Abstract
The challenge of constructing surfaces with nanostructured chemical functionality is central to many areas of biology and biotechnology. This protocol describes the steps required for performing molecular printing using polymer pen lithography (PPL), a cantilever-free scanning probe-based technique that can generate sub-100-nm molecular features in a massively parallel fashion. To illustrate how such molecular printing can be used for a variety of biologically relevant applications, we detail the fabrication of the lithographic apparatus and the deposition of two materials, an alkanethiol and a polymer onto a gold and silicon surface, respectively, and show how the present approach can be used to generate nanostructures composed of proteins and metals. Finally, we describe how PPL enables researchers to easily create combinatorial arrays of nanostructures, a powerful approach for high-throughput screening. A typical protocol for fabricating PPL arrays and printing with the arrays takes 48–72 h to complete, including two overnight waiting steps.
INTRODUCTION
The desire to fabricate molecular patterns with sub-100-nm resolution is increasingly widespread for many applications, including ultrahigh density bioassays, tissue engineering, and studies of cell surface interactions with chemical patterns on the length scale of focal adhesions1,2. The fabrication of these patterns is a very challenging problem, because many of the advances in lithography have been driven by the semiconductor industry, which focuses on the use of a limited set of hard materials. Although photolithography and electron beam lithography (EBL) are staples of conventional nanofabrication, they have attracted limited attention from chemists and biologists, as they rely on harsh processing conditions, which limit their compatibility with many biomaterials, and require complex, time-consuming processing1-3.
A common approach for converting these resist-based techniques (here, ‘resist-based’ refers to lithographic techniques in which a sacrificial polymer layer known as a ‘resist’ is used to transfer patterns onto a desired substrate) into one that is more amenable to patterning molecules is to fabricate a master using photolithography or EBL and mold an elastomeric stamp that can be used to transfer molecules to a surface4-8. This technique, known as microcontact printing (μCP), is well suited for stamping duplicates of sub-micrometer-scale features. The recently developed scanning probe-based technique termed PPL9, which is described in this protocol, is distinct from μCP in two important ways: (i) it uses a rigid backing layer, which enforces planarity across the tip array and enables all of the printing tips to make simultaneous and gentle contact for dot-matrix style printing of 100-nm-scale features, and (ii) it uses a scanning probe platform that enables arbitrary patterns to be written in a mask-free fashion.
Ink-jet printing has also been used to fabricate arrays of biomolecules10,11, but resolution, feature positioning and compatibility with proteins are key limitations of this technique. Indeed, although sub-500-nm resolution has been demonstrated with this approach12, formation of satellite droplets10 hampers the ability to precisely control feature position, and proteins are often too fragile to withstand the severe conditions within an ink-jet printer. Another method for transferring proteins is laser writing, but the resolution of this technique is above the 1-μm scale10.
Since the invention of dip-pen nanolithography (DPN) in 1999, it has been known that a scanning probe can be used as a very-high-resolution molecular patterning tool. However, the main barrier to the widespread application of this capability in bio-engineering is throughput, as it would take prohibitively long to pattern centimeter-scale samples with a single probe. Although early efforts to parallelize cantilever-based DPN for large-scale patterning were successful, they relied on expensive and delicate cantilever arrays13,14. In PPL, however, it is not cantilevers that are used to make hard probes mechanically compliant; instead, the probes themselves are deformable and are held in a plane by a hard transparent slide (e.g., glass)15,16, thus markedly decreasing the cost and complexity of the probe array.
The critical advance introduced by PPL is, therefore, the cantilever-free architecture: nanoscale tips rest on an elastomeric base that adheres to a rigid backing layer, which enforces planarity across the tip array, while the elastomeric base ensures that all tips can be brought into simultaneous contact with the sample. This setup is in contrast to previous experiments in which arrays of deformable polydimethylsiloxane (PDMS) pens without rigid backing layers were used to deposit molecular features17,18; these experiments, however, did not provide the fine and uniform control over contact pressure that is required to pattern large areas with sub-500-nm features. This control in cantilever-free probe arrays is provided by the piezo stages of the scanning probe instrument, which is another key advance when using cantilever-free arrays to pattern surfaces. In the PPL setup, it is the Z piezo that controls the contact pressure and the X and Y piezos that enable researchers to implement arbitrary pattern design and execution. In addition to being the crucial advance in PPL, the cantilever-free architecture has been the focus of considerable research, and now dual-elastomer tips19, hard Si tips20,21, hard Si tips with tunable spring constants22 and opaque tips with nanoscale apertures23 have been developed.
The ability that PPL confers to researchers to quickly redesign and print new patterns means that this technique is poised to fill an important role in the rapid prototyping and small-batch manufacturing of large-scale molecularly patterned surfaces with sub-100-nm resolution. In particular, the crucial attributes of PPL that are most desirable for a lithographic tool for biological applications are listed below. (i) Resolution: by using the molecular transport from the tip to a surface, sub-100-nm features12,24 can be directly deposited with no restriction on how closely features can be placed. (ii) Registration: as feature locations are controlled by precise piezo controllers, sub-nm placement of features relative to each other is easily attainable. (iii) Throughput: arrays as large as 11 million pens have been operated simultaneously to generate highly uniform patterns. (iv) Mask-free: because the features are written individually, arbitrary patterns can be generated in a massively parallel fashion12,25. (v) Combinatorial synthesis: by slightly tilting the pen array to controllably modulate the force applied to each pen, it is possible to synthesize a gradient of feature sizes. (vi) Simplicity: patterning experiments are performed under ambient conditions, thus obviating the need for a clean room or complicated postprocessing2,26. (vii) Materials generality: PPL has already been used to deposit a wide variety of materials onto a wide variety of both hydrophilic and hydrophobic substrates.
Initially, research using PPL focused on the deposition of alkanethiols, such as 16-mercaptohexadecanoic acid (MHA), onto thin films of gold9,19,25,27,28. PPL has also been used to print polymers29-33, lipids34 and proteins35 onto both hydrophilic and hydrophobic surfaces. The case of polymers is of particular interest, as various polymers have been used as matrices to deliver small molecules30,31 and nanoparticles29, as well as reaction vessels to direct the synthesis of nanoparticles32,33. These nanoparticles were used to assemble patterns of single proteins36, which could be used to probe the limits of cell surface interactions. The ability of PPL to deliver multiple reagents simultaneously has also been used to facilitate organic reactions on surfaces, including the Click reaction30 and Staudinger ligation31; furthermore, the controlled application of pressure by PPL tip arrays has been used to induce a force-accelerated Diels-Alder reaction for patterning graphene37. The range of ink compositions and substrate surface energies that are accessible for patterning by PPL is quite large, and the only limit to potential combinations is the need for a relative difference in adhesion between the tip and the surface. Although patterning by PPL is unlikely to work for any ink that strongly associates with PDMS, such as silanes, alternative chemistries are likely to exist that can be used to deliver the functionality present in such PDMS-adhering inks to the target substrate. It is worth mentioning, however, that oil-based inks have not been studied and complications could arise from the permeability of PDMS to most nonpolar solvents.
PPL has also been applied to the generation of biomolecule arrays for cell biology27. Indeed, the use of molecular printing techniques, such as DPN, to generate arrays of biomolecules for studying cell surface interactions has been extensively studied1,26,38-41, and PPL could easily be performed under the conditions optimized for DPN in order to deposit DNA26, proteins39,40 or lipids41. One recent study used PPL to generate combinatorial arrays of the extracellular matrix protein fibronectin for studying the differentiation of mesenchymal stem cells (MSCs) into osteoblasts27. The authors used the deformable nature of the elastomeric pens to generate a gradient of fibronectin feature sizes in order to determine the optimum feature size for MSC adhesion. This ability to tilt the plane of the array with respect to the substrate and thereby systematically vary the feature size is unique to PPL and could be extremely useful for rapid screening. For instance, in the aforementioned recent study27, the optimal feature size for cell binding was unknown, and tilting the PPL array enabled the researchers to address this question in a single experiment without the need to fabricate new masks or masters. After testing a range of feature sizes, the same arrays were used to generate large-area patterns of uniform features to monitor the differentiation of MSCs into osteoblasts using immunofluorescence and reverse-transcription (RT)-PCR27.
The intent of the above discussion was to illustrate the capabilities of PPL and opportunities for using it as a tool in bioengineering. Indeed, given the wide range of inks and substrates that have been used for molecular printing, the toolbox of PPL can be viewed as essentially limitless. The purpose of this protocol is to show researchers how to easily implement PPL to create molecular patterns on surfaces for applications that include label-free biomolecule assays and platforms for studying fundamental cell surface interactions.
Experimental design
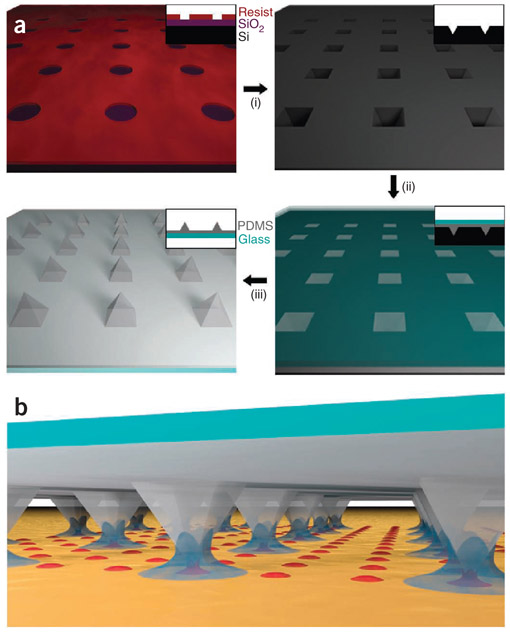
The basic process of fabricating PPL arrays and using them to deposit molecules is depicted schematically in Figure 1. Performing molecular printing with PPL consists of three major stages: master fabrication (i in Fig. 1a), tip array fabrication (ii and iii in Fig. 1a) and patterning of nanostructures (Fig. 1b). The nanostructures generated may consist of molecules (e.g., alkanethiols or proteins) or hard materials (e.g., metals or oxides). After deposition, these structures can also be used to template the assembly of biomolecules, such as proteins26,38 and viruses42. In this protocol, the direct deposition of molecules will be detailed, as well as a method for functionalizing the molecular patterns with proteins. In addition, the deposition of a polymer matrix for synthesizing metallic nanoparticles will be described (Box 1).
Figure 1 ∣.
Polymer pen lithography (PPL) protocol. (a) Schematic of the major steps in the PPL tip array fabrication process, which begins by using photolithography to define patterns on a silicon wafer. These patterns are transferred into the underlying wafer (i) using a series of etching steps; the resulting inverted pyramid array serves as a master for molding PPL tips. These tips are made by pouring h-PDMS pre-polymer into the mold and placing a glass slide on top (ii). These arrays are removed from the master to yield a PPL tip array, which consists of pyramidal h-PDMS pens adhered to a rigid glass backing layer (iii). (b) After coating the arrays in ink, they can be used to deposit patterns by molecular printing. Note that the PPL pens depicted here are not to scale; in a typical array, the pens are 40 μm wide at the base, ~30 μm tall and 150 μm apart.
Box 1 ∣. Deposition of metallic nanoparticles on a silicon substrate.
● TIMING 12–16 h
1. Prepare a hydrophobic Si surface by placing a Si wafer in a desiccator with 4 ml of a 50:50 solution of HMDS:toluene. Pull vacuum on the desiccators until the toluene begins to boil, and then leave the wafer under static vacuum overnight.
2. Prepare the ink for nanoparticle synthesis by stirring 0.5% (wt/vol) PEO-b-P2VP in NANOpure H2O for 12 h; follow this by adding HAuCl4 and stirring for another 24 h. The HAuCl4 can be mixed in molar ratios ranging from 2:1 pyridine:Au to 128:1. For the study used as a representative example in this protocol, a 2:1 pyridine:Au ratio was used.
3. Ink the PPL array by drop-casting 100 μl of ink onto the array, and then spin-coat at 2,000 r.p.m. for 60 s, with a ramp rate of 1,000 r.p.m. s−1.
4. Mount the PPL array in the AFM, level it and design the pattern according to Steps 34–38. For pattern deposition, follow Steps 39–41, but use RH values in the 75–100% range. The patterns shown in Figure 6c,d were printed at RH = 80% with dwell times ranging from 0.3 to 1 s.
5. After polymer patterning, remove the substrate and place it in a tube furnace under flowing Ar at 300 standard cubic centimeters per min. While the substrate is in the tube furnace, subject the samples to the following recipe: ramp to 150 °C, anneal at 150 °C for 1 h, cool to room temperature for 30 min, ramp to 500 °C, anneal for 2 h and then cool to room temperature. In this protocol, the first step serves to reduce and coalesce the nanoparticles within the polymer, and the second step burns off the polymer. SEM images of the final patterns are shown in Figure 6d.
Design of the photomask.
Photomasks for fabricating a Si master comprise a 2D array of circular holes whose diameters are substantially >1 μm and can thus be fabricated by conventional photolithography. Photomask design is quite simple, as only two parameters are varied: tip spacing and hole diameter, where the hole diameter is approximately equal to the pyramid edge length. These photomasks can be designed by graphical drawing programs such as Autodesk AutoCAD and DesignCAD. After design, they are given to a commercial design service, such as HTA Photomask, which will then fabricate a chromium-coated fused silica or quartz mask. Each hole in the photomask corresponds to a pyramidal pen in the tip array, where the center-to-center spacing between holes in the photomask corresponds to the tip-to-tip spacing in the resulting pen array, which, in turn, corresponds to the spacing between patterns generated from each tip on the surface. Typically, hole diameters in the 20–40-μm range are chosen, with the pen pitch in the 40–400-μm range. The tip height, h, is determined by the edge length of the pyramid, l, according to the geometrical relation . In the experiment described in this protocol, the hole diameter was 40 μm and the pen-to-pen pitch was 150 μm.
It is important to note that the pyramids can be defined by using a mask with either square holes or circular holes. If square holes are used, then care must be taken to align the crystal planes of the wafer to the edges of the mask to ensure uniform etching into sharp tips. Slight misalignment between the wafer flat and the square edges will result in asymmetric, line-shaped tips; to avoid this potential problem, we recommend using circular holes (where diameter corresponds to edge length).
Preparation of the master.
The master for PPL tips consists of an array of pyramidal pits etched into a Si wafer, which are prepared using conventional microfabrication techniques. The process begins by spin-coating photoresist onto a Si <100> wafer with a thermal oxide, and then using a mask aligner to selectively expose portions of the resist to define the pen size and spacing. After development of the resist, the patterns are transferred into the oxide, which serves as a hard mask for a subsequent anisotropic etch. This anisotropic etch, which etches the <100> face of silicon ~74 times faster than the <111> face43, defines the pyramidal pits that will be used to mold the elastomer into PPL tips. After etching, the remaining oxide mask is stripped from the wafer, a uniform oxide is regrown and the masters are vapor-coated with a fluorosilane to prevent elastomer adhesion during the tip fabrication process. This fluorosilanization allows the cured tip arrays to be easily peeled from the masters.
Fabrication of the pen arrays.
The pen arrays are fabricated by mixing the hard PDMS (h-PDMS)44 precursor and drop-casting it on the Si masters. Here we use h-PDMS instead of softer PDMS because h-PDMS represents a good compromise between tip deformability, rigidity and tip sharpness. A glass or quartz slide is then placed on the top of the h-PDMS, and the tip arrays are thermally cured. After curing, the tip array is slowly detached from the master. The master can subsequently be reused roughly 20–40 times for making more PPL tip arrays, and the array can be used for many cycles of molecular printing. Masters can no longer be used when too much h-PDMS residue has built up in the bottom of the pyramidal wells, because this residue prevents the formation of sharp tips.
Pattern writing.
The general process for patterning molecules with PPL consists of four key stages: (i) inking the array, (ii) leveling the array, (iii) designing the pattern and (iv) printing the pattern under controlled environmental conditions. After deposition, the patterns can be visualized by a variety of techniques, such as optical microscopy, atomic force microscopy (AFM) and scanning electron microscopy (SEM). The procedure begins by plasma-cleaning the tip array in order to promote ink wetting, followed by drop-casting or spin-coating of the ink. Drop-casting is recommended for volatile solvents, such as ethanol and acetonitrile, because drop-casting allows maximal ink loading on the tip arrays. For aqueous inks and liquid inks, spin-coating is recommended. Pen arrays can also be inked after leveling by inserting the PPL pens into an array of inkwells35, or by pressing the tips onto a ‘stamping pad’ that consists of dried ink on a silicon wafer34. Although these techniques enable nonuniform inking of the pen array, we recommend spin-coating as an inking strategy when uniform ink coverage is desired.
After inking, the PPL tip array is loaded into an AFM instrument, and the array is ‘leveled’ until it is co-planar with the substrate. Leveling is accomplished using an iterative method using either optical or force feedback, wherein the stage is tilted until all of the tips are simultaneously in contact with the surface. As the array is inked before leveling, the process of leveling can lead to undesired material transfer; to avoid ink transfer during this procedure, the relative humidity (RH) in the environmental chamber can be maintained at <10% during the leveling process. After leveling, the environmental chamber is programmed to the desired RH and temperature, and the pattern is designed while the chamber is equilibrating. As pattern design is an entirely computer-based process, it can be carried out at any point before patterning, and patterns can be saved for future use.
Patterns are written in a ‘dot-matrix’ style, in which the pen array is lowered into contact with the surface for a programmed ‘dwell time,’ raised several microns above the surface, moved to the position of the next element and lowered again to print the next feature. This process is repeated until the pattern is fully written. The relevant AFM parameters to control during patterning are the Z piezo extensions during deposition (Zext) and while moving between the elements (Zlift), as well as the XY-stage translation speed. Each pen can write thousands of features before re-inking is necessary, and in general no limit exists to how complex a pattern can be.
MATERIALS
REAGENTS
Reverse osmosis–purified H2O, >18 MΩ·cm (e.g., NANOpure) for cleaning and preparation of solutions
Ethanol (≥99.5%; Sigma-Aldrich, cat. no. 459844) for cleaning glass slides and preparation of solutions
Acetone (≥99.9%; Sigma-Aldrich, cat. no. 270725) for cleaning and photoresist liftoff
2-Propanol (99.9%; Sigma-Aldrich, cat. no. 650447) for cleaning and silicon etching
Shipley S1805 (MicroChem, cat. no. 10018321) photoresist
Developer MF-319 (MicroChem, cat. no. 10018042) for developing the photoresist after the exposure
4-inch <100> Si wafer (Nova Electronic Materials, cat. no. STK8414 ) with 5,000 Å thermal or wet oxide for photolithography ▲ CRITICAL It is extremely important to use a <100> wafer and not a <111> Si wafer, as the PPL master fabrication relies on etching chemistry43 that is selective for the <100> face of Si.
Buffered HF improved (Transene Etchants; pH = 5.0) for SiO2 etching ! CAUTION This reagent is corrosive and dangerous for the environment. It may be hazardous at high concentrations. Manipulate it inside a fume hood, wear a lab coat, use nitrile/latex gloves beneath neoprene gloves, and wear safety goggles while working with it.
Potassium hydroxide (KOH; ≥99.995% (metals basis); Sigma-Aldrich, cat. no. 306568) for Si etching ! CAUTION KOH is highly corrosive and dangerous for the environment. It may be hazardous at high concentrations. Manipulate this reagent inside a fume hood, and use gloves and wear safety goggles while working with it.
Heptadecafluoro-1,1,2,2-tetra(hydrodecyl)trichlorosilane (Gelest, cat. no. SIH5841.0) for hydrophobic coating of Si masters ! CAUTION This reagent is corrosive and dangerous for the environment. It may be hazardous at high concentrations. Manipulate it inside a fume hood, and use gloves and wear safety goggles while handling it.
Toluene (≥99.5%; Sigma-Aldrich, cat. no. 155004) for coating Si masters with hydrophobic silanes
(7–8% vinylmethylsiloxane)-dimethylsiloxane copolymer, trimethylsiloxy-terminated (Gelest, product code VDT-731) for making h-PDMS
Pt divinyltetramethyldisiloxane (Gelest, product code SIP 6831.2) for making h-PDMS
1,3,5,7-Tetramethyl-1,3,5,7-tetravinylcyclotetrasiloxane (Gelest, product no. SIT7900.0) for making h-PDMS
(25–35% methylhydrosiloxane)-dimethylsiloxane copolymer, trimethylsiloxane-terminated (Gelest, product no. HMS-301) for making h-PDMS
16-Mercaptohexadecanoic acid, (MHA; 90%; Sigma-Aldrich, cat. no. 448303) for forming a self-assembled monolayer (SAM) on Au surface
Thiourea (≥99.0%; Sigma-Aldrich, cat. no. T8656) for Au etching
Iron (III) nitrate nonahydrate (99.99%; Sigma-Aldrich, cat. no. 254223) for Au etching
Hydrochloric acid (HCl; 37% (vol/vol); Sigma-Aldrich, cat. no. 320331) for Au etching ! CAUTION HCl is highly corrosive and dangerous for the environment. Manipulate this reagent inside a fume hood, wear a lab coat, use nitrile/latex gloves and wear safety goggles while working with it.
Hexamethyldisilazane (HMDS; >99.9%; Sigma-Aldrich, cat. no. 379212) for modifying the surface energy of a Si wafer to facilitate polymer patterning
(1-Mercapto-11-undecyl)hexa(ethylene glycol), (90%; Sigma-Aldrich, cat. no. 675105) for backfilling Au surface after patterning to prevent nonspecific adsorption of proteins and cells
Cobalt(III) nitrate hexahydrate (Sigma-Aldrich, cat. no. 60832) for mediating protein adsorption to MHA
Solution of PBS (1×; Fisher Scientific, cat. no. BP2438-4) for making protein solutions
Human plasma fibronectin (1 mg ml−1, Millipore, cat. no. FC010), for immobilizing onto MHA patterns
Human anti-fibronectin produced in rabbit (Sigma-Aldrich, cat. no. F3648-5 ML) for immunofluorescence imaging
Goat anti-rabbit IgG (Invitrogen, cat. no. A11036) for immunofluorescence imaging
Poly(ethylene oxide)-b-poly(2-vinyl pyridine) (Polymer Source, PEO-b-P2VP MW = 2,800/1,500 g/mol) for metal nanoparticle patterning
Tetrachloroauric acid (HAuCl4; 99.999% trace metals; Sigma-Aldrich, cat. no. 254169) for metal nanoparticle patterning
Au substrate, prepared by physical vapor deposition (i.e., e-beam or thermal evaporation) onto a Si wafer or glass slide. Use of a Cr or Ti adhesion layer is highly recommended. When possible, substrate should be used within 1 week of deposition
EQUIPMENT
Mask aligner (MA6, Karl Suss)
Spin coater (Cee 200×, Brewer Science)
5 3/4 disposable Pasteur pipette (Fisher Scientific, cat. no. 13-678-20A)
Polystyrene tissue culture dish, 150 × 20 mm style (Falcon, cat. no. 353025)
Glass Petri dishes (VWR International, cat. no. 89000-302)
Glass beaker (250 ml; VWR International, cat. no. 13912-207)
Glass beaker (4 liters; VWR International, cat. no. 89090-524)
Syringes (1 ml; Becton Dickinson, cat. no. 309602, 309586)
Multichannel timer (VWR International, cat. no. 62344-641)
Plasma cleaner (PDC-001, Harrick Plasma)
Wafer tweezers (Ted Pella, cat. no. 5589)
Ceramic top hot plate stirrer with temperature control up to 500 °C (VWR International cat. no. 97042-714)
Magnetic stir bar (VWR International, cat. no. 58949-210)
Optical microscope (Axiovert 200 M; Carl Zeiss)
Glass vial (20 ml; VWR International, cat. no. 66022-060)
Vacuum desiccators (VWR International, cat. no. 89178-332)
Plain glass slides (25 mm × 75 mm × 1 mm; VWR International, cat. no. 48300-025)
Diamond scribes (Structure Probe, cat. no. 06004-AB)
Plastic weigh boats (140 mm × 140 mm × 25 mm; VWR International, cat. no. 89106-770)
Single-edge carbon steel razor blades (Ted Pella, cat. no. 121-82)
Patterning platform (Park XE-PPL, Park Systems)
Portable scale with sensitivity of 0.01 g (PS121, Ohaus)
Double-sided carbon tape (Ted Pella, cat. no. 16073)
Electron-beam evaporator (PVD-75, Kurt J. Lesker) loaded with Au pellets (99.99%; Plasmaterials, 1–3-mm random-size pieces), Cr chips (99.995%; Sigma-Aldrich, cat. no. 374849), and Ti pellets (99.995%; Kurt J. Lesker, part no. EVMTI45EXE-B)
Pistol grip nozzle (Ted Pella, cat. no. 800) and flexible air hose (Ted Pella, cat. no. 803), attached to a dry N2 line in a fume hood. If this is not available, a tank of N2 will suffice
Tube furnace (Lindberg/Blue M 1,100 °C tube furnace, Thermo Scientific)
Mass flow controller (M100B, MKS Instruments)
Mass flow controller power supply/readout (247D, MKS Instruments)
Tank of compressed Ar (ultra-high purity, Airgas)
REAGENT SETUP
Si etchant
Prepare a 30% (wt/vol) KOH solution consisting of 750 g of KOH in 2 liters of NANOpure H2O and 500 ml of 2-propanol. After preparation, the solution can be used for up to 12 h, although 2-propanol will evaporate and must be replenished every 1–2 h. After more than 12 h have passed, we recommend making a fresh solution.
h-PDMS precursor
Mix 500 g of (7–8% (wt/wt) vinylmethylsiloxane)-dimethylsiloxane co-polymer with 20 μl of Pt divinyl tetramethyl disiloxane and 344 μl of 1,3,5,7-tetramethyl-1,3,5,7-tetravinylcyclotetrasiloxane and stir for 3 d. After stirring, this solution is stable at room temperature (19–22 °C) for more than 12 months in a sealed container.
Alkanethiol ink solution
Prepare a 5 mM solution consisting of 14.4 mg of MHA in 10 ml of ethanol. This solution can be used for up to 3 months after storage at room temperature in a sealed glass container.
Au etchant
Mix equal volumes of aqueous solutions of 40 mM thiourea, 27 mM Fe(NO3)3·9H2O and 100 mM HCl. ▲ CRITICAL The etchant solution can be used for 1 d, after which it should be freshly re-made. The component solutions should be stored separately and are stable for ~6 months in sealed containers.
Au substrate
Prepare an Au-coated substrate by placing a clean Si <100> wafer into an e-beam evaporator, and pump the chamber down until the pressure is below ~5 × 10−7 Torr. Once this pressure is reached, first evaporate 2 nm of Cr at 0.2 Å s−1 onto the Si wafer, and then evaporate 20 nm of Au at 0.5 Å s−1. After successful evaporation, let the chamber cool for ~30 min before venting the chamber and removing the sample.
Alkanethiol backfilling solution
Dissolve 93.7 mg of (1-mercapto-11-undecyl)hexa(ethylene glycol) in 20 ml of ethanol to obtain a final concentration of 10 mM. This solution can be used for up to 3 months after storage in a sealed container at 4 °C.
Cobalt nitrate chelating solution
Prepare an ethanolic solution of 10 mM Co(NO3)3 by dissolving 36.6 mg of Co(NO3)3 in 20 ml of ethanol. This solution can be stored in a sealed container at room temperature and is stable for 3 months.
Fibronectin solution
Dilute the as-purchased stock solution from 1 mg ml−1 (1 ml) to 50 μg ml−1 by adding 20 ml of 1× PBS. This solution should be stored at 4 °C and is stable for up to 1 month.
Antibody solutions used for immunofluorescence labeling
Prepare dilute primary antibody solution by diluting 1:100 in 1× PBS from the as-purchased stock solution. Prepare dilute fluorescently labeled secondary antibody solution by diluting 1:250 in 1× PBS from the stock solution. Both dilute solutions should be stored at 4 °C for up to 2 months.
PROCEDURE
Defining the masters ● TIMING 2-4 h
1∣ Use CAD software to design the master and send the CAD file to a commercial design service, such as HTA Photomask, which will fabricate a chromium-coated fused silica or quartz mask for use in a clean room. The pattern used for demonstration purposes here consisted of 40-μm-diameter circular holes with a 150-μm pitch, grouped into squares with a 2-cm edge length. In total, seven arrays of holes were defined over a 4-inch Si wafer, and thus each wafer yields seven masters.
2∣ In a clean room, spin-coat a positive-tone photoresist, such as Shipley S1805, on a 4-inch <100> wafer. Cover the wafer in photoresist, taking care to avoid bubbles, and then spin it at 500 r.p.m. for 5 s at room temperature, followed immediately by 4,000 r.p.m. for 40 s using ramp speeds of 500 r.p.m. s−1 at room temperature.
3∣ Soft-bake the resist-covered wafer on a hot plate set to 115 °C for 80 s and let it cool to room temperature before exposure.
4∣ Expose the resist-coated wafer in a mask aligner for 6 s at 12 mW cm−2. This value represents a slight overexposure, because it is important to completely remove resist from the patterned areas. It is also important to note that the misalignment of the wafer crystal plane relative to the mask pattern will result in rotation of each pyramidal pen with respect to the pen array, but we have not found the exact alignment to be crucial.
5∣ Develop the patterns in the developer MF-319 for 60 s, rinse with NANOpure water for 10 s, and then dry the wafer under a stream of N2. Verify that the resist has been removed in the circular regions using an optical microscope.
∎ PAUSE POINT Photolithography-defined wafers can be stored in a container under ambient conditions for months.
Fabricating the masters ● TIMING 2–5 h
6∣ Expose the photolithography-defined wafers to air plasma at <200 mTorr for 2 min at 30 W to remove any residual photoresist in the patterned regions.
7∣ Fill a polystyrene tissue culture dish with enough buffered HF improved to cover the wafer. Place the photolithography-defined surface in the dish for 6 min to etch the 5,000 Å thermal oxide layer.
! CAUTION HF solutions are mildly corrosive and extremely toxic. Wear full protective clothing, including nitrile/latex gloves beneath neoprene gloves, a lab coat and safety glasses at all times while working with it, and work in a fume hood. Always pour HF solutions into plastic containers, and use Teflon-coated or carbon-coated tweezers to handle wafers.
8∣ Fill a polystyrene tissue culture dish with enough NANOpure H2O to cover the wafer. Place the wafer in the dish for 2 min. After removal from the dish, rinse off any remaining HF with NANOpure H2O, and then dry the wafer under a stream of N2.
9∣ Fill a glass Petri dish with enough acetone to cover the wafer. Place the wafer in the dish for 5 min to remove the patterned photoresist. Remove the wafer and rinse sequentially with acetone, NANOpure H2O and 2-propanol. After rinsing, blow-dry the wafer under a stream of N2.
▲ CRITICAL STEP Native oxides will form quickly on Si wafers exposed to atmospheric water, so if the wafers cannot be immediately transferred to the Si etchant, leave them in acetone until the etchant is prepared at the correct temperature.
10∣ Heat and stir the Si etchant solution in a 4-liter beaker to 75 °C on a hot plate, but do not add the 2-propanol until the solution has reached 75 °C in order to prevent excessive evaporation of the 2-propanol. The uniformity of the etching process is sensitive to the amount of 2-propanol remaining in the Si etchant, so we also advise covering the etchant solution to minimize 2-propanol evaporation. After addition, 2-propanol forms a layer on the top of the H2O. Insert the wafer face-down into the beaker, beneath the 2-propanol layer, and leave it in the etchant for 50–90 min (Fig. 2). The rate of <100> etching is much faster than the rate of <111> etching, making this step a self-terminating process. Care should still be taken, however, because the etchant will remove the oxide that serves as a mask on the unpatterned regions after ~90 min. Remove the Si masters from the etchant solution, rinse with NANOpure H2O and dry them under a stream of N2.
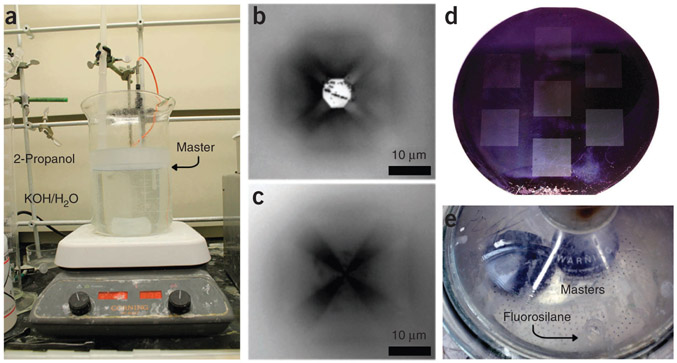
Figure 2 ∣.
Selected steps of the master fabrication process. (a) PPL master in the anisotropic KOH etching solution. During etching, the wafer (labeled ‘Master’) is held face-down in the aqueous KOH phase of the etchant, below the 2-propanol layer. (b) After 25 min of etching, an optical micrograph of a representative tip shows incomplete tip formation. (c) After etching the master for a total of 60 min, the tips form a sharp point. (d,e) Once the master fabrication process has been completed, the etched wafer (d) is placed in a desiccator (e) for overnight fluorosilane coating. In the desiccator, the wafers are placed far away from the vial containing the fluorosilane, so as to avoid splashing onto the wafers while the vacuum is pulling.
! CAUTION The KOH-containing Si etchant solution is highly corrosive and should be used in a fume hood while wearing full protective clothing, including safety glasses, a lab coat and nitrile or latex gloves.
11∣ Examine the Si masters with an optical microscope to ensure complete etching. By adjusting the microscope focal plane, it is possible to see four lines that form the edges of the inverted pyramid converge to a single point when the etching has been completed (Fig. 2c). If the edges converge to a square (Fig. 2b) rather than to a point, reimmerse the wafer in the etching solution as described in Step 10 until etching is complete.
12∣ To remove the remaining oxide that served as a mask for the Si etchant, place the masters (Fig. 2d) in the buffered HF solution for 5 min, as described in Steps 7 and 8.
∎ PAUSE POINT After oxide removal, the wafers can be stored before silanization for years under ambient conditions.
Coating the masters with fluorosilane ● TIMING 12 h
13∣ Expose the master to air plasma for 2 min at 30 W to grow a thin layer of oxide that will react with the fluorinated silane. This step will also remove any organic contamination that has accumulated if the wafers have been stored for long periods.
14∣ In a 20-ml glass vial, add 4 ml of toluene. Use a glass Pasteur pipette to add 8–10 drops of heptadecafluoro-1,1,2,2-tetra(hydrodecyl)trichlorosilane.
15∣ Place the Si masters and the toluene solution on opposite sides of a large (~30-cm diameter) vacuum desiccator (Fig. 2e). Apply vacuum until the toluene solution begins to boil, and then leave the desiccator under static vacuum for 12 h.
▲ CRITICAL STEP The fluorinated silane will rapidly cross-link upon exposure to atmospheric moisture, so the time between Steps 14 and 15 should be as small as possible.
16∣ Remove the master from desiccator and dispose of the toluene solution. Place the master face-up in a glass Petri dish with toluene for 5 min to remove multilayers of the fluorosilane, and then dry it under a stream of N2.
? TROUBLESHOOTING
∎ PAUSE POINT After silane functionalization, the masters can be stored for years under ambient conditions. If possible, they should be stored in an enclosed container to minimize dust contamination.
Fabrication of h-PDMS tip arrays ● TIMING 1–2 h
17∣ Score a glass microscope slide into 2.5 cm × 2.5 cm squares with a glass cutter or a diamond scribe. If smaller arrays (e.g., 1.25 cm × 1.25 cm) are desired, the glass can be further subdivided by scoring the large square into four smaller squares. In our experience, small arrays (1.25 cm × 1.25 cm) are generally easier to level and manipulate in addition to yielding more arrays per master.
▲ CRITICAL STEP If smaller arrays are subdivided from the large 2.5 cm × 2.5 cm arrays, do not break the glass into smaller squares until after the arrays are fabricated. The reason for this instruction is twofold: using larger glass squares makes it easier to handle the arrays and the large squares are commensurate with the 2 cm × 2 cm patterns defined in Step 1.
18∣ Place the glass squares in a 250-ml beaker with 150 ml of 2-propanol, and sonicate them for 5 min to remove glass debris. After sonication, rinse the slides with 2-propanol and dry under a stream of N2.
19∣ Subject the glass slides to air plasma at 30 W for 2 min to increase their hydrophilicity and to promote h-PDMS adhesion.
20∣ Fill a plastic weighing boat with 3.4 g of h-PDMS precursor. Add 1.0 g of (25–35% (wt/wt) methylhydrosiloxane)-dimethylsiloxane copolymer to the h-PDMS precursor.
21∣ Stir the mixture vigorously for ~5 min, using a plastic 1-ml syringe or a plastic spatula.
22∣ Put the weighing boat containing the pre-polymer mixture into a desiccator and connect it to a vacuum line. Keep the desiccator under vacuum for ~15 min to remove trapped air bubbles. After 15 min, there should be no visible bubbles, and the h-PDMS mixture should be transparent.
23∣ Place each master at the bottom of separate, large plastic weighing boats. Use a 1-ml plastic syringe to dispense one drop (~25 μl) of pre-polymer over each master (Fig. 3a).
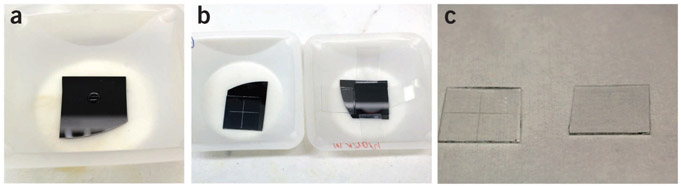
Figure 3 ∣.
Selected steps of the tip array fabrication process. (a,b) A single drop of the h-PDMS precursor is placed on top of the PPL master (a), and a plasma-cleaned glass slide is then placed on the top of the droplet, sandwiching it between the Si master and the glass backing layer (b). (c) After thermal curing, the arrays are removed from the masters, and the extraneous h-PDMS is scraped off of the edges, resulting in tip arrays that can be inked and used for printing. Note that the array on the left-hand side in b and c has been prescored so that it can be broken into four 1 cm × 1 cm tip arrays.
24∣ Place the 2.5 cm × 2.5 cm glass squares over the polymer in the master with the hydrophilic side that had been exposed to air plasma facing the polymer, so that they overlap with the patterned area of the master (Fig. 3b). Apply pressure to the slide by holding a 20-ml vial and pressing down briefly on the array; this step ensures a uniform and thin film of h-PDMS.
▲ CRITICAL STEP Make sure that there are no bubbles trapped between the array and the master. If bubbles are present, move the glass slide move the glass slide around until there are no more bubbles in the area of the array.
25∣ The glass squares may move around during curing (Steps 26–28); to prevent this movement, add glass struts to support the glass squares of the pen arrays. Place glass slides into contact with the sides of the scored glass square and have them rest on the edge of the weighing dish (Fig. 3b, right side).
Curing the h-PDMS tip arrays ● TIMING 24–36 h
26∣ Place the weighing boats containing the tip arrays and masters in an oven at 80 °C for 24–36 h to thermally cure the h-PDMS.
27∣ After curing is complete, allow the polymer to cool for 10 min after removing it from the oven. Gently remove the glass struts and discard them. With a razor, scrape off excess h-PDMS on the back of the glass, as well as the back and sides of the master.
28∣ To remove the h-PDMS pen arrays from the mold, gently insert a razor blade between the h-PDMS and the master. Slowly pry the two apart until the cured h-PDMS separates cleanly from the Si master. Use the razor to remove the h-PDMS from the sections of the array without tips (Fig. 3c). Clean the tip arrays under a stream of N2 gas. If the arrays were scored into smaller square arrays, use two wafer tweezers to break the glass along the scoring lines, and then remove any excess polymer from the sides with a razor.
? TROUBLESHOOTING
Substrate and ink preparation ● TIMING 0.5–3 h
29∣ Prepare an Au-coated substrate by thermal or e-beam evaporation (see Reagent Setup). Typically, a chip of Si <100> or a glass slide is used as the substrate. The thickness of Au ranges from 10 to 50 nm and an adhesion layer of 2–5 nm of Ti or Cr is deposited onto the substrate before the Au. Whenever possible, printing should be performed on substrates that are less than a week old.
30∣ Sonicate the alkanethiol solution until the MHA crystals completely dissolve; this should take ~5 min. Once made, the solution is stable for roughly 1 month.
Inking the pen array ● TIMING 10–20 min
31∣ Place the pen array in a plasma cleaner, and subject it to an air plasma at <200 mTorr for 2 min at 10 W. This will render the h-PDMS hydrophilic, ensuring even dispersal of the ink on the tip array.
32∣ Remove the PPL pen array from the plasma cleaner and check for hydrophilicity.
▲ CRITICAL STEP To ensure good wetting of the ink, check that the h-PDMS is hydrophilic by placing a drop of water on the surface; if it beads up, then subject the pen array to another round of plasma-cleaning. Dry the array under a stream of N2 before proceeding.
33∣ Transfer 100 μl of 5 mM ethanol solution of MHA onto the top of the pen array and let it evaporate; this will take 5–10 min.
34∣ Mount the inked PPL pen array onto the scanning probe system by adhering the backside of the pen array to a stainless steel chip carrier with carbon tape. If you are using a Park XE-150, then hold this chip carrier in place on the scanner head with three magnets.
Leveling the pen array
35∣ Before patterning, the plane of the tip array must be parallel to the plane of the AFM instrument stage. To ensure co-planarity of the array and the substrate, tilt the stage such that it is parallel with the array (Fig. 4). This process, termed leveling, can be performed according to option A, optical leveling, or via option B, force-feedback leveling. The chief difference between the two approaches is that force-feedback leveling requires a scale with a sensitivity of 10 mg or better, but it generally results in a better alignment than that obtained by optical methods. Optical leveling should be used if a scale is not available or if precise alignment is not critical. In contrast, if precise alignment is crucial or the pen array is opaque, and the AFM instrument’s scanner head has enough range in the Z direction to accommodate the height of the scale, then force-feedback leveling should be used. In particular, optical leveling can be used to ensure co-planarity of the array and substrate to ~0.01° for 1-cm-wide arrays, regardless of the density of the pens. Force-feedback leveling can theoretically achieve 1.6 × 10−4° precision25 for 1-cm-wide arrays with ~10,000 pens.
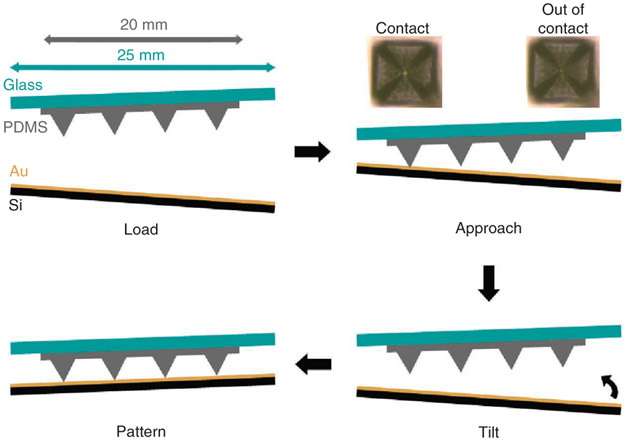
Figure 4 ∣.
Procedure for correcting misalignment between the tip array and substrate by tilting the stage until the PPL array and substrate are co-planar. Typical dimensions of the tip array and glass slide are included in the upper left, and typical optical images of tips that are in and out of contact are included in the upper right as a reference to aid the reader during the leveling procedure. Note that the PPL pens depicted here are not to scale; in a typical array, the pens are ~30-μm tall and 150-μm apart.
(A). Optical leveling ● TIMING 10–20 min
Place the substrate on the AFM instrument stage and center the pen array above it. If you are using a small substrate, make sure that it adheres to the stage by using carbon tape.
Lower the pen array until it is a few hundred micrometers above the substrate, and then monitor the pens using the video feed from an optical microscope. When using Au substrate, the background will become increasingly brighter in the video feed as the array approaches the substrate.
Once the pen array is ~100 μm above the substrate, shadows will appear around each tip. At this point, lower the head in 10-μm increments until the shadows shrink into the tips and vanish. At this point, the pens are within ~20 μm of the substrate.
-
Lower the pen array in 5-μm increments until visible deformation of the pens is observed (Fig. 5a) or until the pen array goes out of focus. If the array becomes defocused, it means pens that are out of the view of the microscope objective are in contact with the substrate.
? TROUBLESHOOTING
Compare the deformation of the pens at each of the four corners in the array. A larger deformation results in a larger square within each pyramid; comparing the extent of pen deformation at each corner gives the deviation of tilting angle.
-
Withdraw the pen array by a safe distance (~100 μm), and correct the stage-array misalignment by changing the tilt angle(s) of the stage.
▲ CRITICAL STEP The safe distance will vary on the basis of the specifics of the microscope stage, and on how extreme the desired tilting angle is. For the Park XE-150, tilting either axis up by 100 μm requires a withdraw distance of over 25 μm.
Repeat Step 35A(iv–vi) until the pens at each corner deform uniformly.
Figure 5 ∣.
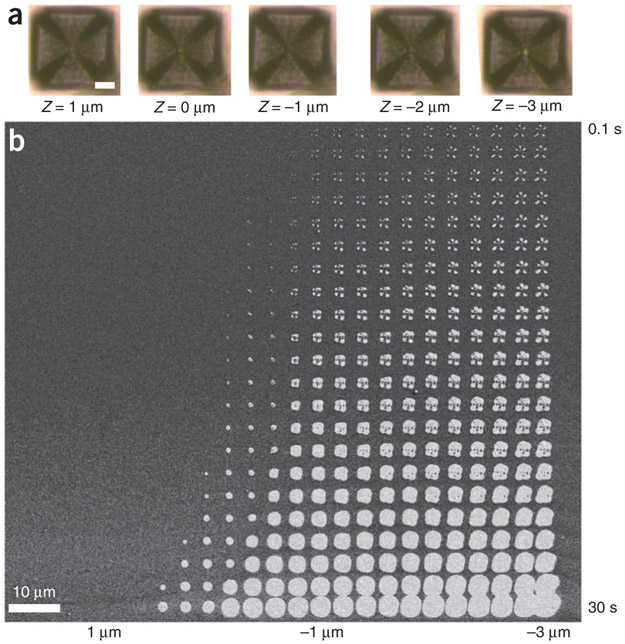
Selecting the correct patterning height for performing PPL. (a) Optical micrographs of a selected tip at a range of Zext values, where negative values indicate that the tips are moving toward the substrate. Note that when moving from Z = 0 to −1 μm, the color of the pyramidal tip center changes from light to dark, and extending the tip further from Z = −2 to −3 μm results in a change from dark black to light as the tip displays the color of the gold substrate. In this pen array, the edge length of the tips is 40 μm, and the scale bar in the leftmost micrograph is 10 μm. The scale bar in the leftmost micrograph applies to all five images, which were taken at the same resolution. (b) A SEM image of a pattern written by depositing MHA onto gold using PPL. The pattern shown here is from a single tip. After deposition, the unpatterned regions of the substrate were etched away, thereby enabling facile pattern characterization. Here, the value of Zext ranges from 1 to −3 μm (left to right), and the dwell time increases logarithmically from 100 ms to 30 s (top to bottom).
(B). Force-feedback leveling ● TIMING 45–60 min
Attach the substrate for the patterning on top of the scale with carbon tape.
Mount the scale together with the substrate on the two-axes tilting stage by using carbon tape in case the scale moves horizontally on the stage.
Make the polymer pen array approach the surface while monitoring the reading from the scale carefully, especially when the pen array is close to the surface.
Stop the pen array from approaching the substrate when there is a force sensed by the scale. Adjust the Z motor slightly (1 micron increments) until the force shown on the scale is 0.10 g. The initial small reading on the scale is to make sure only a very small number of pens are in contact with the surface. If too many pens are in contact, the sensitivity of this method is reduced.
Extend the Z piezo an additional 12 μm toward the surface. Wait until the reading is stable on the scale, and then record the scale output.
-
Withdraw the pen array to a safe height to protect the pen array from damage while adjusting the tilting angle. Then, adjust the value of the Y motor by 100 μm.
▲ CRITICAL STEP The safe withdrawal distance will vary on the basis of the specifics of the microscope stage and on how extreme the desired tilting angle is. For the Park XE-150, tilting either axis up by 100 μm requires a withdraw distance of over 25 μm.
-
Repeat Step 35B(iii–vi), and compare the scale readings to obtain a local maximum in the force. Adjust the Y tilt such that it is at the local force maximum.
? TROUBLESHOOTING
Repeat Step 35B(iii–vi) in a similar manner for the X tilt. After this procedure, the force reading should be close to the global maximum.
Search in a small region of the hypothesized global maximum by making small changes to both tilts (steps of 5–20 μm), using a similar procedure to that described in Step 35B(iii–viii). Once a global maximum is reached, the pen arrays are level with respect to the surface.
36∣ Withdraw the pen arrays from the surface by 100 μm while designing the pattern (Steps 37 and 38 below).
Designing the pattern ● TIMING 5–10 min
37∣ Use commercial software to design the desired pattern. In the Park XEP software, the interface can be found under the ‘lithography’ tab. Note that the pattern can be designed within the software itself, or made in a separate program and then imported as a .bmp image into the AFM software.
38∣ When designing a pattern, two key parameters determine feature size in PPL: dwell time and Z piezo extension (Zext). Dwell time determines how long the pen will remain at each point; extending this time will increase ink diffusion, which will increase the size of the features. The Z piezo extension corresponds to contact force, whereby decreasing Zext will increase contact forces and thus lead to features of increased size. If you are using a Park XE-150, Zext ranges from +12 to −12 μm. We recommend choosing Zext = 0 as the initial contact point (by adjusting the stage height); once the height of the initial contact point is established, Zext can be varied within the pattern to determine the optimal height for future patterns. An example of pen deformation in the range of Zext = 1 to −3 μm is shown in Figure 5a; note that here negative distances mean a higher tip-substrate force, and that Zext = 0 μm corresponds to the contact point.
Printing the desired pattern of MHA ● TIMING 5–60 min
39∣ Set the RH and temperature of the environmental chamber and wait until the desired set points are reached. The best results for MHA are obtained at room temperature with RH in the 40–80% range. The pattern shown in Figure 5b was printed at RH = 60%.
40∣ Bring the pens to the substrate until the pens are at the desired height for patterning, as determined in Step 38.
41∣ Press the ‘Start Lithography’ button in the XEP software to begin the pattern. Once patterning is done, open the environmental chamber, withdraw the pen array and remove the substrate from the AFM instrument. Please note that, if postpatterning functionalization with proteins is desired (Steps 44–49), the Au etching Steps (42 and 43) should not be implemented, and the researcher should skip directly to Step 44. The Au-etching process interferes with MHA functionalization because the carboxylate groups in MHA will bind to the Fe3+ ions in the Au etchant, and thus etching should be avoided if protein functionalization is desired.
∎ PAUSE POINT After deposition, the SAMs of MHA are stable in ambient conditions for over 6 months.
42∣ Mix a fresh batch of Au etchant in a Petri dish, and place the substrate face-up in the dish. The etch rate of Au with the listed concentrations is ~3 nm min−1, so it will take 6–7 min to etch a 20-nm film of Au. During etching, the SAM formed by MHA acts as a mask, preventing dissolution of Au beneath the patterned regions.
43∣ Once etching is complete, remove the patterned substrate, rinse it with NANOpure H2O and blow-dry it under a stream of N2. The resulting patterns can be visualized with optical microscopy or SEM; examples of a pattern etched in this fashion are shown in Figure 5b.
Functionalization of MHA-patterned substrates with fibronectin ● TIMING 12–16 h
▲ CRITICAL Steps 44–49 are optional and should be implemented if the experimenter plans to functionalize MHA patterned onto the substrate. Here, fibronectin is used as a specific example to outline the procedure.
44∣ Backfill the patterned substrate by placing it in a Petri dish containing a 1 mM solution of (1-mercapto-11-undecyl)hexa(ethylene glycol) in ethanol for 1 h. After backfilling, remove the substrate, rinse with ethanol and dry the substrate under a stream of N2.
45∣ Place the substrate in a Petri dish containing an aqueous solution of 10 mM Co(NO3)3 for 5 min. After 5 min, remove the substrate, rinse with NANOpure H2O and dry the substrate under a stream of N2.
46∣ Place the substrate in a Petri dish filled with a solution of 50 μg ml−1 fibronectin in 1× PBS. Incubate the sample in this solution overnight on a shaker at 100 r.p.m. and at 4 °C. Rinse the sample copiously with 1× PBS after fibronectin immobilization and dry it under a stream of N2.
47∣ (Optional) Visualize the patterns by AFM before proceeding to the antibody incubation steps (Steps 48 and 49). This step should only be implemented if the researcher wants to check on pattern quality before going through the time-consuming process of immunofluorescence staining and microscopy. We recommend performing this step as the typical method for pattern verification, as Au etching and optical microscopy cannot be used before protein immobilization.
48∣ To verify the pattern quality using immunofluorescence microscopy, place the fibronectin-modified substrate in a 1:100 solution of primary antibody (human anti-fibronectin produced in rabbit) in 1× PBS on a shaker stirring (100 r.p.m.) overnight at 4 °C. After antibody incubation, rinse the sample with 1× PBS and dry it under flowing N2.
49∣ Place the substrate in a 1:250 solution of fluorescently labeled secondary antibody (here, goat anti-rabbit Alexa Fluor 568) diluted in 1× PBS for 1 h. Rinse the substrate sequentially with 1× PBS and NANOpure H2O, dry it under a stream of N2 and image it with a fluorescence microscope (Fig. 6a,b).
Figure 6 ∣.
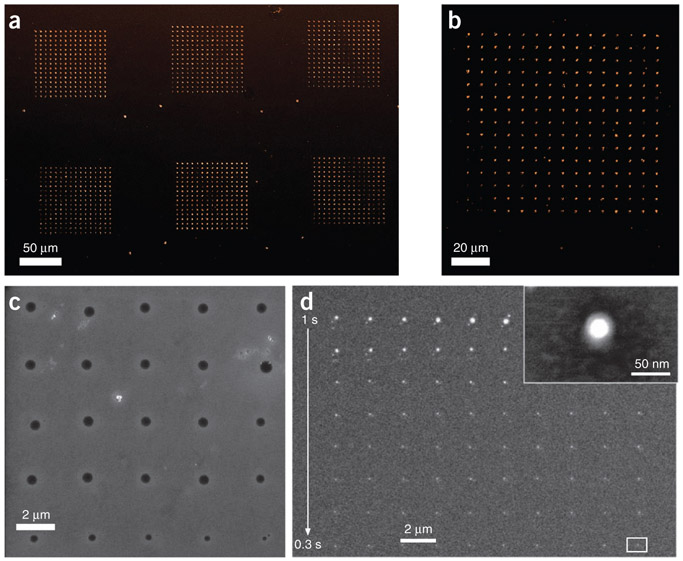
Patterns of soft and hard materials written by PPL. (a) Immunofluorescence image of fibronectin patterns tagged with Alexa Fluor 568 goat anti-rabbit antibody generated by depositing MHA with PPL and then adhering fibronectin to the molecular patterns. (b) A higher-resolution immunofluorescence image of a fibronectin pattern written by a single pen. (c) SEM image of features consisting of block co-polymer mixed with Au salt, deposited by PPL. After annealing, the gold salt is reduced to form nanoparticles and the polymer is burned away. (d) SEM image of the resulting nanoparticles from a pattern written by a single pen, where the inset in the upper right corresponds to the white box in the lower right corner.
? TROUBLESHOOTING
Step 16
Failure can occur here if the fluorosilane is past its shelf life. To test whether the wafers are hydrophobic, place a drop or two of NANOpure water onto the master. The droplet should bead up and easily roll off the master when the master is tilted. Remove any water from the master by drying it under a stream of N2 before proceeding to the next step.
Step 28
Failure can occur here if the h-PDMS is not completely cured; this will leave liquid h-PDMS in the master. Unfortunately, if attempts are made to remove the array before the h-PDMS is fully cured, the master(s) can no longer be used for array fabrication. For this reason, we advise erring on the longer side of the curing time.
Step 35A(iv)
If the misalignment between the array and the stage is severe, the array will go out of focus at all four corners, and thus the tips will not appear to be in contact anywhere. If this is the case, lift the array by 500 μm, and perform a coarse optical leveling. This coarse leveling is done by focusing the camera on the substrate instead of the tip array and iteratively tilting the stage until all four corners of the substrate are in the same focal plane.
Step 35B(vii)
If there is a large misalignment in the X tilt, the local maximum may be too small to be seen. If this is the case, try varying the X tilt until a local maximum can be observed. Coarse leveling with the optical method can also help to address this problem.
● TIMING
Steps 1–5, defining the masters: 2–4 h
Steps 6–12, fabricating the masters: 2–5 h
Steps 13–16, coating the masters with fluorosilane: 12 h
Steps 17–25, fabrication of h-PDMS tip arrays: 1–2 h
Steps 26–28, curing the h-PDMS tip arrays: 24–36 h
Steps 29 and 30, substrate and ink preparation: 0.5–3 h
Steps 31–34, inking the pen array: 10–20 min
Steps 35 and 36, leveling the pen array: 10–60 min
Steps 37 and 38, designing the pattern: 5–10 min
Steps 39–43, printing the desired pattern of MHA: 5–60 min
Steps 44–49, functionalization of MHA-patterned substrates with fibronectin: 12–16 h
Box 1, deposition of metallic nanoparticles on a silicon substrate: 12–16 h
ANTICIPATED RESULTS
Upon successful completion of the experiments described in this protocol, one should obtain (i) patterned features of MHA on a Au substrate, (ii) patterned nanostructures of Au on a Si substrate (Fig. 5b), (iii) patterned nanostructures of fibronectin on an Au substrate (Fig. 6a,b), (iv) patterns of block co-polymer complexed with Au salt (Fig. 6c) and (v) arrays of Au nanoparticles on a Si substrate (Fig. 6d). These ink and substrate combinations demonstrate the full capabilities of PPL: patterns of molecules, transfer of these molecular patterns into the underlying substrate to make ‘hard’ nanostructures (Figs. 5b and 6d) and patterns of soft materials (Fig. 6a-c).
ACKNOWLEDGMENTS
This material is based on the work supported by the US Defense Advanced Research Projects Agency (DARPA) Microsystems Technology Office (MTO) award N66001-08-1-2044; Asian Office of Aerospace Research and Development (AOARD) award FA2386-10-1-4065; Air Force Office of Scientific Research (AFOSR) awards FA9550-12-1-0280 and FA9550-12-1-0141; US National Science Foundation awards DBI-1152139, DBI-1152169 and DMB-1124131; Department of Defense (DoD)/Naval Postgraduate School (NPS)/National Security Science and Engineering Faculty (NSSEF) fellowship awards N00244-09-1-0012 and N00244-09-1-0071; Chicago Biomedical Consortium with support from Searle Funds at The Chicago Community Trust; and Center of Cancer Nanotechnology Excellence (CCNE) initiative of the US National Institutes of Health (NIH) award U54 CA151880. D.J.E. is supported by a DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate Fellowship (NDSEG) 32 CFR 168a. X.L. gratefully acknowledges support from the Ryan Fellowship from Northwestern University. K.A.B. gratefully acknowledges support from Northwestern University’s International Institute for Nanotechnology. B.R. acknowledges the Indo-US Science and Technology Forum (IUSSTF) for a postdoctoral fellowship. L.R.G. acknowledges the NSF for a Postdoctoral Research Fellowship in Biology.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Tran H, Killops KL & Campos LM Advancements and challenges of patterning biomolecules with sub-50 nm features. Soft Matter 9, 6578–6586 (2013). [Google Scholar]
- 2.Schmidt RC & Healy KE Controlling biological interfaces on the nanometer-length scale. J. Biomed. Mater. Res. A 90A, 1252–1261 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Kim D-H, Lee H, Lee YK, Nam J-M & Levchenko A Biomimetic nanopatterns as enabling tools for analysis and control of live cells. Adv. Mater 22, 4551–4566 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, Biebuyck HA & Whitesides GM Patterning self-assembled monolayers: applications in materials science. Langmuir 10, 1498–1511 (1994). [Google Scholar]
- 5.Kumar A & Whitesides GM Features of gold having micrometer to centimeter dimensions can be formed through a combination of stamping with an elastomeric stamp and an alkanethiol ‘ink’ followed by chemical etching. Appl. Phys. Lett 63, 2002–2004 (1993). [Google Scholar]
- 6.Wilbur JL, Kumar A, Kim E & Whitesides GM Microfabrication by microcontact printing of self-assembled monolayers. Adv. Mater 6, 600–604 (1994). [Google Scholar]
- 7.Xia Y & Whitesides GM Soft lithography. Angew. Chem. Int. Ed 37, 550–575 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Qin D, Xia Y & Whitesides GM Soft lithography for micro- and nanoscale patterning. Nat. Protoc 5, 491–502 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Huo FW et al. Polymer pen lithography. Science 321, 1658–1660 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbulovic-Nad I et al. Bio-microarray fabrication techniques—a review. Crit. Rev. Biotechnol 26, 237–259 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Allain LR, Stratis-Cullum DN & Vo-Dinh T Investigation of microfabrication of biological sample arrays using piezoelectric and bubble-jet printing technologies. Anal. Chim. Acta 518, 77–85 (2004). [Google Scholar]
- 12.Park J-U et al. High-resolution electrohydrodynamic jet printing. Nat. Mater 6, 782–789 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Salaita K et al. Massively parallel dip-pen nanolithography with 55,000-pen two-dimensional arrays. Angew. Chem. Int. Ed 45, 7220–7223 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Salaita K, Wang YH & Mirkin CA Applications of dip-pen nanolithography. Nat. Nanotechnol 2, 145–155 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Giam LR & Mirkin CA Cantilever-free scanning probe molecular printing. Angew. Chem.Int. Ed 50, 7482–7485 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Braunschweig AB, Huo F & Mirkin CA Molecular printing. Nat. Chem 1, 353–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilbur JL, Kim E, Xia Y & Whitesides GM Lithographic molding: A convenient route to structures with sub-micrometer dimensions. Adv. Mater 7, 649–652 (1995). [Google Scholar]
- 18.Jung Moo H, Fatih MO & Jun Z A micromachined elastomeric tip array for contact printing with variable dot size and density. J. Micromech. Microeng 18, 015003 (2008). [Google Scholar]
- 19.Xie Z et al. Polymer pen lithography using dual-elastomer tip arrays. Small 8, 2664–2669 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Shim W et al. Hard-tip, soft-spring lithography. Nature 469, 516–521 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Shim W et al. Multifunctional cantilever-free scanning probe arrays coated with multilayer graphene. Proc. Natl. Acad. Sci. USA 109, 18312–18317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichelsdoerfer DJ, Brown KA, Boya R, Shim W & Mirkin CA Tuning the spring constant of cantilever-free tip arrays. Nano Lett. 13, 664–667 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Huo FW et al. Beam pen lithography. Nat. Nanotechnol 5, 637–640 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Brown KA et al. A cantilever-free approach to dot-matrix nanoprinting. Proc. Natl. Acad. Sci. USA 110, 12921–12924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao X, Braunschweig AB & Mirkin CA ‘Force-Feedback’ leveling of massively parallel arrays in polymer pen lithography. Nano Lett. 10, 1335–1340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginger DS, Zhang H & Mirkin CA The evolution of dip-pen nanolithography. Angew. Chem. Int. Ed 43, 30–45 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Giam LR et al. Scanning probe-enabled nanocombinatorics define the relationship between fibronectin feature size and stem cell fate. Proc. Natl. Acad. Sci. USA 109, 4377–4382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong X et al. Materials for the preparation of polymer pen lithography tip arrays and a comparison of their printing properties. J. Pol. Sci. A 51, 1533–1539 (2013). [Google Scholar]
- 29.Huang L et al. Matrix-assisted dip-pen nanolithography and polymer pen lithography. Small 6, 1077–1081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bian S, He J, Schesing KB & Braunschweig AB Polymer pen lithography (PPL)-induced site-specific click chemistry for the formation of functional glycan arrays. Small 8, 2000–2005 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Bian S, Schesing KB & Braunschweig AB Matrix-assisted polymer pen lithography–induced Staudinger ligation. Chem. Commun 48, 4995–4997 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Chai JA et al. Scanning probe block co-polymer lithography. Proc. Natl. Acad. Sci. USA 107, 20202–20206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giam LR et al. Positionally defined, binary semiconductor nanoparticles synthesized by scanning probe block copolymer lithography. Nano Lett. 12, 1022–1025 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Brinkmann F et al. Interdigitated multicolored bioink micropatterns by multiplexed polymer pen lithography. Small 9, 3266–3275 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Zheng Z et al. Multiplexed protein arrays enabled by polymer pen lithography: addressing the inking challenge. Angew. Chem. Int. Ed 48, 7626–7629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai J, Wong LS, Giam L & Mirkin CA Single-molecule protein arrays enabled by scanning probe block co-polymer lithography. Proc. Natl. Acad. Sci. USA 108, 19521–19525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bian S et al. Covalently patterned graphene surfaces by a force-accelerated Diels-Alder reaction. J. Am. Chem. Soc 135, 9240–9243 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Lee K-B, Park S-J, Mirkin CA, Smith JC & Mrksich M Protein nanoarrays generated by dip-pen nanolithography. Science 295, 1702–1705 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Wilson DL et al. Surface organization and nanopatterning of collagen by dip-pen nanolithography. Proc. Natl. Acad. Sci. USA 98, 13660–13664 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee K-B, Lim J-H & Mirkin CA Protein nanostructures formed via direct-write dip-pen nanolithography. J. Am. Chem. Soc 125, 5588–5589 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Sekula S et al. Multiplexed lipid dip-pen nanolithography on subcellular scales for the templating of functional proteins and cell culture. Small 4, 1785–1793 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Cheung CL et al. Fabrication of assembled virus nanostructures on templates of chemoselective linkers formed by scanning probe nanolithography. J. Am. Chem. Soc 125, 6848–6849 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Shikida M, Sato K, Tokoro K & Uchikawa D Comparison of anisotropic etching properties between KOH and TMAH solutions. in Twelfth IEEE International Conference on Micro Electro Mechanical Systems, 1999 (MEMS ′99). 315–320 (1999). [Google Scholar]
- 44.Schmid H & Michel B Siloxane polymers for high-resolution, high-accuracy soft lithography. Macromolecules 33, 3042–3049 (2000). [Google Scholar]