Abstract
Meis genes have been shown to control essential processes during development of the central and peripheral nervous system. Here we have explored the roles of the Meis2 gene during vertebrate inner ear induction and the formation of the cochlea. Meis2 is expressed in several tissues required for inner ear induction and in non-sensory tissue of the cochlear duct. Global inactivation of Meis2 in the mouse leads to a severely reduced size of the otic vesicle. Tissue-specific knock outs of Meis2 reveal that its expression in the hindbrain is essential for otic vesicle formation. Inactivation of Meis2 in the inner ear itself leads to an aberrant coiling of the cochlear duct. By analyzing transcriptomes obtained from Meis2 mutants and ChIPseq analysis of an otic cell line, we define candidate target genes for Meis2 which may be directly or indirectly involved in cochlear morphogenesis. Taken together, these data show that Meis2 is essential for inner ear formation and provide an entry point to unveil the network underlying proper coiling of the cochlear duct.
Keywords: inner ear, cochlea, Meis, organ of corti, mouse
Introduction
Development of the inner ear begins as a thickening of the ectoderm adjacent to the posterior hindbrain termed the otic placode, which can be observed at embryonic day 8 (E 8) in the mouse. In vertebrates, induction of the otic placode requires the interaction with neighboring tissues such as the neural tissue of the hindbrain, the mesoderm and/or the endoderm. Members of the fibroblast growth factor (Fgf) gene family such as Fgf3 have been shown to play a central role during otic placode induction (Schimmang, 2007; Whitfield, 2015). After E8 in the mouse, the otic placode invaginates and forms the otic vesicle, which undergoes a series of morphogenetic steps to form the complex shape of the mature inner ear. Cochlear morphogenesis is initiated at the ventral portion of the otic vesicle that elongates and coils in an anterior–medial direction until it reaches its full one and three−quarters turns. This process is paralleled by cellular differentiation that leads to the formation of sensory cells such as hair cells and auditory neurons, and non-sensory components within the cochlear duct (Basch et al., 2016).
Meis genes are vertebrate orthologs of the Drosophila homolog homothorax (hth) gene which encode for transcription factors belonging to the superclass of TALE (three amino acid loop extension) proteins. Meis genes play key roles during development of the central and peripheral nervous systems, and interact with signaling pathways such as those controlled by Wnt, Fgf and retinoic acid (Schulte and Frank, 2014). The TALE superclass of proteins contains an atypical homeodomain and comprises five separate classes: Meinox, including Prep (Prep1–2 genes) and Meis (Meis1–3 genes), Pbc (Pbx1–4 genes) and the three more distantly related TG-interacting factors, Iroquois, and Mohawk (Schulte and Geerts, 2019). A prominent characteristic of Meinox proteins is their capacity to heterodimerize with the structurally related Pbx proteins. A further group of Meis protein-binding partners participating in these cooperative interactions is the Hox proteins. In this case, Meis proteins often do not interact directly with DNA (Penkov et al., 2013).
In the present work, we have analyzed the role of Meis2 during inner ear induction and cochlear development. We show that during inner ear induction Meis2 is prominently expressed in the hindbrain neighboring the otic placode and to a lesser extent in the periotic mesoderm and endoderm. Specific inactivation in the rhombomeres flanking the otic placode, led to reduced otic vesicles, uncovering a dominant role of hindbrain Meis2 in otic vesicle formation. Inactivation of Meis2 within the otic placode caused morphological defects including improper coiling of the cochlea. Microarray analysis revealed a set of genes that were downregulated in mutant cochleas, representing potential Meis2 target genes required for proper morphogenesis of the cochlear duct. Finally, ChIPseq analysis in an otic cell line with the potential to give rise to sensory and non-sensory cochlear tissue (VOT-E36) allowed the detection of genes whose regulatory regions bind Meis2 directly or indirectly. Our results thus show that Meis2 is essential for otic vesicle formation and cochlear duct morphogenesis.
Results
Meis2 Expression During Inner Ear Development
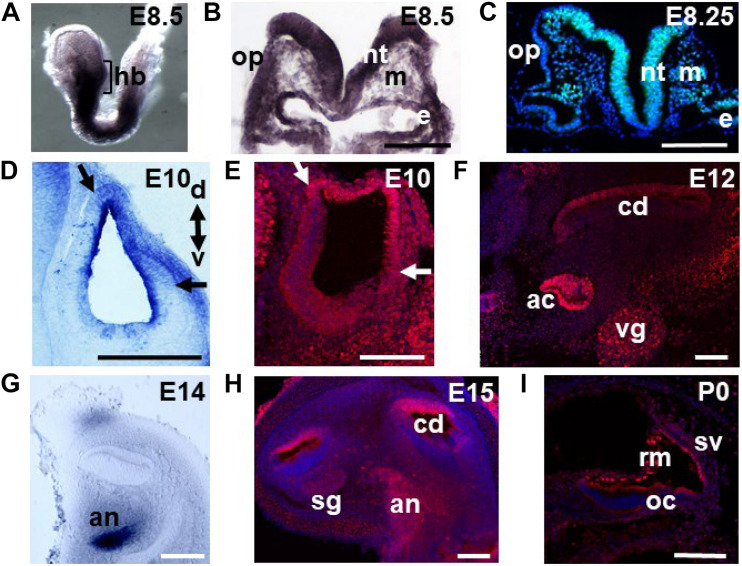
In order to define Meis2 expression throughout inner ear development, we performed whole mount RNA in situ hybridization and immunohistochemistry studies. During inner ear induction around E8–8.5, high levels of mRNA were detected in the developing hindbrain (Figure 1A). Sections of the posterior hindbrain confirmed this expression and revealed Meis2 transcripts in the otic placode and the neighboring mesoderm and endoderm (Figure 1B). Immunohistochemistry confirmed high levels of Meis2 protein in the hindbrain and moderate to low amounts in the endoderm, mesoderm and otic placode (Figure 1C). Upon formation of the otic vesicle Meis2 immunoreactivity was observed in its dorsolateral domain, accompanied by low but detectable Meis2 mRNA (Figures 1D,E).
FIGURE 1.
Meis2 expression throughout inner ear development. Meis2 expression was detected via mRNA in situ hybridization (A,B,D,G) or immunohistochemistry (C,E,F,H,I) at the indicated stages. (A) At embryonic day (E) 8.5 high levels of Meis2 are detected at the level of the hindbrain (hb) and on corresponding sections at the level of the otic placode (op) (B). High levels of Meis2 protein are detected in the neural tube (nt), the endoderm (e), and weaker expression in the mesoderm (m) and the placode itself (C). (D) In the otic vesicle low Meis2 mRNA levels are observed in its dorsal-lateral portion (borders indicated by arrows). The orientations of the sections through the otic vesicles along the dorsal (d)–ventral (v) axis are indicated. (E) Immunohistochemical detection of Meis2 protein in the dorsal-lateral quadrant of the otic vesicle. (F) Presence of Meis2 protein in the roof of the cochlear duct (cd), the vestibular ganglion (vg) and the ampullary cristae (ac). (G) High levels of Meis2 mRNA are detected in the auditory nerve (an). (H) Next to the auditory nerve, Meis2 protein is also observed in the roof of the cochlear duct and to a lesser extent in the spiral ganglion (sg). (I) At P0 Meis2 immunoreactivity is detected in Reissner’s membrane (rm) and the stria vascularis (sv). oc, organ of Corti. Scale bars in (B,C,E–H): 100 μm; in (D): 200 μm and in (I): 75 μm.
During inner ear morphogenesis around E12, Meis2 protein was detected in the roof of the cochlear duct, which will give rise to non-sensory tissue (Figure 1F). Expression in this domain was maintained at E15 and was also detected in the auditory nerve at the protein and shortly before at the mRNA level (Figures 1G,H). Low levels of anti-Meis2 immunoreactivity were also observed in the spiral ganglion (Figure 1H). At postnatal day 0 (P0), Meis2 protein was observed in Reissner’s membrane and the stria vascularis but not in the sensory tissue corresponding to the organ of Corti (Figure 1I). Adjacent to the cochlea, Meis2 protein was also detected in different parts of the vestibular system such as the ampullary cristae, the utriculus, the semicircular canals and the vestibular ganglion (Figure 1F and data not shown).
Meis2 Is Required for Otic Vesicle Formation
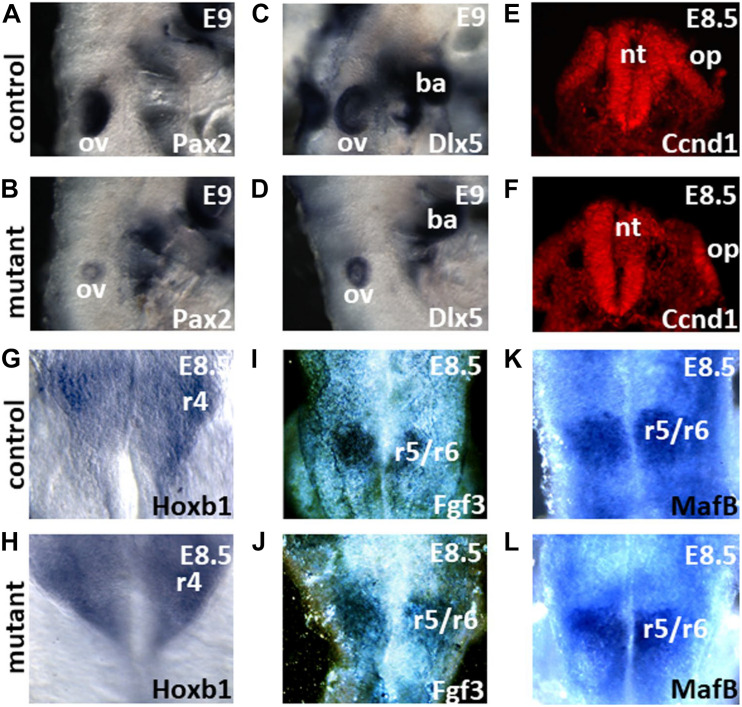
To analyze the requirement of Meis2 for inner ear formation, we inactivated its expression throughout the epiblast by crossing a mouse strain carrying a floxed Meis2 allele with a Sox2-Cre deleter strain (Hayashi et al., 2002). Meis2flox/flox; Sox2Cre/+ embryos showed a severely reduced otic vesicle at E9 as revealed by in situ hybridization with the otic markers Pax2 and Dlx5 (Figures 2A–D), confirming similar observations made in Meis2 null mutants (Machon et al., 2015). Therefore, Meis2 is required for otic vesicle formation.
FIGURE 2.
Global inactivation of Meis2 affects inner ear formation. (A–D) Meis2flox/flox; Sox2Cre/+ mutants show severely reduced otic vesicles (ov) at embryonic day 9 (E9) in comparison to controls as revealed by Pax2 and Dlx5 riboprobes. (E,F) Sections reveal widespread cyclinD1 immunoreactivity in the neural tube (nt) and the otic placode (op) of controls and mutants. Note the reduced size of the otic placode in the mutant. (G–L) Flatmounts of the hindbrain reveal an unaltered expression of Hoxb1, Fgf3 and MafB riboprobes in the indicated rhombomeres (r) in Meis2flox/flox; Sox2Cre/+ mutants in comparison to control embryos. ba, branchial arch.
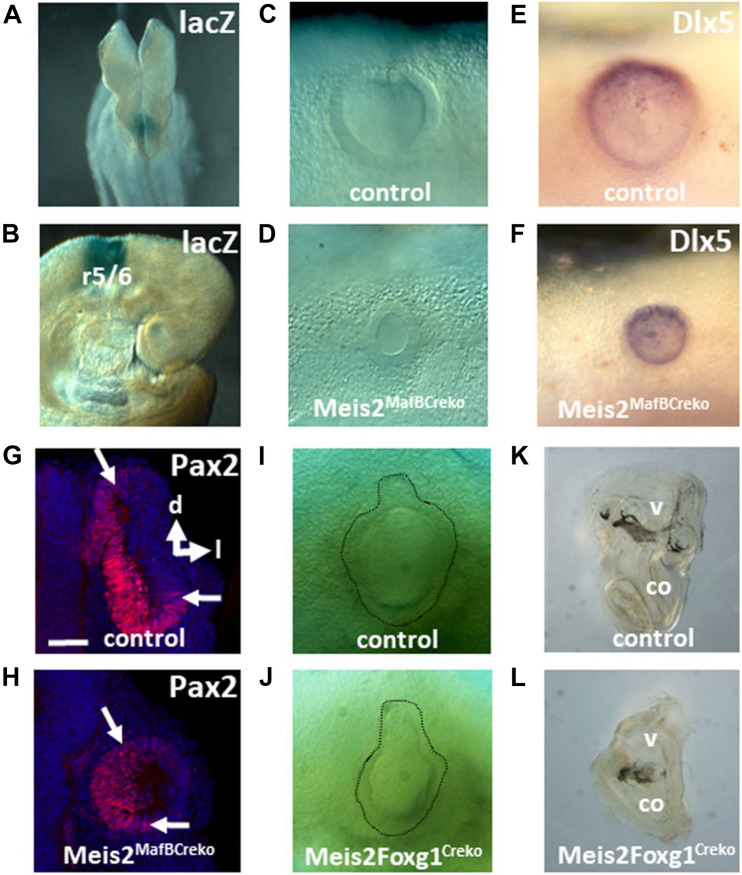
Next, we were interested to determine the tissue-specific requirements for Meis2 during otic induction. Since Meis2 was strongly expressed in the hindbrain during otic development, we sought to find a Cre line with a specific activity in the hindbrain. The MafB gene is expressed in the posterior hindbrain next to the developing otic placode and vesicle (Cordes and Barsh, 1994). Recently, a MafB-Cre line has been employed for lineage tracing of macrophages which are also characterized by endogenous MafB expression (Wu et al., 2016). To assay if Cre activity also occurred in the developing hindbrain, we crossed MafB-Cre transgenic mice with ROSA26 reporter mice (Soriano, 1999). Cre activity was detected by X-gal staining at E8.5 in the posterior hindbrain and in rhombomeres flanking the otic vesicle at E9 (Figures 3A,B and Supplementary Figure 1). This confirmed its utility for the inactivation of floxed genes during inner ear formation. In order to study the requirement for hindbrain Meis2 expression during otic vesicle formation, we analyzed the effects of Meis2 conditional inactivation induced by MafB-Cre at the otic vesicle stage. Meis2flox/flox; MafBCre/+ mutants showed a severely reduced vesicle at E9 comparable to the size observed upon global Meis2 inactivation with a penetrance of 50% (n = 2/4; Figures 3C,D). Therefore Meis2 expression in the hindbrain is required for otic vesicle formation. Staining with the otic markers Dlx5 and Pax2 revealed that their expression was maintained in their corresponding domains in Meis2flox/flox; MafBCre/+ mutants (Figures 3E–H). To analyze the influence of Meis2 expression in neighboring tissue during otic induction, we used a Foxg1-Cre line which next to the otic placode also drives expression in peri-otic mesoderm and endoderm (Zelarayan et al., 2007; Supplementary Figure 1). Conditional inactivation of Meis2 using the Foxg1-Cre line did not affect inner ear formation until E11.5 when a reduced sized otic vesicle with a flask-like shape was observed in Meis2flox/flox; Foxg1Cre/+ mutants (Figures 3I,J). Meis2flox/flox; Foxg1Cre/+ inner ears isolated at P0 and cleared with methylsalicylate revealed no discernible structures such as the cochlea or the semicircular canals (Figures 3K,L).
FIGURE 3.
Effects of tissue-specific inactivation of Meis2 on inner ear formation. (A,B) lacZ ROSA26 reporter staining caused by the MafBCre driver in the posterior hindbrain of E8.25 embryos and in rhombomeres (r) flanking the otic vesicle at E9 (B). (C–H) Meis2flox/flox; MafBCre/+ mutants show reduced sized vesicles (C,D) which maintain expression of Dlx5 (at E9) and Pax2 (at E9,5), as revealed by whole mount RNA in situ hybridization (E,F) and immunohistochemistry (G,H). The orientations of the sections through the otic vesicles along the dorsal (d)–lateral (l) axis are indicated. (I–L) Bright field view of otic vesicles at E11.5 and cleared inner ears at P1 from Meis2flox/flox; Foxg1Cre/+ mutants. Note the flask-shaped morphology of the otic vesicle and the lack of a discernible cochlea (co) in the mutants. v, vestibule.
With the aim to define the molecular changes underlying defective inner ear induction, we analyzed Meis2flox/flox; Sox2Cre/+ mutants which show severely reduced sized otic vesicles with a penetrance of 100% for alterations in gene expression or signaling pathways known to be controlled by Meis genes and related to inner ear formation. Formation of the hindbrain requires Meis gene expression in lower vertebrates and has been shown to activate Fgf3 expression which is redundantly required together with other Fgf family members to induce the otic placode in vertebrates (Maroon et al., 2002; Alvarez et al., 2003; Wright and Mansour, 2003; Ladher et al., 2005; Gutkovich et al., 2010). Moreover, Fgf3 has been shown to participate in the induction of MafB (Zelarayan et al., 2007), and MafB inactivation also leads to reduced sized otic vesicles (Cordes and Barsh, 1994; Wiellette and Sive, 2003; Hernandez et al., 2004). Lastly, Meis proteins together with Fgf signaling have also been shown to induce in the rhombencephalon expression of Hox genes that belong to the paralogous group 1 (PG1) and which are likewise required for in inner ear induction (Rossel and Capecchi, 1999; Pasqualetti et al., 2001; Schulte and Frank, 2014). RNA in situ hybridization with probes corresponding to Fgf3, MafB and the PG1 gene HoxB1 showed no changes in their expression pattern within rhombomeres flanking the developing otic placode in Meis2flox/flox; Sox2Cre/+ mutants (Figures 2G–L).
Alternatively to hindbrain patterning defects, reduced proliferation may cause the smaller sized vesicles. Meis has been shown to regulate cyclinD1 and thereby to control eye size in vertebrates (Bessa et al., 2008; Marcos et al., 2015). CyclinD1 was prominently expressed in the hindbrain and the neighboring otic placode of both controls and Meis2flox/flox; Sox2Cre/+ mutants (Figures 2E,F).
Conditional Inactivation of Meis2 During Inner Ear Development Affects Cochlear Coiling
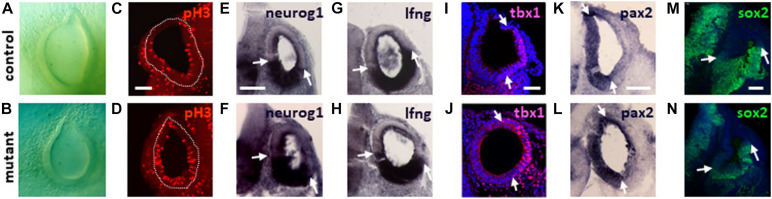
To analyze the function of Meis2 expression in the inner ear, we generated conditional mutant mice using a Cre line driven by Pax2 regulatory sequences which has been used to inactivate floxed alleles throughout inner ear development (Ohyama and Groves, 2004; Supplementary Figure 1). No morphological defects in inner ear formation were found until E10.5 in Meis2flox/flox; Pax2Cre/+ mutants, when a reduced sized otic vesicle was observed in comparison to controls (Figures 4A,B).
FIGURE 4.
Expression of otic vesicle markers in Meis2flox/flox; Pax2Cre/+ mutants. Expression of the indicated markers was detected via mRNA in situ hybridization (E–H,K,L) or immunohistochemistry (C,D,I,J,M,N). (A,B) Bright field image of controls and mutants showing a reduced sized otic vesicle at E10,5. (C,D) Sections at E10 reveal widespread pH3 immunoreactivity in the otic vesicles (circumference indicated by stippled lines) of controls and mutants. (E–L) No changes in the spatial domains of the otic markers (borders indicated by arrows) is observed in the mutants at E9 (E–H) and E10 (I–L). (M,N) At E11 the ventral domain of Sox2 expression is smaller in mutants correlating with the reduced sized dimensions of the otic vesicle at this stage. Scale bars in (C,E,I): 50 μm; in (K): 100 μm and in (M): 75 μm.
The smaller size of the otic vesicle observed in Meis2flox/flox; Pax2Cre/+ mutants may be caused by a lack of proliferation. To examine cell proliferation, we used staining with an antibody against pH3 that labels cells in late G2 and M phase of the cell cycle. Immunoreactivity for pH3 was observed throughout the otic epithelium of both wild-type and Meis2flox/flox; Pax2Cre/+ mutants (Figures 4C,D). To examine potential changes in molecular markers in mutant otic vesicles, we first examined the neurosensory region. This develops in an anterior-medial domain of the otic vesicle and is characterized by the expression of Neurogenin 1 (Ngn1) and Lunatic fringe (Lfng). Complementary to Ngn1 expression, genes like Tbx1 (T-box transcription factor 1) stabilize the neurogenic region (for a review, see Bok et al., 2007). No difference in the expression patterns of these markers was observed in Meis2flox/flox; Pax2Cre/+ mutants when compared to control otic vesicles (Figures 4E–J).
During normal development, Pax2 expression is localized in the medial wall of the otic vesicle (Figure 4K) whereas the ventral portion is characterized by a broad domain of Sox2 at E11.5 from where the cochlear anlage derives (Basch et al., 2016; Figure 4M). Later on, Pax2 and Sox2 label the non-sensory and pro-sensory regions within the developing cochlear duct, respectively (Burton et al., 2004; Mak et al., 2009; Figures 5A,C). Pax2 showed a normal pattern of expression at the otic vesicle stage in Meis2flox/flox; Pax2Cre/+ mutants (Figure 4L) but its domain was severely reduced at E14, pointing to a potential truncation of the cochlear duct (Figure 5B). Sox2 staining at E11.5 revealed a reduced expression domain that correlated with the smaller size of the otic vesicle in Meis2flox/flox; Pax2Cre/+ mutants (Figure 4N). At E14 we observed a small patch of Sox2 expression at the basal portion of the cochlea, confirming the shortening of the cochlear duct (Figure 5D). However, sections through the cochlear duct revealed that the formation of the prosensory region in Meis2flox/flox; Pax2Cre/+ mutants was unaffected (Figures 5E,F). Sox2 staining was also unaffected in the prosensory region of all vestibular sensory epithelia, including the utricular and saccular maculae and posterior, lateral, and anterior cristae (Figures 5C,D).
FIGURE 5.
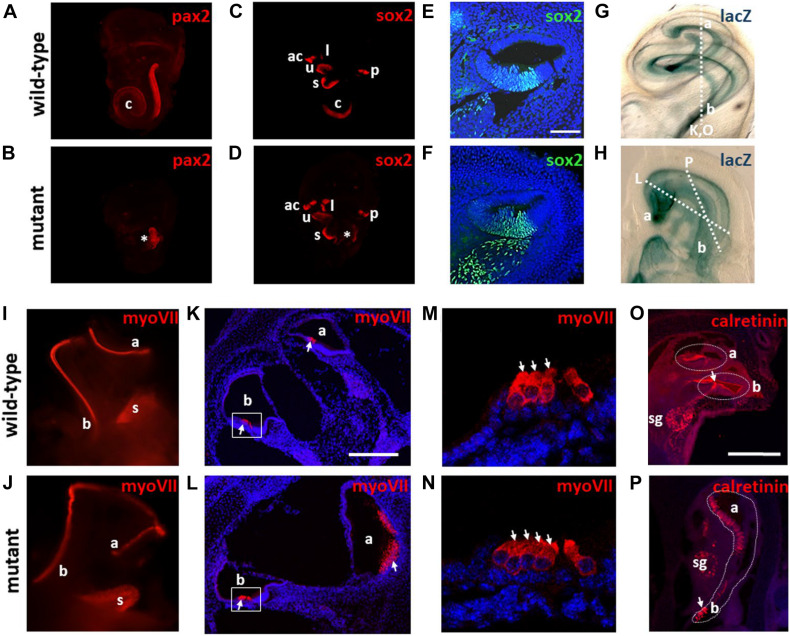
Meis2 is required for cochlear outgrowth and coiling. Expression was monitored by staining of whole mounts or on sections with the corresponding antibodies indicated. (A–D) Staining with Pax2 and Sox2 antibodies reveals a truncation of the cochlear duct (c), indicated by an asterisk in the mutants. Note that the prosensory regions within the vestibular system maintain Sox2 expression in the mutants. (E,F) Sections through the cochlear duct reveal that formation of the prosensory region is unaffected in the mutants. (G,H) lacZROSA26 reporter staining caused by the Pax2-Cre driver reveals the abnormal coiling of the cochlear duct in mutants. The basal (b) and apical (a) portions of the cochlea are indicated. (I–L) Staining with myosin VII antibodies labeling hair cells (indicated by arrows) confirms the abnormal morphology of the cochlear duct in mutants. Note the increased sized patches of hair cells in the apex of mutants. (M,N) High magnification view of the basal region (boxed in K,L) reveals the presence of an extra row of outer hair cells (indicated by arrows) in the mutants. (O,P) Sections stained with calretinin antibodies labeling inner hair cells (arrows) and the spiral ganglion (sg). Note the presence of a normal morphology in the wild-type with the typical appearance of basal and apical turns whereas the mutant shows an abnormal extension along the basal-apical axis. The plane of the sections shown in (K,L,O,P) are indicated in (G,H). ac, anterior cristae; l, lateral cristae; p, posterior cristae; s, sacculus; u, utriculus; Scale bars: 50 μm in (E); 250 μm in (K,O).
To further explore the cochlear abnormality in Meis2flox/flox; Pax2Cre/+ mice, we performed whole-mount β-galactosidase staining of the inner ears from mutants which also carried a ROSA26lacZ reporter (Soriano, 1999). Pax2-Cre is active in the early otic placode and vesicle, and the ROSA26lacZ reporter allows labeling of all inner ear components throughout development (Ohyama and Groves, 2004). Beta-galactosidase staining revealed that in control animals, the cochlea had undertaken its one and three quarter turns (Figure 5G). In contrast, in Meis2flox/flox; Pax2Cre/+ mutants, instead of turning ventrally along the anterior-medial axis, the cochlear duct extended toward the apex but then took a U-turn toward the base. This lead to its termination being positioned half-way from its point of initiation (Figure 5H). This irregular turning was also confirmed by whole mount staining with myosin VII antibodies which label sensory hair cells within the cochlear duct (Figures 5I,J). Sections through control ears confirmed the typical structure of the cochlear duct including one row of inner hair cells and three rows of outer hair cells in controls (Figures 5K,M). In contrast, sections from Meis2flox/flox; Pax2Cre/+ mutant cochleas revealed an extra row of outer hair cells in the basal turn and an enlarged cochlear duct with clusters of hair cells at the apex (Figures 5L,N). Staining with calretinin antibodies that label spiral ganglion neurons and inner hair cells confirmed a normally structured cochlea with the typical appearance of basal and apical turns in control animals, whereas sections of Meis2flox/flox; Pax2Cre/+ mutants revealed an abnormal extension of the cochlear duct along the basal to apical axis (Figures 5O,P). In summary, these data confirm that loss of Meis2 during inner ear development leads to a defective cochlear coiling and extra rows of hair cells.
Direct and Indirect Targets of Meis2 in the Cochlea
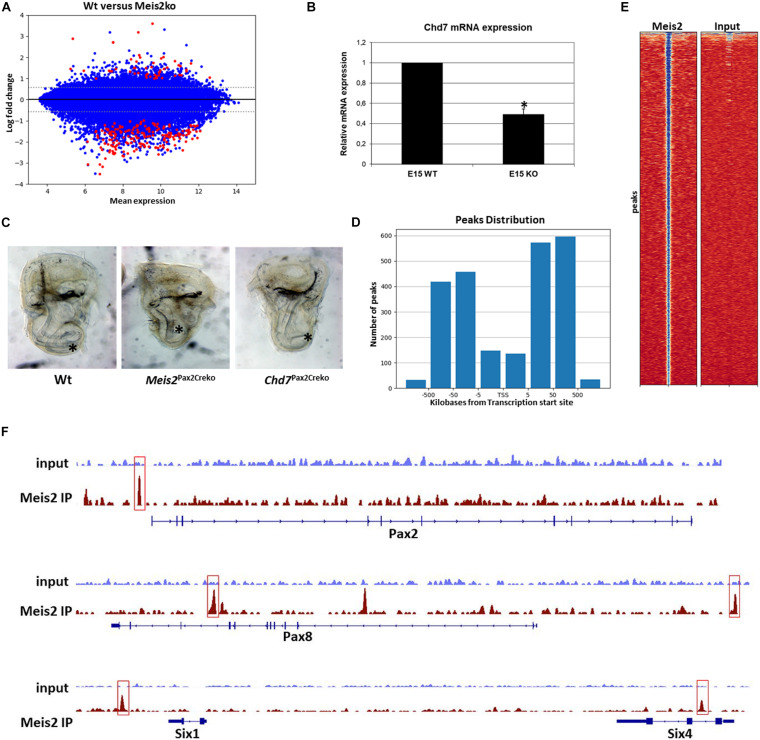
In order to identify potential target genes of Meis2 in the mammalian cochlea, we performed a microarray-based screen for differential gene expression in Meis2flox/flox; Pax2Cre/+ mutant vs. wild-type cochleas (for details, see section “Materials and Methods”). We used whole E15 cochleas when Meis2 expression is detected in the cochlear duct, the spiral ganglion and the auditory nerve (Figure 1E). The results of the microarrays showed that the vast majority of the 255 transcripts differentially expressed were downregulated in mutant cochleas (Figure 6A). Several of the downregulated genes are expressed in the cochlear duct or the spiral ganglion (Table 1). Within this group of genes, the cochlea of mouse mutants for the chromatin remodeling enzyme Chd7 has been described to undergo an abnormal twist at their apex (Hurd et al., 2010). We first confirmed downregulation of Chd7 by qPCR (Figure 6B) and then created a conditional mouse mutants for Chd7 using the Pax2-Cre driver. Cleared whole mounts of inner ears isolated from Chd7flox/flox; Pax2Cre/+ mutants revealed a defective phenotype in cochlear coiling which was very similar to the one observed in Meis2flox/flox; Pax2Cre/+ mutants (Figure 6C). Therefore, Chd7 may be a crucial downstream target of Meis2.
FIGURE 6.
Targets of Meis2. (A) MA plot showing differential gene expression between Meis2flox/flox; Pax2Cre/+ mutant vs. wild-type cochleas at E15. Red dots indicate significantly regulated genes. (B) Significantly reduced expression of Chd7 is observed in the cochlea of Meis2flox/flox; Pax2Cre/+ mutants (KO) by qPCR at E15. Data were normalized to expression levels in wild-type (WT) cochleas. Student’s t-test: *p < 0.0001 (C) Meis2flox/flox; Pax2Cre/+ animals show a similar mutant cochlear phenotype to Chd7flox/flox; Pax2Cre/+ animals. The asterisk indicates the ending of the cochlear duct. (D) Distribution of Meis2 binding sites by their position with respect to the nearest transcription start site (TSS) in VOT-E36 cells. (E) Density plots showing the distribution of Meis2 binding sites vs. input control in VOT-E36 cells. (F) Input control and Meis2 ChIP-seq read profiles from VOT-E36 cells. Detected Meis2 binding sites within or close to the genomic regions of Pax2, Pax8, Six1 and Six4 are boxed.
TABLE 1.
Summary of genes downregulated in the cochlea of Meis2 mutants.
| Gene (MGI reference) | Fold Downregulation | Expression pattern |
| Acidic nuclear phosphoprotein 32 family (5332981) | 4.1x | Mesenchyme |
| THUMP domain containing 3 (1277973) | 3.7x | Otic capsule |
| Itchy, E3 ubiquitin protein ligase (4825634) | 3.4x | Ganglion |
| Zinc finger protein 788 (1914857) | 3.1x | Ganglion |
| Neurofilament, light polypeptide (97313) | 3.1x | Ganglion |
| Trafficking protein particle complex 4 (4480575) | 2.9x | Ganglion |
| Centrosome/spindle pole associated protein (2681832) | 2.7x | Ganglion |
| FK506 binding protein 3 (5335642) | 2.7x | Mesenchyme |
| Chromodomain helicase DNA binding protein 7 (2444748) | 2.6x | Cochlear duct, ganglion and mesenchyme |
| Craniofacial development protein 1 (1344403) | 2.4x | Cochlear duct and ganglion |
| Formin binding protein 1-like (1925642) | 2.4x | Ganglion |
| Ephrin A1 (103236) | 2.4x | Cochlear duct |
| Thymidylate synthase (98878) | 2x | Mesenchyme |
The localization of the genes within the cochlea is annotated according to Mouse Genome Informatics (MGI) using RNA in situ hybridization. The table shows a list of genes which are expressed in the cochlea and down regulated by twofold or higher in Meis2 mutants according to the results obtained by the microarrays.
To identify direct targets of Meis2, we performed ChIPseq analysis (see section “Materials and Methods”) using the VOT-E36 cell line derived from the ventral portion of the otic vesicle, the domain that gives rise to the cochlea (Lawoko-Kerali et al., 2004; Figure 6E). We identified a total of 2,401 Meis2 binding sites in the genome and a collection of 917 genes with a transcription start site closest to any Meis2 binding site (GEO GSE166072). As previously reported for Meis1 (Penkov et al., 2013; Marcos et al., 2015), most Meis2 binding sites were located in regions remote from their closest associated transcription start site (Figure 6D). Meis proteins select two main sequences in DNA: a motif resembling the Pbx-Hox binding sequence (A/TGATNNAT), to which it binds indirectly, and a direct binding site (TGACAG) (Penkov et al., 2013). Transcription factor motif enrichment analysis using Homer and MEME suite software (see section “Materials and Methods”) confirmed these motifs as the most frequently detected with 15.7 and 28.9% of peaks containing at least one of the conserved and/or the PBX-Hox binding sites, respectively. Gene ontology analysis revealed the presence of Meis2 binding peaks in the vicinity of 30 genes related to inner ear development, such as members of the Pax- and Six-gene families (Figure 6F). Interestingly, a Meis2 peak was observed in the vicinity of the Otx1 gene, whose inactivation also leads to defective cochlear coiling (Morsli et al., 1999). A comparison between the observed peaks and differentially expressed genes in the cochlea of Meis2flox/flox; Pax2Cre/+ mutants at E15 revealed no Meis2 binding sites in the vicinity of the Chd7 gene. However, an overlap in the vicinity of 7 genes including Dlg1, Pnn, Nsmce2, Usf3, Zfp945, Lrrcc1, Rybp and Mras was observed. Interestingly, Dlg1 is expressed in the otic vesicle and its loss is associated with a shortening of cochlear length (Iizuka-Kogo et al., 2015), albeit to a lesser degree than that observed in Meis2flox/flox; Pax2Cre/+ mutants. Therefore, loss of Dlg1 expression may, at least in part, contribute to the shortening of the cochlear duct in Meis2flox/flox; Pax2Cre/+ mutants.
Discussion
Meis2 was expressed in all three tissue layers involved in otic induction: the endoderm, mesoderm and the neural tissue of the hindbrain. Correlating with the highest expression levels, the loss of Meis2 in the hindbrain led to a phenotype similar to that observed by global inactivation at E9 (compare Figures 2A–D with Figures 3C–H). Therefore, it may be concluded that Meis2 expression in the hindbrain is likely to be sufficient to account for Meis2 contribution to otic vesicle formation. Genes required for inner ear formation, which are directly or indirectly induced by Meis2 showed no changes in their expression in the hindbrain, and thus so far indicate no major defects in patterning within the hindbrain in Meis2 mutants. However, yet unknown downstream targets for Meis2 within the hindbrain may be involved in inner ear induction. During later stages, otic vesicle development was also affected in Meis2flox/flox; Foxg1Cre/+ mutants that lacked Meis2 expression in the otic placode and the neighboring mesoderm and endoderm. The otic vesicle was reduced at E11 and at the postnatal stage the inner ear showed severe defects (Figures 3I–L). Compared to Meis2flox/flox; Pax2Cre/+ mutants which show Cre activity only in the otic placode (Supplementary Figure 1 and Figure 5), Meis2flox/flox; Foxg1Cre/+ mutants show a more severe inner ear phenotype (Figures 3K,L). Therefore, inactivation of Meis2 expression from the mesoderm and/or endoderm leads to additional defects during inner ear development. Within the inner ear, Meis2 initially showed expression in the dorsal portion of the otic vesicle and was later on found in non-sensory tissue of the cochlea and the peripheral nervous system innervating the inner ear. Very similar patterns of expression for Meis2 have also been described during chicken inner ear development (Sanchez-Guardado et al., 2011).
Meis2flox/flox; Pax2Cre/+ mouse mutants lacking Meis2 throughout inner ear development showed a smaller otic vesicle at E10.5, indicating that initial expression of Meis2 in the otic placode or vesicle is also required for its normal formation. Nevertheless, otic markers related to neurosensory development and formation of the cochlear anlage were unaffected. However, by E13.5 Meis2flox/flox; Pax2Cre/+ mutants a striking defective outgrowth of the cochlear duct became apparent which resulted in its abnormal coiling. The phenotype was similar to that described in mouse mutants lacking β-catenin, required for Wnt signaling, or the transcription factors Sox9 or Tbx1 (Xu et al., 2007; Trowe et al., 2010; Bohnenpoll et al., 2014). However, in mutants lacking Sox9 or Tbx1, the observed inner ear phenotype is caused by the inactivation of these genes in the surrounding mesenchyme, pointing to their indirect influence on morphogenesis. Defective turning of the cochlear duct has also been described in a mouse mutant lacking the chromatin remodeling enzyme Chd7 throughout inner ear development (Hurd et al., 2010). Interestingly, we found that Chd7 expression was reduced in the cochlea of Meis2flox/flox; Pax2Cre/+ mutants, suggesting that these genes may share a common pathway. Indeed, inactivation of Chd7 using the Pax2-Cre line led to a phenotype very similar to Meis2flox/flox; Pax2Cre/+ mutants. Moreover, like Meis2, Chd7 null mutants also show severely reduced sized otic vesicles (Hurd et al., 2007). In spite of this functional relationship, we did not find evidence for the binding of Meis2 within the regulatory regions of Chd7 in the VOT-E36 cell line. Additionally, Chd7 mutants show reduced expression of Ngn1, which was unaltered in Meis2 mutants in the present study. This suggests that Chd7 and Meis2 rather belong to parallel pathways controlling cochlear coiling.
Only very few genes (Hayashi et al., 2002) from the ChIPseq analysis showed binding close to the transcription start site of genes. Similar findings have been recently reported for Meis1 (Penkov et al., 2013; Marcos et al., 2015). The most frequently detected binding motif detected in our studies corresponded to a site to which Meis binds indirectly to DNA. A major group of binding partners that facilitate indirect contact with DNA belong to the Hox gene family. Due to its rostral position in the embryo, the inner ear is a relatively Hox-free tissue, although the expression of PG-1 group genes like Hoxa1 and its control of downstream targets have been reported in the inner ear (Makki and Capecchi, 2010, 2011). Additionally, Meis cooperates with a wide variety of transcription factors containing homeodomains, such as members of the Pax, Dlx, and Otx gene families which are also expressed in the inner ear (Schulte and Geerts, 2019). Nevertheless, only very few genes with Meis2 binding sites showed an overlap with differentially expressed transcripts in the cochlea. Therefore, the VOT-E36 cell line used for the ChIPseq analysis might not reflect an ideal model for in vivo inner ear development. Unfortunately, ChIP analysis from isolated otic vesicles was not feasible in our hands owing to technical limitations due to small-scale tissue samples. Nevertheless, further analysis of the genes showing both differential expression in Meis2flox/flox; Pax2Cre/+ mutants and containing binding sites detected by the ChIPseq analysis may reveal novel regulators of cochlear morphogenesis which are controlled by Meis2.
Materials and Methods
Transgenic Mice
Mice carrying a floxed Meis2 allele (Delgado et al., 2020), a floxed Chd7 allele (Hurd et al., 2007), the Sox2-Cre (Hayashi et al., 2002), Pax2-Cre (Ohyama and Groves, 2004), Foxg1-Cre (Hebert and McConnell, 2000) and MafB-Cre (Wu et al., 2016) transgenes and the ROSA26 Cre reporter strain (Soriano, 1999) have been described previously. With the exception of the Meis2flox/flox; MafBCre/+ mutants which showed a penetrance of 50%, all other phenotypes were fully penetrant and observed in a minimum of n = 3 animals for each experimental condition. Experiments conformed to the institutional and national regulatory standards concerning animal welfare.
β-Galactosidase Staining and in situ Hybridization
β-Galactosidase staining, RNA whole-mount in situ hybridization and sectioning of stained embryos have been described previously (Alvarez et al., 2003). Riboprobes were generated for detection of Meis2 (Delgado et al., 2020), LFng, Ngn1, Dlx5, (Vazquez-Echeverria et al., 2008), Fgf3, Pax2, MafB (Alvarez et al., 2003) and HoxB1 (Vendrell et al., 2013).
Immunohistochemistry
For immunohistochemistry cryostat sections were prepared and processed using standard protocols. The following antibodies were used: Meis-2 (Mercader et al., 2005); Pax2 (PRB-276P) from Covance, Sox2 (sc-17320) from Santa Cruz Biotechnology; myosin VIIA (25-6790) from Proteus; calretinin (7699/3H) from Swant; cyclin D1 (RM-9104-S0) from Thermo Fisher Scientific. For immunofluorescence, cryostat sections were incubated with primary antibodies, and the corresponding secondary antibodies used were donkey anti-goat Alexa Fluor-488, and goat anti-rabbit Alexa Fluor-568 (all from Invitrogen). Some of the sections were counterstained with DAPI. Whole-mount immunolabeling, dehydration, and clearing of inner ears was performed as described previously (MacDonald and Rubel, 2008). Bright-field images were captured with a DFC 490 camera (Leica) on a Labophot-2 microscope (Nikon). Immunofluorescence images were taken with a Nikon Eclipse 90i fluorescence microscope, or Leica SP confocal microscope.
Screening for Differentially Regulated Genes in Meis2flox/flox; Pax2Cre/+ Mutants
RNA was isolated from E15 cochleas of wild type and Meis2flox/flox; Pax2Cre/+ mutants using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA integrity was assessed using Agilent 2100 Bioanalyzer (Agilent). Labeling and hybridizations were performed according to protocols from Affymetrix. Briefly, 100–300 ng of total RNA were amplified and labeled using the WT Expression Kit (Ambion) and then hybridized to Mouse Gene 1.0 ST Arrays (Affymetrix) covering a total of 21,041 gene transcripts. Washing and scanning were performed using the Affymetrix GeneChip System (GeneChip Hybridization Oven 640, GeneChip Fluidics Station 450 and GeneChip Scanner 7G). The robust microarray analysis algorithm was used for background correction, intra- and inter-microarray normalization, and expression signal calculation. The absolute expression signal for each gene was calculated in each microarray and significance analysis of microarrays was applied to calculate differential expression and find the gene probe sets that characterized the highly metastatic samples. The method uses permutations to provide robust statistical inference of the most significant genes and provides > P-values adjusted to multiple testing using false discovery rate. Probe synthesis, hybridizations and microarray data analysis were performed by the Genomics facility of the Centro de Investigación del Cancer (Salamanca, Spain). The microarray data from this screen have been deposited at GEO with accession number GSE149916. Genes twofold or greater were examined for their expression in the cochlea (Visel et al., 2004; Diez-Roux et al., 2011; Agoston et al., 2014; Bult et al., 2019) and are listed in Table 1.
Quantitative PCR
Individual RNA samples were each prepared from the cochleae of two embryonic day (E) 15 mice (i.e., 4 cochleae were used to generate one RNA sample). RNA was extracted following homogenization in TRIzol® Reagent (Invitrogen), and according to manufacturer’s instructions. Each RNA was then reverse transcribed into cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Life Technologies). cDNA samples were amplified in a LightCycler® 480 II (Roche Molecular Diagnostics, Pleasanton, CA, United States) using SYBR® Green PCR Master Mix (Life Technologies); duplicate reactions were carried out with each cDNA sample. The thermocycling conditions consisted of an initial denaturation step of 10 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The sequences of the PCR primers used in this work were: Gapdh, TCCTGCACCACCAACTGCTT and GTGGCAGTGATGGCATGGAC; Chd7, GAATACCCCA CAGAAAGTGCC and TCGCTCTTCACTAGCTGAGCG. Data were analyzed using the Software version LCS480 1.5.0.39. Relative levels of mRNA expression were calculated according to the 2–ΔΔCt method, using Gapdh as the housekeeping gene. The data presented are the results from three independent experiments.
ChIP-Seq
US/VOT-E36 cells (University of Sheffield/ventral otocyst-epithelial cell line clone 36) were used, derived from the otocyst of Immortomice at embryonic day 10.5 (Lawoko-Kerali et al., 2004). The cells were cultured in minimum essential medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 50 U/ml γ-IFN (Immunotools) in a humidified atmosphere with 10% CO2 and at a temperature of 33°C, conditions under which these cells proliferate in the absence of differentiation.
For the ChIP assay, once the cultures had reached 85–90% confluence, the chromatin from the cells in six 75-cm2 flasks was cross-linked in 1% formaldehyde for 10 min at room temperature; next, the reaction was quenched by adding glycine to a final concentration of 125 mM and mixing during an additional 5 min. Following removal of the medium and various washes with phosphate buffer saline, cells were scraped off the tissue culture flasks, pelleted and lysed; the chromatin was then sheared into 200–500 bp fragments by sonication (using pulses of 30 s on and 30 s off in a water-bath sonicator). Any remaining cellular debris was subsequently removed by centrifugation and the sonicated chromatin was pre-cleared for 4 h at 4°C with protein A agarose beads (Roche). After this time, the beads were removed by centrifugation; half the volume of the chromatin sample was immunoprecipitated (IP sample) at 4°C overnight with a 1:1 mixture of an antibody against Meis2 (K846) and K830, an antibody that recognizes both Meis 1a and Meis 2a isoforms (Penkov et al., 2013). A volume that was equivalent to one tenth of the volume used for the IP reaction was also at that time set apart and frozen, constituting the Input control sample. Following immunoprecipitation with the antibodies, beads that had been pre-blocked at 4°C overnight with BSA and a rabbit IgG isotype control (ChromPure Rabbit IgG, Jackson ImmunoResearch Laboratories) were added to each tube; the samples were then incubated at 4°C for 4 h. Afterward, a series of washes using multiple buffers were conducted in order to remove any molecule that had bound non-specifically to the beads; the chromatin was finally eluted from the beads and cross-linking to the antibody reversed by incubating the IP sample in the presence of NaCl at 65°C overnight; the Input sample that had been stored at –20°C was also incubated in the same solution at 65°C. The next day, both the IP and the Input chromatin were purified using a PCR purification kit (Qiagen), following manufacturer’s instructions. The samples were then used to carry out ChIP-seq analysis.
0.5 ng of total DNA for both Input and IP were used to generate barcoded libraries using the NEBNext® UltraTM II DNA Library Prep Kit for Illumina (New England Biolabs). Basically, adapters were ligated to DNA followed by an amplification and clean up. The size of the libraries was checked using the Agilent 2100 Bioanalyzer High Sensitivity DNA chip and their concentration was determined using the Qubit® fluorometer (Thermo Fisher Scientific).
Libraries were sequenced on a HiSeq 2500 (Illumina) and processed with RTA v1.18.66.3. FastQ files for each sample were obtained using bcl2fastq v2.20.0.422 software (Illumina).
Sequencing reads were trimmed for Illumina adapter sequence with cutadapt, aligned to the mouse reference genome (mm10 v92) with bowtie and PCR duplicates were excluded with samtools MarkDuplicates tool.
Peaks were called with HOMER2 and params “-region -localSize 50000 -size 150 -minDist 1000-ntagThreshold 5-regionRes 6.” Peaks were also annotated with HOMER2 and inspected for MOTIFs with Meme (from Meme Suite) and with HOMER2. Data have been deposited in the NCBI GEO database under accession number GSE166072.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE166072, https://www.ncbi.nlm.nih.gov/, GSE149916.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of the University of Valladolid.
Author Contributions
TS and MD designed the research. TS, MD, VV, IL-H, FG, EV, LC, and GG performed the research. MA, MT, and DM contributed unpublished reagents and analytic tools. TS, MD, VV, and EV analyzed the data. TS wrote the first draft of the manuscript and wrote the manuscript. TS, MD, DM, and FG edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Bernice Morrow for providing Tbx1 antibodies and Kiril Schimmang-Alonso for help on designing of figures.
Footnotes
Funding. We acknowledge grants from Mineco BFU2016-76580-P and Consejería de Educación, Junta de Castilla y León (FEDER CSI143P20) (TS).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.679325/full#supplementary-material
Expression domains of Cre lines. MafB-Cre is expressed in the neural tube (nt) corresponding to rhombomeres 5 and 6 of the hindbrain whereas Pax2-Cre is active in the otic placode (op). Next to the otic placode, Foxg1-Cre is also active in the peri-otic mesoderm (m) and endoderm (e).
References
- Agoston Z., Heine P., Brill M. S., Grebbin B. M., Hau A. C., Kallenborn-Gerhardt W., et al. (2014). Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141 28–38. 10.1242/dev.097295 [DOI] [PubMed] [Google Scholar]
- Alvarez Y., Alonso M. T., Vendrell V., Zelarayan L. C., Chamero P., Theil T., et al. (2003). Requirements for FGF3 and FGF10 during inner ear formation. Development 130 6329–6338. 10.1242/dev.00881 [DOI] [PubMed] [Google Scholar]
- Basch M. L., Brown R. M., II, Jen H. I., Groves A. K. (2016). Where hearing starts: the development of the mammalian cochlea. J. Anat. 228 233–254. 10.1111/joa.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J., Tavares M. J., Santos J., Kikuta H., Laplante M., Becker T. S., et al. (2008). meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development 135 799–803. 10.1242/dev.011932 [DOI] [PubMed] [Google Scholar]
- Bohnenpoll T., Trowe M. O., Wojahn I., Taketo M. M., Petry M., Kispert A. (2014). Canonical Wnt signaling regulates the proliferative expansion and differentiation of fibrocytes in the murine inner ear. Dev. Biol. 391 54–65. 10.1016/j.ydbio.2014.03.023 [DOI] [PubMed] [Google Scholar]
- Bok J., Chang W., Wu D. K. (2007). Patterning and morphogenesis of the vertebrate inner ear. Int. J. Dev. Biol. 51 521–533. 10.1387/ijdb.072381jb [DOI] [PubMed] [Google Scholar]
- Bult C. J., Blake J. A., Smith C. L., Kadin J. A., Richardson J. E., Mouse Genome, et al. (2019). Mouse Genome Database (MGD). Nucleic Acids Res. 47 D801–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Q., Cole L. K., Mulheisen M., Chang W., Wu D. K. (2004). The role of Pax2 in mouse inner ear development. Dev. Biol. 272 161–175. 10.1016/j.ydbio.2004.04.024 [DOI] [PubMed] [Google Scholar]
- Cordes S. P., Barsh G. S. (1994). The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79 1025–1034. 10.1016/0092-8674(94)90033-7 [DOI] [PubMed] [Google Scholar]
- Delgado I., Lopez-Delgado A. C., Rosello-Diez A., Giovinazzo G., Cadenas V., Fernandez-de-Manuel L., et al. (2020). Proximo-distal positional information encoded by an Fgf-regulated gradient of homeodomain transcription factors in the vertebrate limb. Sci. Adv. 6:eaaz0742. 10.1126/sciadv.aaz0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., et al. (2011). A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9:e1000582. 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkovich Y. E., Ofir R., Elkouby Y. M., Dibner C., Gefen A., Elias S., et al. (2010). Xenopus Meis3 protein lies at a nexus downstream to Zic1 and Pax3 proteins, regulating multiple cell-fates during early nervous system development. Dev. Biol. 338 50–62. 10.1016/j.ydbio.2009.11.024 [DOI] [PubMed] [Google Scholar]
- Hayashi S., Lewis P., Pevny L., McMahon A. P. (2002). Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119(Suppl. 1), S97–S101. [DOI] [PubMed] [Google Scholar]
- Hebert J. M., McConnell S. K. (2000). Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222 296–306. 10.1006/dbio.2000.9732 [DOI] [PubMed] [Google Scholar]
- Hernandez R. E., Rikhof H. A., Bachmann R., Moens C. B. (2004). vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development 131 4511–4520. 10.1242/dev.01297 [DOI] [PubMed] [Google Scholar]
- Hurd E. A., Capers P. L., Blauwkamp M. N., Adams M. E., Raphael Y., Poucher H. K., et al. (2007). Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome 18 94–104. 10.1007/s00335-006-0107-6 [DOI] [PubMed] [Google Scholar]
- Hurd E. A., Poucher H. K., Cheng K., Raphael Y., Martin D. M. (2010). The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 137 3139–3150. 10.1242/dev.047894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka-Kogo A., Senda T., Akiyama T., Shimomura A., Nomura R., Hasegawa Y., et al. (2015). Requirement of DLG1 for cardiovascular development and tissue elongation during cochlear, enteric, and skeletal development: possible role in convergent extension. PLoS One 10:e0123965. 10.1371/journal.pone.0123965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher R. K., Wright T. J., Moon A. M., Mansour S. L., Schoenwolf G. C. (2005). FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 19 603–613. 10.1101/gad.1273605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawoko-Kerali G., Milo M., Davies D., Halsall A., Helyer R., Johnson C. M., et al. (2004). Ventral otic cell lines as developmental models of auditory epithelial and neural precursors. Dev. Dyn. 231 801–814. 10.1002/dvdy.20187 [DOI] [PubMed] [Google Scholar]
- MacDonald G. H., Rubel E. W. (2008). Three-dimensional imaging of the intact mouse cochlea by fluorescent laser scanning confocal microscopy. Hear. Res. 243 1–10. 10.1016/j.heares.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machon O., Masek J., Machonova O., Krauss S., Kozmik Z. (2015). Meis2 is essential for cranial and cardiac neural crest development. BMC Dev. Biol. 15:40. 10.1186/s12861-015-0093-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A. C., Szeto I. Y., Fritzsch B., Cheah K. S. (2009). Differential and overlapping expression pattern of SOX2 and SOX9 in inner ear development. Gene Exp. Patterns 9 444–453. 10.1016/j.gep.2009.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki N., Capecchi M. R. (2010). Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev. Biol. 341 499–509. 10.1016/j.ydbio.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki N., Capecchi M. R. (2011). Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol. 357 295–304. 10.1016/j.ydbio.2011.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos S., Gonzalez-Lazaro M., Beccari L., Carramolino L., Martin-Bermejo M. J., Amarie O., et al. (2015). Meis1 coordinates a network of genes implicated in eye development and microphthalmia. Development 142 3009–3020. 10.1242/dev.122176 [DOI] [PubMed] [Google Scholar]
- Maroon H., Walshe J., Mahmood R., Kiefer P., Dickson C., Mason I. (2002). Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development 129 2099–2108. 10.1242/dev.129.9.2099 [DOI] [PubMed] [Google Scholar]
- Mercader N., Tanaka E. M., Torres M. (2005). Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132 4131–4142. 10.1242/dev.01976 [DOI] [PubMed] [Google Scholar]
- Morsli H., Tuorto F., Choo D., Postiglione M. P., Simeone A., Wu D. K. (1999). Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development 126 2335–2343. 10.1242/dev.126.11.2335 [DOI] [PubMed] [Google Scholar]
- Ohyama T., Groves A. K. (2004). Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis 38 195–199. 10.1002/gene.20017 [DOI] [PubMed] [Google Scholar]
- Pasqualetti M., Neun R., Davenne M., Rijli F. M. (2001). Retinoic acid rescues inner ear defects in Hoxa1 deficient mice. Nat. Genet. 29 34–39. 10.1038/ng702 [DOI] [PubMed] [Google Scholar]
- Penkov D., Mateos San, Martin D., Fernandez-Diaz L. C., Rossello C. A., Torroja C., et al. (2013). Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 3 1321–1333. 10.1016/j.celrep.2013.03.029 [DOI] [PubMed] [Google Scholar]
- Rossel M., Capecchi M. R. (1999). Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 126 5027–5040. 10.1242/dev.126.22.5027 [DOI] [PubMed] [Google Scholar]
- Sanchez-Guardado L. O., Ferran J. L., Rodriguez-Gallardo L., Puelles L., Hidalgo-Sanchez M. (2011). Meis gene expression patterns in the developing chicken inner ear. J. Comp. Neurol. 519 125–147. 10.1002/cne.22508 [DOI] [PubMed] [Google Scholar]
- Schimmang T. (2007). Expression and functions of FGF ligands during early otic development. Int. J. Dev. Biol. 51 473–481. 10.1387/ijdb.072334ts [DOI] [PubMed] [Google Scholar]
- Schulte D., Frank D. (2014). TALE transcription factors during early development of the vertebrate brain and eye. Dev. Dyn. 243 99–116. 10.1002/dvdy.24030 [DOI] [PubMed] [Google Scholar]
- Schulte D., Geerts D. (2019). MEIS transcription factors in development and disease. Development 146:dev174706. [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21 70–71. 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Trowe M. O., Shah S., Petry M., Airik R., Schuster-Gossler K., Kist R., et al. (2010). Loss of Sox9 in the periotic mesenchyme affects mesenchymal expansion and differentiation, and epithelial morphogenesis during cochlea development in the mouse. Dev. Biol. 342 51–62. 10.1016/j.ydbio.2010.03.014 [DOI] [PubMed] [Google Scholar]
- Vazquez-Echeverria C., Dominguez-Frutos E., Charnay P., Schimmang T., Pujades C. (2008). Analysis of mouse kreisler mutants reveals new roles of hindbrain-derived signals in the establishment of the otic neurogenic domain. Dev. Biol. 322 167–178. 10.1016/j.ydbio.2008.07.025 [DOI] [PubMed] [Google Scholar]
- Vendrell V., Vazquez-Echeverria C., Lopez-Hernandez I., Alonso B. D., Martinez S., Pujades C., et al. (2013). Roles of Wnt8a during formation and patterning of the mouse inner ear. Mech. Dev. 130 160–168. 10.1016/j.mod.2012.09.009 [DOI] [PubMed] [Google Scholar]
- Visel A., Thaller C., Eichele G. (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32 D552–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield T. T. (2015). Development of the inner ear. Curr. Opin. Genet. Dev. 32 112–118. [DOI] [PubMed] [Google Scholar]
- Wiellette E. L., Sive H. (2003). vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development 130 3821–3829. 10.1242/dev.00572 [DOI] [PubMed] [Google Scholar]
- Wright T. J., Mansour S. L. (2003). Fgf3 and Fgf10 are required for mouse otic placode induction. Development 130 3379–3390. 10.1242/dev.00555 [DOI] [PubMed] [Google Scholar]
- Wu X., Briseno C. G., Durai V., Albring J. C., Haldar M., Bagadia P., et al. (2016). Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med. 213 2553–2565. 10.1084/jem.20160600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chen L., Baldini A. (2007). In vivo genetic ablation of the periotic mesoderm affects cell proliferation survival and differentiation in the cochlea. Dev. Biol. 310 329–340. 10.1016/j.ydbio.2007.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelarayan L. C., Vendrell V., Alvarez Y., Dominguez-Frutos E., Theil T., Alonso M. T., et al. (2007). Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev. Biol. 308 379–391. 10.1016/j.ydbio.2007.05.033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression domains of Cre lines. MafB-Cre is expressed in the neural tube (nt) corresponding to rhombomeres 5 and 6 of the hindbrain whereas Pax2-Cre is active in the otic placode (op). Next to the otic placode, Foxg1-Cre is also active in the peri-otic mesoderm (m) and endoderm (e).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE166072, https://www.ncbi.nlm.nih.gov/, GSE149916.