Abstract
Background
Citrulline post-translational modification of proteins is mediated by protein arginine deiminase (PADI) family members and has been associated with autoimmune diseases. The role of PADI-citrullinome in immune response in cancer has not been evaluated. We hypothesized that PADI-mediated citrullinome is a source of neoantigens in cancer that induces immune response.
Methods
Protein expression of PADI family members was evaluated in 196 cancer cell lines by means of indepth proteomic profiling. Gene expression was assessed using messenger RNA data sets from The Cancer Genome Atlas. Immunohistochemical analysis of PADI2 and peptidyl-citrulline was performed using breast cancer tissue sections. Citrullinated 12–34-mer peptides in the putative Major Histocompatibility Complex-II (MHC-II) binding range were profiled in breast cancer cell lines to investigate the relationship between protein citrullination and antigen presentation. We further evaluated immunoglobulin-bound citrullinome by mass spectrometry using 156 patients with breast cancer and 113 cancer-free controls.
Results
Proteomic and gene expression analyses revealed PADI2 to be highly expressed in several cancer types including breast cancer. Immunohistochemical analysis of 422 breast tumor tissues revealed increased expression of PADI2 in ER− tumors (p<0.0001); PADI2 protein expression was positively correlated (p<0.0001) with peptidyl-citrulline staining. PADI2 expression exhibited strong positive correlations with a B cell immune signature and with MHC-II-bound citrullinated peptides. Increased circulating citrullinated antigen–antibody complexes occurred among newly diagnosed breast cancer cases relative to controls (p=0.0012).
Conclusions
An immune response associated with citrullinome is a rich source of neoantigens in breast cancer with a potential for diagnostic and therapeutic applications.
Keywords: antigens, autoimmunity, biomarkers, tumor, immunity, humoral
Introduction
Dysregulated protein citrullination by peptidyl arginine deiminases (PADI) has been associated with autoimmune diseases, with recent interest in its relevance to cancer given the occurrence of autoimmunity as a manifestation of cancer.1 2 PADI comprises a family of enzymes that, in the presence of calcium ions, catalyze the post-translational modification of proteins via the deamination of arginine to citrulline. In total, five PADI family members are known with sequence homology ranging from 70% to 95%.2 To date no enzyme has been identified that can reverse protein citrullination. The role of protein citrullination has been best investigated in the context of rheumatoid arthritis (RA), where elevated protein citrullination, notably of keratins, filaggrin, vimentin, actin, histones, nucleophosmin, and nuclear lamin C, has been shown to elicit an autoimmune response.3 4 Autoimmunity in RA is considered to be principally facilitated through Major Histocompatibility Complex (MHC) class II-mediated presentation of citrullinated peptides that elicit a B cell response.5 6 There is also currently increased interest in MHC-II neoantigens as shaping tumor immunity.7
Comprehensive assessment of the expression of PADI family members in cancer has been limited. PADI4 has been investigated largely with respect to its interactions with histone H3, ING4, p53, and HDAC2.8–11 Moreover, the extent to which protein citrullination in tumors induces an immune response is largely unexplored. Studies aimed at interrogating antitumor immunity against citrulline-peptides of vimentin and α-enolase have demonstrated that these peptides can trigger a CD4+ T cell response,12 13 providing evidence that protein citrullination in tumors may be immunogenic. Given the antigenicity of citrullinated proteins, exploration of PADI family members among cancer types and their impact on citrullination and immune response has potential for the development of tumor vaccines with citrullinated antigens or for identification of citrullinated antigens as biomarkers for cancer detection or prediction of response to immunotherapy.7 14–18
Herein, we evaluated the protein expression of PADI family members among 196 cancer cell lines reflective of 12 common cancer types by proteomic profiling as well as by analysis of messenger RNA (mRNA) expression data sets from The Cancer Genome Atlas (TCGA) for 9721 human tumors consisting of 32 different cancer types. Using mass spectrometry, we further explored the association of PADI2 with the citrullinome of 28 breast cancer cell lines and confirmed the findings of PADI2 expression and citrullination in 422 breast tumors analyzed by immunohistochemistry (IHC). We additionally interrogated whether PADI2-mediated citrullination is associated with a distinct tumor immunophenotype using TCGA gene expression data sets and immunohistochemical analysis of human breast cancer tumors. We further provide evidence for circulating autoantibodies against citrullinated tumor-associated proteins in breast cancer.
Methods
Mass spectrometry analyses
Breast cancer cell line citrullinome analysis
Proteomic analysis was performed as previously described.19–22 For indepth citrullinome analysis, 28 breast cancer cell lines (MCF7, MDA-MB-231, SKBR3, HCC1954, HCC1143, BT474, HCC1500, T47D, ZR75-1, HCC1937, HCC1599, HCC202, HCC1806, MDA-MB-468, HCC2218, HCC70, HCC1187, Hs578T, BT549, MCF10A, MCF12A, MDA-MB-361, HCC1395, CAMA1, HCC38, MDA-MB-436, BT20, and MDA-MB-157) were labeled with 13C6 Lys (#CNLM-2247; Cambridge Isotope Laboratories) in RPMI 1640 containing 10% dialyzed Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin cocktail (Gibco). Stable Isotope Labeling by Amino acid in Cell culture (SILAC labeling)23 was performed to discriminate FBS-derived proteins from cell proteins.
For proteomic analysis of whole cell lysates, −2×107 cells were lysed in 1 mL of Phosphate-Buffered Saline (PBS) containing octyl-glucoside (1% w/v) and protease inhibitors (cOmplete Protease Inhibitor Cocktail, Roche Diagnostics), followed by sonication and centrifugation at 20,000× g with collection of the supernatant and filtration through a 0.22 µm filter. Two milligrams of Whole Cell Extract (WCE) proteins were reduced in Dithiothreitol (DTT) and alkylated with acrylamide before fractionation with Reversed Phase-High Performance Liquid Chromatography (RP-HPLC). A total of 84 fractions were collected at a rate of 3 fractions/min. Mobile phase A consisted of Water (H2O): Acetonitrile (ACN) (95:5, v/v) with 0.1% Trifluoroacetic acid (TFA). Mobile phase B consisted of ACN:H2O (95:5) with 0.1% TFA. Collected fractions from HPLC were dried by lyophilization, followed by insolution digestion with trypsin (Mass Spectrometry Grade, Thermo Fisher Scientific).
Based on the chromatogram profile, 84 fractions were pooled into 24 fractions for Liquid Chromatography-Tandem mass spectrometry (LC-MS/MS) analysis per cell line. In total, 2688 fractions were subjected to Reversed Phase Liquid Chromatography-Tandem mass spectrometry (RPLC-MS/MS) using a nanoflow LC system (EASYnano HPLC System, Thermo Fisher Scientific) coupled online with LTQ Orbitrap ELITE mass spectrometer (Thermo Fisher Scientific). Separations were performed using 75 µm inner diameter (id) × 360 µm od × 25 cm long fused-silica capillary column (Column Technology) slurry packed with 3 µm, 100 A° pore size C18 silica-bonded stationary phase. Following injection of ~500 ng of protein digest onto a C18 trap column (180 µm id × 20 mm; Waters), peptides were eluted using a linear gradient of 0.35% mobile phase B (0.1 formic acid in ACN) per minute for 90 min, then to 95% B for an additional 10 min, all at a constant flow rate of 300 nL/min. Eluted peptides were analyzed by LTQ Orbitrap ELITE in a data-dependent acquisition mode. Each full MS scan (400–1800 m/z) was followed by 20 MS/MS scans (Collision-Induced Dissociation (CID) normalized collision energy of 35%). Acquisition of each full mass spectrum was followed by acquisition of MS/MS spectra for the 20 most intense +2, +3, or +4 ions within a duty cycle; dynamic exclusion was enabled to minimize redundant selection of peptides previously selected for MS/MS analysis. Parameters for MS1 were 60,000 for resolution, 1×106 for automatic gain control target, and 150 ms for maximum injection time. MS/MS was done by CID fragmentation with 3×104 for automatic gain control, 10 ms for maximum injection time, 35 for normalized collision energy, 2.0 m/z for isolation width, 0.25 for activation q-value, and 10 ms for activation time.
MS/MS spectra were searched against the UniProt human proteome database (January 2017) using Sequest HT in Proteome Discoverer V.1.4 pipeline. One fixed modification of propionamide at Cys (71.037114 Da) and three variable modifications, oxidation at Met (15.9949 Da), deamidation at Arg (0.984016 Da), and SILAC 13C6 at Lys (6.0201 Da), were chosen. The mass error allowed was 10 parts per million (ppm) for parent monoisotopic and 0.5 Da for Tandem mass (MS2) fragment monoisotopic ions. Full trypsin was specified as protein cleavage site, with possibility of two missed cleavages allowed. The searched result was filtered with False Discovery Rate (FDR)=0.01, and the peptides with deamidated Arg at C-terminal of the tryptic peptides were considered as false identification and removed as well.
Plasma Ig-bound citrullinome analysis
Clinical subjects for Ig-bound analysis
Plasma samples were collected from 156 women with newly diagnosed breast cancer (0–0.8 years) as cases and 40 age-matched cancer-free women as control. For the cases, only patients who had no documented distant metastasis at the time of sample collection were included in this study. Written informed consent was obtained. The timing of blood draw was after the diagnostic biopsy and prior to neoadjuvant chemotherapy, or definitive surgery in patients who did not receive chemotherapy in the neoadjuvant setting23 (online supplemental tables S4 and S6). The additional 73 healthy control plasmas were obtained from MD Anderson Cancer Center Gynecologic Tissue Bank following institutional review board approval and informed consent (online supplemental table S4).
jitc-2021-002549supp001.pdf (246.3KB, pdf)
Plasma Ig-bound workflow
Plasma immunoglobulin (Ig)-bound proteins were prepared and analyzed as described previously.24 Twenty-six pooled plasma samples from 156 women with newly diagnosed breast cancer and 12 pooled plasma samples from 113 cancer-free subjects as controls were analyzed (online supplemental table S4). Briefly, 100 µL of pooled plasma for each experimental condition was processed with the immuno-depletion column Hu-14 10×100 mm (#5188-6559; Agilent Technologies, Santa Clara, California, USA) to remove the top 14 high abundance proteins: albumin, IgG, IgA, transferrin, haptoglobin, fibrinogen, α1-antitrypsin, α1-acid glycoprotein, apolipoprotein AI, apolipoprotein AII, complement C3, transthyretin, IgM, and α2-macroglobulin. The bound fraction was used for IgG-bound protein analysis as previously described.24 25
LC-High-Definition MSE (HDMSE) data were acquired in resolution mode with SYNAPT G2-S using Waters Masslynx (V.4.1, SCN851). The capillary voltage was set to 2.80 kV, sampling cone voltage to 30 V, source offset to 30 V, and source temperature to 100°C. Mobility used high-purity N2 as the drift gas in the Ion-Mobility Spectrometry (IMS) TriWave cell. Pressures in the helium cell, trap cell, IMS TriWave cell, and transfer cell were 4.50 mbar, 2.47e-2 mbar, 2.90 mbar, and 2.53e-3 mbar, respectively. The IMS wave velocity was 600 m/s, the helium cell DC was 50 V, the trap DC bias was 45 V, the IMS TriWave DC bias was 3 V, and the IMS wave delay was 1000 μs. The mass spectrometer was operated in V-mode with a typical resolving power of at least 20,000. All analyses were performed using positive mode Electrospray Ionization (ESI) using a NanoLockSpray source. The lock mass channel was sampled every 60 s. The mass spectrometer was calibrated with a [Glu1]-fibrinopeptide solution (300 fmol/µL) delivered through the reference sprayer of the NanoLockSpray source. Accurate mass LC-HDMSE data were collected in an alternating low-energy (MS) and high-energy (MSE) mode of acquisition with mass scan range from 50 m/z to 1800 m/z. The spectral acquisition time in each mode was 1.0 s, with a 0.1 s interscan delay. In low-energy HDMS mode, data were collected at constant collision energy of 2 eV in both trap cell and transfer cell. In high-energy HDMSE mode, the collision energy was ramped from 25 eV to 55 eV in the transfer cell only. The RF applied to the quadrupole mass analyzer was adjusted such that ions from 300 m/z to 2000 m/z were efficiently transmitted, ensuring that any ions observed in the LC-HDMSE data less than 300 m/z were known to arise from dissociations in the transfer collision cell.
The acquired LC-HDMSE data were processed and searched against the UniProt human proteome database (January 2017) through ProteinLynx Global Server (Waters Company) with false discovery rate of 4%. The modification search settings and the deamidated Arg, C-terminal miss cleavage assessment were the same with the cell line citrullinome. The plot values are represented as the number of citrullinated proteins relative to the total unique peptides per sample as to adjust for batch effects that occurred during the data acquisition. The Ingenuity Pathway Analysis (IPA) network analysis of plasma IgG-bound was done by calculating the ratio of the spectral counts of the citrullinome in each BC receptor subtype compared with healthy controls with greater than or equal to 1.5.
Plasma autoantibody western blot assay
The recombinant unmodified vimentin (#11234; Cayman Chemical) was incubated with 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.6) containing 100 mM NaCl and 1 mM CaCl2 buffer and stored under room temperature for 1 hour, and the citrullinated form was prepared. Each of the recombinant unmodified vimentin and citrullinated vimentin was loaded 0.1 μg, 0.5 μg, and 1.0 μg to Criterion XT 12% gel and transferred to Polyvinylidene difluoride (PVDF) membrane using Trans-Blot Turbo Transfer System (#1704150; BioRad). From the healthy control (n=8 pooled) and Triple negative breast cancer (TNBC) stage II (n=11 pooled), 2 μL of plasmas was diluted 150-fold with 0.05% casein dissolved in Tris-buffered saline containing 0.01% Tween 20 (TBST) and incubated with the recombinant proteins transferred PVDF membrane for 2 hours, respectively. Then after washing the membrane with TBST, the secondary antibody of ECL anti-human IgG horseradish peroxidase-linked whole Ab (GE Healthcare #NA933) was added and stored for 1 hour in room temperature, followed by Clarity Western ECL (#170-5061; BioRad) detection. The band intensities were read by ImageJ V.1.46r (https://imagej.nih.gov/).
Plasma autoantibody ELISA assay
Clinical subjects for the plasma ELISA assay
Eleven newly diagnosed stage II TNBC patient plasmas and 31 healthy controls were used for autoantibody assays. Case plasmas were derived from a subset of patients in the breast cohort that were used for proteomic analyses of Ig-bound proteins that had sufficient volume. The independent set of healthy control plasmas was obtained from the MD Anderson Cancer Center Gynecologic Tissue Bank as described above (online supplemental tables S4 and S6).
Plasma autoantibody assay method
Concentrations of antivimentin and anticitrullinated vimentin autoantibodies were determined using Luminex bead-based immunoassays on the MAGPIX instrument (Luminex Corporation, Austin, Texas). Samples were analyzed in the same batch in random order. MagPlex Microspheres were conjugated with purified recombinant vimentin (#11234; Cayman Chemical) and recombinant citrullinated vimentin (#21942; Cayman Chemical) at 5 μg/million concentration. The samples were tested at the final dilution of 1:16,250. The acid dissociated autoantibodies were prepared by 5 μL plasma diluted 50-fold with 0.1 M Gly-HCl (pH 3.0), stored under room temperature for 30 min, and exchanged buffer to Reagent Diluent Concentrate 2 buffer (#841380; R&D Systems) using Zeba Spin Column (#89890; Thermo Fisher Scientific). The samples were incubated with MagPlex beads for 2 hours at room temperature on a shaker. After the samples were washed with Phosphate-Buffered Saline with Tween 20 (PBST), PE-conjugated secondary antibody was used to incubate samples for 30 min. The measured fluorescent intensity of each well was read using MAGPIX instrument. The TNBC stage II plasmas in online supplemental table S4 were used for the assay, and anonymous individual subject information is presented in online supplemental table S6.
Cell surface Human Leukocyte Antigen (HLA)-bound peptidyl-citrullinome
A total of 5×108 HCC1954 and TNBC MDA-MB-468 cells were used for culture in peptidyl-citrullinome analysis using the LTQ Orbitrap ELITE as described previously.26 The MS/MS spectra were searched against the UniProt human proteome database (January 2017) using Sequest HT in Proteome Discoverer V.1.4 pipeline. The search was conducted for amino acid lengths from 8 to 34. Two variable modifications, oxidation at Met (15.9949 Da) and deamidation at Arg (0.984016 Da), were chosen. The mass error allowed was 10 ppm for parent monoisotopic and 0.5 Da for MS2 fragment monoisotopic ions. The searched result was filtered with FDR=0.01. The identified peptides with amino acid lengths putatively from 8 to 11 were considered as MHC-I binding peptides and from 12 to 34 as MHC-II binding peptides.
We additionally performed in silico binding affinity predictions between peptides and MHC-II molecules using the well-established prediction tool NetMHCIIpan V.2.3.27 28 Briefly, identified peptide sequences were loaded to the prediction tool NEetMHC-II pan2.3, then the data were sorted by the binding peptide core affinity prediction (IC50, nM) with MHC-II pocket assessed by the artificial intelligent network SSNAlignment, and the percentile rank that is generated by comparing the peptide’s score against the scores of one million random 15 mers selected from Swiss-Prot database.
PADI2 siRNA knockdown
Transient knockdown of PADI2 was performed by transfecting cells with 50 nM of siControl (Silencer Select Negative Control No 1; Thermo Fisher Scientific), siPADI2#1 (s22187; Thermo Fisher Scientific), and siPADI2#2 (s22188; Thermo Fisher Scientific) in Lipofectamine RNAiMAX (Thermo Fisher Scientific). After 72 hours of transfection, total RNA was isolated using RNeasy Mini Kit (Qiagen, Germantown, Maryland). Reverse transcription was performed with 1 μg of total RNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and real-time PCR performed using TaqMan Gene Expression Assays (Hs00247108_m1, PADI2 FAM-MGB; Thermo Fisher Scientific). 18S (Hs99999901_s1, VIC-MGB; Thermo Fisher Scientific) was used as an internal control. All samples were assayed in triplicate.
Proteins were extracted using 8 M urea and 50 mM triethylammonium bicarbonate (TEAB) with protease inhibitor cocktail (cOmplete Protease Inhibitor Cocktail, Millipore Sigma). A total of 100 μg protein was used for Tandem Mass Tag (TMT) labeling per channel. After reduction with TCEP (tris(2-carboxyethyl)phosphine) and alkylation with acrylamide, proteins were digested with Lys-C (Wako) overnight at 37°C. Digested peptides were desalted with MonoSpin C18 column (GL Sciences, Tokyo, Japan) and dried by speedvac. The tryptic peptides were resuspended in 0.1 M TEAB buffer/acetonitrile and reacted with each TMT channel for 1 hour (TMTsixplex Isobaric Label Reagent Set, Thermo Fisher Scientific) and then quenched with hydroxylamine, mixed together, and dried by speedvac. Tryptic peptides were subsequently fractionated under high-pH conditions using MonoSpin L C18 column (GL Sciences) in 0.1% trimethylamine/acetonitrile with step elution of acetonitrile into 10 fractions.
Fractionated peptides were injected into an EASYnano HPLC System (Thermo Fisher Scientific) online coupled with Q Exactive Mass Spectrometer (Thermo Fisher Scientific). The system was equipped with Waters Symmetry C18 nanoAcquity trap column (180 µm × 20 mm, 5 µm) and a C18 analytical column (75 µm × 200 mm, 3 µm) (Column Technology). The separation column temperature was set ambient, and the temperature of the tray compartment in the autosampler was set at 6°C. The parameters for mass spectrometer were the following: spray voltage 2.5 kV, capillary temperature 320°C, Fourier Transform (FT) resolution 70,000, AGC target value 3×106, and 1 microscan with 30 ms injection time. Mass spectra were acquired in a data-dependent mode in the range of 350–1800 m/z. The step gradient of normalized collision energy 20, 25, and 35 was applied to induce fragmentation. Acquisition of each full mass spectrum was followed by acquisition of MS/MS spectra for the 10 most intense +2, +3, or +4 ions within a duty cycle. The acquired LC-MS/MS data were processed by Proteome Discoverer V.1.4 (Thermo Fisher Scientific). Sequest HT was used as a search engine with the parameters including fixed modification of Cys alkylated with acrylamide (+71.03714), Lys with TMT (+229.162932, N-terminal and Lys), and variable modification of Met oxidation (+15.99491) and Arg deamidation (+0.984016). Mass error of 10 ppm was allowed for the parent MS1 and 0.02 Da was allowed for the MS2 fragments. Data were searched against the UniProt human database (2017) and were further filtered with FDR=0.01 and TMT ratios were quantified.
Immunohistochemistry
Clinical subjects for breast cancer tissue microarray analysis
Breast cancer tissue microarrays (TMA; BC081120C, BR20810, and BR20811), including 127 Luminal A, 99 Luminal B, 57 HER2-enriched, and 139 TNBC from a total of 422 patient tissues, were used for assessment of PADI2 expression related to clinical parameters and coexpression with peptidyl-citrulline (table 1).
Table 1.
Citrullinated protein count in breast cancer tissues (n=422) in relation to PADI2 expression and clinical parameters
| Variable | PADI2 expression* | Comparison | P value† | OR (95% CI)‡ | |||
| Low | Low-moderate | Moderate-high | High | ||||
| Age | |||||||
| ≤50 | 77 | 57 | 40 | 60 | Age ≤50 vs >50 | 0.0477 | 1.55 (0.91 to 2.59) |
| >50 | 43 | 45 | 48 | 52 | |||
| Grade | |||||||
| I–II | 93 | 68 | 56 | 50 | Grade I+II vs III | <0.0001 | 8.12 (3.83 to 17.44) |
| III | 11 | 21 | 22 | 48 | |||
| Unknown | 16 | 13 | 10 | 14 | |||
| Stage | |||||||
| I | 12 | 11 | 10 | 10 | Stage I vs II+III+IV | 0.8284 | 1.13 (0.49 to 2.75) |
| II | 90 | 75 | 62 | 85 | Stage I+II vs III+IV | 0.8618 | 1.01 (0.51 to 2.01) |
| III | 17 | 15 | 16 | 17 | |||
| IV | 1 | 1 | 0 | 0 | |||
| Receptor subtype | |||||||
| Luminal A | 61 | 30 | 19 | 17 | HR+ vs HR− | <0.0001 | 6.71 (3.75 to 11.66) |
| Luminal B | 32 | 29 | 17 | 21 | TNBC vs non-TNBC | <0.0001 | 4.33 (2.41 to 8.12) |
| HER2-enriched | 7 | 14 | 14 | 22 | |||
| TNBC | 20 | 29 | 38 | 52 | |||
| Citrullination | |||||||
| Low | 19 | 9 | 9 | 11 | Citrullination | <0.0001 | 4.45 (2.48 to 7.78) |
| (low+low-moderate vs moderate-high+high) | |||||||
| Low-moderate | 77 | 57 | 49 | 42 | Citrullination | <0.0001 | 7.97 (2.5 to 25.95) |
| (high vs rest) | |||||||
| Moderate-high | 21 | 29 | 22 | 40 | |||
| High | 3 | 7 | 8 | 19 | |||
*PADI2 and peptidyl-citrulline (citrullination) staining positivity: low: 0%–24%; low-moderate: 25%–49%; moderate-high: 50%–74%; high: ≥75%.
†P value represents two-sided Χ2 test for trend between PADI2 staining positivity and the respective comparative groups.
‡OR (95% CI) for comparison of high PADI2 (≥75% staining positivity) vs low PADI2 (0%–24% staining positivity) expression.
HR, Hormone Receptor; PADI2, protein arginine deiminase 2; TNBC, Triple Negative Breast Cancer.
IHC workflow
Healthy TMA (BN0001a) and breast cancer tissue sections (HuCAT297, HuCAT298, 2017-16604A TNBC, Fmg0105378 Her2+, and Fmg030209B5 ER+) were purchased from US Biomax (Rockville, Maryland, USA), and healthy mammary gland tissue section was obtained from Zyagen (HP-414). Sections were deparaffinized in xylene, rehydrated in a descending ethanol series, and then treated with 3% hydrogen peroxide for 10 min. Antigen retrieval was conducted in a pressure cooker in 1X ImmunoDNA Retriever with Citrate (Bio SB, Santa Barbara, California, USA) and 0.1% Tween 20 at 121°C for 15 min. Sections were hybridized with 1:2000 times diluted anti-PADI2 monoclonal antibody (66386-1-1g; Proteintech, Rosemont, Illinois, USA), 1:1000 times diluted anticitrulline monoclonal antibody (Clone F95; Millipore Sigma, Burlington, Massachusetts, USA), 1:250 times diluted anti-CD20cy monoclonal antibody (Clone L26; Agilent Technologies), 1:50 times diluted anti-CD19 monoclonal antibody (Clone LE-CD19; Agilent Technologies), and 1:1000 times diluted anti-pan-cytokeratin monoclonal antibody (Clone AE1/AE3+5D3; Abcam, Cambridge, UK) for 16 hours at 4°C. After washing with Tris Buffered Saline (TBS) for 5 min ×3, signal development was performed by Histofine DAB-2V Kit (Nichirei Bioscience, Tokyo, Japan). Images were scanned by MD Anderson Cancer Center (MDACC)-North Campus Research Histology Core Laboratory and analyzed using Aperio Imagescope (Leica Biosystems, Buffalo Grove, Illinois, USA). For PADI2 and citrulline IHC evaluation was independently performed by three operators. Positivity of cancer cells was scored as low (0%–24%), low-moderate (25%–49%), moderate-high (50%–74%), and high (75%–100%), and p values were calculated by two-sided Χ2 test for trend between PADI2 staining positivity and the respective comparative groups. Since BR20810 and BR20811 consisted of duplicate tissues per patient, we derived an average score.
For antigen absorption-based IHC, we used a previously developed method29 with slight modification. Anti-PADI2 antibody was 2000× and mixed with various concentrations of recombinant citrullinated fibrinogen (#18473; Cayman Chemical) and uncitrullinated fibrinogen (#16088; Cayman Chemical) and incubated at 4°C overnight. After centrifugation at 20,000× g for 30 min, the supernatant was used as primary antibody for IHC. Reactivity of the antibody was confirmed using breast invasive ductal carcinoma tissue sections, and positive staining was observed in the presence of unmodified fibrinogen, whereas staining was abrogated in the presence of citrullinated fibrinogen (online supplemental figure S2A–C).
jitc-2021-002549supp003.pdf (931.6KB, pdf)
Gene expression data
TCGA gene expression data, HM450 methylation data, and clinical data were downloaded from cBioPortal.30 Gene expression for the Curtis et al data set31 was obtained through the Oncomine database.32
Immune cell signature analyses
Immune signatures were derived as previously described.33 Briefly, specific immune cell infiltration was computationally inferred using RNA-Seq data based on gene sets overexpressed in one of 24 immune cell types according to Bindea et al.34 Scoring of TCGA cancer samples for each of the immune cell signatures and for expression of antigen presentation MHC class I (APM1) genes (HLA-A/B/C, β2M, TAP1/2, TAPBP) or antigen presentation MHC class II (APM2) genes (HLA-DR/DQ/DP/DM) is described elsewhere.35
Statistical analyses
Unsupervised hierarchical clustering heatmaps were generated using R statistical software. Figures were generated using R statistical software or GraphPad Prism V.8. Spearman’s correlation analyses were performed to assess relationships between continuous variables. Fisher’s exact tests were used to assess relationships between categorical variables. All statistical tests were two-sided unless specified otherwise.
Results
PADI2 is highly expressed in breast cancer among various cancer types
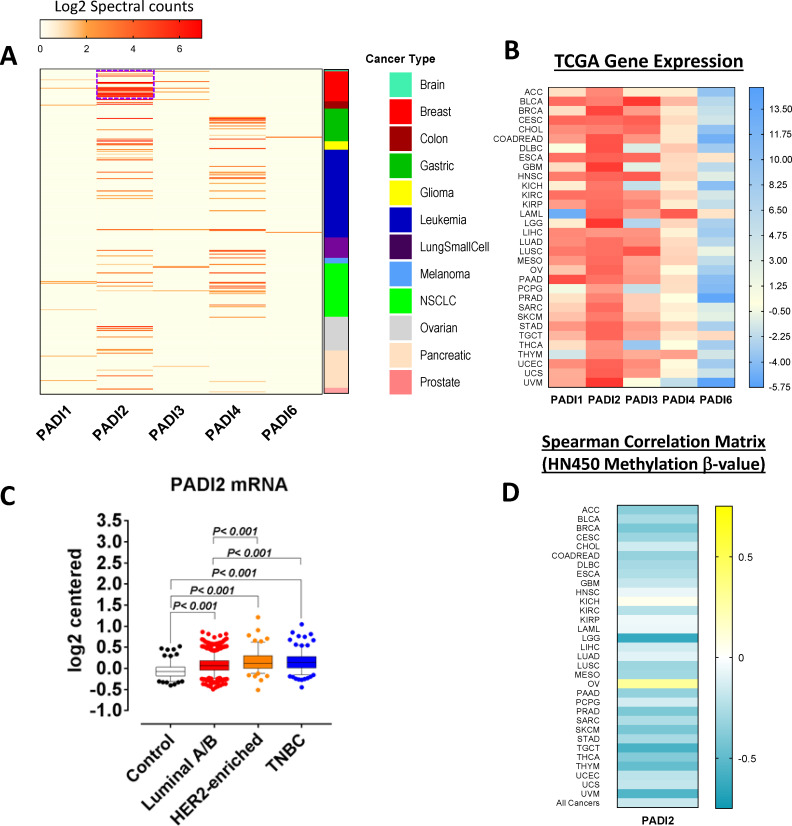
Analysis of proteomic mass spectrometry data for 196 cancer cell lines representing 12 different cancer types revealed that, of the five family members, PADI2 had the highest expression with particularly elevated levels in breast cancer (figure 1A). Using mRNA expression data sets from TCGA, we further interrogated differential expression of PADI family members among 9721 human tumors consisting of 32 different cancer types. Concordant with proteomic data, relative to the other PADI family members, PADI2 exhibited the highest mRNA expression levels (RNA Seq V2 RSEM) among the cancer types (figure 1B). We determined the differential expression of PADI2 among breast cancer subtypes in 1725 breast tumors and 144 normal breast tissues from the Curtis et al cohort.36 Breast tumors exhibited statistically significantly elevated mRNA expression of PADI2 compared with normal breast tissues with an increasing trend from Luminal A/B, HR-/HER2-enriched to TNBC tumors (Dunn’s multiple comparison test, two-sided p<0.001; figure 1C). Spearman’s correlation analyses between PADI2 mRNA expression and HM450 methylation β values further indicated statistically significant inverse associations for most of the cancer types, suggesting that PADI2 gene expression is in part regulated through DNA methylation (figure 1D).
Figure 1.
PADI family gene and protein expression in various cancer types. (A) Proteomic analysis of PADI family protein expression in whole cell lysates from 196 cancer cell lines stratified by cancer type. Values represent spectral counts of the peptides in common as well as unique sequences that distinct PADI family members. The PADI2 protein expression was highest among other family members and relatively enriched in breast cancer. (B) mRNA levels of PADI family members for 9721 TCGA-derived human tumors consisting of 32 different cancer types. Data were obtained from cBioPortal that were normalized based on autoscaled z-score.30 Concordantly, PADI2 gene expression was highest among other PADI family members. (C) Gene expression of PADI family members in the Curtis et al breast cohort36 for 144 normal breast tissues as well as 1725 breast tumors stratified by hormone receptor subtype. Statistical significance was determined by Dunn’s multiple comparison test, and significant elevation of PADI2 was observed in breast cancer compared with normal control. (D) Heatmap depicting Spearman’s correlation coefficients between PADI2 mRNA expression and HM450 methylation ß values for 32 different cancer types as well as all cancers combined. HM450 methylation data were obtained from cBioPortal.30 TCGA abbreviations: ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. mRNA, messenger RNA; PADI, protein arginine deiminase; TCGA, The Cancer Genome Atlas; NSCLC, Non-Small Cell Lung Cancer; TNBC, Triple Negative Breast Cancer.
Proteomics revealed PADI2-mediated citrullinome in breast cancer
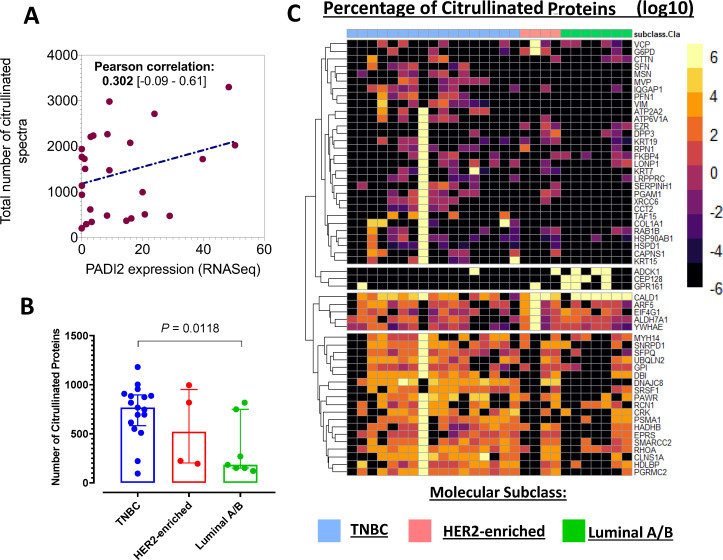
We next examined whether the extent of protein citrullination was reflective of differential expression of PADI2 among the breast cancer subtypes. Global analysis of the protein citrullinome in whole cell lysates of 28 breast cancer cell lines (8 Luminal A/B, 5 Hormone Receptor (HR)-/HER2-enriched, and 15 TNBC) by mass spectrometry with a strict criteria of FDR=0.01 revealed that the total number or citrullinated spectra in the cell lines had positive correlation with PADI2 expression level (Pearson correlation=0.302; figure 2A) and TNBC had significantly higher number of citrullinated proteins compared with Luminal A/B (p=0.0118; figure 2B). The heatmaps in figure 2C demonstrate the discrimination between Luminal A/B, HR-/HER2-enriched and TNBC (analysis of variance, two-sided p<0.05). Of the citrullinated proteins identified in the whole cell lysates, their cellular localization was classified as nucleus (22.8%), cytoplasm (59.6%), plasma membrane (10.5%), and extracellular space (7.0%) based on Ingenuity Pathway Analysis (www.ingenuity.com)37 (online supplemental table S1). We also identified citrullinated vimentin (online supplemental table S1) and citrullinated α-enolase (online supplemental table S2), which are known to occur in autoimmune diseases.38–40
Figure 2.
Hormone receptor specificity of citrullinome in breast cancer cell lines. (A) Scatter plot illustrating the correlation (Pearson correlation, 95% CI) between the total number of citrullinated mass spectra and PADI2 mRNA expression in breast cancer cell lines. (B) Distribution plots illustrating the number of citrullinated proteins in whole cell lysates of breast cancer cell lines stratified by hormone receptor positivity. Statistical significance was determined using Wilcoxon rank-sum test. (C) Heatmap of whole cell lysate citrullinome. Unsupervised heatmaps illustrate the percentage of differentially expressed citrullinated proteins across the hormone receptor subtypes (one-way analysis of variance, two-sided p<0.05). mRNA, messenger RNA; PADI, protein arginine deiminase; TNBC, Triple Negative Breast Cancer.
Positive correlation of PADI2 and citrullinome in TMA analysis
To confirm the association of PADI2 expression with protein citrullination in breast tumor tissues, we assessed PADI2 protein expression and citrullinome by means of IHC in 422 breast tumors (table 1). PADI2 protein expression in the cohort of 422 breast tumors was statistically significantly higher in grade III versus grades I and II tumors (Χ2 test, two-sided p<0.0001), Estrogen Receptor (ER)− (HR-/HER2-enriched and TNBC) versus ER+ (Luminal A/B) (Χ2 test, two-sided p<0.0001), and TNBC versus non-TNBC (Χ2 test, two-sided p<0.0001) (table 1). PADI2 expression was not associated with age or stage in a statistically significant manner (table 1). Staining for peptidyl-citrulline (low+low-moderate vs moderate-high+high) was significantly positively correlated with PADI2 protein expression (OR: 4.45, 95% CI 2.48 to 7.78; Χ2 test, two-sided p<0.0001; table 1, online supplemental figure S3). Staining for PADI2 and peptidyl-citrulline was negative in mammary gland, colon, kidney, liver, lung, stomach, rectum, and esophagus, as well as the tumor’s adjacent normal tissue (online supplemental figure S4).
jitc-2021-002549supp004.pdf (1.6MB, pdf)
jitc-2021-002549supp005.pdf (1.2MB, pdf)
PADI2 is associated with tumor immunophenotype
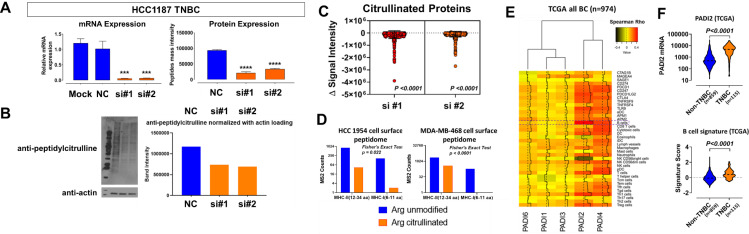
We determined whether PADI2 is a major contributor to protein arginine deamination by knocking down PADI2 with Small interfering RNA (siRNA) in the TNBC HCC1187 cell line and performing global assessment of protein citrullination by immunoblots as well as by mass spectrometry analysis. Knockdown of PADI2 reduced PADI2 mRNA and protein (figure 3A). Immunoblots demonstrated reductions in protein citrullination globally (figure 3B). Mass spectrometry-based analyses similarly demonstrated that citrullinated digested peptides were statistically significantly reduced (paired two-sided t-test, p<0.0001) compared with control (figure 3C). We next evaluated the cell surface HLA-bound peptidome of HR-/HER2-enriched (HCC1954) and TNBC (MDA-MB-468) cell lines as a means to evaluate MHC antigen presentation.26 Specifically, we screened for the presence of citrullinated peptides that were putatively in the MHC class II binding peptide length (12–34 amino acids) or in the MHC class I binding peptide length (6–11 amino acids) with mild acid elution method that can recover class I26 and class II.41 We identified 23 (MHC class II) and 1 (MHC class I) citrullinated peptides in HCC1954 and 126 (MHC class II) and 0 (MHC class I) in MDA-MB-468 (figure 3D, online supplemental table S2), indicating predominance of citrullinated peptides within the MHC class II binding peptide length, compared with the MHC class I binding peptide length (Fisher’s exact test, two-sided p=0.022 for HCC1954 and p<0.0001 for MDA-MB-468) (figure 3D). The citrullinated peptides identified were further searched with the affinity prediction software NetMHCIIpan V.4.1,27 28 considered as unmodified form, and the immunogenicity of actual citrullinated forms was possibly enhanced compared with the predicted result (online supplemental table S2).
Figure 3.
Association between citrullinome and tumor immune response in breast cancer. (A) Relative mRNA and protein expression of PADI2 following siRNA-mediated knockdown of PADI2 in HCC1187 TNBC cell line. Statistical significance was determined by two-sided Student’s t-test: ***p<0.001, ****p<0.0001. (B) Immunoblots for anti-peptidyl-citrulline following siRNA-mediated knockdown of PADI2 in HCC1187 TNBC cell line. Bar plot to the right illustrates densitometry analysis of anti-peptidyl-citrulline normalized against beta-actin. (C) Downregulation of citrullinome following siRNA-mediated knockdown of PADI2 in HCC1187 TNBC cell line. Scatter plots represent the delta in signal intensity of the TMT channels subtracted by siRNA-PADI2-treated cells to si-control; statistical significance was determined by two-sided paired t-test of the citrullinated peptide TMT ratios. (D) Cell surface MHC peptides identified in HCC1954 (HER2-enriched) and MDA-MB-468 (TNBC) breast cancer cell lines. Statistical significance was determined by Fisher’s exact test. The putative MHC class II binding peptide length (12–34 amino acids) containing Arg citrullinated peptides were searched against the NetMHCIIpan V.4.1,27 28 considered as unmodified form, and the binding affinity is presented in online supplemental table S2. (E) Heatmap depicting Spearman’s correlation coefficients between mRNA expression of PADI family members and immune gene signatures in TCGA of all breast cancers (n=974). The broken red line highlights the association between mRNA expression PADI family members and B cell gene-based signatures. PADI2 was strongly positively correlated with B cell gene-based signatures. (F) TCGA-derived gene expression revealed elevated levels of PADI2 and gene signatures of B cells in TNBC (n=115) compared with non-TNBC (Luminal A/B and HER2-enriched combined; n=859) tumors. Statistical significance was determined using two-sided Wilcoxon rank-sum test. mRNA, messenger RNA; TCGA, The Cancer Genome Atlas; TNBC, Triple Negative Breast Cancer; NC, Negative Control; TMT, Tandem Mass Tag; MS2, Tandem mass; MHC, Major Histocompatibility Complex; si, small interfereing.
We next evaluated whether elevated PADI2 expression is associated with a distinct tumor immunophenotype. Using TCGA-derived mRNA expression data sets for 974 breast tumors, we first performed Spearman’s correlation analyses between tumor PADI2 mRNA expression and gene expression profiles of checkpoint blockade-related genes as well as gene expression signatures reflective of immune cell infiltrates.34 Statistically significant positive correlations were observed between PADI2 mRNA and virtually all checkpoint blockade-related genes as well as gene expression signatures reflective of immune cell infiltrates (figure 3E, online supplemental table S3). PADI2 mRNA expression was most positively correlated with a gene signature for B cell infiltrates (p<0.0001, figure 3E; Spearman’s r=0.49 (0.44–0.53), online supplemental table S3). TNBC is considered to be more immunogenic than non-TNBC because of genomic instability and higher rates of mutation.42 We previously reported that TNBC exhibited enhanced immune cell infiltrates compared with non-TNBC tumors.33 The higher expression of PADI2 in TNBC was associated with an enhanced B cell response (p<0.0001) in comparison with non-TNBC (Luminal A/B and HR-/HER2-enriched combined) (figure 3F, online supplemental figure S1). Notably, the association between mutational burden (defined herein as the number of mutation events per case) with mRNA expression of PADI2 (Spearman’s r=0.10 (95% CI −0.05 to 0.26), p=0.17) or B cell gene-based signatures (Spearman’s r=−0.10 (95% CI −0.25 to 0.06), p=0.20) was non-significant in basal-type TNBC tumors (online supplemental figure S1). These findings implicate that the association between PADI2 and B cell gene signatures is independent of mutational burden.
jitc-2021-002549supp002.pdf (1.1MB, pdf)
Citrullinome contributes to B cell tumor immune infiltration and autoantibody elevation in breast cancer
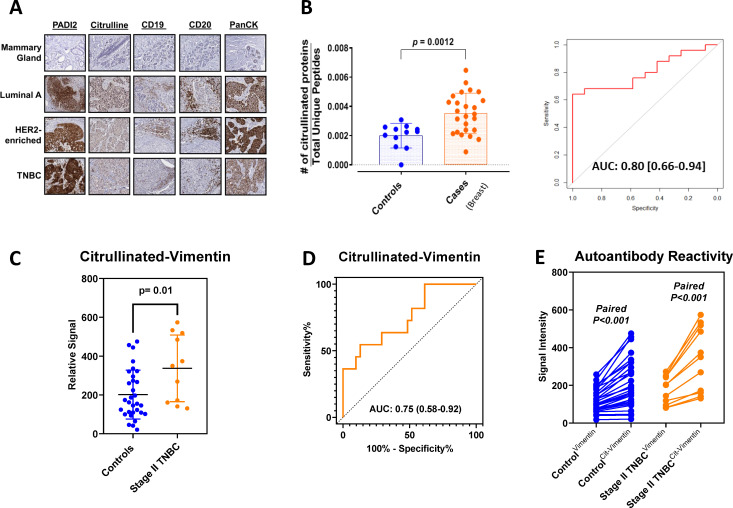
To further confirm the association of PADI2 protein expression with increased tumor protein citrullination and with increased tumor infiltration of B cells, we performed IHC on breast cancer tissue section slides for PADI2, anti-peptidyl-citrulline, pan-cytokeratin, and B cell markers CD19 and CD20 (figure 4A). Our findings confirmed that elevated PADI2 and increased protein citrullination were associated with infiltrating B cells (figure 4A). We therefore aimed to determine whether a tumor B cell response facilitated by PADI2-mediated citrullination would manifest in elevated circulating plasma autoantibodies. We evaluated IgG-bound proteins by mass spectrometry, with 26 plasma pools from 156 patients with breast cancer (10 ER+, 8 HR-/HER2-enriched, 8 TNBC pools) and 11 healthy control pools from 113 cancer-free subjects (online supplemental table S4). The number of IgG-bound citrullinated proteins was statistically significantly higher in plasmas of breast cancer cases compared with controls (Wilcoxon rank-sum test, two-sided p=0.0012, figure 4B, left; Area under the curve (AUC)=0.80, figure 4B, right). IgG-bound citrullinated proteins elevated in patients with breast cancer were further characterized by IPA network analyses. In Luminal A, the top 1 and top 2 networks were cytokeratin complex and estrogen-progesterone-centered networks, whereas in TNBC a cytokeratin complex and an MYC-centered network that included ENO1 were observed (online supplemental figure S5 and table S5). The HR-/HER2-enriched subtype exhibited a cytokeratin complex and an FN1-centered network (online supplemental figure S5 and table S5).
Figure 4.
PADI2-mediated citrullination and B cell tumor infiltration. (A) Representative IHC sections for PADI2, peptidyl-citrulline (citrulline), B cell markers CD19 and CD20, and the tumor marker PanCK in mammary gland and breast tumors stratified by hormone receptor subtype (original magnification ×200). (B) Immunoprecipitated IgG-bound citrullinome identified by mass spectrometry in plasma from subjects with breast cancer. Distribution of citrullinated proteins normalized against total unique peptides (see the Methods section) in 26 pooled plasma samples of newly diagnosed breast cancer consisting of 156 patients and 12 pooled plasma samples corresponding to 113 cancer-free subjects (see online supplemental table S4). Statistical significance was determined by two-sided Student’s t-test: <0.001. The AUC performance of the same cohort. (C) Autoantibody reactivity against citrullinated VIM in individual patient plasma from 11 stage II TNBC cases and 31 healthy controls. The 11 stage II TNBC patient plasmas were the same used for autoantibody reactivity against unmodified and citrullinated VIM by immunoblotting assay (online supplemental figure S6). Statistical significance was determined by two-sided Wilcoxon rank-sum test. (D) Classifier performance (AUC) of citrullinated VIM for distinguishing TNBC cases (n=11) from healthy controls (n=31). (E) Autoantibody reactivity against citrullinated and unmodified VIM in TNBC case (orange) and healthy control (blue) plasmas. Nodes and connecting lines represent matched samples. AUC, Area under the curve; Statistical significance was determined by two-sided paired t-test. IHC, immunohistochemistry; PADI, protein arginine deiminase; PanCK, pan-cytokeratin; TNBC, Triple Negative Breast Cancer; VIM, vimentin.
jitc-2021-002549supp006.pdf (1.3MB, pdf)
jitc-2021-002549supp007.pdf (728.7KB, pdf)
To determine the extent to which autoantibody reactivity is directed against citrulline-containing epitopes, we first used a western blot approach and evaluated differential autoantibody reactivity against citrullinated and non-citrullinated vimentin using plasma pools from patients with newly diagnosed stage II TNBC (n=11 subjects/pool) (online supplemental table S6) and healthy controls (n=8 subjects/pool). We chose to focus on vimentin as our antigen of interest as autoantibody reactivity against vimentin has previously been reported in autoimmune disease and cancer.43–46 The high autoantibody reactivity against citrullinated vimentin was observed in the TNBC stage II patient plasma pool compared with the healthy control pool (online supplemental figure S6). We note that immunoblots using plasma primary autoantibodies can cause considerable background due to reactivity of autoantibodies that are common between healthy control and patients with cancer. Therefore, we additionally developed Luminex autoantibody ELISA assays to test autoantibody reactivity against citrullinated and non-citrullinated vimentin using individual plasmas from patients with newly diagnosed stage II TNBC (n=11) (online supplemental table S6) and healthy controls (n=31). Autoantibody reactivity against citrullinated vimentin was statistically significantly elevated in cases compared with controls (Wilcoxon rank-sum test, two-sided p=0.01, with an AUC of 0.75 (95% CI 0.58 to 0.92); figure 4C, D) and compared with non-citrullinated vimentin among cases (two-sided paired t-test <0.001; figure 4E).
Discussion
We have undertaken a global analysis of PADI family members across cancer types and assessed the extent of citrullinome and its impact on immune response in breast cancer. PADI2-mediated citrullination was found to be strikingly elevated in breast tumor tissue compared with adjacent non-tumor tissue or normal mammary gland tissue and other organ sites. Importantly, we provide compelling evidence that IgG-bound citrullinated proteins are elevated in the plasma of patients with newly diagnosed breast cancer compared with controls, suggesting utility of autoantibodies against citrullinated proteins as cancer biomarkers and utility of citrullinated proteins and peptides as neoantigens.
To date, PADI4 has been the most investigated among family members in the context of cancer. PADI4 is the only PADI member known to encompass a nuclear transport sequence to citrullinate nuclear proteins including histones.47 Our comprehensive analysis revealed that with respect to overall expression levels, PADI2 is preferentially expressed at the protein level in cancer compared with other PADI family members. Expression of PADI2 in breast cancer cell lines was recapitulated in breast cancer TMA. With the anti-PADI2 antibody, 2000-fold dilution was used to ensure PADI2 expression was within the linear range in the tumor tissues; healthy control stains were negative when the same condition was applied. Our proteomic analyses also demonstrated broad protein citrullination among breast cancer cell lines as well as breast tumors. Citrullination is contingent on PADI expression and intracellular Ca2+ levels. In RA, cytosolic Ca2+ levels are increased compared with normal cellular concentrations,48 and in tumors aberrant levels of Ca2+ channels and pumps were expressed,49 which may change the intercellular Ca2+ flux preferable to PADIs. Those providing a rationale for our findings of elevated levels of citrullination by PADI2 in breast cancer. Unsupervised clustering of citrullinated proteins identified both common and distinct signatures between breast cancer molecular subtypes.
In contrast to limited prior studies of protein citrullination in cancer and their impact on immune response, the occurrence of anti-citrulline autoantibodies in RA driven by a B cell response is well documented.5 In our study, PADI2 exhibited statistically significant positive correlations with tumor infiltrating B cells, indicative of similarities with autoimmune disease. MHC analysis of citrullinated peptides in breast cancer cell lines yielded peptides in the MHC-II binding sequence length, supporting a B cell-mediated immune response. Of interest is the occurrence of a substantial fraction of citrullinated proteins as derived from the nuclear compartment, pointing to a similarity between breast cancer and autoimmune diseases, for which elevated levels of circulating antinuclear antibodies are diagnostic markers.50 51 Our proteomic analyses provided strong evidence that IgG-bound citrullinated proteins are statistically significantly elevated in plasmas of patients with newly diagnosed breast cancer as compared with respective controls. Moreover, we provide evidence that the presence of citrulline markedly increases autoantibody reactivity. These findings collectively provide a strong rationale for exploring the utility of circulating autoantibodies against citrullinated proteins for cancer detection.24 25 We recognize that some autoantibodies to citrullinated proteins are likely to be cancer type and subtype specific. Network analysis yielded a cytokeratin complex in common among breast cancer subtypes, whereas distinct features of hormone receptors-centered network were observed in Luminal A and an MYC centered network was observed in TNBC.
Given the current interest in cancer vaccine development and in immunotherapy, our findings point to the importance of citrullination for antigenicity. Interestingly, we identified several citrullinated proteins that have been targeted for vaccine development, including vimentin and α-enolase.12 13 The importance of citrullination of α-enolase in inducing anticancer immunity has been recently demonstrated.13 Our findings suggest broad relevance of citrullination for inducing cancer immunity.
Acknowledgments
We thank all the collaborators in the authors list.
Footnotes
Correction notice: This paper has been updated to amend author name 'Alejandro M Sevillano'.
Contributors: HK, MK, LR, JV, and NP performed the experiments. HK conceived and designed the experiments and wrote the manuscript. HK, MK, and JLi did the IHC image analysis, HK, MK, EI, AS, XM, JV, YC, FH, C-YY, JF, JLo worked on the analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis). FE provided breast cancer patient plasma. The study was supervised by HK, JF, and SH.
Funding: This work was supported by NCI grant U01CA141539, The University of Texas MD Anderson Cancer Center Moonshot Program, The University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment (JFF), and the Little Green Book Foundation.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The study was approved by the Institutional Review Board of MD Anderson Cancer Center.
References
- 1.Malmström V, Catrina AI, Klareskog L. The immunopathogenesis of seropositive rheumatoid arthritis: from triggering to targeting. Nat Rev Immunol 2017;17:60–75. 10.1038/nri.2016.124 [DOI] [PubMed] [Google Scholar]
- 2.Yuzhalin AE. Citrullination in cancer. Cancer Res 2019;79:1274–84. 10.1158/0008-5472.CAN-18-2797 [DOI] [PubMed] [Google Scholar]
- 3.Schellekens GA, de Jong BA, van den Hoogen FH, et al. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest 1998;101:273–81. 10.1172/JCI1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witalison EE, Thompson PR, Hofseth LJ. Protein arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr Drug Targets 2015;16:700–10. 10.2174/1389450116666150202160954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherer HU, Huizinga TWJ, Krönke G, et al. The B cell response to citrullinated antigens in the development of rheumatoid arthritis. Nat Rev Rheumatol 2018;14:157–69. 10.1038/nrrheum.2018.10 [DOI] [PubMed] [Google Scholar]
- 6.Margulies DH. How MHC molecules grab citrullinated peptides to foster rheumatoid arthritis. J Biol Chem 2018;293:3252–3. 10.1074/jbc.H118.001829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alspach E, Lussier DM, Miceli AP, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019;574:696–701. 10.1038/s41586-019-1671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P, Wang D, Yao H, et al. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene 2010;29:3153–62. 10.1038/onc.2010.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Wysocka J, Sayegh J, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004;306:279–83. 10.1126/science.1101400 [DOI] [PubMed] [Google Scholar]
- 10.Guo Q, Fast W. Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J Biol Chem 2011;286:17069–78. 10.1074/jbc.M111.230961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanikawa C, Espinosa M, Suzuki A, et al. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun 2012;3:676. 10.1038/ncomms1676 [DOI] [PubMed] [Google Scholar]
- 12.Brentville VA, Metheringham RL, Gunn B, et al. Citrullinated vimentin presented on MHC-II in tumor cells is a target for CD4+ T-cell-mediated antitumor immunity. Cancer Res 2016;76:548–60. 10.1158/0008-5472.CAN-15-1085 [DOI] [PubMed] [Google Scholar]
- 13.Cook K, Daniels I, Symonds P, et al. Citrullinated α-enolase is an effective target for anti-cancer immunity. Oncoimmunology 2018;7:e1390642. 10.1080/2162402X.2017.1390642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsou P, Katayama H, Ostrin EJ, et al. The emerging role of B cells in tumor immunity. Cancer Res 2016;76:5597–601. 10.1158/0008-5472.CAN-16-0431 [DOI] [PubMed] [Google Scholar]
- 15.Disis ML, Stanton SE. Immunotherapy in breast cancer: an introduction. Breast 2018;37:196–9. 10.1016/j.breast.2017.01.013 [DOI] [PubMed] [Google Scholar]
- 16.Ryan SO, Turner MS, Gariépy J, et al. Tumor antigen epitopes interpreted by the immune system as self or abnormal-self differentially affect cancer vaccine responses. Cancer Res 2010;70:5788–96. 10.1158/0008-5472.CAN-09-4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (time) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derks S, de Klerk LK, Xu X, et al. Characterizing diversity in the tumor-immune microenvironment of distinct subclasses of gastroesophageal adenocarcinomas. Ann Oncol 2020;31:1011–20. 10.1016/j.annonc.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faça VM, Hanash SM. In-depth proteomics to define the cell surface and secretome of ovarian cancer cells and processes of protein shedding. Cancer Res 2009;69:728–30. 10.1158/0008-5472.CAN-08-3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Hanash S. Mass spectrometry based proteomics for absolute quantification of proteins from tumor cells. Methods 2015;81:34–40. 10.1016/j.ymeth.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitteri SJ, JeBailey L, Faça VM, et al. Integrated proteomic analysis of human cancer cells and plasma from tumor bearing mice for ovarian cancer biomarker discovery. PLoS One 2009;4:e7916. 10.1371/journal.pone.0007916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taguchi A, Delgado O, Celiktaş M, et al. Proteomic signatures associated with p53 mutational status in lung adenocarcinoma. Proteomics 2014;14:2750–9. 10.1002/pmic.201400378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama H, Tsou P, Kobayashi M, et al. A plasma protein derived TGFβ signature is a prognostic indicator in triple negative breast cancer. NPJ Precis Oncol 2019;3:10. 10.1038/s41698-019-0082-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama H, Boldt C, Ladd JJ, et al. An autoimmune response signature associated with the development of triple-negative breast cancer reflects disease pathogenesis. Cancer Res 2015;75:3246–54. 10.1158/0008-5472.CAN-15-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ladd JJ, Chao T, Johnson MM, et al. Autoantibody signatures involving glycolysis and splicesome proteins precede a diagnosis of breast cancer among postmenopausal women. Cancer Res 2013;73:1502–13. 10.1158/0008-5472.CAN-12-2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripathi SC, Peters HL, Taguchi A, et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci U S A 2016;113:E1555–64. 10.1073/pnas.1521812113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics 2007;8:238. 10.1186/1471-2105-8-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen KK, Andreatta M, Marcatili P, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018;154:394–406. 10.1111/imm.12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katono K, Sato Y, Jiang S-X, et al. Prognostic significance of MYH9 expression in resected non-small cell lung cancer. PLoS One 2015;10:e0121460. 10.1371/journal.pone.0121460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis C, Shah SP, Chin S-F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6. 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fahrmann JF, Vykoukal J, Fleury A, et al. Association between plasma Diacetylspermine and tumor spermine synthase with outcome in triple-negative breast cancer. J Natl Cancer Inst 2020;112:607–16. 10.1093/jnci/djz182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 35.Chen F, Zhang Y, Bossé D, et al. Pan-urologic cancer genomic subtypes that transcend tissue of origin. Nat Commun 2017;8:199. 10.1038/s41467-017-00289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis C, Shah SP, Chin S-F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krämer A, Green J, Pollard J, et al. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014;30:523–30. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feitsma AL, van der Voort EIH, Franken KLMC, et al. Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis Rheum 2010;62:117–25. 10.1002/art.25059 [DOI] [PubMed] [Google Scholar]
- 39.Lundberg K, Kinloch A, Fisher BA, et al. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum 2008;58:3009–19. 10.1002/art.23936 [DOI] [PubMed] [Google Scholar]
- 40.Kinloch A, Tatzer V, Wait R, et al. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res Ther 2005;7:R1421–9. 10.1186/ar1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capello M, Vykoukal JV, Katayama H, et al. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat Commun 2019;10:254. 10.1038/s41467-018-08109-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwa MJ, Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): where to go from here. Cancer 2018;124:2086–103. 10.1002/cncr.31272 [DOI] [PubMed] [Google Scholar]
- 43.Hong S-H, Misek DE, Wang H, et al. Identification of a specific vimentin isoform that induces an antibody response in pancreatic cancer. Biomark Insights 2006;1:117727190600100–83. 10.1177/117727190600100006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bretherton L, Toh BH, Jack I. Igm autoantibody to intermediate filaments in Mycoplasma pneumoniae infections. Clin Immunol Immunopathol 1981;18:425–30. 10.1016/0090-1229(81)90135-5 [DOI] [PubMed] [Google Scholar]
- 45.Alcover A, Molano J, Renart J, et al. Antibodies to vimentin intermediate filaments in sera from patients with systemic lupus erythematosus. Arthritis Rheum 1984;27:922–8. 10.1002/art.1780270812 [DOI] [PubMed] [Google Scholar]
- 46.Kurki P, Helve T, Virtanen I. Antibodies to cytoplasmic intermediate filaments in rheumatic diseases. J Rheumatol 1983;10:558–62. [PubMed] [Google Scholar]
- 47.Stadler SC, Vincent CT, Fedorov VD, et al. Dysregulation of PAD4-mediated citrullination of nuclear GSK3β activates TGF-β signaling and induces epithelial-to-mesenchymal transition in breast cancer cells. Proc Natl Acad Sci U S A 2013;110:11851–6. 10.1073/pnas.1308362110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Venrooij WJ, van Beers JJBC, Pruijn GJM. Anti-CCP antibodies: the past, the present and the future. Nat Rev Rheumatol 2011;7:391–8. 10.1038/nrrheum.2011.76 [DOI] [PubMed] [Google Scholar]
- 49.Monteith GR, McAndrew D, Faddy HM, et al. Calcium and cancer: targeting Ca2+ transport. Nat Rev Cancer 2007;7:519–30. 10.1038/nrc2171 [DOI] [PubMed] [Google Scholar]
- 50.Friou GJ, Finch SC, Detre KD. Interaction of nuclei and globulin from lupus erythematosis serum demonstrated with fluorescent antibody. J Immunol 1958;80:324–9. [PubMed] [Google Scholar]
- 51.Vlagea A, Falagan S, Gutiérrez-Gutiérrez G, et al. Antinuclear antibodies and cancer: a literature review. Crit Rev Oncol Hematol 2018;127:42–9. 10.1016/j.critrevonc.2018.05.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-002549supp001.pdf (246.3KB, pdf)
jitc-2021-002549supp003.pdf (931.6KB, pdf)
jitc-2021-002549supp004.pdf (1.6MB, pdf)
jitc-2021-002549supp005.pdf (1.2MB, pdf)
jitc-2021-002549supp002.pdf (1.1MB, pdf)
jitc-2021-002549supp006.pdf (1.3MB, pdf)
jitc-2021-002549supp007.pdf (728.7KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.