Abstract
Posttranscriptional modifications of RNA represent an emerging class of regulatory elements in human biology. Improved methods for studying how these elements are controlled and where they occur has the potential to transform our understanding of gene expression in development and disease. Here we describe a chemical method for nucleotide resolution sequencing of N4-acetylcytidine (ac4C), a highly conserved modified nucleobase whose formation is catalyzed by the essential cytidine acetyltransferase enzyme NAT10. This approach enables the sensitive, PCR-amplifiable detection of individual ac4C sites from nanograms of unfractionated cellular RNA. The sensitive and quantitative nature of this assay provides a powerful tool to understand how cytidine acetylation is targeted, profile RNA acetyltransferase dynamics, and validate the sites and stoichiometry of ac4C in novel RNA species.
1. Introduction
To date more than 100 modified nucleotides have been identified in RNA across all domains of life (Boccaletto et al., 2018). The majority of these posttranscriptional modifications have been characterized in abundant ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), where they often are found at highly conserved sites and can influence properties such as ribosome maturation and tRNA stability (Agris, 2004; Sloan et al., 2017). More recently, advances in next-generation sequencing technologies have enabled the mapping of several posttranscriptional modifications in messenger RNAs (mRNAs) as well as in functional non-coding RNAs (e.g., microRNAs) (Alarcon, Lee, Goodarzi, Halberg, & Tavazoie, 2015; Gilbert, Bell, & Schaening, 2016). In these species the presence of posttranscriptional modifications, referred to as the “epitranscriptome,” is capable of influencing diverse functions including translational efficiency, mRNA stability, and RNA-protein interactions (Frye, Harada, Behm, & He, 2018; Schwartz, 2016).
1.1. Cytidine acetylation
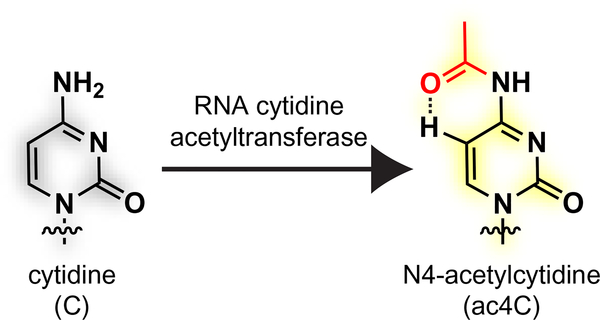
The regulatory potential of RNA modifications has often been viewed as conceptually analogous to the epigenome, where dynamic changes in histone acetylation serve as a hallmark of active transcription (He, 2010; Meier, 2013). Acetylation of RNA also exists, in the form of three modified nucleobases: N6-acetyladenosine and N4-acetyl-2′-O-methylcytidine (ac4Cm), which are found in thermophilic archaea; and N4-acetylcytidine (ac4C), which is found in all domains of life (Boccaletto et al., 2018) (Fig. 1). Historically, ac4C was first identified in the 1960s during the initial sequence determination of yeast and mammalian transfer RNA (tRNAs) (Kowalski, Yamane, & Fresco, 1971; Staehelin, Rogg, Baguley, Ginsberg, & Wehrli, 1968; Zachau, Dutting, & Feldmann, 1966). Subsequently, this modification was also discovered to occur in eukaryotic ribosomal RNA (Thomas, Gordon, & Rogg, 1978) and bacterial tRNAs (Oashi et al., 1972). Recent reports suggest ac4C may also be found in small RNAs and messenger RNAs in human cell lines (Arango et al., 2018; Lan et al., 2018; Tardu, Lin, & Koutmou, 2018), as well as viral RNA (McIntyre et al., 2018). The function of ac4C has been most well studied in bacterial tRNA, where it occurs at the wobble position of the initiator tRNAMet and improves translational fidelity by favoring pairing with the AUG codon, which specifies methionine, over the AUA codon, which specifies isoleucine (Stern & Schulman, 1978; Taniguchi et al., 2018). This effect has been attributed to the ability of ac4C’s preferred pseudo-bicyclic conformation, in which the acetyl group is oriented proximal to C5 of cytidine, to influence Watson-Crick base pairing and mismatch discrimination (Fig. 1) (Kumbhar, Kamble, & Sonawane, 2013; Parthasarathy, Ginell, De, & Chheda, 1978). Less is known about the role of ac4C in eukaryotic systems, although studies in yeast have pointed to a role in rRNA biogenesis and tRNA stability (Ito et al., 2014; Johansson & Bystrom, 2004; Sharma et al., 2015).
Fig. 1.
Posttranscriptional acetylation of RNA cytidine residues forms N4-acetylcytidine (ac4C), shown in the more stable proximal conformation which preserves the native Watson-Crick edge of cytidine.
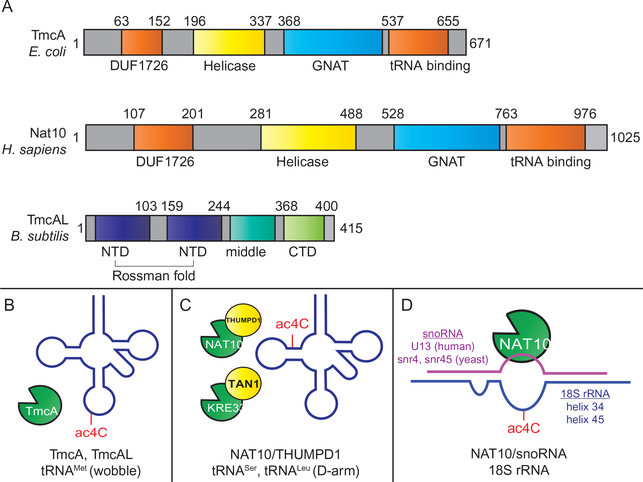
Regulation of ac4C is accomplished via RNA cytidine acetyltransferase enzymes. Thus far, all organisms in which ac4C has been characterized harbor a single RNA acetyltransferase gene. These enzymes can be broadly grouped into three general types (Fig. 2A):
TmcA-type RNA acetyltransferases are found in bacteria where they catalyze acetylation of the wobble base of tRNAMet (Chimnaronk et al., 2009; Ikeuchi, Kitahara, & Suzuki, 2008) (Fig. 2B). These are multi-domain enzymes approximately 600–700 amino acids in length which contain both a Walker-type ATPase/helicase domain and a Gcn5-related N-acetyltransferase (GNAT) domain. Structural and functional studies of these enzymes favor a reaction mechanism in which cytidine acetylation occurs following unwinding of the tRNA anticodon stem by the helicase domain. Genetic studies in E. coli and H. volcanii have found that TmcA is a non-essential gene whose deletion does not cause overt growth defects unless combined with other mutations (El Yacoubi et al., 2009; Ikeuchi et al., 2008).
Nat10/Kre33-type RNA acetyltransferases are eukaryotic enzymes (Nat10, human; Kre33, yeast) that mediate cytidine acetylation in the stem of the D arm of tRNALeu and tRNASer, as well as in helix 34 and helix 45 of 18S ribosomal RNA (Ikeuchi et al., 2008; Ito et al., 2014; Sharma et al., 2015). These are ~1000 amino acid proteins comprised of a helicase domain, a GNAT domain, and multiple predicted RNA-binding domains. Nat10 and its homologs are essential in humans, mice, and yeast, although mutational studies indicate the acetyltransferase activity of the latter may be dispensable (Sharma et al., 2015). Relatively little biochemical data exists for these enzymes. However, all sites of ac4C that have been localized in organisms with Nat10-type acetyltransferases occur within base-paired 5′-CCG-3′ sequences. Furthermore, cellular studies indicate deposition of these sites requires either an adapter protein for tRNA acetylation (Thumpd1, human; Tan1, yeast) (Johansson & Bystrom, 2004) (Fig. 2C), or an adapter antisense snoRNA to guide rRNA acetylation (U13, human; snr4/snr45, yeast) (Sharma et al., 2015; Sharma et al., 2017) (Fig. 2D). Chemical proteomic profiling indicates Nat10 may be among the most abundant and active acetyltransferases in human cell lines (Montgomery, Sorum, Guasch, Nicklaus, & Meier, 2015; Montgomery, Sorum, & Meier, 2014).
TmcAL-type enzymes are a third class of cytidine acetylation catalysts which were recently discovered (Taniguchi et al., 2018) (Fig. 2A–B). These enzymes were found through the study of bacteria that utilize ac4C at the wobble base of tRNAMet but do not harbor TmcA homologs in their genome. TmcAL is distinguished from TmcA and Nat10 in that it does not contain a helicase domain, a GNAT domain, or use acetyl-CoA as a cofactor. Instead, TmcAL uses a two-step mechanism in which acetate and ATP react to form a high energy acetyl adenylate intermediate, which is subsequently acetylates the exocyclic amine of cytidine (Taniguchi et al., 2018). This mechanism is similar to that used by aminoacyl-tRNA synthetases. Bacteria lacking TmcAL display a cold-sensitive phenotype and mistranslate AUA codons, which specify isoleucine, as AUG, which specifies methionine.
Fig. 2.
Cytidine acetylation enzymes, substrates, and adapter molecules. (A) Cytidine acetylation enzymes. (B) Known substrates of TmcA and TmcAL cytidine acetylation enzymes. (C) Known tRNA substrates of Nat10/Kre33-type cytidine acetyltransferases. tRNA acetylation in eukaryotes requires an adapter protein (THUMPD1, human; Tan1, yeast). (D) Known ribosomal RNA substrates of Nat10/Kre33-type cytidine acetyltransferases. rRNA Acetylation requires short nuclear RNA (snoRNA) adapters (human helix 45, U13; yeast helix 45, snr45; yeast helix 34, snr4). Note: Identify of snoRNA guiding Human helix 34 acetylation has not yet been identified.
Dysregulation of the only known human cytidine acetyltransferase, Nat10, has recently been linked to many diseases including Hutchinson-Gilford progeria syndrome (Larrieu, Britton, Demir, Rodriguez, & Jackson, 2014), ovarian cancer (Tan et al., 2013), colorectal cancer (Zhang et al., 2014), and liver cancer (Tschida et al., 2017). The pathophysiological associations and essential role of Nat10 in eukaryotes, as well as recent proposals of pervasive cytidine acetylation in coding RNAs (Arango et al., 2018), have spurred new interest in the development of methods to study the sites, dynamics, and regulation of ac4C.
1.2. Detection, localization, and quantification of N4-acetylcytidine in RNA
Early methods used to identify ac4C in tRNA and rRNA involved partial enzymatic digestions and two-dimensional paper chromatography (Kowalski et al., 1971; Staehelin et al., 1968; Zachau et al., 1966). More recently, ac4C has been localized at precise sites in eukaryotic rRNA using two methods: (1) LC-MS analysis of RNase-digests, and (2) UV-HPLC analysis of nucleosides derived from mung-bean nuclease assays (Ito, Akamatsu, et al., 2014; Sharma et al., 2015; Sharma, Marchand, Motorin, & Lafontaine, 2017). While these digest-based methods have greatly advanced our understanding of ac4C, they suffer from poor sensitivity due to a lack of signal amplification. This leads to significant material requirements which can limit utility and throughput, and has prompted us to explore alternative methods for the analysis of cytidine acetylation (Sinclair et al., 2017), including nucleotide resolution sequencing (Motorin, Muller, Behm-Ansmant, & Branlant, 2007; Schwartz & Motorin, 2017).
Sequencing-based detection relies on the ability of RNA modifications to interfere with reverse transcription. For example, many RNA modifications including N1-methyadenosine (m1A) and N3-methycytidine (m3C) block Watson-Crick hydrogen bonding (Harcourt, Kietrys, & Kool, 2017). This causes reverse transcriptase (RT) enzymes to prematurely terminate or misincorporate non-cognate deoxynucleoside triphosphates (dNTPs) during cDNA synthesis, the latter of which can be detected by PCR amplification and sequencing. In contrast to m1A and m3C, many RNA modifications including ac4C, 5-methylcytidine (5mC), and pseudouridine do not alter the Watson Crick edge, rendering them nearly “invisible” during reverse transcription (Ryvkin et al., 2013) (Fig. 1). These modifications can be instead be detected by antibody-based methods, or by exploiting differences in chemical reactivity to distinguish their RT profile from that of canonical nucleobases. A prototypical example of the latter approach is bisulfite sequencing of 5mC in DNA and RNA (Hayatsu, 2008; Schaefer, 2015).
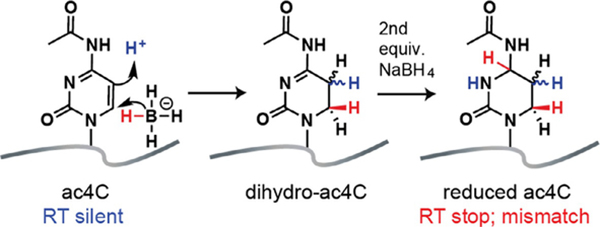
Recently we reported a chemical reaction to enable sequencing-based detection of ac4C in RNA (Thomas et al., 2018). Analogous to bisulfite sequencing, our approach exploits the unique electronic structure of ac4C, which renders its 5,6-double bond susceptible to nucleophilic attack by sodium borohydride (Fig. 3) (Cerutti & Miller, 1967; Miller & Cerutti, 1967). The addition of two equivalents of hydride forms the reduced nucleotide N4-acetyl-3,4,5,6-tetrahydrocytidine. Analysis of model substrates as well as physiological sites in rRNA revealed that chemical reduction of ac4C causes premature termination and misincorporation during reverse transcription (Fig. 3). The ratio of termination (RT stop) to misincorporation was found to vary based on the specific polymerase used, with the thermostable group II intron reverse transcriptase (TGIRT) showing most efficient read through (Thomas et al., 2018) (Fig. 4). This approach is sensitive, requiring only a few hundred nanograms RNA for assessment of individual ac4C sites by Sanger sequencing. Furthermore, the extent of misincorporation was found to vary linearly with ac4C content, indicating the utility of this approach to study RNA acetylation stoichiometry (Fig. 5). Here we describe detailed protocols for the sequencing-based detection of ac4C in RNA. We specify key steps and controls important for assessing ac4C in RNA, and highlight future opportunities for applying this chemistry to better understand the role of cytidine acetylation in fundamental biology and human disease.
Fig. 3.
Chemical reduction of N4-acetylcytidine in RNA to N4-acetyl-3,4,5,6-tetrahydrocytidine (referred to here as “reduced ac4C”). Reduction of ac4C can cause termination (RT stop) or dNTP misincorporation (mismatch) during reverse transcription. Figure is adapted with permission from Thomas, J. M., Briney, C. A., Nance, K. D., Lopez, J. E., Thorpe, A. L., Fox, S. D., et al. (2018). A chemical signature for cytidine acetylation in RNA. Journal of the American Chemical Society. 140, 12667–12,70. Copyright American Chemical Society, 2018.
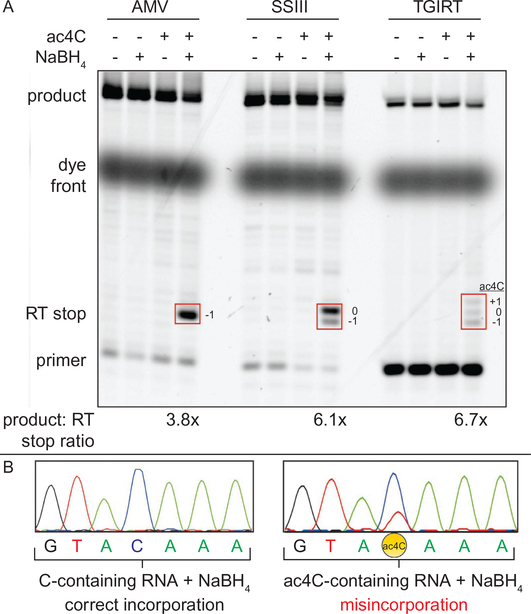
Fig. 4.
Monitoring reduction of ac4C in RNA by RT stop and misincorporation. (A) Comparison of primer extension following NaBH4 reduction of an ac4C-containing RNA. A model in vitro transcribed RNA containing a single site of ac4C was treated with NaBH4 (100mM, 37 °C, 1h). The product of this reaction was incubated with a Cy5 primer, dNTPs, and the specified reverse transcriptase according to the manufacturer’s recommended conditions. Substantial RT stop was observed with AMV, while slightly more product was formed using SuperScript III and TGIRT RT. The position of the RT stop product is specified relative to the ac4C site (+1, 0, −1). (B) Analysis of PCR-amplified full-length cDNAs (sense strand) by Sanger sequencing. Misincorporation of dNTPs is observed only for the ac4C-containing model RNA. All cDNAs were produced via TGIRT RT. Figure is adapted with permission from Thomas, J. M., Briney, C. A., Nance, K. D., Lopez, J. E., Thorpe, A. L., Fox, S. D., et al. (2018). A chemical signature for cytidine acetylation in RNA. Journal of the American Chemical Society. 140, 12667–12670. Copyright American Chemical Society, 2018.
Fig. 5.
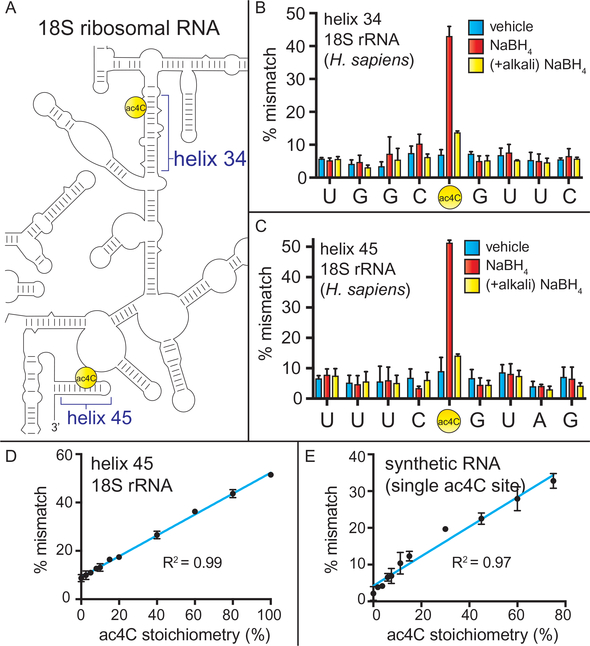
Quantitative sequencing-based detection of ac4C in model substrates and 18S rRNA. (A) Schematic of human rRNA ac4C sites. (B) Sequencing-based detection of ac4C-dependent mismatches in cDNAs generated from human rRNA helix 34 and (C) helix 45 following borohydride treatment and RT. Sequence corresponds to the cDNA sense strand. Vehicle = water; NaBH4 = 100mM sodium borohydride; (+alkali) NaBH4 = alkali pre-treatment (100mM NaCO3, pH10, 60 °C, 1h), precipitation, then borohydride. (D) Relationship between misincorporation signal and stoichiometry of ac4C in rRNA and (E) a synthetic RNA harboring ac4C in an “ACA” sequence context. For all data, error bars indicate the standard deviation (n = 3). Figure is reprinted with permission from Thomas, J. M., Briney, C. A., Nance, K. D., Lopez, J. E., Thorpe, A. L., Fox, S. D., et al. (2018). A chemical signature for cytidine acetylation in RNA. Journal of the American Chemical Society. 140, 12667–12670. Copyright American Chemical Society, 2018.
2. Technical aspects
2.1. Sample preparation and RNA isolation
This protocol utilizes total RNA extracted from tissues or cultured cells by TRIzol extraction. High quality total RNA isolated using alternative methods may also be used. Treatment with DNase is essential to avoid downstream amplification of ribosomal DNA. All reagents used should be free of RNases and care should be taken at all times to avoid contamination of samples with RNases which come from human skin and are prevalent in laboratory environments. It is recommended to wear gloves during all procedures and avoid contact between gloves and the sealing surfaces of microcentrifuge tube lids which can occur when opening.
2.1.1. Materials
TRIzol Reagent (Thermofisher, 15596018)
PBS (Thermofisher, 10010)
Chloroform:isoamyl alcohol 24:1, suitable for nucleic acid purification (Sigma, CO549)
Isopropyl Alcohol (Macron Fine Chemicals, 3032–16)
TURBO DNase kit (2U/μL) (Invitrogen, AM2238)
UltraPure Phenol:Chloroform:isoamyl Alcohol 24:24:1 (Thermofisher, 15593–049)
Molecular biology grade Ethanol, Absolute (200 Proof ) (Fisher Bio-Reagents, BP2818500)
Nuclease free ultra-pure water (Invitrogen 10977)
Sodium Acetate (3M) pH5.5 (Invitrogen, AM9740)
Thermo Scientific NanoDrop 2000 Spectrophotometer
Centrifuge capable accepting 15mL falcon tubes, refrigeration to 4°C and >12,000×g
Tabletop microcentrifuge capable of refrigeration to 4°C and >18,000×g.
2.1.2. RNA isolation
Homogenize freshly washed cell pellets using TRIzol reagent. For homogenization of frozen cell pellets, initially resuspend cells in ice-cold PBS, pellet by centrifugation (200×g, 4°C, 4min), and discard supernatant. Next, add TRIzol reagent (approximately 1mL TRIzol per 15cm2 dish of cells harvested) directly to the freshly washed cell pellet. Vortex and incubate 5min at room temperature.
Add chloroform:isoamyl alcohol solution such that the final proportion of the sample is four parts TRIzol, 1 part chloroform:isoamyl alcohol (4:1 ratio). Once the chloroform-containing solution is added, the sample should become cloudy.
Vortex for 15s, incubate at room temperature for 5min, and centrifuge (12,000×g, 4°C, 15min).
Transfer top aqueous layer, which contains RNA, to a new tube. Care should be taken to avoid touching the middle/interface layer, which contains DNA.
To precipitate RNA, add isopropyl alcohol such that the final proportion of the sample is 1:1 isopropanol:aqueous layer. Mix by vortexing for 15s, incubate at room temperature for 10min, and centrifuge (12,000×g, 4°C, 15min).
Discard the supernatant and add a volume of 75% ethanol equal to the amount of TRIzol used during homogenization (step #1).
Vortex for 5s and centrifuge (7500×g, 4°C, 5min).
Discard supernatant and dry pellet for up to 10min.
Dissolve pellet in 100μL of ultra-pure water and quantify using the NanoDrop 2000 Spectrophotometer
2.1.3. DNase-treatment of total RNA
Dilute RNA to 200μg/mL with ultra-pure water and add 10× Turbo DNase buffer to 1× concentration.
Add 1μL Turbo DNase for every 10μg RNA. Mix gently and incubate at 37°C for 1h.
If DNase treatment reaction is <200μL, add sufficient ultra-pure water to bring the volume to 200μL.
Next you will phenol: chloroform extract and ethanol precipitate the total RNA in order to remove DNase. First, add equal volume phenol: chloroform:isoamyl alcohol to the sample. Vortex and centrifuge at 12,000×g at room temperature for 5min using a tabletop ultracentrifuge.
Collect top aqueous layer and transfer to a clean tube.
Add a 1:1 volume of chloroform to the aqueous layer. Vortex and centrifuge at 12,000×g at room temperature for 5min using a tabletop ultracentrifuge.
Collect top aqueous layer and transfer to a clean tube.
To ethanol precipitate DNase-treated RNA, Add 1:9 volume of 3M sodium acetate and 3:1 volume of 100% Ethanol to the sample. Mix thoroughly.
Store in −80°C for ≥1h.
After −80°C incubation, spin in a microcentrifuge at 18,000×g at 4°C for 30min.
Decant the supernatant, and wash pellet with 500μL of ice-cold 70% ethanol. Spin in a microcentrifuge for an additional 10min at 18,000 ×g at 4°C.
Decant supernatant and air-dry pellet for 10–15min.
Resuspend pellet in 50μL ultra-pure water per 15cm2 dish of cells harvested and heat/incubate at 55°C for 15min.
Quantify RNA using the NanoDrop 2000 Spectrophotometer.
2.2. Chemical manipulation of ac4C in total cellular RNA
As little as 10pg of total RNA can be used to successfully generate cDNA for nucleotide resolution ac4C sequencing; however, for convenient sample handling when material is not limiting, we recommend beginning with approximately 3μg of total RNA from each biological condition to be analyzed.
2.2.1. Materials
Sodium Borohydride (Sigma-Aldrich, 632287)
Hydrochloric acid (Sigma-Aldrich, H1758)
Sodium Bicarbonate (Sigma Aldrich, S5761)
UltraPure 1M Tris-HCI Buffer, pH7.5 (Invitrogen, 15567027)
2.2.2. Reductive RNA treatment
This procedure details the parallel preparation of samples subjected to three sets of conditions: Borohydride treated RNA (+borohydride), borohydride treated deacetylated RNA (+borohydride +alkali) and mock treated RNA (−borohydride). The latter two conditions serve as controls. Sodium borohydride selectively reduces ac4C, converting it to tetrahydroacetycytidine while leaving cytidine unmodified (Fig. 3). Caution should be used when preparing and handling solutions containing NaBH4 as borohydride decomposes in aqueous solutions to produce hydrogen gas which can pressurize reaction vessels. We recommend preparing NaBH4 stocks immediately before use and taking care to vent reactions during or after chemical treatments.
Prepare fresh 1M NaBH4 solution immediately before using in ice cold nuclease-free water. Once dissolved the solution will begin bubbling (release of hydrogen gas). Reactions should be vented in order to prevent pressurization of vessels.
Prepare three separate reactions per sample: (1) For the +borohydride RNA, bring 1μg total RNA to 90μL with nuclease-free water. Add 10μL 1M NaBH4. (2) For the +borohydride +alkali RNA, bring alkali-treated total RNA (prepared in Section 2.2.3) to 90μL with nuclease-free water and add 10μL 1M NaBH4. (3) For the −borohydride RNA, bring 1μg RNA to 100μL in nuclease-free water.
Incubate at 37°C for 60min.
Add 15μL 1M HCl to each sample to quench residual NaBH4. Addition of acid will cause borohydride containing samples to bubble vigorously.
Once bubbling has ceased or 1min, add 30μL 1M Tris pH7.5 to neutralize residual HCL.
Adjust volume to 200μL and ethanol precipitate as described above in Section 2.1.3 steps 8–12.
Resuspend pellet in 25μL ultra-pure water and vortex.
Quantify RNA using the NanoDrop 2000 spectrophotometer
2.2.3. Chemical deacetylation control
Chemical deacetylation via base-pretreatement serves as a convenient control for ac4C dependent misincorporation. A portion of the sample (1μg) is subjected to treatment in mild alkali. This causes hydrolysis of ac4C to cytidine, rendering it non-reactive with borohydride and limiting borohydride-dependent dNTP misincorporation during RT.
To chemically deacetylate ac4C, incubate 1μg of RNA sample in 100μL of 100mM sodium bicarbonate pH9.5 (prepared in nuclease-free water) at 60°C for 1h. This condition promotes substantial conversion (>80%) of ac4C to cytidine without causing excessive degradation of the RNA backbone as assessed by Bioanalyzer.
Adjust the volume of the RNA reaction to 200μL with nuclease-free water. Ethanol precipitate the sample as described above in Section 2.1.2, steps 8–12.
Resuspend pellet in 25μL ultra-pure water and vortex.
Quantify RNA using the NanoDrop 2000 spectrophotometer
2.3. Nucleotide resolution sequencing of individual ac4C sites
2.3.1. Materials
TGIRT-III reverse transcription enzyme (INGEX)
5× TGIRT reaction buffer (user prepared with nuclease-free reagents, 2.25M NaCl, 100mM Tris-HCL pH7.5)
RNasin plus, 40units/μL (Promega, N2611)
Optimized dNTP mix [10mM dTTP, 10mM dCTP, 10mM dATP, 5mM dGTP] (prepared from 100mM dNTPs set, NEB N0446S)
Phusion High-Fidelity PCR Kit (New England Biolabs, E0553S)
Agarose LE, Molecular Biology Grade (Thomas Scientific, C996H59)
100bp DNA Ladder (New England Biolabs, N3231S)
SYBR Safe DNA Gel Stain (Invitrogen, S33102)
UltraPure TBE Buffer, 10× (Invitrogen, 15581044)
UV transilluminator (UVP MultiDoc-It Imager Benchtop UV Transilluminator)
6× DNA gel loading dye (NEB, B7021S)
QIAquick Gel extraction kit (Qiagen, 28704)
Thermocycler
Agarose gel electrophoresis equipment.
2.3.2. Reverse transcription of borohydride-treated RNA
Reverse transcription of sodium borohydride-treated ac4c results in partial incorporation of adenosine in the cDNA strand opposite the reduced ac4C. It should also be noted that reduced ac4C results in partial termination of reverse transcription at or adjacent to its location, often referred to as RT-stops (Fig. 4A). Of the reverse transcriptase enzymes screened by our lab TGIRT-III RT (Ingex) produces the highest proportion of full-length to stop product, as well as the most robust C-to-T signature of the RT enzymes screened by our lab (Fig. 4B). The following protocol describes the use of TGIRT RT for sequence-defined cDNA generation, which we have used to profile ac4C in model substrates and human 18S rRNA (Thomas et al., 2018).
Dilute RNA from Section 2.2.2 to 100pg/μL in water for use as template.
Perform TGIRT RT reactions. We typically carry out RT in 20μL volumes in PCR tubes. Exemplary protocol: combine 4μL 5× TGIRT reaction buffer with 200pg (2μL) of template, 4 pM DNA primer, 2μL 50mM MgCl2 and sufficient ultra-pure water to bring the mixture to 17μL. Note 1: TGIRT reactions are incubated at 57 °C, higher than many other commercially available reverse transcription enzymes. Therefore, it is essential to design the reverse transcription primer to form a stable duplex with the target sequence at this temperature. Note 2: Fluorophore or radiolabeled primers can be substituted at this step to assess RT stop via primer extension assay (Fig. 4)
Heat to 75 °C for 3min and placed on ice for 1min to anneal the primer and RNA.
After cooling, add 0.5μL TGIRT-III, 0.5μL Rnasin Plus and 100μL 100mM DTT. Incubate 20min at room temperature. Initiate the reverse transcription reaction by adding 1μL of optimized dNTP mix. In optimization studies we have found that decreasing the dGTP concentration from 500 to 250μM increases the sensitivity of C-to-T misincorporation. Incubate the reaction for one hour at 57 °C and proceed to PCR amplification step (Section 2.3.3).
Note on additional controls: We recommend performing a reverse transcription reaction in the absence of template to ensure the reagents being used are free from contaminating RNA/DNA. It is also essential to include control reactions that lack RT enzyme for each biological replicate, which allows confirmation that the PCR reaction (Section 2.3.3) is amplifying only cDNA generated through reverse transcription and not contaminating gDNA from other sources.
2.3.3. PCR amplification and purification of cDNAs
In order to PCR amplify cDNA, first prepare 50μL PCR reactions using Phusion Hot Start Flex DNA polymerase according to the manufacturers recommended protocol using 1μL TGIRT reaction product as template. Final reaction conditions used are 1× HF buffer, 200μM dNTPs, 0.5μM forward primer, 0.5μM reverse primers, 1 unit polymerase, and 1μL TGIRT reaction product. Note: A PCR reaction lacking template is recommended to check that PCR reagents are not contaminated with amplifiable genomic DNA. Annealing temperature and number of amplification cycles should be determined empirically for each cDNA to be analyzed. To amplify cDNAs derived from the ac4C-continaing helix 45 of human 18S rRNA, we utilize an annealing temperature of 67.4 °C and amplify for 33cycles (Thomas et al., 2018).
Purifying the PCR product by agarose gel purification/extraction is recommended prior to submission of PCR product for sequencing for best results. Mix PCR product with 6 × loading buffer and run on 2% agarose gel containing 1 × SYBRsafe DNA gel stain until bromophenol blue marker dye runs approximately two-thirds of the way across the gel. 1μg of 100bp dsDNA ladder should be run alongside PCR reaction products.
Visualize PCR product on UV transilluminator and excise bands of expected size with a clean razor blade. Control reactions (minus RT and minus template) should be run alongside and visualized to ensure no PCR product is present in these samples.
Process agarose gel slices using commercially available gel extraction kit, elute DNA and submit for Sanger sequencing with an appropriate primer.
2.3.4. Sanger sequencing-based analysis of cytidine acetylation
To calculate ac4C-dependent misincorporation, open processed Sanger sequencing traces using sequence trace viewing software. Free software which allow peak heights to be calculated include 4peaks (Mac) and Applied Biosystems Sequence Scanner 2.0 (Windows). Only high-quality sequence traces with low background noise and little or no peak ghosting should be analyzed.
Identify sites of ac4C by locating bases that appear as a mixture of C and T in borohydride treated samples (+borohydride). Sites of high ac4C stoichiometry may produce more intense signal for T than C in sequencing chromatograms and may be assigned as T in base called sequences. Sites of ac4C exhibit substantially decreased C-to-T mutation in alkali pretreated controls (+borohydride +alkali) and only background levels of misincorporation (<10%) in untreated controls (−borohydride) (Fig. 5).
Quantify misincorporation at ac4C sites by measuring the height of C and T peaks in the sequencing traces and calculate misincorporation using the equation: percent misincorporation = 100*(T-peak)/((C-peak)+(T-peak)).
To estimate ac4C stoichiometry at a given site, fit calculated misincorporation to a standard curve. A standard curve for ac4C dependent misincorporation can be generated by plotting the average fraction of C-to-T conversion at the site of interest across a series of RNA mixtures prepared from defined ratios of wild-type and Nat10 knockout cells (100:0, 80:20, 60:40, etc.; Fig. 5).
3. Discussion
3.1. Critical parameters and troubleshooting
A critical aspect in sequencing-based detection of ac4C is the use of proper genetic and/or chemical controls. At least three other modified RNA nucleobases are susceptible to reaction with sodium borohydride: dihydrouridine, 7-methylguanosine and N3-methylcytidine (m3C) (Cerutti, Kondo, Landis, & Witkop, 1968; Marchand et al., 2018; Wintermeyer & Zachau, 1970). One way in which ac4C can be distinguished from these other borohydride-reactive modifications is by monitoring the influence of deletion or mutation of cellular cytidine acetyltransferase on misincorporation (Arango et al., 2018). Cytidine acetyltransferases are highly conserved, and can be rapidly identified by searching genomes for homologs to TmcA, Kre33, and Nat10. The selection of genetic controls is further simplified by the fact that in all organisms known to harbor ac4C, a single cytidine acetylation enzyme is known. One caveat to this approach is the fact that in eukaryotes Nat10/Kre33-type cytidine acetyltransferases have been found to be essential for life (Sharma et al., 2015; Wang et al., 2015). In these cases, siRNA knockdown, targeted mutation of the acetyltransferase domain, or conditional knockout may serve as useful controls. Genetic ablation may also be used to target non-essential protein and snoRNA adapters required for tRNA and rRNA acetylation (Fig. 3C and D) (Sharma, Yang, et al., 2017).
In the absence of genetic approaches, chemical deacetylation provides a valuable alternative for attributing a borohydride-dependent misincorporation event to ac4C (Sinclair et al., 2017). These controls take advantage of the fact that the electron-deficient ac4C acetamide is susceptible to cleavage by nucleophiles and alkali conditions, which convert ac4C sites to unmodified cytidine. This property is unique compared to other borohydride-reactive RNA modifications, and defines base-sensitive borohydride-dependent misincorporation as a unique chemical signature of ac4C in RNA (Thomas et al., 2018). An important aspect of these chemical controls, as well as borohydride-treatment itself, is that they create basic conditions that can cause RNA strand scission. This may contribute to background in RT stop assays (Marchand et al., 2018), and is an important consideration for PCR amplification and next-generation sequencing library preparation strategies. Other features of ac4C’s reductive chemistry that distinguish it from other modifications include that its reduction proceeds best in unbuffered solutions of aqueous borohydride, in contrast to the buffered solutions used to reduce dihydrouridine (Cerutti & Miller, 1967). Finally, for Sanger sequencing studies of ac4C in abundant species such as 18S rRNA we have found that purity of the borohydride-treated amplified cDNA is critical for obtaining reproducible quantitative data. Controls that can be used to assess contamination include performing RT in the absence of an RNA template, ensuring the dependence of PCR amplification on the presence RT enzyme, and performing PCR in the absence of template cDNA. In addition, we recommend purifying the final PCR amplicon by agarose gel to obtain the highest quality sequencing results.
3.2. Applications and future directions
Here we have described a sensitive protocol for the nucleotide resolution sequencing-based detection of the RNA modification ac4C. The method described here is immediately applicable to the analysis ac4C’s known rRNA and pre-rRNA sites. The ability to quantitatively assess the acetylation of these sites has several applications, including studying how helix 34 and helix 45 ac4C is affected by stimuli such as small molecules (Larrieu et al., 2014), the metabolic state of the cell (Montgomery et al., 2016), and disease states. In addition, similar to how primer extension assays have facilitated the understanding of snoRNA-dependent ribose methylation (Bachellerie & Cavaille, 1997), the availability of a sensitive and site-specific assay for ac4C should be useful in defining how snoRNAs target cytidine acetylation for deposition. In addition to ac4C, the chemistry described here should also be useful for studies of ac4Cm, which is found in some Archaea (Bruenger et al., 1993; Kawai et al., 1991). One limitation of our method is its ability to interrogate ac4C in more densely-functionalized modification landscapes. The known ac4C sites in eukaryotic tRNASer and tRNALeu lie directly adjacent to dihydrouridine, whose coordinate reduction may severely limit RT readthrough. In the future we anticipate this limitation may be addressed by integrating borohydride-based ac4C detection with recently reported tRNA sequencing methods, exploring the use of more processive polymerases such as HIV and Marathon RT, and by integrating our protocol with deep sequencing methods (Sexton, Wang, Rutenberg-Schoenberg, & Simon, 2017; Zheng et al., 2015; Zhou et al., 2018). Finally, we anticipate nucleotide resolution sequencing will form an essential component of strategies to identify, validate, and assess the stoichiometry of novel ac4C sites in coding RNAs, whose existence is supported by recent RIP-Seq and LC-MS studies in humans and yeast (Arango et al., 2018; Tardu et al., 2018). Overall, our methods provide a foundation for the quantitative and site-specific analysis of ac4C, and as such have the potential to expand our understanding of acetylation as a component of the emerging epitranscriptome.
Acknowledgments
We are grateful to members of the Meier lab for providing helpful discussions and critical feedback on this chapter. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (ZIA-BC011488-06).
References
- Agris PF (2004). Decoding the genome: A modified view. Nucleic Acids Research, 32, 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, & Tavazoie SF (2015). N6-methyladenosine marks primary microRNAs for processing. Nature, 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell, 175, 1872–1886.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie JP, & Cavaille J (1997). Guiding ribose methylation of rRNA. Trends in Biochemical Sciences, 22, 257–261. [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. (2018). MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Research, 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenger E, Kowalak JA, Kuchino Y, McCloskey JA, Mizushima H, Stetter KO, et al. (1993). 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. The FASEB Journal, 7, 196–200. [DOI] [PubMed] [Google Scholar]
- Cerutti P, Kondo Y, Landis WR, & Witkop B (1968). Photoreduction of uridine and reduction of dihydrouridine with sodium borohydride. Journal of the American Chemical Society, 90, 771–775. [DOI] [PubMed] [Google Scholar]
- Cerutti P, & Miller N (1967). Selective reduction of yeast transfer ribonucleic acid with sodium borohydride. Journal of Molecular Biology, 26, 55–66. [DOI] [PubMed] [Google Scholar]
- Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, et al. (2009). RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. The EMBO Journal, 28, 1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Phillips G, Blaby IK, Haas CE, Cruz Y, Greenberg J, et al. (2009). A gateway platform for functional genomics in Haloferax volcanii: Deletion of three tRNA modification genes. Archaea, 2, 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Harada BT, Behm M, & He C (2018). RNA modifications modulate gene expression during development. Science, 361, 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Bell TA, & Schaening C (2016). Messenger RNA modifications: Form, distribution, and function. Science, 352, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt EM, Kietrys AM, & Kool ET (2017). Chemical and structural effects of base modifications in messenger RNA. Nature, 541, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H (2008). Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis–a personal account. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 84, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C (2010). Grand challenge commentary: RNA epigenetics? Nature Chemical Biology, 6, 863–865. [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Kitahara K, & Suzuki T (2008). The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. The EMBO Journal, 27, 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Akamatsu Y, Noma A, Kimura S, Miyauchi K, Ikeuchi Y, et al. (2014). A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. The Journal of Biological Chemistry, 289, 26201–26212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, et al. (2014). Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). The Journal of Biological Chemistry, 289, 35724–35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, & Bystrom AS (2004). The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA, 10, 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai G, Ue H, Yasuda M, Sakamoto K, Hashizume T, McCloskey JA, et al. (1991). Relation between functions and conformational characteristics of modified nucleosides found in tRNAs. Nucleic Acids Symposium Series, 49–50. [PubMed] [Google Scholar]
- Kowalski S, Yamane T, & Fresco JR (1971). Nucleotide sequence of the “denaturable” leucine transfer RNA from yeast. Science, 172, 385–387. [DOI] [PubMed] [Google Scholar]
- Kumbhar BV, Kamble AD, & Sonawane KD (2013). Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochemistry and Biophysics, 66, 797–816. [DOI] [PubMed] [Google Scholar]
- Lan MD, Xiong J, You XJ, Weng XC, Zhou X, Yuan BF, et al. (2018). Existence of diverse modifications in small-RNA species composed of 16–28 nucleotides. Chemistry, 24, 9949–9956. [DOI] [PubMed] [Google Scholar]
- Larrieu D, Britton S, Demir M, Rodriguez R, & Jackson SP (2014). Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science, 344, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Ayadi L, Ernst FGM, Hertler J, Bourguignon-Igel V, Galvanin A, et al. (2018). AlkAniline-Seq: Profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angewandte Chemie (International Ed. in English), 57, 16785–16790. [DOI] [PubMed] [Google Scholar]
- McIntyre W, Netzband R, Bonenfant G, Biegel JM, Miller C, Fuchs G, et al. (2018). Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Research, 46, 5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JL (2013). Metabolic mechanisms of epigenetic regulation. ACS Chemical Biology, 8, 2607–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N, & Cerutti PA (1967). Synthesis of N4-acetyl-3,4,5,6-tetrahydrocytidine and copolymers of cytidylic acid and N4-acetyl-3,4,5,6-tetrahydrocytidylic acid. Journal of the American Chemical Society, 89, 2767. [Google Scholar]
- Montgomery DC, Garlick JM, Kulkarni RA, Kennedy S, Allali-Hassani A, Kuo YM, et al. (2016). Global profiling of acetyltransferase feedback regulation. Journal of the American Chemical Society, 138, 6388–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC, Sorum AW, Guasch L, Nicklaus MC, & Meier JL (2015). Metabolic regulation of histone acetyltransferases by endogenous Acyl-CoA cofactors. Chemistry & Biology, 22, 1030–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC, Sorum AW, & Meier JL (2014). Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation. Journal of the American Chemical Society, 136, 8669–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, & Branlant C (2007). Identification of modified residues in RNAs by reverse transcription-based methods. Methods in Enzymology, 425, 21–53. [DOI] [PubMed] [Google Scholar]
- Oashi Z, Murao K, Yahagi T, Von Minden DL, McCloskey JA, & Nishimura S (1972). Characterization of C + located in the first position of the anticodon of Escherichia coli tRNA Met as N 4 -acetylcytidine. Biochimica et Biophysica Acta, 262, 209–213. [PubMed] [Google Scholar]
- Parthasarathy R, Ginell SL, De NC, & Chheda GB (1978). Conformation of N4-acetylcytidine, a modified nucleoside of tRNA, and stereochemistry of codonanticodon interaction. Biochemical and Biophysical Research Communications, 83, 657–663. [DOI] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, et al. (2013). HAMR: High-throughput annotation of modified ribonucleotides. RNA, 19, 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M (2015). RNA 5-methylcytosine analysis by bisulfite sequencing. Methods in Enzymology, 560, 297–329. [DOI] [PubMed] [Google Scholar]
- Schwartz S (2016). Cracking the epitranscriptome. RNA, 22, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, & Motorin Y (2017). Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biology, 14, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Wang PY, Rutenberg-Schoenberg M, & Simon MD (2017). Interpreting reverse transcriptase termination and mutation events for greater insight into the chemical probing of RNA. Biochemistry, 56, 4713–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, & Lafontaine DL (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Research, 43, 2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Marchand V, Motorin Y, & Lafontaine DLJ (2017). Identification of sites of 2’-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Scientific Reports, 7, 11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, van Nues R, Watzinger P, Kotter P, Lafontaine DLJ, et al. (2017). Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genetics, 13, e1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair WR, Arango D, Shrimp JH, Zengeya TT, Thomas JM, Montgomery DC, et al. (2017). Profiling cytidine acetylation with specific affinity and reactivity. ACS Chemical Biology, 12, 2922–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, & Bohnsack MT (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biology, 14, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin M, Rogg H, Baguley BC, Ginsberg T, & Wehrli W (1968). Structure of a mammalian serine tRNA. Nature, 219, 1363–1365. [DOI] [PubMed] [Google Scholar]
- Stern L, & Schulman LH (1978). The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. The Journal of Biological Chemistry, 253, 6132–6139. [PubMed] [Google Scholar]
- Tan TZ, Miow QH, Huang RY, Wong MK, Ye J, Lau JA, et al. (2013). Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Molecular Medicine, 5, 1051–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Miyauchi K, Sakaguchi Y, Yamashita S, Soma A, Tomita K, et al. (2018). Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nature Chemical Biology, 14, 1010–1020. [DOI] [PubMed] [Google Scholar]
- Tardu M, Lin Q, & Koutmou K (2018). N4-acetylcytidine and 5-formylcytidine are present in Saccharomyces cerevisiae mRNAs. biorxiv. 10.1101/327585. [DOI] [Google Scholar]
- Thomas JM, Briney CA, Nance KD, Lopez JE, Thorpe AL, Fox SD, et al. (2018). A chemical signature for cytidine acetylation in RNA. Journal of the American Chemical Society, 140, 12667–12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Gordon J, & Rogg H (1978). N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. The Journal of Biological Chemistry, 253, 1101–1105. [PubMed] [Google Scholar]
- Tschida BR, Temiz NA, Kuka TP, Lee LA, Riordan JD, Tierrablanca CA, et al. (2017). Sleeping beauty insertional mutagenesis in mice identifies drivers of steatosis-associated hepatic tumors. Cancer Research, 77, 6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. (2015). Identification and characterization of essential genes in the human genome. Science, 350, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W, & Zachau HG (1970). A specific chemical chain scission of tRNA at 7-methylguanosine. FEBS Letters, 11, 160–164. [DOI] [PubMed] [Google Scholar]
- Zachau HG, Dutting D, & Feldmann H (1966). Nucleotide sequences of two serine-specific transfer ribonucleic acids. Angewandte Chemie (International Ed. in English), 5, 422. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hou W, Wang HL, Liu HJ, Jia XY, Zheng XZ, et al. (2014). GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clinical Cancer Research, 20, 4717–4729. [DOI] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, et al. (2015). Efficient and quantitative high-throughput tRNA sequencing. Nature Methods, 12, 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KI, Clark WC, Pan DW, Eckwahl MJ, Dai Q, & Pan T (2018). Pseudouridines have context-dependent mutation and stop rates in high-throughput sequencing. RNA Biology, 15, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]