SUMMARY
Given the immense antigenic load present in the microbiome, we hypothesized that microbiota mimotopes can be a persistent trigger in human autoimmunity via cross-reactivity. Using antiphospholipid syndrome (APS) as a model, we demonstrate cross-reactivity between non-orthologous mimotopes expressed by a common human gut commensal, Roseburia intestinalis (R. int), and T and B cell autoepitopes in the APS autoantigen β2-glycoprotein I (β2GPI). Autoantigen-reactive CD4+ memory T cell clones and an APS-derived, pathogenic monoclonal antibody cross-reacted with R. int mimotopes. Core-sequence-dependent anti-R. int mimotope IgG titers were significantly elevated in APS patients and correlated with anti-β2GPI IgG autoantibodies. R. int immunization of mice induced β2GPI-specific lymphocytes and autoantibodies. Oral gavage of susceptible mice with R. int induced anti-human β2GPI autoantibodies and autoimmune pathologies. Together, these data support a role for non-orthologous commensal-host cross-reactivity in the development and persistence of autoimmunity in APS, which may apply more broadly to human autoimmune disease.
INTRODUCTION
Commensal microorganisms chronically colonize host barrier sites and impact a broad range of human physiology (Cho and Blaser, 2012). Commensal-host interactions with the mucosal and systemic immune systems have evolved over millennia (O’Hara and Shanahan, 2006). Microbial antigens are continuously sampled by the mucosal immune system and presented to adaptive immune cells that can mount local and systemic responses (Hand et al., 2012; Hegazy et al., 2017; Ladinsky et al., 2019; Zeng et al., 2016). Commensal-host immune interactions are implicated in the development of autoimmunity via bystander activation, cross-reactivity, dual antigen receptors, and epitope spreading (Ost and Round, 2018; Ruff and Kriegel, 2015). Because the particular antigens presented by innate immune cells to antigen-specific T cells are dictated by polymorphisms in major histocompatibility (MHC) genes, host-commensal cross-reactivity may contribute to the development and persistence of human autoimmunity in genetically predisposed individuals. Given the enormous antigenic load of the human gut microbiome, which is encoded by over 9.8 million non-redundant genes (Li et al., 2014; Qin et al., 2010), we hypothesized that chronic exposure of the gut immune system to non-orthologous, commensal mimotopes generates and sustains human autoimmune disease via cross-reactivity.
We utilized antiphospholipid syndrome (APS) as a model of systemic autoimmunity with well-characterized autoepitopes to test for commensal mimotope cross-reactivity. The common autoantigen β2-glycoprotein I (β2GPI), also known as apolipoprotein H, is targeted in the majority of APS patients (Ruiz-Irastorza et al., 2010). It contains five domains and circulates at high levels in human plasma (Lozier et al., 1984; Polz and Kostner, 1979). T cell-dependent autoantibodies against β2GPI lead to autoimmune clotting events and obstetric complications (Garcia and Erkan, 2018; Giannakopoulos and Krilis, 2013). Widespread thrombotic events can be lethal and occur also in patients with other systemic rheumatic diseases such as lupus (Garcia and Erkan, 2018; Giannakopoulos and Krilis, 2013; Rauch etal., 2018). A thoroughly characterized CD4+ T cell epitope in domain V (DV) of β2GPI is p276-290 (KVSFFCKNKEKKCSY), which is restricted by the human leukocyte antigen polymorphism HLA-DRB4*0103 (serotype DR53) (Arai et al., 2001; Kuwana et al., 2005). The major B cell autoepitope resides in domain I (DI) of β2GPI, the arginine-rich R39-R43 (RGGMR) sequence, and is strongly associated with thrombosis (de Laat et al., 2005, 2006; Ioannou et al., 2007; Iverson et al., 1998; Mahler et al., 2016; Pericleous et al., 2015). Focusing on commensals with mimotopes to both T cell (p276-290) and B cell (R39-R43) epitopes within β2GPI, we identified the prevalent, immunogenic human gut commensal Roseburia intestinalis (R. int) as a cross-reactive trigger in APS patients. We provide in vitro and in vivo evidence supporting T and B cell cross-reactivity between human β2GPI autoepitopes and R. int mimotopes. We show that human, gut-tropic, β2GPI-reactive memory CD4+ Th1 cell clones cross-react with R. int and mimotope peptides. Further, an APS-derived, pathogenic autoepitope-specific antibody binds to a mimotope in a bacterial DNA methyltransferase expressed by R. int (R. int DNMT). Consistent with these findings, APS patients have significantly elevated levels of anti-R. int DNMT IgG, which positively correlate with anti-β2GPI IgG and are dependent on the core sequence based on mutagenesis studies. Moreover, immunization of BALB/c mice with R. int induces autoepitope-specific cross-reactivity to human β2GPI. Finally, oral gavage of R. int into the spontaneous APS mouse model, (NZW x BXSB)F1 mice, induced significantly elevated anti-human β2GPI IgG autoantibodies and thrombotic events. These proof-of-concept studies support cross-reactivity between non-orthologous commensal mimotopes and autoepitopes in genetically susceptible individuals as a potential mechanism sustaining chronic autoimmunity in humans.
RESULTS
APS Patients Exhibit Signs of Gut Inflammation with Systemic Adaptive Immune Responses to β2-Glycoprotein I Mimotope-Expressing Roseburia intestinalis
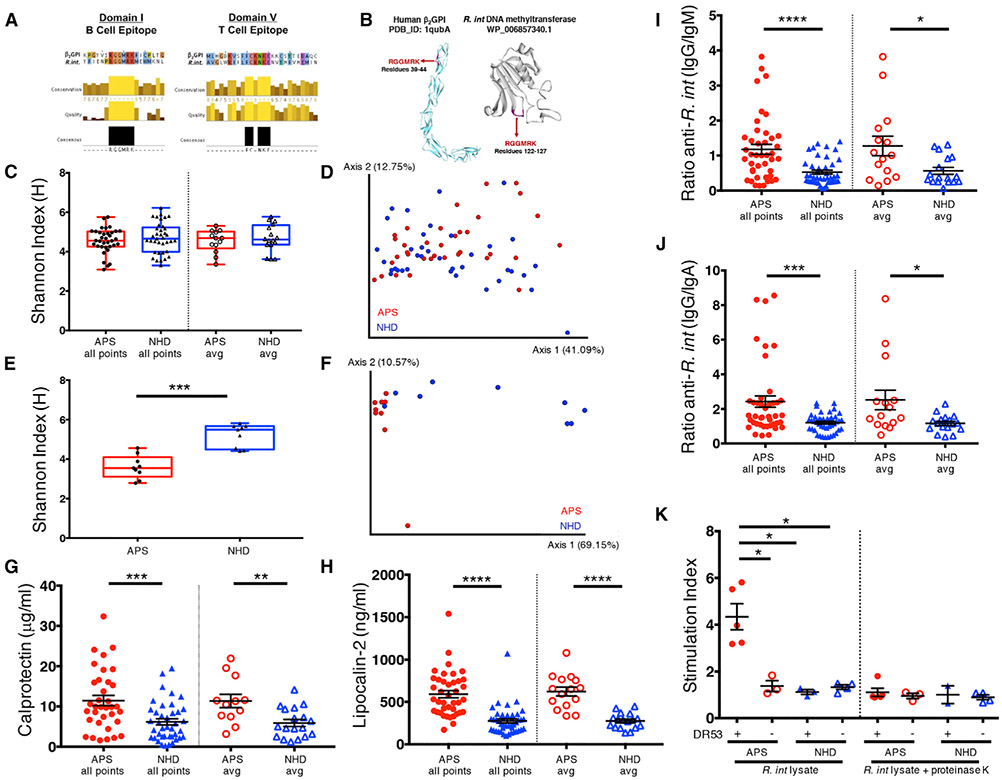
We aligned the HLA-DRB4*0103 (DR53)-restricted β2GPI-immunodominant CD4+ T cell epitope p276-290 (KVSFFCKNKEKKCSY) (Arai et al., 2001; Kuwana et al., 2005) and the major APS B cell autoepitope in β2GPI, R39-R43 (RGGMR) (de Laat et al., 2005; Ioannou et al., 2007; Iverson et al., 1998) to bacterial proteins in non-redundant protein databases (Altschul et al., 1990). Candidate sequences were cross-referenced to identify human commensal gut bacteria containing homology to both of these well-defined T and B cell autoepitopes. Excluding pathogens that are expected to represent mainly transient stimuli, we identified R. int as a common, human colonic bacterium containing proteins with highly homologous amino acid sequences to the CD4+ T cell β2GPI-immunodominant epitope p276-290 (GenBank: EEU99424.1) in DV and the core region (R39-R43) of the major B cell autoepitope in DI of β2GPI (NCBI Reference Sequence: WP_118597735.1) (Figure 1A). Further, the B cell mimotope was predicted by in silico modeling to be exposed, indicating a potential antibody-binding site within R. int DNMT (Figure 1B).
Figure 1. APS Patients Exhibit Signs of Gut Inflammation with Systemic Adaptive Immune Responses to β2-Glycoprotein I-Mimotope-Expressing Roseburia intestinalis.
(A) Clustal Omega alignment of β2GPI B cell domain I epitope (left panel) aligned to Roseburia intestinalis L1-82 (R. int) mimotope (WP_118597735.1). Clustal Omega alignment of HLA-DRB4*0103-restricted immunodominant T cell domain V epitope within β2GPI (p276-290, KVSFFCKNKEKKCSY) (right panel) aligned to R. int mimotope (EEU99424.1).
(B) In silico modeling of the core domain I epitope R39-R43 exposed on human β2GPI (PDB_ID: 1qubA) and R. int mimotope (SWISS-MODEL, WP_118597735.1). (C–F) 16S rRNA sequencing (C and D) and IgA-seq (E and F) was performed as described in STAR Methods.
(C) Alpha diversity between APS (n = 35) and NHD (n = 37) by Shannon-Weiner diversity index for all time points and the average of visits from each patient.
(D) Principal-coordinate analysis of weighted UniFrac distances shows no difference (PERMANOVA 999 permutations, p = 0.164).
(E) IgA-coated fraction of fecal bacteria have decreased Shannon-Weiner diversity in APS (n = 9) compared to NHD (n = 9), p = 0.0003.
(F) IgA-coated fraction of fecal bacteria have significantly different weighted UniFrac diversity (PERMANOVA 999 permutations, p = 0.001).
(G) Elevated fecal calprotectin in APS (all time points n = 38, p = 0.0002; average n = 13, p = 0.002) compared to NHD (all time points n = 40, average n = 18).
(H) Elevated plasma lipocalin-2 in APS (all time points n = 42, p = < 0.0001; average n = 15, p = < 0.0001) compared to NHD (all time points n = 43, average n = 17).
(I) Ratio of plasma anti-R. int IgG to anti-R. int IgM is increased in APS (all time points n = 42, p < 0.0001; average n = 15, p = 0.024) compared to NHD (all time points n = 43; average n = 17, p = 0.04).
(J) Ratio of plasma anti-R. int IgG to anti-R. int IgA is increased in APS (all time points n = 42, p = 0.0008; average n = 15, p = 0.022) compared to NHD (all time points n = 43; average n = 17).
(K) Peripheral blood mononuclear cells from HLA-DRB4*01 (DR53)+ APS (n = 5) proliferate more in response to R. int lysate compared to HLA-DRB4*01 (DR53)− APS (n = 3, p = 0.036), NHD HLA-DRB4*01 (DR53)+ (n = 3, p = 0.036), and NHD HLA-DRB4*01 (DR53)− (n = 4, p = 0.0016) donors, respectively. Treatment of lysates with proteinase K abrogated the response. Counts per minute (CPM) as measured by 3[H]-thymidine incorporation was used to calculate proliferation with averages of triplicates shown as single points. Stimulation index was calculated as CPM of stimulated PBMCs divided by CPM of unstimulated PMBCs. All points, all time points; avg, average; APS, antiphospholipid syndrome; NHD, normal healthy donors. Two-tailed Mann-Whitney U test was performed unless noted. Error bars represent ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We enrolled 15 subjects positive for persistent moderate-to-high-titer β2GPI IgG autoantibodies (11 APS patients and 4 individuals at risk for APS, defined as APS hereafter) and 20 healthy donors (defined as normal healthy donors [NHD] hereafter). Study subjects donated fecal samples, whole blood, and plasma at up to three longitudinal visits separated by 1 month each to capture microbial and immune marker variability over time. Health status, medications, 24-h diet histories, and APS-relevant HLA and autoantibody status were collected over time (Tables 1 and S1).
Table 1.
Cohort Information
| Subject ID | Age | Sex | Diagnosis | Anti-β2GPI | Anti-domain I β2GPI |
Arterial Events |
Venous Events |
Pregnancy comorbidities |
R. int | Anti-R. int DNMT IgG |
|---|---|---|---|---|---|---|---|---|---|---|
| APS01 | 27 | F | APS + SLE | positive | positive | No | no | HELLP, miscarriages | + | +++ |
| APS02 | 41 | F | APS + SLE | positive | negative | CVA | no | no | + | + |
| APS03 | 61 | M | APS | positive | positive | CVA | no | no | − | + |
| APS04 | 56 | F | APS | positive | positive | CVA | no | no | + | + |
| APS05 | 49 | F | APS | positive | positive | TIA, CVA | PE × 2, DVT × 2 | no | ++ | + |
| APS06 | 66 | F | APS | positive | negative | stroke | cortical vein thrombosis | no | + | + |
| APS07 | 38 | F | triple-positive | positive | positive | NA | NA | no | + | + |
| APS08 | 47 | F | triple-positive | positive | negative | NA | NA | no | + | + |
| APS09 | 55 | F | APS | positive | positive | TIAs | no | placental blood clots | + | + |
| APS10 | 47 | M | APS | positive | positive | No | DVT and PE | no | − | ++ |
| APS11 | 70 | F | single-positive | positive | negative | NA | NA | no | + | + |
| APS12 | 60 | M | APS | positive | positive | No | DVT and PE | no | − | + |
| APS13 | 45 | M | APS | positive | positive | No | DVT | no | +++ | ++ |
| APS14 | 50 | F | triple-positive + SLE | positive | negative | NA | NA | no | + | + |
| APS15 | 40 | M | APS | positive | negative | stroke | DVT and PE | no | + | ++ |
| NHD01 | 42 | F | healthy | negative | negative | no | no | no | +++ | + |
| NHD02 | 50 | F | healthy | negative | negative | no | no | no | − | + |
| NHD03 | 49 | F | healthy | negative | negative | no | no | no | + | − |
| NHD04 | 23 | M | healthy | negative | negative | no | no | no | + | − |
| NHD05 | 40 | M | healthy | negative | negative | no | no | no | + | − |
| NHD06 | 29 | M | healthy | negative | negative | no | no | no | + | − |
| NHD07 | 23 | F | healthy | negative | negative | no | no | no | − | + |
| NHD08 | 32 | F | healthy | negative | negative | no | no | no | + | ++ |
| NHD09 | 60 | F | healthy | negative | negative | no | no | no | + | + |
| NHD10 | 48 | F | healthy | negative | negative | no | no | no | + | − |
| NHD11 | 42 | M | healthy | negative | negative | no | no | no | + | + |
| NHD12 | 45 | F | healthy | negative | negative | no | no | no | + | + |
| NHD13 | 21 | F | healthy | negative | negative | no | no | no | + | + |
| NHD14 | 50 | F | healthy | negative | negative | no | no | no | + | + |
| NHD15 | 31 | F | healthy | negative | negative | no | no | no | + | + |
| NHD16 | 55 | F | healthy | negative | negative | no | no | no | + | + |
| NHD17 | 29 | F | healthy | negative | negative | no | no | no | + | + |
| NHD18 | 29 | F | healthy | negative | negative | no | no | no | + | + |
| NHD19 | 47 | F | healthy | negative | negative | no | no | no | ++ | ++ |
| NHD20 | 55 | F | healthy | negative | negative | no | no | no | ++ | + |
Study cohort information. APS, antiphospholipid syndrome or at risk (single- or triple-positive, i.e., anti-β2GPI, aCL, LA; see Table S1); SLE, systemic lupus erythematosus; NHD, normal healthy donor; anti-β2GPI, anti-β2GPI IgG autoantibodies (negative < 40.0 chemiluminescent units (CU), positive > 40.0 CU); anti-domain I β2GPI, anti-β2GPI domain I-specific IgG autoantibodies (negative < 20 CU, positive > 20 CU); R. int, R. int qPCR relative levels; anti-R. int DNMT IgG, anti-R. int DNA methyltransferase IgG; values were determined to be + when between −1 and 1 SD from the mean, ++ > 1 SD, +++ > 2 SD, − < −1 SD; HELLP, hemolysis elevated liver enzymes low platelets; CVA, cerebrovascular accident; TIA, transient ischemic attack; DVT, deep vein thrombosis; PE, pulmonary embolism; NA, not assessed.
To determine overall gut microbial community structure, sequencing of the 16S ribosomal RNA (rRNA) V4 region was performed on fecal DNA (Cullen et al., 2015; Kozich et al., 2013). Compared to NHDs on average and across time, overall taxonomy, alpha diversity as measured by Shannon-Weiner diversity index, and beta diversity as measured by principal-coordinate analysis of weighted UniFrac distances were not different in APS patients (Figures 1C, 1D, and S1A-S1E). Similarly, the relative abundance of Roseburia at the genus level was not significantly different in APS patients (Figure S1F), supporting its broad prevalence in human gut microbiomes. Next, we determined IgA coating of the gut microbiota using fluorescence-activated cell sorting (FACS). IgA coating varied across time (Figure S1G) with a trend toward being increased in APS patients compared to NHDs (Figure S1H). Furthermore, sorting of IgA-coated fractions followed by 16S rRNA sequencing, so called IgA-seq (Palm et al., 2014), was performed on a limited cohort. IgA-coated fractions differed in taxonomy with significantly lower alpha diversity and different beta diversity in APS patients compared to NHDs (Figures 1E, 1F, and S2). IgA coating of the genus Roseburia was not different by IgA-coating index (ICI) (Figure S3), but this method does not capture species-specific alterations in IgA coating. Using R. int species-specific qPCR primers (Figure S4A), R. int was detected in 13 of 15 APS patients and positive in 86.7% (n = 44) of all visits. Similarly, R. int was present in 18 of 20 healthy donors and positive in 88.6% (n = 48) of all visits (Figure S4B). These data support that while gut microbial community structure is not significantly different in APS compared to NHD, gut mucosal IgA targets are contracted in APS patients and that R. int is widely prevalent in the gut microbiome of our cohort.
We next examined if subclinical intestinal and corresponding systemic inflammation is present in APS patients, with the hypothesis that the contracted mucosal IgA response may associate with a more pronounced systemic IgG response toward certain gut microbiota. Using the surrogate fecal marker of inflammation, calprotectin (Berstad et al., 2000; Konikoff and Denson, 2006), and a marker of intestinal-associated systemic inflammation, lipocalin-2 (Chassaing et al., 2012; Vijay-Kumar et al., 2010), we compared signs of inflammation between APS and NHD. For fecal calprotectin measurements, we excluded individuals taking proton pump inhibitors and non-steroidal anti-inflammatory drugs because these interventions are associated with increased fecal calprotectin levels (Poullis et al., 2003; Tibble et al., 1999). Fecal calprotectin was significantly elevated in APS compared to NHD and did not change across time within individuals (Figures 1G and S4C). Moreover, plasma lipocalin-2 was significantly elevated in APS patients and did not differ across time (Figures 1H and S4D). These data support that chronic, subclinical intestinal and peripheral inflammation is present in APS patients, possibly allowing for cross-reactive immune responses to commensal bacteria to spread systemically.
To determine if R. int is recognized outside of the gut, we determined the highest expression of both R. int-mimotope-containing genes. Both genes displayed variable expression by qPCR throughout a 24-h growth curve, with maximal expression during the late-exponential growth phase (Figure S4E). Using whole R. int or the soluble fraction of lysates from cultures grown to late-exponential phase, we examined peripheral antibody and peripheral blood mononuclear cell (PBMC) responses to R. int. Using bacterial FACS, we determined plasma antibody coating of R. int. APS patients had significantly higher ratios of anti-R. int IgG than anti-R. int IgM or IgA, respectively (Figures 1I and 1J). These ratios did not change over time (Figures S4F and S4G). Moreover, PBMCs from HLA-DRB4*01 (DR53)-positive APS subjects but not HLA-DRB4*01 (DR53)-negative APS or NHDs vigorously proliferated in response to R. int proteins (Figure 1K). These data extend recent data that R. int is capable of eliciting a peripheral immune response in humans (Hegazy et al., 2017). Furthermore, these data support that T cells from APS patients respond systemically to R. int in an HLA-dependent manner. Together with an IgG isotype-skewed response toward R. int, these results suggest adaptive T and B cell reactivity against this species in APS patients as occurs also toward other Roseburia species in autoimmune-prone humans (Paun et al., 2019). To investigate mimotope cross-reactivity in APS, we examined memory CD4+ T cells specific for the β2GPI-immuno-dominant epitope p276-290.
β2GPI-Reactive Memory CD4+ Th1 Cells from APS Patients Cross-React with the Corresponding R. int Mimotope
To investigate human CD4+ T cell cross-reactivity with R. int, we cloned β2GPI-specific T cells from HLA-DRB4*01+ APS PBMCs using a T cell library method (Geiger et al., 2009). We sorted CD4+, CD45RA−, CD45RO+, CD25−, β7 integrin+ memory T cells from patients into CCR6+/− populations. Integrin β7 is a marker of mucosa-homing lymphocytes, and CCR6 is enriched in Th17 cells linked with human autoimmunity (Cao et al., 2015; Farstad et al., 1996; Geiger et al., 2009). We identified β2GPI-reactive T cells in both the CCR6− and CCR6+ populations (Figures S5A and S5B). β2GPI p276-290-reactive gut homing, CCR6− memory T cells but not CCR6+ clones cross-reacted with the R. int p276-290 mimotope (Figures 2A). The cross-reactive clones displayed a Th1-like phenotype, as shown by secretion of IFN-γ and GM-CSF (Figures 2B and 2C), which has also been linked to a pathogenic Th1 response in human autoimmunity (Noster et al., 2014). We noted increased IL-2 and IL-4 in β2GPI p276-290-stimulated clones but not mimotope clones, with no significant increase in IL-10 or IL-17A, consistent with the lack of CCR6 expression in cross-reactive clones (Figures S5C-S5F). These findings support peptide cross-reactivity between the disease-relevant T cell-β2GPI DV peptide and the R. int mimotope. Cross-reactivity of these particular clones occurred within the Th1 and not Th17 pool, the former having been previously linked to APS (Benagiano et al., 2017; Salem et al., 2015).
Figure 2. β2GPI-Reactive Memory CD4+ T Cells from APS Patients Cross-React with the Corresponding R. int Mimotope.

(A–C) β2GPI p276-290 (DV epitope)-specific CD4+, CCR6−, β7+ memory T cells (n = 2, p < 0.001) cross-react with R. int mimotope peptide (DV mimotope) in (A) proliferation assays (n = 2, p < 0.0087). These clones secrete (B) GM-CSF (n = 10, p < 0.023) and (C) IFN-γ (n = 8, p < 0.001) in the culture supernatant 72 h post-antigen stimulation. (D–F) Representative single cell-sorted β2GPI p276-290, DRB4*0103 MHC class II tetramer-positive CD4+, CD45RA−, CD45RO+, and CCR6− T cell clones that proliferate (D) significantly (n = 3, p < 0.05) in response to whole, heat-killed R. int, DV tetramer epitope (n = 2, p < 0.005), and β2GPI (n = 2, p < 0.0042). Phytohemagglutinin (PHA) plus rhIL-2 serve as positive control. Clones secrete (E) GM-CSF (n = 4, p < 0.002) and (F) IFN-γ (n = 3, p < 0.0001). CPM measured by 3[H]-thymidine incorporation with averages of triplicates shown. Tetramer proliferation was determined using ATP luminescence. Fold increase determined by dividing the average of triplicates from individual clones by the average of the unstimulated (“no antigen”) clones in relative light units (RLU). Unpaired, two-tailed Student’s t test. Error bars represent ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p< 0.0001.
To further investigate APS CD4+ T cell cross-reactivity, we synthesized a DRB4*0103 MHC class II tetramer specific for β2GPI p276-290. To limit disulfide bond formation when producing the tetramer, we substituted cytosine at position 13 with alanine (C13A, KVSFFCKNKEKKASY). Tetramer-specific CD4+, CD45RA−, CD45RO+, CD25−, and CCR6+/− memory T cells were isolated by FACS. Tetramer-positive cells were expanded and tested for reactivity to both the tetramer peptide and to whole heat-killed R. int, which we used to determine if physiologically processed peptides presented on MHC II would be sufficient to induce cross-reactivity similar to PBMCs shown in Figure 2A. We identified tetramer-positive clones that showed significant responses to R. int (Figure 2D). Tetramer-positive, R. int cross-reactive clones showed a similar Th1-like (IFN-γ+ and GM-CSF+) phenotype consistent with the phenotype of clones identified using the T cell library method (Figures 2E, 2F, and S5G-S5J). These data from two different approaches confirmed that APS-derived cross-reactive memory CD4+ T cells have the capacity to produce proinflammatory cytokines and are polarized toward a Th1 response in vitro.
A Pathogenic, APS-Derived β2GPI R39-R43-Specific Autoantibody Cross-Reacts with R. int DNMT
To test autoantibody cross-reactivity, we cloned two APS-derived, lupus anticoagulant (LA)-inducing autoantibodies, previously demonstrated to bind to β2GPI DI, as full-length human IgG1 antibodies (Dienava-Verdoold et al., 2011; Pelkmans et al., 2013). Clone P1-117 binds to a discontinuous DI-DII epitope containing the major R39-R43 epitope, whereas clone P2-6 binds to an unrelated DI epitope outside of the R39-R43 sequence (Dienava-Verdoold et al., 2011; Pelkmans et al., 2013). We confirmed P1-117 DI interaction by ELISA and bio-layer interferometry (Figures 3A and S6A-S6C). Supporting cross-reactivity of an R39-R43-specific mAb with R. int, we found that P1-117, but not P2-6, bound to R. int lysates similarly to serum from mice immunized with R. int (Figure 3B). Finally, we show that P1-117 bound to the R. int DNMT mimotope compared to a mutant version with RGGMR substituted with alanines (R. int DNMTΔ122-126) (Figures 3C and S6D). To confirm the specificity of these interactions using bio-layer interferometry, we screened recombinant human insulin (rhInsulin) as an unrelated human autoantigen, which is typically employed in polyreactivity assays (Tiller et al., 2007). Compared to β2GPI or R. int DNMT, we found very weak interactions between rhInsulin and P1-117, as reflected by a KD value of 5,800 nM (Figure S6E).
Figure 3. A Pathogenic, APS-Derived β2GPI R39-R43-Specific Autoantibody Cross-Reacts with R. int DNA Methyltransferase.
APS-derived monoclonal antibodies P1-117 and P2-6 were cloned as full-length human IgG1 and purified. Full-length mature rhβ2GPI, a mutant version containing alanines in positions R39-R43 (rhβ2GPIΔ39-43), R. int DNA methyltransferase (R. int DNMT, WP_118597735.1), and a mutant version containing alanines in positions R122-R126 (R. int DNMTΔ122-126) were expressed and purified.
(A) Representative ELISA titration curve showing P1-117 binds to R39-R43 within domain I (DI) of β2GPI.
(B) Representative R. int ELISA showing P1-117, P2-6 negative control (binding domain I outside R39-R43), R. int-immunized, and sham-immunized mouse sera, respectively. X-axis shows serum dilutions (1:100 serum = 10 μg/mL antibody) in 2-fold dilutions.
(C) Representative ELISA titration curve of P1-117 and P2-6 binding to R. int DNMT or R. int DNMTΔ122-126, respectively.
(D) Human trophoblast cells were treated with media, P1-117 (50 μg/mL) or IgG isotype control (50 μg/mL). After 48 h, cell migration was measured (n = 4, *p < 0.05).
(E and F) Human endometrial endothelial cells (red) were plated on Matrigel overnight to allow tube formation after which trophoblast cells (green) were added with media alone, P1-117 (50 μg/mL) or IgG isotype control (50 μg/mL), and then co-cultured for 48 h.
(E) Representative images from each experiment (n = 3) are shown.
(F) Number of tubes counted per field (n = 3, *p < 0.05). Panels A–C, two-way ANOVA with Bonferroni correction. D, one-way ANOVA with Tukey’s multiple comparison test. E, Friedman test with Dunn’s multiple comparison test. Error bars represent ± SEM. *p < 0.05, ***p < 0.001, ****p < 0.0001.
To further validate the pathogenic nature of LA-inducing P1-117, we performed in vitro assays testing the ability of P1-117 to induce human placental trophoblast abnormalities, which is one of the mechanisms how β2GPI autoantibodies mediate obstetric complications in pregnant women with APS such as pre-eclampsia and abortions (Abrahams et al., 2017; Garcia and Erkan, 2018; Giannakopoulos and Krilis, 2013). P1-117 reduced trophoblast migration by 67.5% ± 3.6% when compared to media control, and by 60.0% ± 5.9% when compared to IgG isotype control (Figure 3D). This reduction in the ability of trophoblast cells to migrate was not due to the P1-117 mAb affecting cell viability (P1-117 viability 96.6% ± 1.67). Using an in vitro model of spiral artery transformation (Alvarez et al., 2015), the presence of P1-117 disrupted trophoblast-endothelial interactions and tube stability, resulting in a 35.7 ± 2.8% decrease in the number of tube-like structures when compared to media control, and also a significant 40.1 ± 2.4% decrease in tube-like structures when compared to IgG isotype control (Figures 3E and 3F). Together, these results support that P1-117 is pathogenic and cross-reactive with R. int, interacting with the RGGMR core epitope in both β2GPI and R. int DNMT.
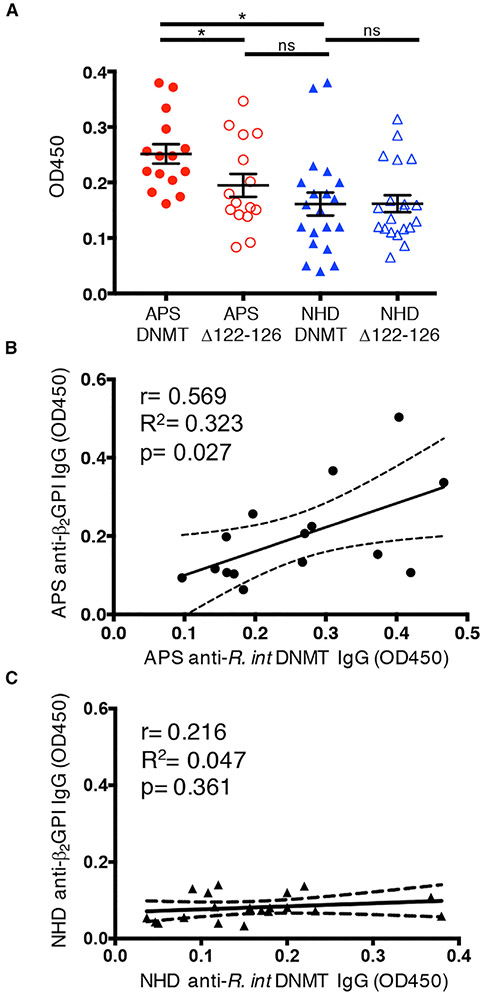
Moreover, APS patients had elevated levels of anti-R. int DNMT IgG compared to NHDs (Figure 4A). When comparing the mutant R. int DNMTΔ122-126 with wild-type R. int DNMT, anti-DNMT IgG levels in APS patients were decreased (Figure 4A). We did not observe a significant loss of binding in healthy donors (Figure 4A), suggesting that APS patients but not healthy subjects had antibodies specific for the mimotope. Anti-R. int DNMT and anti-R.int DNMTΔ122-126 IgG levels remained consistent across time (Figures S7A and S7B). Furthermore, plasma anti-R. int DNMT IgG levels significantly correlated with anti-β2GPI IgG levels in APS patients but not in NHDs (Figures 4B and 4C). Providing additional support for epitope specificity, significant correlation to anti-β2GPI IgG was lost when comparing anti-β2GPI IgG with anti-R. int DNMTΔ122-126 IgG levels (Figure S7C). These data reveal a robust correlation between anti-R. int DNMT IgG and anti-β2GPI IgG titers ex vivo, which is lost when the RGGMR mimic sequence is mutated in R. int DNMT.
Figure 4. APS Patients Have Significantly Elevated Levels of Anti-R. int DNMT IgG that Positively Correlate with Anti-β2GPI IgG Autoantibodies.
ELISAs of R. int DNMT and R. int DNMTΔ122-126 plasma diluted 1:1000 and probed for IgG.
(A) Averages from 2-3 time points are shown as individual points. APS patients have significantly higher anti-R. int DNMT IgG (n = 15, p = 0.0112) compared to NHD (n = 20). APS patient plasma contains significantly less anti-R. int DNMTΔ122-126 IgG compared to wild-type DNMT (n = 15, p = 0.0103). APS plasma anti-R. int DNMTΔ122-126 IgG values were not significantly different from NHD plasma anti-R. int DNMT IgG or anti-R. int DNMTΔ122-126 IgG, respectively (n = 20 each).
(B) Average APS anti-R. int DNMT IgG levels (x axis) significantly correlate with increasing anti-β2GPI IgG (y axis). Pearson r = 0.569, R2 = 0.323, p = 0.0271 two-tailed.
(C) Average NHD R. int DNMT IgG levels (x axis) do not correlate with average anti-β2GPI IgG (y axis). Pearson r = 0.216, R2 = 0.047, p = 0.361 two-tailed. Two-tailed Mann-Whitney U test performed for inter-group comparison and two-tailed Wilcoxon signed-rank test performed for intra-group comparison. APS, antiphospholipid syndrome; NHD, normal healthy donor. Error bars represent ± SEM. *p < 0.05, ns = not significant.
Immunization of BALB/c Mice with R. int Induces Human β2GPI Cross-Reactivity
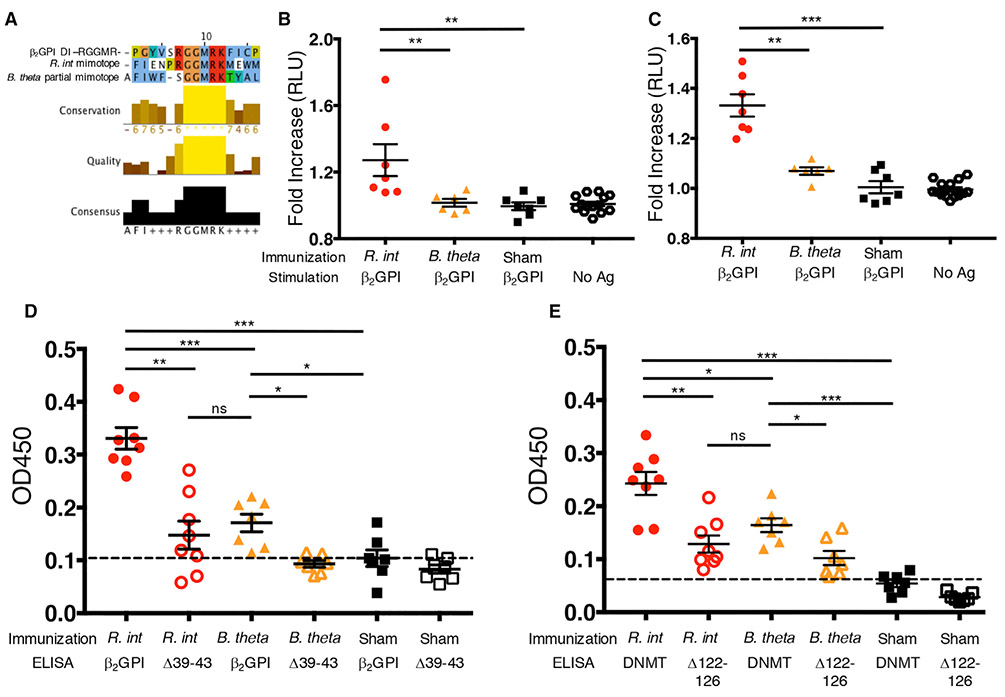
To determine if R. int is able to induce cross-reactivity in vivo, we immunized BALB/c mice as prior immunization studies in this strain were shown to induce T cell and antibody responses to both DI and DV β2GPI domains (Salem et al., 2015). As a control, we chose another prevalent human gut bacterium, Bacteroides thetaiotaomicron (B. theta), which contained no DV mimotope and only a partial RGGMR epitope in a non-DNMT protein (Figure 5A). Lymphocytes from spleens or peripheral lymph nodes of R. int-immunized mice proliferated significantly more to recombinant human β2GPI (rhβ2GPI) than B. theta- or sham-immunized mice (Figures 5B and 5C), supporting peripheral cross-reactive lymphocyte responses in vivo.
Figure 5. Immunization of BALB/c Mice with R. int Induces Human β2GPI Cross-Reactivity.
(A) Clustal Omega alignment of RGGMR DI epitope in β2GPI, Roseburia intestinalis L1-82 (R. int) and Bacteroides thetaiotaomicron VPI-5482 (B. theta). B. theta lacks T cell mimotope homology but contains a partial B cell mimotope (GGMR) homology.
(B and C) Proliferative response to immunization was determined using ATP luminescence in triplicates. Fold increase was determined by averaging triplicates from stimulated populations and dividing by the average of the unstimulated (“no antigen”) cells in RLU. Splenocytes (B) and peripheral lymphocytes isolated from inguinal and axillary lymph nodes (C) were restimulated ex vivo with rhβ2GPI.
(B) Splenocytes from R. int-immunized mice (n = 7) respond significantly more to rhβ2GPI than sham-immunized mice (n = 7, p = 0.0017) and B. Theta-immunized mice (n = 6, p = 0.0047).
(C) Peripheral lymphocytes from R. int-immunized mice (n = 7) respond significantly more to rhβ2GPI than sham-immunized mice (n = 7, p = 0.0006) and B. theta-immunized mice (n = 6, p = 0.0012).
(D and E) Mice were immunized s.c. with lysates from R. int, B. theta, or no lysates (sham) in IFA followed by three s.c. IFA boosts. Representative ELISA using 1:100 diluted serum. Values represent the average OD450 of individual mice in triplicates.
(D) R. int-immunized mice have significantly more serum anti-β2GPI IgG (n = 8) compared to B. theta-immunized mice (n = 7, p = 0.0003) and sham-immunized mice (n = 7, p = 0.0003). R. int-immunized and B. theta-immunized sera lose significantly binding to rhβ2GPI39-43 compared to rhβ2GPI.
(E) R. int-immunized mice have significantly more serum anti-R. int DNMT IgG (n = 8) compared to B. theta-immunized mice (n = 7, p = 0.0205) and sham-immunized mice (n = 7, p = 0.0003). R. int-immunized and B. theta-immunized sera lose significantly binding to R. int DNMTΔ122-126 compared to R. int DNMT. Two-tailed Mann-Whitney U test performed for inter-group comparison and two-tailed Wilcoxon signed-rank test performed for intra-group comparison. Error bars represent ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant.
Next, we immunized BALB/c mice with R. int in incomplete Freund’s adjuvant (IFA) only over a 48-day time course to allow for full development of a humeral response to the bacteria. Mice immunized with R. int lysates had markedly elevated titers of serum anti-rhβ2GPI IgG autoantibodies compared to B. theta- or sham-immunized controls (Figure 5D). Supporting epitope-specific cross-reactivity, we observed a significant decrease in binding to rhβ2GPIΔ39-43 (Figure 5D). Consistent with these findings, anti-R. int DNMT IgG levels were markedly elevated in R. int-immunized mice compared to B. theta- or sham-immunized control mice (Figure 5E). Lastly, we observed a significant loss of binding to the RGGMR epitope-mutant recombinant R. int DNMTΔ122-126 (Figure 5E), which supports mimotope specificity within the bacterial protein besides autoepitope specificity within the autoantigen β2GPI.
We noted that B. theta immunization induced a slight but significant rise in anti-rhβ2GPI and anti-R. int DNMT IgG titers compared to IFA alone, and reduced binding to rhβ2GPIΔ39-43 and R. int DNMTΔ122-126 (Figures 5D and 5E). This could be explained by the presence of a partial RGGMR sequence (4 out of the 5 core amino acids) expressed by B. theta (Figure 5A). However, compared to R. int-immunized mice, B. theta binding to β2GPI and DNMT was significantly decreased, possibly owing to the lack of T cell help in the absence of DV β2GPI-related T cell cross-reactivity (Figures 5A-5C). Taken together, in vivo immunization studies support that R. int mimotopes are recognized in vivo and generate cross-reactive T and B cells systemically that target β2GPI with other partially homologous commensal antigens likely contributing in vivo to the overall levels of autoantibodies in APS.
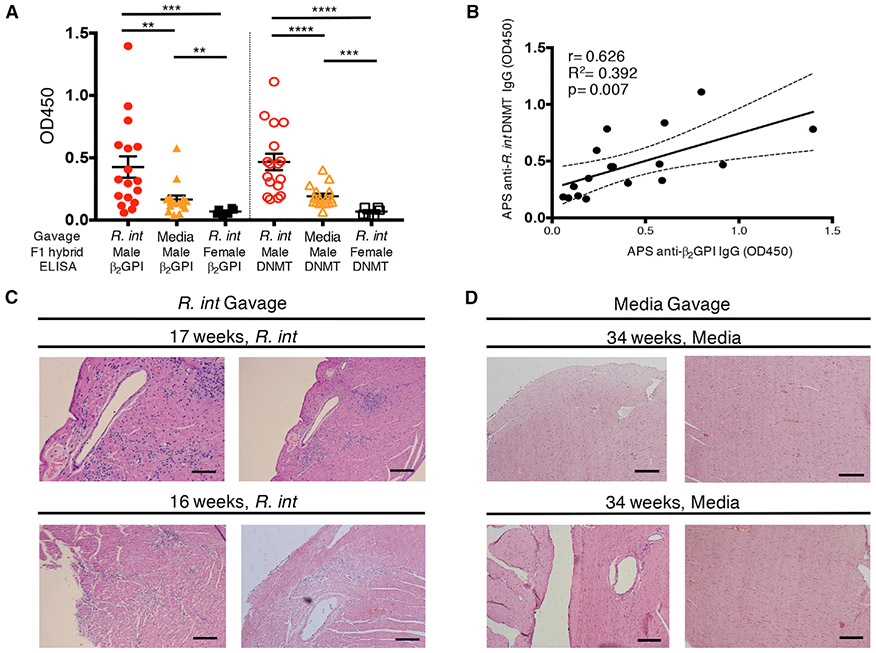
Gavage of (NZW × BXSB)F1 Mice with R. intestinalis Induces Anti-Human β2GPI IgG and Lethal Thromboses
To determine if R. int gut colonization would impact a spontaneous APS mouse model, we chose the (NZW × BXSB)F1 hybrid (F1 male mice), with deaths occurring by 30 weeks of age because of anti-β2GPI-autoantibody-mediated coronary microthrombi and subsequent myocardial infarctions (Hashimoto et al., 1992; Takemura et al., 1989). This model is characterized by an autoimmune gene duplication on the Y chromosome, rendering males susceptible to disease. This innate genetic predisposition is mirrored also functionally in human APS and thus represents a useful model to test adaptive immune stimuli from gut commensals (Giannakopoulos and Krilis, 2013). All mice were pre-treated with vancomycin for 2 weeks prior to gavage with R. int for several reasons. First, vancomycin has been shown by our lab to protect F1 male mice from developing autoantibodies and APS by depleting a pathobiont identified in this model (Figure S8) (Manfredo Vieira et al., 2018). Secondly, R. int is a human gut-adapted commensal that does not normally colonize a murine colon. After vancomycin pre-treatment to open the colonic niche for R. int, we repeatedly gavaged mice with fresh cultures of R. int or media alone and measured autoantibodies at 16 weeks of age when anti-β2GPI IgG peaks (Figure 6A) (Manfredo Vieira et al., 2018). R. int-gavaged (n = 17) but not control-gavaged F1 male (n = 16) or R. int-gavaged female (n = 6) mice developed high-titer anti-β2GPI IgG autoantibodies as well as anti-R. int DNMT IgG antibodies (Figure 6A). Autoantibodies were markedly above low titers detectable in control-gavaged F1 males at 16 weeks of age that are likely induced by a recolonizing microbiota. Human anti-β2GPI IgG positively correlated with anti-R. int DNMT IgG, similarly to what we observed in APS patient plasma (Figures 6B and 4B). Importantly, R. int-gavaged F1 males showed extensive myocardial as well as subendocardial inflammation with large lymphocytic infiltrates and necrosis, which were not seen in controls that survived beyond 30 weeks of age (Figures 6C, 6D, S9A, and S9B). Additionally, areas of hemorrhage were noted in one mouse, which can be associated with chronic ischemia from old myocardial infarction, a finding seen in prior studies (Hashimoto et al., 1992; Takemura et al., 1989). Furthermore, R. int-gavaged—but not control—animals suffered from prominent lymphocytic infiltrates in the lungs (Figures S9C and S9D). The histopathologic findings developed at the time of premature deaths of R. int-gavaged mice, which occurred as early as 16 weeks of age (Figures 6C, S9A, and S9C). Overall, these results support that colonization of a genetically prone host with a primary human isolate of mimotope-containing R. int is sufficient to elicit cross-reactive, pathogenic T and B cell responses in vivo, leading to APS-related morbidity and mortality.
Figure 6. Gavage of (NZW × BXSB)F1 Male Mice with R. intestinalis Induces Anti-Human β2GPI IgG Autoantibodies and Thromboses.
(NZW × BXSB)F1 mice were gavaged weekly with R. intestinalis L1-82 (R. int) or media after two weeks of vancomycin treatment.
(A and B) Sera from 16-week-old mice were tested for anti-human β2GPI IgG or anti-R. int DNMT IgG by ELISA at 1:100 dilution.
(A) R. int gavage of male mice (n = 17) induces significantly elevated anti-human β2GPI IgG compared to males gavaged with media (n = 16, p = 0.006) and females gavaged with R. int (n = 6, p = 0.0002). R. int gavage of male mice induces significantly elevated anti-R. int DNMT IgG compared to males gavaged with media (p < 0.0001) and females gavaged with R. int (p < 0.0001).
(B) Anti-human β2GPI IgG positively correlates with anti-R. int DNMT in male R. int-gavaged mice at 16 weeks of age. Pearson r = 0.626, R2 = 0.392, p = 0.007 two-tailed.
(C) Shown are representative micrographs of H&E-stained myocardium from two R. int-gavaged males that died spontaneously because of autoimmunity at 17 weeks (upper panel) and 16 weeks (lower panel) of age, respectively. R. int-gavaged males show widespread myocardial and subendocardial lymphocytic infiltrates.
(D) Shown are representative micrographs of H&E-stained myocardium from two male mice gavaged with media that survived beyond 34 weeks at which point they were euthanized (representative images of two separate mice, upper and lower). Surviving mice show unremarkable myocardium without any evidence of inflammation or myocardial infarction. Scale bar, 500 μm. Two-tailed Mann-Whitney U test performed for inter-group comparison and two-tailed Wilcoxon signed-rank test performed for intra-group comparison. Error bars represent ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001.
DISCUSSION
Antigens from the microbiota are continuously recognized by the adaptive immune system during homeostasis. This process generates a pool of memory T cells and IgG+ memory B cells that can potentially cross-react with antigens from pathogens or self (Birnbaum et al., 2014; Blank et al., 2002; Greiling et al., 2018; Horai et al., 2015; Su et al., 2013; Szymula et al., 2014; Tai et al., 2016; Varrin-Doyer et al., 2012). The gut microbiota is thought to contribute to the pathogenesis of autoimmune diseases by this mechanism (Dehner et al., 2019; Ruff and Kriegel, 2015), which is supported by recent murine studies cited above and an obvious case of ortholog cross-reactivity in humans (Greiling et al., 2018). Since the number of microbiota-derived proteins exceeds by 100-fold the number of eukaryotic proteins in the host, non-orthologous commensal cross-reactivity is likely to impact the development and maintenance of human autoimmune diseases via T and B cell cross-reactivity.
We utilized the systemic autoimmune disorder APS as a paradigm. Infectious agents are the main factors associated with catastrophic APS, which still leads to exceedingly high mortality despite current treatment (Asherson et al., 2001; Cervera et al., 2015). Besides triggers leading to acute thrombotic events, additional factors induce and sustain chronic autoimmunity in APS patients. We identified the common human gut commensal R. int as a chronic driver of β2GPI autoreactivity based on non-orthologous mimotopes to major autoantigenic epitopes. APS patients had signs of subclinical intestinal inflammation together with peripheral adaptive immune recognition of R. int. The latter was reflected by circulating anti-R. int antibodies polarized to the IgG isotype, indicating a mature, class-switched B cell response to R. int in APS patients compared to controls. APS CD4+ memory T cells, specific for the HLA-DRB4*0103-restricted β2GPI epitope p276-290, cross-reacted with a mimotope derived from R. int and had a Th1-like phenotype. Moreover, we demonstrated that an APS-derived monoclonal antibody with LA activity exhibits pathogenic features by functionally impairing human trophoblasts. We were able to show that this functional mAb, which targets the pathogenic DI epitope R39-R43, bound to both R. int lysates as well as the mimotope within the R. int protein DNMT. Ex vivo, APS patient plasma also bound to the mimotope within R. int DNMT, a finding that positively correlated with anti-β2GPI IgG autoantibody titers. Finally, we also demonstrated T and B cell cross-reactivity in vivo with progression to full-blown clinical disease in a genetically susceptible background. These findings support a role for R. int-mediated cross-reactivity in the development and maintenance of pathogenic autoantibodies in genetically predisposed individuals. In non-predisposed individuals, commensal mimotopes likely provide a source of antigen-specific tolerance in homeostatic settings. This hypothesis is partially supported by a recent study showing that commensal-specific diabetogenic T cells are recruited to the gut and were capable of suppressing colitis (Hebbandi Nanjundappa et al., 2017). In a genetically predisposed host, this mechanism of tolerance may break down and support the induction and maintenance of pathogenic autoreactive lymphocytes. Additional insults act then as a tipping point that allow for pathogenic cross-reactivity to progress to tissue damage and overt autoimmune pathology. These data are consistent with the current “second hit hypothesis” in APS, which states that additional environmental hits, such as oxidative stress or lipopolysaccharide (LPS), lead to acute thrombotic events following systemic anti-β2GPI responses in vivo (Agaret al., 2011; Fischetti et al., 2005; Laplante et al., 2011).
Following this model, exposed, cross-reactive epitopes on gut commensals, that naturally turn over in the gut and release immunogenic antigens such as DNMT, may induce autoantibodies against the unexposed, cryptic epitopes in β2GPI years before a second, innate immune-driven event occurs (“second hit hypothesis”). Such “second hits” then lead to exposure of the cryptic epitopes within β2GPI, which facilitates binding of pathogenic autoantibodies to β2GPI DI and subsequent thrombotic episodes. This scenario is plausible given the immense antigenic variability of the microbiota and the various second hits that can occur in humans such as oxidative events during stress or trauma, a burst of LPS in the circulation during gramnegative infections, exposure of phospholipids during excessive apoptosis or viral infections (Giannakopoulos and Krilis, 2013; Passam et al., 2011; Ruff et al., 2015). The gut microbiota likely contributes also to these second hits if one considers that phospholipids, LPS, or oxidative stress can also be derived from commensals. This is particularly likely in the setting of heightened gut inflammation, as we noted in APS patients, or a disturbed gut barrier present in a spontaneous model for lupus-related APS (Manfredo Vieira et al., 2018).
In summary, a complex interplay between antigen-specific responses against chronically colonizing commensals in a human host together with a genetic predisposition and intermittent “second hits” from the environment are likely drivers of persistent autoreactive T and B cells with subsequent waves of tissue damage. Defining the molecular interactions between the host immune system and the microbiota will be essential for a better understanding of the pathogenesis of chronic immune diseases. This study dissected a molecular process of host-microbiota interactions in the multistep pathogenesis of APS that may be applicable more broadly to other autoimmune diseases. In addition, similar processes may apply to microbiota effects on antitumor immunity, where a cross-reactive tissue-directed host response would be desirable (Routy et al., 2018). Evolutionarily, commensal cross-reactive immune responses may be tolerated by the host to allow for tumor surveillance and enhanced pathogen clearance under inflammatory conditions. The trade-off would be a heightened risk of developing and sustaining autoimmunity in susceptible individuals as we have demonstrated here.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Martin Kriegel (martin.kriegel@yale.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
15 patients positive for anti-β2GPI IgG autoantibodies (11 APS patients and 4 individuals at risk for developing APS, ages 27-70, total 44 samples, see Table 1) and 20 normal healthy donors (NHD, ages 21–60, total 48 samples) were recruited from two institutions: Yale University, New Haven, CT, and Hospital for Special Surgery, New York, NY. Inclusion criteria were: Persistent positivity (defined as positive for β2GPI autoantibodies ± classification of antiphospholipid syndrome (APS), primary or associated with systemic lupus erythematosus, in accordance with the revised Sapporo criteria, with positive anti-β2GPI IgG defined as titers above 40 chemiluminescent units (Miyakis et al., 2006) and ages of 18-75. Exclusion criteria were as follows: ongoing chronic infection, antibiotic or probiotic use in the last 90 days, major gastrointestinal surgery in the last 5 years, gastrointestinal bleeding history, inflammatory bowel disease, bulimia or anorexia nervosa, morbid obesity defined as body mass index (BMI) greater than 40, uncontrolled diabetes mellitus, concurrent autoimmune disease with the exception of systemic lupus erythematosus, malignancy in the past year, current pregnancy, and known excessive alcohol use. Subjects with APS and healthy donors completed up to three monthly study visits for the collection of detailed health and diet histories, whole blood, serum, plasma, and fecal microbiota sampling. DNA was extracted from blood using DNeasy blood and tissue kit (Qiagen). HLA typing was performed as previously published for HLA-DRB4*01 (serotype DR53) (Olerup and Zetterquist, 1992). Further study details are listed at ClinicalTrials.gov identifiers NCT01787305 and NCT02394964. Metadata about participants, including gender, age, BMI, dietary and medical history, are described in Tables 1 and S1. A majority of our cohort were female. When samples sizes were sufficient for statistical power, we found no differences between APS males and females and NHD males and females. All human subject protocols were approved by the Yale Human Investigations Committee and in accordance with the Declaration of Helsinki. A signed document of informed consent was obtained from all study subjects.
Animals
Specific-pathogen-free BALB/cJ, NZW/LacJ, and BXSB/mpJ mice were purchased from Jackson laboratory and further housed and bred in the Yale Amistad Animal Facility under specific-pathogen-free conditions. 10- to 12-week-old BALB/cJ male and female littermate mice were used for immunization studies. There were no significant differences between male and female BALB/cJ responses to immunizations. 4-6-week -old (NZW x BXSB)F1 male and female hybrids were used for oral gavage study and bred as previously described (Manfredo Vieira et al., 2018). Female (NZW x BXSB)F1 mice do not develop disease and were used as a negative control for the effect of R. intestinalis. Mice were provided with water and a standard laboratory diet ad libitum (2018 Harlan Teklad) except if noted otherwise. They were supplied with hardwood chips as bedding and housed in a temperature-controlled, air-conditioned room on a 12-hr light-dark cycle. Animal care and handling was approved by the Yale Institutional Animal Care and Use Facility and in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Bacterial Strains, Culture Conditions, and Lysate Preparation
Human stool isolates of Roseburia intestinalis L1-82 (R. int) and Bacteroides thetaiotaomicron VPI-5482 (B. theta) were grown in Gut Microbiota Medium (Goodman et al., 2011) anaerobically at 37°C to an OD600 value of 1.5-1.7 for all experiments. All cultures were confirmed to be pure by PCR amplification of the 16S rRNA region (forward primer AGAGTTTGATCCTGGCTCAG; reverse primer GACGGGCGGTGWGTRCA; 95°C for 5 min; 30 cycles of 95°C for 10 s, 60°C for 20 s, 72°C for 15 s; 72°C for 10 min) (Turner et al., 1999), followed by Sanger sequencing.
Bacterial lysates were prepared from pelleted monocultures of bacteria, washed three times with PBS, bead-beaten using 0.1-mm glass beads for 2 minutes, centrifuged at 10,000 g x 5 min, and quantified by bicinchoninic acid (BCA) using a BSA standard curve (Thermo Fisher).
METHOD DETAILS
Identification of Commensal Epitopes with Homology to β2GPI Autoepitopes
Beta 2-glycoprotein I (β2GPI) epitope mimics were identified by searching non-redundant protein database in NCBI BLAST for the major epitopes associated with β2GPI. The major B cell epitope, previously identified as spanning residues R39-R43 (RGGMR), and 6-8 overlapping sequences from the human HLA-DRB4*01 (serotype DR53)-restricted immunodominant epitope p276-290 (KVSFFCKNKEKKCSY) were used as query sequences with searches limited to bacteria. Results were manually annotated to exclude known human pathogens and further cross-referenced with the Pathosystems Resource Integration Center (PATRIC) and the Human Microbiome Project for previous identification as a human commensal (Human Microbiome Project Consortium, 2012a, 2012b; Wattam et al., 2017). Commensal protein sequences were aligned using Clustal Omega (Sievers et al., 2011). The R. intestinalis DNA methyltransferase (WP_118597735.1) structural model was prepared by homology modeling with SWISS-MODEL (ExPASy) (Arnold et al., 2006; Biasini et al., 2014; Guex et al., 2009; Kiefer et al., 2009).
Stool Sample Collection
Stool samples were collected by subjects at home in sterile containers and shipped overnight on ice to the laboratory, at which time they were aliquoted and stored at −80°C. Fecal DNA was extracted according to the HMP protocol (Aagaard et al., 2013). 80 to 200 mg of human stool was combined with 1 ml of Qiagen Bead Solution and 1-mm ceramic beads (BioSpec) and bead-beaten twice for 1 minute with a 2-minute rest on ice in between using a mini-beadbeater (Biospec). Samples were centrifuged and supernatant was transferred to a Qiagen Garnet Bead tube, heated for 10 minutes at 65°C, then heated for 10 minutes at 95°C, followed by further processing per the Qiagen DNeasy Power Soil DNA Isolation Kit protocol.
16S rRNA Sequencing
Dual-indexed 16S rRNA high-throughput sequencing was performed on isolated fecal DNA or IgA+ and IgA− sorted bacterial DNA as previously described (Kozich et al., 2013; Palm et al., 2014). The V4 region of the 16S rRNA gene was PCR-amplified, pooled, normalized, and sequenced using the Illumina MiSeq platform with 2 x 250 bp paired-end reads. Analysis of 16S rRNA sequencing reads was performed as previously described (Cullen et al., 2015). The following minor modifications were included. QIIME analysis was performed using version 2 core distribution 2018.11.0, with denoising performed using DADA2 (trimLeft = 0 nucleotides for forward and reverse; truncLen = 251 nucleotides forward and 250 nucleotides reverse) (Callahan et al., 2016; Caporaso et al., 2010). Denoised and filtered amplicon sequencing variants were rarefied to a depth of 7091 sequences per sample for analysis of overall composition and 5528 sequences per sample for IgA-seq.
Fecal Calprotectin and Plasma Lipocalin-2 Assays
Calprotectin was measured in a single frozen stool sample from human subjects using the Legend Max™ ELISA kit (BioLegend). Stool samples were aliquoted into pre-weighed bead-containing tubes and stored at −80°C until sample preparation. 100 mg of stool samples were diluted with 1 ml Tris pH 7.4 followed by bead-beating for 5 s. Samples were centrifuged at 10,000 x g for 10 minutes at 4°C. The supernatant was carefully transferred to a new microcentrifuge tube avoiding the pellet. The supernatant was centrifuged again using the same settings as above. The resulting supernatant was transferred to a new tube and diluted 1:20 with a dilution buffer supplied in the Legend Max™ ELISA kit. Lipocalin-2 was measured in previously frozen human plasma from all subjects using the Legend Max™ ELISA kit (BioLegend). Experimental samples were assayed with the standards and controls included with the kit according to the manufacturer’s instructions.
Plasma Anti-R. int Ig Bacterial Flow Cytometry and Sorting of Fecal IgA-Coated Bacteria
1 ml of fresh R. int L1-82 cultures were pelleted (8,000 x g, 1 min, room temperature) and frozen prior to use. Pellets were resuspended in 1 ml PBS containing 1% (w/v) BSA (Sigma-Aldrich, staining buffer) and washed twice with staining buffer. Pellets were resuspended in 1 ml of staining buffer and pooled. 100 μl-aliquots were incubated with 1 μl of plasma overnight at 4°C. Samples were pelleted and washed twice with staining buffer. Samples were then pelleted and resuspended in blocking buffer (staining buffer containing 20% Normal Mouse Serum from MP Biomedical), incubated for 20 min on ice, and then stained with 100 μl staining buffer containing PE-conjugated anti-human IgA (1:20; Miltenyi Biotec clone IS11-8E10), FITC-conjugated anti-human IgM (1:20; Jackson ImmunoResearch polyclonal), and PerCP-conjugated anti-human IgG (1:20; Jackson ImmunoResearch polyclonal) for 30 min on ice. Samples were then washed twice with 1-ml staining buffer before flow cytometric analysis.
Fecal samples were prepared as previously described (Palm et al., 2014). Briefly, approximately 100 mg of frozen human fecal material were placed in 2-ml microtubes (Sarstedt AG & Co.) containing 1-mm ceramic beads (Big D Lysing Matrix, MP Biomedicals) and incubated in 1-ml phosphate buffered saline (PBS) per 100 mg fecal material on ice for 30 minutes. Fecal pellets were homogenized by bead beating for 5 s (MiniBeadBeater-16, BioSpec products, Inc.) and then centrifuged (50 x g, 15 min, 4°C) to remove large particles. Fecal bacteria in the supernatants were removed (100 μl/sample), washed with 1 ml PBS containing 1% (w/v) BSA (Sigma-Aldrich, staining buffer) and centrifuged for 5 min (8,000 x g, 4°C) before resuspension in 1 ml staining buffer. After an additional wash, bacterial pellets were resuspended in 100 μl blocking buffer (staining buffer containing 20% Normal Mouse Serum from MP Biomedical), incubated for 20 min on ice, and then stained with 100 μl staining buffer containing PE-conjugated anti-human IgA (1:20; Miltenyi Biotec clone IS11-8E10) for 30 min on ice. Samples were then washed twice with 1-ml staining buffer before flow cytometric analysis or cell separation via FACS (FACS Aria, BD Biosciences). For each separated sample, 2 million IgA-positive bacteria were collected, pelleted (10,000 x g, 5 min, 4°C), and frozen along with the IgA-negative samples at −80°C for future use. IgA coating percentages were determined by dividing the total lgA+ bacteria by the total number of bacteria analyzed by FACS. IgA coating index (ICI) was determined as previously described (Palm et al., 2014). Heat map of ICI was generated with “heatmap.2” and “vegan” R packages.
Gene Expression Analysis
Quantitative real-time PCRs of genus, species, and total bacterial load as well as R. intestinalis genes containing autoepitope mimics, were performed on a QuantStudio 6 (Applied Biosystems). RNA was isolated from tissue sections and stored in Trizol (Thermo Fisher Scientific) at −80°C. RNA was then quantified and purified with a DNA-free DNA Removal Kit (Thermo Fisher Scientific). cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Relative load and expression were determined using 20 ng of stool or isolated bacterial DNA, 250 nM forward and reverse primers, and Power SYBR green PCR Master Mix (Thermo Fisher) in a total reaction volume of 20 μl in either duplicates or triplicates. Samples were heated at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 s, 55°C for 1 min.
Primer Pairs
R. intestinalis species-specific (CBL07561.1) forward CTTGTGACAGATGATGAAGATCGTG, reverse GCAGATCAGTCCTTTTCCATGTGTT, length 114 nt, efficiency 96.2%; universal 16S rRNA forward CGGCAACGAGCGCAACCC, reverse CCATTGTAGCACGTGTGTAGC, length 146 nt, efficiency 89% (Denman and McSweeney, 2006); R. intestinalis DNMT-specific (WP_118597735.1) forward TGGACGAATCATCCGAACCC, reverse CCCTCGAACCTTTCAGTCCC, length 115 nt, efficiency 99.1%; and R. intestinalis T cell mimic-specific (EEU99424.1) forward AGAAAATCCGTCAAAGACTGGGA, reverse CGCCAAAGACCCACTGCATAG, length 52 nt, efficiency 87%.
All commensal-specific and gene-specific primers were validated by determining qPCR efficiency using isolated bacterial DNA ranging from 20 ng to approximately 2.5 pg using 2-fold dilutions. Each primer pair had a single peak during melt curve analysis and a single band by gel electrophoresis. Specificity was tested by using primers to amplify 20 ng of unrelated bacteria (B. theta, Eubacterium rectale, E. coli, Staphylococcus epidermidis) and fecal DNA previously shown to contain or not contain Roseburia at the genus level by 16S rRNA sequencing. Replicates were averaged and bacterial load was quantified using the delta-delta-Ct method per the formula: 2 ^ -((Ct of experimental sample – Ct of experimental sample 16S) – (Ct of control bacteria – Ct of control bacteria 16S)). If replicates were not concordant, i.e. one well failed to amplify, the assay was repeated with a new sample.
Production and Purification of Proteins and Antibodies
Double-stranded DNA sequences corresponding to mature human β2GPI (apolipoprotein H, NP_000033.2) or mature human β2GPI with alanine replacing the R39-R43 (rhβ2GPIΔ39-43) were purchased as gBlocks (Integrated DNA Technologies). These sequences were cloned into a custom pcDNA3.4 expression vector (Thermo Fisher) containing an N-terminal human CD5 signal peptide, a C-terminal HRV3C protease cleavage site, and a C-terminal murine IgG2a. These proteins were expressed in EXPI293F mammalian expression system (Thermo Fisher) and purified over a HiTrap protein A column (GE Healthcare). EXPI 293F cells were cultured as recommended by the manufacturer. Briefly, EXPI293F cells were thawed and incubated in EXPI 293 expression medium (Thermo Fisher) in 125-ml polycarbonate, disposable, sterile, vent-cap Erlenmeyer shaker flasks. EXPI293F cells were cultured at 37°C at 8% CO2 on an orbital shaker set to 125 rpm. Transfection was carried out per the manufacturer’s instructions. Briefly, 7.5 x 107 viable cells in 25.5 ml of Expi293 Expression medium were transfected with 30 μg of purified plasmid DNA. Purified plasmid DNA was diluted in 1.5 ml of Opti-MEM reduced serum medium. 81 μl of ExpiFectamine 293 reagent were diluted into 1.5ml of Opti-MEM reduced serum medium, gently mixed and allowed to incubate for 5 minutes at room temperature. After the 5-minute incubation the diluted plasmid DNA and ExpiFectamine 293 reagent were mixed and incubated for 20 min at room temperature. Following this 20-minute incubation step, the DNA-ExpiFectamine mix was added to 25.5 ml of EXPI293F cells and incubated as above. After 20 hours of culture, 150 μl of ExpiFectamine 293 transfection enhancer 1 and 1.5 ml of enhancer 2 were added to the cultures. Cultures supernatant was collected after 72 hours of further incubation and centrifuged at 800 x g for 5 minutes. Culture supernatant was further passed through a 0.2-μM filter before being purified over a HiTrap protein A column (GE Healthcare). Following purification, the mIgG2a tag was cleaved by HRV3C protease overnight at 4°C (Thermo Fisher). Cleaved protein was separated from mIgG2a by purification over a HiTrap protein A column. Cleaved, tagless recombinant proteins were validated to be devoid of contaminating mIgG2a by SDS-PAGE, western blot, and ELISA, respectively. Heavy and light chains of the previously identified anti-human β2GPI clones P1-117 (HQ129860.1 and HQ129861.1) and P2-6 (HQ129864.1 and HQ129865.1) were obtained as gBlocks and cloned into a pcDNA 3.1 expression vector containing full length human IgG1. Antibodies were expressed and purified as above. Purified products were confirmed to be >95% pure by SDS-PAGE and western blot. Serum-derived human β2GPI was purchased from Haematologic Technologies. Recombinant R. int DNMT and R. int DNMTΔ122-126 fused with C-terminal 6xHis-tag were purchased from GenScript technologies and shown to be >90% pure and <1.0 EU endotoxin.
Trophoblast Migration and Trophoblast-Endothelial Cell Co-Cultures
The human first trimester extravillous trophoblast telomerase-transformed cell line Sw.71 was used for trophoblast experiments (Straszewski-Chavez et al., 2009). The trophoblast cell line was cultured in DMEM (Gibco-Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Hyclone), 10 mM Hepes, 0.1 mM MEM non-essential amino acids, 1mM sodium pyruvate and 100 nM penicillin/streptomycin (Gibco-Invitrogen) and maintained at 37°C, 5% CO2. Trophoblast cells were treated in serum-free OptiMEM (Gibco) with media alone, anti-β2GPI IgG1 mAb P1-117 (50 μg/ml), or an isotype control human IgG1 (Southern Biotech) (50 μg/ml). Trophoblast migration was measured using a two-chamber colorimetric assay from EMD Millipore as described (Gysler et al., 2016; Mulla et al., 2010). Trophoblast viability was measured using the CellTiter 96™ assay (Promega) as described (Mulla et al., 2009). A three-dimensional in vitro system was used to study trophoblast-endothelial cell interactions as a model for spiral artery transformation (Alvarez et al., 2015). Human endometrial endothelial cells (HEECs) (Krikun et al., 2004; Schatz et al., 2000) were cultured in flasks coated with 2% gelatin in Endothelial Basal Medium-2 (Lonza) supplemented with 2% FBS and maintained at 37°C/5% CO2. HEECs were stained with the red fluorescent linker dye PKH-26 (Sigma) and seeded into 24-well tissue culture plates over undiluted reduced growth factor Matrigel (Corning). Cells were cultured overnight until tube-like structures were observed. Media was then removed and replaced with trophoblast Sw.71 cells stained with the green fluorescent linker dye PKH-67 (Sigma) in OptiMEM media alone, or with P1-117 (50 μg/ml) or control IgG (50 μg/ml). The trophoblast-endothelial co-culture was then incubated for 48 hrs. Over this time, the trophoblast cells are observed by fluorescent microscopy to invade the Matrigel, invade and co-localize with the endothelial cells, and eventually replace them to take on the vessel tube-like structures (Aldo et al., 2007; Alvarez et al., 2015). After 48 hrs, five fields per well were recorded by fluorescent microscopy (Carl-Zeiss Observer Z1) using OpenLab software (Perkin Elmer) and the number of tubes per field counted.
Immunization Scheme with Bacterial Lysates for In Vivo Lymphocyte Proliferation
10- to 12-week-old male and female BALB/c mice were immunized s.c. initially with 100 μg of R. intestinalis, B. theta, or no lysate (sham) in complete Freund’s adjuvant (CFA). 14 days post-initial immunization, mice were immunized s.c. with 100 μg of lysate or sham in incomplete Freund’s adjuvant (IFA). A final s.c. immunization was performed seven days later with 100 μg of lysate or sham per mouse in IFA. Mice were euthanized five days after the third immunization.
Immunization Scheme with Bacterial Lysates for In Vivo Serum Antibody Responses
10- to 12-week-old male and female BALB/c mice were immunized s.c. with 100 μg of bacterial lysates or sham in IFA. Mice were immunized every 14 days as above for a total of four immunizations. Mice were euthanized five days after the final immunization.
Oral Gavage of (NZW x BXSB)F1 Mice with R. intestinalis
4-6-week-old (NZW x BXSB)F1 hybrids were given orally vancomycin for two weeks as previously described (Manfredo Vieira et al., 2018). Mouse fecal pellets were collected after two weeks of vancomycin and confirmed to be devoid of Enterococcus gallinarum as previously described (Manfredo Vieira et al., 2018). After a 48-hour period without vancomycin, mice were orally gavaged with 100 μl containing 109 CFU of freshly cultured R. int L1-82, which was washed in culture media without short-chain fatty acids (SCFAs) two times, or with culture media alone without SCFAs as control. Aliquots for gavage were prepared anaerobically. Gavages were performed weekly until 20 weeks of age. Blood and serum were collected at 16 weeks of age when autoantibodies peak in the (NZW x BXSB)F1 model (Manfredo Vieira et al., 2018). Mice were followed until APS-related deaths or 34 weeks of age.
Histology
Hearts and lungs of (NZW x BXSB)F1 mice were dissected at the time of euthanasia and fixed in 10% neutral formalin. After paraffin embedding, sectioning, and staining with hematoxylin and eosin (H&E), histologic features of each sample were assessed in a blinded fashion using an Olympus microscope (Olympus DP71). Representative pictures were taken and analyzed using the cellSens software (Olympus).
Lymph Node and Spleen Proliferation Assays
Peripheral lymph nodes (inguinal and axillary) and spleens from BALB/c mice were harvested in RPMI 1640 medium (Life Technologies) supplemented with 2 mM L-glutamine, 2 mM Hepes, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, penicillin (50 U/ml), streptomycin (50 U/ml, Lonza), and 10% fetal bovine serum. Tissues were dissociated into single cell suspensions and filtered through a 40-μm cell strainer and washed into RPMI. For spleens, red blood cells were lysed with 1 ml of red blood cell lysing buffer (Sigma) for 1 minute, then washed three times with PBS. Cells were plated in 96-well plates at 25,000 cells per well, then stimulated with rhβ2GPI (50 μg/ml) and incubated for 72 hours at 37°C. After that, cells were transferred to white opaque 96-well plates (Greiner Bio-one 655073). Proliferation was measured using the CellTiter-Glo Luminscent Cell Viability Assay (Promega) or ATPlite 1step luminescence kit (Perkin Elmer) following the manufacturers’ instructions.
Immunoassays
Enzyme-linked immunosorbent assay (ELISA) for β2GPI, R. intestinalis DNMT, and bacterial lysate were performed with 96-well high-binding, hydrophobic, positively charged plates (Corning 3369) coated with 5 μg/ml for recombinant serum-derived or recombinant β2GPI, or 25 μg/ml of the soluble fraction of bacterial lysates in NaHPO4 (pH 7.6) overnight at 4°C, washed three times with PBS with 0.01% Tween-20, blocked with protein-free blocking buffer (Pierce 37572) for 1 hour at room temperature with shaking, and incubated with primary antibody diluted in protein-free blocking buffer at room temperature with shaking for 2 hours. Wells were washed 4 times and incubated with pre-adsorbed horseradish peroxidase-conjugated secondary antibody sheep anti-human IgG 1:6,000 (Thermo Fisher 31412) or rabbit anti-mouse IgG 1:1,000 (Thermo Fisher A16166) diluted in protein-free blocking buffer for 30 minutes, followed by five washes and colorimetric development with TMB Substrate Buffer (Thermo Fisher). Reactions were stopped after 15 minutes with 2M H2SO4. Previously frozen plasma samples were used to determine the presence of anti-domain I β2GPI IgG by chemiluminescent immunoassay (CIA) (Inova Diagnostics). The anti-domain I β2GPI IgG CIA assay was performed by the manufacturer.
Human T Cell Cloning and Proliferation Assays
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by Lymphoprep (STEMCELL Technologies) gradient centrifugation 24-hour post-blood draw. PBMCs were immunomagnetically separated using the following kits (STEMCELL Technologies) per the manufacturers’ instructions: monocytes using the EasySep Human CD14 Positive Selection kit, and CD4+ T cells using the EasySep Human CD4+ T Cell Negative Isolation kit. Total PBMCs and selected cells were frozen at −80°C in 90% heat-inactivated human AB serum with 10% dimethyl sulfoxide and transferred to liquid nitrogen within 24 hours. Human autologous monocytes were used as antigen-presenting cells for all T cell library assays. Cell numbers were assessed using the Neubauer counting chamber.
Peripheral Blood Mononuclear Cell (PBMC) Proliferation
PBMCs were thawed and plated in 96-well round bottom plates (Corning) at a concentration of 25,000 cells per well in X-Vivo 15 media (Lonza). PMBCs were incubated with the soluble fraction of mechanically lysed, heat-killed R. int L1-82 lysates at a concentration of 1 mg/ml with or without treatment of Proteinase K at 50 μg/ml (Promega) followed by heat inactivation. PBMCs were cultured for 5 days and proliferation was measured by [3H]-thymidine incorporation 16 hours before harvest. Stimulation index was calculated as proliferation of stimulated PBMCs divided by proliferation of unstimulated PBMCs.
T Cell Library and Tetramer Assay
Viable CCR6− memory (VVD−, CD45RA−, CD45RO+, CD25−, CCR6−) CD4+ T cells and CCR6+ memory (CD45RA−, CD45RO+, CD25−, CCR6+) CD4+ T cells were sorted (antibodies from BioLegend) on a FACS Aria machine (BD Biosciences). CD25+ cells were excluded from sorting and analysis. Tetramer assays were performed by sorting into CD45RA−, CD45RO+, CD25−, CCR6±, tetramer± populations.
The T cell library assays were performed as previously described (Geiger et al., 2009). CCR6− and CCR6+ memory CD4+ T cells from APS patients were sorted and cultured in 96-well round-bottom plates (Corning) at 2,000 cells per well in X-Vivo 15 media (Lonza) or as single-cell clones in complete media (RPMI 1640 with 1% hepes, 1% glutamine, 1% pyruvate, 1% non-essential amino acids, 1% pen/strep, and 5% heat-inactivated human serum). Cells were stimulated with phytohemagglutinin (PHA, 1 μg/ml) (Roche) and rhIL-2 (30 U/ml) (R&D) in the presence of irradiated (45 Gy) allogeneic PBMCs (25,000 per well). IL-2 was added on days 3, 6, and 10, respectively. After 14 days of maintenance and expansion, T cell cultures were washed and split equally into two “mirror” 96-well plates. Library screening was carried out by stimulation of 250,000 T cells per well with irradiated (45 Gy) autologous monocytes (~25,000 per well). Monocytes were then pulsed for 3 hours with 100 μg/ml native β2GPI. Wells responding to β2GPI were expanded and restimulated with irradiated monocytes pulsed with DV peptide (KVSFFCKNKEKKCSY) or commensal mimotope peptide (DV mimotope, RIFLFCRNKENVYHF). Negative controls contained unpulsed monocytes only. After 72 hours, culture supernatants were removed for cytokine measurement. Cell proliferation was measured either by measuring [3H]-thymidine incorporation on a scintillation β-counter (Perkin Elmer) or via non-radioactive ATP using the ATP lite kit (Perkin Elmer).
Tetramer cell assays were performed using a similar approach: Tetramer-positive T cells were expanded as above, plated at 50,000 cells/well, and re-stimulated with 10,000 autologous monocytes per well pulsed with native β2GPI (100 μg/ml) or 100 μg/ml DV tetramer peptide (KVSFFCKNKEKKASY) as controls as well as whole heat-killed R. intestinalis (10 bacterial cells/1 monocyte). Proliferation was measured as described above. Supernatants were stored at −80°C prior to cytokine analysis.
Cytokine Immunoassays
Cytokine concentrations from supernatants of memory CD4+ T cell clones were measured using a bead-based immunoassay for human Tfh, Th1, and Th17 cytokines with custom LEGENDplex (BioLegend) and Luminex (Millipore) panels, respectively. Supernatants of activated T cell clones stimulated as detailed above were collected and LEGENDplex or Luminex panels were run according to the manufacturers’ protocols.
Bio-Layer Interferometry and Label-free Determination of KD Calculations
Label-free kinetic assays were determined by bio-layer interferometry on a BLItz instrument (ForteBio). Manufacturer’s instructions were followed unless otherwise noted. Sensors were soaked in and reagents diluted in kinetics buffer (ForteBio). Measurements consisted of 5 steps: Initial baseline 30 s, loading 180 s, baseline 120 s, association 300 s, dissociation 300 s. Purified anti-β2GPI mAb P1-117 was immobilized onto anti-human IgG Fc capture biosensors at a concentration of 10 μg/ml (ForteBio). Sensorgrams were fit globally to a 1:1 binding model by BLItz Pro v1.1.0.28. The equilibrium dissociation constant (KD) and association (ka) and dissociation (kd) rate constants were calculated from a minimum of 4 molar concentrations ranging from 200 nM to 25 nM with a 0 nM control.
QUANTIFICATION AND STATISTICAL ANALYSIS
Plotting of data and statistical analyses were performed using Graphpad Prism 7 software. Unless otherwise stated, statistical significance was determined by the unpaired two-tailed Mann-Whitney U test, and differences were considered statistically significant if the p-value was < 0.05. P-values are represented using * for p < 0.05, ** for p < 0.01, *** for p < 0.001, and **** for p < 0.0001. Sample sizes and relevant statistical information can be found in corresponding figure legends. Data are shown as means ± SEM unless otherwise noted. Shannon-Weiner diversity index was determined using the “vegan” R package (Oksanen et al., 2017; R Core Team, 2017). Weighted UniFrac distances were calculated in QIIME 2, core distribution 2018.11.0, using the Silva 132 reference database (Quast et al., 2013) and visualized using Emperor (Caporaso et al., 2010; Lozupone and Knight, 2005; Vázquez-Baeza et al., 2013).
DATA AND SOFTWARE AVAILABILITY
Data generated by 16S rRNA high-throughput sequencing of human fecal and IgA-seq samples have been deposited in the European Nucleotide Archive. The accession number for the data reported in this paper is ENA: PRJEB32067.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-conjugated anti-human IgA | Miltenyi Biotec, Palm et al., 2014 | Cat# 130-113-476 Clone IS11-8E10 RRID:AB_2733861 |
| FITC-conjugated anti-human IgM | Jackson Immuno Research Laboratories Inc. | Cat# 709-096-073 RRID:AB_2340515 |
| PerCP-conjugated anti-human IgG | Jackson Immuno Research Laboratories Inc. | Cat# 109-125-098 RRID:AB_2337683 |
| Human IgG1 isotype control | Southern Biotech | Cat# 0151K-14 RRID:AB_2794083 |
| anti-human IgG-HRP | Thermo Fisher Scientific | Cat# 31412 RRID:AB_228265 |
| anti-mouse IgG-HRP | Thermo Fisher Scientific | Cat# A16166 RRID:AB_2534837 |
| Bacterial and Virus Strains | ||
| Roseburia intestinalis | Goodman et al., 2011 | Strain: L1-82 |
| Bacteroides thetaiotaomicron | Goodman et al., 2011 | Strain: VPI-5482 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DV epitope peptide, KVSFFCKNKEKKCSY, >90% pure | GenScript | Custom peptide |
| DV mimotope peptide, RIFLFCRNKENVYHF, >90% pure | GenScript | Custom peptide |
| DV tetramer peptide, KVSFFCKNKEKKASY, >90% pure | GenScript | Custom peptide |
| HRV3C protease | Thermo Fisher | Cat# 88946 |
| Recombinant R. int DNMT- C-terminal 6xHIS >90% pure and <1.0 EU endotoxin, WP_118597735.1 | GenScript | Custom order |
| Recombinant R. int DNMTΔ122-126- C-terminal 6xHIS >90% pure and <1.0 EU endotoxin, WP_118597735.1 with alanine substitution as indicated | GenScript | Custom order |
| Critical Commercial Assays | ||
| DNeasy blood and tissue kit | Qiagen | Cat# 69504 |
| DNeasy PowerSoil Kit | Qiagen | Cat# 12888-100 |
| LEGEND MAX™ Human MRP8/14 (Calprotectin) ELISA Kit | Biolegend | Cat# 439707 |
| LEGEND MAX™ Human NGAL (Lipocalin-2) ELISA Kit | Biolegend | Cat# 443407 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | Cat# 4368577 |
| HiTrap protein A column | GE Healthcare | Cat# 29048576 |
| Two-chamber colorimetric assay | EMD Millipore Gysler et al., 2016; Mulla et al., 2010 | Cat# ECM220 |
| CellTiter 96 assay | Promega; Mulla et al., 2009 | Cat# G3582 |
| CellTiter-Glo Luminscent Cell Viability Assay | Promega | Cat# G7570 |
| ATPlite 1step luminescence kit | Perkin Elmer | Cat# 6016736 |
| ELISA plate, 96-well high-binding, hydrophobic, positively charged | Corning | Cat# 3369 |
| TMB Substrate Kit | Thermo Fisher | Cat# 34021 |
| Anti-domain I β2GPI IgG chemiluminescent immunoassay, QUANTA Flash® (B2) GPI Domain 1 | Inova Diagnostics | Cat# 701188 |
| Lymphoprep | Stemcell Technologies | Cat# 07851 |
| EasySep Human CD14 Positive Selection kit | Stemcell Technologies | Cat# 17858 |
| EasySep Human CD4+ T Cell negative isolation kit | Stemcell Technologies | Cat# 17952 |
| LEGENDplex human cytokine kit | Biolegend | Custom |
| Luminex human cytokine kit | Millipore | Custom |
| Anti-Human Fc Capture (AHC) Biosensors | ForteBio | Cat# 18-5060 |
| PKH26 red fluorescent cell linker kit | Sigma-Aldrich | Cat# PKH26GL |
| PKH67 green fluorescent cell linker kit | Sigma-Aldrich | Cat#PKH67GL |
| Deposited Data | ||
| 16S rRNA sequencing data from all samples (fecal total and IgA-coated bacteria) | This manuscript | ENA: PRJEB32067 |
| Expi293 Expression System Kit | Thermo Fisher Scientific | Cat# A14635 |
| Experimental Models: Cell Lines | ||
| EXPI293F cell line (human female) | Thermo Fisher Scientific | Cat# A14527 RRID:CVCL_D615 |
| Human first trimester extravillous trophoblast telomerase-transformed Swan.71; Sw.71 (human female) | Straszewski-Chavez et al., 2009; kindly provided by Dr. Gil Mor (Yale) | RRID:CVCL_D855 |
| Human endometrial endothelial cells (HEECs) (human female) | Krikun et al., 2004; kindly provided by Dr. Gil Mor (Yale) | N/A |
| Experimental Models: Organisms/Strains | ||
| BALB/cJ mice | Jackson Laboratory | 000651 |
| NZW/LacJ mice | Jackson Laboratory | 001058 |
| BXSB/mpJ mice | Jackson Laboratory | 000740 |
| Oligonucleotides | ||
| Universal 16S rRNA: 8F AGAGTTTGATCCTGGCTCAG | Turner et al., 1999 | Yale Oligo Synthesis Resource |
| Universal 16S rRNA: 1391R GACGGGCGGTGWGTRCA | Turner et al., 1999 | Yale Oligo Synthesis Resource |
| R. intestinalis species-specific forward (CBL07561.1, ROI_02380) CTTGTGACAGATGATGAAGATCGTG | This manuscript | Yale Oligo Synthesis Resource |
| R. intestinalis species-specific reverse (CBL07561.1, ROI_02380) GCAGATCAGTCCTTTTCCATGTGTT | This manuscript | Yale Oligo Synthesis Resource |
| Universal 16S rRNA qPCR forward CGGCAACGAGCGCAACCC | Denman and McSweeney, 2006 | Yale Oligo Synthesis Resource |
| Universal 16S rRNA qPCR reverse CCATTGTAGCACGTGTGTAGC | Denman and McSweeney, 2006 | Yale Oligo Synthesis Resource |
| R. intestinalis DNMT-specific forward (WP_118597735.1) TGGACGAATCATCCGAACCC | This manuscript | Yale Oligo Synthesis Resource |
| R. intestinalis DNMT-specific reverse (WP_118597735.1) CCCTCGAACCTTTCAGTCCC | This manuscript | Yale Oligo Synthesis Resource |
| R. intestinalis T cell mimic-specific forward (EEU99424.1) AGAAAATCCGTCAAAGACTGGGA | This manuscript | Yale Oligo Synthesis Resource |
| R. intestinalis T cell mimic-specific reverse (EEU99424.1) CGCCAAAGACCCACTGCATAG | This manuscript | Yale Oligo Synthesis Resource |
| Recombinant DNA | ||
| Mature human β2GPI (NP_000033.2) gBlock | Integrated DNA Technologies | Custom gBlock |
| Human β2GPI with alanine replacing the R39-R43 (rhβ2GPIΔ39-43) gBlock | Integrated DNA Technologies | Custom gBlock |
| Customized pcDNA3.4 expression vector, containing N-terminal human CD5 signal peptide, a C-terminal HRV3C protease cleavage site, and a C-terminal murine IgG2a | Thermo Fisher Scientific, custom cloning | A14697, custom cloning |
| Human anti-β2GPI P1-117 heavy chain, GenBank accession HQ129860.1 | Integrated DNA Technologies | Custom gBlock |
| Human anti-β2GPI P1-117 light chain, GenBank accession HQ129861.1 | Integrated DNA Technologies | Custom gBlock |
| Human anti-β2GPI P2-6 heavy chain, GenBank accession HQ129864.1 | Integrated DNA Technologies | Custom gBlock |
| Human anti-β2GPI P2-6 light chain, GenBank accession HQ129865.1 | Integrated DNA Technologies | Custom gBlock |
| pcDNA 3.1, full-length human IgG1 | This paper; kindly provided by Dr. Lieping Chen (Yale) | N/A |
| Software and Algorithms | ||
| SWISS-MODEL | Arnold et al., 2006; Biasini et al., 2014; Guex et al., 2009; Kiefer et al., 2009 | https://swissmodel.expasy.org/ |
| Clustal Omega | Sievers et al., 2011 | https://www.ebi.ac.uk/rools/msa/clustalo/ |
| Quantitative Insights Into Microbial Ecology (QIIME) core distribution 2018.11.0 | Caporaso et al., 2010 | https://qiime2.org/ |
| DADA2 | Callahan et al., 2016 | https://benjjneb.github.io/dada2/tutorial.html |
| OpenLab | Perkin Elmer, Agilent | https://www.agilent.com/en/products/software-informatics/openlab-software-suite |
| cellSens | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/ |
| BLItz Pro v1.1.0.28 | Forte Bio | https://www.blitzmenow.com/blitz_pro.html |
| Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ‘Vegan’ R package | Oksanen et al., 2017 | http://vegan.r-forge.r-project.org/ |
| Other | ||
| Serum-derived human β2GPI | Haematologic technologies Inc | Cat# B2GI-0001 |
| Normal mouse serum | Jackson ImmunoResearch laboratories Inc | Cat# 015-000-120 |
| Three-dimensional in vitro system to study trophoblast-endothelial cell interactions as a model for sprial artery transformation | Alvarez et al., 2015 | Custom |
| X-Vivo 15 media | Lonza | Cat# BE02-053Q |
| Growth Factor Reduced Matrigel | Corning | Cat# 356231 |
| Endothelial Basal Medium-2 | Lonza | Cat# CC-3156 |
| 24-well tissue culture plates; Nunc | Thermo Fisher Scientific | Cat# 142475 |
ACKNOWLEDGMENTS
The authors would like to thank all study subjects for their participation, all current and former lab members for helpful discussions, Lieping Chen and Jun Wang for antibody expression vectors, Lesley Devine for assistance with FACS sorting, Roger Albesa and Inova Diagnostics for performing the anti-domain I β2GPI CIA assay, David Schatz for the use of the bio-layer interferometer, Noah Palm for use of an anaerobic chamber and advice on IgA-seq, Emine Guven-Maiorov (NIH) for in silico structural modeling, and John Sterpka for technical assistance. The authors also thank Kristin DeFrancesco and Irene Matos for patient recruitment at the Yale Center for Clinical Investigation. This work was supported by grants from the National Institutes of Health (NIH) (K08AI095318, R01AI118855, T32AI07019, and T32DK007017-39), Yale Rheumatic Diseases Research Core (NIH P30 AR053495), Women’s Health Research at Yale, O’Brien Center at Yale (NIH P30DK079310), Arthritis National Research Foundation, Arthritis Foundation, and Lupus Research Alliance (to M.A.K.) as well as the American Heart Association (15GRNT24480140) and Lupus Research Institute (to V.M.A.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2019.05.003.
DECLARATION OF INTERESTS
M.A.K. received salary, consulting fees, honoraria, and research funds from Roche, Bristol-Meyers Squibb, AbbVie, and Cell Applications and is an employee of Roche. M.A.K. and S.M.V. hold an international patent on the use of antibiotics and commensal vaccination to treat autoimmunity and received royalties. The remaining authors declare no conflict of interests.
REFERENCES
- Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, Patel S, Cutting M, Madden T, Hamilton H, et al. (2013). The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 27, 1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams VM, Chamley LW, and Salmon JE (2017). Emerging treatment models in rheumatology: antiphospholipid syndrome and pregnancy: pathogenesis to translation. Arthritis Rheumatol. 69, 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agar C, de Groot PG, Mörgelin M, Monk SD, van Os G, Levels JH, de Laat B, Urbanus RT, Herwald H, van der Poll T, et al. (2011). Beta(2)-glycoprotein I: a novel component of innate immunity. Blood 117, 6939–6947. [DOI] [PubMed] [Google Scholar]
- Aldo PB, Krikun G, Visintin I, Lockwood C, Romero R, and Mor G (2007). A novel three-dimensional in vitro system to study trophoblast-endothelium cell interactions. Am. J. Reprod. Immunol 58, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez AM, Mulla MJ, Chamley LW, Cadavid AP, and Abrahams VM (2015). Aspirin-triggered lipoxin prevents antiphospholipid antibody effects on human trophoblast migration and endothelial cell interactions. Arthritis Rheumatol. 67, 488–497. [DOI] [PubMed] [Google Scholar]
- Arai T, Yoshida K, Kaburaki J, Inoko H, Ikeda Y, Kawakami Y, and Kuwana M (2001). Autoreactive CD4(+) T-cell clones to beta2-glycoprotein I in patients with antiphospholipid syndrome: preferential recognition of the major phospholipid-binding site. Blood 98, 1889–1896. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, and Schwede T (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Asherson RA, Cervera R, Piette JC, Shoenfeld Y, Espinosa G, Petri MA, Lim E, Lau TC, Gurjal A, Jedryka-Góral A, et al. (2001). Catastrophic antiphospholipid syndrome: clues to the pathogenesis from a series of 80 patients. Medicine (Baltimore: ) 80, 355–377. [DOI] [PubMed] [Google Scholar]
- Benagiano M, Gerosa M, Romagnoli J, Mahler M, Borghi MO, Grassi A, Della Bella C, Emmi G, Amedei A, Silvestri E, et al. (2017). beta2 glyco-protein I recognition drives Th1 inflammation in atherosclerotic plaques of pa-tients with primary antiphospholipid syndrome. J. Immunol 198, 2640–2648. [DOI] [PubMed] [Google Scholar]
- Berstad A, Arslan G, and Folvik G (2000). Relationship between intestinal permeability and calprotectin concentration in gut lavage fluid. Scand. J. Gastroenterol 35, 64–69. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42 (web server issue), W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, and Garcia KC (2014). Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank M, Krause I, Fridkin M, Keller N, Kopolovic J, Goldberg I, Tobar A, and Shoenfeld Y (2002). Bacterial induction of autoantibodies to beta2-glycoprotein-I accounts for the infectious etiology of antiphospholipid syndrome. J. Clin. Invest 109, 797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Goods BA, Raddassi K, Nepom GT, Kwok WW, Love JC, and Hafler DA (2015). Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci. Transl. Med 7, 287ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera R, Serrano R, Pons-Estel GJ, Ceberio-Hualde L, Shoenfeld Y, de Ramón E, Buonaiuto V, Jacobsen S, Zeher MM, Tarr T, et al. (2015). Morbidity and mortality in the antiphospholipid syndrome during a 10-year period: a multicentre prospective study of 1000 patients. Ann. Rheum. Dis 74, 1011–1018. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, and Vijay-Kumar M (2012). Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 7, e44328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, and Blaser MJ (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet 13, 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, and Goodman AL (2015). Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat B, Derksen RH, Urbanus RT, and de Groot PG (2005). IgG antibodies that recognize epitope Gly40-Arg43 in domain I of beta 2-glycoprotein I cause LAC, and their presence correlates strongly with thrombosis. Blood 105, 1540–1545. [DOI] [PubMed] [Google Scholar]
- de Laat B, Derksen RH, van Lummel M, Pennings MT, and de Groot PG (2006). Pathogenic anti-beta2-glycoprotein I antibodies recognize domain I of beta2-glycoprotein I only after a conformational change. Blood 107, 1916–1924. [DOI] [PubMed] [Google Scholar]
- Dehner C, Fine R, and Kriegel MA (2019). The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr. Opin. Rheumatol 31, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman SE, and McSweeney CS (2006). Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol 58, 572–582. [DOI] [PubMed] [Google Scholar]
- Dienava-Verdoold I, Boon-Spijker MG, de Groot PG, Brinkman HJ, Voorberg J, Mertens K, Derksen RH, and de Laat B (2011). Patient-derived monoclonal antibodies directed towards beta2 glycoprotein-1 display lupus anticoagulant activity. J. Thromb. Haemost 9, 738–747. [DOI] [PubMed] [Google Scholar]
- Farstad IN, Halstensen TS, Lien B, Kilshaw PJ, Lazarovits AI, and Brandtzaeg P (1996). Distribution of beta 7 integrins in human intestinal mucosa and organized gut-associated lymphoid tissue. Immunology 89, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti F, Durigutto P, Pellis V, Debeus A, Macor P, Bulla R, Bossi F, Ziller F, Sblattero D, Meroni P, et al. (2005). Thrombus formation induced by antibodies to β2-glycoprotein I is complement dependent and requires a priming factor. Blood 106, 2340–2346. [DOI] [PubMed] [Google Scholar]
- Garcia D, and Erkan D (2018). Diagnosis and management of the antiphospholipid syndrome. N. Engl. J. Med 378, 2010–2021. [DOI] [PubMed] [Google Scholar]
- Geiger R, Duhen T, Lanzavecchia A, and Sallusto F (2009). Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J. Exp. Med 206, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulos B, and Krilis SA (2013). The pathogenesis of the antiphospholipid syndrome. N. Engl. J. Med 368, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, and Gordon JI (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 108, 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiling TM, Dehner C, Chen XG, Hughes K, Iñiguez AJ, Boccitto M, Ruiz DZ, Renfroe SC, Vieira SM, Ruff WE, et al. (2018). Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med 10, eaan2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, and Schwede T (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, S162–S173. [DOI] [PubMed] [Google Scholar]
- Gysler SM, Mulla MJ, Guerra M, Brosens JJ, Salmon JE, Chamley LW, and Abrahams VM (2016). Antiphospholipid antibody-induced miR-146a-3p drives trophoblast interleukin-8 secretion through activation of Toll-like receptor 8. Mol. Hum. Reprod 22, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagán AJ, Pepper M, Maynard CL, Elson CO 3rd, and Belkaid Y (2012). Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Kawamura M, Ichikawa K, Suzuki T, Sumida T, Yoshida S, Matsuura E, Ikehara S, and Koike T (1992). Anticardiolipin antibodies in NZW x BXSB F1 mice. A model of antiphospholipid syndrome. J. Immunol 149, 1063–1068. [PubMed] [Google Scholar]
- Hebbandi Nanjundappa R, Ronchi F, Wang J, Clemente-Casares X, Yamanouchi J, Sokke Umeshappa C, Yang Y, Blanco J, Bassolas-Molina H, Salas A, et al. (2017). A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell 171, 655–667.e17. [DOI] [PubMed] [Google Scholar]
- Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, et al. (2017). Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 153, 1320–1337.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai R, Zárate-Bladés CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, Jittayasothorn Y, Chan CC, Yamane H, Honda K, et al. (2015). Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 43, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012a). A framework for human microbiome research. Nature 486, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012b). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou Y, Pericleous C, Giles I, Latchman DS, Isenberg DA, and Rahman A (2007). Binding of antiphospholipid antibodies to discontinuous epitopes on domain I of human beta(2)-glycoprotein I: mutation studies including residues R39 to R43. Arthritis Rheum. 56, 280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson GM, Victoria EJ, and Marquis DM (1998). Anti-beta2 glycoprotein I (beta2GPI) autoantibodies recognize an epitope on the first domain of beta2GPI. Proc. Natl. Acad. Sci. USA 95, 15542–15546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, and Schwede T (2009). The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikoff MR, and Denson LA (2006). Role of fecal calprotectin as a biomarker of intestinal inflammation in inflammatory bowel disease. Inflamm. Bowel Dis 12, 524–534. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, and Schloss PD (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol 79, 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikun G, Mor G, Alvero A, Guller S, Schatz F, Sapi E, Rahman M, Caze R, Qumsiyeh M, and Lockwood CJ (2004). A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology 145, 2291–2296. [DOI] [PubMed] [Google Scholar]
- Kuwana M, Matsuura E, Kobayashi K, Okazaki Y, Kaburaki J, Ikeda Y, and Kawakami Y (2005). Binding of beta 2-glycoprotein I to anionic phospholipids facilitates processing and presentation of a cryptic epitope that activates pathogenic autoreactive T cells. Blood 105, 1552–1557. [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, Lee C, Irie K, Pinker EY, Narushima S, et al. (2019). Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363, eaat4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante P, Amireault P, Subang R, Dieudé M, Levine JS, and Rauch J (2011). Interaction of beta2-glycoprotein I with lipopolysaccharide leads to Toll-like receptor 4 (TLR4)-dependent activation of macrophages. J. Biol. Chem 286, 42494–42503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol 32, 834–841. [DOI] [PubMed] [Google Scholar]
- Lozier J, Takahashi N, and Putnam FW (1984). Complete amino acid sequence of human plasma beta 2-glycoprotein I. Proc. Natl. Acad. Sci. USA 81, 3640–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, and Knight R (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol 71, 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler M, Albesa R, Zohoury N, Bertolaccini ML, Ateka-Barrutia O, Rodriguez-Garcia JL, Norman GL, and Khamashta M (2016). Autoantibodies to domain 1 of beta 2 glycoprotein I determined using a novel chemiluminescence immunoassay demonstrate association with thrombosis in patients with antiphospholipid syndrome. Lupus 25, 911–916. [DOI] [PubMed] [Google Scholar]
- Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E, Greiling T, Ruff W, et al. (2018). Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakis S, Lockshin MD, Atsumi T, Branch DW, Brey RL, Cervera R, Derksen RH, DE Groot PG, Koike T, Meroni PL, et al. (2006). International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J. Thromb. Haemost 4, 295–306. [DOI] [PubMed] [Google Scholar]
- Mulla MJ, Brosens JJ, Chamley LW, Giles I, Pericleous C, Rahman A, Joyce SK, Panda B, Paidas MJ, and Abrahams VM (2009). Antiphospholipid antibodies induce a pro-inflammatory response in first trimester trophoblast via the TLR4/MyD88 pathway. Am. J. Reprod. Immunol 62, 96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulla MJ, Myrtolli K, Brosens JJ, Chamley LW, Kwak-Kim JY, Paidas MJ, and Abrahams VM (2010). Antiphospholipid antibodies limit trophoblast migration by reducing IL-6 production and STAT3 activity. Am. J. Reprod. Immunol 63, 339–348. [DOI] [PubMed] [Google Scholar]
- Noster R, Riedel R, Mashreghi MF, Radbruch H, Harms L, Haftmann C, Chang HD, Radbruch A, and Zielinski CE (2014). IL-17 and GM-CSF expression are antagonistically regulated by human T helper cells. Sci. Transl. Med 6, 241ra80. [DOI] [PubMed] [Google Scholar]
- O’Hara AM, and Shanahan F (2006). The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Guillaume F, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, et al. (2017). Vegan: community Ecology Package. [Google Scholar]
- Olerup O, and Zetterquist H (1992). HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 39, 225–235. [DOI] [PubMed] [Google Scholar]
- Ost KS, and Round JL (2018). Communication between the microbiota and mammalian immunity. Annu. Rev. Microbiol 72, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passam FH, Giannakopoulos B, Mirarabshahi P, and Krilis SA (2011). Molecular pathophysiology of the antiphospholipid syndrome: the role of oxidative post-translational modification of beta 2 glycoprotein I. J. Thromb. Haemost 9, 275–282. [DOI] [PubMed] [Google Scholar]
- Paun A, Yau C, Meshkibaf S, Daigneault MC, Marandi L, Mortin-Toth S, Bar-Or A, Allen-Vercoe E, Poussier P, and Danska JS (2019). Association of Hla-dependent islet autoimmunity with systemic antibody responses to intestinal commensal bacteria in children. Sci. Immunol 4, eaau8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Kelchtermans H, de Groot PG, Zuily S, Regnault V, Wahl D, Pengo V, and de Laat B (2013). Variability in exposure of epitope G40-R43 of domain I in commercial anti-beta2-glycoprotein I IgG ELISAs. PLoS One 8, e71402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericleous C, Ruiz-Limon P, Romay-Penabad Z, Marin AC, Garza-Garcia A, Murfitt L, Driscoll PC, Latchman DS, Isenberg DA, Giles I, et al. (2015). Proof-of-concept study demonstrating the pathogenicity of affinity-purified IgG antibodies directed to domain I of beta2-glycoprotein I in a mouse model of anti-phospholipid antibody-induced thrombosis. Rheumatology 54, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz E, and Kostner GM (1979). The binding of beta 2-glycoprotein-I to human serum lipoproteins: distribution among density fractions. FEBS Lett. 102, 183–186. [DOI] [PubMed] [Google Scholar]
- Poullis A, Foster R, Mendall MA, Shreeve D, and Wiener K (2003). Proton pump inhibitors are associated with elevation of faecal calprotectin and may affect specificity. Eur. J. Gastroenterol. Hepatol 15, 573–574. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glöckner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Rauch J, Salem D, Subang R, Kuwana M, and Levine JS (2018). beta2-glycoprotein I-reactive T cells in autoimmune disease. Front. Immunol. 9, 2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, and Kroemer G (2018). The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol 15, 382–396. [DOI] [PubMed] [Google Scholar]
- Ruff WE, and Kriegel MA (2015). Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol. Med 21, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff WE, Vieira SM, and Kriegel MA (2015). The role of the gut microbiota in the pathogenesis of antiphospholipid syndrome. Curr. Rheumatol. Rep 17, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Irastorza G, Crowther M, Branch W, and Khamashta MA (2010). Antiphospholipid syndrome. Lancet 376, 1498–1509. [DOI] [PubMed] [Google Scholar]
- Salem D, Subang R, Okazaki Y, Laplante P, Levine JS, Kuwana M, and Rauch J (2015). beta2-glycoprotein I-specific T cells are associated with epitope spread to lupus-related autoantibodies. J. Biol. Chem 290, 5543–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz F, Soderland C, Hendricks-Muñoz KD, Gerrets RP, and Lockwood CJ (2000). Human endometrial endothelial cells: isolation, characterization, and inflammatory-mediated expression of tissue factor and type 1 plasminogen activator inhibitor. Biol. Reprod 62, 691–697. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straszewski-Chavez SL, Abrahams VM, Alvero AB, Aldo PB, Ma Y, Guller S, Romero R, and Mor G (2009). The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 30, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, and Davis MM (2013). Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 38, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, and Deshmukh US (2014). T cell epitope mimicry between Sjogren’s syndrome antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin. Immunol 152, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai N, Peng J, Liu F, Gulden E, Hu Y, Zhang X, Chen L, Wong FS, and Wen L (2016). Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med 213, 2129–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura G, Fujiwara H, Yoshida H, Wu DJ, Matsuda M, Ishida M, Kawamura A, Fujiwara T, and Kawai C (1989). High frequency of spontaneous acute myocardial infarction due to small coronary artery disease in dead (NZWxBXSB)F1 male mice. Am. J. Pathol 135, 989–999. [PMC free article] [PubMed] [Google Scholar]
- Tibble JA, Sigthorsson G, Foster R, Scott D, Fagerhol MK, Roseth A, and Bjarnason I (1999). High prevalence of NSAID enteropathy as shown by a simple faecal test. Gut 45, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, and Wardemann H (2007). Autoreactivity in human IgG+ memory B cells. Immunity 26, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S, Pryer KM, Miao VP, and Palmer JD (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol 46, 327–338. [DOI] [PubMed] [Google Scholar]
- Varrin-Doyer M, Spencer CM, Schulze-Topphoff U, Nelson PA, Stroud RM, Cree BA, and Zamvil SS (2012). Aquaporin 4-specific T cells in neuromyelitis optica exhibit a Th17 bias and recognize Clostridium ABC transporter. Ann. Neurol 72, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, and Knight R (2013). EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, and Gewirtz AT (2010). Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, and Núñez G (2016). Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated by 16S rRNA high-throughput sequencing of human fecal and IgA-seq samples have been deposited in the European Nucleotide Archive. The accession number for the data reported in this paper is ENA: PRJEB32067.