Abstract
There is considerable interest in the development of libraries of scaffold-diverse macrocycles as a source of ligands for difficult targets, such as protein–protein interaction surfaces. A classic problem in the synthesis of high-quality macrocyclic libraries is that some linear precursors will cyclize efficiently while some will not, depending on their conformational preferences. We report here a powerful quality control method that can be employed to readily distinguish between scaffolds that do and do not cyclize efficiently during solid-phase synthesis of thioether macrocycles without the need for tedious liquid chromatography/mass spectrometry analysis. We demonstrate that this assay can be employed to identify linear impurities in a DNA-encoded library of macrocycles. We also use the method to establish a useful quality control protocol for re-synthesis of putative macrocyclic screening hits.
Keywords: DNA-encoded libraries, macrocycles, peptidomimetics
Introduction
Libraries of macrocycles are of interest as a potential source of high affinity ligands for difficult to target proteins.[1] Indeed, powerful methods for the creation of huge peptide libraries such as ribosome display and phage display have been adapted to make macrocyclic peptides,[2] which have proven to be a rich source of protein ligands. Unfortunately, most[3] macrocyclic peptides are not cell permeable, which limits their applicability to extracellular targets. As a result, there has been considerable interest in the development of synthetic libraries of non-peptidic macrocycles with the appropriate physicochemical properties to passively cross membranes.[4] DNA-encoded libraries (DELs) of non-peptidic macrocycles[5] are of particular interest, since this technology allows the synthesis of libraries as large or larger than those created by phage display. This area was pioneered by Liu and co-workers. They employed DNA-templated chemistry[6] and a ring-closing Wittig olefination to create libraries of up to 256,000 macrocycles from which ligands for several protein targets have been mined.[7] More recently, Gillingham and co-workers created a library of DNA-encoded macrocycles using amide bond formation to close the ring.[8]
An important feature of both of these libraries is that they are scaffold-diverse, which is likely to be important in order to provide a general source of ligands for a variety of different protein targets.[9] However, scaffold diversity brings along with it the problem of disparate efficiencies of macrocyclization of the various molecules in the library, since efficient ring closure depends on the linear precursor accessing a conformation that brings the two ends into close proximity. The same issue has also been encountered with some long peptides that adopt a fold that hinders the close approach of the N- and C-termini.[10] While variable yields are tolerable in parallel synthesis strategies where reaction products can be purified,[11] this is not the case for libraries created by split and pool chemistry,[12] which produces an intractable mixture of molecules. Indeed, to the best of our knowledge, it is not known what percentage of the molecules in existing macrocyclic DELs are linear impurities.
We recently developed methodology to create DELs of scaffold-diverse macrocyclic PICCOs[13] (peptoid-inspired conformationally constrained oligomers) by solid-phase split and pool synthesis.[14] PICCOs are made using peptoid-like chemistry[15] in which amines are stitched together with carboxylic acid-containing building blocks that also have a good leaving group elsewhere in the molecule (Scheme 1). Because the carboxylate building blocks employed in the library synthesis have structural features that provide considerable conformational constraint, we anticipated that the aforementioned issue of variable cyclization yields could be significant in the construction of macrocyclic PICCO libraries.
Scheme 1.
Representation of the fluorescence-based assay for monitoring the efficiency on on-resin macrocyclization via thioether formation.
To address this question, we report here a simple but powerful assay for assessment of the efficiency of on-resin cyclization capable of monitoring this process on millions of individual beads. The strategy is shown schematically in Scheme 1 for macrocycles closed via thioether bond formation. We demonstrate that this method is useful for monitoring the progression of an on-resin cyclization reaction at the level of a single compound or a library without the requirement for tedious analysis of reaction products by liquid chromatography/mass spectrometry (LC/MS) after release from the bead.
Results and Discussion
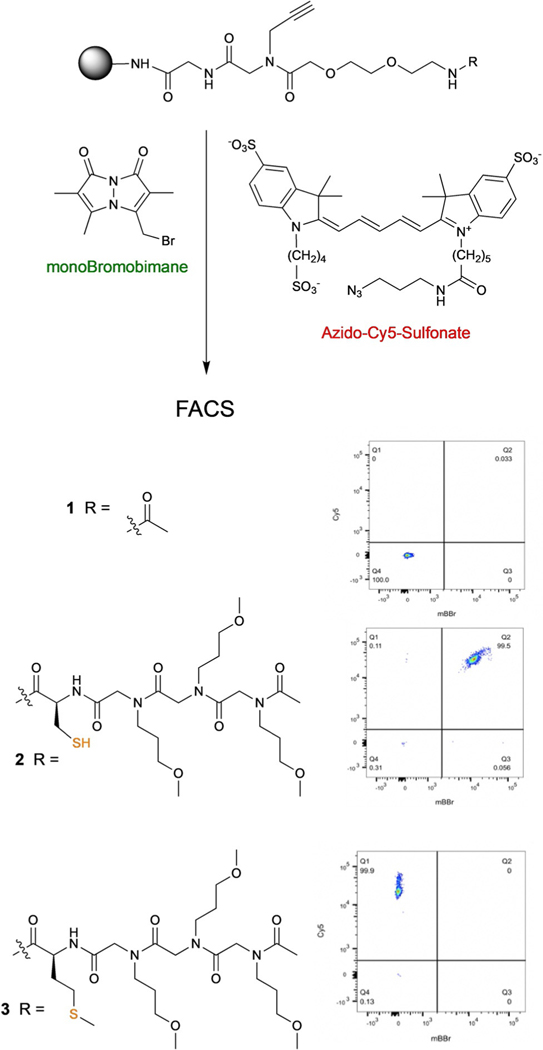
We have created one bead one compound (OBOC) DELs of macrocyclic thioethers.[14] The cyclization reaction involves deprotection of the invariant cysteine followed by incubation in the presence of a mild base, resulting in nucleophilic displacement of the chloride by the thiol (Scheme 1). After staining with a thiol-reactive green dye, beads displaying mostly macrocyclic molecules would have little green color, while those with significant amounts of uncyclized material would fluoresce brightly at this wavelength. To account for the fact that not all beads display the same amount of compound, we also envisioned attaching a red dye to a conserved alkyne in the linker as a normalization marker (Scheme 1). Since the libraries are constructed on 10 μm TentaGel beads, which are about the size of a red blood cell, we imagined that measurement of the ratio of green/red fluorescence on each bead using a flow cytometer would provide a high-throughput method for the determination of the fraction of linear molecules on large numbers of beads.
To establish appropriate conditions for this assay, the control compounds shown in Figure 1 were synthesized on 10 μm TentaGel beads. All contain an alkyne unit in the linker. Compound 1 has no sulfur atom, while compound 2 contains a cysteine and compound 3 contains a methionine. 2 and 3 serve as models for uncyclized and cyclized library members, respectively. We experimented with a variety of thiol-reactive and azide-containing dyes and found that most of them produced an unacceptable level of background staining, even on beads that displayed compound 1 and in the absence of a copper catalyst for Click chemistry. We eventually found that this undesirable absorption of dye by the TentaGel beads could be circumvented by using either a turn-on fluorescent probe or highly hydrophilic dyes, which are far less prone to be trapped by the TentaGel resin. For thiol labeling, we turned to monoBromobimane (mBBr) (Figure 1), a green dye whose fluorescence increases dramatically when the bromide is displaced by a thiol. For conjugation to the conserved alkyne, allowing normalization of the density of reactive sites on the bead, we settled on azido-Cy5-(SO3)3−2 (Figure 1). Different dye concentrations and reaction times were assessed to determine empirically the appropriate amount of staining to be in the linear range of the FACS instrument (Supporting Information, Section 2D). Under these conditions, beads displaying compounds 1 or 3 acquired little or no green fluorescence when treated with mBBr, whereas most of the beads displaying cysteine-containing compound 2 became intensely fluorescent (Figure 1). Beads subjected to the Click reaction with azido-Cy5-(SO3)3–2 fluoresced intensely in the red channel. Only a modest level of variability in the degree of staining of the 10 μm TentaGel beads with azido-Cy5-(SO3)3–2 was observed, presumably reflecting a relatively homogenous population of beads with respect to the number of reactive sites. This is in contrast to larger TentaGel beads, in which this value can vary by over 20-fold from bead to bead.[16] Therefore, some of the experiments below employed this normalization dye, while some did not.
Figure 1.
Structures of control compounds and FACS analysis of beads displaying these molecules after staining. Beads displaying compound 1 were not stained with either dye, whereas beads displaying compounds 2 or 3 were stained with both dyes.
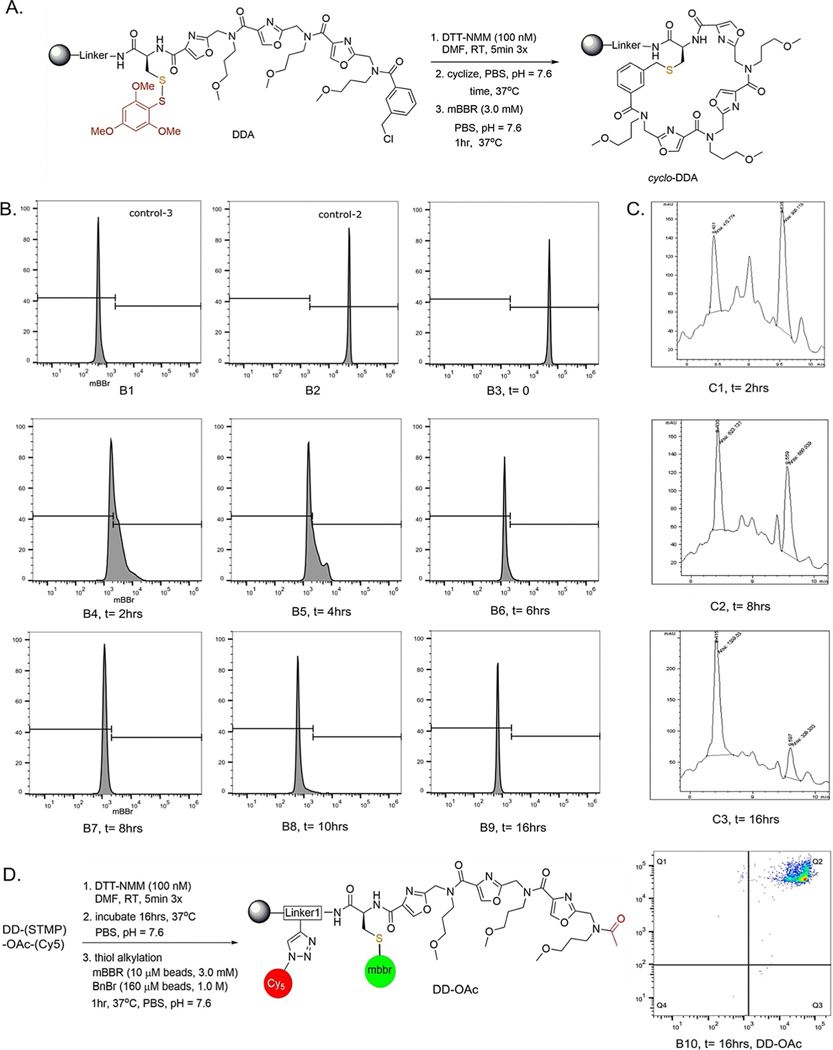
With these conditions in hand, we asked if this method was suitable for following the progress of a macrocyclization reaction of a single compound on resin. Compound DDA (Figure 2A), was subjected to standard macrocyclization conditions. Specifically, the thiol was exposed by treatment with DTT (100 mM) and N-methyl morpholine (100 mM) in DMF for five minutes, three times. After buffer exchange (Supporting Information, Section 2B) the beads were then incubated in a buffered aqueous solution. At various times afterward, an aliquot of 10 μm beads was removed from the reaction and stained with mBBr for an hour. Following a wash, the fluorescence intensity of these beads was analyzed using a flow cytometer. As shown in Figure 2B, B4, the green fluorescence intensity on beads treated with mBBr dye two hours after exposure of the thiol was reduced greatly in comparison to beads displaying the linear control molecule 2 (Figure 2B, B2), indicating a fast cyclization rate. Indeed, for this molecule the reaction appeared almost complete in two hours. With additional time, the level of staining decreased slowly and by 16 h following thiol exposure, about 99% of the beads were 50-fold less fluorescent than the control linear model (B4–B6).
Figure 2.
Monitoring the progress of macrocylization of DDA. A) Reaction conditions used for the experiment. B) Flow cytometer histograms showing the level of staining of 10 μm TentaGel beads with mBBr at the times indicated (post-deprotection). B1 shows beads displaying 3 (the methionine control) and B2 shows beads displaying 2 (the cysteine control; see Figure 1). B3–B10 show mBBr staining of beads displaying DDA. C) Progress of DDA cyclization on 160 μm TentaGel ebads as monitored by LC-mS analysis after alkylation of free thiol with benzylbromide and release of the compound from the bead. D) mBBr staining and FACS analysis of 10 μm TentaGel beads displaying DD-OAc, a molecule that cannot cyclize. Even 16 h post-deprotection, the thiol is robustly alkylated, by mBBr, showing that oxidation of the sulfur over this time period is not significant.
LC/MS analysis of the reaction generally corroborated this interpretation of the flow data. Scaffold DDA was synthesized on larger 160 μm TentaGel RAM beads with a linker that facilitates ionization in the mass spectrometer. At various times following exposure of the thiol, an aliquot of the beads was treated with excess benzyl bromide for two hours to freeze the cyclization reaction prior to release of the compounds from the beads (90% TFA, 2.5% TIPS, 2.5% thioanisole, 5.0% DCM). The ratio of linear and macrocyclic compounds was then assessed by LC/MS. Using this assay, the reaction was 24% complete after two hours, 48% complete after eight hours and reached 81% conversion after sixteen hours. This is slower than the same reaction on the smaller TentaGel resin, but this is perhaps not surprising, since it is impossible to conduct identical experiments on the two resins. The compounds are attached to the bead by different linkers and there is likely a significant difference in the fraction of sites buried in the polystyrene core versus the hydrophilic PEGylated coating on the surface of the beads.
To investigate if signal was lost due to thiol oxidation rather than macrocyclization, acetylated scaffold DD-STMP was incubated at 37°C for 16 h in PBST buffer (Figure 2D) after thiol deprotection. Staining with mBBr dye produced almost identical fluorescence intensity as linear standard (B10 vs. B2). This demonstrates thiol oxidation does not occur to a substantial degree in the time frame of the experiment. Similarly, benzylated product was observed in mass analysis from 160 μm beads after treatment with benzyl bromide (Supporting Information, Section 4C). We conclude that the dye staining/flow cytometry assay is suitable for monitoring the progress of a macrocyclization reaction.
Presumably, this method will be useful for monitoring other thioether bond-forming reactions on-resin. To conform this, we also applied the mBBr-staining protocol to examine the macrocyclization of analogues of DDD and DDA in which the terminal unit was an acrylamide, and the ring closure was the result of a Michael addition to the double bond. As shown in section 11 of the Supplementary Information, the staining assay showed clearly that this reaction also provides PICCO macrocycles, though the cyclization reaction appears to be somewhat slower than displacement of a primary chloride.
A set of 64 molecules with the general structure shown in Figure 3A were prepared by parallel solid-phase synthesis on both 10 & 160 μm TentaGel resin in a microtiter plate format. Azido-Cy5-(SO3)3−2 was added to the conserved alkyne in the linker for 10 μm beads prior to thiol deprotection. The 64 compounds represent different scaffolds with distinct conformational preferences. Each compound was subjected to the cyclization reaction conditions for eight hours, followed by staining the 10 μm beads with mBBr to assess the degree of cyclization on-resin. The analogous 160 μm beads were treated identically for eight hours, after which the compounds were cleaved from resin and processed for LC/MS analysis (Supporting Information, Section 9). Figure 3B shows a summary of the results from the fluorescence analysis (for all of the primary data see Supporting Information Sections 5 & 9). Most of the compounds in this small library cyclized efficiently, with at least 95% of the beads being located in a quadrant of the flow plot indicating a low level of staining with mBBr and thus a small amount of linear starting material. The primary data for four of these compounds is shown in Figure 3D. In contrast, a few of the compounds (highlighted in yellow in Figure 3B) provided a different result. Eight hours after thiol exposure, the level of mBBr staining suggested that the beads displayed a substantial amount of linear material, but less than that displayed by the linear control. The most reasonable interpretation of these data is that these compounds cyclize more slowly, and only partial conversion is realized after eight hours. Indeed, when beads displaying four of these compounds were stained 16 hours after thiol exposure, the level of staining had decreased to the level expected of a high yield macrocyclization reaction (Figure 3E). Again, the LC/MS data from the analogous reactions on the 160 μm TentaGel RAM beads largely corroborate the interpretation of the flow data, with some variability in rate. Eight hours after thiol exposure, the compounds were released from the resin by treatment with TFA. The majority of the scaffolds (50/64) appeared to have completed cyclization (>90%) while others progressed more slowly (Supporting Information, Table T5). In general, the scaffolds that cyclize more slowly have acid B (Figure 3A) at the terminus. In these reactions the thiol must attack a secondary carbon rather than a primary benzylic or allylic center, presumably explaining the slower rate of ring closure.
Figure 3.
Analysis of the efficiency of macrocyclization of 64 PICCO scaffolds created by parallel solid-phase synthesis. A) Structures of the building blocks employed (box) and the protocol employed for macrocylization. Diamine K was always employed following acid B, while amine L was always used following all of the other acids. This is because acylation of N-alkylated alanines is difficult.[19] Cy5-azide dye was attached to alkyne handle of the linker, the thiol protection was removed and the beads were stained with mBBr eight hours later. Bead fluorescence was then analyzed using a flow cytometer. B) Summary of ring-closure status of 64 scaffolds from FACS & LCMS analysis. Cells in yellow indicate scaffolds that had between 5–40% linear material after 8 hours while red indicates scaffolds that had more than 40% linear material present. The rest of the scaffolds did not show any detectable linear material. C) FACS plots for the control molecules 2 and 3 (see Figure 1). D) FACS plots for selected scaffolds that showed a high degree of cyclization after eight hours. E) Selected scaffolds that showed incomplete cyclization after eight hours (top row of FACS plots) but completed macrocyclization by 16 h (bottom row of FACS plots).
Interestingly, in the LC/MS-based analysis of some of the molecules synthesized on 160 μm beads, we saw peaks representing deletions of one of the PICCO units, reflecting incomplete addition of one of the acid building blocks to the terminal amine during synthesis. Yet even these shorter chains cyclized with high efficiency (see Supporting Information section 8, molecules DBA, DCA).
We next turned to applying this method to monitor the on-resin macrocyclization of DNA-encoded compounds. In this platform, less than 1% of the molecules on the bead have a DNA encoding tag appended to the invariant linker. Nonetheless, we have found that some reactions are affected by the presence of the DNA.
Azido-headpiece DNA was clicked to a modified version of compound-1 (Supporting Information, Figure S10) and a full length test-DNA encoding tag was ligated. After three acylations and aminations the linear precursor was subjected to CuAAC reaction to link Cy5-azide dye. Encoding DNA tags from these beads could not be amplified (data not shown) indicating that a late stage CuAAC reaction is not suitable for this assay. Therefore, we turned to a strain-promoted Click reaction. For this, 1,3-diazidopropane was clicked to the invariant alkyne during azido-HDNA addition via CuAAC. A full-length test-DNA tag was ligated onto the headpiece and chemistry was carried out to construct the linear precursor to cyclization. Resin was suspended in CRB buffer, and a cyclooctyne-containing dye (Cy5-DBCO; 0.11 mM, DMSO) was added and incubated at 37°C, 16 h to label the azide in the linker. We found that the conditions for mBBr labeling described above for beads lacking DNA resulted in a significantly lower labeling efficiency for encoded beads. Reoptimization showed that a higher concentration of dye (9 mM) and a longer reaction time (two hours) provided a level of labeling commensurate to what had been observed with the beads lacking encoding tags and to retain sufficient DNA tags for PCR amplification.
With these conditions set, we assessed the utility of the fluorescent method to monitor macrocyclic thioether formation on beads with encoding tags. Four compounds (DDD, DDA, DDE and EBB) were constructed on both 10 and 160 μm TentaGel beads with DNA tags and subjected to the standard cyclization conditions (Supporting Information, Section 6). The degree of cyclization was determined using the flow assay (10 μm) and by mass analysis after release from the beads (160 μm). In the latter case, any free thiol was first quenched by the addition of excess benzyl bromide (1.0 M for 1 h at 37°C) prior to TFA-mediated release of the compound from the resin to prevent cyclization during the processing and analysis period. Two of these molecules, DDD and DDA, were shown to be excellent cyclization substrates in the absence of the DNA tag (Figure 3B), and we anticipated that compound DDE would also likely cyclize well given the presence of the highly reactive bromomethyl benzene at the chain terminus. In contrast, we hypothesized that compound EBB might cyclize more slowly given the presence of a secondary bromide at the terminus. As shown in Figure 4, flow analysis suggested that compounds DDD, DDA and DDE cyclized in almost quantitative yield in 16 h, whereas the production of the thioether from EBB was still in progress, as anticipated. This interpretation was corroborated via mass analysis of the material synthesized on the 160 μm beads (Supporting Information, Section 6A). We conclude that this assay is suitable for tracking the cyclization of DNA-encoded compounds on-resin.
Figure 4.
Staining in the presence of DNA encoding tags to monitor on-resin cyclization. The FACS plots show the level of staining of beads eight hours after removal of the thiol protecting group.
Finally, we proceeded to employ the fluorescent labeling/flow cytometry assay for analysis of macrocyclization in the context of a DEL synthesis. As shown in Figure 5C, seven carboxylic acids and 34 amines (Supporting Information, Section 7) were employed to create an OBOC DEL by split and pool synthesis on 25 mg of 10 μm TentaGel beads. Library synthesis began by attaching azido-headpiece DNA[17] (<1%) and 1,3-diazidopropane (>50%) at once via a copper-catalyzed reaction to the alkyne unit in the linker. After 3 cycles of split & pool chemistry and consecutive enzymatic ligations of encoding DNA tags, 110,000 diverse linear precursor were prepared. About 4% of the library members were capped with acid H, and I (Figure 5C & D) at position 3. Since these units lack a leaving group, they cannot cyclize and thus function as internal linear control molecules in the library. Finally, the beads were labeled with Cy5-DBCO.
Figure 5.
Monitoring the progress of macrocyclization in the context of an OBOC DEL. A) Flow cytometry plot of library beads stained with Cy5 and mBBr as described in the text. The plots shown are for aliquots of library beads stained 8, 12, and 16 hours following exposure of the thiol group. B) Collection of beads with the highest level of staining by mBBr. The first two plots are beads displaying control molecules 3 (methionine) and 2 (cysteine), respectively. These are provided for comparison. The third plot is a blow-up of the stained library beads 16 h after deprotection of the thiol. The rectangle represents the gate set to collect the beads (ca. 6500) with a level of green fluorescence significantly higher than that of the methionine control. The encoding tags on these beads were amplified and deep sequenced. C) Building blocks used to create the library (left) and the general structure of the library and protocol for the experiment. Note that acids H and I were used only at position X3 in amounts such that about 4% of the beads would display molecules unable to cyclize.
About 5 mg of beads were treated so as to remove the STMP protecting group, then allowed to incubate at 37°C in buffered aqueous solution. Aliquots (ca. 10000 beads) were removed 8, 12, and 16 h later, stained with mBBr, and then analyzed using a flow cytometer. As shown in Figure 5A, almost all of the beads had substantial amounts of thiol remaining after 8 h. Four hours later, substantial ring closure was reflected by the lower average level of green fluorescence. By 16 h post-deprotection, almost all of the beads evinced a low level of fluorescence in the green channel, indicating that macrocyclization was complete for the vast majority of compounds in the library. A fluorescence based gate was established to collect beads (ca. 6500) that, after the 16 hour incubation, had higher fluorescence intensity than the DNA-linked control 3 (Figure 5B). The DNA encoding tags on these beads were deep sequenced to identify acid building blocks contained in these difficult-to-cyclize scaffolds. As anticipated, the vast majority of the sequences returned from these tags revealed that building blocks H, I, B, or F were in position X3 (Supporting Information section 9). Molecules terminating in units H and I cannot cyclize, while the analyses described above show that molecules containing B and F are slower to cyclize. There was no significant enrichment of particular building blocks in positions X1 and X2. These data show clearly that linear PICCO molecules terminating in a reactive, primary alkyl chloride cyclize with a high degree of efficiency, demonstrating unequivocally that high quality libraries of PICCO macrocycles can be created by solid-phase synthesis.
This result was somewhat surprising to us. We had anticipated that linear chains of some of the constrained (relative to peptides or peptoids) PICCO molecules might prove difficult to cyclize, but that is clearly not the case, at least for the ring sizes analyzed here. This is likely due to the fact that PICCOs are oligomers of tertiary amides, which can adopt either the cis or trans conformation with almost equal facility and which interconvert rapidly (seconds) relative to the time course of the cyclization reaction (hours). This likely facilitates the population of a linear conformer that allows the molecules to cyclize efficiently.
Conclusion
We have developed a flow cytometer-based assay to analyze the on-resin ring-closure process for OBOC DNA-encoded thioether macrocycles. We have demonstrated this powerful methodology is able to monitor the chemistry that occurs on hundreds of thousands or even millions of individual 10 μm TentaGel beads. This makes it possible to quality control even very large DELs of macrocycles, something that is not currently possible using standard techniques. While this study focused on monitoring the efficiency of thioether formation by staining unreacted sulfhydryls with a thiol-reactive dye, it should be possible to adapt this approach to almost any ring-closing chemistry so long as one of the reacting partners is unique in the molecule. It also seems reasonable to suggest that this Scheme could be adapted to identify difficult-to-cyclize molecules in DELs produced by solution-phase split and pool synthesis, where each molecule is affixed to a DNA tag and the compounds are part of an intractable mixture.[17a] In this case, the entire library could be treated, for example, with a thiol-reactive biotinylating agent. The biotinylated molecules could be pulled out of solution with immobilized streptavidin and the corresponding encoding tags deep sequenced to identify linear scaffolds. Indeed, Neri and co-workers have used a similar strategy to remove products of incomplete acylation from a DEL created by solution-phase synthesis.[18] Therefore, we believe that this basic assay format will be of general utility in the quality assessment of macrocyclic DELs.
Supplementary Material
Acknowledgements
We thank Dr. Ofelia Utset for administrative support. This work was supported by a SBIR grant from the NIH (GM130164).
Footnotes
[**] A previous version of this manuscript has been deposited on a preprint server (A version of this manuscript was posted on ChemRxiv https://doi.org/10.26434/chemrxiv.13491312.v1).
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.202100230.
Conflict of interest
T.K. is a major shareholder in Deluge Biotechnologies, which is commercializing DNA-encoded libraries of macrocycles.
Contributor Information
Animesh Roy, Deluge Biotechnologies, 6671 W. Indiantown Rd., Suite 50-325, Jupiter, FL 33458 (USA).
Eric Koesema, Deluge Biotechnologies, 6671 W. Indiantown Rd., Suite 50-325, Jupiter, FL 33458 (USA).
Thomas Kodadek, Dr., Department of Chemistry, The Scripps Research Institute, 130 Scripps Way, Jupiter, FL 33458 (USA).
References
- [1] a).Driggers EM, Hale SP, Lee J, Terrett NK, Nat. Rev. Drug Discovery 2008, 7, 608–624; [DOI] [PubMed] [Google Scholar]; b) Whitty A, Viarengo LA, Zhong M, Org. Biomol. Chem 2017, 15, 7729–7735; [DOI] [PubMed] [Google Scholar]; c) Bauer RA, Wurst JM, Tan DS, Curr. Opin. Chem. Biol 2010, 14, 308–314; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Dandapani S, Marcaurelle LA, Nat. Chem. Biol 2010, 6, 861–863. [DOI] [PubMed] [Google Scholar]
- [2] a).Reid PC, Goto Y, Katoh T, Suga H, Methods Mol. Biol 2012, 805, 335–348; [DOI] [PubMed] [Google Scholar]; b) Vinogradov AA, Yin Y, Suga H, J. Am. Chem. Soc 2019, 141, 4167–4181; [DOI] [PubMed] [Google Scholar]; c) Heinis C, Rutherford T, Freund S, Winter G, Nat. Chem. Biol 2009, 5, 502–507. [DOI] [PubMed] [Google Scholar]
- [3] a).Bockus AT, McEwan CM, Lokey RS, Curr. Top. Med. Chem 2013, 13, 821–836; [DOI] [PubMed] [Google Scholar]; b) Fouch8 M, Sch-fer M, Berghausen J, Desrayaud S, Blatter M, Pi8chon P, Dix I, Martin Garcia A, Roth H-J, ChemMedChem 2016, 11, 1048–1059. [DOI] [PubMed] [Google Scholar]
- [4] a).Villar EA, Beglov D, Chennamadhavuni S, Porco JA, Kozakov D, Vajda S, Whitty A, Nat. Chem. Biol 2014, 10, 723–731; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Over B, Matsson P, Tyrchan C, Artursson P, Doak BC, Foley MA, Hilgendorf C, Johnston SE, Lee MD, Lewis RJ, McCarren P, Muncipinto G, Norinder U, Perry MWD, Duvall JR, Kihlberg J, Nat. Chem. Biol 2016, 12, 1065–1074; [DOI] [PubMed] [Google Scholar]; c) Pye CR, Hewitt WM, Schwochert J, Haddad TD, Townsend CE, Etienne L, Lao Y, Limberakis C, Furukawa A, Mathiowetz AM, Price DA, Liras S, Lokey RS, J. Med. Chem 2017, 60, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Connors WH, Hale SP, Terrett NK, Curr. Opin. Chem. Biol 2015, 26, 42–47. [DOI] [PubMed] [Google Scholar]
- [6].Li X, Liu DR, Angew. Chem. Int. Ed 2004, 43, 4848–4870; Angew. Chem. 2004, 116, 4956–4979. [DOI] [PubMed] [Google Scholar]
- [7] a).Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR, J. Am. Chem. Soc 2010, 132, 11779–11791; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Usanov DL, Chan AI, Maianti JP, Liu DR, Nat. Chem 2018, 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stress CJ, Sauter B, Schneider LA, Sharpe T, Gillingham D, Angew. Chem. Int. Ed 2019, 58, 9570–9574; Angew. Chem. 2019, 131, 9671–9675. [DOI] [PubMed] [Google Scholar]
- [9].Dickson P, Kodadek T, Org. Biomol. Chem 2019, 17, 4676–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jagasia R, Holub JM, Bollinger M, Kirshenbaum K, Finn MG, J. Org. Chem 2009, 74, 2964–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] a).Mortensen KT, Osberger TJ, King TA, Sore HF, Spring DR, Chem. Rev 2019, 119, 10288–10317; [DOI] [PubMed] [Google Scholar]; b) Nielsen TE, Schreiber SL, Angew. Chem. Int. Ed 2008, 47, 48–56; Angew. Chem. 2008, 120, 52–61; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Isidro-Llobet A, Hadje Georgiou K, Galloway WRJD, Giacomini E, Hansen MR, M8ndez-Abt G, Tan YS, Carro L, Sore HF, Spring DR, Org. Biomol. Chem 2015, 13, 4570–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12] a).Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ, Nature 1991, 354, 82–84; [DOI] [PubMed] [Google Scholar]; b) Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH, Nature 1991, 354, 84–86. [DOI] [PubMed] [Google Scholar]
- [13].Kodadek T, McEnaney PJ, Chem. Commun 2016, 52, 6038–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Koesema E, Roy A, Paciaroni NG, Kodadek T, ChemRxiv 2020, submitted. [Google Scholar]
- [15].Zuckermann RN, Kerr JM, Kent SBH, Moos WH, J. Am. Chem. Soc 1992, 114, 10646–10647. [Google Scholar]
- [16].Doran TM, Gao Y, Mendes K, Dean S, Simanski S, Kodadek T, ACS Comb. Sci 2014, 16, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a).Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJ, Israel DI, Jiang J, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer JA, Myers P, OQKeefe H, Oliff MC, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang L, van Vloten K, Wagner RW, Yao G, Zhao B, Morgan BA, Nat. Chem. Biol 2009, 5, 647–654; [DOI] [PubMed] [Google Scholar]; b) MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM, ACS Comb. Sci 2015, 17, 518–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Franzini RM, Biendl S, Mikutis G, Samain F, Scheuermann J, Neri D, ACS Comb. Sci 2015, 17, 393–398. [DOI] [PubMed] [Google Scholar]
- [19].Gao Y, Kodadek T, Chem. Biol 2013, 20, 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.