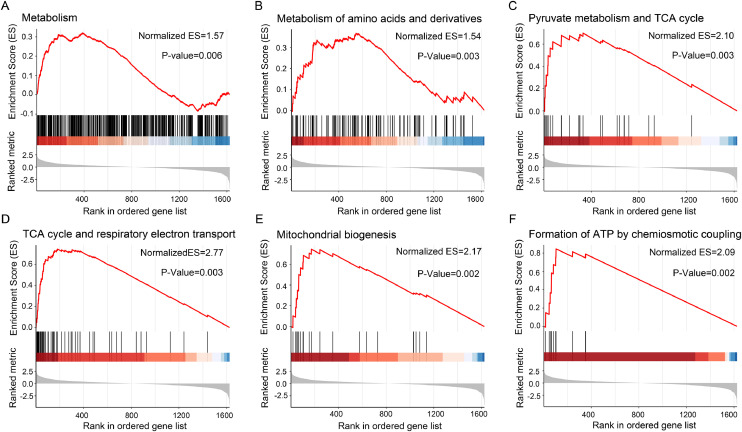
Gene-set enrichment analysis (GSEA) allows detection of modest but coordinated changes in expression of genes involved in a common biological function, rather than concentrating on specific gene changes. Here we used GSEA to identify pathways associated with SDHB loss using expression data derived from patients with HNPGLs compared to normal controls. The results showed that pathways involving metabolism and the mitochondrial respiratory chain were enriched in HNPGLs, as characterized by GSEA using the Reactome database (all with P-values ≤ 0.05) (Fig. 2A–F, Table S3).
Fig. 2.
Gene set enrichment analysis (GSEA) of the proteomic dataset in HNPGLs. GSEA demonstrated that the gene sets involving metabolism (A), metabolism of amino acids and derivatives (B), pyruvate metabolism and TCA cycle (C), TCA cycle and respiratory electron transport (D), mitochondrial biogenesis (E) and formation of ATP by chemiosmotic coupling (F) were enriched in the tumors compared to nerve controls. Normalized enrichment score (ES) and P-value are shown as indicated. The top portion of the plot showed the running ES. The middle portion of the plot showed where the members of the gene set appear in the ranked list of genes. The lower portion of the plot shows the value of the ranking metric as you move down the list of ranked genes. The ranking metric measures a gene's correlation with a phenotype.
A protein interaction network was generated using the STRING database to identify potential binding partners for the host-specific proteins (Fig. 3). There, a PPI ''hot spot'' was shown in NDUFA2, NDUFA4, and NDUFA10 with neighbors (UQCRC1, NNT, ITPR3, SLC25A3, RAB5C, GFAP, IDH3A, UGGT1, and LMAN1) (Fig. 3A). NDUFA2 was identified as the most significantly up-regulated protein in HNPGLs, and NDUFA4 and NDUFA10 were also highly upregulated in the tumors (Table 2). The circle of the protein interaction network on the right in Fig. 3A showed enrichment of DEPs in the oxidative phosphorylation and metabolic pathways. We also showed that the proteins NDUFA2, NDUFA4, and NDUFA10 interacted strongly with pathogenic genes for HNPGLs, including SDHA, SDHAF2, SDHB, SDHC, and SDHD (Fig. 3B).
Fig. 3.
Protein-protein interaction (PPI) network analysis in HNPGLs compared to nerve controls.A PPI network of all the significantly deregulated proteins (left), and proteins involved in metabolism pathways (right). Circular nodes denoted proteins, and their connecting edges denoted protein-protein interactions. The node color was made proportional to the fold-change of the corresponding modulated protein. B PPI network between NDUFA2, NDUFA4, NDUFA10 and SDHx (SDHA, SDHAF2, SDHB, SDHC and SDHD). A protein–protein interaction with combined score > 0.7 was considered to be high-confidence.
Western blotting analysis of SDHB, NDUFA2, NDUFA4, NDUFA10, and HIF-1α
We used western analysis to confirm our proteomic findings of three significant candidates (NDUFA2, NDUFA4, and NDUFA10) along with two proteins of interest, SDHB and HIF-1α (Fig. 4). There was an appreciable amount of SDHB in nerve controls, whereas low levels of this protein were observed in SDHB-mutated HNPGL samples. The samples T622 and T679, with no detectable mutations in the SDHB gene, did not show SDHB deficiency (Fig. 4, Table 1). The expression of HIF-1α in HNPGLs showed almost unchanged compared with normal controls, although previous studies considered that up-regulation of HIF-1α may contribute to tumor cell proliferation and invasion during PGL tumorigenesis. The blots showed significantly increased levels of NDUFA2 by 4.07-fold, NDUFA4 by 13.8-fold and NDUFA10 by 5.67-fold in HNPGLs relative to control nerves, confirming their differential expression patterns determined by proteomics analysis. The three proteins belong to the complex I and complex IV subunits of the respiratory electron transport chain. Similar observations were found in a subset of pheochromocytomas (PCCs) and PGLs in which SDHB loss led to up-regulated activities of complex I components, including NDUFS2, NDUFS3, and NDUFB6 [33]. The hyperactivation of complex I might function as a compensatory mechanism to maintain electron input and mitochondrial membrane potential (MMP). As an inhibitor of complex I, metformin can suppress its activity to baseline level [33].
Fig. 4.
The expression of SDHB, NDUFA2, NDUFA4, NDUFA10 and HIF-1α in HNPGLs compared with nerve controls by Western Blotting. The left panel showed representative blots of the five target proteins for each tumor. The band intensities of proteins in each tumor are normalized to β-actin to obtain a gray scale value. The average gray scale values of the proteins in HNPGL group are expressed relative to those in the nerve controls to obtain relative average gray scale values, which were listed in the right panel. Data are presented as the mean ± SD. Full-length gels are presented in Supplementary Fig. S3.
Establishment of a HNPGL cell line with SDHB mutation
HNPGL (jugular PGL) tissue, which has been characterized as containing a missense mutation (c.649 G > A) in SDHB from a 56-year-old HNPGL patient, was used for in vitro isolation and primary culture (Fig. 5A, upper panel). At the first passage, the cells were immortalized by the transfection with lentivirus construct EF1α-SV40-IRES-puromycin. After 8–10 days of puromycin selection, characterization of the cell line was performed (Fig. 5A, lower panel). The cell line was designated as PGL-626.
Fig. 5.
The establishment of a HNPGL cell line.A Morphology of the primary paraganglioma cells (x50; upper panel) and the immortalized PGL cell line (PGL-626; x100; lower pane). B Immunofluorescence staining of the PGL markers (red), including S100, synaptophysin, chromogranin A and vimentin. Scale bar=25 μm. C The PGL-626 cells were indentified with a SDHB mutation (c.649C > T), consistent with the mutation in corresponding tumor sample. D Western blot analysis of the SDHB protein in PGL-626 cells compared to other cell types, including human schwann cell (HSC), 293T and SHY5Y. Full-length gels are presented in Supplementary Fig. S4.
To assure that the cells purified and expanded from the HNPGL were PGL cells, the primary cells were stained for paraganglia cell markers, including S100, synaptophysin, chromogranin A, and vimentin [23].The paraganglia cell markers showed positive expression after immortalization (Fig. 5B). It was reported that missense mutations in SDHB gene might increase the degradation of its product in tumor cells [34]. Accordingly, Western blot analysis demonstrated the deficiency of SDHB in the cell line with a missense mutation (c.649 G > A) (Fig. 5C). Immunofluorescence analyses showed that NDUFA4 and NDUFA10 were co-expressed in the cytoplasm, as well as in mitochondria of PGL-626 cells. However, NDUFA2 was expressed at a high level in the nuclei of the tumor cells (Fig. 6). The target proteins SDHB, SDHC, and SDHD were also expressed in the cytoplasm of the HNPGL cells. As expected, SDHB exhibited lower fluorescence intensity compared to the other two target proteins (Fig. 6).
Fig. 6.
The expression of NDUFA2, NDUFA4, NDUFA10, SDHB, SDHC and SDHD in PGL626 cell culture by Immunofluorescence. NDUFA2 was highly expressed in nucleoplasm, while NDUFA4 and NDUFA10 showed a stripe linear pattern of distribution in the cytoplasm of PGL-626 cells. SDHB, SDHC and SDHD were expressed in the cytoplasm of PGL626 cells. Scale bar =25 μm.
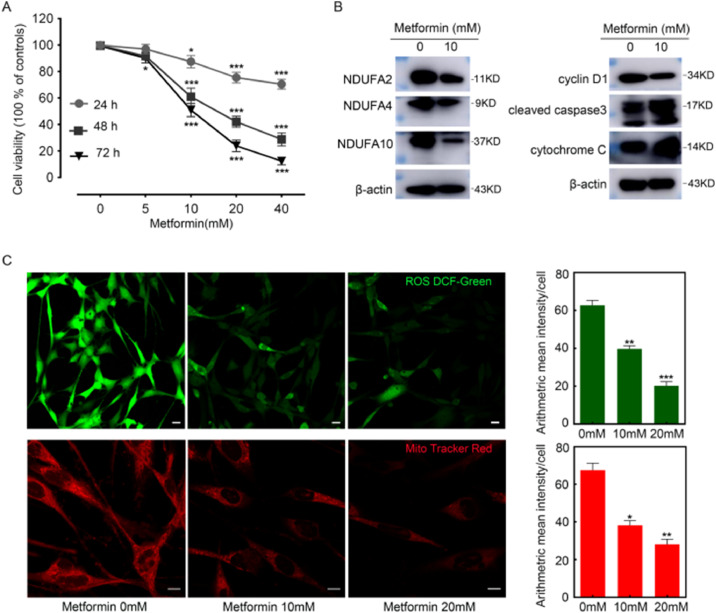
Metformin inhibited the proliferation of cultured PGL-626 cells
In the PGL-626 cell line, metformin inhibited cell proliferation in a time- and dose-dependent manner by MTT assays (Fig. 7A). The results were in agreement with the observation of Florio et al. that metformin could inhibit PGL cell proliferation [35]. Previous studies have confirmed that metformin inhibits mitochondrial function by inducing ROS degradation [36]. Therefore, we investigated the oxidative status of metformin-treated PGL-626 cells. Intracellular ROS levels were monitored with the fluorescent probe 2ʹ,7ʹ-dichlorofluorescin diacetate (DCFDA). As shown in Fig. 7C, metformin (10 mM and 20 mM, 48 h) treatment caused significant downregulation of ROS, as demonstrated by the green fluorescence in cells. A statistical analysis further confirmed the antioxidant effects of metformin. Similarly, as shown in Fig. 7C (lower panel), metformin treatment (10 mM and 20 mM, 48 h) induced significantly reduced MMP. Interestingly, metformin treatment rendered the down-regulation of the key proteins NDUFA2, NDUFA4, and NDUFA10 (Fig. 7B). Also, western blot analysis suggested the activation of apoptosis-related proteins cleaved-caspase 3 and cytochrome c in PGL-626 cells upon metformin treatment (Fig. 7B).
Fig. 7.
Inhibitory effects of metformin on the proliferation of PGL cells in vitro. A Viabilities of PGL-626 treated with different doses of metformin for 24, 48 and 72 h. B Western blot analysis of NDUFA2, NDUFA4, NDUFA10, cyclin D1, cleaved caspase 3 and cytochrome C in response to metformin at doses of 10 mM for 48 h. C Intracellular ROS stained by DCFDA (green) and MMP evidenced by mito-tracker red (red) in PGL-626 were significantly suppressed by metformin (10 and 20 mM) for 24 h. Representative confocal images and histograms of fluorescence intensity were shown in the left and right panel. respectively. *P < 0.05, **P < 0.01 and ***P = 0.001 compared with controls. Data were presented as mean ± SE, n = 3. Scale bar =50 μm. Full-length gels are presented in Supplementary Fig. S5 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Effects of metformin on metabolite profiles in HNPGL cells
To further investigate the effects of metformin on the metabolism of PGL-626 cells, we performed an untargeted metabolomic analysis of the tumor cells before and after metformin treatment. We characterized the metabolic features of HNPGL cells using a non-targeted metabolic profiling strategy based on liquid chromatography-mass spectrometry (LC-MS). The results of principal component analysis (PCA) show that there was a significant difference in cell metabolism between the metformin-treated group and blank control group at several different time points, especially 48 h (Fig. 8A). In order to identify the critical endogenous metabolites, we analyzed the profile data of metabolic footprints by PCA and orthogonal projections to latent structures discriminant analysis (OPLS-DA) to obtain the variable importance value (VIP), which can directly reflect the contribution rate to metabolic profile changes. The higher the VIP value and the greater distance from the center represent a greater contribution to the difference between groups, and can identify which biomarkers are the focus (Table S4). To determine endogenous metabolites, we performed a structural analysis of potential biomarkers based on the information in the VIP diagram (VIP>1, P-value <0.05). Metabolites (n = 653) were filtered and identified from the metabolomic data analysis. Among these, 189 metabolites, including 49 negative metabolites, were significantly different in metformin-treated cells compared to blank controls (Table S5). Thirty-one metabolites were upregulated and 158 were downregulated in the metformin-treated cells (Fig. 8B, Table S5).
Fig. 8.
Untargeted metabolomics analyses of PGL-626 cell culture treated with metformin.A Cluster analysis of metformin-treated group and non-treated control based on the deregulated metabolites using PCA analysis. The first two principal components (PC1 and PC2) from the PCA are plotted for each sample. The percentage variation in the plotted principal components was marked on the axes. Each spot represents one sample, and each group of cells was indicated by a different color. B Heatmap analysis of the deregulated metabolite patterns in the treatment group relative to the control. The heatmap showed the 189 significantly changed metabolites between the two groups. Among these metabolites, 31 and 158 proteins were found to be up- and down-regulated, respectively. Each column represented one sample, and each row represented one metabolite. The color bar showing green to red indicated the Z-score of metabolites, red represented a high z-score, and green represents a low z-score. C KEGG pathway enrichment analysis based on the deregulated metabolites. The results showed that the top 20 frequently enriched pathways were associated with protein digestion and absorption. The number of metabolites in each pathway was showed in the top of each histogram (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
As a clinical study showed [37], significant differences in metabolites were observed before and after surgery in PCC/PGL, including increased plasma concentrations of some amino acids/biogenic amines (creatinine, histidine, ornithine, sarcosine, tyrosine), and decreased levels of some glycerophospholipids (PCaaC38:1, PCaaC 42:0, PCaeC40:2, PCaeC42:5, PCaeC44:5, PCaeC44:6), sphingomyelins (SMC24:1, SMC 26:1), and hexose in plasma. The amino acids/biogenic amines (creatinine, histidine, tyrosine) were significantly decreased in PGL-626 cells treated with 10 mM metformin (Fig. 8B). An increase in urine concentration of 5-hydroxyindoleacetic acid has been reported to be associated with poor prognosis for PCC/PGL patients [38]. Interestingly, 5-hydroxyindoleacetic acid decreased most significantly among metabolites in metformin-treated cells relative to non-treated controls. Metabolic pathways involving catecholamine and serotonin synthesis play a key role in the tumorigenesis of HNPGLs [39,40]. Here, the levels of intermediate metabolites of the serotonin synthesis pathway, such as alpha, beta-didehydrotryptophan, tryptophanol, and 5-hydroxy-L-tryptophan were decreased by metformin treatment (Fig. 8B).
In the GO and KEGG analyses we identified significant expression of metabolites and enzymes (Table S5) mainly enriched in central carbon metabolism in cancer, sphingolipid signaling pathway, biosynthesis of amino acids, protein digestion and absorption, carbon metabolism, and other metabolic pathways (Fig. 8C). Among these pathways, the term “central carbon metabolism in cancer” has been reported to be strongly associated with metformin inhibition of tumors [41]. In addition, glutamine metabolism might be involved in the pathogenesis of SDHx-related PCCs/PGLs [38]. Here, we found that D-glutamine and D-glutamate metabolic-pathway-related metabolites were enriched with metformin treatment. About 16 metabolites were involved in the protein digestion and absorption pathway including L-glutamate, L-tyrosine, L-tryptophan, L-serine, L-valine, and L-glutamine. Eleven metabolites were involved in central carbon metabolism in cancer pathways, including L-glutamate, L-tyrosine, L-aspartic acid, L-isoleucine, L-glutamine, L-histidine, and L-tryptophan, suggesting that deregulation of these pathways might play a role in the inhibitory effects of metformin on PGL-626 cells.
Discussion
HNPGLs are rare neoplasms that with difficult treatment paradigms in neurotology, and little is known about the molecular pathways that drive HNPGLs tumorigenesis. Germline mutations in SDHx are the cause of nearly all familial HNPGLs [3]. The frequency of SDHB mutations in tumors was reported to be 87.1% in Dutch patients, with hotspot mutations, including deletion of exon 3 (33%) and the c.423+1G > A mutation (20%) [42]. In our series, the frequency of SDHB mutation identified in HNPGLs was estimated to be 28.6% (4/14). With the advent of mass spectrometers that are able to analyze complex protein mixtures within a reasonable timeframe, the systematic analysis of all proteins in a tumor becomes feasible. It is very likely that such studies will provide sights into how biological systems operate leading to HNPGL pathologies. In current study, the proteomics analysis of DEPs between the SDH-mutated tumors and normal tissue samples were carried out, and further subjected to a series of bioinformatic analysis. We also attempt to investigate the effects of the mitochondrial complex I inhibitor metformin on the proliferation, ROS, MMP and metabolic status of a HNPGL cell line with a SDHB mutation. A workflow was shown in the supplementary Fig.S1.
The possible molecular mechanisms underlying the tumorigenesis of PCCs/PGLs has been investigated by several studies [23,43]. Fishbein et al. [43] reported that kinase and hypoxia signaling pathways, Krebs cycle/electron transport, and Wnt signaling were deregulated in PCCs/PGLs, based on careful manual review of somatic and germline alterations in DNA and RNA. Cama et al. [23] demonstrated that the NOTCH signaling pathway was the most significantly deregulated in HNPGLs through gene-centric copy number variant (CNV) analysis of 24 patients. In our study, we first applied proteomics analysis of HNPGLs with SDHB mutations compared to nerve controls. GO and pathway enrichment analysis of proteomic data showed that the oxidative phosphorylation, citrate cycle, carbon metabolism, and metabolic pathways were deregulated in this tumor. GSEA analysis also indicated activation of pathways related to metabolism, citric acid cycle, and respiratory electron transport. Our study further confirmed previous reports on PCCs/PGLs, which exhibited distinctive changes in the TCA cycle, hypoxia signaling pathway, mitochondrial electron transport chain, and abnormalities in metabolic pathways [33,44].
The oxidative phosphorylation (OXPHOS) joints electron transport through respiratory chain complexes including complex I, II, III, and IV to ATP synthesis. The TCA cycle is composed by a set of enzymes primarily linking the product of the oxidation of pyruvate to acetyl-CoA with the generation of NADH for the oxidation by the mitochondrial respiratory chain. SDH, also known as Mitochondrial complex II, is one such protein involved in both TCA and OXPHOS [44,45]. Under normal conditions, succinate is converted to fumarate bySDH in TCA cycle. Mutations in the SDHx genes abolish SDH's ability to convert succinate to fumarate, contributing to succinate accumulation in tumor cells. Accumulation of succinate results in an over-reduced ubiquinone pool, leading to a reverse electron transfer to complex I, where electrons escape as ROS [46]. Excessive ROS levels have been shown to stabilize HIF and induce the pseudohypoxia pathway in SDH-mutated PGLs/PCCs [47]. In addition, SDH mutations can lead to reprogramming of cancer-related cell metabolism such as enhanced glycolysis (Warburg effect), and the main driver of the Warburg effect is HIF which induces expression of GLUT1/3, hexokinase 2, PKM2 and LDH-A [44,45]. A diagram showing different aspects of oxidative phosphorylation and metabolism pathways in HNPGLs was presented as Supplementary Fig.S2. Consistent with these views, our bioinformatics analysis of proteomics data revealed high levels of enrichment of OXPHOS and metabolism pathways in HNPGLs. Our findings may also highlight the potential role of OXPHOS and metabolism pathways as therapeutic targets in clinical settings. However, it appeared that the expression levels of HIF-1α were unchanged in HNPGLs, except in one case where it was overexpressed (#T559), compared with normal controls. A possible explanation may be that the tissue vascularization during tumorigenesis leads to cellular adaptation to hypoxic microenvironment.
It has been showed that the activity of SDH or respiratory chain enzyme complex II is low in SDH-related tumors and associated with increased activities of respiratory chain complexes I, III, and IV; all these factors suggest a compensatory response to the lack of SDH activity [48]. In agreement with their results, we demonstrated that that expression of complex I (NDUFA2 and NDUFA10) and complex IV (NDUFA4) subunits of the respiratory electron transport chain was dysregulated in HNPGLs. In addition, proteomics analysis showed a significantly increased level of UQCRC, a complex III subunit, by 2.97-fold (P-value =0.033) in HNPGLs relative to control nerves (Table S2).
Metformin acts directly on mitochondrial complex I, suppressing ROS production and restricting the cell's ability to cope with energetic stress [36]. In this study, metformin was shown to inhibit the proliferation of PGL cells in a time- and dose-dependent manner, which is in line with previously reported results [35]. Inhibition of electron transport at complex I by metformin should reduce the MMP, as proton pumping is linked to electron transport [36]. In our study, PGL cells’ intracellular ROS and MMP were significantly decreased after metformin treatment, and this may correlate with the inhibition of ADP production, as indicated in the data obtained from metabolomic experiments (Table S5). Interestingly, western blot showed significant down-regulation of the candidate proteins NDUFA2, NDUFA4, and NDUFA10 in metformin-treated cells compared with non-treated controls, suggesting that the drug can reverse the deregulated signaling associated with the tumorigenesis of HNPGLs. Metformin targets central carbon metabolism and mitochondrial metabolism pathways, thus playing a key role in controlling tumor growth [41]. Notably, our results showed that the activity of the central carbon metabolism pathway was altered in HNPGL cells after metformin treatment.
However, the potential of metformin in cancer therapeutics is particularly associated its functions in multiple signaling pathways such as p38 MAPK, AMPK, mTOR, IGF, HER-2, and NF-kappa B pathways [49]. Metformin has also been identified as a key regulator in autophagy, cytoskeleton remodeling and even epigenetic changes [49]. Future experiments should strive to confirm if these pathways are affected by metformin in treatment of HNPGLs. Also, there are several limitations in our study. The study involved only 14 HNPGL patients because of the low prevalence of the tumor, so more cases need to be analyzed. LC- tandem MS-based methodology is not sensitive to small and highly charged peptides, hydrophobic membrane proteins, and proteins expressed at a low level, suggesting that we may have missed some valuable information.
Conclusions
The deregulation of the oxidative phosphorylation-respiratory electron transport and metabolic pathways, which is characterized by the significant up-regulation of mitochondrial complex I subunits NDUFA2 and NDUFA10 and complex IV subunit NDUFA4, is involved in the development of HNPGLs, Metformin exerted inhibitory effects on PGL cells, possible through reduced ROS production, decreased MMP, and altered cellular metabolism. One of the interesting finding was that the drug could also reverse the deregulation of the key proteins NDUFA2, NDUFA4, and NDUFA10.
CRediT authorship contribution statement
Zhigang Wang: Investigation, Methodology, Project administration. Hongsai Chen: Investigation, Funding acquisition, Methodology, Formal analysis. Lu Xue: Data curation, Formal analysis, Investigation. Weiwei He: Methodology, Resources. Wenying Shu: Data curation, Formal analysis. Hao Wu: Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Zhaoyan Wang: Funding acquisition, Investigation, Methodology, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81800898 to Hongsai Chen, Grant No. 81901959 to Mengling Chang, Grant No.81670919 and 81870713 to Zhaoyan Wang, and Grant No. 81970872 to Hao Wu), Shanghai Sailing Program (19YF1430500 to Mengling Chang), and Shanghai Key Laboratory of Translational Medicine on Ear and Nose Diseases (14DZ2260300).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101146.
Contributor Information
Hao Wu, Email: wuhao622@sina.cn.
Zhaoyan Wang, Email: wzyent@126.com.
Appendix. Supplementary materials
References
- 1.Lotti L.V., Vespa S., Pantalone M.R., Perconti S., Esposito D.L., Visone R., V eronese A., Paties C.T., Sanna M., Verginelli F., Nauclér C.S., Mariani-Costantini R. A developmental perspective on Paragangliar Tumori- genesis. Cancers. 2019;11:1–21. doi: 10.3390/cancers11030273. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H., Zhu W., Li X., Xue L., Wang Z., Wu H. Genetic and epigenetic patterns in patients with the head- and-neck paragangliomas associate with differential clinical characteristics. J. Cancer Res. Clin. Oncol. 2017;143:953–960. doi: 10.1007/s00432-017-2355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amar L., Bertherat J., Baudin E., Ajzenberg C., Bressac-de Paillerets B., Chabre O., Chamontin B., Dele mer B., Giraud S., Murat A., Niccoli-Sire P., Richard S., Rohmer V., Sadoul J.L., Strompf L., Schlumberger M., Bertagna X., Plouin P.F., Jeunemaitre X., Gimenez-Roqueplo AP. Genetic testing in pheochromocytoma or functional paraganglioma. J. Clin. Oncol. 2005;23:8812–8818. doi: 10.1200/JCO.2005.03.1484. [DOI] [PubMed] [Google Scholar]
- 4.Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., Richard C.W., Cornelisse C.J., Devilee P., Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 5.Zhu W.D., Wang Z.Y., Chai Y.C., Wang X.W., Chen D.Y., Wu H. Germline mutations and genotype-phenotype associations in head and neck paraganglioma patients with negative family history in China. Eur. J. Med. Genet. 2015;58:433–438. doi: 10.1016/j.ejmg.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Baysal BE. Clinical and molecular progress in hereditary paraganglioma. J. Med. Genet. 2008;45:689–694. doi: 10.1136/jmg.2008.058560. [DOI] [PubMed] [Google Scholar]
- 7.Heesterman B.L., Bayley J.P., Tops C.M., Hes F.J., van Brussel B.T., Corssmit E.P., Hamming J.F., van der Mey A.G., Jansen JC. High prevalence of occult paragangliomas in asymptomatic carriers of SDHD and SDHB gene mutations. Eur. J. Hum. Genet. 2013;21:469–470. doi: 10.1038/ejhg.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller U. Pathological mechanisms and parent-of-origin effects in hereditary paraganglioma/pheochro mocytoma (PGL/PCC) Neurogenetics. 2011;12:175–181. doi: 10.1007/s10048-011-0280-y. [DOI] [PubMed] [Google Scholar]
- 9.Hensen E.F., Jordanova E.S., van Minderhout I.J., Hogendoorn P.C., Taschner P.E., van der Mey A.G., Devilee P., Cornelisse CJ. Somatic loss of maternal chromosome 11 causes parent-of-origin-dependent inheritance in SDHD-linked paraganglioma and phaeochromocytoma families. Oncogene. 2004;23:4076–4083. doi: 10.1038/sj.onc.1207591. [DOI] [PubMed] [Google Scholar]
- 10.Pigny P., Vincent A., Cardot Bauters C., Bertrand M., de Montpreville V.T., Crepin M., Porchet N., Caron P. Paraganglioma after maternal transmission of a succinate dehydrogenase gene mutation. J. Clin. Endocrinol. Metab. 2008;93:1609–1615. doi: 10.1210/jc.2007-1989. [DOI] [PubMed] [Google Scholar]
- 11.Hartzell L.D., McKelvey K.D., Van Hemert R.L., Dornhoffer J. Cerebellopontine angle tumor in a patient with a maternally inherited SDHD gene mutation. Int. Tinnitus J. 2008;14:97–100. [PubMed] [Google Scholar]
- 12.Yeap P.M., Tobias E.S., Mavraki E., Fletcher A., Bradshaw N., Freel E.M., Cooke A., Murday V.A., Davidson H.R., Perry C.G., Lindsay RS. Molecular analysis of pheochromocytoma after maternal transmission of SDHD mutation elucidates mechanism of parent-of-origin effect. J. Clin. Endocrinol. Metab. 2011;96:E2009–E2013. doi: 10.1210/jc.2011-1244. [DOI] [PubMed] [Google Scholar]
- 13.Richter S., Klink B., Nacke B., de Cubas A.A., Mangelis A., Rapizzi E., Meinhardt M., Skondra C., Mannelli M., Robledo M., Menschikowski M., Eisenhofer G. Epigenetic mutation of the succinate dehydrogenase C promoter in a patient with two paragangliomas. J. Clin. Endocrinol. Metab. 2016;101:359–633. doi: 10.1210/jc.2015-3856. [DOI] [PubMed] [Google Scholar]
- 14.Bernardo-Castiñeira C., Valdés N., Sierra M.I., Sáenz-de-Santa-María I., Bayón G.F., Perez R.F., Fernández A.F., Fraga M.F., Astudillo A., Menéndez R., Fernández B., Del Olmo M., Suarez C., Chiara MD. SDHC promoter methylation, a novel pathogenic mechanism in parasympathetic paragangliomas. J. Clin. Endocrinol. Metab. 2018;103:295–305. doi: 10.1210/jc.2017-01702. [DOI] [PubMed] [Google Scholar]
- 15.Roy D., Tiirikainen M. Diagnostic power of DNA methylation classifiers for early detection of cancer. Trends Cancer. 2020;6:78–81. doi: 10.1016/j.trecan.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letouzé E., Martinelli C., Loriot C., Burnichon N., Abermil N., Ottolenghi C., Janin M., Menara M., Nguyen A.T., Benit P., Buffet A., Marcaillou C., Bertherat J., Amar L., Rustin P., De Reyniès A., Gimenez-Roqueplo A.P., Favier J. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Moosavi B., Berry E.A., Zhu X.L., Yang W.C., Yang GF. The assembly of succinate dehydrogenase: a key enzyme in bioenergetics. Cell Mol. Life Sci. 2019;76:4023–4042. doi: 10.1007/s00018-019-03200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cervera A.M., Apostolova N., Crespo F.L., Mata M., McCreath KJ. Cells silenced for SDHB expression display characteristic features of the tumor phenotype. Cancer Res. 2008;68:4058–4067. doi: 10.1158/0008-5472.CAN-07-5580. [DOI] [PubMed] [Google Scholar]
- 19.Ke Q., Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol. Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 20.Movafagh S., Crook S., Vo K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: new developments in an old debate. J. Cell Biochem. 2015;116:696–703. doi: 10.1002/jcb.25074. [DOI] [PubMed] [Google Scholar]
- 21.Chua Y.L., Dufour E., Dassa E.P., Rustin P., Jacobs H.T., Taylor C.T., Hagen T. Stabilization of hypoxia-induc- ible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J. Biol. Chem. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verginelli F., Perconti S., Vespa S., Schiavi F., Prasad S.C., Lanuti P., Cama A., Tramontana L., Esposito D.L., Guarnieri S., Sheu A., Pantalone M.R., Florio R., Morgano A., Rossi C., Bologna G., Marchisio M., D'Argenio A., Taschin E., Visone R., Opocher G., Veronese A., Paties C.T., Rajasekhar V.K., Söderberg-Nauclér C., Sanna M., Lotti L.V., Mariani-Costantini R. Paragangliomas arise through an autonomous vasculo-angio- neurogenic program inhibited by imatinib. Acta Neuropathol. 2018;135:779–798. doi: 10.1007/s00401-017-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cama A., Verginelli F., Lotti L.V., Napolitano F., Morgano A., D'Orazio A., Vacca M., Perconti S., Pepe F., Romani F., Vitullo F., di Lella F., Visone R., Mannelli M., Neumann H.P., Raiconi G., Paties C., Moschetta A., Tagliaferri R., Veronese A., Sanna M., Mariani-Costantini R. Integrative genetic, epigenetic and pathological analysis of paraganglioma reveals complex dysregulation of NOTCH signaling. Acta Neuropathol. 2013;126:575–594. doi: 10.1007/s00401-013-1165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jara J.A., López-Muñoz R. Metformin and cancer: between the bioenergetic disturbances and the antifo- late activity. Pharmacol. Res. 2015;101:102–108. doi: 10.1016/j.phrs.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Griess B., Klinkebiel D., Kueh A., Desler M., Cowan K., Fitzgerald M., Teoh-Fitzgerald M. Association of SOD3 promoter DNA methylation with its down-regulation in breast carcinomas. Epigenetics. 2020;15:1325–1335. doi: 10.1080/15592294.2020.1777666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mira E., Carmona-Rodríguez L., Pérez-Villamil B., Casas J., Fernández-Aceñero M.J., Martínez-Rey D., Martín-González P., Heras-Murillo I., Paz-Cabezas M., Tardáguila M., Oury T.D., Martín-Puig S., Lacalle R.A., Fabriás G., Díaz-Rubio E., Mañes S. SOD3 improves the tumor response to chemotherapy by stabilizing endothelial HIF-2α. Nat. Commun. 2018;9:575. doi: 10.1038/s41467-018-03079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmona-Rodríguez L., Martínez-Rey D., Fernández-Aceñero M.J., González-Martín A., Paz-Cabezas M., Rodríguez-Rodríguez N., Pérez-Villamil B., Sáez M.E., Díaz-Rubio E., Mira E., Mañes S. SOD3 induces a HIF-2α-dependent program in endothelial cells that provides a selective signal for tumor infiltration by T cells. J. Immunother. Cancer. 2020;8:1–17. doi: 10.1136/jitc-2019-000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L.D., Sun H.F., Bai Y., Gao S.P., Jiang H.L., Jin W. Significant prognostic values of nuclear genes encoding mitochondrial complex I subunits in tumor patients. Neoplasma. 2016;63:548–558. doi: 10.4149/neo_2016_408. [DOI] [PubMed] [Google Scholar]
- 29.Ugalde C., Janssen R.J., van den Heuvel L.P., Smeitink J.A., Nijtmans LG. Differences in assembly or stabi- lity of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 2004;13:659–667. doi: 10.1093/hmg/ddh071. [DOI] [PubMed] [Google Scholar]
- 30.Li L., Li Y., Huang Y., Ouyang Y., Zhu Y., Wang Y., Guo X., Yuan Y., Gong K. Long non-coding RNA MIF- AS1 promotes gastric cancer cell proliferation and reduces apoptosis to upregulate NDUFA4. Cancer Sci. 2018;109:3714–3725. doi: 10.1111/cas.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gimenez-Roqueplo A.P., Favier J., Rustin P., Mourad J.J., Plouin P.F., Corvol P., Rötig A., Jeunemaitre X. The R22X mutation of the SDHD gene in hereditary paraganglioma abolishes the enzymatic activity of complex II in the mitochondrial respiratory chain and activates the hypoxia pathway. Am. J. Hum. Genet. 2001;69:1186–1197. doi: 10.1086/324413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douwes Dekker P.B., Hogendoorn P.C., Kuipers-Dijkshoorn N., Prins F.A., van Duinen S.G., Taschner P.E., van der Mey A.G., Cornelisse CJ. SDHD mutations in head and neck paragangliomas result in destabilization of complex II in the mitochondrial respiratory chain with loss of enzymatic activity and abnormal mitochondrial morphology. J. Pathol. 2003;201:480–486. doi: 10.1002/path.1461. [DOI] [PubMed] [Google Scholar]
- 33.Pang Y., Lu Y., Caisova V., Liu Y., Bullova P., Huynh T.T., Zhou Y., Yu D., Frysak Z., Hartmann I., Taïeb D., Pacak K., Yang C. Targeting NAD+/PARP DNA repair pathway as a novel therapeutic approach to SDHB-mutated cluster I pheochromocytoma and paraganglioma. Clin. Cancer Res. 2018;24:3423–3432. doi: 10.1158/1078-0432.CCR-17-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C., Matro J.C., Huntoon K.M., Ye D.Y., Huynh T.T., Fliedner S.M., Breza J., Zhuang Z., Pacak K. Missense mutations in the human SDHB gene increase protein degradation without altering intrinsic enzymatic function. FASEB J. 2012;26:4506–4516. doi: 10.1096/fj.12-210146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Florio R., De Lellis L., Veschi S., Verginelli F., di Giacomo V., Gallorini M., Perconti S., Sanna M., Mariani- Costantini R., Natale A., Arduini A., Amoroso R., Cataldi A., Cama A. Effects of dichloroacetate as single agent or in combination with GW6471 and metformin in paraganglioma cells. Sci. Rep. 2018;8:13610. doi: 10.1038/s41598-018-31797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheaton W.W., Weinberg S.E., Hamanaka R.B., Soberanes S., Sullivan L.B., Anso E., Glasauer A., Dufour E., Mutlu G.M., Budigner G.S., Chandel NS. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erlic Z., Kurlbaum M., Deutschbein T., Nölting S., Prejbisz A., Timmers H., Richter S., Prehn C., Weismann D., Adamski J., Januszewicz A., Reincke M., Fassnacht M., Robledo M., Eisenhofer G., Beuschlein F., Kroiss M. Metabolic impact of pheochromocytoma/paraganglioma: targeted metabolomics in patients before and after tumor removal. Eur. J. Endocrinol. 2019;181:647–657. doi: 10.1530/EJE-19-0589. [DOI] [PubMed] [Google Scholar]
- 38.Imperiale A., Moussallieh F.M., Roche P., Battini S., Cicek A.E., Sebag F., Brunaud L., Barlier A., Elbayed K., Loundou A., Bachellier P., Goichot B., Stratakis C.A., Pacak K., Namer I.J., Taïeb D. Metabolome profiling by HRMAS NMR spectroscopy of pheochromocytomas and paragangliomas detects SDH deficiency: clinical and pathophysiological implications. Neoplasia. 2015;17:55–65. doi: 10.1016/j.neo.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauss M., Nicholas G.G., Abt A.B., Harrison T.S., Seaton JF. Malignant catecholamine-secreting carotid body paraganglioma. Otolaryngol. Head Neck Surg. 1983;91:315–321. doi: 10.1177/019459988309100323. [DOI] [PubMed] [Google Scholar]
- 40.Osinga T.E., Korpershoek E., de Krijger R.R., Kerstens M.N., Dullaart R.P., Kema I.P., van der Laan B.F., van der Horst-Schrivers A.N., Links TP. Catecholamine-synthesizing enzymes are expressed in parasympathetic head and neck paraganglioma tissue. Neuroendocrinology. 2015;101:289–295. doi: 10.1159/000377703. [DOI] [PubMed] [Google Scholar]
- 41.Liu X., Romero I.L., Litchfield L.M., Lengyel E., Locasale JW. Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab. 2016;24:728–739. doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hes F.J., Weiss M.M., Woortman S.A., de Miranda N.F., van Bunderen P.A., Bonsing B.A., Stokkel M.P., Morre- au H., Romijn J.A., Jansen J.C., Vriends A.H., Bayley J.P., Corssmit EP. Low penetrance of a SDHB mutation in a large Dutch paraganglioma family. BMC Med. Genet. 2010;11:92. doi: 10.1186/1471-2350-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fishbein L., Leshchiner I., Walter V., Danilova L., Robertson A.G., Johnson A.R., Lichtenberg T.M., Murray B.A., Ghayee H.K., Else T., Ling S., Jefferys S.R., de Cubas A.A., Wenz B., Korpershoek E., Amelio A.L., Makowski L., Rathmell W.K., Gimenez-Roqueplo A.P., Giordano T.J., Asa S.L., Tischler A.S., Cancer Genome Atlas Research Network. Pacak K., Nathanson K.L., Wilkerson MD. Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31:181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vicha A., Taieb D., Pacak K. Current views on cell metabolism in SDHx-related pheochromocytoma and paraganglioma. Endocr. Relat. Cancer. 2014;21:R261–R277. doi: 10.1530/ERC-13-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eijkelenkamp K., Osinga T.E., Links T.P., van der Horst-Schrivers A. Clinical implications of the oncometabolite succinate in SDHx-mutation carriers. Clin. Genet. 2020;97:39–53. doi: 10.1111/cge.13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szeto S., Reinke S.N., Sykes B.D., Lemire BD. Ubiquinone-binding site mutations in the Saccharomyces cerevisiae succinate dehydrogenase generate superoxide and lead to the accumulation of succinate. J. Biol. Chem. 2007;282:27518–27526. doi: 10.1074/jbc.M700601200. [DOI] [PubMed] [Google Scholar]
- 47.Selak M.A., Armour S.M., MacKenzie E.D. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Rao J.U., Engelke U.F., Rodenburg R.J. Genotype-specific abnormalities in mitochondrial function associate with distinct profiles of energy metabolism and catecholamine content in pheochromocytoma and paraganglioma. Clin. Cancer Res. 2013;19:3787–3795. doi: 10.1158/1078-0432.CCR-12-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B., Luo J., Yu T., Zhou L., Lv H., Shang P. Anticancer mechanisms of metformin: a review of the current evidence. Life Sci. 2020;254 doi: 10.1016/j.lfs.2020.117717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.