Abstract
A fundamental step in many data-analysis techniques is the construction of an affinity matrix describing similarities between data points. When the data points reside in Euclidean space, a widespread approach is to from an affinity matrix by the Gaussian kernel with pairwise distances, and to follow with a certain normalization (e.g. the row-stochastic normalization or its symmetric variant). We demonstrate that the doubly-stochastic normalization of the Gaussian kernel with zero main diagonal (i.e., no self loops) is robust to heteroskedastic noise. That is, the doubly-stochastic normalization is advantageous in that it automatically accounts for observations with different noise variances. Specifically, we prove that in a suitable high-dimensional setting where heteroskedastic noise does not concentrate too much in any particular direction in space, the resulting (doubly-stochastic) noisy affinity matrix converges to its clean counterpart with rate m−1/2, where m is the ambient dimension. We demonstrate this result numerically, and show that in contrast, the popular row-stochastic and symmetric normalizations behave unfavorably under heteroskedastic noise. Furthermore, we provide examples of simulated and experimental single-cell RNA sequence data with intrinsic heteroskedasticity, where the advantage of the doubly-stochastic normalization for exploratory analysis is evident.
1. Introduction
1.1. Affinity matrix constructions
Given a dataset of points in Euclidean space, a useful approach for encoding the intrinsic geometry of the data is by a weighted graph, where the vertices represent data points, and the edge-weights describe similarities between them. Such a graph can be described by an affinity (or adjacency/similarity) matrix, namely a nonnegative matrix whose (i, j)’th entry holds the edge-weight between vertices i and j. To measure the similarity between pairs of data points, one can employ the Gaussian kernel with pairwise (Euclidean) distance. In particular, given data points , we consider the matrix given by
| (1) |
for i, j = 1, … , n, where ε is the kernel width parameter. For many applications it is a common practice to normalize K, so to equip the resulting affinity matrix with a useful interpretation and favourable properties. Two such normalizations, which are closely related to each other, are the row-stochastic and the symmetric normalizations:
| (2) |
| (3) |
where r = [r1, … , rn], and diag(r) is a diagonal matrix with r on its main diagonal.
Notably, the matrix W(r) is row-stochastic, i.e., the sum of every row of W(r) is 1, which allows for a useful interpretation of W(r) as a transition-probability matrix (in the sense of a Markov chain). An important characteristic of the row-stochastic affinity matrix W(r) is its relation to the heat kernel and the Laplace-Beltrami operator on a manifold [4, 12, 24, 46, 53]. Specifically, under the “manifold assumption” – where the points x1, … , xn are uniformly sampled from a smooth low-dimensional Riemannian manifold embedded in the Euclidean space – W(r) approximates the heat kernel on the manifold, and the matrix L(r) = I−W(r) (known as the random-walk graph Laplacian) approximates the Laplace-Beltrami operator. This property of the row-stochastic normalization establishes the relation between W(r) and the intrinsic local geometry of the data, thereby justifying the use of W(r) as an affinity matrix.
The affinity matrix W(s) (obtained by the symmetric normalization) is closely-related to W(r), and in particular, since W(s) = [diag(r)]−1/2W(r)[diag(r)]1/2, W(s) shares the spectrum of W(r), and their eigenvectors are related through the vector r. Even though W(s) is not a proper transition-probability matrix, it enjoys symmetry, which is advantageous in various applications.
We also mention that the row stochastic and symmetric normalizations can be used in conjunction with a kernel with variable width, i.e., when a different value of ε is taken for each row or column of K (see for instance [6] and references therein). We further discuss one such variant in the example in Section 3.2.
The matrices W(r) and W(s) (or equivalently, their corresponding graph Laplacians I − W(r) and I − W(s)) are used extensively in data processing and machine learning, notably in non-linear dimensionality reduction (or manifold learning) [4, 12, 38, 33], community detection and spectral-clustering [44, 39, 42, 55, 20, 43, 29], image denoising [9, 40, 37, 31, 47], and in signal processing and supervised-learning over graph domains [45, 13, 23, 15, 7].
1.2. The doubly-stochastic normalization
In this work, we focus on the doubly-stochastic normalization of K:
| (4) |
where d = [d1, … , dn] > 0 is a vector chosen such that W(d) is doubly-stochastic, i.e., such that the sum of every row and every column of W(d) is 1. The problem of finding d such that W(d) has prescribed row and column sums is known as a matrix scaling problem, and the entries of d are often referred to as scaling factors. Matrix scaling problems have a rich history, with a long list of applications and generalizations [2, 27]. Since the scaling factors are defined implicitly, their existence and uniqueness are not obvious, and depend on the zero-pattern of the matrix to be scaled. For the particular zero-pattern of K, existence and uniqueness are established by the following lemma.
Lemma 1 (Existence and uniqueness). Suppose that , n > 2, is symmetric with zero main diagonal and strictly positive off-diagonal entries. Then, there exist scaling factors d1, … , dn > 0 such that for all i = 1, … , n, and moreover, are unique.
The proof can be found in Appendix A, and is based on the simple zero-pattern of A and on a lemma by Knight [30]. On the computational side, the scaling factors d can be obtained by the classical Sinkhorn-Knopp algorithm [48] (known also as the RAS algorithm), or by more recent techniques based on optimization (see [1] and references therein). We detail a lean variant of the Sinkhorn-Knopp algorithm adapted to symmetric matrices (see [30]) in Algorithm 1 below, and briefly discuss its convergence and computational complexity in Remark 1.
| Algorithm 1 Sinkhorn-Knopp algorithm for symmetric matrices [30] | |
|---|---|
By definition, W(d) is a symmetric transition-probability matrix. Hence, it naturally combines the two favorable properties that W(r) and W(s) hold separately. It is worthwhile to point-out that W(d) is in fact the closest symmetric and row-stochastic matrix to K in KL-divergence [8, 58], and interestingly, it can also be obtained by iteratively re-applying the symmetric normalization (3) indefinitely (see [57]). Another appealing interpretation of the doubly-stochastic normalization is through the lens of optimal transport with entropy regularization [14], summarized by the following proposition.
Proposition 2 (Optimal transport interpretation). W(d) from (4) is the unique solution to
| (5) |
where 1 is a column vector of n ones, and is the negative entropy.
The proof of Proposition 2 follows very closely with the proof of Lemma 2 in [14], with the additional use of Lemma 1 (to account for the constraint Wi,i = 0), and is omitted for the sake of brevity. In the optimal transport interpretation of the problem (5), each point xi holds a unit mass that should be distributed between all the other points xj ≠ xi, while minimizing the transportation cost between the points (measured by the pair-wise distances ∥xi − xj∥2). The outcome of this process is constrained so that each point ends up holding a unit mass. In this context, the matrix W describes the distribution of the masses from all points to all other points, and is therefore required to be doubly-stochastic. The negative entropy regularization term εH(W) controls the “fairness” of the mass allocation, such that each mass is distributed more evenly between the points for large values of ε.
The optimization problem (5) can also be interpreted as an optimal graph construction. In this context, the term can be considered as accounting for the regularity of the data (as a multivariate signal) with respect to the weighted graph represented by W, while the negative entropy term εH(W) controls the approximate sparseness of W. Since the solution to (5) is a symmetric matrix, W(d) can be thought of as describing the undirected weighted graph that optimizes the “smoothness” of the dataset, under the constraints of prescribed entropy (or approximate sparseness), no self-loops, and stochasticity (i.e., so that W(d) is a transition-probability matrix).
In the context of manifold learning, the relation between the doubly-stochastic normalization and the heat kernel (or the Laplace-Beltrami operator) on a Riemannian manifold has been recently established in [36]. That is, under the manifold assumption (and under certain conditions) W(d) is expected to approximate the heat kernel on the manifold, and therefore to encode the local geometry of the data much like W(r). The doubly-stochastic normalization was also demonstrated to be useful for spectral clustering in [3], where it was shown to achieve the best clustering performance on several datasets. Last, we note that several other constructions of doubly-stochastic affinity matrices have appeared in the literature [56, 58], typically involving a notion of closeness to K other than KL-divergence (e.g. Frobenius norm).
Remark 1 (Computational complexity of Algorithm 1). It is evident that the computational complexity of each iteration in Algorithm 1 is dominated by the matrix-vector multiplication Kd(τ), and is therefore . As for the number of iterations required, in [30] it was shown that if the matrix to be scaled is fully indecomposable, then the scaling factors in the Sinkhorn-knopp algorithm admit a linear convergence whose rate is equal to the squared subdominant eigenvalue of the resulting doubly-stochastic matrix (see Theorem 4.4 in [30]). In the proof of Lemma 1 in Appendix A we show that K from (1) is indeed fully indecomposable, hence the number of iterations in Algorithm 1 is expected to be , where λ2{W(d)} is the subdominant eigenvalue of W(d).
1.3. Robustness to noise
When considering real-world datasets, it is desirable to construct affinity matrices that are robust to noise. Specifically, suppose that we do not have access to the points x1, … , xn (which are non-random in our setting), but rather to their noisy observations , given by
| (6) |
where are pairwise independent noise vectors satisfying
| (7) |
for all i = 1, … , n, where 0 is the zero column vector in , and Σi is the covariance matrix of ηi. We then define , , , , and {} analogously to W(r), W(s), W(d), K , and {di}, respectively, when replacing {xi} in (1) with {}. For the noise model described above, we say that the noise is homoskedastic if Σ1 = Σ2 = … = Σn, and heteroskedastic otherwise.
The influence of homoskedastic noise on kernel matrices (such as K) was investigated in [16], and the results therein imply that and are robust to high-dimensional homoskedastic noise. Specifically, in the high-dimensional setting considered in [16], converges to a biased version K where all the off-diagonal entries of admit the same multiplicative bias. Such bias can therefore be corrected by applying either the row-stochastic or the symmetric normalizations (see [17]). However, this is not the case in the more general setting of heteroskedastic noise.
Heteroskedastic noise is a natural assumption for many real-world applications. For example, heteroskedastic noise arises in certain biological, photon-imaging, and Magnetic Resonance Imaging (MRI) applications [10, 41, 22, 19], where observations are modeled as samples from random variables whose variances depend on their means, such as in binomial, negative-binomial, multinomial, Poisson, or Rice distributions. In natural image processing, heteroskedastic noise occurs due to the spatial clipping of values in an image [18]. Additionally, heteroskedastic noise is encountered when the experimental setup varies during the data collection process, such as in spectrophotometry and atmospheric data acquisition [11, 49]. Generally, many modern datasets are inherently heteroskedastic as they are formed by aggregating observations collected at different times and from different sources. Last, we mention that heteroskedastic noise can be considered as a natural relaxation to the popular manifold assumption. In particular, heteroskedastic noise arises whenever data points are sampled from the high-dimensional surroundings of a low-dimensional manifold embedded in the ambient space, where the size of the sampling neighborhood (in the ambient space) around the manifold is determined locally by the manifold itself. See Figure 4 and the corresponding example in Section 3.1.2.
Figure 4:
Typical array of clean and noisy data points for n = 1000, m = 2, and additive noise sampled uniformly from a sphere whose radius depends on the angle of the corresponding clean point (on the unit circle) according to (18).
1.4. Contributions
Our main contribution is to establish the robustness of the doubly-stochastic normalization of the Gaussian kernel (with zero main diagonal) to high-dimensional heteroskedastic noise. In particular, we prove that in the high-dimensional setting where the number of points n is fixed, the dimension m is increasing, and the noise does not concentrate too much in specific direction in space, converges to W(d) with rate m−1/2. See Theorem 3 in Section 2. An intuitive justification of the robustness of the doubly-stochastic normalization to heteroskedastic noise, and also why zeroing-out the main diagonal of K is important, can be found in Section 2, equations (9)-(10). The proof of Theorem 3, see Appendix B, relies on a perturbation analysis of the doubly-stochastic normalization.
We demonstrate the robustness of to heteroskedastic noise in several examples (see Section 3). In Section 3.1.1 we corroborate Theorem 3 numerically, and exemplify that W(r) and W(s) suffer from inherent point-wise bias due to heteroskedastic noise (see Figures 1-3). In Section 3.1.2 we demonstrate the robustness of the leading eigenvectors of W(d) to heteroskedastic noise whose characteristics depend locally on the manifold of the clean data (see Figures 4-6). In Section 3.2 we apply the doubly-stochastic normalization for both simulated and experimental single-cell RNA sequence data with inherent heteroskedasticity, showcasing its ability to accurately recover the underlying structure of the data despite the noise (see Figures 7,8,9,10).
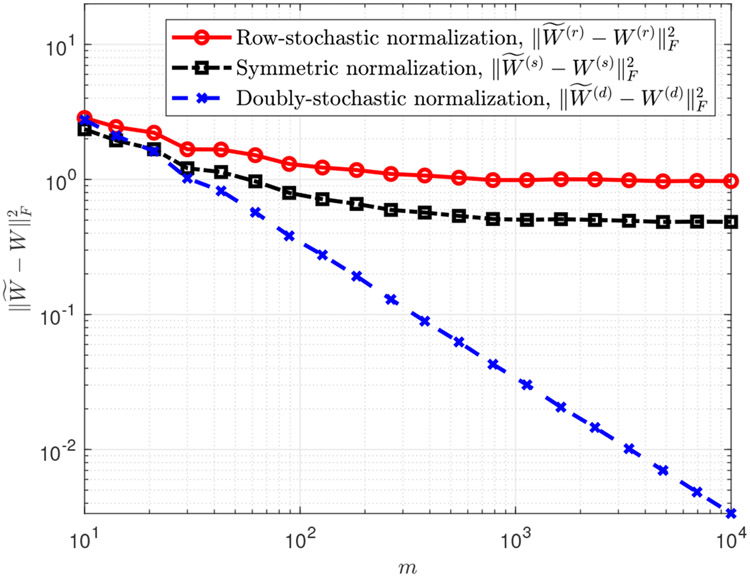
Figure 1:
Squared Frobenius loss (averaged over 10 trials) between clean and noisy affinity matrices from different normalizations, versus the dimension m. The dataset is the unit circle embedded in different dimensions (see (14), with n = 103 and heteroskedastic noise simulated according to (15)-(17)
Figure 3:
First row of the clean and noisy affinity matrices obtained using different normalizations. The dataset is the unit circle (see (14), with n = 103, m = 104, and heteroskedastic noise simulated according to (15)-(17).
Figure 6:
Two-dimensional embedding using the second and third eigenvectors (corresponding to the second- and third-largest eigenvalues) of affinity matrices obtained from different normalizations. The dataset is the unit circle, with n = 103, m = 500, and heteroskedastic noise generated according to (18).
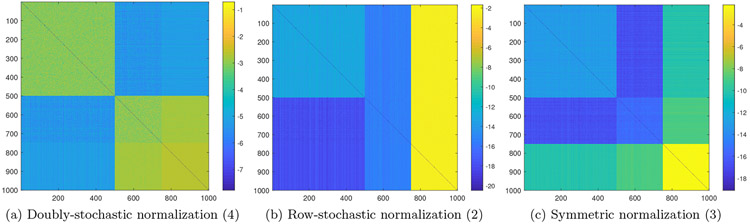
Figure 7:
Entries of the affinity matrices obtained from different normalizations in logarithmic scale (from left to right: , , ), for single-cell RNA sequence data simulated according to (19)-(21), with n = 1000, m = 4000.
Figure 8:
Two-dimensional visualization from t-SNE with different perplexity values (Figures 8a,8b,8c), and from t-SNE modified to use the doubly-stochastic affinity matrix (Figure 8d). The dataset is a simulated single-cell RNA sequence data (see (19)-(21) with n = 1000, m = 4000.
Figure 9:
Entries of the affinity matrices obtained from different normalizations in logarithmic scale (from left to right: , , ), computed from 1000 CD14 cells and 1000 CD34 cells from the purified PBMC dataset [61].
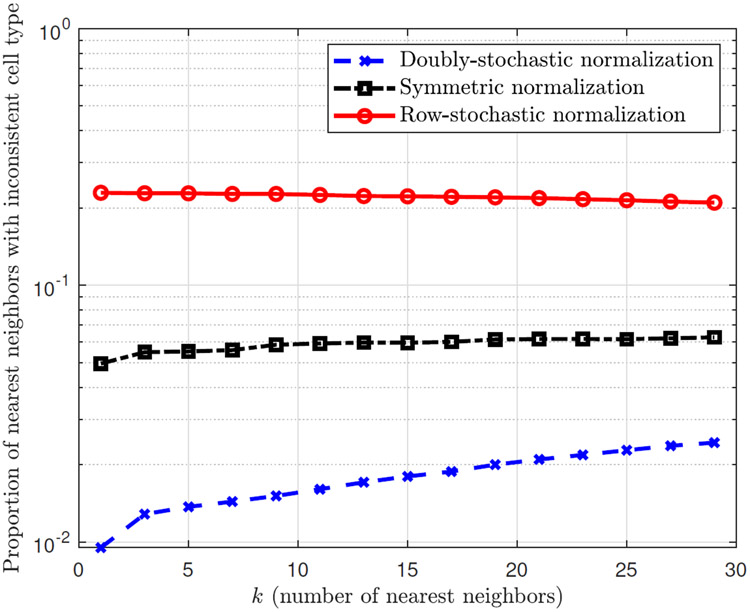
Figure 10:
Inconsistency between the type of a cell and the types of its nearest neighbours, according to each of the affinity matrices , , , and using 1000 CD14 cells and 1000 CD34 cells from the purified PBMC dataset [61]. The x-axis is the number of nearest neighbors chosen for each cell (according to the largest entries in each row of a given affinity matrix), and the y-axis is the proportion of these nearest neighbors with a cell type that is different from the type of the reference cell (whose nearest neighbors are considered), averaged over all cells in the dataset and over 20 randomized trials (of sampling from the CD14 cells and the CD34 cells).
2. Main result
We now place ourselves in the high-dimensional setting where the dimension m is increasing while the number of points n and the kernel parameter ε are fixed. Formally, let , , , , K(m), , W(d),(m), and be the same as xi, , ηi, Σi, K, , W(d), and , respectively, and consider a sequence of each of the former quantities (with superscript (m)) in m = M, M + 1, … , ∞, where M is a positive integer. Our main result is as follows, where stands for order in probability [35] (or stochastic boundedness).
Theorem 3 (Convergence of to W(d),(m)). Suppose that and for all i = 1, …, n and m ≥ M, where Cη is a universal constant (independent of m). Then,
| (8) |
In other words, under the conditions in Theorem 3, it follows that for any probability p > 0 there exist a constant C′ and an integer M′ (both of which may depend on n, p, ε, and Cη) such that for all m ≥ M′ we have . The proof of Theorem 3 is detailed in Appendix B. For simplicity of the presentation, we omit the superscript (m) from all quantities in the rest of this section, as it should be clear that all quantities associated with Theorem 3 are sequences in the dimension m (where n and ε are fixed).
We now provide some remarks on the conditions in Theorem 3. Evidently, the constant 1 in the condition ∥xi∥ ≤ 1 is arbitrary and can be replaced with any other constant (since can always be normalized appropriately). Additionally, note that even though the quantities ∥Σi∥2 are required to decrease with m, the expected noise magnitudes (which are equal to Tr{Σi}) can remain constant, and can possibly be large compared to the magnitudes of the clean data points ∥xi∥2. For example, if we have Σi = m−1Im for all i, where Im is the m×m identity matrix, then it follows that , asserting that the magnitude of the noise is greater or equal to that of the clean data points (under the condition ∥xi∥ ≤ 1). In this regime of non-vanishing high-dimensional noise, the condition ∥Σi∥2 ≤ Cηm−1 guarantees that the noise spreads-out in Euclidean space, and does not concentrate too much in any particular direction (observe that ∥Σi∥2 is the largest singular value of Σi, and is therefore the variance of the noise in the direction where it is largest). Hence, the condition ∥Σi∥2 ≤ Cηm−1 is primarily a convenience for considering noise that has bounded magnitude regardless of the ambient dimension (since ), and whose variance in any particular direction is not too large. In many situations, the data can be normalized appropriately to satisfy this condition, see Remark 2 below and the discussion in Section 3.2.3. Clearly, the setup of Theorem 3 accommodates for heteroskedastic noise, and importantly, the ratios between the noise magnitudes for different data points can be arbitrary.
The main reason behind the robustness of the doubly-stochastic normalization to high-dimensional heteroskedastic noise, is that it is invariant to the type of bias introduced by heteroskedastic noise. Specifically, our analysis in the proof of Theorem 3 (see Appendix B) shows that for i ≠ j,
| (9) |
where stands for convergence in probability, and correspondingly,
| (10) |
for all i, j (since ). Crucially, in (10) is biased by symmetric diagonal scaling, which is precisely the type of bias corrected automatically by the doubly-stochastic normalization (4).
Equations (9) and (10) also highlight why zeroing-out the main diagonal of the Gaussian kernel (see Eq. (1) is important. Without it, the entries on the main diagonal of would be 1, while the off-diagonal entries of would be small due to the bias in the noisy pairwise distances (9). Thus, would be close to the identity matrix, which would render any normalization (row-stochastic, symmetric, or doubly-stochastic) ineffective.
Remark 2. Consider an alternative setting for high-dimensionality where the noise is only required to have bounded variance in each direction, i.e., ∥Σi∥2 ≤ Cη for some universal constant Cη. This assumption holds, for instance, in the standard model where ηi has bounded variance in each coordinate and is uncorrelated between different coordinates. In addition, as the dimension m increases, suppose that the newly-added clean data coordinates are determined by a latent variable that is sampled from some underlying distribution. Specifically, suppose that each clean observation xi is given by
| (11) |
where Fi is a bounded function, and y1, … , ym are i.i.d samples from some latent “coordinate” random variable Y (which can be multivariate or reside in a non-Euclidean space). In this case, one has
| (12) |
| (13) |
which is due to Hoeffding’s inequality [25] (for sums of independent and bounded random variables). Evidently, a natural distance between xi and xj in this setting is as it does not depend on the ambient dimension m and allows for a constant kernel parameter ε to be used for all m. This suggests that the noisy observations should be normalized by , which places us in the setting of Theorem 3 since , and ∥Σi/m∥2 ≤ Cηm−1.
3. Examples
3.1. Example 1: The unit circle embedded in high-dimensional space
In our first example, we sampled n = 103 points uniformly from the unit circle in , and embedded them in , for m ∈ [10, 104], using randomly-generated orthogonal transformations. In more details, we first sampled angles θ1, … , θn independently and uniformly from [0, 2π]. Then, for each embedding dimension m, we generated a random orthogonal matrix (i.e., such that ), and computed the data points {xi} as
| (14) |
Note that as a result, the magnitude of all points is constant, with ∥xi∥ = 1 for all 1 ≤ i ≤ n and embedding dimension m.
3.1.1. Gaussian noise with arbitrary variances
We begin by demonstrating Theorem 3 numerically. Towards that end, we created the noise as follows. For every embedding dimension m, we set (so that the noise is uncorrelated between coordinates), and generated the noise standard-deviations σi,j according to
| (15) |
where , were sampled (independently) from the uniform distribution over [0.05, 0.5]. Therefore, the noise magnitudes satisfy
| (16) |
for all 1 ≤ i ≤ n, and can take any values in that range. Importantly, the noise magnitudes can vary substantially between data points, which is key in our setting. Then, {ηi[j]}i,j were sampled (independently) according to
| (17) |
where ηi[j] stands for the j’th entry of ηi. Once we generated the noisy data points according to (6), we formed the clean and noisy kernel matrices K and with ε = 0.1, and computed W(d), using Algorithm 1 with δ = 10–12. Last, we also evaluated W(r), W(s) and , using K and , respectively, according to (2) and (3).
The behavior of the errors , , as a function of m can be seen in Figure 1. It is evident that for m > 100 the error for the doubly-stochastic normalization is substantially smaller than that for the row-stochastic normalization or for the symmetric normalization. Additionally, the error for the doubly-stochastic normalization decreases linearly in logarithmic scale, while the errors for the row-stochastic and the symmetric normalizations reach saturation and never fall below a certain value. In this experiment, the slope of versus log m (between m = 102 and m = 104) was −0.9996, matching the slope suggested by the upper bound in Theorem 3 (which implies a slope of −1 for the squared Frobenius norm).
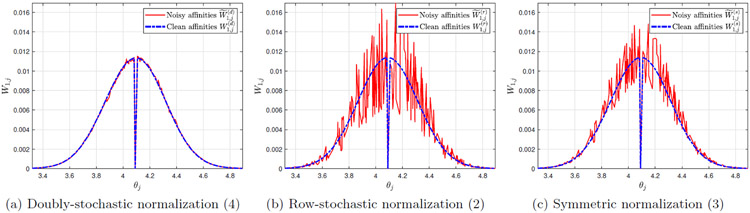
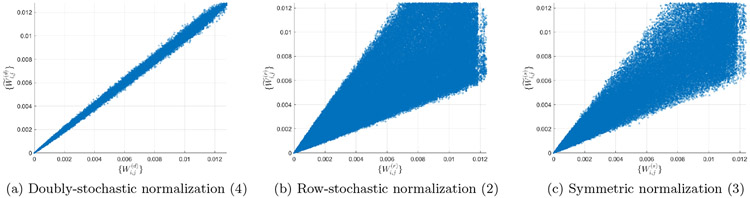
In Figure 2 we depict the noisy affinities , , versus their corresponding clean affinities , , for m = 104. It can be observed that the noisy affinities from the doubly-stochastic normalization concentrate near their corresponding clean affinities, while the noisy affinities from the row-stochastic and symmetric normalizations deviate substantially from their clean counterparts, particularly for larger affinity values.
Figure 2:
Entries of the noisy affinity matrices (y-axis) versus the corresponding entries in the clean affinity matrices (x-axis), using different normalizations. The dataset is the unit circle (see (14), with n = 103, m = 104, and heteroskedastic noise simulated according to (15)-(17).
Last, in Figure 3 we visually demonstrate the first row of the clean and noisy affinity matrices W(d), W(r), W(s) and , , , using m = 104. Note that we only display about a quarter of all the entries, since all the other entries are vanishingly small. It can be seen that the clean row-stochastic, clean symmetric, and clean doubly-stochastic affinities are all very similar, and resemble a Gaussian. This is explained by the fact that both W(d) and W(r) are expected to approximate the heat kernel on the unit circle (see [12, 36] and other related references given in the introduction), which is close to the Gaussian kernel with geodesic distance (for sufficiently small ε). Additionally, since the sampling density on the circle is uniform, diag(r) (from (2)) is close to a multiple of the identity, and hence W(s) is expected to be close to W(r) (recall that W(s) = [diag(r)]−1/2W(r)[diag(r)]1/2). Indeed, we found that .
Importantly, the doubly-stochastic normalization recovers the true affinities with high accuracy, with an almost perfect match between the corresponding clean and noisy affinities. On the other hand, there is an evident discrepancy between the corresponding clean and noisy affinities from the row-stochastic normalization and from the symmetric normalization.
3.1.2. Noise sampled uniformly from a ball with smoothly varying radius
Next, we proceed by demonstrating the robustness of the leading eigenvectors from the doubly-stochastic normalization under heteroskedastic noise, and in particular, in the presence of noise whose magnitude depends on the local geometry of the clean data. Specifically, we simulated heteroskedastic noise whose magnitude varies smoothly according to the angle θi of each point xi on the circle (see (14), according to
| (18) |
where stands for the uniform distribution over , which is a ball with radius r in (centered at the origin). That is, every noisy observation is sampled uniformly from a ball whose center is xi and its radius is ρ(θi) from (18). Consequently, the maximal noise magnitude varies smoothly between 0.01 (for θ = π/2, 3π/2) and 1 (for θ = 0, π). A typical array of clean and noisy points arising from the noise model (18) for dimension m = 2 can be seen in Figure 4.
We generated the noisy data points according to (6) for dimension m = 500, and formed the noisy kernel matrix with ε = 0.1. We next computed using Algorithm 1 with δ = 10−12, and evaluated , using according to (2) and (3).
Figure 5 displays the five leading (right) eigenvectors of W(d), W(r), W(s), denoted by , , , respectively, and the five leading (right) eigenvectors of , , , denoted by , , , respectively. It can be seen that the leading eigenvectors from the doubly-stochastic normalization are almost unaffected by the noise, and approximate sines and cosines, which are the eigenfunctions of the Laplace-Beltrami operator on the circle. As sines and cosines are advantageous for expanding periodic functions, it is natural to employ the eigenvectors of for the purposes of regression, interpolation, and classification over the dataset. It is important to mention that other useful bases and frames can potentially be constructed from (see [13, 23]). On the other hand, the eigenvectors obtained from and are strongly biased due to the heteroskedastic noise, and exhibit undesired effects such as discontinuities and localization. Specifically, as evident from Figure 5, the leading eigenvectors of are discontinuous at θ = 0 and θ = π, and the leading eigenvectors of are localized around θ = π/2 and θ = 3π/2 (i.e., their values are close to 0 around θ = 0 and θ = π). Clearly, this behaviour of the leading eigenvectors of and does not reflect the geometry of the clean data, but rather the characteristics of the noise (since the noise variance is smallest at θ = π/2, 3π/2 and largest at θ = 0, π).
Figure 5:
Eigenvectors corresponding to the five largest eigenvalues of the clean and noisy affinity matrices obtained from different normalizations. The top row corresponds to clean affinity matrices (from left to right: W(d), W(r), W(s)) and the bottom row corresponds to noisy affinity matrices (from left to right: , , ). The dataset is the unit circle, with n = 103, m = 500, and heteroskedastic noise generated according to (18).
In Figure 6 we illustrate the two-dimensional embedding of the noisy data points using the second and third eigenvectors of , , and (corresponding to their second- and third-largest eigenvalues). That is, the x-axis and y-axis values for each embedding are given by the entries of and for the doubly-stochastic normalization, and for the row-stochastic normalization, and and for the symmetric normalization (see also [4, 12]). It is clear that the embedding due to the doubly-stochastic normalization reliably represents the intrinsic structure of the clean dataset – a unit circle with uniform density, whereas the embeddings due to the row-stochastic and the symmetric normalizations are incoherent with the geometry and density of the clean points.
3.2. Example 2: Single-cell RNA sequence data
Single-cell RNA sequencing (scRNA-seq) is a revolutionary technique for measuring target gene expressions of individual cells in large and heterogeneous samples [50, 34]. Due to the method’s high resolution (single-cell level) it allows for the discovery of rare cell populations, which is of paramount importance in immunology and developmental biology. A typical scRNA-seq dataset is an m × n nonnegative matrix corresponding to n cells and m genes, where its (i, j)’th entry is an integer called the read count, describing the expression level of i’th gene in the j’th cell. Importantly, the total number of read counts (or in short total reads) per cell (i.e., column sums) may vary substantially within a sample [28]. We next exemplify the advantage of using the doubly-stochastic normalization for exploratory analysis of scRNA-seq data.
3.2.1. Simulated data
We begin with a simple prototypical example where the gene expression levels of cells are measured in two different batches, such that the number of total reads (per cell) within each batch is constant, but is substantially different between the batches. Therefore, the noise variance (modeled by the variance of the multinomial distribution, to be described shortly) differs between the observations in the two batches, giving rise to heteroskedastic noise. Such a scenario can arise naturally in scRNA-seq, either from the intrinsic read count variability common to such datasets, or when two datasets from two independent experiments are merged for unified analysis.
We consider a simulated dataset which includes only two cell types, denoted by p1, , with m = 4000 genes. The prototypes p1 and p2 were created by first sampling their entries uniformly (and independently) from [0, 1], and then normalizing them so that they sum to 1. That is,
| (19) |
Next, each noisy observation was drawn from a multinomial distribution using either p1 or p2 as the probability vector, and normalized to sum to 1, as described next. First, we generated a batch containing 500 observations of p1 and 250 observations of p2, each with 1000 multinomial trials. Second, we added a batch containing 250 observations of p2 only, each with 104 multinomial trials. To summarize, the total number of observations is n = 1000, given explicitly by
| (20) |
Therefore, the dataset consists of 500 (normalized) multinomial observations of p1, followed by 500 (normalized) multinomial observations of p2. While all observations of p1 are with 103 multinomial trials, the observations of p2 are split between 250 observations with 103 multinomial trials, and 250 observations with 104 multinomial trials. Evidently, we can write
| (21) |
where ηi is a zero-mean noise vector (arising from the multinomial sampling) satisfying that is significantly smaller (by a factor of 10 roughly) for 751 ≤ i ≤ 1000 compared to 1 ≤ i ≤ 750.
Using the noisy observations , we formed the noisy kernel matrix of (1) with ε = 2 · 10−5, computed the corresponding matrix using Algorithm 1 with δ = 10−12, and evaluated the matrices , according to (2) and (3). Our methodology for choosing ε was to take it to be the smallest possible such that Algorithm 1 converges within the desired tolerance (specifically, in this experiment we set a maximum of 106 iterations for the algorithm). We note that if ε is too small, then becomes too sparse (approximately), and the doubly-stochastic normalization may become numerically ill-posed.
Figure 7 illustrates the values (in logarithmic scale) of the obtained affinity matrices , , . It is evident that the affinity matrix from the doubly-stochastic normalization accurately describes the relationships between the data points. That is, indicates the similarities within the two groups of cell types (i.e., p1 and p2), but also the dissimilarities between them, regardless of batch association. On the other hand, the affinity matrices from the row-stochastic and the symmetric normalizations are not loyal to the grouping according to cell types, but rather to batch association. In particular, and highlight the observations from the second batch (observations 751–1000) as being most similar to all other observations. Clearly, the fundamental issue here is the heteroskedasticity of the noise, and specifically, the fact that the noise in the last 250 observations is considerably smaller than the noise in all the other observations.
One of the main goals of exploratory analysis of scRNA-seq data is to identify different cell types. Towards that end, non-linear dimensionality reduction techniques are often employed, among which t-distributed stochastic neighbor embedding (t-SNE) [33] is perhaps the most prominent [32, 51, 54, 21]. For its operation, t-SNE employs an affinity matrix which is a close variant of the row-stochastic normalization (2), where the kernel width parameter ε in (1) is allowed to vary between different rows of K, and the resulting row-stochastic matrix is symmetrized by averaging it with its transpose. The different kernel widths are determined by a parameter called the perplexity, which is related to the entropy of each row of the resulting affinity matrix.
Even though the affinity matrix employed by t-SNE is a modification of the standard row-stochastic normalization, and uses a different value of ε for each row, it is still expected to suffer from the inherent bias observed in Figure 9b. Specifically, note that the order of the entries in each row of (when sorted by their values) does not depend on ε, and only on the noisy pair-wise distances , which are strongly biased by the magnitudes of the noise, as evident from Figure 9b.
In Figures 8a,8b,8c we demonstrate the two-dimensional visualization obtained from t-SNE for the dataset , using typical perplexity values of 10, 30, 100. We used MATLAB’s standard implementation of t-SNE, activating the option of forcing the algorithm to be exact (i.e., without approximating the affinity matrix). All other parameters of t-SNE were set to their default values suggested by the code (we also mention that the default suggested perplexity is 30).
In Figure 8d we display the two-dimensional visualization obtained from t-SNE when replacing its default affinity matrix construction with the doubly-stochastic matrix (obtained using ε = 2 · 10−5), while leaving all other aspects of t-SNE unchanged. Since the optimization procedure in t-SNE is affected by randomness, we ran the experiment several times to verify that the results we exhibit are consistent.
While there are only two types of cell in the data (p1 and p2), no clear evidence of this fact can be found in the visualizations by t-SNE (Figures 8a,8b,8c). Furthermore, the visualizations by t-SNE do not provide any noticeable separation between the cell types. On the other hand, the visualization obtained by modifying the t-SNE to employ the doubly-stochastic affinity matrix (Figure 8d) allows one to easily identify and distinguish between the two cell types.
3.2.2. Experimental data
In our second example for scRNA-seq, we use an experimental dataset of purified Peripheral Blood Mononuclear Cells (PBMC) [61], which includes 94654 cells and 32733 genes. This dataset is particularly advantageous for our purposes since each cell in the experiment was labeled according to a known cell type (with 10 different types in total). While this particular dataset does not include different experimental batches (as in the previous simulated example), there is nonetheless inherent variability in the read counts associated with different cell types. To demonstrate the advantage of the doubly-stochastic normalization over the row-stochastic or symmetric normalizations, we focus on the two cell types in the data that have the largest difference in their read counts (on average), which are the CD14 and CD34 cells. Specifically, the CD34 cells have roughly four times more read counts on average than the CD14 cells.
We randomly sampled n1 = 103 cells out of all CD14 cells, sampled n2 = 103 cells out of all CD34 cells, and concatenated their gene expressions (using all genes) into a matrix of size 32733 × (n1 + n2). We then normalized each column of this matrix to sum to 1 (which is a standard procedure used in scRNA-seq for normalizing the read count of each cell, see also Section 3.2.1), and denoted the resulting columns by . That is, are the normalized gene expressions of the sampled CD14 cells, and are the normalized gene expressions of the sampled CD34 cells. We then formed the kernel matrix of (1) with ε = 5 · 10−4, computed the corresponding matrix using Algorithm 1 with δ = 10−12, and evaluated the matrices , according to (2) and (3). We mention that other values of ε produce similar results to what we report next.
Figure 9 illustrates the values (in logarithmic scale) of the obtained affinity matrices , , . It is evident that the affinity matrices form the doubly-stochastic and the symmetric normalizations accurately reflect the structure of the data, as they assign large affinities between cells of the same type and small affinities between cells of different type. The row-stochastic normalization, on the other hand, assigns large affinities between CD14 cells and CD34 cells, which is clearly a bias from the fact that the CD34 cells are less noisy compared to the CD14 cells (due to the difference between their read counts).
Aside from the qualitative differences between the affinity matrices depicted in Figure 9, it is of interest to quantitatively assess their accuracy. Even though we do not have access to the corresponding clean affinity matrices W(d), W(r), W(s), we can make use of the logical reasoning that the nearest neighbors of any given reference cell, defined as the cells with largest affinities to that reference cell, should belong to the same cell type as the reference cell. Following this logic, for each cell i we first found its k nearest neighbors, which are given by the k indices with largest entries in the i’th row of a given affinity matrix (, , or ). Then, as a measure of error, for each cell i we found the proportion of its k nearest neighbors that do not share its cell type. We averaged this proportion for all cells i = 1, … , n1 + n2, and furthermore averaged these results over 20 randomized trials of sampling from the CD14 and CD34 cells.
Figure 10 depicts the resulting proportions of inconsistent cell types as a function of the number of nearest neighbors k for each of the affinity matrices , , and . It is evident that according to our measure of cell type consistency, the affinity matrix from the doubly-stochastic normalization provides the lowest proportion of incorrectly determined near neighbors for all choices of k, establishing the advantage of the doubly-stochastic normalization. In particular, the proportion of the nearest neighbor (i.e., k = 1) with inconsistent cell type using the doubly-stochastic normalization is about 5 times less than that of the symmetric normalization, and about 20 times less than that of the row-stochastic normalization.
3.2.3. Validity of the asymptotic model for scRNA-seq
We conclude our scRNA-seq example with a brief discussion on the validity of Theorem 3 in an asymptotic setting of scRNA-seq, where the number of cells n and the kernel width ε are fixed, and the number of genes m is increasing. Suppose that the gene expression levels for each cell are sampled from a multinomial variable [52], where the number of multinational trials may differ between cells. That is, let ~ Multinomial(ri, pi), where ri is the read count for the i’th cell, pi[j] is the underlying proportion for the expression level of the j’th gene in the i’th cell, and for all i. When preprocessing scRNA-seq data, an important first step is to normalize the gene expression levels by the number of read counts. Hence, we define , so that . In this case, the matrix is equal to the covariance matrix of the multinomial divided by . Using Theorem 1 in [5] (which provides an inequality on the eigenvalues of the covariance of a multinomial),
| (22) |
To consider an asymptotic setting, we think of an experimental setup where an increasing number of genes is sequenced. It is clear that in this experimental setup the number of read counts ri for each cell needs to be controlled appropriately as a function of m. Since , it is evident that the conditions in Theorem 3 hold if ri = Ω(m), a condition which was recently identified as important for large-scale scRNA-seq experiments [60].
4. Summary and discussion
In this work, we investigated the robustness of the doubly-stochastic normalization to heteroskedastic noise, both from a theoretical perspective and from a numerical one. Our results imply that the doubly-stochastic normalization is advantageous over the popular row-stochastic and symmetric normalizations, particularly when the data at hand is high-dimensional and suffers from inherent heteroskedasticity. Moreover, our experiments suggest that incorporating the doubly-stochastic normalization into various data analysis, visualization, and processing techniques for real-world datasets can be worthwhile. The doubly-stochastic normalization is particularly appealing due to is simplicity, solid theoretical foundation, and resemblance to the row-stochastic/symmetric normalizations (which proved useful in countless applications).
The results reported in this work naturally give rise to several possible future research directions. On the theoretical side, it is of interest to characterize the convergence rate of to W(d) also in terms of the number of points n and the covariance matrices {Σi} explicitly. As a particular simpler case, one may consider the high-dimensional setting where both n and m tend to infinity, while the quantity n/m is fixed (or tends to a fixed constant). On the practical side, it is of interest to investigate how to best incorporate the affinity matrix from the doubly-stochastic normalization into data analysis and visualization techniques. To that end, it is desirable to derive a method for picking the kernel parameter ε automatically, or in more generality, to determine how to make use of a variable kernel width (similarly to [59]) while retaining the robustness to heteroskedastic noise.
5. Acknowledgements
We would like to thank Boaz Nadler for his useful comments and suggestions. B.L, R.R.C, and Y.K. acknowledge support by NIH grant R01GM131642. R.R.C and Y.K acknowledge support by NIH grant R01HG008383. Y.K. acknowledges support by NIH grant 2P50CA121974. R.R.C acknowledges support by NIH grant 5R01NS10004903.
Appendix
Appendix A. Proof of Lemma 1
We first recall the definition of a fully indecomposable matrix [2]. A matrix B is called fully indecomposable if there are no permutation matrices P and Q such that
| (23) |
with B1 square. We now proceed to show that A from Lemma 1 is fully indecomposable. Since the only zeros in A are on its main diagonal, there is only one zero in every row and every column of A. Consequently, any permutation of the rows and columns of A would retain this property, namely have a single zero in every row and every column. Therefore, if n > 2, it is impossible to find P and Q such that (23) would hold for B = A, since there cannot be a block of zeros in PAQ whose number of rows or columns is greater than 1. Hence, A is fully indecomposable, and the existence and uniqueness of d = [d1, … , dn] > 0 follows from Lemma 4.1 in [30].
Appendix B. Proof of Theorem 3
Throughout this proof we omit the superscript (m) from the quantities , , , , K(m), , W(d),(m), , and it should be noted that the resulting notation corresponds to sequences in the dimension m where n and ε are fixed.
Let us define
| (24) |
for i, j = 1, …, n. By the definition of W(d) in (4), for i ≠ j we can write
| (25) |
Analogously, we define and by replacing {xi} and {di} in (24) with {} and {}, respectively, and we have that .
Let ⨀ denote the Hadamard (element-wise) product, u = [u1, … , un]T, and . We can write
| (26) |
We begin by bounding the quantity , which is the subject of the following Lemma.
Lemma 4. For all i ≠ j,
| (27) |
Proof. Let us write
| (28) |
According to (7) and the conditions in Theorem 3, for i ≠ j we have
| (29) |
| (30) |
| (31) |
| (32) |
Therefore,
| (33) |
where we used the inequality (a + b + c)2 ≤ 3(a2 + b2 + c2). Consequently, Chebyshev’s inequality yields that for any p > 0
| (34) |
which implies
| (35) |
Using the above for i ≠ j, a first-order Taylor expansion of exp(y) around y = 0 gives
| (36) |
and by (28) we have
| (37) |
where we used Hi,j = e⟨xi,xj⟩/ε ≤ e∥xi∥∥xj∥/ε ≤ e1/ε in the last equality. □
Using Lemma 4 and applying the union bound on the off-diagonal entries of , we obtain
| (38) |
Continuing, we bound the quantities maxi,j{uiuj} and from (26). Towards that end, we have the following result.
Proposition 5. Under the conditions of Theorem 3, {Hi,j}i≠j and are upper- and lower-bounded by positive constants independent of m.
Proof. Observe that for i ≠ j and for all m,
| (39) |
Since the set of n × n matrices satisfying the above is compact, and using the fact that {ui} > 0 can be uniquely determined by H (from Lemma 1 applied to H), there must exist constants cu, Cu independent of m such that
| (40) |
for all i and m. □
Consequently, Proposition 5 together with Lemma 4 guarantee that
| (41) |
Next, we turn to bound the quantity from (26). From Lemma 1 applied to H and , it follows that u and are unique. Additionally, by Lemma 4 it is clear that . Therefore, we also have that (as otherwise we have a contradiction to the uniqueness of u and ). Since is doubly-stochastic, we have
| (42) |
Let us define the multivariate functions , where , , as
| (43) |
To bound the error , we expand fi(A, v) around (H, u) using a first-order Taylor expansion. Towards that end, Proposition 5 can be used to verify that the second-order partial derivatives of fi in the vicinity of (H, u) are bounded by constants independent of m. In particular,
| (44) |
for all i, j, k, m, ℓ, where is a ball of radius 1 in Euclidean space around (H, u):
| (45) |
The choice of the radius of the ball is arbitrary, and is only required to guarantee that the point is included in for sufficiently large m. Therefore, by (42) and (44), the first-order Taylor expansion of fi(A, v) around (H, u) gives
| (46) |
where
| (47) |
and we used the fact that is doubly-stochastic). Next, using that fi(H, u) = 1, denoting , and multiplying both hand sides of (46) by ui (ui is bounded according to Proposition 5), we can write
| (48) |
where e = [e1, … , en]T. Consequently, since Hi,i = 0, writing (48) in matrix form gives
| (49) |
where In is the n × n identity matrix. In order to bound the vector e, we must be able to invert the matrix
| (50) |
which is the subject of the following Lemma.
Lemma 6. Under the conditions of Theorem 3, the matrix G from (50) is invertible for all m, and ∥G−1∥2 ≤ CG for some constant CG independent of m.
Proof. Notice that G is similar to the matrix
| (51) |
Therefore, G is invertible if In + W(d) is invertible. Since W(d) is symmetric and doubly-stochastic, its largest eigenvalue is exactly 1, and λmin{W(d)} ≥ −1. Moreover, since Wi,j > 0 for all i ≠ j, we have that {(W(d))2}i,j > 0 for all i, j. Therefore, by Lemma 8.4.3 in [26] W(d) has only one eigenvalue with maximal absolute-value (which is 1). Hence, λmin{W(d)} > −1, and we obtain that
| (52) |
The fact that ∥G−1∥2 is bounded by some constant independent of m is established by Proposition 5 (since the set of all possible matrices G that satisfy (39) and (40) is compact). □
Using (49) together with Lemma 6 and Proposition 5, we have that
| (53) |
where we used the inequality . From (38), (53), and the fact that , , it follows that
| (54) |
Consequently,
| (55) |
where we used Proposition 5 to bound ∥u∥. Overall, substituting (55), (38), and (41) into (26), we arrive at the required result
| (56) |
References
- [1].Allen-Zhu Zeyuan, Li Yuanzhi, Oliveira Rafael, and Wigderson Avi. Much faster algorithms for matrix scaling. In 2017 IEEE 58th Annual Symposium on Foundations of Computer Science (FOCS), pages 890–901. IEEE, 2017. [Google Scholar]
- [2].Bapat Ravi B, Bapat Ravindra B, Raghavan TES, et al. Nonnegative matrices and applications, volume 64. Cambridge University Press, 1997. [Google Scholar]
- [3].Beauchemin Mario. On affinity matrix normalization for graph cuts and spectral clustering. Pattern Recognition Letters, 68:90–96, 2015. [Google Scholar]
- [4].Belkin Mikhail and Niyogi Partha. Laplacian eigenmaps for dimensionality reduction and data representation. Neural computation, 15(6):1373–1396, 2003. [Google Scholar]
- [5].Bénasséni Jacques. A new derivation of eigenvalue inequalities for the multinomial distribution. Journal of Mathematical Analysis and Applications, 393(2):697–698, 2012. [Google Scholar]
- [6].Berry Tyrus and Harlim John. Variable bandwidth diffusion kernels. Applied and Computational Harmonic Analysis, 40(1):68–96, 2016. [Google Scholar]
- [7].Bronstein Michael M, Bruna Joan, LeCun Yann, Szlam Arthur, and Vandergheynst Pierre. Geometric deep learning: going beyond euclidean data. IEEE Signal Processing Magazine, 34(4):18–42, 2017. [Google Scholar]
- [8].Brown Jack B, Chase Phillip J, and Pittenger Arthur O. Order independence and factor convergence in iterative scaling. Linear algebra and its applications, 190:1–38, 1993. [Google Scholar]
- [9].Buades Antoni, Coll Bartomeu, and Morel J-M. A non-local algorithm for image denoising. In 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), volume 2, pages 60–65. IEEE, 2005. [Google Scholar]
- [10].Cao Yuanpei, Zhang Anru, and Li Hongzhe. Multi-sample estimation of bacterial composition matrix in metagenomics data. arXiv preprint arXiv:1706.02380, 2017. [Google Scholar]
- [11].Cochran Robert N and Horne Frederick H. Statistically weighted principal component analysis of rapid scanning wavelength kinetics experiments. Analytical Chemistry, 49(6):846–853, 1977. [Google Scholar]
- [12].Coifman Ronald R and Lafon Stéphane. Diffusion maps. Applied and computational harmonic analysis, 21(1):5–30, 2006. [Google Scholar]
- [13].Coifman Ronald R and Maggioni Mauro. Diffusion wavelets. Applied and Computational Harmonic Analysis, 21(1):53–94, 2006. [Google Scholar]
- [14].Cuturi Marco. Sinkhorn distances: Lightspeed computation of optimal transport. In Advances in neural information processing systems, pages 2292–2300, 2013. [Google Scholar]
- [15].Defferrard Michaël, Bresson Xavier, and Vandergheynst Pierre. Convolutional neural networks on graphs with fast localized spectral filtering. In Advances in neural information processing systems, pages 3844–3852, 2016. [Google Scholar]
- [16].El Karoui Noureddine et al. On information plus noise kernel random matrices. The Annals of Statistics, 38(5):3191–3216, 2010. [Google Scholar]
- [17].El Karoui Noureddine, Wu Hau-Tieng, et al. Graph connection laplacian methods can be made robust to noise. The Annals of Statistics, 44(1):346–372, 2016. [Google Scholar]
- [18].Foi Alessandro. Clipped noisy images: Heteroskedastic modeling and practical denoising. Signal Processing, 89(12):2609–2629, 2009. [Google Scholar]
- [19].Foi Alessandro. Noise estimation and removal in mr imaging: The variance-stabilization approach. In 2011 IEEE International symposium on biomedical imaging: from nano to macro, pages 1809–1814. IEEE, 2011. [Google Scholar]
- [20].Fortunato Santo. Community detection in graphs. Physics reports, 486(3–5):75–174, 2010. [Google Scholar]
- [21].Habib Naomi, Li Yinqing, Heidenreich Matthias, Swiech Lukasz, Avraham-Davidi Inbal, Trombetta John J, Hession Cynthia, Zhang Feng, and Regev Aviv. Div-seq: Single-nucleus rna-seq reveals dynamics of rare adult newborn neurons. Science, 353(6302):925–928, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hafemeister Christoph and Satija Rahul. Normalization and variance stabilization of single-cell rna-seq data using regularized negative binomial regression. Genome Biology, 20(1):1–15, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hammond David K, Vandergheynst Pierre, and Gribonval Rémi. Wavelets on graphs via spectral graph theory. Applied and Computational Harmonic Analysis, 30(2):129–150, 2011. [Google Scholar]
- [24].Hein Matthias, Audibert Jean-Yves, and Von Luxburg Ulrike. From graphs to manifolds–weak and strong pointwise consistency of graph laplacians. In International Conference on Computational Learning Theory, pages 470–485. Springer, 2005. [Google Scholar]
- [25].Hoeffding Wassily. Probability inequalities for sums of bounded random variables. In The Collected Works of Wassily Hoeffding, pages 409–426. Springer, 1994. [Google Scholar]
- [26].Horn Roger A. and Johnson Charles R.. Matrix Analysis. Cambridge University Press, 2 edition, 2012. [Google Scholar]
- [27].Idel Martin. A review of matrix scaling and sinkhorn’s normal form for matrices and positive maps. arXiv preprint arXiv:1609.06349, 2016. [Google Scholar]
- [28].Kim Tae, Zhou Xiang, and Chen Mengjie. Demystifying” drop-outs” in single cell umi data. bioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kluger Yuval, Basri Ronen, Chang Joseph T, and Gerstein Mark. Spectral biclustering of microarray data: coclustering genes and conditions. Genome research, 13(4):703–716, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Knight Philip A. The sinkhorn–knopp algorithm: convergence and applications. SIAM Journal on Matrix Analysis and Applications, 30(1):261–275, 2008. [Google Scholar]
- [31].Landa Boris and Shkolnisky Yoel. The steerable graph laplacian and its application to filtering image datasets. SIAM Journal on Imaging Sciences, 11(4):2254–2304, 2018. [Google Scholar]
- [32].Linderman George C, Rachh Manas, Hoskins Jeremy G, Steinerberger Stefan, and Kluger Yuval. Fast interpolation-based t-sne for improved visualization of single-cell rna-seq data. Nature methods, 16(3):243–245, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].van der Maaten Laurens and Hinton Geoffrey. Visualizing data using t-sne. Journal of machine learning research, 9(November):2579–2605, 2008. [Google Scholar]
- [34].Macosko Evan Z, Basu Anindita, Satija Rahul, Nemesh James, Shekhar Karthik, Goldman Melissa, Tirosh Itay, Bialas Allison R, Kamitaki Nolan, Martersteck Emily M, et al. Highly parallel genomewide expression profiling of individual cells using nanoliter droplets. Cell, 161(5):1202–1214, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mann Henry B and Wald Abraham. On stochastic limit and order relationships. The Annals of Mathematical Statistics, 14(3):217–226, 1943. [Google Scholar]
- [36].Marshall Nicholas F and Coifman Ronald R. Manifold learning with bi-stochastic kernels. IMA Journal of Applied Mathematics, 84(3):455–482, 2019. [Google Scholar]
- [37].Meyer François G and Shen Xilin. Perturbation of the eigenvectors of the graph laplacian: Application to image denoising. Applied and Computational Harmonic Analysis, 36(2):326–334, 2014. [Google Scholar]
- [38].Nadler Boaz, Lafon Stéphane, Coifman Ronald R, and Kevrekidis Ioannis G. Diffusion maps, spectral clustering and reaction coordinates of dynamical systems. Applied and Computational Harmonic Analysis, 21(1):113–127, 2006. [Google Scholar]
- [39].Ng Andrew Y, Jordan Michael I, and Weiss Yair. On spectral clustering: Analysis and an algorithm. In Advances in neural information processing systems, pages 849–856, 2002. [Google Scholar]
- [40].Pang Jiahao and Cheung Gene. Graph laplacian regularization for image denoising: Analysis in the continuous domain. IEEE Transactions on Image Processing, 26(4):1770–1785, 2017. [DOI] [PubMed] [Google Scholar]
- [41].Salmon Joseph, Harmany Zachary, Deledalle Charles-Alban, and Willett Rebecca. Poisson noise reduction with non-local pca. Journal of mathematical imaging and vision, 48(2):279–294, 2014. [Google Scholar]
- [42].Sarkar Purnamrita, Bickel Peter J, et al. Role of normalization in spectral clustering for stochastic blockmodels. The Annals of Statistics, 43(3):962–990, 2015. [Google Scholar]
- [43].Shaham Uri, Stanton Kelly, Li Henry, Basri Ronen, Nadler Boaz, and Kluger Yuval. Spectralnet: Spectral clustering using deep neural networks. In International Conference on Learning Representations, 2018. [Google Scholar]
- [44].Shi Jianbo and Malik Jitendra. Normalized cuts and image segmentation. IEEE Transactions on pattern analysis and machine intelligence, 22(8):888–905, 2000. [Google Scholar]
- [45].Shuman David I, Narang Sunil K, Frossard Pascal, Ortega Antonio, and Vandergheynst Pierre. The emerging field of signal processing on graphs: Extending high-dimensional data analysis to networks and other irregular domains. IEEE signal processing magazine, 30(3):83–98, 2013. [Google Scholar]
- [46].Singer Amit. From graph to manifold laplacian: The convergence rate. Applied and Computational Harmonic Analysis, 21(1):128–134, 2006. [Google Scholar]
- [47].Singer Amit, Shkolnisky Yoel, and Nadler Boaz. Diffusion interpretation of nonlocal neighborhood filters for signal denoising. SIAM Journal on Imaging Sciences, 2(1):118–139, 2009. [Google Scholar]
- [48].Sinkhorn Richard and Knopp Paul. Concerning nonnegative matrices and doubly stochastic matrices. Pacific Journal of Mathematics, 21(2):343–348, 1967. [Google Scholar]
- [49].Tamuz Omer, Mazeh Tsevi, and Zucker Shay. Correcting systematic effects in a large set of photometric light curves. Monthly Notices of the Royal Astronomical Society, 356(4):1466–1470, 2005. [Google Scholar]
- [50].Tang Fuchou, Barbacioru Catalin, Wang Yangzhou, Nordman Ellen, Lee Clarence, Xu Nanlan, Wang Xiaohui, Bodeau John, Tuch Brian B, Siddiqui Asim, et al. mrna-seq whole-transcriptome analysis of a single cell. Nature methods, 6(5):377, 2009. [DOI] [PubMed] [Google Scholar]
- [51].Tirosh Itay, Izar Benjamin, Prakadan Sanjay M, Wadsworth Marc H, Treacy Daniel, Trombetta John J, Rotem Asaf, Rodman Christopher, Lian Christine, Murphy George, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell rna-seq. Science, 352(6282):189–196, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].William Townes F, Hicks Stephanie C, Aryee Martin J, and Irizarry Rafael A. Feature selection and dimension reduction for single-cell rna-seq based on a multinomial model. Genome biology, 20(1):1–16, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Trillos Nicolás García, Gerlach Moritz, Hein Matthias, and Slepčev Dejan. Error estimates for spectral convergence of the graph laplacian on random geometric graphs toward the laplace–beltrami operator. Foundations of Computational Mathematics, pages 1–61, 2019. [Google Scholar]
- [54].Villani Alexandra-Chloé, Satija Rahul, Reynolds Gary, Sarkizova Siranush, Shekhar Karthik, Fletcher James, Griesbeck Morgane, Butler Andrew, Zheng Shiwei, Lazo Suzan, et al. Single-cell rna-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science, 356(6335):eaah4573, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Von Luxburg Ulrike. A tutorial on spectral clustering. Statistics and computing, 17(4):395–416, 2007. [Google Scholar]
- [56].Wang Fei, Li Ping, König Arnd Christian, and Wan Muting. Improving clustering by learning a bi-stochastic data similarity matrix. Knowledge and information systems, 32(2):351–382, 2012. [Google Scholar]
- [57].Zass Ron and Shashua Amnon. A unifying approach to hard and probabilistic clustering. In Tenth IEEE International Conference on Computer Vision (ICCV’05) Volume 1, volume 1, pages 294–301. IEEE, 2005. [Google Scholar]
- [58].Zass Ron and Shashua Amnon. Doubly stochastic normalization for spectral clustering. In Advances in neural information processing systems, pages 1569–1576, 2007. [Google Scholar]
- [59].Zelnik-Manor Lihi and Perona Pietro. Self-tuning spectral clustering. In Advances in neural information processing systems, pages 1601–1608, 2005. [Google Scholar]
- [60].Jinye Zhang Martin, Ntranos Vasilis, and Tse David. Determining sequencing depth in a single-cell rna-seq experiment. Nature communications, 11(1):1–11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zheng Grace XY, Terry Jessica M, Belgrader Phillip, Ryvkin Paul, Bent Zachary W, Wilson Ryan, Ziraldo Solongo B, Wheeler Tobias D, McDermott Geoff P, Zhu Junjie, et al. Massively parallel digital transcriptional profiling of single cells. Nature communications, 8(1):1–12, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]