Abstract
The toxicokinetic (TK) parameters fraction of the chemical unbound to plasma proteins and metabolic clearance are critical for relating exposure and internal dose when building in vitro-based risk assessment models. However, experimental toxicokinetic studies have only been carried out on limited chemicals of environmental interest (~1000 chemicals with TK data relative to tens of thousands of chemicals of interest). This work evaluated the utility of chemical structure information to predict TK parameters in silico; development of cluster-based read-across and quantitative structure-activity relationship models of fraction unbound or fub (regression) and intrinsic clearance or Clint (classification and regression) using a dataset of 1487 chemicals; utilization of predicted TK parameters to estimate uncertainty in steady-state plasma concentration (Css); and subsequent in vitro–in vivo extrapolation analyses to derive bioactivity-exposure ratio (BER) plot to compare human oral equivalent doses and exposure predictions using androgen and estrogen receptor activity data for 233 chemicals as an example dataset. The results demonstrate that fub is structurally more predictable than Clint. The model with the highest observed performance for fub had an external test set RMSE/σ=0.62 and R2=0.61, for Clint classification had an external test set accuracy = 65.9%, and for intrinsic clearance regression had an external test set RMSE/σ=0.90 and R2=0.20. This relatively low performance is in part due to the large uncertainty in the underlying Clint data. We show that Css is relatively insensitive to uncertainty in Clint. The models were benchmarked against the ADMET Predictor software. Finally, the BER analysis allowed identification of 14 out of 136 chemicals for further risk assessment demonstrating the utility of these models in aiding risk-based chemical prioritization.
1. Introduction
Human health risk assessment associated with environmental chemical exposure is limited by the tens of thousands of chemicals with little or no experimental in vivo toxicity data 1. The wealth of in vitro toxicity data generated over the last decade has emerged as a promising alternative to animal testing and has enabled better insight into potential mechanism(s) of toxicity 1-5. However, in vitro toxicity data suffers from a drawback in that it cannot account for the toxicokinetic (TK) factors such as bioavailability, plasma protein binding and intrinsic clearance which are required for the transformation of an in vitro active concentration to a relevant in vivo oral equivalent dose (OED) below which significant in vitro bioactivity is not expected to occur. However, these parameters can be measured, and TK models can be built using them, yielding estimates of steady-state plasma concentration (Css). The OED can then be calculated as the ratio of an in vitro potency value (e.g. an AC50) to the Css value 6-10.
Incorporation of toxicokinetic and exposure information can be used in chemical prioritization and can facilitate the addition of a risk context to high-throughput in vitro screening results 6-8, 11-13. Two key experimental TK parameters that are required for relating oral dose to an internal steady state plasma concentration are fraction unbound in plasma (fub) and intrinsic clearance (Clint). Although these parameters can be measured experimentally in vitro 7, 8, 14, the protocols are not high-throughput, primarily due to the need to develop chemical-specific analytical methods. As a result, in vitro TK data are available only for fraction of environmental chemicals of interest (~1000 to date), which in turn limit the ability to provide bioactivity exposure ratio (BER) estimates for most environmental chemicals.
In the absence of experimental data, in silico approaches such as read-across 15-20 and quantitative structure-activity relationship (QSAR) models 21, 22 can potentially be used to predict fub and Clint. Several in silico models that have been derived for predicting fub23-29 as well as Clint30-32. Some of these models have been published in the peer reviewed literature, whilst others have been implemented into commercial software tools, such as ADMET Predictor (Simulations Plus Inc., Lancaster, CA). Most of these models were derived using data generated for pharmaceutical chemicals and their relevance for environmental chemicals is unclear.
Here, we derive new in silico models for fub and Clint using data extracted from published literature collected for 1486 environmental chemicals 7-9, 33. This study aimed at (1) evaluating the suitability of chemical structure information for predicting these parameters in silico, (2) exploring the utility of read-across and QSAR modeling techniques for developing predictive models for the two in vitro TK parameters, (3) evaluating the implications of variability in experimental and predicted TK parameters, and physicochemical properties on the uncertainty in resultant OED estimates, and (4) integration of IVIVE methods along with high-throughput exposure predictions using the EPAs ExpoCast tool 34, 35 to facilitate rapid risk-assessment and chemical prioritization.
2. Workflow
The overall workflow in this study comprised three main steps (Supplemental Figure S1). First, experimental data along with fingerprints and molecular descriptors were used to develop QSAR models. Second, the predictions from the models developed in this work were compared with the predictions from the commercially available ADMET Predictor package. Last, the predictions from this work were used to calculate OEDs using IVIVE methods implemented in the HTTK package and compared with the human exposure predictions from ExpoCast 34, 35 to facilitate high-throughput risk-assessment. The methods are described in detail in the following section. Additional analysis was performed where unsupervised clustering of chemicals with human in vitro TK parameters data was used to determine whether structurally related chemicals had similar TK parameters. The clusters derived using unsupervised clustering were used along with experimental data to derive cluster-based read-across predictions. These analyses do not directly impact the main findings of this study and are discussed in supplemental information (1-Supplemental-Methods&Results.docx).
3. Methods
3.1. Dataset
The data used in this analysis was obtained from published literature and available through the high-throughput toxicokinetic (HTTK) R package 7, 33, 36-47. The dataset consists of 1486 chemicals that span a variety of use classes including pharmaceuticals, food-use chemicals, pesticides and industrial chemicals 48 of which 1139 chemicals had experimental human in vitro fub data and 642 chemicals that had experimental human hepatic in vitro Clint data. An external dataset of 1,814 chemicals tested in a battery of 18 ER and 11 AR related assays 49, 50 was also utilized in this study for model validation. Before developing any models, the chemicals from this external ER-AR dataset were removed to ensure there is no training bias in the predictions for external validation on this set. All the structures were curated and the sdf file format was obtained from the DSSTox database 51, 52. The distribution of experimental values for fub and Clint are shown in Supplemental Figure S2. Since the data were non-normally distributed, they were appropriately transformed before any analysis was conducted. The details of the transformation and the transformed data distribution are presented in the results section and Supplemental Figures S2 and S3. A complete list of chemicals with CAS registry numbers (CASRN) and experimental data for both parameters and the chemical structure sdf file are included as supplemental information (2-fub_data.csv, 3-clint_data.csv, and 5-QSARreadyStructures.sdf).
3.2. Molecular descriptors
The chemicals used in this study were characterized using two structure-based fingerprints PubChem 53 and ToxPrint chemotypes 54; two physicochemical descriptors (acid dissociation constant, acidic and basic pKa and logarithm of water-octanol partition coefficient, logP) computed using the OPERA software 55; 12 molecular descriptors calculated using the Chemistry Development Kit (CDK) 56, 57 implemented in KNIME the KNIME analytics platform 58 (version 2.11.3); and 1875 descriptors (1444 1D, 2D and 431 3D descriptors) calculated using PaDEL software59. PubChem fingerprints were generated in the KNIME analytics platform 58 (version 2.11.3). ToxPrints were generated within the publicly available Chemotyper application (version1.0.r12976, https://chemotyper.org). The fingerprint and descriptor generation require an sdf file format which was obtained from the DSSTox database 60. The final descriptor selection and preparation for both fub and Clint datasets was done as follows:
PubChem fingerprints and ToxPrints were combined to generate one combined fingerprint. Feature selection was performed to remove features with less than 80% variation across the chemical set, and only one feature from a pair of features was retained if the Pearson correlation coefficient between them was more than 80%,
The OPERA pKa acidic (pKa_a) and pKa basic (pKa_b) predictions were used to infer a pKa value as the lower of the 2 values. In case, only one them exists then that was used as the pKa,
All continuous descriptors were normalized to have mean = 0 and standard deviation = 1,
A supervised recursive feature elimination algorithm was used to select 10 descriptors from PaDEL and CDK descriptors combined, and
All chemicals for which the fingerprints/descriptors could not be calculated were dropped from the analysis.
A complete list of all the fingerprints and descriptors used in the final models is provided in table S6 of supplementary information (1-Supplemental-Methods&Results.docx).
3.3. QSAR modeling
3.3.1. Data Preparation
Fraction unbound in plasma
Chemicals with a fub value equal to 0 (below limit of detection) were set at a default of 0.005, based on the assumptions in the HTTK package 47, and those with a value of 1 were set to 0.99 (upper limit of detection). The final dataset after removing the chemicals from the external ER-AR dataset comprised 1003 chemicals that had defined Pubchem fingerprints and Toxprints. Before modeling, fub was transformed using the log-odds ratio form 25:
| (2) |
Supplemental Figure S3 shows the distribution of the transformed fub values. This transformed dataset was divided into an 80% training dataset (802 chemicals) and a 20% external test dataset (201 chemicals). Regression QSAR models were developed for fub, details of which are provided below.
Intrinsic Clearance
The final dataset after removing the chemicals from the external ER-AR dataset comprised 524 chemicals that had defined Pubchem fingerprints and Toxprints. Each chemical was assigned a clearance value of low, medium or high based on the following scheme: Clint values less than or equal to 0.9 were set as low, values between 0.9 and 50 were set as medium and values greater than 50 were set as high. These thresholds were defined by Ekins et al. 30. All Clint values are in units of uL/min/million cells. Two types of QSAR models were developed for Clint: (1) classification models to predict low, medium and high clearance, and (2) regression models using data for chemicals in the medium clearance group to avoid the effect of outliers (low or high clearance values) on the models. For regression models, the clearance data from the medium bins was log transformed before modeling. Supplemental Figure S4 shows the distribution of clearance data for classification and regression models. For both classification and regression models, the transformed dataset was then divided into two parts where 80% of the data (419 chemicals for classification and 269 chemicals for regression) was used as the training set and 20% of the data (105 chemicals for classification and 68 chemicals for regression) was used as an external test set.
Once the classification (low, medium, high) and regression (medium bin) QSAR models for intrinsic clearance were run, the results were combined as follows. Chemicals predicted to have low clearance were assigned a default clearance value equal to the median clearance value for training set chemicals in the low clearance bin, and the chemicals predicted to have high clearance were assigned a default value equal to the median clearance value for training set chemicals in the high clearance bin. Thus, the models can predict if a chemical exhibits low, medium or high clearance, and provides a quantitative value in each case.
3.3.2. Algorithm
Regression models for fub and Clint were developed using the lasso regression 61, support vector machine (SVM) 62, 63, random forest (RF) 64, 65 and neural network multiple layer perceptron 65, 66 algorithms. Classification models for Clint were developed using the logistic regression 65, 67, support vector machine (SVM) 62, 63, random forest (RF) 64, 65 and neural network multiple layer perceptron 65, 66 algorithms. The datasets were randomly split into a training set (80% chemicals) and an external test set (20% chemicals). The training set was used to build the models using 5-fold cross-validation with hyper-parameter tuning where the model is developed over a grid of parameter values and the values with the best model performance are selected as the final algorithm parameters. The final models were then evaluated on the external test set. Detailed discussion of the machine learning algorithms and the hyperparameters tuned for each model are available in supplemental information (1-Supplemental-Methods&Results.docx).
For both fub and Clint, several models were developed in an additive fashion where a new set of descriptors were added incrementally to observe any improvements in model predictivity. The first set of models, referred to as the baseline models, were developed using structural information encoded by PubChem fingerprints and ToxPrints. The subsequent models expanded on the baseline models in terms of the descriptor space. In the second set of models, two physicochemical descriptors (LogP and pKa) were added to the fingerprints. In the third set of models, 10 additional physicochemical descriptors from PaDEL and CDK were added. Finally, the two best performing models for each endpoint were combined to develop a consensus model 68, where the final prediction was the average prediction from the two best performing models. The performance of each regression model was evaluated in terms of MAE (mean absolute error), RMSE (root mean square error), RMSE/σ (RMSE / standard deviation of the endpoint distribution) and the variance explained (R2). The performance of each classification model was evaluated in terms of accuracy, F1-score (harmonic mean of positive predictive value and sensitivity) for each class and R2. Different additive models were compared based on the improvement in performance relative to their coverage. Note that the number of chemicals used in developing each additive model was dependent on generation of valid set of descriptors for each model.
3.4. Prediction of TK parameters and comparison with ADMET predictor
Model predictions from this work were compared with those of the ADMET Predictor package on an external dataset of 1,814 chemicals tested in a battery of 18 ER and 11 AR related assays 49, 50. ADMET predictions were obtained for these chemicals using data previously generate by Sipes et al 12. Overall, 472 chemicals had predicted fraction unbound and 410 chemicals had predicted Clint from the models developed in this work and the ADMET predictor. The residuals (difference between the observed experimental value and the predicted value) from both models (this work and ADMET predictor) were compared across both TK parameters to identify any chemicals or regions where both the models performed poorly. The experimental data for selected chemicals was then re-evaluated to account for potential experimental errors and data anomalies. Note that the chemicals in this set were not used in training the models in this work but it is unknown if those chemicals were present in the training dataset for the ADMET predictor.
3.5. Prediction and validation of Css: Calculation of in silico Css and comparison with in vitro Css
The HTTK R package 47 was used to predict steady-state concentration in plasma (Css) using the R software environment 69. Css is interpreted as the steady-state concentration of a chemical in the plasma given a constant 1 mg/kg/day oral dose rate and has units of μM/mg/kg/day. The default parameters of the population simulator in HTTK, httkpop (using calc_mc_Css) were used 47, 70 to calculate Css. The httkpop function returns the upper 95th percentile of Css in the population – corresponding to individuals for whom the same 1 mg/kg/day exposure produces plasma concentrations higher than 95% of the population. This is intended to be a conservative estimate, calculated with a simple steady-state model that estimates clearance from passive renal filtration and well-stirred hepatic metabolism. The omission of other routes of clearance acts to make Css higher (more conservative when comparing to estimated exposure). The calculation of Css for each chemical requires chemical-specific physicochemical properties (LogP and pKa), in addition to fub and Clint. Experimental fub and Clint data was available for 709 chemicals from the HTTK package. So, in vitro Css (using experimental fub, Clint and the default physicochemical properties from the HTTK package) and in silico Css (using predicted fub, Clint and the physicochemical properties calculated from the OPERA tool) could be calculated for 709 chemicals. The HTTK package LogP was obtained from the DSSTox database 51 or predicted using the EPA’s estimation program interface (EPI) suite (http://www.epa.gov/tsca-screening-tools/epi-suitetmestimation-program-interface) or using OPERA71. pKa was taken from the literature when available or taken from predictions produced by Strope et al 72. The in vitro and in silico Css calculations were compared to get an estimate of variance in Css values based on the models developed in this work.
3.6. Comparison of human oral equivalent doses (OEDs) and exposure predictions: Bioactivity-exposure ratio plot
A set of OEDs for each chemical in the dataset described in Section 3.4 was calculated by dividing in vitro potency values (ACC: activity concentration at cut-off) for a series of in vitro assays by the corresponding Css values. The assays that were used measured activities in the estrogen and androgen receptors (ER and AR), and have been combined to provide definitive estimates of agonist and antagonist activity for these receptors 50, 73. The OEDs for each chemical were then compared with the median estimated daily exposure taken from the EPAs ExpoCast estimates 34, 35. For each chemical, the following analysis was done:
The response (ACC value) in any ER or AR related assay was retained if the agonist or antagonist model AUC value 50, 73 was greater than 0.1, indicating activity in the specific target and mode (ER, AR, agonist, antagonist). The lowest ACC value across all assays (the most potent assay) was then used to calculate a conservative estimate of OED and is referred to as ACC hereafter,
The ACC was divided by the chemical’s in silico Css value to obtain an OED,
If the chemical had an in vitro Css value, ACC was divided by the in vitro Css value to obtain an overall conservative estimate of OED for the chemical based on in vitro Css,
An estimate of variance in the in silico values based on the Css prediction analysis from the analysis in Section 3.5 was incorporated to obtain an overall conservative estimate of OED for the chemical based on in silico Css. The ACC was divided by the in silico Css plus twice the standard deviation of the residuals between in silico and in vitro Css values,
Finally, all estimates of OEDs were compared with the exposure estimates.
The software for data analysis and model development was developed using the functions implemented in the scikit-learn module 74 of Python 2.7 75 and is available as supplemental information (code.zip).
4. Results and Discussion
4.1. QSAR modeling
Fraction Unbound in Plasma
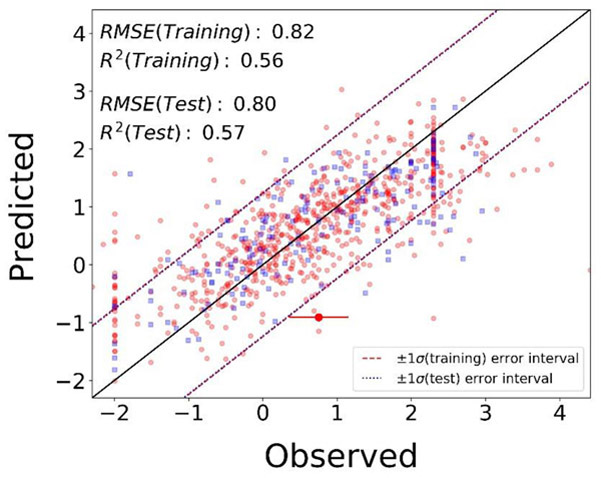
Feature selection on combined PubChem fingerprints and Toxprints resulted in 80 substructural features that were used for baseline model development. Subsequent models expanded the baseline feature set (80 features) with additional physicochemical descriptors. Across all sets of models, the best predictive performance was achieved when using RF, SVM or Lasso algorithms. Consequently, the consensus models were developed by averaging the predictions of best two models. Supplemental Table S2 summarizes the model details and the performance metrics for all the models developed. The coverage for all the baseline models was 1003 chemicals (the entire dataset). Adding two physicochemical descriptors LogP and pKa(from OPERA) to the baseline models resulted in the best consensus model performance metrics with MAE = 0.61, RMSE = 0.82, RMSE/σ = 0.66 and R2 = 0.56 for 5-fold internal cross-validation, and MAE = 0.61, RMSE = 0.80, RMSE/σ = 0.65 and R2 = 0.57 for the external test set validation with over 10% loss in coverage at 886 chemicals. Figure 1 shows the observed versus predicted transformed Fub values as evaluated in 5-fold internal cross-validation (red dots) and external validation (blue squares). Note that the RMSEs for all the models are well within one standard deviation (=1.24) of the endpoint distribution (Figure S3), which provides a context to the error rates.
Figure 1.
Observed versus predicted fraction unbound (transformed scale) for the random forest and support vector machine consensus model (highlighted in red in Supplemental Table S2) for 5-fold internal cross-validation (red dots) and external test set validation (blue squares). The root-mean-squared-error and R-squared for the 5-fold internal cross-validation are 0.82 and 0.56, respectively, and for external test set validation are 0.80 and 0.57, respectively. The black solid line indicates the line of perfect fit, where the predicted values would equal the experimental values. The red dashed lines indicate an error margin of ±1 standard deviation of the training dataset and the blue dotted lines indicate an error margin of ±1 standard deviation of the test dataset. The uncertainty in the observed data is indicated as a red error bar (coefficient of variation for uncertainty = 0.4) on an example chemical.
Intrinsic Clearance
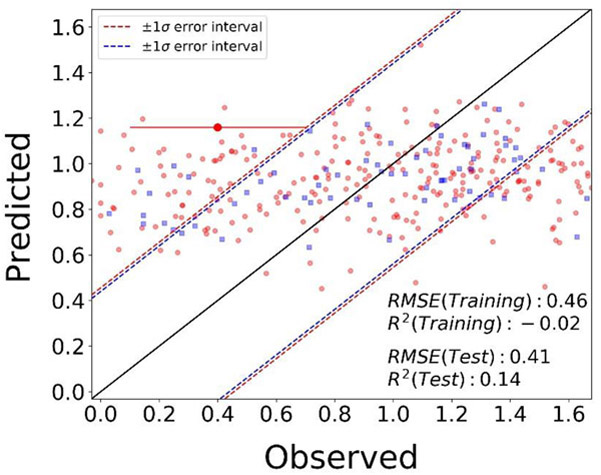
Feature selection on PubChem fingerprints and Toxprints resulted in 79 substructural features that were used for baseline model development. Subsequent models expanded the baseline feature set (of 79 features) with additional physicochemical descriptors. As expected from the unsupervised clustering analysis results (see supplemental file Supplemental-Methods&Results.docx for the methods used), use of structural descriptors did not yield highly predictive models. Supplemental Tables S4 and S5 summarize the model details and the performance metrics for all the classification and regression models, respectively. Adding LogP and pKa as descriptors to the classification baseline models using the support vector algorithm resulted in the best consensus model performance metrics with accuracy = 66.47% and F1-score = [0.43, 0.77, 0.18] for 5-fold internal cross-validation, and accuracy = 73.26% and F1-score = [0.41, 0.83, 0.25] for the external test set validation. The regression models were all poorly performing with the random forest baseline model being the best one with MAE = 0.39, RMSE = 0.46, RMSE/σ = 1.01 and R2 = −0.02 for 5-fold internal cross-validation, and MAE = 0.33, RMSE = 0.41, RMSE/σ = 0.92 and R2 = 0.14 for the external test set validation. Figure 2 shows the observed versus predicted transformed clearance values as evaluated in 5-fold internal cross-validation (red dots) and external test set validation (blue squares).
Figure 2.
Observed versus predicted medium intrinsic clearance (transformed scale) for the random forest model (highlighted in red in Supplemental Table S5) for (a) 5-fold internal cross-validation (red dots), and (b) external test set validation (blue squares). The root-mean-squared-error and R-squared for the 5-fold internal cross-validation are 0.46 and −0.02, respectively, and for external test set validation are 0.41 and 0.14, respectively. The black solid line indicates the line of perfect fit, where the predicted values would equal the experimental values. The red dashed lines indicate an error margin of ±1 standard deviation of the training dataset and the blue dotted lines indicate an error margin of ±1 standard deviation of the test dataset. The uncertainty in the observed data is indicated as a red error bar (coefficient of variation for uncertainty = 0.3) on an example chemical.
Overall, fub models performed better than Clint models. Unsupervised clustering analysis (described in supplemental file 1-Supplemental-Methods&Results.docx) show that: (i) fub values are more tightly bounded across different clusters as compared to Clint, and (ii) the mean value of fub are more distinct across clusters as compared to Clint. A new evaluation of uncertainty in experimental Clint values identified a median coefficient of variation of 0.31 [Wambaugh 2019, submitted]. Therefore, there is significant uncertainty in the experimental values on which the QSAR modeling was performed.
4.2. Prediction of TK parameters and comparison with ADMET predictor
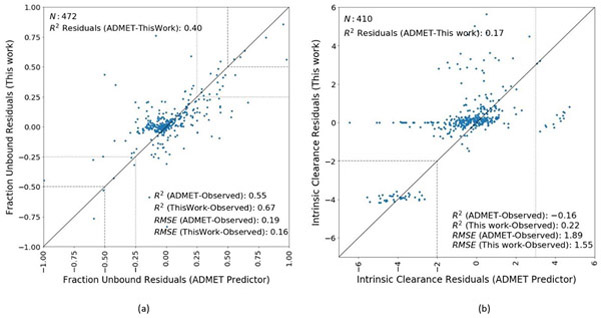
Figure 3(a) shows the plot of the residuals between the current model and those of ADMET Predictor for fraction unbound for 472 chemicals. The RMSE and R2 for ADMET Predictor were 0.19 and 0.55, respectively, and the RMSE and R2 for this work were comparatively better for this dataset at 0.16 and 0.67, respectively. The residuals from the two models had low correlation (R2 = 0.40). In general, the ADMET predictor over-predicts fub while the consensus model from this work under-predicts fub as compared to the experimental data. The dotted lines and the dashed lines highlight the chemicals for which the absolute residuals were greater than 0.25 and 0.50 for both predictors (outliers), respectively. The outliers with experimental data from Wetmore et al. 2012 and 2015 were cross-checked for any potential measurement errors7, 46. In general, the experimental data appeared to be reliable with the predicted fraction unbound values lower than the measured data. It is speculated that the ADMET predictor looks specifically at predicted binding to AAG and albumin, but does not consider additional binding to lipoprotein complexes, whereas the experimental measures capture these interactions.
Figure 3.
Residual comparison plots between the models developed in this work with those of the ADMET predictor. The black line on the plots is the line of perfect fit, where the residuals (observed minus predicted) from both the predictors are the same. (a). Plot of fraction unbound residuals. The dotted lines highlight the regions where the absolute residuals were greater than 0.25 for both the predictors. The dashed lines highlight the regions where the absolute residuals were greater than 0.50 for both the predictors. (b) Plot of intrinsic clearance residuals. The dashed lines highlight the region where the residuals were less than −2 log-units for both the predictors. The dotted line highlights the region where the residuals from the ADMET predictor were greater than 3 log-units.
Figure 3(b) shows the plot of the residuals for Clint for 410 chemicals on the log10-transformed dataset. For the purposes of plotting on a logarithmic-scale, the chemicals with an observed clearance value of zero were defaulted to 10−3 (i.e. log-transformed clearance value = −3). The models developed in this work worked better on this dataset with explained variance of 0.22 as compared to the negative R2 values from the ADMET predictor. However, the RMSEs were 1.89 and 1.55 for ADMET and this work, respectively. The residuals from the two models were quite uncorrelated (R2 = 0.17). The dashed lines highlight the region where the residuals for both predictors were less than −2 log-units and dotted line highlights the region where the residuals from the ADMET predictor were greater than 3 log-units (outliers). In general, the residuals from this work are more centered around zero, indicating lesser bias, as compared to the ADMET predictor which has a wider range of residuals. The outliers with experimental data from Wetmore et al. 2012 and 2015 were cross-checked for any potential measurement errors (details described in the supplementary file 1-Supplemental-Methods&Results.docx)7, 46. In general, the discrepancies in primary hepatocyte data can be accounted due to non-P450 related clearance (ADMET predictor uses only P450 metabolism in their calculations), non-metabolic degradation and sensitivity/detection issues at 1μM measurements. The poor predictions from the models developed in this work could be due to the general inability of chemical structural descriptors to adequately model Clint. A complete list of all the chemicals highlighted in the residual plots, experimental and predicted data, and data evaluation results are provided in supplemental information (4-ERAR-FubClintData.xlsx).
4.3. Prediction and validation of Plasma Css: Calculation of in silico Css and comparison with in vitro Css
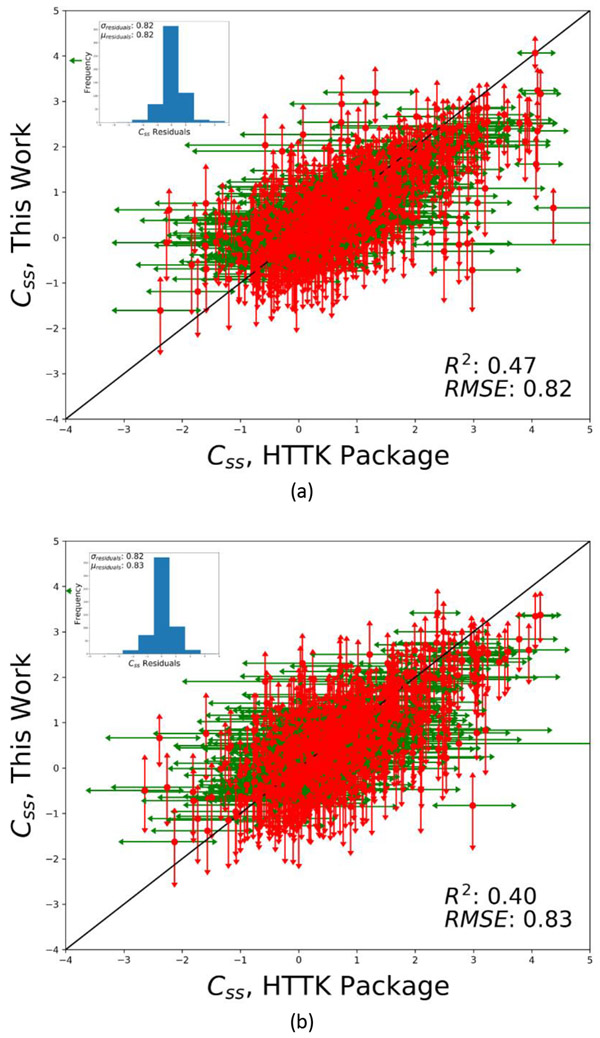
Figure 4 shows a plot of Css values calculated using the models in this work (in silico Css) against Css calculated using the data in HTTK package (in vitro Css) for the ER-AR dataset and the entire set of chemicals with in vitro fub and clint data from the HTTK package. The Css units are log10 mg/kg. The observed RMSE and the R2 values for the ER-AR dataset were 0.82 and 0.47, and for the big dataset were 0.83 and 0.40, respectively. These plots incorporate the variability in Css calculations owing to the underlying experimental variability in the fub and Clint data as calculated using the HTTK package and are shown as error bars in the plot. The results of this analysis demonstrate that Css calculations tend to be relatively stable given the uncertainty in (predicted) clearance values. The standard deviation of the residuals from this analysis were further used in the BER analysis to arrive at a conservative estimate of dose for hazard assessment. Note that the analysis excludes all chemicals that were in the training dataset for development of the fub and Clint models.
Figure 4.
Comparison of Css calculated using the models in this work (in silico Css) calculations with Css calculated using the data in HTTK package (in vitro Css) for (a) the ER-AR dataset and (b) the entire set of chemicals with in vitro fub and clint data from the HTTK package. The plots incorporate the variability in Css calculations owing to the underlying experimental variability in the fub and Clint data. The standard deviation of residuals from this analysis were used in the BER analysis (for the ER-AR dataset) to derive a conservative estimate of dose for hazard assessment. The Css units are log10 mg/kg. Note that the analysis excludes all chemicals that were in the training dataset for development of the fub and clint models.
4.4. Comparison of human oral equivalent doses (OEDs) and exposure predictions: Bioactivity-exposure ratio plot
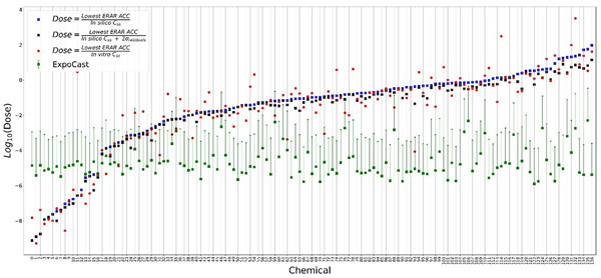
Figure 5 shows the range of OEDs derived for each chemical, based on ER-AR in vitro assay data, using Css values calculated based on the fraction unbound and clearance predictions from this work, along with exposure predictions. A total of 136 chemicals were active for ER or AR and had Css predictions. The OED value from the lowest assay potency (ACC) and using the in silico Css estimate is represented as a blue square dot. The exposure predictions are represented as green squares with error bars indicating the 95% upper confidence interval of the median exposure estimate. The red solid circles indicate OEDs derived using in vitro Css values but are only shown for chemicals that had in vitro fraction unbound and clearance data. The black squares indicate a conservative estimate of OEDs derived using in silico Css predictions and incorporating the standard error (=2σ of the residuals) due to uncertainty in Css predictions for the ER-AR dataset (Figure 4(a)).
Figure 5.
Bioactivity-exposure ratio (BER) plot to compare human oral equivalent doses (OEDs) and exposure predictions. Lowest and a conservative estimate of ToxCast derived OEDs (visualized as blue and black squares, respectively) were compared with the exposure estimates (visualized as median exposure value along with a 95% upper confidence interval in green). The plot is ordered by lowest ToxCast OED estimate. This analysis allows for the generation of a BER plot that compares hazard to exposure estimates within a high-throughput risk assessment framework to aid chemical screening and risk-prioritization.
About 92% (121/136) of the chemicals have a hazard point estimate (lowest OED value from in vitro assays, blue squares) higher than the upper 95% confidence interval (CI) of the exposure estimate. After adjusting for uncertainty, (black squares), about 91% (120/136) of chemicals have an OED higher than the upper 95% CI exposure estimate. For chemicals with in vitro OED estimates, 85% (120/136) have an OED higher than the upper 95% CI exposure estimate, leaving 16 that would be prioritized for follow-up based on an overlap between the OED and the exposure estimate. Of these, the in silico estimates identified 14 of the 16. In general, the chemicals on the left side of the plot with overlapping exposure and OED estimates are naturally occurring hormones or pharmaceuticals that are intended to occur at levels that are bioactive. A complete list of chemicals and the corresponding data are available in supplemental information (3.ERAR_BERPlot-Data.csv).
Overall the novelty of this work lies in (1) the development of open-source QSAR models for fub and Clint using a simple descriptor space and a rich chemical dataset, allowing prediction of pharmacokinetics for thousands of chemicals for which experimental data is not available, (2) incorporation of uncertainty due to the source of physicochemical properties in Css calculations in the estimation of OEDs to allow for a conservative comparison with exposure predictions resulting in higher confidence, and (3) extending the ability to prioritize large numbers of data-poor chemicals using in silico predictions in an effort to ease the transition from hazard-based prioritization to exposure related risk assessment. The model of fub shows higher predictivity than does that for Clint, but the Clint model developed here has similar predictivity to the commercial model it was compared to. The model for Css combines fub, Clint and physicochemical parameters, which are also typically the result of QSAR models. A recent analysis using new experimental fub and Clint measurements characterizes the uncertainty in the observed data and estimates the coefficient of variation for uncertainty as 0.4 for fub and 0.3 for Clint data [Wambaugh submitted 2019]. Both this new analysis and that shown here indicates that Css is less sensitive to uncertainty in Clint than to the other inputs (fub, pKa, logP). A final important point is illustrated by Figure 5 and the associated discussion. The uncertainties associated with using the in silico Css values are on the same order of magnitude as the uncertainties in the exposure predictions, but the BER (minimum OED, uncertainty included/maximum exposure) are often large compared with the individual component uncertainties. This means that in general, classifications of chemicals into those with BER > or < 1 are relatively accurate. 16 of 18 chemicals with exposure ratio<1 using in vitro TK parameter values were identified with in silico TK parameter values. This indicates that the models developed here for fub and Clint can provide useful input for efforts to prioritize thousands of chemicals lacking the appropriate experimental data.
Supplementary Material
Acknowledgments:
This work was supported in part by an appointment to the ORISE participant research program supported by an interagency agreement between the US EPA and DOE.
Footnotes
Disclaimer: The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Supporting information: Detailed information on methods and some relevant results to aid in the understanding of the manuscript are provided in the supplemental file Supplemental-Methods&Results.docx. The datasets, final model details and codes are provided as Supplemental.zip.
References
- 1.Council NR, Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press: Washington, DC, 2007; p 216. [Google Scholar]
- 2.Attene-Ramos MS; Miller N; Huang R; Michael S; Itkin M; Kavlock RJ; Austin CP; Shinn P; Simeonov A; Tice RR; Xia M, The Tox21 robotic platform for the assessment of environmental chemicals – from vision to reality. Drug Discovery Today 2013, 18 (15-16), 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins FS; Gray GM; Bucher JR, Toxicology. Transforming environmental health protection. Science 2008, 319 (5865), 906–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H; Xia M; Austin CP; Huang R, Paradigm shift in toxicity testing and modeling. AAPS J 2012, 14 (3), 473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dix DJ; Houck KA; Martin MT; Richard AM; Setzer RW; Kavlock RJ, The ToxCast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicological Sciences 2007, 95 (1), 5–12. [DOI] [PubMed] [Google Scholar]
- 6.Thomas RS; Philbert MA; Auerbach SS; Wetmore BA; Devito MJ; Cote I; Rowlands JC; Whelan MP; Hays SM; Andersen ME; Meek ME; Reiter LW; Lambert JC; Clewell HJ; Stephens ML; Zhao QJ; Wesselkamper SC; Flowers L; Carney EW; Pastoor TP; Petersen DD; Yauk CL; Nong A, Incorporating New Technologies Into Toxicity Testing and Risk Assessment: Moving From 21st Century Vision to a Data-Driven Framework. Toxicological Sciences 2013, 136 (1), 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wetmore BA; Wambaugh JF; Ferguson SS; Sochaski MA; Rotroff DM; Freeman K; Clewell HJ; Dix DJ; Andersen ME; Houck KA; Allen B; Judson RS; Singh R; Kavlock RJ; Richard AM; Thomas RS, Integration of Dosimetry, Exposure, and High-Throughput Screening Data in Chemical Toxicity Assessment. Toxicological Sciences 2012, 125 (1), 157–174. [DOI] [PubMed] [Google Scholar]
- 8.Rotroff DM; Wetmore BA; Dix DJ; Ferguson SS; Clewell HJ; Houck KA; LeCluyse EL; Andersen ME; Judson RS; Smith CM; Sochaski MA; Kavlock RJ; Boellmann F; Martin MT; Reif DM; Wambaugh JF; Thomas RS, Incorporating Human Dosimetry and Exposure into High-Throughput In Vitro Toxicity Screening. Toxicological Sciences 2010, 117 (2), 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wetmore BA; Wambaugh JF; Ferguson SS; Li L; Clewell HJ 3rd; Judson RS; Freeman K; Bao W; Sochaski MA; Chu TM; Black MB; Healy E; Allen B; Andersen ME; Wolfinger RD; Thomas RS, Relative impact of incorporating pharmacokinetics on predicting in vivo hazard and mode of action from high-throughput in vitro toxicity assays. Toxicol Sci 2013, 132 (2), 327–46. [DOI] [PubMed] [Google Scholar]
- 10.Blaauboer BJ, Biokinetic modeling and in vitro-in vivo extrapolations. J Toxicol Environ Health B Crit Rev 2010, 13 (2-4), 242–52. [DOI] [PubMed] [Google Scholar]
- 11.Judson R; Houck K; Martin M; Knudsen T; Thomas RS; Sipes N; Shah I; Wambaugh J; Crofton K, In Vitroand Modelling Approaches to Risk Assessment from the U.S. Environmental Protection Agency ToxCast Programme. Basic & Clinical Pharmacology & Toxicology 2014, 115 (1), 69–76. [DOI] [PubMed] [Google Scholar]
- 12.Sipes NS; Wambaugh JF; Pearce R; Auerbach SS; Wetmore BA; Hsieh J-H; Shapiro AJ; Svoboda D; DeVito MJ; Ferguson SS, An Intuitive Approach for Predicting Potential Human Health Risk with the Tox21 10k Library. Environmental Science & Technology 2017, 51 (18), 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell SM; Chang X; Wambaugh JF; Allen DG; Bartels M; Brouwer KLR; Casey WM; Choksi N; Ferguson SS; Fraczkiewicz G; Jarabek AM; Ke A; Lumen A; Lynn SG; Paini A; Price PS; Ring C; Simon TW; Sipes NS; Sprankle CS; Strickland J; Troutman J; Wetmore BA; Kleinstreuer NC, In vitro to in vivo extrapolation for high throughput prioritization and decision making. Toxicology in Vitro 2018, 47, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters NJ; Jones R; Williams G; Sohal B, Validation of a Rapid Equilibrium Dialysis Approach for the Measurement of Plasma Protein Binding. Journal of Pharmaceutical Sciences 2008, 97 (10), 4586–4595. [DOI] [PubMed] [Google Scholar]
- 15.Patlewicz G; Ball N; Booth ED; Hulzebos E; Zvinavashe E; Hennes C, Use of category approaches, read-across and (Q)SAR: General considerations. Regulatory Toxicology and Pharmacology 2013, 67 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 16.Patlewicz G, Read-across approaches - misconceptions, promises and challenges ahead. ALTEX 2014, 31 (4), 387–396. [DOI] [PubMed] [Google Scholar]
- 17.Patlewicz G; Roberts DW; Aptula A; Blackburn K; Hubesch B, Workshop: Use of “read-across” for chemical safety assessment under REACH. Regulatory Toxicology and Pharmacology 2013, 65 (2), 226–228. [DOI] [PubMed] [Google Scholar]
- 18.Ball N, Toward Good Read-Across Practice (GRAP) guidance. ALTEX 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enoch SJ, Chemical Category Formation and Read-Across for the Prediction of Toxicity. In Recent advances in QSAR studies: Methods and applications., Springer; Netherlands: 2010; pp 209–219. [Google Scholar]
- 20.Patlewicz G; Helman G; Pradeep P; Shah I, Navigating through the minefield of read-across tools: A review of in silico tools for grouping. Computational Toxicology 2017, 3, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ECHA, 2008. Guidance on information requirements and chemical safety assessment. Chapter R.6: QSARs and grouping of chemicals.
- 22.Dudek A; Arodz T; Galvez J, Computational Methods in Developing Quantitative Structure-Activity Relationships (QSAR): A Review. Combinatorial Chemistry & High Throughput Screening 2006, 9 (3), 213–228. [DOI] [PubMed] [Google Scholar]
- 23.Basant N; Gupta S; Singh KP, Predicting binding affinities of diverse pharmaceutical chemicals to human serum plasma proteins using QSPR modelling approaches. SAR and QSAR in Environmental Research 2016, 27 (1), 67–85. [DOI] [PubMed] [Google Scholar]
- 24.Ghafourian T; Amin Z, QSAR models for the prediction of plasma protein binding. Bioimpacts 2013, 3 (1), 21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingle BL; Veber BC; Nichols JW; Tornero-Velez R, Informing the Human Plasma Protein Binding of Environmental Chemicals by Machine Learning in the Pharmaceutical Space: Applicability Domain and Limits of Predictability. Journal of Chemical Information and Modeling 2016, 56 (11), 2243–2252. [DOI] [PubMed] [Google Scholar]
- 26.Sun L; Yang H; Li J; Wang T; Li W; Liu G; Tang Y, In Silico Prediction of Compounds Binding to Human Plasma Proteins by QSAR Models. ChemMedChem 2017. [DOI] [PubMed] [Google Scholar]
- 27.Votano JR; Parham M; Hall LM; Hall LH; Kier LB; Oloff S; Tropsha A, QSAR modeling of human serum protein binding with several modeling techniques utilizing structure-information representation. J Med Chem 2006, 49 (24), 7169–81. [DOI] [PubMed] [Google Scholar]
- 28.Yamazaki K; Kanaoka M, Computational prediction of the plasma protein-binding percent of diverse pharmaceutical compounds. J Pharm Sci 2004, 93 (6), 1480–94. [DOI] [PubMed] [Google Scholar]
- 29.Zhu XW; Sedykh A; Zhu H; Liu SS; Tropsha A, The Use of Pseudo-Equilibrium Constant Affords Improved QSAR Models of Human Plasma Protein Binding. Pharmaceut Res 2013, 30 (7), 1790–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekins S; Obach RS, Three-dimensional quantitative structure activity relationship computational approaches for prediction of human in vitro intrinsic clearance. J Pharmacol Exp Ther 2000, 295 (2), 463–73. [PubMed] [Google Scholar]
- 31.Pirovano A; Brandmaier S; Huijbregts MAJ; Ragas AMJ; Veltman K; Hendriks AJ, QSARs for estimating intrinsic hepatic clearance of organic chemicals in humans. Environmental Toxicology and Pharmacology 2016, 42, 190–197. [DOI] [PubMed] [Google Scholar]
- 32.Yamagata T; Zanelli U; Gallemann D; Perrin D; Dolgos H; Petersson C, Comparison of methods for the prediction of human clearance from hepatocyte intrinsic clearance for a set of reference compounds and an external evaluation set. Xenobiotica 2016, 47 (9), 741–751. [DOI] [PubMed] [Google Scholar]
- 33.Wambaugh JF; Wetmore BA; Pearce R; Strope C; Goldsmith R; Sluka JP; Sedykh A; Tropsha A; Bosgra S; Shah I; Judson R; Thomas RS; Setzer RW, Toxicokinetic Triage for Environmental Chemicals. Toxicological Sciences 2015, 147 (1), 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wambaugh JF; Wang A; Dionisio KL; Frame A; Egeghy P; Judson R; Setzer RW, High Throughput Heuristics for Prioritizing Human Exposure to Environmental Chemicals. Environmental Science & Technology 2014, 48 (21), 12760–12767. [DOI] [PubMed] [Google Scholar]
- 35.Cohen Hubal EA; Richard A; Aylward L; Edwards S; Gallagher J; Goldsmith M-R; Isukapalli S; Tornero-Velez R; Weber E; Kavlock R, Advancing Exposure Characterization for Chemical Evaluation and Risk Assessment. Journal of Toxicology and Environmental Health, Part B 2010, 13 (2-4), 299–313. [DOI] [PubMed] [Google Scholar]
- 36.Paixão P; Gouveia LF; Morais JAG, Prediction of the human oral bioavailability by using in vitro and in silico drug related parameters in a physiologically based absorption model. International Journal of Pharmaceutics 2012, 429 (1-2), 84–98. [DOI] [PubMed] [Google Scholar]
- 37.McGinnity DF, Evaluation of Fresh and Cryopreserved Hepatocytes as in Vitro Drug Metabolism Tools for the Prediction of Metabolic Clearance. Drug Metabolism and Disposition 2004, 32 (11), 1247–1253. [DOI] [PubMed] [Google Scholar]
- 38.Tonnelier A; Coecke S; Zaldívar J-M, Screening of chemicals for human bioaccumulative potential with a physiologically based toxicokinetic model. Arch Toxicol 2011, 86 (3), 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata Y, Prediction of Hepatic Clearance and Availability by Cryopreserved Human Hepatocytes: An Application of Serum Incubation Method. Drug Metabolism and Disposition 2002, 30 (8), 892–896. [DOI] [PubMed] [Google Scholar]
- 40.Obach RS; Lombardo F; Waters NJ, Trend Analysis of a Database of Intravenous Pharmacokinetic Parameters in Humans for 670 Drug Compounds. Drug Metabolism and Disposition 2008, 36 (7), 1385–1405. [DOI] [PubMed] [Google Scholar]
- 41.Obach RS, Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: An examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos 1999, 27 (11), 1350–9. [PubMed] [Google Scholar]
- 42.Ito K; Houston JB, Comparison of the Use of Liver Models for Predicting Drug Clearance Using in Vitro Kinetic Data from Hepatic Microsomes and Isolated Hepatocytes. Pharmaceut Res 2004, 21 (5), 785–792. [DOI] [PubMed] [Google Scholar]
- 43.Naritomi Y, Utility of Hepatocytes in Predicting Drug Metabolism: Comparison of Hepatic Intrinsic Clearance in Rats and Humans in Vivo and in Vitro. Drug Metabolism and Disposition 2003, 31 (5), 580–588. [DOI] [PubMed] [Google Scholar]
- 44.Lau YY, Development of a Novel in Vitro Model to Predict Hepatic Clearance Using Fresh, Cryopreserved, and Sandwich-Cultured Hepatocytes. Drug Metabolism and Disposition 2002, 30 (12), 1446–1454. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt W, General approach for the calculation of tissue to plasma partition coefficients. Toxicology in Vitro 2008, 22 (2), 457–467. [DOI] [PubMed] [Google Scholar]
- 46.Wetmore BA; Wambaugh JF; Allen B; Ferguson SS; Sochaski MA; Setzer RW; Houck KA; Strope CL; Cantwell K; Judson RS; LeCluyse E; Clewell HJ; Thomas RS; Andersen ME, Incorporating High-Throughput Exposure Predictions With Dosimetry-AdjustedIn VitroBioactivity to Inform Chemical Toxicity Testing. Toxicological Sciences 2015, 148 (1), 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearce RG; Setzer RW; Strope CL; Sipes NS; Wambaugh JF, httk: R Package for High-Throughput Toxicokinetics. J Stat Softw 2017, 79 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richard AM; Judson RS; Houck KA; Grulke CM; Volarath P; Thillainadarajah I; Yang C; Rathman J; Martin MT; Wambaugh JF; Knudsen TB; Kancherla J; Mansouri K; Patlewicz G; Williams AJ; Little SB; Crofton KM; Thomas RS, ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chemical Research in Toxicology 2016, 29 (8), 1225–1251. [DOI] [PubMed] [Google Scholar]
- 49.Huang R; Sakamuru S; Martin MT; Reif DM; Judson RS; Houck KA; Casey W; Hsieh JH; Shockley KR; Ceger P; Fostel J; Witt KL; Tong W; Rotroff DM; Zhao T; Shinn P; Simeonov A; Dix DJ; Austin CP; Kavlock RJ; Tice RR; Xia M, Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci Rep 2014, 4, 5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Judson RS; Magpantay FM; Chickarmane V; Haskell C; Tania N; Taylor J; Xia M; Huang R; Rotroff DM; Filer DL; Houck KA; Martin MT; Sipes N; Richard AM; Mansouri K; Setzer RW; Knudsen TB; Crofton KM; Thomas RS, Integrated Model of Chemical Perturbations of a Biological Pathway Using 18In VitroHigh-Throughput Screening Assays for the Estrogen Receptor. Toxicological Sciences 2015, 148 (1), 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richard AM; Williams CR, Distributed structure-searchable toxicity (DSSTox) public database network: a proposal. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2002, 499 (1), 27–52. [DOI] [PubMed] [Google Scholar]
- 52.Young D; Martin T; Venkatapathy R; Harten P, Are the Chemical Structures in Your QSAR Correct? QSAR & Combinatorial Science 2008, 27 (11-12), 1337–1345. [Google Scholar]
- 53.PubChem. https://pubchem.ncbi.nlm.nih.gov/help.html.
- 54.Yang C; Tarkhov A; Marusczyk J; Bienfait B; Gasteiger J; Kleinoeder T; Magdziarz T; Sacher O; Schwab CH; Schwoebel J; Terfloth L; Arvidson K; Richard A; Worth A; Rathman J, New Publicly Available Chemical Query Language, CSRML, To Support Chemotype Representations for Application to Data Mining and Modeling. Journal of Chemical Information and Modeling 2015, 55 (3), 510–528. [DOI] [PubMed] [Google Scholar]
- 55.Mansouri K; Grulke CM; Judson RS; Williams AJ, OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminform 2018, 10 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chemistry Development Kit. https://cdk.github.io/.
- 57.Steinbeck C; Han Y; Kuhn S; Horlacher O; Luttmann E; Willighagen E, The Chemistry Development Kit (CDK): An Open-Source Java Library for Chemo- and Bioinformatics. Journal of Chemical Information and Computer Sciences 2003, 43 (2), 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berthold MR; Cebron N; Dill F; Gabriel TR; Kötter T; Meinl T; Ohl P; Thiel K; Wiswedel B, KNIME - the Konstanz information miner: version 2.0 and beyond. ACM SIGKDD Explorations Newsletter 2009, 11 (1), 26. [Google Scholar]
- 59.Yap CW, PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. Journal of Computational Chemistry 2011, 32 (7), 1466–1474. [DOI] [PubMed] [Google Scholar]
- 60.Grulke CM; Williams AJ; Thillanadarajah I; Richard AM, EPA’s DSSTox database: History of development of a curated chemistry resource supporting computational toxicology research. Computational Toxicology 2019, 12, 100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tibshirani R, Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2011, 73 (3), 273–282. [Google Scholar]
- 62.Smola AJ; Schölkopf B, A tutorial on support vector regression. Statistics and Computing 2004, 14 (3), 199–222. [Google Scholar]
- 63.Cortes C; Vapnik V, Support-Vector Networks. Mach Learn 1995, 20 (3), 273–297. [Google Scholar]
- 64.Breiman L, Random forests. Mach Learn 2001, 45 (1), 5–32. [Google Scholar]
- 65.Schapire RE; Freund Y, Foundations of Machine Learning. Adapt Comput Mach Le 2012, 23–52. [Google Scholar]
- 66.McCulloch WS; Pitts W, A logical calculus of the ideas immanent in nervous activity. The Bulletin of Mathematical Biophysics 1943, 5 (4), 115–133. [PubMed] [Google Scholar]
- 67.Agresti A, Categorical data analysis. 3rd ed.; Wiley: Hoboken, NJ, 2013; p xvi, 714 p. [Google Scholar]
- 68.Pradeep P; Povinelli RJ; White S; Merrill SJ, An ensemble model of QSAR tools for regulatory risk assessment. Journal of Cheminformatics 2016, 8 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hornik K The R FAQ. https://CRAN.R-project.org/doc/FAQ/R-FAQ.html.
- 70.Ring CL; Pearce RG; Setzer RW; Wetmore BA; Wambaugh JF, Identifying populations sensitive to environmental chemicals by simulating toxicokinetic variability. Environ Int 2017, 106, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mansouri K; Grulke CM; Judson RS; Williams AJ, OPERA models for predicting physicochemical properties and environmental fate endpoints. Journal of Cheminformatics 2018, 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strope CL; Mansouri K; Clewell HJ; Rabinowitz JR; Stevens C; Wambaugh JF, High-throughput in-silico prediction of ionization equilibria for pharmacokinetic modeling. Sci Total Environ 2018, 615, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kleinstreuer NC; Ceger P; Watt ED; Martin M; Houck K; Browne P; Thomas RS; Casey WM; Dix DJ; Allen D; Sakamuru S; Xia M; Huang R; Judson R, Development and Validation of a Computational Model for Androgen Receptor Activity. Chemical Research in Toxicology 2016, 30 (4), 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedregosa F; Varoquaux G; Gramfort A; Michel V; Thirion B; Grisel O; Blondel M; Prettenhofer P; Weiss R; Dubourg V; Vanderplas J; Passos A; Cournapeau D; Brucher M; Perrot M; Duchesnay E, Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011, 12, 2825–2830. [Google Scholar]
- 75.Python Software Foundation. Python Language Reference, version 2.7. Available at http://www.python.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.