Abstract
The outbreak of COVID-19 caused by SARS-CoV-2 is spreading worldwide, with the pathogenesis mostly unclear. Both virus and host-derived microRNA (miRNA) play essential roles in the pathology of virus infection. This study aims to uncover the mechanism for SARS-CoV-2 pathogenicity from the perspective of miRNA. We scanned the SARS-CoV-2 genome for putative miRNA genes and miRNA targets and conducted in vivo experiments to validate the virus-encoded miRNAs and their regulatory role on the putative targets. One of such virus-encoded miRNAs, MR147-3p, was overexpressed that resulted in significantly decreased transcript levels of all of the predicted targets in human,i.e., EXOC7, RAD9A, and TFE3 in the virus-infected cells. The analysis showed that the immune response and cytoskeleton organization are two of the most notable biological processes regulated by the infection-modulated miRNAs. Additionally, the genomic mutation of SARS-CoV-2 contributed to the changed miRNA repository and targets, suggesting a possible role of miRNAs in the attenuated phenotype of SARS-CoV-2 during its evolution. This study provided a comprehensive view of the miRNA-involved regulatory system during SARS-CoV-2 infection, indicating possible antiviral therapeutics against SARS-CoV-2 through intervening miRNA regulation.
Keywords: COVID-19, SARS-CoV-2, virus-encoded miRNA, host miRNA, virus infection, immune response
Introduction
The current pandemic of COVID-19, caused by a novel virus, SARS-CoV-2, has led to over 24 000 000 infections and 830 000 fatalities in over 100 countries since its emergence in late 2019 (data collected on August 30, 2020). SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA betacoronavirus. Human coronaviruses cause respiratory tract infections that can be mild, such as some cases of the common cold. However, over the past two decades, highly pathogenic human coronaviruses have emerged, including SARS-CoV in 2002 and 2003, which infected 8000 people worldwide and with a fatality rate of approximately 10% and MERS-CoV in 2012 with 2500 confirmed cases and a death rate of 37%[1]. Compared to MERS and SARS, SARS-CoV-2 appears to spread more efficiently, although with relatively lower mortality of 2% to 3%[1–2].
The SARS-CoV-2 gains entry to cells by engaging the angiotensin-converting enzyme 2 (ACE2) as receptor with the spike (S) protein, and then using the host serine protease TMPRSS2 for S priming[3]. Pneumonia is the common symptom of SARS-CoV-2 infection[4]; however, damages to the liver[5] and spleen[6], as well as gastrointestinal symptoms[7] were also reported. The SARS-CoV-2 infection causes increased secretion of cytokines, and the leading death cause of SARS-CoV-2 is ARDS, for which one of the main mechanisms is the cytokine storm, the deadly uncontrolled systemic inflammatory response resulting from the release of large amounts of proinflammatory cytokines and chemokines by immune cells[4]. Nevertheless, the mechanism of virus invasion and release, and the causes of these exuberant inflammatory responses in SARS-CoV-2 infection remain mostly unknown.
MicroRNAs (miRNAs) are widely found in plants, animals, and some viruses and involved in a variety of biological processes. In most cases, miRNAs interact with the 3′ untranslated region (3 ′-UTR) of target mRNAs to induce mRNA degradation and translational repression. However, except for the role as a repressor, increasing evidence has shown that miRNAs could activate gene expression by interacting with other regions, including the 5′-UTR and enhancers[8–9]. As crucial regulators of gene expression, miRNA-targeted therapeutics have been proposed for the treatment of cancers, virus infection, and other diseases. For example, antimiR targeted at has-miR-122 is in trials for treating hepatitis[10].
It has shown that both DNA and RNA viral genomes can encode miRNAs[11–12]. The virus-derived miRNAs can be expressed in host cells and participate in the lifecycle and cellular consequences of infection. To date, over 300 viral encoded miRNAs have been described, and many of them were involved in the host immune response, cell cycle, and vesicle trafficking[13]. Viral miRNAs can target a large number of host genes involved in regulating cell proliferation, apoptosis, and host immunity[11]. The miR-HA-3p encoded by H5N1 accentuates the production of antiviral cytokines, resulting in a high level of cytokine production and high mortality[14]. Also, viral miRNAs target the virus genes, which also help the virus to remain hidden from the host immune response. For example, miR-BART2 encoded by EBV lie antisense to the EBV DNA polymerase gene BALF5 and inhibit DNA polymerase expression by inducing cleavage within its 3′-UTR[15].
Furthermore, viruses have evolved mechanisms to degrade, boost, or hijack cellular miRNAs to benefit the viral life cycle[16]. The non-translated region of North American eastern equine encephalitis virus genome base pairs with the miR-142-3p, resulting in a miR-142-3p mediated innate immune suppression[17]. Moreover, recent studies indicate the virus-encoded miRNAs, host-encoded miRNAs, and miRNA targets together form a novel regulatory system between the virus and the host, which contributes to the outcome of infection[18–19]. Hence, it is crucial to comprehensively understand the role of the regulatory system in SARS-CoV-2 infection. Here, we showed that the virus-encoded miRNAs might participate in the process of virus infection and host immunity, and the attenuated phenotype of mutant SARS-CoV-2 might partly be attributed by the mutation-introduced alteration of the miRNA repository and targets.
Materials and methods
Computational identification of virus-encoded miRNAs
The complete genome sequence of SARS-CoV-2 (accession number: MN908947) was obtained from the National Center for Biotechnology Information (NCBI). The genome sequence of SARS-CoV, MERS-CoV-2, and HCoV-NL63 were obtained from NCBI with accession numbers of NC_004718.3, NC_005831.2, and NC_019843.3, respectively. The VMir software package (v2.2)[20] was used to identify potential pre-miRNAs within the virus genome. The parameters of window and step sizes were set at 500 and 1 nucleotide (nt), respectively. The minimum free energy (MFE) of the predicted hairpin structures was predicted with RNAFold algorithm from the Vienna package[21]. Then HuntMi software was used to further distinguish miRNA hairpins from pseudo hairpins with a virus-based model[22]. Maturebayes web server[23] was used to identify the mature miRNAs within the pre-miRNAs based on sequence and secondary structure information of their miRNA precursors.
Prediction of miRNA targets and functional annotation
Human 3′- and 5′-UTR, and the enhancer sequence for five tissues were downloaded from the UTRdb database[24] and EnhancerAtlas database[25], respectively. Human miRNA sequence was downloaded from the TargetScan database. The targets of virus-encoded miRNAs and human miRNAs were predicted with TargetFinder[26] and miRanda[27]. The miRNA targets on human genes were extracted from the TargetScanHuman 7.2 database[28], with context++ score <–0.4 and percentile >0.99 as previously applied [29]. Then UTR and enhancer associated genes were extracted from annotations in UTRdb database[24] and EnhancerAtlas database[25]. Genes were functionally annotated using Gene Ontology (GO) terms using R packages, and the function enrichment tests were conducted with GOstats[30].
Virus variation processing
The genomic variations were downloaded from China National Center for Bioinformation; data were collected until Mar 19, 2020. The mutations were obtained based on sequence alignment against the reference genome MN908947.3, as we used for miRNA scanning.
SARS-CoV-2 infection of Vero E6 cell line
The African green monkey kidney Vero E6 cell line was purchased from the Cell Resources Center of Shanghai Institute of Life Science, Chinese Academy of Sciences (China) and cultured in DMEM medium (Cat. No. 12430-054, Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco) at 37 °C with 5% CO2 atmosphere.
BetaCoV/JS03/human/2020 (EPI_ISL_411953), a SARS-CoV-2 virus strain, was isolated from nasopharyngeal swab of a 40-year-old female confirmed with COVID-19 by reverse transcriptase PCR (RT-PCR) in December 2019. The virus was propagated in Vero E6 cells, and the viral titer was determined by the 50% tissue culture infective dose based on microscopic observation of cytopathic effects. All the in vitro SARS-CoV-2 infection experiments were performed in a bio-safety level-3 laboratory of Jiangsu Provincial Center for Diseases Control and Prevention, Jiangsu, China.
Vero E6 cells were seeded into 12-well plates with a density of 4×104 cells/well for incubation in DMEM medium supplemented with 10% FBS for 16 hours in an incubator with 5% CO2 at 37 °C for cells to reach 80% confluent. Six wells were infected with the SARS-CoV-2 virus with multiplicity of infection of 0.05 at 37 °C with a 5% CO2 atmosphere for 48 hours. Then the infected cells of each well were harvested for RNA extraction and quantitation. RNA was extracted from cells using the RNeasy Plus Mini Kit (Cat. No. 74134, Qiagen, Germany) according to the manufacturer's instructions.
Cell culture and transfection
Human pulmonary epithelial BEAS-b2 cells were maintained in DMEM (Cat. No. 8120360, Gibco) supplemented with 10% FBS (Cat. No. FSP500, ExCell Bio, China), penicillin-streptomycin, and L-glutamine at 37 °C. The transfections were carried out according to standard procedures using PolyJet Transfection Reagent (Cat. No. SL100688, Signagen, China).
Real-time RT-PCR analysis
Total RNA was extracted from cells using TRIzol reagent (Life Technology, USA). RNA integrity was verified by gel electrophoresis. RNA was reverse transcribed using the HiScript II Q Select RT SuperMix for qPCR (+gDNA wiper) (Cat. No. R233-01, Vazyme, China) according to the manufacturer's directions. The reverse transcribed cDNA products were subjected to quantitative real-time PCR using the Hieff qPCR SYBR Green Master Mix (Low Rox Plus) (Cat. No. 11202ES08, Yeasen, China) according to the manufacturer's directions. Reverse transcription and RT-qPCR for miRNA, as well as internal reference U6 were performed using miRcute Plus miRNA First-Strand cDNA Kit (Cat. No. KR211, TIANGEN, China) and miRcute Plus miRNA qPCR Kit (Cat. No. FP411, TIANGEN) according the manufacturer's instructions. Briefly, The RNA mixture was incubated for 60 minutes at 42 °C and 5 minutes at 95 °C for first-strand cDNA synthesis. For qPCR, after an initial denaturation step at 95 °C for 15 minutes, the amplifications were carried out with 40 cycles at a melting temperature of 94 °C for 15 seconds, and an annealing and extending temperature of 60 °C for 34 seconds. The primers used for virus-encoded miRNAs and host genes were listed in Supplementary Table 1 , available online. The hits of miRNA primers at the green monkey genome were provided in the Supplementary Note, available online. The Ct values of test and control samples were compared for three independent repeats, relative expression of test was calculated with 2−ΔΔCt method.
Virus MR147-3p construct and transfection
According to the virus MR147-3p sequence shown in Supplementary Table 1 , oligonucleotides encoding MR147-3p precursor were subcloned into the EcoRI and AgeI restriction sites of pPG/miR/EGFP (GenePharma, China), and verified by DNA sequencing. BEAS-2B cells were transfected with control plasmid pPG/miR/EGFP or pPG-MR147-3p-EGFP. The transfection samples were collected 24 hours later. Each transfection included three repeats.
Statistical analysis
Data are presented as the mean±SD. P-values less than 0.05 indicated statistical significance. Differences between two samples were analyzed by Student's t test or Wilcoxon signed-rank test.
Results
Identification of SARS-CoV-2 encoded miRNAs
A total of 808 potential pre-miRNAs in the SARS-CoV-2 genome were detected with VMir Analyzer[20], an ab initio prediction program explicitly designed to identify pre-miRNAs in viral genomes. Then the candidate pre-miRNA sequences were further filtered using HuntMi[22] with the virus model, and the sequences with MFE larger than −20 kcal/mol were removed. A total of 45 candidate virus pre-miRNAs were finally obtained (Fig. 1A, Supplementary Table 2 , available online), with 30 in the direct orientation and 15 in the reverse orientation (Fig. 1B). The average length of the candidate pre-miRNA sequence was 78 nt with an average MFE of −28.1 kcal/mol. Finally, the sequence and position of mature miRNA within the candidate pre-miRNA were identified with MatureBayes, resulting in 90 putative mature miRNAs.
Figure 1.
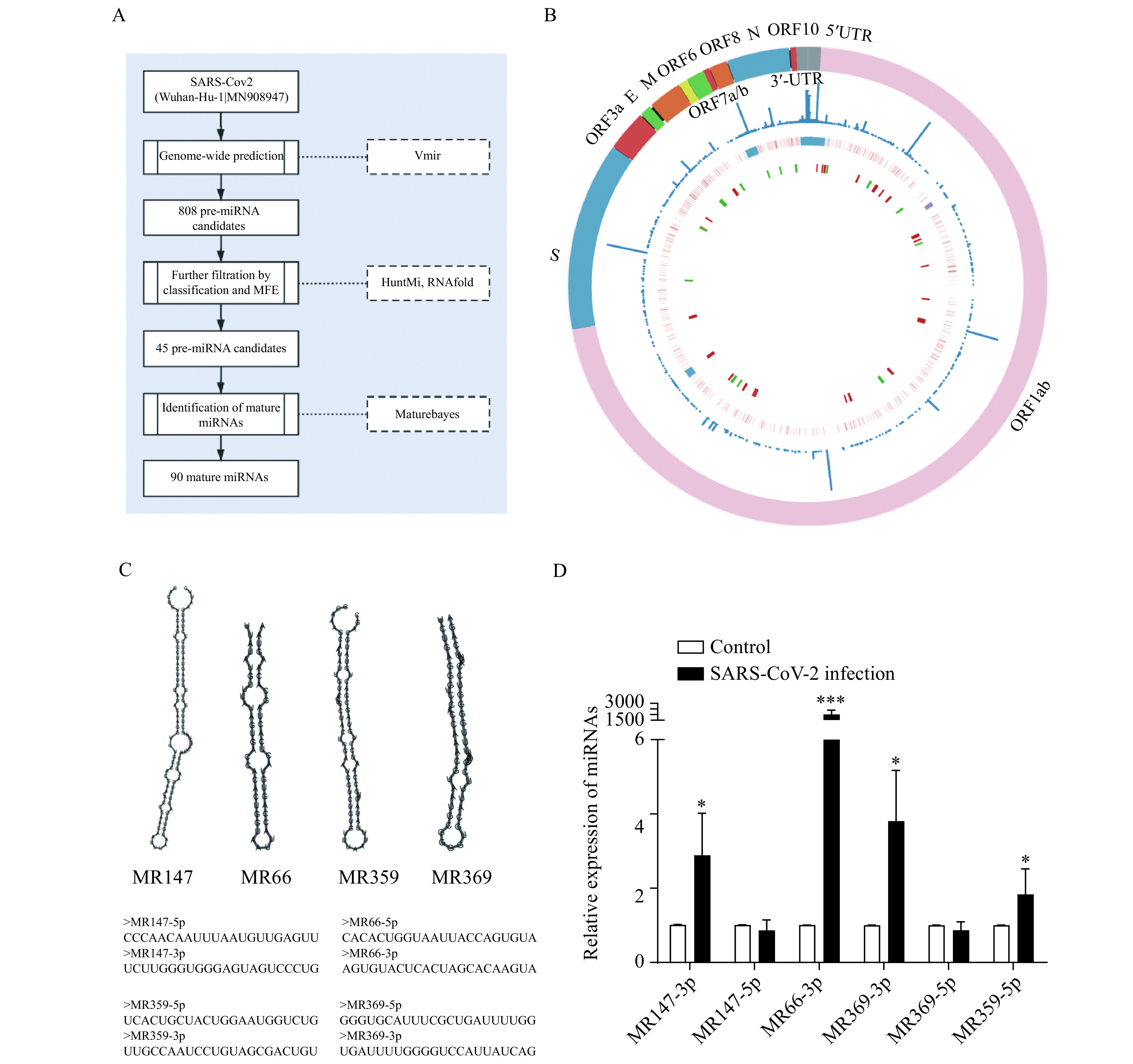
Overview of the putative SARS-CoV-2 encoded miRNAs.
A: The schematic workflow for virus miRNA identification. B: The innermost cycle demonstrated the distribution of the candidate pre-miRNAs across the SARS-CoV-2 genome with red and green representing pre-miRNAs in the direct direction and reserve direction, respectively. The middle layer showed the mutation in the genome with red denoting SNPs, and blue denoting deletions. Outside of it is the frequency of the variations. C: The hairpin structure of the example pre-miRNAs putatively encoded by SARS-CoV-2. D: RT-qPCR analysis of the candidate SARS-CoV-2-encoded miRNAs expression levels in Vero E6 cells infected with SARS-CoV-2 (n=3, data are presented as mean±SD). Two-sided Student's t-test, *P<0.05,***P<0.001. MFE: minimum free energy.
To validate the existence of the candidate virus-encoded miRNAs, we performed qPCR in the SARS-CoV-2-infected Vero E6 cells for six miRNAs, including MR147-3p, MR147-5p, MR369-3p, MR369-5p, MR66-3p, and MR359-5p (Fig. 1C). Four of them were absent in the controls but significantly increased in the infected cells (Fig. 1D).
Virus miRNAs regulate host gene expression
Forty candidate miRNAs were predicted to bind at the 3′-UTR of 73 genes (Supplementary Table 3 , available online). Gene ontology analysis revealed that the target genes were enriched in the apoptosis-related functions, such as Notch binding, single-stranded DNA endodeoxyribonuclease activity, and deoxyribonuclease activity (Fig. 2A). Apoptosis is a powerful mechanism to curtail viral spread[31]. However, viruses have also evolved sophisticated molecular strategies to subvert host cell apoptotic defenses. A couple of studies have investigated the mechanism by which viruses modulate apoptosis signalling[13,32]. CHAC1 and RAD9A were targeted by the virus-encoded miRNAs, MD2-5p and MR147-3p, respectively (Fig. 2B). CHAC1 is a proapoptotic enzyme and a regulator of Notch signalling. RAD9A is a cell cycle checkpoint protein and regulates cell death and promotes apoptosis[33]. FOXO3, which was predicted to be targeted by MR359-5p, is required for antioxidant responses and autophagy, and altered expression of FOXO3 is observed in hepatitis C infection and fatty liver[34]. The suppressive role of the candidate SARS-CoV-2-encode miRNAs on these genes suggested their possible role in reducing the host cell apoptosis to subvert host defense.
Figure 2.
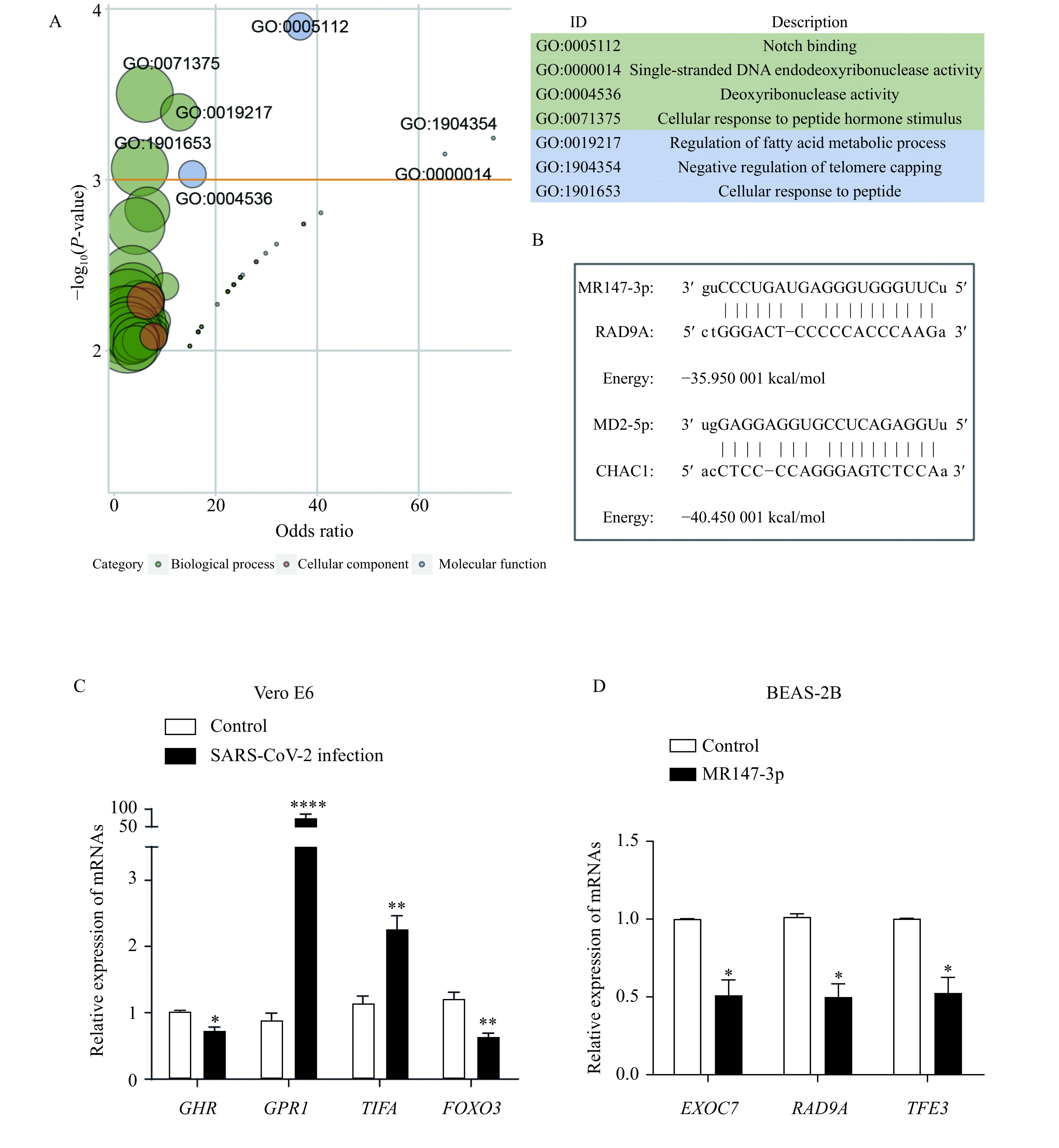
SARS-CoV-2 encoded candidate miRNAs targeting at the 3′-UTR of genes.
A: The GO functions enriched by the putative virus-miRNAs regulated genes. GO IDs with P-value <0.001 were labeled, and the size of bubbles is proportion to the number of genes annotated to the corresponding GO terms. B: Diagram examples of the binding of the putative virus-encoded miRNAs with 3′-UTR. C: The expression of the target genes of putative virus-encoded miRNAs in Vero E6 cells infected with SARS-CoV-2 ( n=3, data are presented as mean±SD). D: The expression of genes targeted by MR147-3p in BEAS-2B cell transfected with MR147-3p (n=3, data are presented as mean±SD). *P<0.05,**P<0.01, and****P<0.0001. GO: Gene Ontology.
Eleven candidate virus miRNAs were predicted to bind at the 5′-UTR of 13 target genes (Supplementary Table 3 ). For example, MR385-3p was predicted to bind at the 5′-UTR of TGFBR3, a gene widely expressed on cells of the innate and adaptive immune system and promote Th1 differentiation and regulation of regulatory T-cell activation and survival[35]. MD2-5p was targeted at ENOX1, which encodes a protein that presents in the cytoskeletal compartment of endothelial cells and suppression of it in human or mouse endothelial cells inhibits migration and the ability to form tubule-like structures in matrigel[36].
miRNAs can also target the enhancer regions to activate their expression[9]. Targets of the candidate virus-encoded miRNAs on the enhancer sequence of five tissues (lung, gut, spleen, liver, and heart) were searched. Target prediction revealed that the lung had got the most significant number of genes targeted by the candidate miRNAs on the enhancer, followed by spleen and gut (Supplementary Fig. 1 , Supplementary Table 4 , available online). Functional annotation of these targeted genes revealed enrichment in distinct functions in these tissues (Supplementary Fig. 1B –E ). Significant enrichment in cell skeleton-related and immune response-related functions was observed in the lung (Supplementary Fig. 1B ) and spleen (Supplementary Fig. 1C ), respectively.
The four validated miRNAs above targeted the UTR sequences of six genes in the Vero E6 cell (Supplementary Table 5 , available online). We then verified the expression of them in the virus-infected cells (two genes that encode putative proteins were not tested, i.e., LOC103242070 and LOC103217999). MR359-5p targeted at 3′-UTR of the same genes (i.e., FOXO3, GHR, and TIFA) in the green monkey genome as in human. Two of these three genes were significantly downregulated in the infected cells (Fig. 2C). Besides, GPR1 (G-protein-coupled receptor 1), which was targeted by MR359-5p at the 5′-UTR, was highly upregulated in the infected cells (Fig. 2C). G-protein-coupled receptors (GPCRs) constitute the largest group of cell-surface proteins that participate in a wide variety of physiological functions. GPCRs are involved in facilitating viral propagation and thereby contributing to viral pathogenesis[37–38], and GPCR antagonists have broad antiviral activity[39].
To further validate the regulatory role of these miRNAs on human genes, the construct of pGCMV/EGFP/MR147-3p was transfected into the pulmonary epithelial BEAS-2B cells. The increased MR147-3p levels relative to empty vector were observed (Supplementary Fig. 2 , available online). Real-time qRT-PCR showed that overexpression of MR147-3p resulted in significantly decreased transcript levels of all of the predicted targets in human, i.e., EXOC7, RAD9A, and TFE3 (Fig. 2D). EXOC7 is a component of the exocyst complex, a protein complex that guides membrane addition and polarized exocytosis[40]. TFE3 is a transcription factor that promotes the expression of genes downstream of transforming growth factor beta signalling and also plays a crucial role in the regulation of lipid and glucose metabolism[41].
Virus genome hijacks host miRNAs to regulate the immune response
Several studies have reported that the virus genome can hijack host miRNAs to modulate host biological processes[38]. A total of 28 human miRNAs were predicted to target the SARS-Cov-2 genome (Supplementary Table 6 , available online). Since these miRNAs were hijacked by the virus genome, we supposed that the genes originally targeted by these miRNAs could be affected. There were over 800 human genes predicted to be regulated by these miRNAs, and a significant enrichment at the immune system process was observed (Fig. 3A and B). Among the hijacked miRNAs, miR-146 is one of the critical modulators of the immune response. Studies in mice have shown that miR-146b−/− mice displayed enlarged spleens and increased myeloid cell populations in both spleen and bone marrow, and spontaneously developed intestinal inflammation[42]. Another miRNA, miR-939, was proposed to regulate proinflammatory genes. Increasing miR-939 levels should restore homeostasis by decreasing inflammatory protein synthesis[43]. Our result posed a possibility that the hijack of human miRNAs by the SARS-Cov-2 genome might contribute to the abnormal immunity activation in patients with COVID-19.
Figure 3.
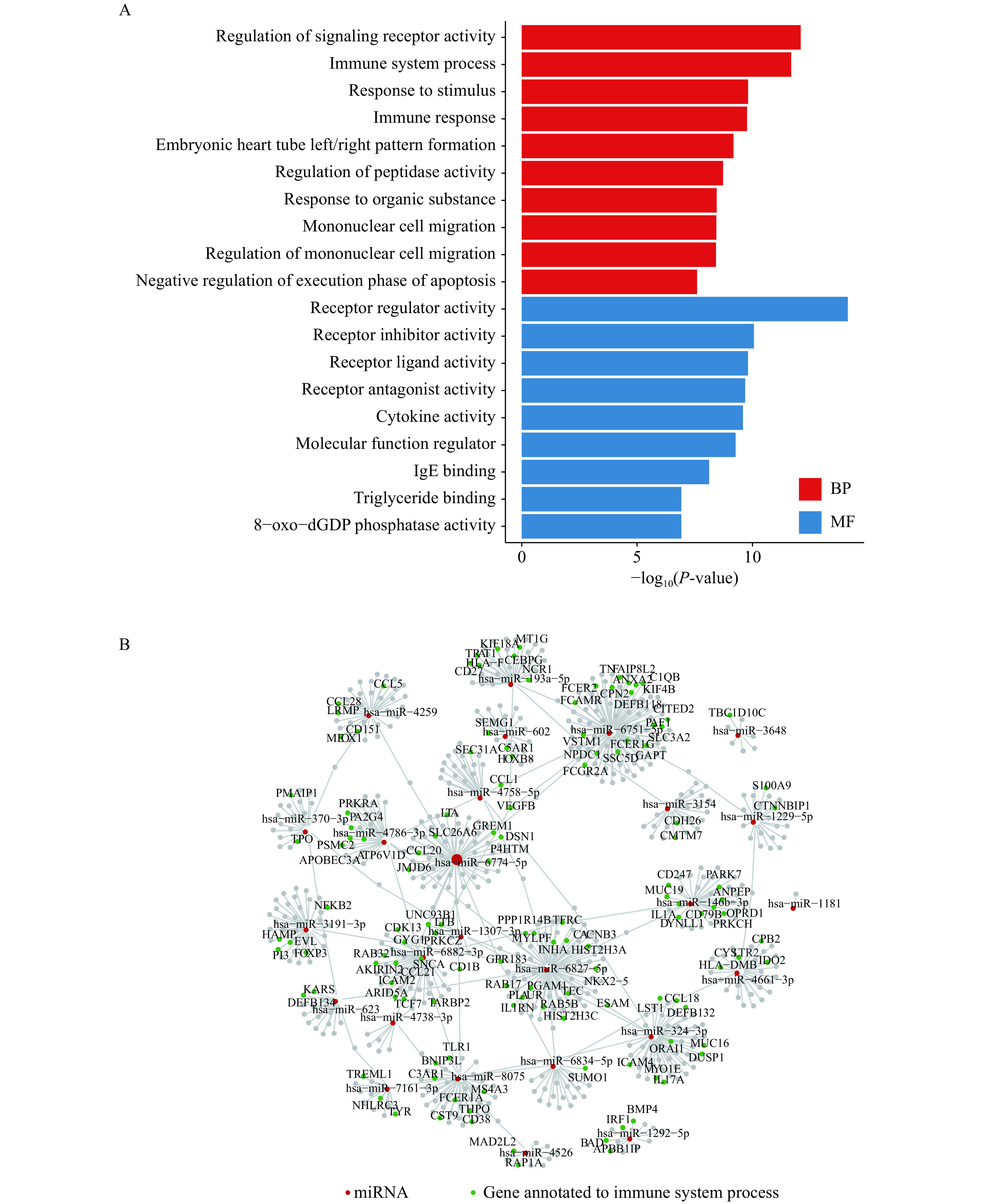
Genes targeted by the virus-genome-hijacked miRNAs.
A: The barplot for the enriched functions of genes regulated by the host miRNAs attracted by the virus miRNAs. B: The network of the regulation between the hijacked human miRNAs and genes. The red nodes represent the miRNAs. The greens are genes annotated to the immune system processes, and greys are annotated to other functions. BP: biological process; MF: molecular function.
Virus genomic region targeted by virus and host miRNAs
We then explored the targets of both virus and host cell-derived miRNAs on the SARS-CoV-2 genome. There were 27 targets of the putative virus miRNAs at the virus genome (Fig. 4A). Forty-three targets were predicted for these miRNAs, and most of them were bound to the ORFlab region (Fig. 4B), the longest ORF in the SARS-CoV-2 genome. There were two miRNAs target at the 5′-UTR of the virus genome, and two at the S gene. Twenty-eight human miRNAs were predicted to have 30 targets on the SASRS-CoV-2 genome (Fig. 4B). Most of the human miRNAs were bound to the ORF1ab, followed by seven miRNAs targeted at the N genes of the virus, and two at the 5′-UTR. The hsa-miR-4661-3p was predicted to target at the genomic region 25296 to 25320 within S genes (21563 to 25384), which is the potential 3′-UTR of the S gene transcript (Fig. 4C), suggesting a possible repressor role on the S gene expression. Since the transcriptome landscape of the SARS-Cov-2 is mostly unknown, further research in characterizing its genomic and transcriptomic features may enlighten the role of miRNAs in regulating the virus gene expression and infection.
Figure 4.
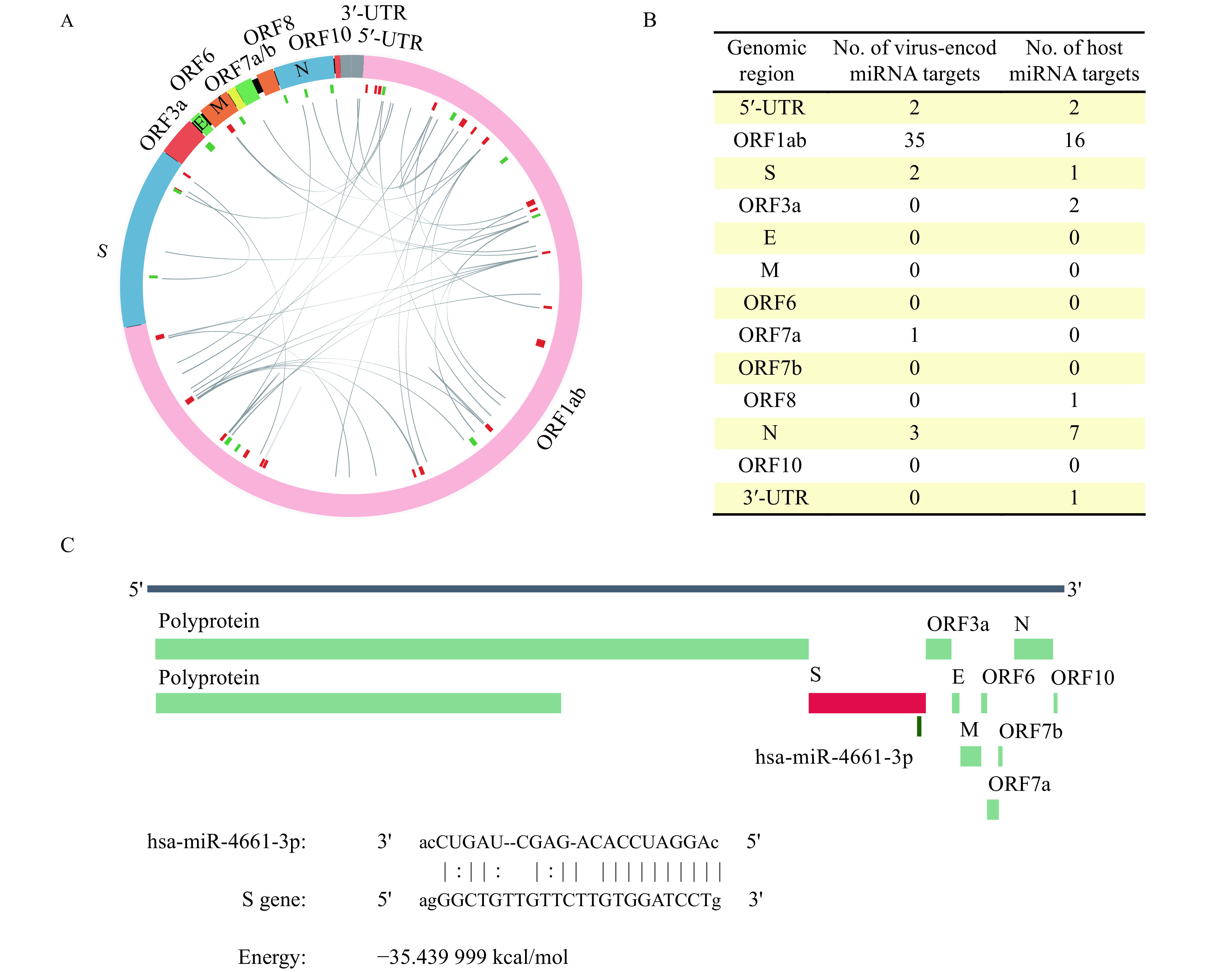
Virus genomic region targeted by virus and host-derived miRNAs.
A: The circos plot for interactions between the putative virus miRNA and virus genomic region. B: The distribution of viruses and host miRNA targets on the virus genome. C: The target of hsa-miR-4661-3p at S gene.
Mutations in virus genome affect miRNA function
Mutations accumulated since the SARS-CoV-2 emerged, and studies have implicated that the variations on the virus genome may lead to an attenuated phenotype of SARS-CoV-2[44]. Therefore, we explored whether miRNAs encoded by either virus or human contributed to the fitness of SARS-CoV-2 to human. A total of 810 mutations on the SARS-CoV-2 genome were collected from China National Center for Bioinformation, including 646 SNPs, 19 indels, and 145 deletions (Fig. 1B). There were 33 candidate pre-miRNAs overlaps the mutant region, including 30 pre-miRNAs overlapped with SNPs and four located in the deletion region. Compared with the wild-type pre-miRNAs, the MFE of the mutant pre-miRNAs were significantly increased (paired Wilcox test P-value <0.001, Fig. 5A), indicating an overall decreased stability of the mutant-genome encoded pre-miRNAs.
Figure 5.
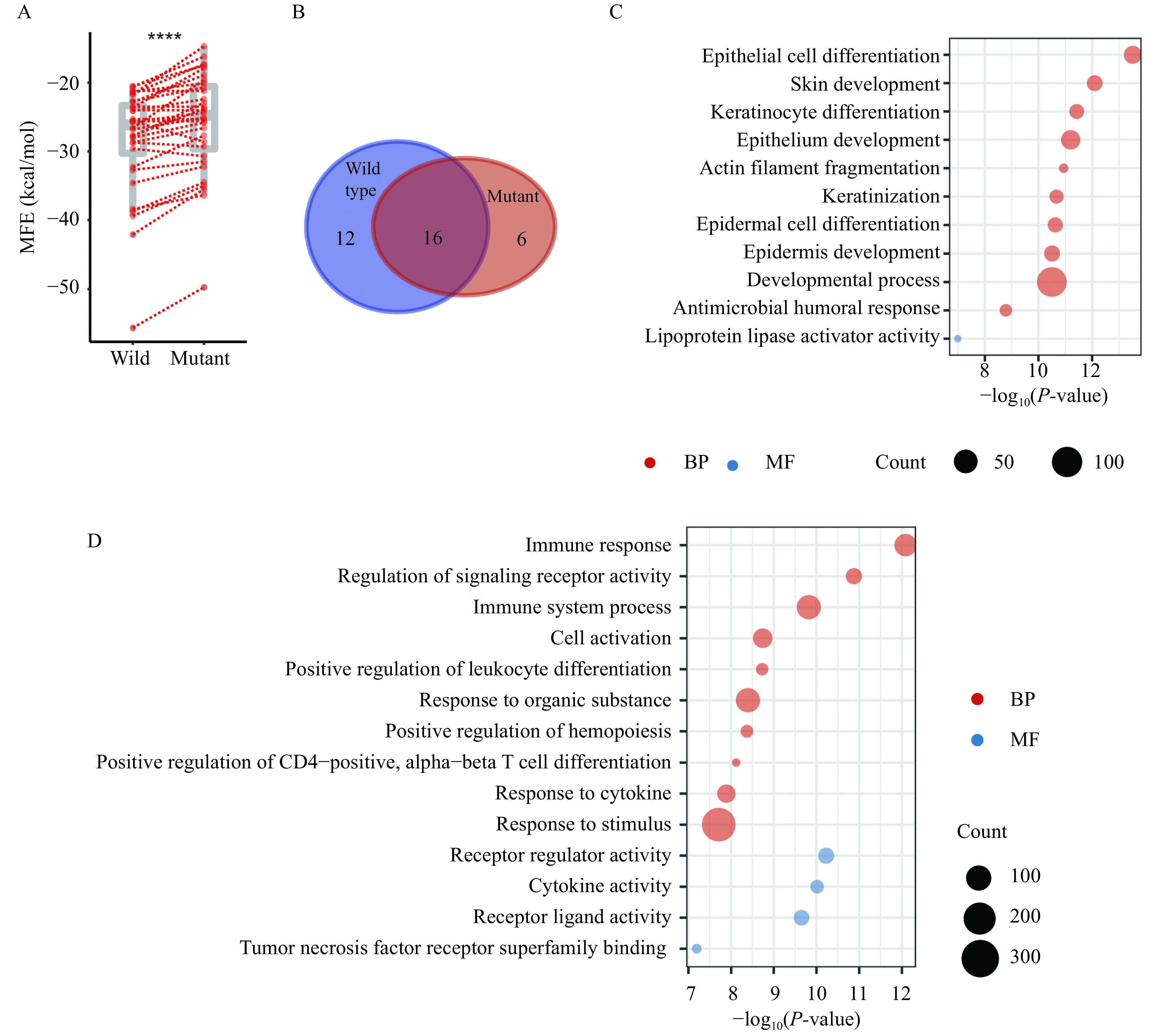
The effect of genome mutation on miRNA repositories and function.
A: The comparison between the MFE of hairpin folding of candidate miRNAs derived from the wild type and mutated SARS-CoV-2 genome (n=33). B: The overlap of the putative miRNAs binding to the wild type and mutant genome. C: The functional annotation of genes targeted by miRNAs specifically binding to the mutant SARS-CoV-2 genome. D: The functional annotation of genes targeted by miRNAs specifically binding to the wild-type SARS-CoV-2 genome. Two-sided Wilcoxon signed-rank test, ****P<0.0001. MFE: minimum free energy; BP: biological process; MF: molecular function.
Next, we investigated the alteration of host miRNA targets on the mutant virus genome. Twenty-two human miRNAs can bind to the mutant genome, with 16 overlapped with those binding to the wild type genome (Fig. 5B). The genes targeted by the newly recruited miRNAs were annotated to the function of epithelial cell differentiation and actin filament fragmentation (Fig. 5C). In contrast, the genes targeted by the miRNAs specifically binding to the wild type genome were annotated to the immune response-related progress (Fig. 5D). The two miRNAs discussed, hsa-miR-939-5p and hsa-miR-146b-3p, which were involved in maintaining homeostasis by decreasing inflammation, failed to be hijacked by the mutant virus genome. Taken together, we speculate that the mutant type of virus might relieve the high level of immunity response, even the cytokine storm. Nevertheless, it enhances the invasion of the virus by intensifying the regulation of the epithelial differential and actin filament fragmentation, through attracting different batches of host miRNAs.
miRNA view of the difference between SARS-CoV-2 with other human coronaviruses
Compared with MERS-CoV and SARS-CoV, SARS-CoV-2 appears to have high transmission rates, but low mortality[1–2]. To explore whether and how miRNAs implicated in this difference between them, we predicted the miRNAs encoded by MERS-CoV, SARS-CoV, as well as HCoV-NL63, a coronavirus that causes mild symptoms in human and uses the same cell acceptor with SARS-CoV and SARS-CoV-2 (ACE2). The largest putative virus-encoded miRNA repository (212 miRNAs) was detected from the MERS-CoV genome (Table 1), followed by SARS-CoV with 168 miRNAs and SARS-CoV-2 with 90 miRNAs. HCoV-NL63 encodes the least number of miRNAs (64 miRNAs). Additionally, the number of targets on the virus genome bound by the host miRNAs follows the same trend (Table 1). Targets and functional analysis also revealed a dramatic difference between these virus (Supplementary Fig. 3 and 4 , available online). These results implicated that differences in miRNA repositories and miRNA-target interaction might be involved in the pathological difference among the four coronaviruses.
Table 1. The number of miRNAs encoded by virus genome and genomic targets bound by host miRNAs for four human coronaviruses.
| Features | SARS-CoV-2 | SARS-CoV | MERS-CoV | HCoV-NL63 |
| Genome length | 29 903 | 29 751 | 30 119 | 27 553 |
| Vmir predict | 808 | 827 | 862 | 740 |
| Pre-miRNA candidates | 45 | 84 | 106 | 32 |
| Mature miRNAs | 90 | 168 | 212 | 64 |
| Hits by human miRNAs | 30 | 39 | 70 | 21 |
Discussion
In this study, we revealed a systematic view of the possible role of the candidate virus-encoded miRNAs as well as the host-derived miRNAs in the SARS-CoV-2 infection (Fig. 6). The analysis revealed that the immune response was the function most affected by the virus-infection introduced miRNA change in repository and targets, which might contribute to the immune evasion of the virus as well as the abnormal immunity activation in the host. The SARS-CoV-2-encoded miRNAs were also implicated in the cytoskeleton dynamics facilitating the virus envision, trafficking within the cell, and release. The virus-encoded miRNAs were also predicted to be able to repress the expression of genes involved apoptosis by binding at the 3′-UTR of host genes. We validated the virus-encoded miRNAs and their targets in the Vero E6 and human cell lines. Additionally, the genomic mutation-introduced alterations on the coding ability of virus miRNAs and the targets of both virus and host miRNAs might contribute to the attenuated phenotype of SARS-CoV-2 during its evolution.
Figure 6.
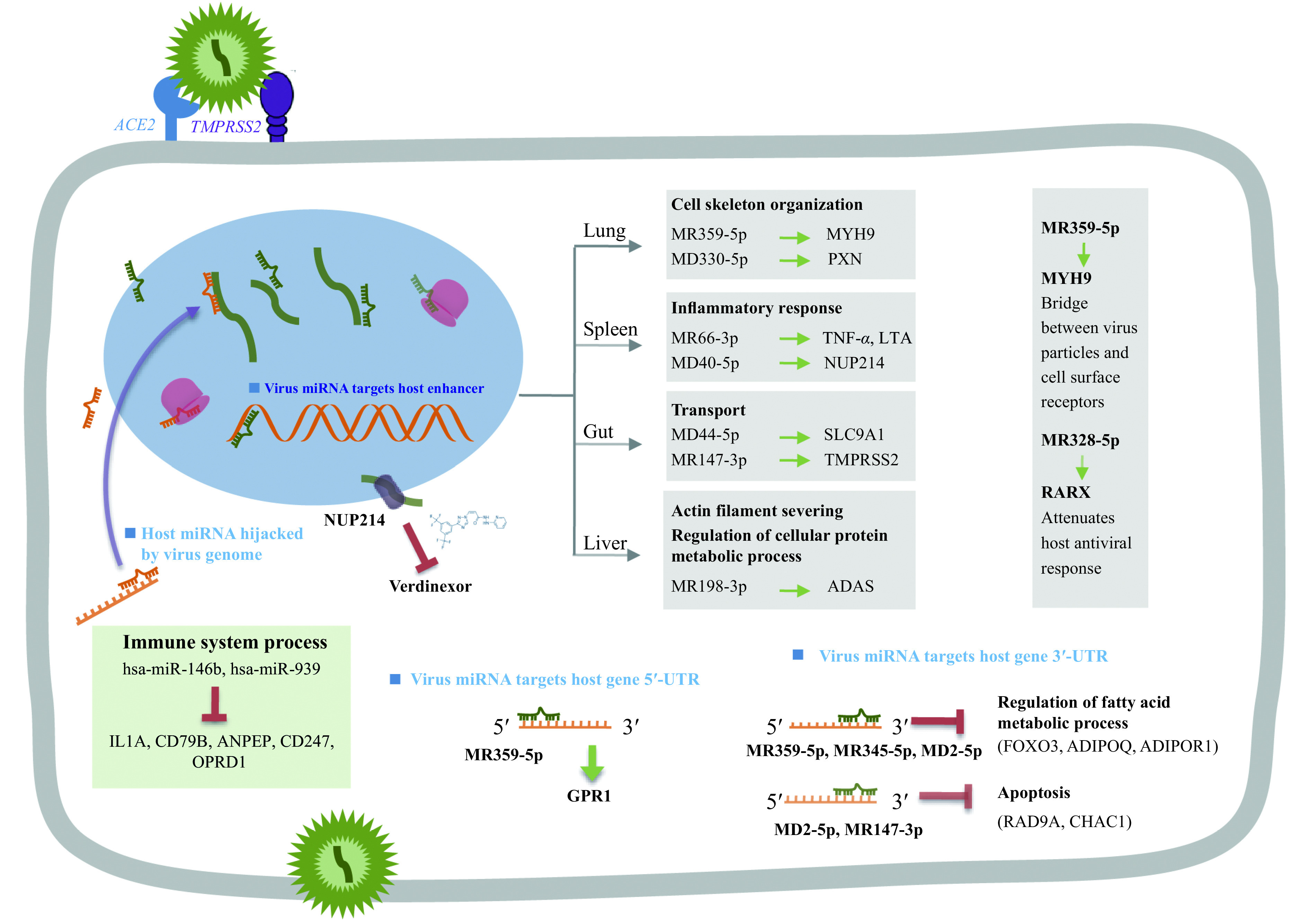
A graphic summary of the findings in this study.
The SARS-CoV-2 encoded miRNAs are able to target the regulatory regions (3′-UTR, 5′-UTR or enhancer) of host genes, and the host miRNAs might be hijacked by the virus genome. The involved miRNAs could target genes that participate in variety of biological process that induced by virus infection.
Two virus miRNAs were predicted to target at potent drug targets, which were MR147-3p that bind to the enhancer of TMPRSS2 and MD40-5p that bind to the enhancer of NUP214 (Supplementary Table 4 , available online). TMPRSS2 enhances SARS-CoV-2 infection, together with ACE2[3]. NUP214 is predicted as a drug target of Verdinexor, a selective inhibitor of nuclear export for virus RNP under clinical trial. Animal experiments have proved the efficiency of Verdinexor in reducing lung viral loads and proinflammatory cytokine expression in influenza A virus-infected mice[45]. We have quantified the expression of TMPRSS2 in the MR147-3p transfected cells, and elevated average expression was observed, but the variation between different replicates was large (data do not show). Since the mechanisms of miRNAs mediated RNA activation are largely unknown, for example, how the miRNAs being imported back into the nucleus[9], more studies are needed to explore the regulation of gene expression of miRNAs by binding enhancer.
In this work, we did exhaustive in silico analysis of the SARS-CoV-2 genome from the perspective of miRNA and aided by simple validations for the virus miRNAs and their targets. Though several interesting phenomena have been observed, more detailed and sophisticated experiments are needed to validate them. With further experiments to validate the role of candidate miRNAs in vivo, there is a reasonable prospect to develop antiviral therapeutics against SARS-CoV-2 through targeting the candidate miRNAs.
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) grant No. 81671983 and 81871628 to X.L., NSFC grant No. 81703306, China Postdoctoral Science Foundation (2017M611867), and Postdoctoral Science Foundation of Jiangsu Province (1701119C) to Z.L., NSFC grant No. 81902027, and Natural Science Foundation of Jiangsu Province to J.W. (BK20171045).
Footnotes
The authors reported no conflict of interests.
CLC number:R511 , Document code: A
Contributor Information
Ying Chi, Email: chiying@jscdc.cn.
Xingyin Liu, Email: xingyinliu@njmu.edu.cn.
References
- 1.Wang C, Horby PW, Hayden FG, et al A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, et al A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, Kleine-Weber H, Schroeder S, et al SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang CL, Wang YM, Li XW, et al Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Shi L, Wang FS Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5(5):428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng ZQ, Diao B, Wang RS, et al. The novel severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) directly decimates human spleens and lymph nodes[EB/OL].[2020-03-31]. https://www.medrxiv.org/content/10.1101/2020.03.27.20045427v1.
- 7.Chen NS, Zhou M, Dong X, et al Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao M, Li J, Li W, et al MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017;14(10):1326–1334. doi: 10.1080/15476286.2015.1112487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramchandran R, Chaluvally-Raghavan P. miRNA-mediated RNA activation in mammalian cells[M]//Li LC. RNA Activation. Singapore: Springer, 2017: 81-89.
- 10.Bandiera S, Pfeffer S, Baumert TF, et al miR-122-A key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Kincaid RP, Sullivan CS Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XH, Zou XP An overview of RNA virus-encoded microRNAs. ExRNA. 2019;1(1):37. doi: 10.1186/s41544-019-0037-6. [DOI] [Google Scholar]
- 13.Guo X, Huang YJ, Qi Y, et al Human cytomegalovirus miR-UL36-5p inhibits apoptosis via downregulation of adenine nucleotide translocator 3 in cultured cells. Arch Virol. 2015;160(10):2483–2490. doi: 10.1007/s00705-015-2498-8. [DOI] [PubMed] [Google Scholar]
- 14.Li XH, Fu Z, Liang HW, et al H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2. Cell Res. 2018;28(2):157–171. doi: 10.1038/cr.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth S, Pfuhl T, Mamiani A, et al Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcinowski L, Tanguy M, Krmpotic A, et al Degradation of cellular miR-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo . PLoS Pathog. 2012;8(2):e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trobaugh DW, Gardner CL, Sun CQ, et al RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506(7487):245–248. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahamtan A, Inchley CS, Marzban M, et al The role of microRNAs in respiratory viral infection: friend or foe? Rev Med Virol. 2016;26(6):389–407. doi: 10.1002/rmv.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernier A, Sagan SM The diverse roles of microRNAs at the host-virus interface. Viruses. 2018;10(8):440. doi: 10.3390/v10080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundhoff A. Computational prediction of viral miRNAs[M]//van Rij RP. Antiviral RNAi: Concepts, Methods, and Applications. New York: Humana Press, 2011: 143-152.
- 21.Gruber AR, Lorenz R, Bernhart SH, et al The Vienna RNA Websuite. Nucleic Acids Res. 2008;36(S1):W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudyś A, Szcześniak MW, Sikora M, et al HuntMi: an efficient and taxon-specific approach in pre-miRNA identification. BMC Bioinformatics. 2013;14(1):83. doi: 10.1186/1471-2105-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gkirtzou K, Tsamardinos I, Tsakalides P, et al MatureBayes: a probabilistic algorithm for identifying the mature miRNA within novel precursors . PLoS One. 2010;5(8):e11843. doi: 10.1371/journal.pone.0011843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignone F, Grillo G, Licciulli F, et al UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33(S1):D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao TS, Qian J EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020;48(D1):D58–D64. doi: 10.1093/nar/gkz980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fahlgren N, Carrington JC. miRNA target prediction in plants[M]//Meyers BC, Green PJ. Plant MicroRNAs: Methods and Protocols. New York: Humana Press, 2010: 51-57.
- 27.John B, Enright AJ, Aravin A, et al Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal V, Bell GW, Nam JW, et al Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu YJ, Wang LQ, Gu JX, et al Identification of microRNA differentially expressed in three subtypes of non-small cell lung cancer and in silico functional analysis. Oncotarget. 2017;8(43):74554–74566. doi: 10.18632/oncotarget.20218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falcon S, Gentleman R Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–257. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 31.Kvansakul M Viral infection and apoptosis. Viruses. 2017;9(12):356. doi: 10.3390/v9120356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols DB, De Martini W, Cottrell J Poxviruses utilize multiple strategies to inhibit apoptosis. Viruses. 2017;9(8):215. doi: 10.3390/v9080215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu K, Miyashita T, Hang HY, et al Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis . Nat Cell Biol. 2000;2(1):1–6. doi: 10.1038/71316. [DOI] [PubMed] [Google Scholar]
- 34.Zhang KB, Li L, Qi YJ, et al Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice . Endocrinology. 2012;153(2):631–646. doi: 10.1210/en.2011-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raman C, Ren C, Boas S, et al TCR signaling strength dependent regulation of T cell proliferation, survival and Th differentiation by TGF-βR3 (betaglycan) https://www.jimmunol.org/content/198/1_Supplement/201.8 J Immunol. 2017;198(S1):201.8. [Google Scholar]
- 36.Venkateswaran A, Sekhar KR, Levic DS, et al The NADH oxidase ENOX1, a critical mediator of endothelial cell radiosensitization, is crucial for vascular development. Cancer Res. 2014;74(1):38–43. doi: 10.1158/0008-5472.CAN-13-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang N, Huang HJ, Tan BH, et al Leucine-rich repeat-containing G protein-coupled receptor 4 facilitates vesicular stomatitis virus infection by binding vesicular stomatitis virus glycoprotein. J Biol Chem. 2017;292(40):16527–16538. doi: 10.1074/jbc.M117.802090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodhi A, Montaner S, Gutkind JS Viral hijacking of G-protein-coupled-receptor signalling networks. Nat Rev Mol Cell Biol. 2004;5(12):998–1012. doi: 10.1038/nrm1529. [DOI] [PubMed] [Google Scholar]
- 39.Cheng H, Lear-Rooney CM, Johansen L, et al Inhibition of Ebola and Marburg virus entry by G protein-coupled receptor antagonists. J Virol. 2015;89(19):9932–9938. doi: 10.1128/JVI.01337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lira M, Arancibia D, Orrego PR, et al The Exocyst component Exo70 modulates dendrite arbor formation, synapse density, and spine maturation in primary hippocampal neurons. Mol Neurobiol. 2019;56(7):4620–4638. doi: 10.1007/s12035-018-1378-0. [DOI] [PubMed] [Google Scholar]
- 41.Pastore N, Vainshtein A, Klisch TJ, et al TFE3 regulates whole‐body energy metabolism in cooperation with TFEB. EMBO Mol Med. 2017;9(5):605–621. doi: 10.15252/emmm.201607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Testa U, Pelosi E, Castelli G, et al miR-146 and miR-155: two key modulators of immune response and tumor development. Non-Coding RNA. 2017;3(3):22. doi: 10.3390/ncrna3030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald MK, Ramanathan S, Touati A, et al Regulation of proinflammatory genes by the circulating microRNA hsa-miR-939. Sci Rep. 2016;6(1):30976. doi: 10.1038/srep30976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holland LA, Kaelin EA, Maqsood R, et al An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January to March 2020) J Virol. 2020;94(14):e00711–20. doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perwitasari O, Johnson S, Yan XZ, et al Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo . J Virol. 2014;88(17):10228–10243. doi: 10.1128/JVI.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


