Abstract
Mitochondria are the key organelles that supply cellular energy. As the most active organ in the body, the energy required to maintain the mechanical function of the heart requires a high quantity of high-quality mitochondria in cardiomyocytes. MicroRNAs (miRNAs) are single-stranded noncoding RNAs, approximately 22 nt in length, which play key roles in mediating post-transcriptional gene silencing. Numerous studies have confirmed that miRNAs can participate in the occurrence and development of cardiac diseases by regulating mitochondrial function-related genes and signaling pathways. Therefore, elucidating the crosstalk that occurs between miRNAs and mitochondria is important for the prevention and treatment of cardiac diseases. In this review, we discuss the biogenesis of miRNAs, the miRNA-mediated regulation of major genes involved in the maintenance of mitochondrial function, and the effects of miRNAs on mitochondrial function in cardiac diseases in order to provide a theoretical basis for the clinical prevention and treatment of cardiac disease and the development of new drugs.
Keywords: microRNAs, mitochondrial function, cardiac disease, cardiac apoptosis, cardiac hypertrophy, diabetic cardiomyopathy, heart failure
Introduction
Cardiovascular diseases (CVDs) refer to a group of diseases that affect cardiac and vascular tissues and are the most common causes of morbidity and mortality worldwide. By 2030, an estimated 23.6 million people will die due to CVD-related complications annually, and cardiac diseases and stroke are the two leading causes of CVD-associated death (World Health Organization, 2019). Cardiac diseases include multiple disorders, such as pathological cardiac remodeling (Schirone et al., 2017), diabetic cardiomyopathy (Paolillo et al., 2019), heart failure (Rossignol et al., 2019), and myocarditis (Ammirati et al., 2020). Recent studies have indicated that microRNAs (miRNAs) and mitochondria are two important factors involved in the pathophysiological processes underlying cardiac diseases. The targeted regulation of genes by miRNAs is closely associated with mitochondrial function, and miRNAs play important roles in the occurrence and development of cardiac diseases (Borralho et al., 2015; Song et al., 2019). Therefore, a broad understanding of the crosstalk that occurs between miRNAs and mitochondrial function is vital for the prevention and treatment of cardiac diseases.
Mitochondria serve as the cellular “energy factory,” supplying intracellular energy to all cells, which synthesize energy-rich phosphate bonds in the form of adenosine triphosphate (ATP) via oxidative phosphorylation (OXPHOS) and are deeply integrated into multiple cellular metabolisms and signaling pathways (Tahrir et al., 2019). As the most metabolically active organ of the body, the heart requires enormous amounts of energy to maintain systolic and diastolic functions, which requires a high content of cardiac mitochondria. Under normal physiological conditions, 90% of the energy required by the heart is derived from mitochondrial OXPHOS, and normal mitochondrial function is a necessary prerequisite for the maintenance of the heart pumping function. Recently, increasing data have shown that mitochondrial dysfunction is involved in the occurrence of cardiac diseases, such as cardiac hypertrophy, myocardial fibrosis (Chen et al., 2019), diabetic cardiomyopathy (Gollmer et al., 2020), and heart failure (Kumar et al., 2019). Accumulating evidence has also suggested that targeting mitochondrial function may represent a promising potential therapeutic strategy for addressing heart failure (Brown et al., 2017). Therefore, reviewing the factors that affect mitochondrial function and cardiac regulatory pathways is important for targeting the clinical treatment of cardiac diseases.
MiRNAs are a class of small, endogenous, single-stranded, noncoding RNAs, and mature miRNAs in eukaryotic cells are approximately 22 nt in length. MiRNAs can inhibit mRNA translation or regulate transcriptional degradation by binding to the 3′untranslated region (UTR) of the target gene, regulating gene expression (Akhtar et al., 2016). The first miRNA, lin-4, was discovered in 1993 (Lee et al., 1993), and miRNA research has continued for more than 20 years. MiRNAs are among the best-studied class of noncoding RNA and are viewed as important regulators of cell differentiation, growth, proliferation, and apoptosis (Colpaert and Calore, 2019), and then participated in a variety of cardiovascular diseases, such as atherosclerosis (Neth et al., 2013), myocardial remodeling (Zhou et al., 2013) and acute myocardial infarction (Zhang et al., 2020). Recent experimental and clinical data have revealed that the differential expression of miRNAs affects mitochondrial function through the regulation of relevant signaling pathways and mitochondrial function-related proteins. Many miRNAs are thought to be involved in the development of CVDs, such as miR-20b, which promotes cardiac hypertrophy by targeting the inhibition of mitofusin 2 (MFN2)-mediated intercellular Ca2+ interactions in cardiomyocytes (Qiu et al., 2020). The knockout of miR-181c upregulated the expression of mitochondrial calcium uptake 1 (MICU1), and the regulation of mitochondrial calcium uptake in the myocardium has been shown to protect against heart injury (Banavath et al., 2019). Therefore, understanding the crosstalk between miRNAs and mitochondrial function is important for the study of cardiac diseases and the development of potential drug interventions.
We reviewed the existing miRNA research associated with mitochondrial function and cardiac disease (myocardial apoptosis, cardiac hypertrophy, diabetic cardiomyopathy, and heart failure) found in an English database and attempted to summarize identified miRNAs thought to be involved in the regulation of mitochondrial function. In particular, we focused on the miRNA-mediated regulation of target genes and related signaling pathways involved in mitochondrial function and are thought to affect cardiac homeostasis. We also attempted to identify existing gaps in the current research, including the challenges encountered in miRNA research. We aimed to provide a better understanding of the potential mechanisms that link miRNAs and mitochondria in the development of cardiac diseases and to provide novel targets for the early diagnosis, prevention, and treatment of cardiac diseases.
MicroRNAs Biogenesis
Canonical MicroRNAs
With the advent of deep sequencing technology, a large number of noncoding miRNAs have been identified, and our understanding of complex miRNAs has increased. The biogenesis of miRNAs occurs in both the nucleus and cytoplasm through multiple enzymatic cleavage steps (Kim Y.-K. et al., 2016). MiRNAs are encoded by genomic DNA and are transcribed by RNA polymerase II (POL II) or POL III, which produce longer primary miRNA transcripts, known as pri-miRNAs, which can be up to 1,000 nt in length (O’Donnell et al., 2005; Cai et al., 2004). Subsequently, the stem-loop structure of pri-miRNAs is modified by the RNase class III enzyme DROSHA to form precursor-miRNAs (pre-miRNAs) by truncating the stem-loop, while the RNA-binding protein DGCR8 assisted DROSHA in acting on miRNAs to form a hairpin-loop structure pre-miRNA that is approximately 70–100 nt in length. DGCR8 and DROSHA are collectively known as the microprocessor complex, resulting in the RNase III-mediated cleavage of 2-nt-3′ overhangs (Lee et al., 2003; Denli et al., 2004; Lund et al., 2004; Han et al., 2006; Ha and Kim, 2014; O’Brien et al., 2018). These processes all occur in the nucleus.
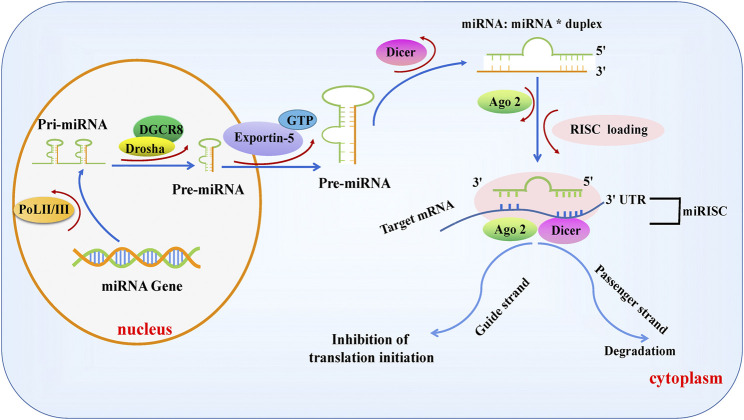
Pre-miRNAs are transported into the cytoplasm by Exportin-5, which depend on Ran gtpase (a GTP-binding nuclear protein) (Yi et al., 2003), and the terminal loops of pre-miRNAs are removed by the enzymatic cleavage of the RNase III Dicer to form a 22-nt miRNA: miRNA* duplex with incomplete complementarity (Hutvágner et al., 2001; Ketting et al., 2001). One of the stranded molecules is introduced into the RNA-induced silencing complex (RISC) containing argonaute proteins (AGOs), known as a guiding strand. At the same time, we see the degradation of the passenger strand. miRNA-loaded RISCs (miRISCs) bind to reverse complementary sequences within the 3’ UTR of the target gene mRNA, inhibiting the translation or degradation of target gene mRNAs and inducing gene silencing. Figure 1 shows the canonical synthesis process for miRNAs (Martinez et al., 2002; Pratt and MacRae, 2009; Winter et al., 2009; Hammond, 2015; Wilson et al., 2015; Bartel, 2018).
FIGURE 1.
Biogenesis of canonical microRNAs. MiRNA genes are transcribed into a single-stranded primary miRNA by POL II or POL III, followed by the modification of the stem-loop structure of pri-MiRNAs by DROSHA, resulting in the formation of a 70–100 nt pre-miRNA with the assistance of the RNA-binding protein DGCR8. Subsequently, pre-miRNAs are exported to the cytoplasm by Ran GTPase and Exportin-5 and cleaved by Dicer to form mature miRNA double strands, one of which, known as the guide strand, is introduced into the RISC complex containing AGO. The other strand, called the passenger chain, is degraded. The miRNA-loaded RISC combines with the reverse complementary sequence within the 3′UTR of the target mRNA, suppressing the translation or degradation of the target mRNA.
Non-Canonical MicroRNAs
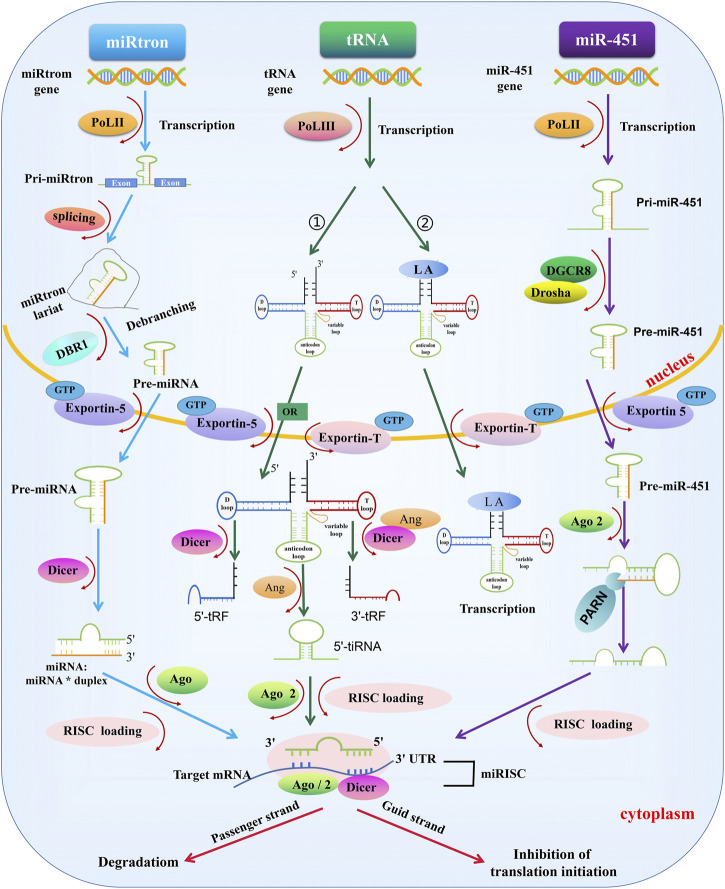
In addition to the canonical pathway, some miRNAs are produced through alternate biogenesis pathways and are similar in structure and function to the miRNAs. These are called non-canonical miRNAs: e.g., miRtrons, transfer RNAs (tRNAs), and miR-451 (Martinez et al., 2002; Abdelfattah et al., 2014). Although non-canonical miRNAs may skip one or more steps of the canonical synthesis pathway, Dicer appears to be essential for non-canonical miRNAs, except for miR-451 (Okamura et al., 2007; Ruby et al., 2007; Cheloufi et al., 2010; Abdelfattah et al., 2014; Stavast and Erkeland, 2019) (Figure 2).
FIGURE 2.
Biogenesis of non-canonical miRNAs. miRtrons: miRtron are primarily localized in the introns of host genes and modified introns, which are formed by the spliceosome and lariat debranching enzyme 1 (DBR1) treatment in the nucleus, can act as pre-miRNAs—which are processed independently of DROSHA/DGCR8. The remaining miRtron biosynthesis process is the same as canonical miRNAs. tRNA: 1) tRNA starts from POL III in the early stage. Next, the pre-tRNA is exported to the cytoplasm by Exportin-5 or Exportin-T. In the cytoplasm, the pre-tRNA adopts a cloverleaf-shaped secondary structure, composed of a D-loop, an anticodon loop, a T-loop, and a variable loop. Pre-tRNA is cleaved by Dicer or angiogenin (ANG): the h D-loop is cleaved by Dicer, the T-loop is cleaved by Dicer and ANG, and the anticodon loop is cleaved by ANG. Some tRNA fragments are loaded into RISC containing AGO2 protein and regulate gene expression. 2) tRNAs transcribed and synthesized by POL III can be stabilized by LA and transported to the cytoplasm under the action of Exportin-T and Ran GTPase to maintain the stability of tRNAs for translation. MiR-451: MiR-451 in transcribed by POL II. After modification by DROSHA/DGCR8 treatment, miR-451 is exported to the cytoplasm by Exportin-5 and Ran GTPase, cleaved by an unknown endonuclease at the 3′end, and loaded into the RISC-AGO complex to exert its biological function, a process independent of Dicer enzymatic cleavage.
MiRtrons
MiRtrons were the first identified miRNAs synthesized through non-canonical pathways in Drosophila melanogaster and Caenorhabditis elegans. MiRtrons are coded by the introns of host genes (Okamura et al., 2007; Wen et al., 2015). Through splice cleavage events, the intron lariat is debranched by lariat debranching enzyme 1 (DBR1), and the modified intron can serve as a pre-miRNA, independent of DROSHA/DGCR8 (Westholm and Lai, 2011). The next step in the synthesis process is consistent with the canonical miRNAs synthesis pathway. (Babiarz et al., 2008).
MicroRNAs From Transfer RNAs
The structure of tRNAs differs from that of other miRNAs. A mature tRNA is processed by POL III during the early stage. The pre-tRNA adopts a cloverleaf-shaped secondary structure, composed of a D-loop, an anticodon loop, a T-loop, and a variable loop (Oberbauer and Schaefer, 2018). Next, the pre-tRNA is exported to the cytoplasm by Exportin-5 or Exportin-T, where the pre-tRNA is cleaved by Dicer or angiogenin (ANG): Dicer cleaves the D-loop, both Dicer and ANG cleave the T-loop, and ANG cleaves anticodon loop. The cleaved tRNA interacts with AGO to form a mature miRNA complex involved in a variety of biological functions (Cao et al., 2020). In addition, the lupus autoantigen (LA) is a conserved nuclear RNA-binding factor. The 3′-oligo (U) of POL III is a sequence-specific signal for the termination POL III transcription and a high-affinity binding site for LA, and LA is closely involved in tRNA synthesis associated with POL III transcription. tRNAs that are transcribed and synthesized by POL III can be stabilized by LA and transported to the cytoplasm by Exportin-T and Ran gtpase, which maintain the stability of tRNAs for translation. LA also plays a major role in the inhibition of Dicer-mediated 5′-tRNA-derived RNA fragment cleavage (Okamura et al., 2007; Bayfield and Maraia, 2009; Blewett and Maraia, 2018; Stavast and Erkeland, 2019).
MiR-451
Unlike other miRNAs, the biosynthesis pathway of miR-451 is independent of Dicer but requires the AGO2 slicer activity to achieve miRNA maturation (Cheloufi et al., 2010; Cifuentes et al., 2010). MiR-451 originates from the transcription of its host gene by POL II, generating pri-miR-451, which is modified by DROSHA/DGCR8 in the nucleus and transformed into pre-miR-451. Following export to the cytoplasm by Exportin-5 under the action of Ran gtpase (Stavast and Erkeland, 2019), pre-miR-451 cannot be cleaved by Dicer because the stem-loop structure is too short. Instead, pre-miR-451 is cleaved by an unknown endonuclease at the three’ end, loaded into AGO2, and subsequently integrated into a RISC complex to perform its function (Abdelfattah et al., 2014). The specificity of miR-451 has encouraged the exploration of Dicer-independent miRNAs.
Increasing investigations have revealed additional Dicer-independent miRNAs, such as miR-1225 and miR-1228, the biogenesis of which are not only independent of Dicer but also DGCR8, Exportin-5, and AGO2 (Havens et al., 2012). In addition, some miRNAs are synthesized through non-canonical pathways, such as snoRNA-derived miRNAs (sno-miR-28) and miRNAs derived from endogenous short hairpin RNAs (miR-320 and miR-484). A more in-depth description of the biosynthetic pathways associated with non-canonical miRNAs can be found elsewhere (Okamura et al., 2007; Stavast and Erkeland, 2019). The biosynthetic pathways that generate miRNAs are very complex, and the mechanisms of miRNA synthesis and the interactions that occur among components require further study. Additional miRNA synthetic pathways likely exist that have not yet been discovered.
MicroRNAs Regulate Core Mitochondrial Signals
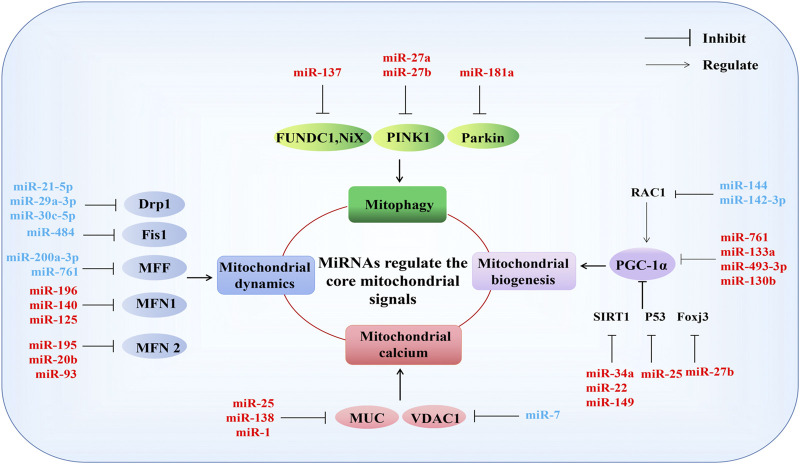
MiRNAs are known to serve as primary regulators of mitochondrial function (Song et al., 2019). Mitochondrial dysfunction can occur due to mitochondrial structural damage, reduced ATP synthesis, the overproduction of reactive oxygen species (ROS), disturbances in calcium ions, mitochondrial DNA (mtDNA) damage, mitochondrial dynamics, biosynthesis (Bose and Beal, 2016), and mitophagy abnormalities (Chistiakov et al., 2018). MiRNAs typically participate in these pathophysiological processes by regulating the expression of mitochondria-related genes (Figure 3).
FIGURE 3.
MiRNAs regulate the core mitochondrial signals. MiRNAs participate in mitochondrial function by regulating genes directly related to mitophagy, mitochondrial dynamics, mitochondrial calcium, and mitochondrial biogenesis. Mitochondrial function-related genes: mitophagy (FUNDC1, NiX, PINK1, Parkin); mitochondrial dynamics (Drp1, Fis1, MFF, MFN1, MFN2); mitochondrial calcium (MUC, VDAC1), mitochondrial biogenesis (PGC-1α). See text for detailed explanations. Red and blue represent negative and positive regulation of mitochondrial function, respectively. T represents inhibitory effect, and arrows represent involvement in regulation.
Mitochondrial Biogenesis
Mitochondrial biogenesis is a key pathway for intracellular mitochondrial self-renewal and repair. Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) family members and related transcription coactivators are the primary regulators of mitochondrial biogenesis, and PGC-1α can be activated in response to growth signals or energy requirements (Puigserver and Spiegelman, 2003; Zhang and Xu, 2016). Previous evidence has suggested that miR-34a, miR-22, and miR-149 negatively regulate mitochondrial biogenesis by targeting the reduction of Sirtuin 1 (SIRT 1) expression to inhibit the PGC-1α activity (Mohamed et al., 2014; Mao et al., 2019; Xiong et al., 2019). MiR-761, miR-133a, miR-493–3p, and miR-130b negatively regulate mitochondrial biogenesis by directly targeting the inhibition of PGC-1α expression (Xu et al., 2015; Nie et al., 2016; Jiang et al., 2017; Lemecha et al., 2018). MiR-27b and miR-25 inhibit mitochondrial biogenesis by downregulating the expression of forkhead box J3 (Foxj3) and P53, respectively (Wang et al., 2016; Shen et al., 2016). Additionally, miR-144 and miR-142-3p promote mitochondrial biogenesis by targeting the rac family small gtpase 1(Rac1) to activate PGC-1α (Tao et al., 2020; Xia et al., 2020).
Mitochondrial Dynamics
Mitochondrial dynamics describe how the mitochondria change their shape, distribution, size, and function through the dynamic balance of continuous fission and fusion to meet the energy needs of host cells and respond to environmental changes. In mammals, these dynamic processes are primarily regulated by the dynamin gtpase family. Dynamin-related protein 1 (Drp1), mitochondrial fission factor (MFF), and mitochondrial fission protein 1(Fis1) regulate mitochondrial fission, MFN1 and MFN2 are responsible for mitochondrial outer membrane (OMM) fusion, and optic atrophy 1 (OPA1) mediates mitochondrial inner membrane (IMM) fusion (Tilokani et al., 2018). MiR-21-5p, miR-29a-3p, and miR-30c-5p have been reported to negatively regulate mitochondrial fission Drp1 targeting (Chowdhury et al., 2017; Zhang et al., 2017; Xie et al., 2020). MiR-484 participates in mitochondrial network regulation by targeting Fis1, improving mitochondrial fission (Wang et al., 2012). In addition, miR-200a-3p and miR-761 can improve mitochondrial dysfunction by targeting the negative regulation of MFF, enhancing mitochondrial activity and ATP production (Long et al., 2013; Lee et al., 2017). MFN2-mediated mitochondrial fusion is inhibited by miR-195, miR-20b, and miR-93 (Feng et al., 2019; Purohit et al., 2019; Qiu et al., 2020). Furthermore, miR-196, miR-140, and miR-125 negatively regulate mitochondrial fusion by inhibiting MFN1 (Li J. et al., 2014; Li X. et al., 2014; Ma et al., 2017). However, the direct regulation of OPA1 by miRNAs has not yet been reported and requires further study.
Mitochondrial Calcium
The ability of mitochondria to act as Ca2+ buffers plays a significant role in the regulation of intracellular Ca2+ signaling. Mitochondria are the primary store of intracellular calcium, and calcium homeostasis serves as the physiological basis of many cellular activities, such as the regulation of OXPHOS, the modulation of cytosolic Ca2+ signals, cell death, secretion, and the production of ROS. The voltage-dependent anion channel (VDAC) in the OMM permits Ca2+entry into mitochondria, and the mitochondrial Ca2+ uniporter complex (MCU) in the IMM serves as a driving force for the electron transport chain (Jaquenod De Giusti et al., 2018). MiR-25, miR138, and miR-1 have been reported to regulate mitochondrial calcium by targeting MUC, disrupting mitochondrial calcium homeostasis, and miR-7 overexpression improves the release of mitochondrial calcium by targeting VDAC1 (Chaudhuri et al., 2016; Hong Z. et al., 2017; Zaglia et al., 2017).
Mitophagy
Mitophagy is a specific type of autophagy mode used to clear dysfunctional or redundant mitochondria and serves as an important regulatory mechanism that ensures the quantity and quality of mitochondria, which is necessary for the maintenance of intracellular homeostasis (Vigié and Camougrand, 2017). BCL2-interacting protein three like (BNIP3L, also called Nix), FUN14-domain containing 1 (FUNDC1), PTEN-induced putative kinase 1 (PINK1), and Parkin are key factors that promote the occurrence of mitophagy (Sun T. et al., 2018). MiR-137 negatively regulates mitophagy by targeting FUNDC1 and Nix (Li W. et al., 2014). MiR-27a/b represses mitophagy by downregulating the expression of PINK1 (Kim J. et al., 2016). Additionally, the miR-181a targeting of Parkin has also been found to downregulate mitophagy (Li W. et al., 2014; Cheng et al., 2016; Kim J. et al., 2016).
In summary, the miRNA-mediated regulation of mitochondria-related genes is important for the maintenance of normal mitochondrial function. The heart is the most active organ, and its high demand for energy is met by mitochondrial OXPHOS, which indirectly indicates the importance of miRNA-mediated mitochondrial functional regulation in cardiac disease.
MicroRNAs Regulate Mitochondrial Function in Cardiac Diseases
Mitochondria account for nearly one-third of the total volume of cardiomyocytes. The homeostasis of mitochondrial function is essential for the maintenance of the normal physiological function of the heart. Abnormal mitochondrial function and quality have been associated with a wide variety of pathological processes that underlie cardiac diseases (Nakamura and Sadoshima, 2018; Picca et al., 2018; Zhou and Tian, 2018). Recent studies have demonstrated that miRNAs regulate mitochondrial functions and participate in cardiac diseases through the direct or indirect targeting of mitochondria-related genes. Table 1 highlights the regulation of mitochondria-related genes by miRNAs in several models of cardiac disease.
TABLE 1.
MiRNAs involved in cardiac disease. Overview of the mitochondrial functions regulated by miRNAs in several different cardiac phenotypes (myocardial apoptosis, cardiac hypertrophy, diabetic cardiomyopathy, and heart failure).
| miRNA | Targets | Model conditions | References |
|---|---|---|---|
| Myocardial apoptosis | |||
| miR-23a | PGC-1α | Dox | Du et al. (2019) |
| miR-15b-5p | Bmpr1 | Wan et al. (2019) | |
| miR-140 | MFN 1 | Li J. et al. (2014) | |
| miR-29b | Bax | Jing et al. (2018) | |
| miR-140 | YES1 | HR/IR | Yang et al. (2019) |
| miR-138 | HIF1-α | Liu et al. (2019) | |
| miR-181c | MICU1 | Banavath et al. (2019) | |
| miR-30a | P53 | Forini et al. (2014); Zhang C. et al. (2019) | |
| miR-486 | P53 | Sun et al. (2017) | |
| miR-361 | PHB1 | Wang et al. (2015a) | |
| miR-539 | PHB2 | Wang et al. (2014) | |
| miR-421 | PINK1 | Wang et al. (2015c) | |
| miR-145 | BNIP3 | IR and AR | Li et al. (2012) |
| miR-324-5p | MTFR1 | Wang et al. (2015b) | |
| miR-484 | Fis 1 | Wang et al. (2012) | |
| miR-761 | MFF | Long et al. (2013) | |
| miR-499 | CnAα/CnBβ | Wang et al. (2011) | |
| miR-410 | HMGB1 | LAD and/or HR | Yang et al. (2018) |
| miR-762 | ND2 | Yan et al. (2019) | |
| miR-143 | C-epsilon | Hong H. et al. (2017) | |
| miR-203 | PTP1 | Zhang J. et al. (2019) | |
| miR-24 | CHOP | Wang and Qian, (2014) | |
| miR-145 | PDCD4 | Xu et al. (2017) | |
| miR-558 | ULK1 | ISO | Guo et al. (2019) |
| miR-208 | SOCS2 | Sepsis | Ouyang et al. (2020) |
| Cardiac hypertrophy | |||
| miR-106 | MFN 2 | Ang Ⅱ/ TAC | Guan et al. (2016) |
| miR-195-5p | Wang et al. (2019) | ||
| miR-20 | Sun et al. (2019) | ||
| miR-20b | Qiu et al. (2020) | ||
| miR-153-3p | MFN 1 | ISO/ PAH | Wang et al. (2020) |
| miR-140 | Joshi et al. (2016) | ||
| miR-29a-3p | Drp 1 | Neuropeptide Y | Xie et al. (2020) |
| miR-376b-3p | MFF | NA | Sun T. et al. (2018) |
| miR-485–5p | MAPL | PE | Zhao et al. (2017) |
| miR-146a | DLST | Ang Ⅱ/TAC | Heggermont et al. (2017) |
| miR-21 | Mt-cytb | SHR | Li H. et al. (2016) |
| miR-1 | MUC | TAC/Chronic exercise | Zaglia et al. (2017) |
| miR-142–3p | HS2B1 | Ang Ⅱ/ AB | Liu et al. (2018) |
| miR-28 | VDAC-1 | PE | Kar and Bandyopadhyay, (2018) |
| Diabetic cardiomyopathy | |||
| Let-7b-5 | Mt-cytb | STZ | Li et al. (2019) |
| miR-92a-2-5 | Mt-cytb | Li et al. (2019) | |
| miR-1 | Junctin | Yildirim et al. (2013) | |
| miR-141 | Slc25a3 | Baseler et al. (2012) | |
| miR-30c | PGC-1β | Palmitate and db/db mice | Yin et al. (2019) |
| miR-1 | Fxra | HG | Cheng et al. (2018) |
| miR-144 | Rac-1 | HG and STZ | Tao et al. (2020) |
| Heart failure | |||
| miR-195 | SIRT 3 | TAC/MI | Zhang et al. (2018) |
| miR-1a-3p | ND1, COXⅠ | ISO | He et al. (2019) |
| miR-29 | PGC-1α | HF patients | Caravia et al. (2018) |
| miR-665 | GLP1R | LCA | Lin et al. (2019) |
| miR-152 | FLRX5 | TAC | LaRocca et al. (2020) |
| miR-181c | COXⅠ | Plasmid miR-181c in vivo | Das et al. (2014) |
| miR-696 | — | TAC | Wang et al. (2017) |
| miR-532 | |||
| MiR-690 | |||
| miR-345–3p | |||
Cardiomyocyte Apoptosis
Apoptosis refers to programmed death that is regulated by various genes and serves to maintain intracellular homeostasis. Abnormal apoptosis in cardiomyocytes has previously been linked to mitochondrial dysfunction caused by the dysregulation of miRNAs (Makhdoumi et al., 2016). Here, we describe the specific regulation of mitochondrial function by miRNAs in several representative models of myocardial apoptosis.
The cardiomyocyte apoptosis model induced by doxorubicin (Dox) is an antitumor antibiotic that can inhibit many types of tumors; however, its clinical applications are limited due to significant heart toxicity (Shi et al., 2018). In both in vivo and in vitro Dox-induced cardiomyocyte apoptosis models, miR-23a mimicking was shown to promote mitochondrial fission, cause mitochondrial membrane potential loss, and exacerbate cardiomyocyte apoptosis by targeting the downregulation of PGC-1α and the activation of Drp1 (Du et al., 2019). The overexpression of miR-15b-5p can silence bone morphogenetic protein receptor 1 (Bmpr1) signaling, causing mitochondrial ROS (mtROS) aggregation and resulting in mitochondrial damage, accelerating apoptosis (Wan et al., 2019). MiR-140 inhibits MFN1, causing mitochondrial fission abnormalities and promoting Dox-induced cardiomyocyte apoptosis (Li J. et al., 2014). However, high expression levels of miR-29b can inhibit Bax, reverse the loss of mitochondrial membrane potential, prevent the opening of the mitochondrial permeability transition pore (MPTP) and cardiomyocyte apoptosis (Jing et al., 2018). These data suggest that the regulation of mitochondrial function by miRNAs can have opposing effects, and the in-depth study of their mechanisms of action might result in a therapeutic response to the cardiotoxicity associated with Dox.
Myocardial hypoxia and ischemia/reperfusion injury (HR/IR) models: HR and IR injury models can induce cardiomyocyte apoptosis and myocardial infarction, and miRNA-mediated mitochondrial regulation is involved in these processes. The upregulation of miR-140 and miR-138 improve mitochondrial morphology and reverse IR-induced myocardial apoptosis and myocardial infarction in mice by inhibiting the expression of YES1 and hypoxia-inducible factor 1α (HIF1α) and downregulating the mitochondrial fission proteins Drp1 and Fis1, respectively (Liu et al., 2019; Yang et al., 2019). In addition, the nuclear-encoded miR-181c can be transported to the mitochondria to form an AGO/mt-COX1 complex, which regulates mtROS. The activation of the transcription factor Sp1 can directly upregulate the transcription of MICU1 by binding to the MICU1 promoter region, clarifying the regulatory role of the miR-181c→mt-COX 1→ROS→Sp1→MICU1 signaling pathway in the regulation of mitochondrial calcium uptake in the model; however, the article did not identify the specific target of miR-181c (Banavath et al., 2019). Similarly, in both in vivo and in vitro models induced by anoxia reoxygenation (AR), HR, or IR, miR-30a, and miR-486 can target p53 transcription, restore the mitochondrial membrane potential and ATP production, improve mitochondrial damage, and inhibit myocardial apoptosis (Forini et al., 2014; Sun et al., 2017; Zhang C. et al., 2019). MiR-361,miR-539, and miR-421 inhibit prohibitin 1 (PHB1), PHB2, and PINK1, respectively, to intensify mitochondrial fragmentation and promote H2O2-induced cardiomyocyte apoptosis and myocardial infarction (Wang et al., 2014; Wang et al., 2015a; Wang et al., 2015c).
In both in vitro and in vivo IR and AR-induced models, miR-145 can improve the mitochondrial structure, reduce the production of mtROS, and reverse myocardial apoptosis by targeting BNIP3 (Li et al., 2012). The upregulation of miR-324-5p, miR-484, and miR-761 can improve mitochondrial over fission and inhibit myocardial apoptosis by inhibiting mitochondrial fission regulator 1 (MTFR1), Fis1, and MFF, respectively (Wang et al., 2012; Long et al., 2013; Wang et al., 2015b). The downregulation of miR-499 can inhibit the α and β isoforms of the calcineurin catalytic subunit (CnAα and CnAβ) to activate Drp1, accelerate mitochondrion fission, and promote the occurrence of apoptosis (Wang et al., 2011).
In the left anterior descending artery (LAD)-ligation or HR-mediated myocardial apoptosis models, miR-410 can cause abnormal mitophagy and promote apoptosis by inhibiting high-mobility group box one protein (HMGB1) (Yang et al., 2018). MiR-762 and miR-143 target NADH dehydrogenase 2 (ND2) and protein kinase C-epsilon (PKCε), respectively, resulting in the depletion of the mitochondrial membrane potential and decreased ATP activity, which disrupts mitochondrial function and accelerates myocardial apoptosis (Hong H. et al., 2017; Yan et al., 2019). However, miR-203 can inhibit protein tyrosine phosphatase 1 (PTP1) expression, promote ATP synthesis, restore the mitochondrial membrane potential, and improve myocardial apoptosis (Zhang J. et al., 2019). MiR-24 can inhibit the expression of C/EBP-homologous protein (CHOP) and improve myocardial apoptosis (Wang and Qian, 2014). The overexpression of miR-145 inhibited programmed cell death 4 (PDCD4) expression, improved mitochondrial function, and inhibited myocardial apoptosis in the above experimental models (Xu et al., 2017). In addition, in the apoptotic model induced by isoprenaline (ISO) treatment, miR-558 was able to promote apoptosis by targeting unc-51-like autophagy activating kinase 1 (ULK1), promoting the loss of mitochondrial membrane potential and mtROS overproduction (Guo et al., 2019). In a sepsis-induced mouse model of myocardial apoptosis, the downregulation of miR-208 improved mitochondrial swelling and cell apoptosis by targeting activation of the suppressor of cytokine signaling 2 (SOCS2) (Ouyang et al., 2020).
These data show that the regulation of mitochondrial function by miRNAs has been extensively studied in myocardial apoptosis, especially in models of IR-mediated myocardial apoptosis, which laid a solid foundation for the possibility of using miRNAs as biomarkers in myocardial IR injury-induced cardiomyopathy, although the functions and mechanisms of miRNAs in other induced myocardial apoptosis models require further study.
Cardiac Hypertrophy
Cardiac hypertrophy is a compensatory response that allows for the maintenance of cardiac function and efficiency under conditions of cardiac overload, which is manifested as the enlargement of the heart and the individual component cardiomyocytes. Physiological hypertrophy often occurs during pregnancy and in athletes, associated with enhanced cardiac contractile function, with no interstitial or alternative fibrosis or cell death, and is usually reversible and does not progress to heart failure. Pathological cardiac hypertrophy is mediated by persistent neuroendocrine hormones and stimulated by mechanical forces and is accompanied by cell death, fibrous remodeling, and mitochondrial dysfunction, which ultimately leads to heart failure (Nakamura and Sadoshima, 2018). Cumulative evidence has demonstrated that miRNA-targeted mitochondrial function and morphological regulation play indispensable roles in cardiac hypertrophy (Jusic et al., 2020).
Mitochondrial Fusion-Related MicroRNAs in Cardiac Hypertrophy
MFN1/2 and OPA1 are the main contributors to the process of mitochondrial fusion and are important for maintaining the balance of mitochondrial dynamics. MiR-106 can downregulate the expression of MFN2, exacerbating cardiac hypertrophy induced by transverse aortic constriction (TAC) or angiotensin II (Ang II) pressure overload, which is associated with mitochondrial dysfunction, including mitochondrial cristae defects, the considerable depolarization of the mitochondrial membrane, and mtROS production, suggesting that high miR-106 expression levels promote mitochondrial dysfunction in cardiac hypertrophy (Guan et al., 2016). The same regulatory pathway was also associated with miR-195-5p and miR-20. MiR-195-5p was upregulated in vitro and in vivo Ang Ⅱ-induced hypertrophic models, and the high expression of miR-195-5p inhibited MFN2 activity, leading to abnormal mitochondrial fusion. MiR-20 is upregulated in hypertrophic cells, causing mitochondrial dysfunction by repressing MFN2 expression, and a similar expression phenomenon has been detected in clinical hypertrophic cardiomyopathy patients (Sun et al., 2019; Wang et al., 2019). Recently, miR-20b was found to be upregulated in an Ang II-induced primary cardiomyocyte hypertrophy model and a TAC-induced hypertrophy model in KM mice, resulting in mitochondrial fragmentation and a sharp decrease in fusion ability. MFN2 plays an important role connecting the sarcoplasmic reticulum to mitochondrial calcium transport, which weakens the mitochondrial buffering capacity and leads to increased cytosolic Ca2+, decreased mitochondrial Ca2+, and disturbed calcium homeostasis. This phenomenon can be reversed by inhibiting the miR-20b-targeted upregulation of MFN2, showing that the hypertrophic effects of miR-20b can be achieved by the targeted downregulation of MFN2 (Qiu et al., 2020). Additionally, studies have demonstrated that both miR-153-3p and miR-140 are involved in ISO-induced cardiac hypertrophy and ventricular hypertrophy in patients with pulmonary arterial hypertension (PAH), downregulating the expression of MFN1 mRNA and leading to excessive mitochondrial fission (Joshi et al., 2016; Wang et al., 2020). However, the role played by miRNAs in the regulation of OPA1 in cardiac hypertrophy has not yet been reported and represents an unexplored field for researchers looking to study the regulatory effects of miRNAs on mitochondrial fusion.
Mitochondrial Fission-Related MicroRNAs in Cardiac Hypertrophy
Drp1/Dynamin I (Dnm1), MFF, and Fis1 are the key genes involved in the regulation of mitochondrial fission. In vitro, miR-29a-3p was shown to improve cardiac hypertrophy induced by neuropeptide Y by inhibiting the expression of Drp1 and reducing the expression of the hypertrophy marker gene atrial natriuretic peptide (Xie et al., 2020). In the norepinephrine (NA)-induced cardiac hypertrophy model, the overexpression of miR-376b-3p and the knockdown of MFF attenuated NA-induced mitochondrial fission and cardiac hypertrophy, which suggested that miR-376b-3p could improve mitochondrial fission and reverse cardiac hypertrophy by inhibiting the expression of MFF by binding to its 3’UTR. These results indicated that miR-376b-3p is a novel regulator that affects mitochondrial morphology (Sun Y. L et al., 2018). MiR-485-5p can improve mitochondrial fission and reverse cardiac hypertrophy by activating MFN2 and causes the downregulation of mitochondrial anchored protein ligase (MAPL) in phenylephrine (PE)-induced cardiac hypertrophy models (Zhao et al., 2017). However, whether miRNAs are involved in the growth underlying cardiac hypertrophy by targeting other mitochondrial mitotic proteins, such as Dnm1 and Fis1, requires further exploration.
In hypertrophic cardiac diseases, miRNAs can regulate mitochondrial function through other signaling pathways. For example, the overexpression of miR-146a and the knockout of dihydrolipoyl succinyltransferase (DLST) can both lead to the downregulation of tricarboxylic acid cycle efficiency, causing impaired energy production, similar to the mechanism identified in Ang II- and TAC-induced cardiac hypertrophy and in clinical patients, suggesting that miR-146a and its target DLST are important regulators of the metabolic pathways involved in cardiac hypertrophy (Heggermont et al., 2017). In a spontaneous hypertensive rat (SHR) model, the high expression of miR-21 can rescue mitochondrial dysfunction and cardiac hypertrophy induced by the gene silencing of mtDNA-encoded cytochrome b (mt-Cytb) (Li H. et al., 2016). MiR-1 has been identified as a muscle-specific miRNA, and MCU is a prerequisite for mitochondrial functional homeostasis, ATP synthesis, and cell metabolism. Recently, miR-1 was shown to effectively bind to the 3′UTR of MCU, affecting mitochondrial Ca2+ uptake and leading to cardiac hypertrophy. This finding suggests a new role for miR-1/MCU signaling in mitochondrial calcium regulation and cardiac hypertrophy (Zaglia et al., 2017). MiR-142-3p overexpression was shown to silence the energy metabolism regulatory factor Src homology 2 (HS2B1), and protect mitochondrial function by restoring oxygen consumption rate and membrane potential, thus reversing Ang Ⅱ- and abdominal aorta ligation (AB)- mediated cardiac hypertrophy (Liu et al., 2018). MiR-28 can target the mitochondrial gene VDAC1, activate PPARγ, improve mitochondrial function, and induce cardiac hypertrophy mediated by the α1-adrenergic agonist PE in vitro (Kar and Bandyopadhyay, 2018).
Although increasing evidence has suggested that miRNAs participate in the development of cardiac hypertrophy by regulating the expression of mitochondria-related genes, more attention has been focused on the pathways involved in mitochondrial fusion and fission. However, the contributions of mitophagy regulation by miRNAs in cardiac hypertrophy should not be ignored and should be further explored.
Diabetic Cardiomyopathy
Diabetic cardiomyopathy (DCM) is a specific cardiomyopathy, independent of hypertension, coronary heart disease, valvular heart disease, and other cardiac diseases, that occurs in diabetic patients and is associated with ventricular pathological remodeling. DCM is the primary mediator of the increased incidence of heart failure and mortality in diabetic patients (Yin et al., 2019). The regulation of mitochondrial function by miRNAs is actively involved in the development and progression of DCM (Hathaway et al., 2018). In db/db mice and palmitate-induced DCM, the downregulation of miR-30c can inhibit the mitochondrial membrane potential of PPARα by activating PGC-1 β, inducing mitochondrial biogenesis and intensifying the occurrence of DCM (Yin et al., 2019). In the past, let-7 downregulation was shown to improve STZ-induced DCM by regulating the Akt/mTOR signal, but its key target has not been studied in-depth (Li J. et al., 2016).
Recently, both let-7b-5 and miR-92a-2-5p were found to inhibit mtROS overproduction, activate the mitochondrial respiratory chain, prevent lipid deposition, and ameliorate DCM by regulating mt-cytb (Li et al., 2019). In an STZ-induced DCM model, the expression of miR-1 was downregulated, and Junctin expression was upregulated, which induced mitochondrial swelling and the loss of the mitochondrial cistern and granule matrix. Junctin is a downstream target of miR-1, but the specific mechanism of Junctin regulation in DCM remains unclear (Yildirim et al., 2013). However, in a high-glucose (HG)-induced DCM model, the overexpression of miR-1 inhibited Fxra, resulting in the loss of the mitochondrial membrane potential and aggravating DCM damage (Cheng et al., 2018). Therefore, the elucidation of the specific role played by miR-1 and its mechanism of action in DCM is important for the prevention and treatment of DCM. MiR-141 was upregulated in the hearts of STZ-induced FVB mice, which inhibited the translation of mitochondrion phosphate carrier (Slc25a3) without affecting the transcription level, resulting in decreased ATP synthetase activity (Baseler et al., 2012). Recently, the overexpression of miR-144 was found to specifically inhibit Rac-1, regulating the AMPK-PGC-1 α signal, enhancing mitochondrial biogenesis, upregulating the copy number of mtDNA, and restoring the mitochondrial membrane potential, which improved the DCM induced by HG and STZ both in vitro and in vivo (Tao et al., 2020).
MiRNAs are involved in DCM by affecting mitochondrial function through a wide range of signaling pathways. However, little research has been performed to examine the direct targeting of mitochondrial functional proteins, especially those involved in mitochondrial dynamics and mitophagy.
Heart Failure
The pathological factors that contribute to heart failure are very complex and are often the cumulative effect of many diseases, including coronary heart disease, hypertension, and ischemic heart disease. Mitochondrial function is an important target for the treatment of heart failure, and the regulation of mitochondrial function by miRNAs contributes to the occurrence and development of heart failure (Pinti et al., 2017). MiR-696, miR-532, miR-690, and miR-345-3p are highly expressed in the mitochondria of C57BL/6 mice with TAC-induced heart failure. Enrichment analysis of these four miRNAs pathways by TarBase showed that these pathways were closely related to mitochondrial genesis, fission, and energy metabolism, which further revealed that the effects of miRNAs on mitochondrial function were closely related to heart failure (Wang et al., 2017). The mitochondrial deacetylase Sirtuin 3 (SIRT3) plays a key role in the maintenance of mitochondrial function by regulating mitochondrial acetylation. In patients with heart failure and TAC/MI- and Ang II-induced heart failure models, the high expression of miR-195 can downregulate SIRT3 activity, aggravating the excessive acetylation of pyruvate dehydrogenase complex (PDC) and ATP synthetase in patients—resulting in disordered mitochondrial energy metabolism (Zhang et al., 2018). Treatment with an miR-1a-3p agomir can improve the symptoms of heart failure in ISO-induced HF mice by increasing the expression of mitochondrial NADH dehydrogenase 1 (ND1) and mitochondrial cytochrome c oxidase I (COX Ⅰ) (He et al., 2019). In addition, in miR-29-deficient mice, cardiac dysfunction, abnormal cardiac metabolism, and the activation of PGC-1 α led to mitochondrial biological disorders. The expression of miR-29 can inhibit PGC-1 α, control mitochondrial biogenesis, and improve heart failure (Caravia et al., 2018). In a heart failure model induced by left coronary artery ligation (LCA), the high expression of miR-665 can reduce mitochondrial ATP enzymatic activity and damage cardiac function, whereas the inhibition of miR-655 can upregulate glucagon-like peptide 1 receptor (GLP1R) and cyclic adenosine monophosphate (cAMP) signaling to stabilize heart failure (Lin et al., 2019). In a TAC-induced heart failure model in C57BL/6 mice, the high expression of miR-152 resulted in the destruction of the mitochondrial ultrastructure and induced cardiac metabolic disorder, which was mediated by the targeted inhibition of FLRX5 (a key factor involved in the regulation of mitochondrial homeostasis). By contrast, the silencing of miR-152 could reverse the above pathological process and improve the heart failure caused by stress overload (LaRocca et al., 2020). In addition, nuclear-encoded miR-181c could be shifted toward mitochondrial targeting to inhibit the expression of the mitochondrial gene COX1, resulting in abnormal mitochondrial metabolism, the loss of OXPHOS, and heart failure (Das et al., 2014). These data suggested that miRNAs primarily contribute to heart failure by regulating mitochondrial metabolic pathways. Therefore, contributions of miRNAs on other mitochondrial functions associated with heart failure require additional study.
Challenges and Development
Since the first miRNA was identified in 1993, research on miRNAs has received a great deal of attention, and miRNAs are the best-studied noncoding RNAs, especially relative to cardiovascular disease (Colpaert and Calore, 2019). One of the most characteristic features of the mitochondria is that it has its own genome, and mtDNA can participate in the synthesis, transcription, and replication of mitochondrial proteins (Gustafsson et al., 2016). Consequently, the discovery of mitochondrial genome-encoded miRNAs, which play key roles in cardiomyopathy, continues (Song et al., 2019). However, some challenges remain to be overcome, which provides avenues for future research:
1) The biosynthetic pathway of miRNAs is very complex, involving interactions among multiple enzymes and complex components. The roles played by each component in the miRNA synthesis pathway remain controversial, and how the passenger strand is degraded remains unclear (Jaé and Dimmeler, 2020). In addition to canonical synthetic pathways, non-canonical synthetic pathways are gradually being uncovered. Although these synthesis pathways bypass one or more steps of the canonical pathway, mature miRNAs synthesized by these pathways function similarly to those synthesized by canonical pathways (Ruby et al., 2007). Therefore, exploring and discovering the biosynthetic pathways used to generate additional miRNAs and elucidating the mechanisms of these miRNAs in cardiac disease remains a difficult and urgent problem to be solved.
2) The regulation of mitochondrial function by miRNAs has received increasing interest, and many mitochondrial-encoded miRNAs and nuclear-encoded miRNAs have been shown to translocate to the mitochondria to perform their biological functions (Fan et al., 2019). However, the extraction of uncontaminated mitochondria remains a challenge; therefore, the biosynthetic pathway and mode of action associated with mitochondrial miRNAs remain unclear. In addition, how nuclear-encoded miRNAs translocate to the mitochondria to perform their functions and the mediators of this process have yet to be determined. Thus, whether the mitochondrial genome can encode miRNAs and how the crosstalk between nuclear-encoded miRNAs and the mitochondria affects the development of cardiac disease remains to be further studied.
3) Generally, a single miRNA can target multiple genes, and multiple miRNAs can regulate the same gene, exerting different biological functions (Selbach et al., 2008). Both miR-106a and miR-20b can induce mitochondrial dysfunction and promote cardiac hypertrophy by inhibiting the expression of MFN2 (Guan et al., 2016; Qiu et al., 2020). MiR-140 improves I/R-induced myocardial apoptosis by inhibiting the expression of Drp1 and Fis1 through the targeting of YES1 (Li J. et al., 2014). However, in the Dox-induced cardiomyocyte apoptosis model, miR-140 was found to mediate cardiomyocyte apoptosis by inhibiting the expression of MFN1 (Li J. et al., 2014). Due to the large number of gene pools associated with mitochondrial function and the complex pathogenic factors that underlie cardiac diseases, research has both increased our understanding of the specific mechanisms and targets through which miRNAs regulated mitochondrial activity in cardiac diseases and introduced new challenges—which can be used to guide our future experimental studies.
4) MiRNA is also widely involved in other cardiac diseases, such as coronary artery disease (Hinkel et al., 2014), myocardial fibrosis (Yuan et al., 2017), and congenital heart disease (Smith et al., 2015). However, there is a lack of research on miRNA involved in these diseases by regulating mitochondrial function. Therefore, it is an urgent need to actively explore and study the specific regulatory mechanism of miRNA regulating mitochondrial function in more cardiac diseases.
Conclusion
Cardiovascular diseases are among the leading causes of death, and mitochondria represent promising potential therapeutic targets for cardiovascular diseases. The emergence of miRNAs has been a major breakthrough in the study of cardiac disease. In recent years, the miRNA-mediated regulation of the mitochondrial network has received much attention and has been verified in cardiac diseases. This review summarizes the biogenesis of miRNAs, their regulatory roles in mitochondrial functions, and the current research regarding miRNAs in cardiac disease, indicating that the regulation of mitochondrial function by miRNAs plays an important role in cardiomyocyte apoptosis, cardiac hypertrophy, diabetic cardiomyopathy, and cardiac failure. Based on the multiple targets downstream of miRNAs and the complexity of these diseases, a great deal of controversy remains regarding the miRNA-mediated regulation of mitochondria. Therefore, the continued vigorous exploration of miRNAs that regulate mitochondrial function remains necessary to elucidate downstream targets, especially those involved in mitophagy and mitochondrial dynamics. This research will improve our understanding of the biological functions of miRNAs and clarify the crosstalk that occurs between miRNAs and mitochondria. Understanding these networks will provide a theoretical basis for discovering new targets for drugs designed to prevent and treat cardiac disease and provide a new research avenue for clinical cardiac disease.
Acknowledgments
We thank SC and other members from SC laboratory for helpful discussions and for providing professional opinions.
Author Contributions
ZQ: consulting, collecting supporting materials, and writing the manuscript. WQ: collecting materials and architecture of figures. CY: providing professional opinions on writing reviews. FY: conducting beneficial discussions on the manuscript. XN: providing valuable opinions and suggestions on figures. LL: giving opinions and modifying forms on the table. TL: reviewing manuscripts and modifying word order. SC: providing ideas, critically reviewing the manuscript, and providing financial support. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 82060729 and U1812403-4-4), the Fund of High-Level Innovation Talents (No. 2015-4029), and the base of International Scientific and Technological Cooperation of Guizhou Province (No. (2017) 5802).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abdelfattah A. M., Park C., Choi M. Y. (2014). Update on Non-canonical MicroRNAs. Biomol. Concepts. 5 (4), 275–287. 10.1515/bmc-2014-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M. M., Micolucci L., Islam M. S., Olivieri F., Procopio A. D. (2016). Bioinformatic Tools for MicroRNA Dissection. Nucleic Acids Res. 44 (1), 24–44. 10.1093/nar/gkv1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammirati E., Veronese G., Bottiroli M., Wang D. W., Cipriani M., Garascia A., et al. (2020). Update on Acute Myocarditis. Trends. Cardiovasc. Med. S1050-1738 (20), 30079–7. 10.1016/j.tcm.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz J. E., Ruby J. G., Wang Y., Bartel D. P., Blelloch R. (2008). Mouse ES Cells Express Endogenous ShRNAs, SiRNAs, and Other Microprocessor-Independent, Dicer-Dependent Small RNAs. Genes Dev. 22 (20), 2773–2785. 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banavath H. N., Roman B., Mackowski N., Biswas D., Afzal J., Nomura Y., et al. (2019). MiR-181c Activates Mitochondrial Calcium Uptake by Regulating MICU1 in the Heart. J. Am. Heart Assoc. 8 (24), e012919. 10.1161/jaha.119.012919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2018). Metazoan MicroRNAs. Cell 173 (1), 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseler W. A., Thapa D., Jagannathan R., Dabkowski E. R., Croston T. L., Hollander J. M., et al. (2012). MiR-141 as a Regulator of the Mitochondrial Phosphate Carrier (Slc25a3) in the Type 1 Diabetic Heart. Am. J. Physiology-Cell Physiol. 303 (12), C1244–C1251. 10.1152/ajpcell.00137.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield M. A., Maraia R. J. (2009). Precursor-Product Discrimination by La Protein During tRNA Metabolism. Nat. Struct. Mol. Biol. 16 (4), 430–437. 10.1038/nsmb.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewett N. H., Maraia R. J. (2018). La Involvement in tRNA and Other RNA Processing Events Including Differences Among Yeast and Other Eukaryotes. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1861 (4), 361–372. 10.1016/j.bbagrm.2018.01.013 [DOI] [PubMed] [Google Scholar]
- Borralho P. M., Rodrigues C. M. P., Steer C. J. (2015). MicroRNAs in Mitochondria: An Unexplored Niche. Adv. Exp. Med. Biol. 887, 31–51. 10.1007/978-3-319-22380-3_3 [DOI] [PubMed] [Google Scholar]
- Bose A., Beal M. F. (2016). Mitochondrial Dysfunction in Parkinson's Disease. J. Neurochem. 139, 216–231. 10.1111/jnc.13731 [DOI] [PubMed] [Google Scholar]
- Brown D. A., Perry J. B., Allen M. E., Sabbah H. N., Stauffer B. L., Shaikh S. R., et al. (2017). Mitochondrial Function as a Therapeutic Target in Heart Failure. Nat. Rev. Cardiol. 14 (4), 238–250. 10.1038/nrcardio.2016.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C. H., Cullen B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10 (12), 1957–1966. 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Cowan D. B., Wang D. Z. (2020). tRNA-Derived Small RNAs and Their Potential Roles in Cardiac Hypertrophy. Front. Pharmacol. 11, 572941. 10.3389/fphar.2020.5729410.3389/fphar.2020.572941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravia X. M., Fanjul V., Oliver E., Roiz-Valle D., Morán-Álvarez A., Desdín-Micó G., et al. (2018). The microRNA-29/PGC1α Regulatory axis Is Critical for Metabolic Control of Cardiac Function. Plos Biol. 16 (10), e2006247. 10.1371/journal.pbio.2006247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A. D., Choi D. C., Kabaria S., Tran A., Junn E. (2016). MicroRNA-7 Regulates the Function of Mitochondrial Permeability Transition Pore by Targeting VDAC1 Expression. J. Biol. Chem. 291 (12), 6483–6493. 10.1074/jbc.M115.691352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Dos Santos C. O., Chong M. M. W., Hannon G. J. (2010). A Dicer-Independent miRNA Biogenesis Pathway that Requires Ago Catalysis. Nature 465 (7298), 584–589. 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Guan Y., Ma Y., Quan D., Zhang J., Wu S., et al. (2019). Danshenol A Alleviates Hypertension-Induced Cardiac Remodeling by Ameliorating Mitochondrial Dysfunction and Suppressing Reactive Oxygen Species Production. Oxidative Med. Cell Longevity 2019, 1–18. 10.1155/2019/2580409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Liu L., Lao Y., Liao W., Liao M., Luo X., et al. (2016). MicroRNA-181a Suppresses Parkin-Mediated Mitophagy and Sensitizes Neuroblastoma Cells to Mitochondrial Uncoupler-Induced Apoptosis. Oncotarget 7 (27), 42274–42287. 10.18632/oncotarget.9786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Zhao W., Zhang X., Sun L., Yang H., Wang Y., et al. (2018). Downregulation of microRNA-1 A-ttenuates G-lucose-induced A-poptosis by R-egulating the L-iver X R-eceptor α in C-ardiomyocytes. Exp. Ther. Med. 16 (3), 1814–1824. 10.3892/etm.2018.6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov D. A., Shkurat T. P., Melnichenko A. A., Grechko A. V., Orekhov A. N. (2018). The Role of Mitochondrial Dysfunction in Cardiovascular Disease: A Brief Review. Ann. Med. 50 (2), 121–127. 10.1080/07853890.2017.1417631 [DOI] [PubMed] [Google Scholar]
- Chowdhury S. R., Reimer A., Sharan M., Kozjak-Pavlovic V., Eulalio A., Prusty B. K., et al. (2017). Chlamydia Preserves the Mitochondrial Network Necessary for Replication via MicroRNA-Dependent Inhibition of Fission. J. Cel. Biol. 216 (4), 1071–1089. 10.1083/jcb.201608063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D., Xue H., Taylor D. W., Patnode H., Mishima Y., Cheloufi S., et al. (2010). A Novel MiRNA Processing Pathway Independent of Dicer Requires Argonaute2 Catalytic Activity. Science 328 (5986), 1694–1698. 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpaert R. M. W., Calore M. (2019). MicroRNAs in Cardiac Diseases. Cells 8 (7), 737. 10.3390/cells8070737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Bedja D., Campbell N., Dunkerly B., Chenna V., Maitra A., et al. (2014). MiR-181c Regulates the Mitochondrial Genome, Bioenergetics, and Propensity for Heart Failure In Vivo . PLoS One 9 (5), e96820. 10.1371/journal.pone.0096820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A. M., Tops B. B. J., Plasterk R. H. A., Ketting R. F., Hannon G. J. (2004). Processing of Primary MicroRNAs by the Microprocessor Complex. Nature 432 (7014), 231–235. 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- Du J., Hang P., Pan Y., Feng B., Zheng Y., Chen T., et al. (2019). Inhibition of miR-23a Attenuates Doxorubicin-Induced Mitochondria-dependent Cardiomyocyte Apoptosis by Targeting the PGC-1α/Drp1 Pathway. Toxicol. Appl. Pharmacol. 369, 73–81. 10.1016/j.taap.2019.02.016 [DOI] [PubMed] [Google Scholar]
- Fan S., Tian T., Chen W., Lv X., Lei X., Zhang H., et al. (2019). Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res. 79 (6), 1069–1084. 10.1158/0008-5472.CAN-18-2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Gao L., Zhang D., Tian X., Kong L., Shi H., et al. (2019). MiR-93 Regulates Vascular Smooth Muscle Cell Proliferation, and Neointimal Formation Through Targeting Mfn2. Int. J. Biol. Sci. 15 (12), 2615–2626. 10.7150/ijbs.36995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forini F., Kusmic C., Nicolini G., Mariani L., Zucchi R., Matteucci M., et al. (2014). Triiodothyronine Prevents Cardiac Ischemia/Reperfusion Mitochondrial Impairment and Cell Loss by Regulating MiR30a/p53 Axis. Endocrinology 155 (11), 4581–4590. 10.1210/en.2014-1106 [DOI] [PubMed] [Google Scholar]
- Gollmer J., Zirlik A., Bugger H. (2020). Mitochondrial Mechanisms in Diabetic Cardiomyopathy. Diabetes Metab. J. 44 (1), 33–53. 10.4093/dmj.2019.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Wang L., Liu Z., Guo X., Jiang Y., Lu Y., et al. (2016). MiR-106a Promotes Cardiac Hypertrophy by Targeting Mitofusin 2. J. Mol. Cell Cardiol. 99, 207–217. 10.1016/j.yjmcc.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Guo X., Wu X., Han Y., Tian E., Cheng J. (2019). LncRNA MALAT1 Protects Cardiomyocytes from Isoproterenol‐induced Apoptosis through Sponging miR‐558 to Enhance ULK1‐mediated Protective Autophagy. J. Cel Physiol. 234 (7), 10842–10854. 10.1002/jcp.27925 [DOI] [PubMed] [Google Scholar]
- Gustafsson C. M., Falkenberg M., Larsson N.-G. (2016). Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160. 10.1146/annurev-biochem-060815-014402 [DOI] [PubMed] [Google Scholar]
- Ha M., Kim V. N. (2014). Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cel Biol 15 (8), 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Hammond S. M. (2015). An Overview of MicroRNAs. Adv. Drug Deliv. Rev. 87, 3–14. 10.1016/j.addr.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.-H., Nam J.-W., Heo I., Rhee J.-K., et al. (2006). Molecular Basis for the Recognition of Primary MicroRNAs by the Drosha-DGCR8 Complex. Cell 125 (5), 887–901. 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- Hathaway Q. A., Pinti M. V., Durr A. J., Waris S., Shepherd D. L., Hollander J. M. (2018). Regulating MicroRNA Expression: at the Heart of Diabetes Mellitus and the Mitochondrion. Am. J. Physiology-Heart Circulatory Physiol. 314 (2), H293–H310. 10.1152/ajpheart.00520.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens M. A., Reich A. A., Duelli D. M., Hastings M. L. (2012). Biogenesis of Mammalian MicroRNAs by a Non-canonical Processing Pathway. Nucleic Acids Res. 40 (10), 4626–4640. 10.1093/nar/gks026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Ding C., Yin P., He L., Xu Q., Wu Z., et al. (2019). MiR-1a-3p Mitigates Isoproterenol-Induced Heart Failure by Enhancing the Expression of Mitochondrial ND1 and COX1. Exp. Cel Res. 378 (1), 87–97. 10.1016/j.yexcr.2019.03.012 [DOI] [PubMed] [Google Scholar]
- Heggermont W. A., Papageorgiou A.-P., Quaegebeur A., Deckx S., Carai P., Verhesen W., et al. (2017). Inhibition of MicroRNA-146a and Overexpression of Its Target Dihydrolipoyl Succinyltransferase Protect Against Pressure Overload-Induced Cardiac Hypertrophy and Dysfunction. Circulation 136 (8), 747–761. 10.1161/CIRCULATIONAHA.116.024171 [DOI] [PubMed] [Google Scholar]
- Hinkel R., Ng J. K. M., Kupatt C. (2014). Targeting MicroRNAs for Cardiovascular Therapeutics in Coronary Artery Disease. Curr. Opin. Cardiol. 29 (6), 586–594. 10.1097/HCO.0000000000000107 [DOI] [PubMed] [Google Scholar]
- Hong H., Tao T., Chen S., Liang C., Qiu Y., Zhou Y., et al. (2017). MicroRNA-143 Promotes Cardiac Ischemia-Mediated Mitochondrial Impairment by the Inhibition of Protein Kinase Cepsilon. Basic Res. Cardiol. 112 (6), 60. 10.1007/s00395-017-0649-7 [DOI] [PubMed] [Google Scholar]
- Hong Z., Chen K.-H., DasGupta A., Potus F., Dunham-Snary K., Bonnet S., et al. (2017). MicroRNA-138 and MicroRNA-25 Down-Regulate Mitochondrial Calcium Uniporter, Causing the Pulmonary Arterial Hypertension Cancer Phenotype. Am. J. Respir. Crit. Care Med. 195 (4), 515–529. 10.1164/rccm.201604-0814oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan J., Pasquinelli A. E., Bálint E., Tuschl T., Zamore P. D. (2001). A Cellular Function for the RNA-Interference Enzyme Dicer in the Maturation of the Let-7 Small Temporal RNA. Science 293 (5531), 834–838. 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- Jaé N., Dimmeler S. (2020). Noncoding RNAs in Vascular Diseases. Circ. Res. 126 (9), 1127–1145. 10.1161/CIRCRESAHA.119.315938 [DOI] [PubMed] [Google Scholar]
- Jaquenod De Giusti C., Roman B., Das S. (2018). The Influence of MicroRNAs on Mitochondrial Calcium. Front. Physiol. 9, 1291. 10.3389/fphys.2018.01291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Teague A. M., Tryggestad J. B., Chernausek S. D. (2017). Role of microRNA-130b in Placental PGC-1α/TFAM Mitochondrial Biogenesis Pathway. Biochem. Biophysical Res. Commun. 487 (3), 607–612. 10.1016/j.bbrc.2017.04.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X., Yang J., Jiang L., Chen J., Wang H. (2018). MicroRNA-29b Regulates the Mitochondria-Dependent Apoptotic Pathway by Targeting Bax in Doxorubicin Cardiotoxicity. Cell Physiol. Biochem. 48 (2), 692–704. 10.1159/000491896 [DOI] [PubMed] [Google Scholar]
- Joshi S. R., Dhagia V., Gairhe S., Edwards J. G., McMurtry I. F., Gupte S. A. (2016). MicroRNA-140 Is Elevated and Mitofusin-1 Is Downregulated in the Right Ventricle of the Sugen5416/Hypoxia/Normoxia Model of Pulmonary Arterial Hypertension. Am. J. Physiol.-Heart Circ. Physiol. 311 (3), H689–H698. 10.1152/ajpheart.00264.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusic A., Devaux Y. EU-CardioRNA COST Action (CA17129) (2020). Mitochondrial Noncoding RNA-Regulatory Network in Cardiovascular Disease. Basic Res. Cardiol. 115 (3), 23. 10.1007/s00395-020-0783-5 [DOI] [PubMed] [Google Scholar]
- Kar D., Bandyopadhyay A. (2018). Targeting Peroxisome Proliferator Activated Receptor α (PPAR α) for the Prevention of Mitochondrial Impairment and Hypertrophy in Cardiomyocytes. Cel Physiol. Biochem. 49 (1), 245–259. 10.1159/000492875 [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J., Plasterk R. H. (2001). Dicer Functions in RNA Interference and in Synthesis of Small RNA Involved in Developmental Timing in C. elegans . Genes Dev. 15 (20), 2654–2659. 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Fiesel F. C., Belmonte K. C., Hudec R., Wang W.-X., Kim C., et al. (2016). MiR-27a and MiR-27b Regulate Autophagic Clearance of Damaged Mitochondria by Targeting PTEN-Induced Putative Kinase 1 (PINK1). Mol. Neurodegeneration 11 (1), 55. 10.1186/s13024-016-0121-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-K., Kim B., Kim V. N. (2016). Re-evaluation of the Roles of DROSHA, Exportin 5, and DICER in microRNA Biogenesis. Proc. Natl. Acad. Sci. USA 113 (13), E1881–E1889. 10.1073/pnas.1602532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. A., Kelly D. P., Chirinos J. A. (2019). Mitochondrial Dysfunction in Heart Failure with Preserved Ejection Fraction. Circulation 139 (11), 1435–1450. 10.1161/CIRCULATIONAHA.118.036259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca T. J., Seeger T., Prado M., Perea-Gil I., Neofytou E., Mecham B. H., et al. (2020). Pharmacological Silencing of MicroRNA-152 Prevents Pressure Overload-Induced Heart Failure. Circ. Heart Fail. 13 (3), e006298. 10.1161/CIRCHEARTFAILURE.119.006298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75 (5), 843–854. 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., et al. (2003). The Nuclear RNase III Drosha Initiates MicroRNA Processing. Nature 425 (6956), 415–419. 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- Lee H., Tak H., Park S. J., Jo Y. K., Cho D. H., Lee E. K. (2017). MicroRNA-200a-3p Enhances Mitochondrial Elongation by Targeting Mitochondrial Fission Factor. BMB Rep. 50 (4), 214–219. 10.5483/bmbrep.2017.50.4.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemecha M., Morino K., Imamura T., Iwasaki H., Ohashi N., Ida S., et al. (2018). MiR-494-3p Regulates Mitochondrial Biogenesis and Thermogenesis through PGC1-α Signalling in Beige Adipocytes. Sci. Rep. 8 (1), 15096. 10.1038/s41598-018-33438-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yan G., Li Q., Sun H., Hu Y., Sun J., et al. (2012). MicroRNA-145 Protects Cardiomyocytes against Hydrogen Peroxide (H2O2)-Induced Apoptosis through Targeting the Mitochondria Apoptotic Pathway. Plos. One. 7 (9), e44907. 10.1371/journal.pone.0044907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Dai B., Fan J., Chen C., Nie X., Yin Z., et al. (2019). The Different Roles of miRNA-92a-2-5p and Let-7b-5p in Mitochondrial Translation in Db/db Mice. Mol. Ther. - Nucleic Acids 17, 424–435. 10.1016/j.omtn.2019.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Y., Jiao J., Wang J., Li Y., Qin D., et al. (2014). Mitofusin 1 Is Negatively Regulated by MicroRNA 140 in Cardiomyocyte Apoptosis. Mol. Cell Biol. 34 (10), 1788–1799. 10.1128/MCB.00774-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang X., Zhuang H., Chen H.-g., Chen Y., Tian W., et al. (2014). MicroRNA-137 Is a Novel Hypoxia-Responsive MicroRNA that Inhibits Mitophagy via Regulation of Two Mitophagy Receptors FUNDC1 and NIX. J. Biol. Chem. 289 (15), 10691–10701. 10.1074/jbc.M113.537050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang F. S., Wu Z. Y., Lin J. L., Lan W. B., Lin J. H. (2014). MicroRNA-19b Targets Mfn1 to Inhibit Mfn1-Induced Apoptosis in Osteosarcoma Cells. neo 61 (3), 265–273. 10.4149/neo_2014_034 [DOI] [PubMed] [Google Scholar]
- Li H., Zhang X., Wang F., Zhou L., Yin Z., Fan J., et al. (2016). MicroRNA-21 Lowers Blood Pressure in Spontaneous Hypertensive Rats by Upregulating Mitochondrial Translation. Circulation 134 (10), 734–751. 10.1161/CIRCULATIONAHA.116.023926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ren Y., Shi E., Tan Z., Xiong J., Yan L., et al. (2016). Inhibition of the Let-7 Family MicroRNAs Induces Cardioprotection Against Ischemia-Reperfusion Injury in Diabetic Rats. Ann. Thorac. Surg. 102 (3), 829–835. 10.1016/j.athoracsur.2016.02.016 [DOI] [PubMed] [Google Scholar]
- Lin B., Feng D. G., Xu J. (2019). microRNA‐665 Silencing Improves Cardiac Function in Rats with Heart Failure through Activation of the cAMP Signaling Pathway. J. Cel. Physiol. 234 (8), 13169–13181. 10.1002/jcp.27987 [DOI] [PubMed] [Google Scholar]
- Liu B.-l., Cheng M., Hu S., Wang S., Wang L., Tu X., et al. (2018). Overexpression of MiR-142-3p Improves Mitochondrial Function in Cardiac Hypertrophy. Biomed. Pharmacother. 108, 1347–1356. 10.1016/j.biopha.2018.09.146 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zou J., Liu X., Zhang Q. (2019). MicroRNA-138 A-ttenuates M-yocardial I-schemia R-eperfusion I-njury through I-nhibiting M-itochondria-mediated A-poptosis by T-argeting HIF1-α. Exp. Ther. Med. 18 (5), 3325–3332. 10.3892/etm.2019.7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B., Wang K., Li N., Murtaza I., Xiao J.-Y., Fan Y.-Y., et al. (2013). MiR-761 Regulates the Mitochondrial Network by Targeting Mitochondrial Fission Factor. Free Radic. Biol. Med. 65, 371–379. 10.1016/j.freeradbiomed.2013.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Güttinger S., Calado A., Dahlberg J. E., Kutay U. (2004). Nuclear Export of MicroRNA Precursors. Science 303 (5654), 95–98. 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- Ma C., Zhang C., Ma M., Zhang L., Zhang L., Zhang F., et al. (2017). MiR-125a Regulates Mitochondrial Homeostasis Through Targeting Mitofusin 1 to Control Hypoxic Pulmonary Vascular Remodeling. J. Mol. Med. 95 (9), 977–993. 10.1007/s00109-017-1541-5 [DOI] [PubMed] [Google Scholar]
- Makhdoumi P., Roohbakhsh A., Karimi G. (2016). MicroRNAs Regulate Mitochondrial Apoptotic Pathway in Myocardial Ischemia-Reperfusion-Injury. Biomed. Pharmacother. 84, 1635–1644. 10.1016/j.biopha.2016.10.073 [DOI] [PubMed] [Google Scholar]
- Mao G.-X., Xu X.-G., Wang S.-Y., Li H.-F., Zhang J., Zhang Z.-S., et al. (2019). Salidroside Delays Cellular Senescence by Stimulating Mitochondrial Biogenesis Partly through a miR-22/SIRT-1 Pathway. Oxidative Med. Cell Longevity 2019, 1–13. 10.1155/2019/5276096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska A., Urlaub H., Lührmann R., Tuschl T. (2002). Single-Stranded Antisense siRNAs Guide Target RNA Cleavage in RNAi. Cell 110 (5), 563–574. 10.1016/s0092-8674(02)00908-x [DOI] [PubMed] [Google Scholar]
- Mohamed J. S., Hajira A., Pardo P. S., Boriek A. M. (2014). MicroRNA-149 Inhibits PARP-2 and Promotes Mitochondrial Biogenesis via SIRT-1/PGC-1α Network in Skeletal Muscle. Diabetes 63 (5), 1546–1559. 10.2337/db13-1364 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Sadoshima J. (2018). Mechanisms of Physiological and Pathological Cardiac Hypertrophy. Nat. Rev. Cardiol. 15 (7), 387–407. 10.1038/s41569-018-0007-y [DOI] [PubMed] [Google Scholar]
- Neth P., Nazari-Jahantigh M., Schober A., Weber C. (2013). MicroRNAs in Flow-Dependent Vascular Remodelling. Cardiovasc. Res. 99 (2), 294–303. 10.1093/cvr/cvt096 [DOI] [PubMed] [Google Scholar]
- Nie Y., Sato Y., Wang C., Yue F., Kuang S., Gavin T. P. (2016). Impaired Exercise Tolerance, Mitochondrial Biogenesis, and Muscle Fiber Maintenance in miR‐133a-Deficient Mice. FASEB j. 30 (11), 3745–3758. 10.1096/fj.201600529R [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J., Hayder H., Zayed Y., Peng C. (2018). Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 9, 402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005). c-Myc-Regulated MicroRNAs Modulate E2F1 Expression. Nature 435 (7043), 839–843. 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- Oberbauer V., Schaefer M. (2018). tRNA-Derived Small RNAs: Biogenesis, Modification, Function and Potential Impact on Human Disease Development. Genes 9 (12), 607. 10.3390/genes9120607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J. W., Duan H., Tyler D. M., Lai E. C. (2007). The Mirtron Pathway Generates MicroRNA-Class Regulatory RNAs in Drosophila. Cell 130 (1), 89–100. 10.1016/j.cell.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H., Tan Y., Li Q., Xia F., Xiao X., Zheng S., et al. (2020). MicroRNA-208-5p Regulates Myocardial Injury of Sepsis Mice via Targeting SOCS2-Mediated NF-Κb/hif-1α Pathway. Int. Immunopharmacol. 81, 106204. 10.1016/j.intimp.2020.106204 [DOI] [PubMed] [Google Scholar]
- Paolillo S., Marsico F., Prastaro M., Renga F., Esposito L., De Martino F., et al. (2019). Diabetic Cardiomyopathy. Heart Fail. Clin. 15 (3), 341–347. 10.1016/j.hfc.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Picca A., Mankowski R. T., Burman J. L., Donisi L., Kim J.-S., Marzetti E., et al. (2018). Mitochondrial Quality Control Mechanisms as Molecular Targets in Cardiac Ageing. Nat. Rev. Cardiol. 15 (9), 543–554. 10.1038/s41569-018-0059-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinti M. V., Hathaway Q. A., Hollander J. M. (2017). Role of MicroRNA in Metabolic Shift During Heart Failure. Am. J. Physiology-Heart Circulatory Physiol. 312 (1), H33–H45. 10.1152/ajpheart.00341.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt A. J., MacRae I. J. (2009). The RNA-Induced Silencing Complex: A Versatile Gene-Silencing Machine. J. Biol. Chem. 284 (27), 17897–17901. 10.1074/jbc.R900012200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Spiegelman B. M. (2003). Peroxisome Proliferator-Activated Receptor-γ Coactivator 1α (PGC-1α): Transcriptional Coactivator and Metabolic Regulator. Endocr. Rev. 24 (1), 78–90. 10.1210/er.2002-0012 [DOI] [PubMed] [Google Scholar]
- Purohit P. K., Edwards R., Tokatlidis K., Saini N. (2019). MiR-195 Regulates Mitochondrial Function by Targeting Mitofusin-2 in Breast Cancer Cells. RNA Biol. 16 (7), 918–929. 10.1080/15476286.2019.1600999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Cheng R., Liang C., Yao Y., Zhang W., Zhang J., et al. (2020). MicroRNA-20b Promotes Cardiac Hypertrophy by the Inhibition of Mitofusin 2-Mediated Inter-organelle Ca2+ Cross-Talk. Mol. Ther. - Nucleic Acids 19, 1343–1356. 10.1016/j.omtn.2020.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P., Hernandez A. F., Solomon S. D., Zannad F. (2019). Heart Failure Drug Treatment. The Lancet 393 (10175), 1034–1044. 10.1016/S0140-6736(18)31808-7 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P. (2007). Intronic MicroRNA Precursors that Bypass Drosha Processing. Nature 448 (7149), 83–86. 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirone L., Forte M., Palmerio S., Yee D., Nocella C., Angelini F., et al. (2017). A Review of the Molecular Mechanisms Underlying the Development and Progression of Cardiac Remodeling. Oxidative Med. Cell Longevity 2017, 1–16. 10.1155/2017/3920195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread Changes in Protein Synthesis Induced by MicroRNAs. Nature 455 (7209), 58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Shen L., Chen L., Zhang S., Du J., Bai L., Zhang Y., et al. (2016). MicroRNA-27b Regulates Mitochondria Biogenesis in Myocytes. PloS one 11 (2), e0148532. 10.1371/journal.pone.0148532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Deng H., Zhang J., Zhang Y., Zhang X., Cui G. (2018). Mitochondria-Targeting Small Molecules Effectively Prevent Cardiotoxicity Induced by Doxorubicin. Molecules 23 (6), 1486. 10.3390/molecules23061486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T., Rajakaruna C., Caputo M., Emanueli C. (2015). MicroRNAs in Congenital Heart Disease. Ann. Transl. Med. 3 (21), 333. 10.3978/j.issn.2305-5839.2015.12.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Hu X.-Q., Zhang L. (2019). Mitochondrial MiRNA in Cardiovascular Function and Disease. Cells 8 (12), 1475. 10.3390/cells8121475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavast C., Erkeland S. (2019). The Non-canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 8 (11), 1465. 10.3390/cells8111465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Su Q., Li L., Wang X., Lu Y., Liang J. (2017). MiR-486 Regulates Cardiomyocyte Apoptosis by P53-Mediated BCL-2 Associated Mitochondrial Apoptotic Pathway. BMC. Cardiovasc. Disord. 17 (1), 119. 10.1186/s12872-017-0549-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Li C., Liu J., Wang Z., Liu Y., Luo C., et al. (2019). Expression Profile of microRNAs in Hypertrophic Cardiomyopathy and Effects of microRNA-20 in Inducing Cardiomyocyte Hypertrophy Through Regulating Gene MFN2. DNA Cel Biol. 38 (8), 796–807. 10.1089/dna.2019.4731 [DOI] [PubMed] [Google Scholar]
- Sun T., Li M.-Y., Li P.-F., Cao J.-M. (2018). MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells 7 (8), 104. 10.3390/cells7080104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. L., Li S. H., Yang L., Wang Y. (2018). MiR-376b-3p Attenuates Mitochondrial Fission and Cardiac Hypertrophy by Targeting Mitochondrial Fission Factor. Clin. Exp. Pharmacol. Physiol. 45 (8), 779–787. 10.1111/1440-1681.12938 [DOI] [PubMed] [Google Scholar]
- Tahrir F. G., Langford D., Amini S., Mohseni Ahooyi T., Khalili K. (2019). Mitochondrial Quality Control in Cardiac Cells: Mechanisms and Role in Cardiac Cell Injury and Disease. J. Cel. Physiol. 234 (6), 8122–8133. 10.1002/jcp.27597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Huang X., Xu M., Yang L., Hua F. (2020). MiR‐144 Protects the Heart from Hyperglycemia‐induced Injury by Regulating Mitochondrial Biogenesis and Cardiomyocyte Apoptosis. FASEB j. 34 (2), 2173–2197. 10.1096/fj.201901838R [DOI] [PubMed] [Google Scholar]
- Tilokani L., Nagashima S., Paupe V., Prudent J. (2018). Mitochondrial Dynamics: Overview of Molecular Mechanisms. Essays. Biochem. 62 (3), 341–360. 10.1042/EBC20170104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigié P., Camougrand N. (2017). Mitophagie et contrôle qualité des mitochondries. Med. Sci. (Paris) 33 (3), 231–237. 10.1051/medsci/20173303008 [DOI] [PubMed] [Google Scholar]
- Wan G.-x., Cheng L., Qin H.-l., Zhang Y.-z., Wang L.-y., Zhang Y.-g. (2019). MiR-15b-5p Is Involved in Doxorubicin-Induced Cardiotoxicity via Inhibiting Bmpr1a Signal in H9c2 Cardiomyocyte. Cardiovasc. Toxicol. 19 (3), 264–275. 10.1007/s12012-018-9495-6 [DOI] [PubMed] [Google Scholar]
- Wang L., Qian L. (2014). MiR-24 Regulates Intrinsic Apoptosis Pathway in Mouse Cardiomyocytes. Plos. One. 9 (1), e85389. 10.1371/journal.pone.0085389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-X., Jiao J.-Q., Li Q., Long B., Wang K., Liu J.-P., et al. (2011). MiR-499 Regulates Mitochondrial Dynamics by Targeting Calcineurin and Dynamin-Related Protein-1. Nat. Med. 17 (1), 71–78. 10.1038/nm.2282 [DOI] [PubMed] [Google Scholar]
- Wang K., Long B., Jiao J.-Q., Wang J.-X., Liu J.-P., Li Q., et al. (2012). MiR-484 Regulates Mitochondrial Network Through Targeting Fis1. Nat. Commun. 3, 781. 10.1038/ncomms1770 [DOI] [PubMed] [Google Scholar]
- Wang K., Long B., Zhou L. Y., Liu F., Zhou Q. Y., Liu C. Y., et al. (2014). CARL lncRNA Inhibits Anoxia-Induced Mitochondrial Fission and Apoptosis in Cardiomyocytes by Impairing MiR-539-Dependent PHB2 Downregulation. Nat. Commun. 5, 3596. 10.1038/ncomms4596 [DOI] [PubMed] [Google Scholar]
- Wang K., Liu C.-Y., Zhang X.-J., Feng C., Zhou L.-Y., Zhao Y., et al. (2015a). MiR-361-Regulated Prohibitin Inhibits Mitochondrial Fission and Apoptosis and Protects Heart from Ischemia Injury. Cell. Death Differ. 22 (6), 1058–1068. 10.1038/cdd.2014.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Zhang D.-l., Long B., An T., Zhang J., Zhou L.-Y., et al. (2015b). NFAT4-Dependent MiR-324-5p Regulates Mitochondrial Morphology and Cardiomyocyte Cell Death by Targeting Mtfr1. Cell. Death Dis. 6 (12), e2007. 10.1038/cddis.2015.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Zhou L.-Y., Wang J.-X., Wang Y., Sun T., Zhao B., et al. (2015c). E2F1-Dependent MiR-421 Regulates Mitochondrial Fragmentation and Myocardial Infarction by Targeting Pink1. Nat. Commun. 6, 7619. 10.1038/ncomms8619 [DOI] [PubMed] [Google Scholar]
- Wang X., Yan M., Zhao L., Wu Q., Wu C., Chang X., et al. (2016). Low-Dose Methylmercury-Induced Genes Regulate Mitochondrial Biogenesis via miR-25 in Immortalized Human Embryonic Neural Progenitor Cells. Ijms 17 (12), 2058. 10.3390/ijms17122058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Song C., Zhou X., Han X., Li J., Wang Z., et al. (2017). Mitochondria Associated MicroRNA Expression Profiling of Heart Failure. Biomed. Res. Int. 2017, 1–10. 10.1155/2017/4042509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Qin D., Shi H., Zhang Y., Li H., Han Q. (2019). MiR-195-5p Promotes Cardiomyocyte Hypertrophy by Targeting MFN2 and FBXW7. Biomed. Res. Int. 2019, 1–10. 10.1155/2019/1580982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Zhai M., Xu S., Ponnusamy M., Huang Y., Liu C.-Y., et al. (2020). NFATc3-Dependent Expression of MiR-153-3p Promotes Mitochondrial Fragmentation in Cardiac Hypertrophy by Impairing Mitofusin-1 Expression. Theranostics 10 (2), 553–566. 10.7150/thno.37181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Ladewig E., Shenker S., Mohammed J., Lai E. C. (2015). Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates. Plos. Comput. Biol. 11 (9), e1004441. 10.1371/journal.pcbi.1004441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm J. O., Lai E. C. (2011). Mirtrons: MicroRNA Biogenesis via Splicing. Biochimie 93 (11), 1897–1904. 10.1016/j.biochi.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. C., Tambe A., Kidwell M. A., Noland C. L., Schneider C. P., Doudna J. A. (2015). Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis. Mol. Cel 57 (3), 397–407. 10.1016/j.molcel.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R. I., Diederichs S. (2009). Many Roads to Maturity: MicroRNA Biogenesis Pathways and Their Regulation. Nat. Cel. Biol. 11 (3), 228–234. 10.1038/ncb0309-228 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2019). Cardiovascular Disease. Available at: https://www.who.int/cardiovascular_diseases/about _cvd/en/ (Accessed on October 30, 2019). [Google Scholar]
- Xia P. P., Zhang F., Chen C., Wang Z. H., Wang N., Li L. Y., et al. (2020). Rac1 Relieves Neuronal Injury Induced by Oxygenglucose Deprivation and Re-Oxygenation via Regulation of Mitochondrial Biogenesis and Function. Neural Regen. Res. 15 (10), 1937–1946. 10.4103/1673-5374.280325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Hu J., Zhang X., Li C., Zuo Y., Xie S., et al. (2020). Neuropeptide Y Induces Cardiomyocyte Hypertrophy via Attenuating miR-29a-3p in Neonatal Rat Cardiomyocytes. Ppl 27 (9), 878–887. 10.2174/0929866527666200416144459 [DOI] [PubMed] [Google Scholar]
- Xiong H., Chen S., Lai L., Yang H., Xu Y., Pang J., et al. (2019). Modulation of MiR-34a/SIRT1 Signaling Protects Cochlear Hair Cells Against Oxidative Stress and Delays Age-Related Hearing Loss Through Coordinated Regulation of Mitophagy and Mitochondrial Biogenesis. Neurobiol. Aging 79, 30–42. 10.1016/j.neurobiolaging.2019.03.013 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhao C., Sun X., Liu Z., Zhang J. (2015). MicroRNA-761 Regulates Mitochondrial Biogenesis in Mouse Skeletal Muscle in Response to Exercise. Biochem. Biophysical Res. Commun. 467 (1), 103–108. 10.1016/j.bbrc.2015.09.113 [DOI] [PubMed] [Google Scholar]
- Xu H., Cao H., Zhu G., Liu S., Li H. (2017). Overexpression of MicroRNA-145 Protects Against Rat Myocardial Infarction Through Targeting PDCD4. Am. J. Transl. Res. 9 (11), 5003–5011. [PMC free article] [PubMed] [Google Scholar]
- Yan K., An T., Zhai M., Huang Y., Wang Q., Wang Y., et al. (2019). Mitochondrial MiR-762 Regulates Apoptosis and Myocardial Infarction by Impairing ND2. Cel. Death Dis. 10 (7), 500. 10.1038/s41419-019-1734-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Li T., Dong Z., Mi R. (2018). MicroRNA‐410 Is Involved in Mitophagy after Cardiac Ischemia/reperfusion Injury by Targeting High‐mobility Group Box 1 Protein. J. Cel. Biochem. 119 (2), 2427–2439. 10.1002/jcb.26405 [DOI] [PubMed] [Google Scholar]
- Yang S., Li H., Chen L. (2019). MicroRNA‐140 Attenuates Myocardial Ischemia‐reperfusion Injury through Suppressing Mitochondria‐mediated Apoptosis by Targeting YES1. J. Cel. Biochem. 120 (3), 3813–3821. 10.1002/jcb.27663 [DOI] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I. G., Cullen B. R. (2003). Exportin-5 Mediates the Nuclear Export of Pre-MicroRNAs and Short Hairpin RNAs. Genes Dev. 17 (24), 3011–3016. 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S. S., Akman D., Catalucci D., Turan B. (2013). Relationship Between Downregulation of MiRNAs and Increase of Oxidative Stress in the Development of Diabetic Cardiac Dysfunction: Junctin as a Target Protein of MiR-1. Cell. Biochem. Biophys. 67 (3), 1397–1408. 10.1007/s12013-013-9672-y [DOI] [PubMed] [Google Scholar]
- Yin Z., Zhao Y., He M., Li H., Fan J., Nie X., et al. (2019). MiR-30c/PGC-1β Protects against Diabetic Cardiomyopathy via PPARα. Cardiovasc. Diabetol. 18 (1), 7. 10.1186/s12933-019-0811-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Chen H., Ge D., Xu Y., Xu H., Yang Y., et al. (2017). MiR-21 Promotes Cardiac Fibrosis After Myocardial Infarction Via Targeting Smad7. Cell. Physiol. Biochem. 42 (6), 2207–2219. 10.1159/000479995 [DOI] [PubMed] [Google Scholar]
- Zaglia T., Ceriotti P., Campo A., Borile G., Armani A., Carullo P., et al. (2017). Content of Mitochondrial Calcium Uniporter (MCU) in Cardiomyocytes Is Regulated by Microrna-1 in Physiologic and Pathologic Hypertrophy. Proc. Natl. Acad. Sci. USA 114 (43), E9006–E9015. 10.1073/pnas.1708772114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu H. (2016). Translational Regulation of Mitochondrial Biogenesis. Biochem. Soc. Trans. 44 (6), 1717–1724. 10.1042/BST20160071C [DOI] [PubMed] [Google Scholar]
- Zhang J.-J., Liu W.-Q., Peng J.-J., Ma Q.-L., Peng J., Luo X.-J. (2017). Mir-21-5p/203a-3p Promote Ox-LDL-Induced Endothelial Cell Senescence Through Down-Regulation of Mitochondrial Fission Protein Drp1. Mech. Ageing Dev. 164, 8–19. 10.1016/j.mad.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ji R., Liao X., Castillero E., Kennel P. J., Brunjes D. L., et al. (2018). MicroRNA-195 Regulates Metabolism in Failing Myocardium Via Alterations in Sirtuin 3 Expression and Mitochondrial Protein Acetylation. Circulation 137 (19), 2052–2067. 10.1161/CIRCULATIONAHA.117.030486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Liao P., Liang R., Zheng X., Jian J. (2019). Epigallocatechin Gallate Prevents Mitochondrial Impairment and Cell Apoptosis by Regulating Mir-30a/P53 Axis. Phytomedicine 61, 152845. 10.1016/j.phymed.2019.152845 [DOI] [PubMed] [Google Scholar]
- Zhang J., Pan J., Yang M., Jin X., Feng J., Wang A., et al. (2019). Upregulating MicroRNA-203 Alleviates Myocardial Remodeling and Cell Apoptosis Through Downregulating Protein Tyrosine Phosphatase 1B in Rats with Myocardial Infarction. J. Cardiovasc. Pharmacol. 74 (5), 474–481. 10.1097/FJC.0000000000000733 [DOI] [PubMed] [Google Scholar]
- Zhang L., Ding H., Zhang Y., Wang Y., Zhu W., Li P., et al. (2020). Circulating MicroRNAs: Biogenesis and Clinical Significance in Acute Myocardial Infarction. Front. Physiol. 11, 1088. 10.3389/fphys.2020.01088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ponnusamy M., Liu C., Tian J., Dong Y., Gao J., et al. (2017). MiR-485-5p Modulates Mitochondrial Fission Through Targeting Mitochondrial Anchored Protein Ligase in Cardiac Hypertrophy. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1863 (11), 2871–2881. 10.1016/j.bbadis10.1016/j.bbadis.2017.07.034 [DOI] [PubMed] [Google Scholar]
- Zhou B., Tian R. (2018). Mitochondrial Dysfunction in Pathophysiology of Heart Failure. J. Clin. Invest. 128 (9), 3716–3726. 10.1172/JCI120849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Liu Y., Prater K., Zheng Y., Cai L. (2013). Roles of MicroRNAs in Pressure Overload- and Ischemia-Related Myocardial Remodeling. Life Sci. 93 (23), 855–862. 10.1016/j.lfs.2013.08.023 [DOI] [PubMed] [Google Scholar]