Abstract
Objectives
The aim of this study was to investigate the effects of exosomes isolated from human bone marrow mesenchymal stem cells (BMSCs) on osteoarthritis (OA) and a competitive endogenous RNA (ceRNA) network.
Methods
Exosomes were isolated from human BMSCs and characterized by transmission electron microscopy (TEM), Nanosight (NTA), and western blotting. Chondrocytes were treated with interleukin-1β (IL-1β) and then transfected with exosomes. Cell viability and apoptosis were determined using Cell Counting Kit-8 (CCK-8) and flow cytometry, respectively. Cells with IL-1β and exosomes were sequenced, and differentially expressed lncRNAs (DE-lncRNAs) and miRNAs (DE-miRNAs) were identified. Thereafter, a ceRNA network (LYRM4-AS1-GRPR-miR-6515-5p) was chosen for further validation.
Results
TEM, NTA, and western blotting showed that exosomes were successfully isolated, and PKH67 staining showed that exosomes could be taken up by IL-1β-induced chondrocytes. Compared with the control group, IL-1β significantly decreased cell viability and promoted apoptosis (P < 0.05), while exosomes reversed the changes induced by IL-1β. For MMP3, AKT, and GRPR, IL-1β upregulated their expression, while exosomes downregulated their expression. For PTEN, there was no significant difference in PTEN expression between the control and IL-1β groups; however, exosomes markedly upregulated PTEN expression. By sequencing, 907 DE-lncRNAs and 25 DE-miRNAs were identified, and a ceRNA network was constructed. The dual-luciferase reporter gene indicated that LYRM4-AS1, miR-6515-5, and GRPR interacted with each other. The results of cell experiments showed that LYRM4-AS1 regulated the growth of IL-1β-induced chondrocytes by GRPR/miR-6515-5p.
Conclusion
Exosomes may alleviate OA inflammation by regulating the LYRM4-AS1/GRPR/miR-6515-5p signaling pathway.
Keywords: osteoarthritis, exosomes, GRPR, miR-6515-5p, lncRNA LYRM4-AS1, IL-1β-induced chondrocytes
Introduction
Osteoarthritis (OA) is a painful joint disease characterized by progressive breakdown of articular cartilage, sclerosis of the subchondral bone, and abnormal bone growth (osteophyte) (Guilak et al., 2018). OA remains a major global public health problem causing increasing morbidity and disability and affecting approximately 240 million people worldwide, particularly adults over the age of 45 (Jonsson et al., 2016; Lankhorst et al., 2017; Nelson, 2018). The pathogenesis of OA is related to many factors, such as aging, inflammation, obesity, trauma, and heredity, but its basic molecular mechanisms are still not fully understood. At present, the standard drugs for OA treatment are mainly non-steroidal anti-inflammatory drugs (NSAIDs), opioids, and steroid injections, which can alleviate the symptoms of OA (Poulet and Staines, 2016). However, long-term use of these drugs may lead to a series of adverse effects, including gastrointestinal, renal, and cardiovascular complications (Benyamin et al., 2008; Harirforoosh et al., 2013). Therefore, the discovery of novel pharmacological and physiological pathways may provide potential targets for new drugs to improve the management of OA.
Bone marrow mesenchymal stem cells (BMSCs), a type of pluripotent cell, are the most commonly used stem cells in cell therapy and tissue repair (Fu et al., 2019). Previous studies have shown that BMSC transplantation can facilitate cartilage repair and wound and fracture healing (Chen et al., 2016; Wang et al., 2018), and has significant therapeutic effects on renal damage, articular cartilage injury, and myocardial damage (Zhang et al., 2015; Li L. et al., 2018; Yang et al., 2018). In addition, increasing evidence has indicated that BMSCs can produce and release a series of bioactive factors during the repair process, such as chemokines, cytokines, and growth factors, which directly stimulate target cells to emit functional active mediators in a paracrine manner to play their protective and endogenous regenerative roles (Francois et al., 2013; Oh et al., 2018). However, there are some limitations in BMSC transplantation, such as the survival time of transplanted cells in vivo and the immune rejection reaction after transplantation.
Recently, exosomes, as media for cell-to-cell communication, have attracted increasing attention. Exosomes are small (30–300 nm) carriers of proteins, lipids, and nucleic acids (DNA and RNA) (Qin and Xu, 2014). Exosomes isolated from BMSCs have been reported to display prominent and typical functions in BMSCs (Baglio et al., 2015). He et al. (2020) showed that BMSC-derived exosomes could effectively promote cartilage repair, extracellular matrix synthesis, and knee pain relief in an OA rat model. Another study reported that lncRNA PVT1 in BMSC-derived exosomes promoted osteosarcoma growth and metastasis via miR-183-5p/ERG (Zhao et al., 2019). LncRNAs have been reported to play important roles in many biological processes, including proliferation, apoptosis, metabolism, and metastasis (Qu et al., 2016; Li X. et al., 2018). Additionally, lncRNAS can serve as a competitive endogenous RNA (ceRNA) to facilitate mRNA expression by sponging microRNAs (miRNAs). A study by Li Z. et al. (2019) showed that exosomal ZFAS1 knockdown induced apoptosis in esophageal squamous cell carcinoma cells and suppressed their proliferation and invasion via the STAT3/miR-124 axis. In MSCs, exosomal lncRNAs have also been reported as novel regulators of osteogenesis (Tye et al., 2018). However, the specific mechanisms of lncRNAs in OA remain unclear, and ceRNA networks in OA have not been fully elucidated.
In addition, chondrocytes play an important role in the development of OA. Inflammation and inflammatory cytokines are closely associated with the occurrence and progression of OA (Goldring and Otero, 2011). Interleukin-1β (IL-1β), a pro-inflammatory cytokine, has been reported to have a potential effect on the destruction of articular cartilage and has been used to establish the inflammatory environment of OA (Feng et al., 2017). Therefore, in this study, IL-1β was used to treat chondrocytes to construct an OA cell model, and the effects of exosomes isolated from human BMSCs on OA were explored. Subsequently, inflammatory chondrocytes and chondrocytes treated with exosomes were sequenced, and a ceRNA network was selected for further verification. These findings provide novel therapeutic targets and pathways for treating OA.
Materials and Methods
Culture of Chondrocytes
Chondrocytes were purchased from CHI Scientific Inc. (Jiangsu, China). The chondrocytes were cultured in DMEM/F12 medium containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, United States), 100 U/mL penicillin (Gibco), and 100 μg/mL streptomycin (Gibco), and maintained in an incubator with 5% CO2 at 37°C. The chondrocytes were passaged upon reaching 80–90% confluency.
The chondrocytes were identified by type II collagen immunohistochemistry staining. Briefly, the chondrocytes were fixed with 4% paraformaldehyde for 20 min, and then were incubated with 0.5% Triton X-100 (Sigma-Aldrich, United States) for 15 min. After that, 3% H2O2 was added, and incubated for 15 min. After washing with PBS, the chondrocytes were incubated with anti-collagen II antibody (1:100, Proteintech, Chicago, United States) overnight. Afterward, the cells were treated with HRP-labeled secondary antibody (1:2000, Jackson ImmunoResearch Laboratories, Inc.) for 1 h. After washing, the cells were stained with diaminobenzidine (DAB, Beyotime Biotechnology) for 5 min, and then were redyed with hematoxylin (Sigma-Aldrich). Finally, the chondrocytes were observed under a microscope under a 100 × magnification.
Isolation and Identification of Exosomes From BMSCs
Human BMSCs were purchased from Cyagen Biosciences Inc. (Guangzhou, China). Human BMSCs were cultured in α-MEM (Gibco) with 5% UltraGRO-Advanced (Gibco) and incubated for 48 h. The cell supernatant (100 mL, approximately 107 cells) was collected for exosome isolation. Exosomes were isolated from the BMSC supernatant by differential centrifugation at 4°C (Bobrie et al., 2012). Briefly, the obtained BMSC supernatant was centrifuged at 300 g for 10 min, and then the supernatant was transferred to a new tube. After centrifugation at 10,000 g for 30 min, the supernatant was transferred to a new tube and centrifuged at 100,000 g for 70 min. The sediment was resuspended in 1 mL PBS and then centrifuged at 10,000 g for 60 min. The exosomes were resuspended in 200 μL PBS and kept either at −80°C for long-term preservation or at −20°C for short-term preservation.
The concentration of exosomes was measured using a BCA Protein Concentration Assay Kit (BOSTER) following the manufacturer’s protocol. Based on the method of Zhu et al. (2018), the morphology and ultrastructure of the exosomes were visualized using transmission electron microscopy (TEM, JEOL Ltd., United States). Exosome size distribution was determined using a Nanosight NS300 particle size analyzer (NTA; Malvern Panalytical, Malvern, UK) as described by the method of Soares Martins et al. (2018). Additionally, according to the method described by Yin et al. (2019), the protein levels of CD63, CD9, and HSP70, which are specific proteins of exosomes, were evaluated by western blotting with anti-CD63 antibody (1:2,000, ABclonal), anti-CD9 antibody (1:2,000, Abcam), and anti-HSP70 antibody (1:2,000, Proteintech).
Establishment of a Chondrocyte Inflammatory Model
Chondrocytes were seeded in a 6-well plate. After the cells adhered to the plate, they were treated with 10 ng/mL IL-1β for 24 h. Then, the inflammatory chondrocytes were treated with different concentrations of exosomes (0, 1, 5, 10, 20, and 50 μg/mL). Cells without treatment served as blank controls. After culturing for 24, 48, and 72 h, cell viability and cell apoptosis were assessed.
Cellular Uptake of Exosomes in Chondrocytes
Cellular uptake of exosomes in chondrocytes was explored by labeling with PKH67 (green fluorescent cell linker for general cell membrane labeling) using a commercial kit (PKH67GL-1KT; Sigma-Aldrich, United States), according to the manufacturer’s instructions. Briefly, Diluent C (200 μL) was added to the exosomes, and then 300 μL of Diluent C and 4 μL PKH67 dye were added. The mixture was incubated at room temperature for 5 min. Subsequently, 1% bovine serum albumin (600 μL BSA; Sigma-Aldrich, United States) was added to bind excess dye. The mixture was centrifuged at 120,000 g for 60 min, and the sediment (PKH67-labeled exosomes) was resuspended in PBS for use.
The chondrocytes were seeded into a 24-well plate at a density of 1 × 104 cells/well and cultured overnight. Next, 10 ng/mL IL-1β was added to the cells for 24 h, and then 10 μg/mL PKH67-labeled exosomes were added. After incubation for 24 h, the chondrocytes were fixed with 4% paraformaldehyde for 15 min. After washing, the cells were stained with 4, 6-diamidino-2-phenylindole (DAPI) and observed under a laser scanning confocal microscope (TCS SP8, Leica Microsystems, Inc., United States) at a magnification of 400×.
Whole Transcriptome Sequencing
Inflammatory cells treated with PBS and 20 μg/mL exosomes were used as the IL-1β and IL-1β + Exos groups. The chondrocytes in the IL-1β and IL-1β + Exos groups were sequenced by Yanzai Biotechnology (Shanghai) Co., Ltd. (Shanghai, China), as previously described (Park and Kim, 2016). The total RNA was extracted from the cells with different treatments using mirVanaTM miRNA Isolation Kit (Thermo Fisher Scientific), and used for whole transcriptome sequencing. DESeq was used to identify the differentially expressed genes (DEGs) between inflammatory cells and inflammatory cells treated with exosomes, including differentially expressed lncRNAs (DE-lncRNAs) and differentially expressed miRNAs (DE-miRNAs). The thresholds for screening DE-lncRNAs and DE-miRNAs were log2Fold change (FC) > 2, P-value < 0.05, and log2Fold change (FC) > 1, P-value < 0.05, respectively. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on these DE-lncRNAs and DE-miRNAs. Next, ceRNA networks were analyzed, and a ceRNA network (ENST00000663198.1 [LYRM4-AS1]—ENSG00000126010 (GRPR)—hsa-miR-6515-5p) was selected for further study. Additionally, six DE-lncRNAs (three upregulated and downregulated) were chosen for real-time quantitative PCR (RT-qPCR) verification, and the sequences of the six DE-lncRNAs are shown in Table 1.
TABLE 1.
The sequences of all primers.
| Primer | Sequence (5′–3′) | |
| ENST00000652428 (LINC00310-209) | F | CCGTGGAATGTCTTTGGC |
| R | TGCGTGCTGGAGGATGAA | |
| ENST00000622968 (AC118344.2-201) | F | TCTATTCTGCTGCTCCATT |
| R | CGCCCTCGTATCTTGTAT | |
| ENST00000654466.1 (AC025171.1-205) | F | TCGCAAACAGCATTACAT |
| R | TTTCCAAGTTCCGAAGAG | |
| ENST00000572856.1 (DLGAP1-AS2-201) | F | CACAGGCTACCACCACTC |
| R | CTTCTTCATGCACGCTCT | |
| ENST00000656992.1 (UBA6-AS1-209) | F | CATACTGCCCAAGATAAT |
| R | CTCTGTTCTGTTCCGTTG | |
| ENST00000626826.1 (HELLPAR-201) | F | CAGCCCTCTACTTACCCA |
| R | TGTCAAGTGTTCTTACGG | |
| ENST00000663198.1 (LYRM4-AS1) | F | CCCAACTGTAACGACCAC |
| R | TTAACATCAAACTAAGGCAC | |
| IL-1β | F | ATGATGGCTTATTACAGTGGCAA |
| R | GTCGGAGATTCGTAGCTGGA | |
| IL-6 | F | CCTGAACCTTCCAAAGATGGC |
| R | TTCACCAGGCAAGTCTCCTCA | |
| GAPDH | F | TGACAACTTTGGTATCGTGGAAGG |
| R | AGGCAGGGATGATGTTCTGGAGAG | |
| miR-6515-5p | F | GCCGTTGGAGGGTGTGGAA |
| R | GTCGTATCCAGTGCAGGGTCCGAG | |
| GTATTCGCACTGGATACGACGATGTC | ||
| U6 | F | CTCGCTTCGGCAGCACA |
| R | AACGCTTCACGAATTTGCGT |
Dual-Luciferase Reporter Gene Assay
The sequences of LYRM4-AS1, hsa-miR-6515-5p mimics, and GRPR 3-untranslated region (3-UTR) were synthesized by Yanzai Biotechnology (Shanghai) Co., Ltd. (Shanghai, China). The psiCHECK2 vector (Yanzai Biotechnology (Shanghai) Co., Ltd.) and pGL3-basic vector (Yanzai Biotechnology (Shanghai) Co., Ltd.) were used to construct the psiCHECK2-LYRM4-AS1 reporter plasmid (psiCHECK2-LYRM4-AS1) and the 3′-UTR GRPR reporter plasmid (pGL3-GRPR). Next, psiCHECK2 (500 ng) or psiCHECK2-LYRM4-AS1 (500 ng), and pGL3-basic vector (500 ng) or pGL3-GRPR (500 ng) were co-transferred into 293T cells with has-miR-6515-5p mimics (100 nM) or negative control (NC) mimics (100 nM) using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s recommendations. A dual luciferase reporter system (Promega, Madison, WI, United States) was used to detect luciferase reactivity.
Cell Transfection
Small interfering (si)-lncRNA LYRM4-AS1 or si-negative control (NC), and miR-6515-5p inhibitor or NC inhibitor were prepared and purchased from Yanzai Biotechnology Co., Ltd. (Shanghai, China). Cell transfection was performed as described previously (Lin et al., 2017). Briefly, chondrocytes were seeded in a 24-well plate. After the cells reached a density of 70%, the cell culture medium was changed to serum-free medium, and the cells were transfected with 100 nM si-lncRNA LYRM4-AS1 or si-NC, and 100 nM miR-6515-5p inhibitor or NC inhibitor using Lipofectamine 3000 (Thermo Fisher Scientific), according to the manufacturer’s instructions. After transfection for 6 h, the medium was replaced with complete medium, and IL-1β (10 ng/mL) was added to the cells. After treatment for another 48 h, total RNA of the cells with different treatments was isolated, and the expression levels of lncRNA LYRM4-AS1 and miR-6515-5p were determined using RT-qPCR to assess the transfection efficiency. The primer sequences of the lncRNA LYRM4-AS1 and miR-6515-5p are shown in Table 1.
Cell Viability and Cell Apoptosis Assays
Cell viability of the chondrocytes with different treatments was measured using the Cell Counting Kit-8 (CCK-8, Beyotime Biotechnology, Shanghai, China) following the manufacturer’s protocols. The cells were treated with different treatments, and 10 μL of CCK-8 was added. After incubation for 2 h, the absorbance was measured at 450 nm using a microplate reader (Multiskan MK3; Thermo Fisher Scientific).
The Annexin V-FITC/PI apoptosis assay kit (Beyotime Biotechnology) was used to assess cell apoptosis of the chondrocytes with different treatments in accordance with the manufacturer’s recommendations. The cells were harvested and centrifuged at 1000 g for 5 min. After washing with PBS, the cells were resuspended in 1 × binding buffer (100 μL). Next, 5 μL of FITC-Annexin V and 5 μL of PI (50 μg/mL) were added. After incubation at 25°C in the dark for 15 min, 400 μL of 1 × binding buffer was added, and the images were acquired by flow cytometry. The apoptosis rate was calculated using CellQuest software (Becton, Dickinson and Company, NJ, United States).
RT-qPCR
Total RNA was extracted from the cells with different treatments using TRIzol reagent (Thermo Fisher Scientific) and 2 times volume of isopropanol for RNA precipitate (Thermo Fisher Scientific) according to the manufacturer’s instructions. The purity and concentration of the total RNA were evaluated using a microplate reader. Stem ring method was used to determine the levels of miR-6515-5p (Wang et al., 2021), and U6 was served as a reference gene. Total RNA was reverse transcribed into cDNA using a cDNA Reverse Transcription Kit (Takara Bio Inc.) according to the manufacturer’s protocol. The RT-qPCR reaction conditions were as follows: pre-denaturation at 95°C for 3 min; 40 cycles at 95°C for 10 s and 60°C for 30 s; melt curve at 60–95°C with an increment of 0.5°C for 10 s. The sequences of all primers are listed in Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a housekeeping gene, and the relative mRNA expression of Interleukin-1β (IL-1β) and IL-6 was calculated using the 2–ΔΔCt method (Xu et al., 2019).
Western Blot
Total protein was isolated from the chondrocytes with different treatments using RIPA protein lysis buffer, and protein concentrations were determined using a BCA assay kit (BOSTER). The protein samples (20 μg) were separated by 10% SDS-PAGE and then transferred to PVDF membranes. After blocking with 5% skim milk at 37°C for 2 h, the membranes were incubated with anti-matrix metallopeptidase 13 (MMP13) antibody (1:4,000; Proteintech Group, Inc., Rosemont, IL, United States), anti-AKT serine/threonine kinase 1 (AKT1) antibody (1:2,000, Proteintech Group, Inc.), anti-phosphatase and tensin homolog (PTEN) antibody (1:4,000, Bioss), anti-gastrin-releasing peptide receptor (GRPR) antibody (1:2,000, Abcam), and anti-GAPDH antibody (1:2,000, Abcam) at 4°C overnight. The membranes were then incubated with the secondary antibody (goat anti-rabbit IgG-HRP, 1:1,000; Jackson ImmunoResearch Laboratories Inc., PA, United States). After incubation at 37°C for 2 h, the Millipore ECL system (Shanghai Tanon Technology Co., Ltd., Shanghai, China) was used to visualize the protein bands.
Statistical Analysis
Data are expressed as the mean ± standard deviation (SD). GraphPad Prism 5 (GraphPad Software, San Diego, CA) was used to perform all statistical analyses. For the comparison of two groups, Student’s t-test was used. One-way analysis of variance (ANOVA) and Tukey’s test were used to compare the differences between more than two groups. Statistical significance was set at p < 0.05.
Results
Identification of Exosomes Isolated Form Human BMSCs
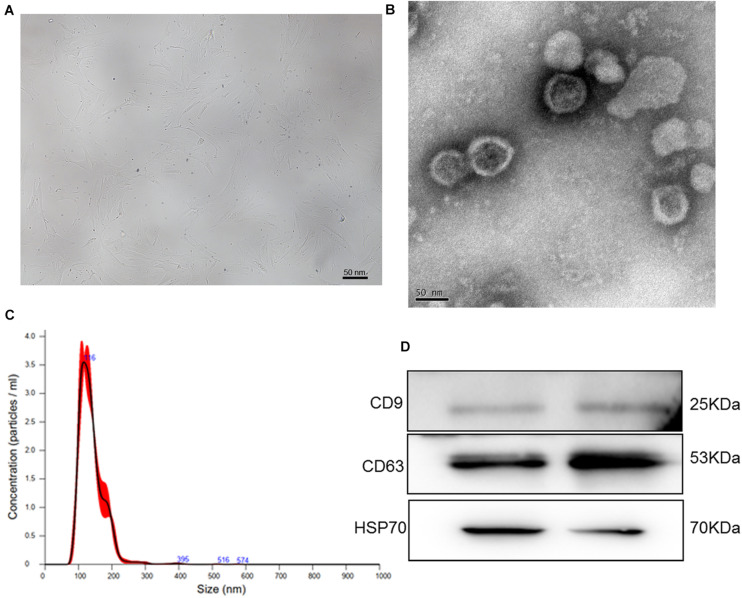
To isolate exosomes from human BMSCs, human BMSCs were cultured for the first time. After the cells reached 80% confluence, BMSCs displayed proliferation ability and showed a relatively homogeneous population of spindle cells (Figure 1A). Thereafter, exosomes were extracted from human BMSCs and identified by TEM, NTA, and western blotting. TEM showed that exosomes exhibited a cup-shaped or nearly round morphology with a diameter of approximately 100 nm (Figure 1B). NTA revealed that the major peak of particle size was approximately 116 nm (Figure 1C), which was in accordance with the size distribution of exosomes reported previously (Tkach and Thery, 2016). Furthermore, western blotting showed that CD9, CD63, and HSP70, which are exosome markers, were expressed in the exosomes (Figure 1D). These results indicate that exosomes were successfully isolated from human BMSCs using the ultracentrifugation method.
FIGURE 1.
(A) The morphology of human bone marrow mesenchymal stem cells (BMSCs). (B) Transmission electron microscopy image of exosomes isolated from human BMSCs. (C) The size distribution of exosomes determined by nanoparticle tracking analysis. (D) The images of exosome-specific CD9, CD63, and HSP70 proteins examined by western blot.
Establishment of a Chondrocyte Inflammatory Model and Cellular Uptake of Exosomes
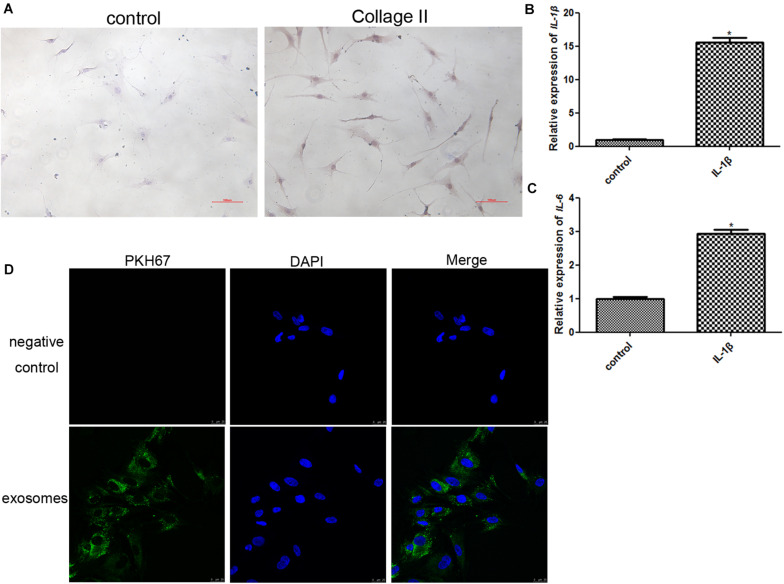
In order to identify the chondrocytes, the expression of collagen II was determined. As shown in Figure 2A, type II collagen immunocytochemical staining confirmed the cellular phenotype of obtained chondrocytes, and showed that the cells were stained dark yellow-brown. Therefore, the positive results of collagen II staining replied that the obtained cells were chondrocytes.
FIGURE 2.
(A) Identification of chondrocytes using immunocytochemical staining of collagen II (100×). (B) The expression level of IL-1β after chondrocytes were induced by IL-1β. *P < 0.05 vs. control group. n = 3. (C) The expression level of IL-6 after chondrocytes were induced by IL-1β. *P < 0.05 vs. control group. n = 3. (D) PKH67-labeled exosomes could be taken up by chondrocytes induced by IL-1β after co-culture.
The chondrocytes were treated with 10 ng/mL IL-1β for 24 h to construct an inflammatory cell model, and the expression of IL-1β and IL6 was determined to assess the model. After IL-1β treatment, the expression of IL-1β and IL6 was significantly upregulated compared to that in the control group (P < 0.05, Figures 2B,C). These results implied that inflammatory chondrocytes were successfully established by treatment with IL-1β.
In addition, PKH67 was used to label exosomes (green fluorescence), and PKH67-labeled exosomes were co-cultured with inflammatory chondrocytes for 24 h. After co-culture, most inflammatory chondrocytes displayed intracellular green fluorescence (Figure 2D). These results indicated that exosomes isolated from human BMSCs could be taken up by chondrocytes induced by IL-1β.
The Effects of Exosomes on the Growth of Chondrocytes Treated With IL-1β and on the Expression of MMP13, AKT1, PTEN, and GRPR
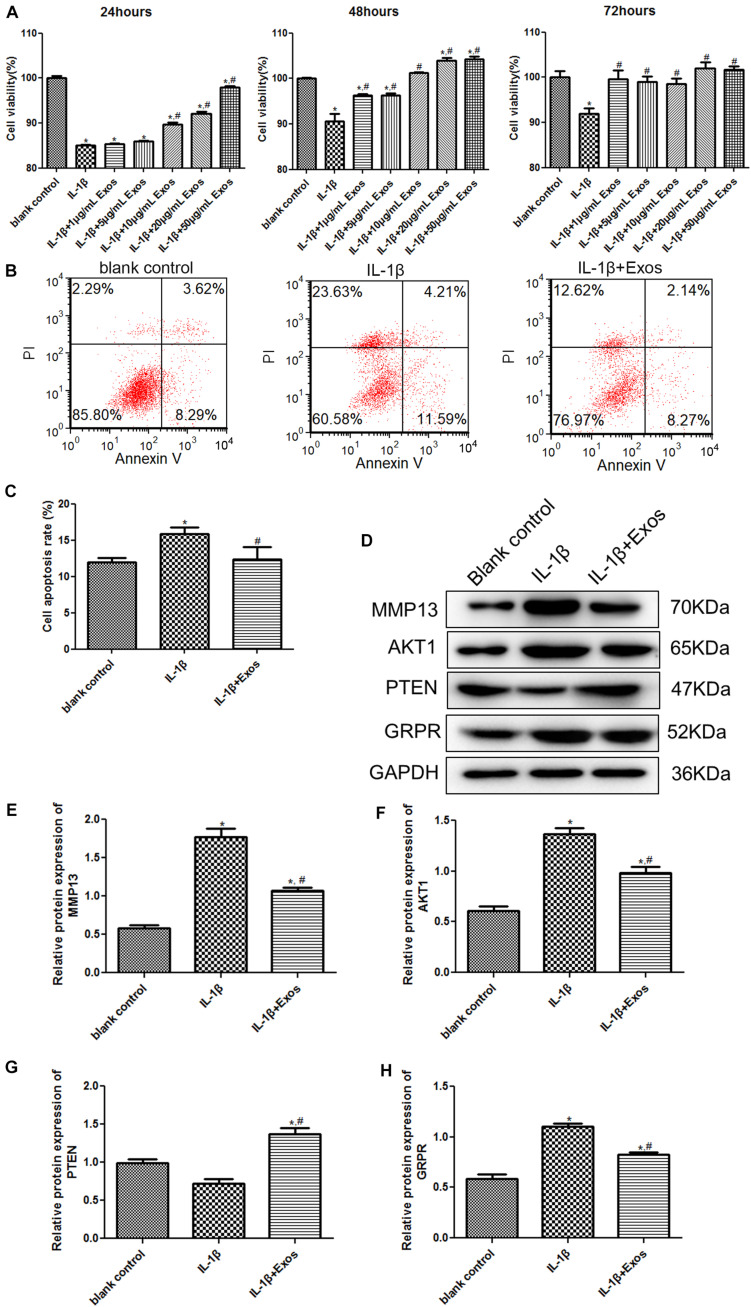
To understand the roles of exosomes in chondrocytes, cell viability and apoptosis were determined. After culturing for 24, 48, and 72 h, there was no significant difference in the cell viability between the normal cells and normal cells treated with exosomes (P > 0.05, Supplementary Figure 1). Therefore, we explored the effects of exosomes on chondrocytes. Compared with the blank control group, the cell viability of chondrocytes was significantly inhibited after IL-1β treatment (P < 0.05, Figure 3A). After treatment with 1 and 5 μg/mL exosomes for 24 h, the cell viability of chondrocytes was similar to that of the IL-1β group (P > 0.05), whereas after treatment with 10, 20, and 50 μg/mL exosomes for 24 h, the cell viability was significantly higher than that of the IL-1β group (P < 0.05). When the inflammatory cells were treated with different concentrations of exosomes for 48 h, the cell viability was significantly increased compared with the IL-1β group (P < 0.05), and when the exosome concentrations were 20 and 50 μg/mL, the cell viability was higher than that in the blank control and IL-1β groups (P < 0.05). Therefore, treatment with 20 μg/mL exosomes for 48 h was selected for subsequent experiments. For cell apoptosis, the cell apoptosis rate in the IL-1β group (15.87 ± 0.49%) was markedly higher than that in the blank control group (11.96 ± 0.34%, P < 0.05, Figures 3B,C); while after treatment with 20 μg/mL exosomes, the cell apoptosis (12.32 ± 1.02%) was restored to a similar level to that of the blank control group (P > 0.05, Figures 3B,C). Taken together, IL-1β treatment suppressed the viability of chondrocytes and promoted their apoptosis, whereas exosomes could properly restore the changes in cell vitality and apoptosis induced by IL-1β.
FIGURE 3.
(A) Cell viability of chondrocytes treated with different concentrations of exosomes for 24, 48, and 72 h using the Cell Counting Kit-8 (CCK-8) assay. n = 3. (B) The images of cell apoptosis acquired by flow cytometry. (C) The cell apoptosis rate of chondrocytes treated with exosomes analyzed by CellQuest software. n = 3. (D) The protein bands acquired by western blot. The protein expressions of MMP13 (E), AKT1 (F), PTEN (G), and GRPR (H) after chondrocytes were treated with exosomes determined by western blot. n = 3. *P < 0.05 vs. blank control group; #P < 0.05 vs. IL-1β group.
To further investigate the molecular mechanisms by which exosomes affect cell viability and apoptosis, the expression of MMP13, AKT1, PTEN, and GRPR was examined by western blotting. Compared with the blank control group, the expression of MMP3 and AKT1 was significantly upregulated after IL-1β treatment, while after exosome treatment, their expression was down-regulated compared to the IL-1β group (Figures 3D–F). For PTEN, there was no significant difference in its expression between the blank control group and the IL-1β group (P > 0.05); however, its expression after exosome treatment was significantly upregulated (Figures 3D,G). In addition, the change trend of GRPR expression was similar to that of MMP13 and AKT expression (Figures 3D,H).
Analyses of Whole Transcriptome Sequencing and RT-qPCR Verification
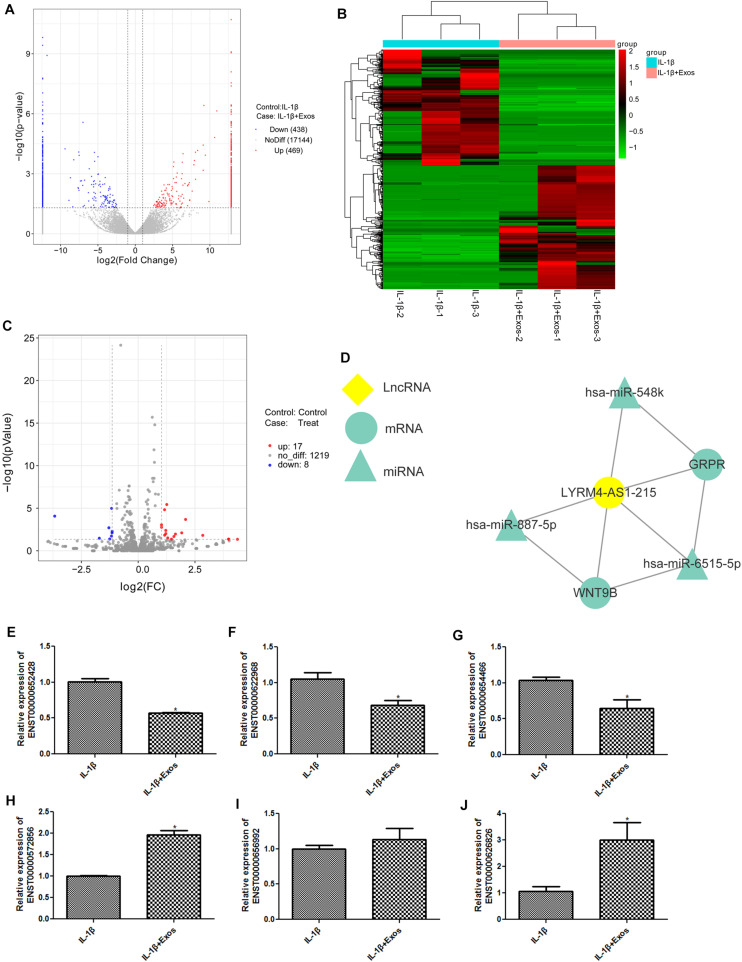
The cells in the IL-1β and IL-1β + Exos groups were sequenced, and DEGs between inflammatory cells and inflammatory cells treated with exosomes were analyzed. A total of 907 DE-lncRNAs were identified, including 438 downregulated lncRNAs and 469 upregulated lncRNAs (Figure 4A). The heatmap distribution of the DE-lncRNA expression is shown in Figure 4B. GO and KEGG analyses were then performed on the DE-lncRNAs (Supplementary Figures 2A,B). Supplementary Figure 1A displays the top10 GO terms in cellular component (CC), molecular function (MF), and biological process (BP), including membrane-bounded organelle, SMAD2-SMAD3 protein complex and cytoplasm in CC; SMAD binding, RNA polymerase II transcription factor activity, and formate-tetrahydrofolate ligase activity in MF; and import across plasma membrane, amino acid transmembrane import, methionine metabolic process, and osteoblast differentiation in BP. These DE-lncRNAs were also enriched in the relaxin signaling pathway, signaling pathways regulating pluripotency of stem cells, protein digestion and absorption, cell cycle, D-arginine and D-ornithine metabolism, IL-17 signaling pathway, and TGF-beta signaling pathway (Supplementary Figure 2B).
FIGURE 4.
(A) The volcano plot of differentially expressed lncRNAs (DE-lncRNAs). (B) The heat map of DE-lncRNAs. (C) The volcano plot of differentially expressed miRNAs (DE-miRNAs). (D) A ceRNA network about lncRNA LYRM4-AS1 was built. (E) The expression of lncRNA ENST00000652428. (F) The expression of lncRNA ENST00000622968. (G) The expression of lncRNA ENST00000654466. (H) The expression of lncRNA ENST00000572856. (I) The expression of lncRNA ENST00000656992. (J) The expression of lncRNA ENST000006268268. n = 3. *P < 0.05 vs. IL-1β group.
Subsequently, 25 DE-miRNAs were screened with the threshold of log2FC > 1 and P-value < 0.05, including 17 upregulated and 8 downregulated miRNAs (Figure 4C and Table 2). These DE-miRNAs were associated with the intrinsic components of the plasma membrane, cell, and plasma membrane region in CC, and DNA binding transcription factor activity, ion binding, protein binding in MF, anatomical structure morphogenesis, regulation of primary metabolic processes, regulation of nitrogen compound metabolic processes, regulation of cellular metabolic processes, and nervous system development in BP (Supplementary Figure 1C). Additionally, these DE-miRNAs were related to the MAPK signaling pathway, TNF signaling pathway, terpenoid backbone biosynthesis, Rap1 signaling pathway, cAMP signaling pathway, and calcium signaling pathway (Supplementary Figure 1D).
TABLE 2.
The differentially expressed miRNAs (DE-miRNAs) between inflammatory cells and inflammatory cells treated with exosomes.
| ID | logFC | P-value | Regulation |
| hsa-miR-4804-5p | 4.369282269 | 0.044934589 | Up regulation |
| hsa-miR-5188 | 3.985399519 | 0.043346613 | Up regulation |
| hsa-miR-6802-3p | 3.978271057 | 0.042313985 | Up regulation |
| hsa-miR-124-5p | 2.842255887 | 0.015303299 | Up regulation |
| hsa-miR-3065-5p | 2.086145606 | 0.000207661 | Up regulation |
| hsa-miR-206 | 1.916945659 | 0.007803674 | Up regulation |
| hsa-miR-6515-5p | 1.639455774 | 0.010792166 | Up regulation |
| hsa-miR-4504 | 1.572682075 | 0.020863067 | Up regulation |
| hsa-miR-449a | 1.46759187 | 0.042832155 | Up regulation |
| hsa-miR-133a-3p | 1.30080826 | 0.030564227 | Up regulation |
| hsa-miR-483-3p | 1.259195958 | 3.6597E-06 | Up regulation |
| hsa-miR-548k | 1.217801224 | 0.009536832 | Up regulation |
| hsa-let-7c-3p | 1.214944911 | 0.004027594 | Up regulation |
| hsa-miR-7976 | 1.165499887 | 0.014562738 | Up regulation |
| hsa-miR-585-3p | 1.161558169 | 1.48771E-05 | Up regulation |
| hsa-miR-548e-3p | 1.028993871 | 0.001738362 | Up regulation |
| hsa-miR-4746-5p | 1.026294946 | 0.000885642 | Up regulation |
| hsa-miR-7704 | −3.673672131 | 8.48286E-05 | Down regulation |
| hsa-miR-147a | −1.71092359 | 0.033242609 | Down regulation |
| hsa-miR-376b-5p | −1.290335472 | 0.001974939 | Down regulation |
| hsa-miR-191-3p | −1.254283585 | 0.041073194 | Down regulation |
| hsa-miR-887-5p | −1.174667937 | 0.018450622 | Down regulation |
| hsa-miR-29c-3p | −1.169956008 | 1.03332E-05 | Down regulation |
| hsa-miR-376c-5p | −1.149515746 | 0.007776615 | Down regulation |
| hsa-miR-154-3p | −1.14467181 | 0.005211388 | Down regulation |
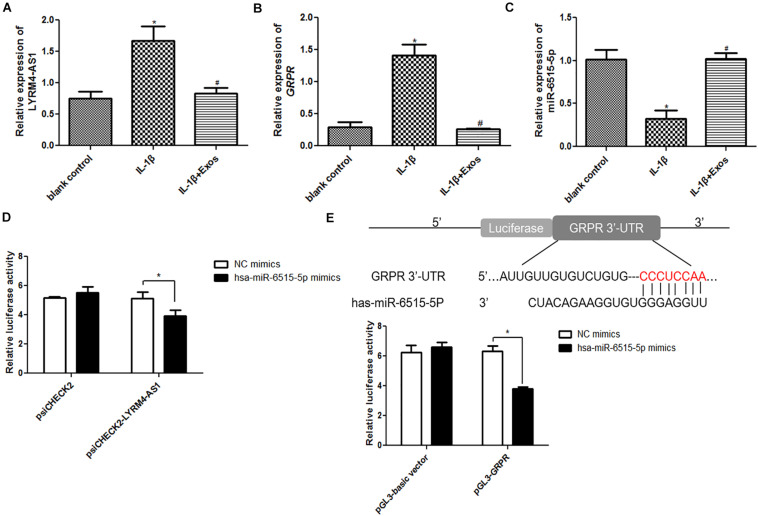
Based on the expression of DE-lncRNAs, DE-miRNAs, and mRNAs obtained by sequencing, we found that the expression of lncRNA LYRM4-AS1 was in line with the forecast, and then a ceRNA network of lncRNA LYRM4-AS1 was built (Figure 4D). It was found that lncRNA LYRM4-AS1 interacted with miR-887-5p, miR-6515, and miR-548k. Because miR-887-5p was downregulated, miR-6515-5p and miR-548k were considered for further study. A previous study showed that the GRP pathway plays an important role in chronic arthritis (Oliveira et al., 2011). Therefore, we selected the lncRNA LYRM4-AS1—mRNA GRPR—miR-6515-5p for further experimental validation.
In addition, three upregulated DE-lncRNAs (ENST 00000572856, ENST00000656992, and ENST00000626826) and three downregulated DE-lncRNAs (ENST00000652428, ENST00000622968, and ENST00000654466) were selected to determine their expression in inflammatory cells and inflammatory cells following exosome treatment. It is clear that the expression of ENST00000652428, ENST00000622968, and ENST00000654466 was significantly downregulated in the IL-1β + Exos group, whereas the expression of ENST00000572856 and ENST00000626826 were both significantly upregulated in the IL-1β + Exos group, compared with the IL-1β group (P < 0.05, Figures 4E–H,J). However, there was no significant difference in ENST00000656992 expression between the two groups (P > 0.05, Figure 4I). The results showed that the consistency rate between sequencing analyses and RT-qPCR results was 83.33%, which indicated a high relative reliability of the sequencing results.
Interaction Among lncRNA LYRM4-AS1, miR-6515-5p, and GRPR
The expression of lncRNA LYRM4-AS1, miR-6515-5p, and GRPR was first determined in the different groups. Compared with the blank control group, lncRNA LYRM4-AS1 and mRNA GRPR expression was significantly upregulated in the IL-1β group (P < 0.05), while exosome treatment downregulated their expression and restored their levels to levels similar to those of the blank control group (P > 0.05, Figures 5A,B). However, for miR-6515-5p, its expression was significantly down-regulated in the IL-1β group compared to the blank control group (P < 0.05), and its expression markedly returned to a level similar to that of the blank control group (P > 0.05, Figure 5C). These results indicated that the expression trends of lncRNA LYRM4-AS1, mRNA GRPR, and miR-6515-5p were consistent with the sequencing results.
FIGURE 5.
The relative expression of lncRNA LYRM4-AS1 (A), mRNA GRPR (B), and miR-6515-5p (C) in the blank control group, IL-1β group, and IL-1β + Exo group. n = 3. *P < 0.05 vs. blank control group; #P < 0.05 vs. IL-1β group. (D) LncRNA LYRM4-AS1 interacted with miR-6515-5p. n = 3. *P < 0.05 vs. negative control (NC) mimics. (E) GRPR directly binds with miR-6515-5p. n = 3. *P < 0.05 vs. NC mimics.
Next, the interaction between lncRNA LYRM4-AS1, miR-6515-5p, and GRPR was analyzed. There was no significant difference between NC mimics and miR-6515-5p mimics in the psiCHECK2 plasmid (P > 0.05), whereas in the psiCHECK2-LYRM4-AS1 plasmid, the relative luciferase activity in the miR-6515-5p mimic group was significantly lower than that in the NC mimics group (P < 0.05, Figure 5D). Additionally, in the pGL3-GRPR plasmid, the relative luciferase activity in the miR-6515-5p mimics group was lower than that in the NC mimics group (P < 0.05, Figure 5E). These results implied that lncRNA LYRM4-AS1, miR-6515-5, and GRPR mRNA could interact with each other.
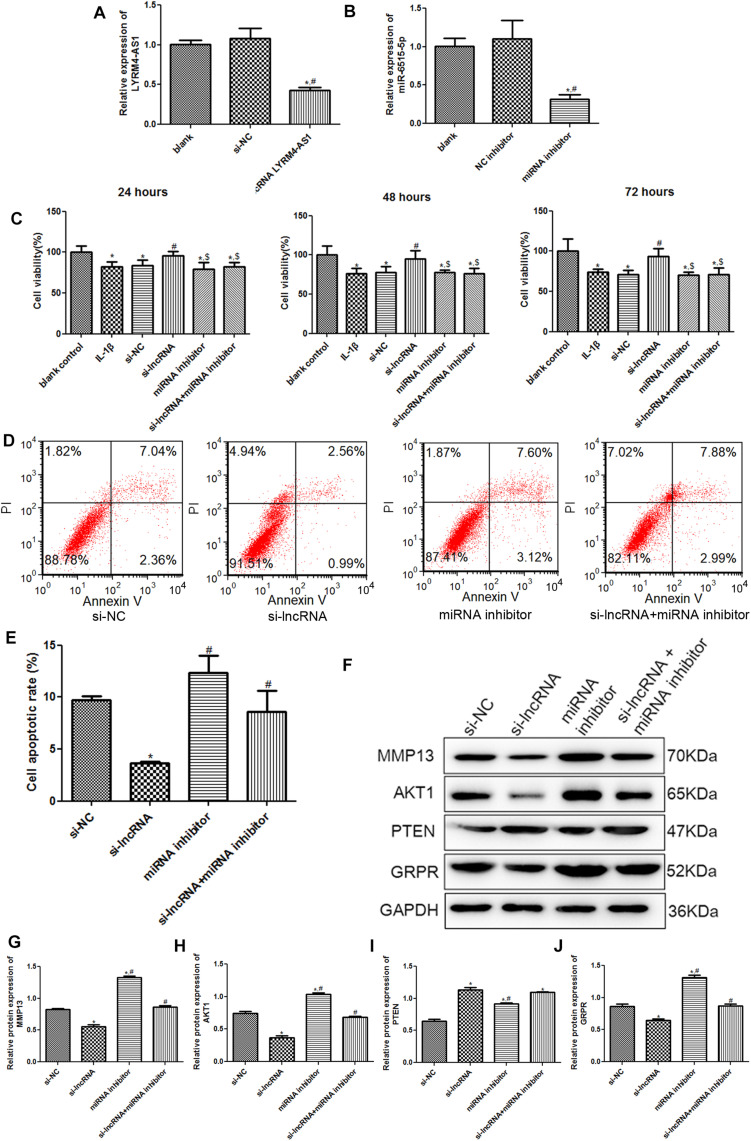
The Effects of lncRNA LYRM4-AS1 on the Growth of Inflammatory Chondrocytes and on the Expression of Related Proteins
To further investigate the roles of lncRNA LYRM4-AS1 in cartilage repair, chondrocytes with lncRNA LYRM4-AS1 interference and miR-6515-5p knockdown were constructed. There was no significant difference in the lncRNA LYRM4-AS1 expression between the blank and si-NC groups (P > 0.05); however, after transfection with si-lncRNA LYRM4-AS1, its expression was significantly decreased (P < 0.05, Figure 6A). miR-6515-5p expression was markedly downregulated after transfection with miR-6515-5p inhibitor compared to that in the blank and NC inhibitor groups (P < 0.05, Figure 6B). These results indicate that chondrocytes with lncRNA LYRM4-AS1 interference and miR-6515-5p knockdown were successfully established.
FIGURE 6.
(A) The relative expression of lncRNA LYRM4-AS1 after transfection. n = 3. *P < 0.05 vs. blank group; #P < 0.05 vs. si-NC group. (B) The relative expression of miR-6515-5p after transfection. n = 3. *P < 0.05 vs. blank group; #P < 0.05 vs. NC inhibitor group. (C) After cell transfection, the cell viability of chondrocytes was examined using CCK-8 assay. n = 3. *P < 0.05 vs. blank control group; #P < 0.05 vs. IL-1β group. $P < 0.05 vs. si-lncRNA LYRM4-AS1 group. (D) The images of cell apoptosis acquired by flow cytometry. (E) The cell apoptosis rate of chondrocytes after transfection analyzed by CellQuest software. n = 3. *P < 0.05 vs. si-NC group; #P < 0.05 vs. si-lncRNA LYRM4-AS1 group. (F) The protein bands of MMP13, AKT1, PTEN, and GRPR detected by western blot. The relative protein expression of MMP13 (G), AKT1 (H), PTEN (I), and GRPR (J) in different groups. n = 3. *P < 0.05 vs. si-NC group; #P < 0.05 vs. si-lncRNA group.
Subsequently, cell viability and apoptosis were detected. After culturing for 24, 48, and 72 h, there was no significant difference in cell viability between the IL-1β and si-NC groups (P > 0.05, Figure 6C). Compared with the blank control group, IL-1β induction significantly inhibited the viability of chondrocytes (P < 0.05). When lncRNA LYRM4-AS1 was knocked down, the cell viability of IL-1β-induced chondrocytes was evidently increased compared with that of the IL-1β group (P < 0.05), and was restored to a level similar to that of the blank control group (P > 0.05). However, in the cells with both lncRNA LYRM4-AS1 and miR-6515-5p knockdown, cell viability was significantly lower than that in the si-lncRNA LYRM4-AS1 group (P < 0.05) and was similar to that of the IL-1β group (P > 0.05, Figure 6C). Next, apoptosis in different groups was analyzed. Compared with the si-NC group, apoptosis was significantly reduced in the si-lncRNA LYRM4-AS1 group (P < 0.05), while in the cells with both lncRNA LYRM4-AS1 and miR-6515-5p knocked down, the cell apoptosis was significantly increased compared with the si-lncRNA LYRM4-AS1 group (P < 0.05, Figures 6D,E). These results indicated that lncRNA LYRM4-AS1 interference could enhance cell viability and suppress cell apoptosis in IL-1β-induced chondrocytes, while miR-6515-5p knockdown reversed the changes in cell viability and apoptosis caused by lncRNA LYRM4-AS1 interference.
Finally, the protein expression of MMP13, AKT1, PTEN, and GRPR in different groups was examined by western blotting (Figure 6F). Compared to the si-NC group, lncRNA LYRM4-AS1 interference significantly downregulated the expression of MMP3, AKT1, and GRPR (P < 0.05); however, miR-6515-5p knockdown markedly reversed the expression changes of MMP3, AKT1, and GRPR induced by lncRNA LYRM4-AS1 interference (Figures 6G,H,J). The expression of PTEN after lncRNA LYRM4-AS1 interference was significantly higher than that in the si-NC group. After miR-6515-5p knockdown, its expression was also higher than that in the si-NC group but was lower than that in the si-lncRNA group (Figure 6I). In addition, there was no significant difference in PTEN expression between the si-lncRNA and si-lncRNA + miRNA inhibitor groups (P > 0.05, Figure 6I).
Discussion
OA, one of the most prevalent chronic diseases, is a degenerative joint disease involving cartilage and many surrounding tissues, and seriously reduces health and quality of life (Litwic et al., 2013). Since the occurrence and development of OA involves complex molecular mechanisms associated with inflammation and degeneration of articular cartilage, there are still many obstacles in the establishment of improved therapies for OA (Wei and Bai, 2016). In our study, exosomes were successfully isolated from human BMSCs and used to treat IL-1β-induced chondrocytes. It was found that exosome treatment significantly increased the viability of chondrocytes induced by IL-1β and inhibited their apoptosis. Then, the cells in the IL-1β and IL-1β + Exos groups were sequenced, and a total of 907 DE-lncRNAs (438 downregulated and 469 upregulated) and 25 DE-miRNAs (8 downregulated and 17 upregulated) were identified. Afterward, a ceRNA network on lncRNA LYRM4-AS1 was constructed, and lncRNA LYRM4-AS1—mRNA GRPR—miR-6515-5p was chosen for further study. The dual-luciferase reporter gene indicated that LYRM4-AS1, miR-6515-5, and GRPR interacted with each other. Additionally, LYRM4-AS1 knockdown enhanced cell viability and suppressed the apoptosis of IL-1β-induced chondrocytes, while miR-6515-5p inhibition reversed the changes caused by LYRM4-AS1 knockdown. These results suggest that exosomes could exert a protective effect on OA in vitro, and LYRM4-AS1/GRPR/miR-6515-5p may be a novel pathway for OA therapy.
Exosomes, a subset of extracellular vesicles generated by all cell types, have been reported to play essential roles in intercellular communication (Meldolesi, 2018). Previous studies have demonstrated the great therapeutic potential of MSC-derived exosomes in joint disease (Li Z. et al., 2018) and the importance of MSC paracrine secretion in a variety of diseases (Linero and Chaparro, 2014). In this study, TEM, NTA, and western blot results showed that exosomes were successfully isolated from human BMSCs, and then exosomes were used to treat IL-1β-induced chondrocytes. It is clear that IL-1β significantly inhibited the viability of chondrocytes and induced cell apoptosis, while exosomes were able to restore the changes in cell vitality and apoptosis induced by IL-1β. A study by Wang et al. (2017) indicated that exosomes isolated from embryonic MSCs could remit OA by balancing the degradation and synthesis of chondrocyte ECM. Another study reported that MSC-derived exosomes could promote cell viability and inhibit cell apoptosis of inflammatory chondrocytes, and may be a novel cell-free therapeutic approach for OA treatment (Qi et al., 2019). Therefore, we speculated that exosomes isolated from BMSCs could protect against OA by regulating cell viability and apoptosis of chondrocytes.
Thereafter, the protein expression levels of MMP3, AKT, GRPR, and PTEN were determined. It was found that the expression of MMP3, AKT, and GRPR was up-regulated after IL-1β induction, and after exosome treatment, the expression was downregulated. Interestingly, there was no significant difference in PTEN expression between the control and IL-1β groups; however, exosomes markedly upregulated PTEN expression. MMP3, a member of the MMP family, has been reported to be highly expressed in OA and rheumatoid arthritis (Tong et al., 2017; Lerner et al., 2018). Zeng et al. (2019) showed that high expression of MMP3 was observed in OA synovial cells, and curcumin could control cell proliferation and induce cell apoptosis by inhibiting MMP3 expression, eventually alleviating OA inflammation. AKT, a serine/threonine protein kinase, is an effector of the PI3K/AKT signaling pathway (Malemud, 2015) and is involved in multiple biological processes, including cell growth, apoptosis, migration, and metabolism (Zhang et al., 2018). PTEN is an important negative regulator of the PI3K/AKT signaling pathway, and loss of PTEN can lead to the activation of the PI3K/AKT signaling pathway (Xi et al., 2020). A study by Zhang et al. (2018) indicated that miRNA-130a could regulate cell growth and relieve OA via the PTEN/PI3K/AKT signaling pathway. Additionally, GRPR is a member of the G-protein coupled receptor superfamily and affects cell growth and invasion (Elshafae et al., 2016). Combined with our results, it can be inferred that exosomes isolated from BMSCs may mediate cell viability and apoptosis induced by IL-1β by downregulating MMP3, AKT, and GRPR expression and upregulating PTEN, thus alleviating OA.
To further investigate the specific molecular mechanisms of exosomes, the cells in the IL-1β and IL-1β + Exos groups were sequenced. A total of 907 DE-lncRNAs, including 438 downregulated and 469 upregulated, and 25 DE-miRNAs, including 8 downregulated and 17 upregulated, were identified. These DE-lncRNAs were involved in SMAD binding, osteoblast differentiation, relaxin signaling pathway, IL-17 signaling pathway, and TGF-beta signaling pathway. Additionally, these DE-miRNAs were enriched in the regulation of nitrogen compound metabolic process, MAPK signaling pathway, TNF signaling pathway, Rap1 signaling pathway, cAMP signaling pathway, and calcium signaling pathway. A previous study has shown that SMAD/TGF-β signaling plays an important role in inflammation in temporomandibular joint OA (Li W. et al., 2019). IL-17A can induce osteoblast differentiation (Jo et al., 2018) and calcium-phosphate complexes can serve as potential catabolic mediators in cartilage (Jung et al., 2018). It has been reported that relaxin and cAMP are associated with anti-fibrogenic effects on OA synoviocytes (Qadri et al., 2018; Ko et al., 2019). Additionally, the activation of the p38MAPK pathway contributes to the overexpression of pro-inflammatory cytokines and chemokines and plays important roles in the progression of OA (Chen et al., 2018). Taken together, we speculated that exosomes may reduce inflammation of IL-1β-induced chondrocytes via the relaxin signaling pathway, IL-17 signaling pathway, TGF-beta signaling pathway, MAPK signaling pathway, TNF signaling pathway, cAMP signaling pathway, and calcium signaling pathway. Further studies should be performed to verify the role of these pathways in OA.
Exosomes contain mRNA, miRNA, lncRNA, and other signal substances, and can carry out information exchange between cells by targeting proximal cells in an autocrine and paracrine manner and targeting distal cells via the circulatory system (Colombo et al., 2014). LncRNAs, a type of non-coding RNA with a length of more than 200 bases, are regulators that do not encode proteins. MiRNAs are multifunctional non-coding RNAs with 22–25 bases and can regulate mRNA expression. Additionally, ceRNAs can sponge miRNAs to affect the expression of target mRNAs. Previous studies have shown that lncRNAs and miRNAs have regulatory effects on chondrocyte growth and inflammation, and play essential roles in the progression of OA (Swingler et al., 2019; Xie et al., 2020). Shen et al. (2018) found that lncRNA SNHG5 could serve as a ceRNA to compete with miR-26a and regulate SOX2 expression, thus participating in OA pathogenesis. In our study, by sequencing, we found a ceRNA network (lncRNA LYRM4-AS1—mRNA GRPR—miR-6515-5p) associated with the development of OA. A dual-luciferase reporter gene assay was used to confirm that LYRM4-AS1, miR-6515-5, and GRPR could interact with each other. The results of cell experiments showed that LYRM4-AS1 could mediate chondrocyte viability and apoptosis by regulating miR-6515-5p and GRPR. LYRM4-AS1 is related to integration hotspots in the human genome (Yang-Chun et al., 2020) and has not been explored in OA. GRPR, a gastrin-releasing peptide receptor, has been reported to be involved in the healing process in the synovium in patients with OA (Grimsholm et al., 2008). Xu et al. (2020) demonstrated that miR-6515-5p was highly expressed in P2 × 7R gene-modified stem cell-derived exosomes and reduced inflammation-mediated impairment of periodontal ligament stem cells through indirect binding to the GREM-1 protein. Together with our results, we speculate that exosomes isolated from human BMSCs may regulate chondrocyte growth by the LYRM4-AS1/GRPR/miR-6515-5p signal axis, reducing inflammation in OA.
However, this study has some limitations. First, the roles of exosomes and the LYRM4-AS1/GRPR/miR-6515-5p signal axis in OA need to be further investigated in vivo. In addition, experiments on the effects of relaxin signaling pathway, IL-17 signaling pathway, TGF-beta signaling pathway, MAPK signaling pathway, TNF signaling pathway, cAMP signaling pathway, and calcium signaling pathway in OA should be carried out. In conclusion, exosomes isolated from human BMSCs could exert a protective effect on OA, and the possible molecular protective mechanisms of exosomes in OA may be associated with MMP3, AKT, PTEN, and GRPR expression and the LYRM4-AS1/GRPR/miR-6515-5p signaling pathway. In addition, exosomes may alleviate OA inflammation through the relaxin signaling pathway, IL-17 signaling pathway, TGF-beta signaling pathway, MAPK signaling pathway, TNF signaling pathway, cAMP signaling pathway, and calcium signaling pathway. Our findings provide a theoretical basis for the application of exosomes in the treatment of OA and provide a basis for novel LYRM4-AS1/GRPR miR-6515-5p therapeutic targets and pathways for OA treatment.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XW and ZW designed the experiments. XW, ZL, YC, XC, and CC performed the experiments and analyzed the experimental results. ZW obtained the funding and supervised the experiments. XW drafted the manuscript and ZW revised the manuscript. All authors have reviewed and approved the submitted version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Featured Clinical Discipline Project of Shanghai Pudong New District (PWYts2018-2), the Construction of Key Discipline Group of Sanitary System of Shanghai Pudong New District (PWZxq2017-12), the Mechanism of Prevotella Coptic Regulating the Progression of Senile Osteoporosis (PKJ2020-Y44), and Shanghai Municipal Key Specialty Construction Fund of Shanghai Municipal Health Bureau (no. ZK2019B05).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.644380/full#supplementary-material
References
- Baglio S. R., Rooijers K., Koppers-Lalic D., Verweij F. J., Perez Lanzon M., Zini N., et al. (2015). Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 6:127. 10.1186/s13287-015-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyamin R., Trescot A. M., Datta S., Buenaventura R., Adlaka R., Sehgal N., et al. (2008). Opioid complications and side effects. Pain Physician 11(2 Suppl) S105–S120. [PubMed] [Google Scholar]
- Bobrie A., Colombo M., Krumeich S., Raposo G., Thery C. (2012). Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 1:18397. 10.3402/jev.v1i0.18397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shou K., Gong C., Yang H., Yang Y., Bao T. (2018). Anti-inflammatory effect of geniposide on osteoarthritis by suppressing the activation of p38 MAPK signaling pathway. Biomed. Res. Int. 2018:8384576. 10.1155/2018/8384576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang S., Du X., Bai B. (2016). Parathyroid hormone-induced bone marrow mesenchymal stem cell chondrogenic differentiation and its repair of articular cartilage injury in rabbits. Med. Sci. Monit. Basic Res. 22 132–145. 10.12659/msmbr.900242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G. A., Théry C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30 255–289. [DOI] [PubMed] [Google Scholar]
- Elshafae S. M., Hassan B. B., Supsavhad W., Dirksen W. P., Camiener R. Y., Ding H., et al. (2016). Gastrin-releasing peptide receptor (GRPr) promotes EMT, growth, and invasion in canine prostate cancer. Prostate 76 796–809. 10.1002/pros.23154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Li X., Lin J., Zheng W., Hu Z., Xuan J., et al. (2017). Oleuropein inhibits the IL-1beta-induced expression of inflammatory mediators by suppressing the activation of NF-kappaB and MAPKs in human osteoarthritis chondrocytes. Food Funct. 8 3737–3744. 10.1039/c7fo00823f [DOI] [PubMed] [Google Scholar]
- Francois S., Mouiseddine M., Allenet-Lepage B., Voswinkel J., Douay L., Benderitter M., et al. (2013). Human mesenchymal stem cells provide protection against radiation-induced liver injury by antioxidative process, vasculature protection, hepatocyte differentiation, and trophic effects. Biomed. Res. Int. 2013:151679. 10.1155/2013/151679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Liu G., Halim A., Ju Y., Luo Q., Song A. G. (2019). Mesenchymal stem cell migration and tissue repair. Cells 8:784. 10.3390/cells8080784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring M. B., Otero M. (2011). Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23 471–478. 10.1097/BOR.0b013e328349c2b1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsholm O., Rantapää-Dahlqvist S., Dalén T., Forsgren S. (2008). Observations favouring the occurrence of local production and marked effects of bombesin/gastrin-releasing peptide in the synovial tissue of the human knee joint–comparisons with substance P and the NK-1 receptor. Neuropeptides 42 133–145. 10.1016/j.npep.2007.12.008 [DOI] [PubMed] [Google Scholar]
- Guilak F., Nims R. J., Dicks A., Wu C. L., Meulenbelt I. (2018). Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 7 40–50. 10.1016/j.matbio.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harirforoosh S., Asghar W., Jamali F. (2013). Adverse effects of nonsteroidal antiinflammatory drugs: an update of gastrointestinal, cardiovascular and renal complications. J. Pharm. Pharm. Sci. 16 821–847. 10.18433/j3vw2f [DOI] [PubMed] [Google Scholar]
- He L., He T., Xing J., Zhou Q., Fan L., Liu C., et al. (2020). Bone marrow mesenchymal stem cell-derived exosomes protect cartilage damage and relieve knee osteoarthritis pain in a rat model of osteoarthritis. Stem Cell Res. Ther. 11:276. 10.1186/s13287-020-01781-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Wang S. E., Lee Y. L., Kang S., Lee B., Han J., et al. (2018). IL-17A induces osteoblast differentiation by activating JAK2/STAT3 in ankylosing spondylitis. Arthritis Res. Ther. 20:115. 10.1186/s13075-018-1582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson H., Olafsdottir S., Sigurdardottir S., Aspelund T., Eiriksdottir G., Sigurdsson S., et al. (2016). Incidence and prevalence of total joint replacements due to osteoarthritis in the elderly: risk factors and factors associated with late life prevalence in the AGES-Reykjavik Study. BMC Musculoskelet. Disord. 17:14. 10.1186/s12891-016-0864-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y. K., Han M. S., Park H. R., Lee E. J., Jang J. A., Kim G. W., et al. (2018). Calcium-phosphate complex increased during subchondral bone remodeling affects earlystage osteoarthritis. Sci. Rep. 8:487. 10.1038/s41598-017-18946-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. H., Kang Y. M., Yang J. H., Kim J. S., Lee W. J., Kim S. H., et al. (2019). Regulation of MMP and TIMP expression in synovial fibroblasts from knee osteoarthritis with flexion contracture using adenovirus-mediated relaxin gene therapy. Knee 26 317–329. 10.1016/j.knee.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Lankhorst N. E., Damen J., Oei E. H., Verhaar J. A. N., Kloppenburg M., Bierma-Zeinstra S. M. A., et al. (2017). Incidence, prevalence, natural course and prognosis of patellofemoral osteoarthritis: the Cohort Hip and Cohort Knee study. Osteoarthritis Cartilage 25 647–653. 10.1016/j.joca.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Lerner A., Neidhofer S., Reuter S., Matthias T. (2018). MMP3 is a reliable marker for disease activity, radiological monitoring, disease outcome predictability, and therapeutic response in rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 32 550–562. 10.1016/j.berh.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Li L., Duan X., Fan Z., Chen L., Xing F., Xu Z., et al. (2018). Mesenchymal stem cells in combination with hyaluronic acid for articular cartilage defects. Sci. Rep. 8:9900. 10.1038/s41598-018-27737-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhao S., Yang H., Zhang C., Kang Q., Deng J., et al. (2019). Potential novel prediction of TMJ-OA: MiR-140-5p regulates inflammation through smad/TGF-beta signaling. Front. Pharmacol. 10:15. 10.3389/fphar.2019.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lei Y., Wu M., Li N. (2018). Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int. J. Mol. Sci. 19:2958. 10.3390/ijms19102958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Qin X., Bian W., Li Y., Shan B., Yao Z., et al. (2019). Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J. Exp. Clin. Cancer Res. 38:477. 10.1186/s13046-019-1473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang Y., Xiao K., Xiang S., Weng X. (2018). Emerging role of exosomes in the joint diseases. Cell Physiol. Biochem. 47 2008–2017. 10.1159/000491469 [DOI] [PubMed] [Google Scholar]
- Lin Y., Jin Y., Xu T., Zhou S., Cui M. (2017). MicroRNA-215 targets NOB1 and inhibits growth and invasion of epithelial ovarian cancer. Am. J. Transl. Res. 9 466–477. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Linero I., Chaparro O. (2014). Paracrine effect of mesenchymal stem cells derived from human adipose tissue in bone regeneration. PLoS One 9:e107001. 10.1371/journal.pone.0107001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwic A., Edwards M. H., Dennison E. M., Cooper C. (2013). Epidemiology and burden of osteoarthritis. Br. Med. Bull. 105 185–199. 10.1093/bmb/lds038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malemud C. J. (2015). The PI3K/Akt/PTEN/mTOR pathway: a fruitful target for inducing cell death in rheumatoid arthritis? Future Med. Chem. 7 1137–1147. 10.4155/fmc.15.55 [DOI] [PubMed] [Google Scholar]
- Meldolesi J. (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol. 28 R435–R444. 10.1016/j.cub.2018.01.059 [DOI] [PubMed] [Google Scholar]
- Nelson A. E. (2018). Osteoarthritis year in review 2017: clinical. Osteoarthritis Cartilage 26 319–325. 10.1016/j.joca.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E. J., Lee H. W., Kalimuthu S., Kim T. J., Kim H. M., Baek S. H., et al. (2018). In vivo migration of mesenchymal stem cells to burn injury sites and their therapeutic effects in a living mouse model. J. Control Release 279 79–88. 10.1016/j.jconrel.2018.04.020 [DOI] [PubMed] [Google Scholar]
- Oliveira P. G., Grespan R., Pinto L. G., Meurer L., Brenol J. C., Roesler R., et al. (2011). Protective effect of RC-3095, an antagonist of the gastrin-releasing peptide receptor, in experimental arthritis. Arthritis Rheum. 63 2956–2965. 10.1002/art.30486 [DOI] [PubMed] [Google Scholar]
- Park S. T., Kim J. (2016). Trends in next-generation sequencing and a new era for whole genome sequencing. Int. Neurourol. J. 20(Suppl. 2) S76–S83. 10.5213/inj.1632742.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet B., Staines K. A. (2016). New developments in osteoarthritis and cartilage biology. Curr. Opin. Pharmacol. 28 8–13. 10.1016/j.coph.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Qadri M. M., Jay G. D., Ostrom R. S., Zhang L. X., Elsaid K. A. (2018). cAMP attenuates TGF-beta’s profibrotic responses in osteoarthritic synoviocytes: involvement of hyaluronan and PRG4. Am. J. Physiol. Cell Physiol. 315 C432–C443. 10.1152/ajpcell.00041.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Liu D. P., Xiao D. W., Tian D. C., Su Y. W., Jin S. F. (2019). Exosomes derived from mesenchymal stem cells inhibit mitochondrial dysfunction-induced apoptosis of chondrocytes via p38, ERK, and Akt pathways. In Vitro Cell Dev. Biol. Anim. 55 203–210. 10.1007/s11626-019-00330-x [DOI] [PubMed] [Google Scholar]
- Qin J., Xu Q. (2014). Functions and application of exosomes. Acta Pol. Pharm. 71 537–543. [PubMed] [Google Scholar]
- Qu L., Ding J., Chen C., Wu Z. J., Liu B., Gao Y., et al. (2016). Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29 653–668. 10.1016/j.ccell.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Shen H., Wang Y., Shi W., Sun G., Hong L., Zhang Y. (2018). LncRNA SNHG5/miR-26a/SOX2 signal axis enhances proliferation of chondrocyte in osteoarthritis. Acta Biochim. Biophys. Sin. 50 191–198. 10.1093/abbs/gmx141 [DOI] [PubMed] [Google Scholar]
- Soares Martins T., Catita J., Martins Rosa I., da Cruz E Silva O. A. B., Henriques A. G. (2018). Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One 13:e0198820. 10.1371/journal.pone.0198820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingler T. E., Niu L., Smith P., Paddy P., Le L., Barter M., et al. (2019). The function of microRNAs in cartilage and osteoarthritis. Clin. Exp. Rheumatol. 37(Suppl. 120) 40–47. [PubMed] [Google Scholar]
- Tkach M., Thery C. (2016). Communication by extracellular vesicles: where we are and where we need to go. Cell 164 1226–1232. 10.1016/j.cell.2016.01.043 [DOI] [PubMed] [Google Scholar]
- Tong Z., Liu Y., Chen B., Yan L., Hao D. (2017). Association between MMP3 and TIMP3 polymorphisms and risk of osteoarthritis. Oncotarget 8 83563–83569. 10.18632/oncotarget.18745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye C. E., Boyd J. R., Page N. A., Falcone M. M., Stein J. L., Stein G. S., et al. (2018). Regulation of osteogenesis by long noncoding RNAs: an epigenetic mechanism contributing to bone formation. Connect. Tissue Res. 59 35–41. 10.1080/03008207.2017.1412432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li L., Hu X., Liu T., Jiang W., Wu R., et al. (2021). Effects of atrial fibrillation-derived exosome delivery of mir-107 to human umbilical vein endothelial cells. DNA Cell Biol. 40 568–579. 10.1089/dna.2020.6356 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang C., Gou W., Xu X., Wang Y., Wang A., et al. (2018). The optimal time to inject bone mesenchymal stem cells for fracture healing in a murine model. Stem Cell Res. Ther. 9:272. 10.1186/s13287-018-1034-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yu D., Liu Z., Zhou F., Dai J., Wu B., et al. (2017). Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res. Ther. 8:189. 10.1186/s13287-017-0632-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Bai L. (2016). Recent advances in the understanding of molecular mechanisms of cartilage degeneration, synovitis and subchondral bone changes in osteoarthritis. Connect. Tissue Res. 57 245–261. 10.1080/03008207.2016.1177036 [DOI] [PubMed] [Google Scholar]
- Xi Y., Ma J., Chen Y. (2020). PTEN promotes intervertebral disc degeneration by regulating nucleus pulposus cell behaviors. Cell Biol. Int. 44 583–592. 10.1002/cbin.11258 [DOI] [PubMed] [Google Scholar]
- Xie F., Liu Y.-L., Chen X.-Y., Li Q., Zhong J., Dai B.-Y., et al. (2020). Role of MicroRNA, LncRNA, and exosomes in the progression of osteoarthritis: a review of recent literature. Orthop. Surg. 12 708–716. 10.1111/os.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Ao R., Zhi Z., Jia J., Yu B. (2019). miR-21 and miR-19b delivered by hMSC-derived EVs regulate the apoptosis and differentiation of neurons in patients with spinal cord injury. J. Cell. Physiol. 234 10205–10217. 10.1002/jcp.27690 [DOI] [PubMed] [Google Scholar]
- Xu X. Y., Tian B. M., Xia Y., Xia Y. L., Li X., Zhou H., et al. (2020). Exosomes derived from P2X7 receptor gene-modified cells rescue inflammation-compromised periodontal ligament stem cells from dysfunction. Stem Cells Transl. Med. 9 1414–1430. 10.1002/sctm.19-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. C., Zhu X. L., Wang J., Wan F., Zhang H. Q., Lin Y., et al. (2018). Bone marrow mesenchymal stem cells attenuate the progression of focal segmental glomerulosclerosis in rat models. BMC Nephrol. 19:335. 10.1186/s12882-018-1137-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Chun F., Sen-Yu W., Yuan Z., Yan-Chun H. (2020). Genome-wide profiling of human papillomavirus DNA integration into human genome and its influence on PD-L1 expression in chinese uygur cervical cancer women. J. Immunol. Res. 2020:6284960. 10.1155/2020/6284960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Zeng A., Zhang Z., Shi Z., Yan W., You Y. (2019). Exosomal transfer of miR-1238 contributes to temozolomide-resistance in glioblastoma. EBioMedicine 42 238–251. 10.1016/j.ebiom.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J. J., Wang H. D., Shen Z. W., Yao X. D., Wu C. J., Pan T. (2019). Curcumin inhibits proliferation of synovial cells by downregulating expression of matrix metalloproteinase-3 in osteoarthritis. Orthop. Surg. 11 117–125. 10.1111/os.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu D., Li S., Chang L., Zhang Y., Liu R., et al. (2015). Bone marrow mesenchymal stem cell transplantation retards the natural senescence of rat hearts. Stem Cells Transl. Med. 4 494–502. 10.5966/sctm.2014-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Huang E., Zhou H., Li B., Shao C., et al. (2018). MicroRNA-130a regulates chondrocyte proliferation and alleviates osteoarthritis through PTEN/PI3K/Akt signaling pathway. Int. J. Mol. Med. 41 3699–3708. 10.3892/ijmm.2018.3551 [DOI] [PubMed] [Google Scholar]
- Zhao W., Qin P., Zhang D., Cui X., Gao J., Yu Z., et al. (2019). Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging (Albany NY) 11 9581–9596. 10.18632/aging.102406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Li Q., Niu X., Zhang G., Ling X., Zhang J., et al. (2018). Extracellular vesicles secreted by human urine-derived stem cells promote ischemia repair in a mouse model of hind-limb ischemia. Cell Physiol. Biochem. 47 1181–1192. 10.1159/000490214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.