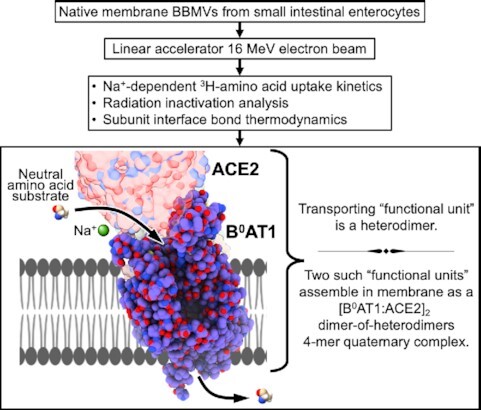
Abstract
The SARS-CoV-2 receptor, angiotensin-converting enzyme-2 (ACE2), is expressed at levels of greatest magnitude in the small intestine as compared with all other human tissues. Enterocyte ACE2 is coexpressed as the apical membrane trafficking partner obligatory for expression and activity of the B0AT1 sodium-dependent neutral amino acid transporter. These components are assembled as an [ACE2:B0AT1]2 dimer-of-heterodimers quaternary complex that putatively steers SARS-CoV-2 tropism in the gastrointestinal (GI) tract. GI clinical symptomology is reported in about half of COVID-19 patients, and can be accompanied by gut shedding of virion particles. We hypothesized that within this 4-mer structural complex, each [ACE2:B0AT1] heterodimer pair constitutes a physiological “functional unit.” This was confirmed experimentally by employing purified lyophilized enterocyte brush border membrane vesicles exposed to increasing doses of high-energy electron radiation from a 16 MeV linear accelerator. Based on radiation target theory, the results indicated the presence of Na+-dependent neutral amino acid influx transport activity functional unit with target size molecular weight 183.7 ± 16.8 kDa in situ in intact apical membranes. Each thermodynamically stabilized [ACE2:B0AT1] heterodimer functional unit manifests the transport activity within the whole ∼345 kDa [ACE2:B0AT1]2 dimer-of-heterodimers quaternary structural complex. The results are consistent with our prior molecular docking modeling and gut–lung axis approaches to understanding COVID-19. These findings advance understanding the physiology of B0AT1 interaction with ACE2 in the gut, and thereby contribute to translational developments designed to treat or mitigate COVID-19 variant outbreaks and/or GI symptom persistence in long-haul postacute sequelae of SARS-CoV-2.
Keywords: ACE2, B0AT1, neutral amino acid transport, transporter, membrane, intestine, radiation inactivation, 6M17, sodium-dependent transport
Graphical Abstract
Graphical Abstract.
Introduction
Infection by SARS-CoV-2 requires its receptor binding domain to bind the ectodomain of angiotensin-converting enzyme-2 (ACE2). ACE2 can be stabilized on the surface of plasma membranes by the sodium-dependent neutral amino acid transporter, B0AT1, forming an [ACE2:B0AT1]2 dimer-of-heterodimers quaternary 4-mer structural complex.1 Pfizer/BioNTech exploited plasmid constructs of this [ACE2:B0AT1]2 structure overexpressed in cultured cell membranes2 as being crucial to their successful preclinical testing of mRNA candidates encoding SARS-CoV-2 spike protein efficacious vaccine epitopes. Following clinical trials, their ACE2:B0AT1-screened choice of BNT162b2 mRNA2 was approved by US FDA for emergency use authorization delivery by lipid nanoparticles as the country's first publicly deployed COVID-19 vaccine.
B0AT1 (literature aliases: NBB, B, B0, B(0)AT1) was originally discovered and functionally characterized by Stevens and coworkers3–17 as being the major sodium-coupled neutral amino acid transport system in small intestine villus epithelial cell apical brush border membranes.7,17 These seminal studies were obligatory to subsequently assigning the functional properties to an SLC6A19 gene expression product by Broer, Verrey, and colleagues, and in implicating ACE2 as indispensable in epithelial cell trafficking/chaperoning18 and apical membrane expression of B0AT1.18–29 Following recommendations made by Halvor Christensen at a 1994 membrane transport symposium in Stowe, Vermont, the Stevens' NBB (Neutral Brush Border) term3,7 was changed to B and then to B0, in order to conform to the then-evolving transporter nomenclature convention.30 This alluded back to Christensen's pioneering Blastocyst classification categories in which the uppercase refers to sodium dependency and the “0” superscript refers to the zwitterion net zero charge of neutral amino acid substrates.30 Ultimately, the NBB/B/B0Amino acid Transporter (AT) various interchangeable appellations in the literature3–16,30,31 were eventually consolidated into the current designation “B0AT1.”17,20,32
The small intestine is the human body's site of greatest magnitude expression of both B0AT1 and ACE2.33–41 In the mucosa, B0AT1 is the central player in villus enterocyte neutral amino acid transport that supplies nutritional amino nitrogen. Its amino acid substrates signal enteroendocrine and goblet cell physiological activities, and steer gut barrier integrity and inflammasome events.17,25,33,34,42
Literature reviews/meta-studies published during the period spanning 1990–2010 presaged various pleiotropic physiological roles for B0AT1 interactions with ACE2, including the remarkably prescient concept of governing coronavirus infectivity.17,21,24 In early 2020, Yan and coworkers in Zhou's group1 utilized 2.9 Å resolution cryo-electron microscopy to determine that two B0AT1 subunits stabilize two ACE2 subunits in cell membranes as the thermodynamically favored atomic structure [ACE2:B0AT1]2 multimeric complex that can bind the SARS-CoV-2 spike (PDB ID: 6M17 and PDB ID: 6M18).
SARS-CoV-2 hijacks ACE2 as its receptor in both small intestinal enterocytes and lung pneumocytes.33,34,43–47 Pulmonary symptoms are the hallmark of severe COVID-19, while about half of COVID-19 patients manifest extrapulmonary gastrointestinal (GI) tropism with gut clinical symptomology accompanied by virion particles shed in feces and RNA in toilet aerosols in the active phase, and intestinal symptoms persist in long-haul postacute sequelae of SARS-CoV-2 (PASC).48–53 The main risk factor decisive for organ-based clinical outcomes of lung versus intestine in COVID-19 is the nature of ACE2 interplay with two particular membrane-bound metalloproteinases—TMPRSS2 and ADAM17—that are expressed in both organs.54 These metalloproteinases are responsible for launching the pernicious events of SARS-CoV-2 tropism via their specific cleavage sites on ACE2.33,34,55,56 Lung cells do not express B0AT1, thus permitting ready access of TMPRSS2 and ADAM17 to pneumocyte monomer ACE2 cleavage sites, resulting in unconstrained lung pathology.33,34 However, for enterocytes that can express the [ACE2:B0AT1]2 complex, our molecular docking studies55,56 predicted that the B0AT1 subunits sterically interfere with TMPRSS2 and ADAM17 access to the cleavage sites of gut ACE2. Thus, the degree to which B0AT1 is expressed and trafficked by ACE2 is likely a pivotal factor that governs gut COVID-19 severity in a given patient. Consequently, the structure–function relationship coupling B0AT1 with ACE2 is important to understanding involvement of the intestine in COVID-19 and why some patients are spared yet others are affected. This relationship is poorly understood.
The present study addresses this knowledge gap, in order to provide insights that may lead to developing new therapies and treatments for COVID-19 in current or future outbreaks. Our approach was to exploit radiation inactivation analysis and electron flux density target theory utilizing high-energy ionizing electrons from a 16 MeV linear accelerator. As empirically established by us57,58 and others,59–70 this technique reveals membrane in situ structure–function relationships, accurately identifying the molecular size of “functional units” entwined within physical structures of complex multi-subunit biological systems such as channels, transporters, enzymes, and receptors. We report that sodium-dependent carrier-mediated B0AT1 activity in situ in small intestinal enterocyte purified apical brush border membrane vesicles (BBMVs) occurs via an apparent physiological “functional unit” of target size molecular weight (mw) ∼184 kDa representing the [ACE2:B0AT1] heterodimer components within the ∼345 kDa [ACE2:B0AT1]2 dimer-of-heterodimer complex.
Methods
Small intestinal epithelium isolated apical BBMVs were prepared using New Zealand white rabbit ileum mucosa, lyophilized, and reconstituted for use in radiation inactivation experiments as previously described by us.3,4,7,15,57,58,71 Briefly, rapidly isolated mucosal scrapings were obtained from 1 m of ileum proximal to the ileocecal junction and treated with 10 mm MgCl2, followed by a series of differential centrifugations and progressively diluted washes using 300–10 mmd-mannitol in 1 mm HCl/Tris pH 7.6 buffer. The final pellets were suspended in distilled water using a glass homogenizer. BBMVs (15 mg protein/mL in 100 μL) aliquoted into individual glass ampules were snap frozen in liquid N2 and then lyophilized under 20 μm Hg vacuum for 12 h and subsequently stored vacuum-sealed at −10°C until needed for radiation inactivation experiments. For postirradiation uptake assays, the lyophilized BBMVs were reconstituted and equilibrated at 22°C with 100 μL of buffer containing 200 mmd-mannitol in 10 mm HEPES/Tris pH 7.5 followed by 3 passes through a 22-gauge needle.
Lyophilized BBMVs vacuum-sealed in glass ampules were stable for several months at ∼22°C room temperature, such that when reconstituted they displayed >90% of original fresh transport activity and the usual BBMV characteristics observed for fresh BBMVs. As we have published previously,4,7,15,57,58 we measured radiotracer labeled l-amino acid or d-glucose time course uptake peak overshoots in zero-trans sodium-containing uptake media, and >95% right-side-out sealed spherical compartments ∼1000 Å diameter, with <5% nonsealed pieces of membrane observed in electron micrographs.4,7,57,58 Transmission electron microscopy cross-sections were prepared using glutaraldehyde/OsO4/uranyl acetate-treated centrifuged pellets of reconstituted lyophilized BBMVs. BBMVs were enriched ∼15-fold in each of the apical membrane markers γ-glutamyl transpeptidase, leucine aminopeptidase, and alkaline phosphatase, relative to mucosal cell scrapings of the starting tissue. On the other hand Na+/K+ ATPase activity representing basolateral membrane contamination was decreased by ∼70% as previously reported.4,7,15,57,58 Alkaline phosphatase (EC 3.1.3.1) activity was employed as a radiation inactivation target size mw internal calibration standard. For each radiation dose, 0.02 mL of reconstituted irradiated BBMV suspension containing 100 mm NaSCN was incubated at 22°C with 1.0 mL 0.9 M diethanolamine pH 9.8, 1.0 mL 30 mmp-nitrophenylphosphate (pNPP) in media lacking K+ ions, with the p-nitrophenol product quantified colorimetrically at 405 nm.62,66
Lyophilized BBMVs in thin wall glass ampules under vacuum were irradiated with a high-energy electron beam (16 MeV in a 10 cm uniform beam) delivered by a linear accelerator (Addenbrooke's Hospital, Cambridge, England) over the range of 5–180 kGy in increments of 20 kGy/min or less to prevent sample heating. Samples were fitted in an aluminum block cooled by a dry-ice streaming system. The accelerator was calibrated using Perspex dosimetry. The irradiated vesicles were stored in their vacuum-sealed ampules at −10°C until required for assays. Following postirradiation, BBMVs were reconstituted with 200 mmd-mannitol pH 7.5 buffer as described above, and the vesicles were then allowed to equilibrate for 30 min before transport measurements were made.
Influx initial rates were measured at 22°C in reconstituted BBMVs, defined as the 5 s initial uptake of zero-trans (ie, substrate outside but not inside) unidirectional carrier-mediated sodium-dependent portion of total uptake of radiolabeled 0.1 mm [3H]-l-alanine or 1 mm [3H]-L-serine, as described by us.3,15,58 The external vesicle uptake buffer contained either 100 mm NaSCN or 100 mm KSCN in 100 mmd-mannitol pH 7.5. Sodium-dependent carrier-mediated transport activity was calculated from the total radiotracer uptake in Na+ media minus diffusion uptake as measured in K+ media replacing Na+ in the presence of unlabeled 100 mml-methionine or 100 mml-alanine. A rapid mix/rapid filtration apparatus was employed with ice-cold 200 mmd-mannitol stop buffer to arrest uptake, as described by us.3,15,58 Uptake measurements were replicated N = 6 times.
Radiation inactivation target size mws were obtained by measuring postirradiation remaining activity of zero-trans unidirectional sodium-dependent initial influx rates in reconstituted lyophilized BBMVs at various radiation doses:
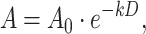 |
(1) |
where A = activity remaining, A0 = control initial activity, D = radiation dose in kGy units, and k = rate constant dependent on target mw. It has been empirically established by us57,58 and others59–70 that for activity of biological systems in lyophilized preparations irradiated by high-energy electron beams, the "functional unit" radiation target size is calculated by
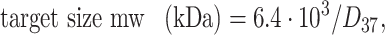 |
(2) |
where D37 = radiation dose (in kGy units) at which activity = A0.e−1 (ie, 37% of control activity). In practice, target sizes were computed by nonlinear regressions constrained to 100% activity at zero dose radiation, fitting the raw data using the R package “investr” with objects of class “nls” using the function:
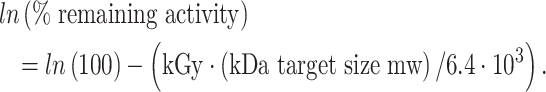 |
(3) |
Atomic coordinates for PDB ID: 6M18, 6M17, or 6M1D were employed for the molecular structure assemblage of ACE2 subunits with B0AT1 subunits as the [ACE2:B0AT1]2 dimer-of-heterodimers quaternary complex determined by Yan and coworkers in Zhou's group1 using 2.90 Å resolution cryo-electron microscopy. In accordance with our previous studies of B0AT1 structures,55,56 molecular modeling of subunit interactions and interface residues' contact distances were executed using ChimeraX software,72 meeting default probe criteria of 1.4 Å or being buried within a 15 Å2 area cutoff. Thermodynamics of chain molecular internal and interface energies were computed using PDBePISA.73 Molecular structures and their membrane location were generated using PyMOL v2.4.0,74 PDBEditor,75 ChimeraX,72 and Orientations of Proteins in Membranes (OPM) database transmembrane server.76
Results
Figure 1 shows cross-section electron micrographs of the reconstituted lyophilized small intestinal purified apical BBMVs, which were ∼100 nm diameter. In Figure 1A and B, note the sealed right-side-out BBMVs populated by 100–150 Å protruding knobs from the membrane surface lipid rafts. Such sealed vesicles are essential for measuring uptake of radiotracer substrates across the purified membrane proteophospholipid components that partition a defined space trapping the radiotracer.
Figure 1.
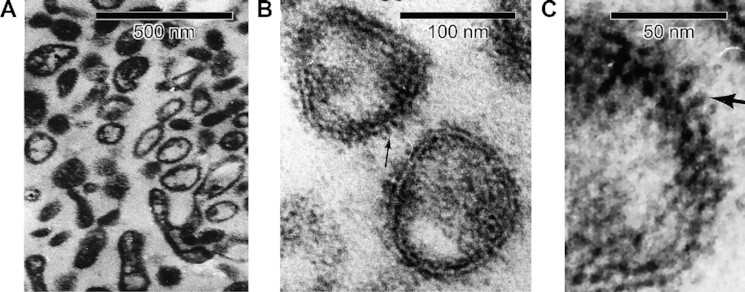
Cross-section Electron Micrographs of Reconstituted Lyophilized Small Intestinal Purified Apical Membrane BBMVs.(A) Wide field view of intact BBMV vesicles employed for radiation inactivation of B0AT1 functional unit activity, with right-side-out orientation of fuzzy glycocalyx. (B) BBMV sealed lipid bilayers showing protruding 100–150 Å knobs (arrow example). (C) Close-up view of reconstituted lyophilized BBMV, showing 100–150 Å protruding glycoprotein knobs from membrane surface lipid rafts (arrow example).
The reconstituted lyophilized intestinal BBMV zero-trans uptake kinetics exhibited a singular saturable carrier-mediated sodium-dependent radiolabeled neutral amino acid unidirectional influx pathway attributable to known characteristics of B0AT1,3–18,20–23,26,28,32 as shown in Eadie–Hofstee plot of Figure 2. The B0AT1 transport activity data were obtained in the BBMVs according to eqn (4), as solved by nonlinear regression using the R package “investr” with objects of class “nls” employing the function:
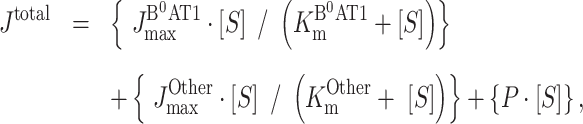 |
(4) |
where J represents influx initial rates, Jmax is the maximal influx rate of a given transport carrier with its kinetics fitting the Michaelis–Menten relationship, [S] is the extravesicular radiolabeled l-alanine concentration (mm), Km is the apparent Michaelis–Menten affinity constant for a given transport carrier, and P is the passive diffusion permeability coefficient. The computed value of P = 1.1 × 10−7 L/mg protein/5 s was also independently empirically verified by measuring total 0.1 mm [3H]-l-alanine uptake in media with K+ replacing Na+ and containing 100 mm unlabeled l-alanine and/or 100 mml-methionine. Based on nonlinear regression analyses, the B0AT1 component within the 95% CI (confidence interval) shown in Figure 2 yielded JB0AT1max = 5.2 ± 0.4 nmol/mg protein/5 s, and KmB0AT1 = 6.9 ± 0.8 mml-alanine. Total influx included an apparent very minor additional non-B0AT1 saturable component, denoted “Other,” which was fitted in Figure 2 by JOthermax = 0.33 ± 0.06 nmol/mg protein/5 s, and KmOther = 1.2 ± 0.3 mm (inset Figure 2B). This “Other” activity contributed <5% to maximal sodium-dependent uptake as compared with >95% of Na+-dependent active attributable to B0AT1. It could be speculated that “Other” might potentially represent systems ASCT2, SNAT2, or the [rBAT:b0,+AT1] heterodimer complex.77 However, unlike B0AT1, ASCT2 is an amino acid exchanger/antiporter77 that mechanistically would be principally unresponsive to the zero-trans initial rate unidirectional sodium-coupled uptake assay conditions employed in the present study (see the Methods section). Furthermore, ASCT2 is reportedly expressed in small intestine at levels ∼2.4% of B0AT1 expression,26,78 with ASCT2 prominence dominating ascending colon compared with small intestine. SNAT217 is a highly unlikely candidate because it is primarily a basolateral membrane transport system that is expressed only transiently during the early development phase of life mainly in the neonatal duodenum, not in adult ileum as in our apical BBMV preparation. A [rBAT:b0,+AT1] heterodimer complex17 would run in reverse under the zero-trans initial uptake experimental conditions, thus likely precluding its activity. Thus “Other” activity was dropped from subsequent consideration in the ensuing analyses, and was discounted as a relevant factor in the present study.
Figure 2.
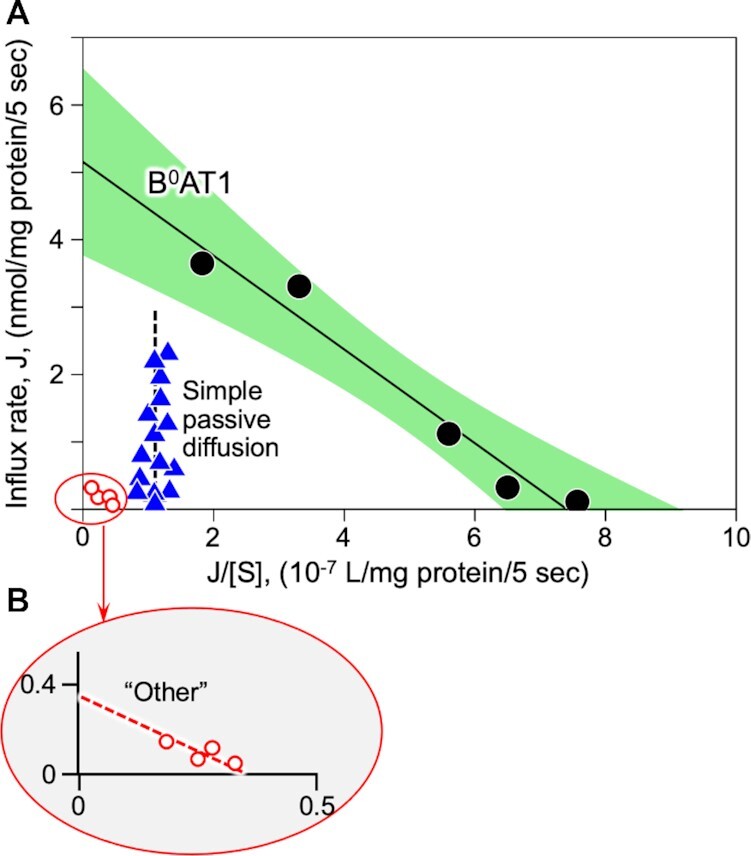
Eadie–Hofstee Plot of Initial Rate Radiotracer Amino Acid Influx Transport Kinetics.(A)Employing multivariate nonlinear analyses of zero-trans unidirectional [3H]-l-alanine initial influx rates measured in reconstituted lyophilized intestinal BBMVs, a single linear component B0AT1 (black circles) was derived by subtracting simple passive diffusion (blue triangles) from total l-alanine uptake in Na+ media. The B0AT1 component fit saturable kinetics per eqn (1) (see the Methods section), defining >95% of the Na+-dependent carrier-mediated uptake, as represented by the solid line within green 95% CI. The computed passive diffusion permeability coefficient, P, of eqn (1) (abscissa intercept of vertical dashed line) was also independently empirically verified by measuring uptake in K+ media replacing Na+ in the presence of 100 mml-alanine and/or 100 mm unlabeled methionine. (B) An apparent additional trivial carrier-mediated component, labeled “Other” (red open circles), contributed <5% of total Na+-dependent maximum uptake activity, and was dropped from subsequent considerations.
Figure 3 shows B0AT1 transport activities and internal calibration standard alkaline phosphatase enzymatic activities remaining in reconstituted BBMVs exposed to increasing doses of high-energy electron irradiation. Based on eqns (1)–(3), nonlinear regression analyses of the radiation target theory relationships57–70 yielded target size mw and D37 value for B0AT1 = 183.7 ± 16.8 kDa (D37 = 34.8 ± 3.3 kGy; P < 0.001). For alkaline phosphatase hydrolysis of pNPP in Na+ media lacking K+, analyses yielded target size mw = 57.4 ± 1.8 kDa (D37 = 111.5 ± 3.5 kGy; P < 0.001).
Figure 3.
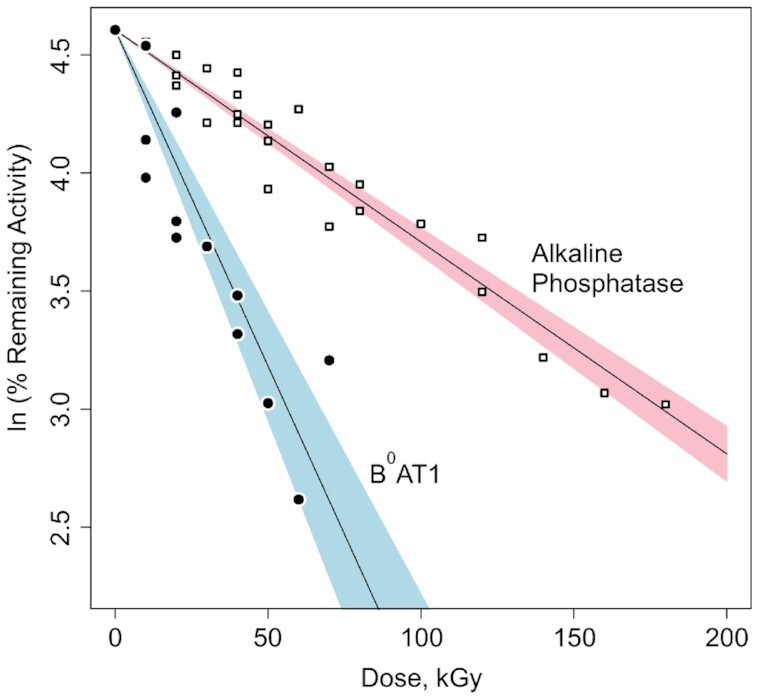
Radiation Inactivation of B0AT1 Transport and Alkaline Phosphatase Activities. At increasing electron irradiation doses, carrier-mediated sodium-dependent zero-trans unidirectional initial influx rates of [3H]-l-alanine or [3H]-L-serine uptake in intestinal BBMVs via B0AT1 (filled circles) were measured along with native alkaline phosphatase activity serving as the internal standard (open squares). Based on ln of % remaining activity at each dose compared with zero dose, nonlinear regression analyses (see the Methods section, eqns (2)–(4)) yielded target size mws. B0AT1 = 183.7 ± 16.8 kDa (blue: 95% CI for alanine and serine uptake, with D37 = 34.8 ± 3.3 kGy; P < .001). Alkaline phosphatase = 57.4 ± 1.8 kDa (pink: 95% CI for pNPP hydrolysis in Na+ media lacking K+, with D37 = 111.5 ± 3.5 kGy; P < .001).
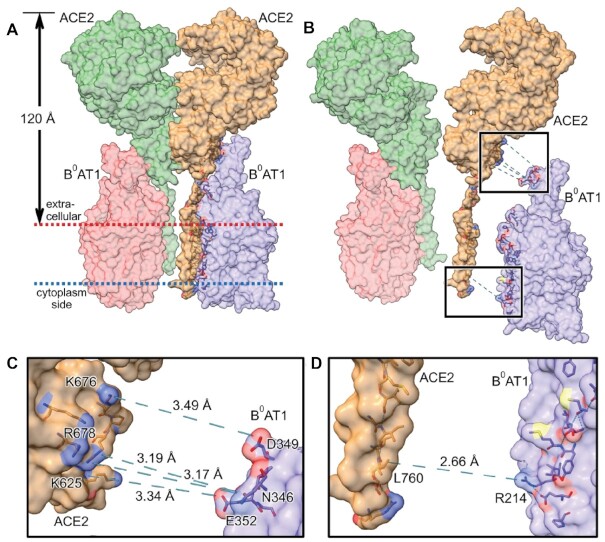
Atomic coordinates for PDB ID: 6M181 represent the thermodynamically favored assembly of the dimer-of-heterodimers complex putatively embedded in the intestinal epithelial cell apical brush border membrane surface. Figure 4 shows this [ACE2:B0AT1]2 quaternary complex total mw = 345.45 kDa assembled as a dimer of 2 [ACE2:B0AT1] heterodimers. Employing the OPM database transmembrane server,76 we calculated that transmembrane hydrophobic residues of all chains secure the complex within a BBMV membrane thickness of 30.2 Å, and that the anchored structure protrudes 120 Å from the membrane surface (Figure 4A). Figure 4B, the exploded view of panel 4A, emphasizes the zones of contact bonds connecting the subunits of the internal heterodimer [ACE2:B0AT1] interface residues in the regions of extracellular milieu (upper box) and membrane anchors (lower box); for graphic simplicity, only the right side [ACE2:B0AT1] exploded pairing is shown (tan color ACE2_chain_B with purple B0AT1_chain_A), although the same relationships hold for the Figure 4B left side unexploded pairing of green ACE2_chain_D complexed with pink B0AT1_chain_C. Employing PDBePISA, ChimeraX, PyMOL, and OPM,72–74,76 we computed the interface contact amino acid residues as being the same whether for heterodimer ACE2_chain_B paired with B0AT1_chain_A (shown exploded), or for heterodimer ACE2_chain_D paired with B0AT1_chain_C. Figure 4C is an enlarged exploded view of the upper box of panel 4B, showing bond distances between specific contact residues. Figure 4D shows an enlarged exploded view of lower box of panel 4B, revealing bond distance measured between specified contact residues. The interface bonding computations are summarized in the data of Table 1. These results indicate that within the [ACE2:B0AT1]2 dimer-of-heterodimers complex, each of the separate heterodimer [ACE2:B0AT1] chain pairing combinations yielded bonds with statistically significant (P = .037 for pairing of [ACE2_chain_B:B0AT1_chain_A]; and P = .040 for pairing of [ACE2_chain_D:B0AT1_chain_C]) negative free energy minimization ΔiG = −20.8 kcal/mol over an interface surface area of 1260.8 Å2, unlike the nonsignificant difference in the homodimer bond pairing of ACE2_chain_A:ACE2_chain_D residue contacts (P = .935; positive ΔiG = +3.8 kcal/mol).
Figure 4.
[ACE2:B0AT1]2 Dimer-of-Heterodimers Complex in Intestinal BBMVs. (A) PDB ID: 6M181 is shown embedded in intestinal epithelial cell apical membrane surface. The [ACE2:B0AT1]2 hetero-4-mer complex total mass is 345.45 kDa assembled as a dimer of 2 [ACE2:B0AT1] heterodimers. Transmembrane hydrophobic residues of all chains anchor the complex within the membrane thickness of 30.2 Å (between red and blue dotted line boundaries), as determined using OPM database transmembrane server.76 The anchored structure protrudes 120 Å from the membrane surface. (B) Exploded view of panel A emphasizing contact bonds for 1 of the 2 internal heterodimer [ACE2:B0AT1] bonding interfaces in the regions of extracellular milieu (upper box) and membrane anchors (lower box). Interface contact residues were the same whether for heterodimer ACE2_chain_B paired with B0AT1_chain_A (shown exploded), or heterodimer ACE2_chain_D paired with B0AT1_chain_C. (C) Enlarged exploded view of upper box of panel B, showing bond distances between contact residues. (D) Enlarged exploded view of lower box of panel B, showing bond distance between contact residues. Key: B0AT1_chain_A, purple; B0AT1_chain_C, pink; ACE2_chain_B, tan; ACE2_chain_D, green.
Table 1.
Interface Bonds within the [ACE2:B0AT1]2 Dimer-of-Heterodimers Complex
| Chain Pairing | ΔiG (kcal/mol) | P-value | Interface Surface Area (Å2) | Hydrogen Bonds Between Contact Residues (Distance Å) |
|---|---|---|---|---|
| [ACE2:B0AT1] (chain_B:chain_A) | −20.8 | .037 | 1260.8 | ACE2_LEU760/B0AT1_ARG214 (2.66 Å)ACE2_ARG678/B0AT1_ASN346 (3.17 Å)ACE2_ARG678/B0AT1_ASN346 (3.19 Å)ACE2_LYS625/B0AT1_GLU352 (3.34 Å)ACE2_LYS676/B0AT1_ASP349 (3.49 Å) |
| [ACE2:ACE2] (chain_B:chain_D) | +3.8 | .935 | 1276.9 | N/A |
| [B0AT1:B0AT1] (chain_A:chain_C) | N/A | N/A | null | N/A |
| [ACE2:B0AT1] (chain_D:chain_C) | −20.8 | .040 | 1263.1 | ACE2_LEU760/B0AT1_ARG214 (2.66 Å)ACE2_ARG678/B0AT1_ASN346 (3.17 Å)ACE2_ARG678/B0AT1_ASN346 (3.19 Å)ACE2_LYS625/B0AT1_GLU352 (3.34 Å)ACE2_LYS676/B0AT1_ASP349 (3.49 Å) |
Solvation-free energies were calculated for each isolated chain, and also for the interfaces between contact residues of chain combinations within the [ACE2:B0AT1]2 dimer-of-heterodimers complex described in Figure 4. The ΔiG values represent solvation-free energy gain (kcal/mol) upon formation of a given interface, with P ≤ .05 representing statistical significance. Shown are the distances between specific residues responsible for interface contact hydrogen bonds between paired chains shown in Figure 4. There were null interactions between the B0AT1 chains.
Intestinal-type alkaline phosphatase (EC 3.1.3.1) was chosen as the radiation inactivation target size internal calibration standard (Figure 3; target size mw = 57.4 ± 1.8 kDa), grounded on various mammalian orthologs exhibiting the same fundamental structural arrangement running as a single ∼55 kDa monomer Western blot band.79 It has been previously demonstrated62 that the radiation inactivation target size mw of intestinal alkaline phosphatase monomer can be identified independent from the homodimer state when the postirradiation enzyme activity is assayed under conditions of using the Na+ salt of pNPP substrate hydrolysis in the absence of K+ at alkaline pH,62,66 as described earlier in the Methods section. Rat intestinal-type alkaline phosphatase atomic coordinates (PDB ID: 4KJG) indicate a homodimer assembly of 2 identical noncovalently associated independent 54.4 kDa monomer chains in the absence of Na+, as shown in Figure 5. Further in Figure 5, the effect of binding Na+ ion in the absence of K+ is revealed as shown by the 54.8 kDa monomer structure from atomic coordinates of human alkaline phosphatase PDB ID: 3MK1, with release of p-nitrophenol product.80
Figure 5.
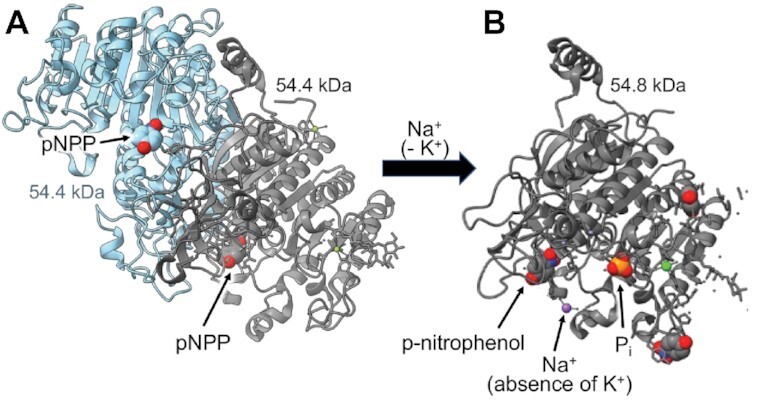
Alkaline Phosphatase (EC 3.1.3.1) With and Without Na+. (A) Rat intestine alkaline phosphatase homodimer assembled as two 54.4 kDa monomers without Na+ (PDB ID: 4KJG), shown with pNPP substrate in each binding site. (B) Alkaline phosphatase 54.8 kDa monomer activity assayed under conditions of Na+ ion (purple) in the absence of K+ (PDB ID: 3MK1), shown with pH 9.8 reaction products p-nitrophenol and inorganic phosphate (Pi).
Discussion
The main finding of this study is that sodium-dependent carrier-mediated B0AT1 activity in situin small intestinal enterocyte purified apical BBMVs occurs via an apparent physiological “functional unit” of target size mw = 183.7 ± 16.8 kDa representing a thermodynamically stabilized [ACE2:B0AT1] heterodimer, determined by high-energy electron radiation inactivation analysis. This finding is consistent with predictions in the literature grounded in prior biochemical, immunohistochemical, molecular modeling, and cryo-EM techniques. Two of these heterodimer functional units behave within the physical structure of an [ACE2:B0AT1]2 dimer-of-heterodimers 4-mer complex, with PDB ID: 6M18 atomic coordinates measured by Yan et al.1 Notably, these data are consistent with our prior molecular docking modeling55,56 and gut–lung axis studies,33,34,81 and prescient antecedent literature review17 that putatively implicated the B0AT1 subunit as a major player with ACE2 in SARS-CoV-2 gastrointestinal tropism in COVID-19.
Previous experimental evidence demonstrated that posttranslational SLC6A19 gene expression of B0AT1 and its sodium-dependent neutral amino acid transporter activity obligatorily engages the accessory protein ACE2 as its chaperone for intracellular trafficking to epithelial cell apical brush border membranes, whereby the mature B0AT1 protein subunit colocalizes with ACE2 within the membrane.18,20,22,24–28,82–85 Pharmacologic manipulation of ACE2 expression demonstrated concomitant parallel changes in B0AT1 amino acid transporter protein expression and uptake activity18,20,22,24–28,82–85; however, the converse does not hold, such that ACE2 can be expressed independent of trafficking B0AT1.
The individual molecular masses of B0AT1, ACE2, and [ACE2:B0AT1]2 physical structures have each been determined previously based on molecular biology, biochemistry, cell transfection/expression, tissue immunofluorescence microscopy colocalization, and epithelial membrane isolation techniques.18,20,22,24–28,82–84 Western blots yielded a single band for each component, reflecting the appropriate molecular weights of each individual cloned monomer (denaturing conditions) or aggregate multimer complex (native gel conditions). The B0AT1 monomer subunit band on SDS-PAGE23,86,87 is ∼75 kDa, with predicted mw = 71.2 kD from 634 amino acids expressed by the SLC6A19 gene (accession NP_001034811.1). ACE2 monomer single bands generally range from ∼110 kDa (glycosylated) to ∼92 kDa (deglycosylated), with predicted mw = 92.5 kDa from 805 amino acids expressed by the ACE2 gene (accession XP_002719891.1).88–91 In mouse intestinal purified brush border membranes, B0AT1 and ACE2 coimmunoprecipitation coupled with digitonin native PAGE yielded a band at 376 or 488 kDa, representing the intact [ACE2:B0AT1]2 dimer-of-heterodimers 4-mer complex.18 Collectively, these biochemical findings are consistent with the 2.9 Å resolution cryo-EM (PDB ID: 6M18) atomic structure1 mw ∼345 kDa (replete with hydrogen atoms) for 2 [ACE2:B0AT1] heterodimers assembled as a [ACE2:B0AT1]2 dimer-of-heterodimers ternary complex shown in Figure 4. As further shown in Figure 4A, the membrane-anchored [ACE2:B0AT1]2 complex protrudes 120 Å outward from the extracellular surface. This is consistent with the well-known phenomenon reported for a wide variety of integral membrane-bound protein multimer ectodomains anchored by lipid rafts in epithelial cell membranes,1,17,76,92 and is in agreement with the results in Figure 1B and C electron micrographs showing 100–150 Å protruding knobs on the BBMVs employed in the present study.
While such biochemical and physical techniques are useful to identify purified individual polypeptides and their physical characteristics, the unique value of radiation inactivation analysis is to reveal structure–function relationships and biological behaviors especially in situ in oligomeric protein assemblies of any form—whether crude samples, intact cells, membranes, or purified molecules. Ionizing radiation inactivation target theory has been used extensively to assess the physiological behavior “functional unit” molecular masses of a diverse variety of complex multi-subunit oligomeric polypeptide structures residing in situ in biological systems, such as channels, transporters, enzymes, and receptors,59–70 including our prior work with intestinal integral membrane-bound proteins in BBMVs.57,58 The literature is replete with evidence of radiation inactivation accurately assigning known biological activities as a “functional unit” whether as a single polypeptide or as an oligomeric assembly of many individual polypeptide subunits.59–70 The technique exploits the loss of measured biological activity surviving a random hit by a high-energy electron from a linear accelerator, with the probability of being knocked out by deposition of the electron's 60 eV (1500 kcal/mol) ionizing energy directly correlated with the mw “target size” of the functioning entity, as described in the Methods section and extensively discussed elsewhere.59–70 In the case of biological activity of a multimer composed of subunits, a single electron hitting any one of the subunit members within the collective assembly will completely abolish functional activity as the consequence of transferring its ionizing energy to other subunits of the complex via bonds of contact interface amino acid residues.59–70 Thus, for a heterodimer with subunits paired by 1 or more bond of interface contact residues, and in accordance with radiation inactivation target theory,59–70 an electron direct hit to either one of the subunits will nullify biological activity, even if only one of the subunit entities is responsible for the actual biological activity.
The data of Figure 3 fit the simple exponential relationship of eqns (1)–(3) for the inactivation of membrane in situ B0AT1 transport activity. The computed values in Table 1 summarizing the structures of Figure 4 indicate that [ACE2:B0AT1] heterodimer pairings are thermodynamically stabilized (ΔiG = −20.8 kcal/mol) via interface contact bonds 2.66–3.49 Å involving 5 specific residue pairings within the hetero 4-mer complex. However, this is in contrast to atomic modeling attempts (Table 1 and Figure 4) to examine [B0AT1:B0AT1] or [ACE2:ACE2] homodimer pairings that each lack residues with bonds able to transfer electron hit energy into the adjoining subunits (ΔiG = +3.8 kcal/mol in the case of [ACE2:ACE2]; and null interfacings between the B0AT1 subunits). Thus, a high-energy electron direct hit to any ACE2 subunit will transfer its energy to a B0AT1 subunit, resulting in annihilating measurable B0AT1 transport activity. Based on eqns (2) and (3), the above arguments collectively indicate that the high-energy electron irradiation “sees” a functional unit target mw ∼184 kDa for B0AT1 transport activity, which is consistent with radiation target theory describing a multimeric functional unit59–70 composed of the [ACE2:B0AT1] heterodimer.
The radiation inactivation target size results (Figure 3) were validated by internal calibration exploiting endogenous alkaline phosphatase activity in the reconstituted BBMVs. As shown in Figures 3 and 5 for the K+-independent activity of pNPP hydrolysis assayed in the presence of Na+, the data revealed the internal alkaline phosphatase radiation target mw = 57.4 ± 1.8 kDa, consistent with prior studies predicting ∼55 kDa monomer subunits on Western blots.62,66,79,80
The present study employed native intestinal BBMV membranes. We posit that it would be beneficial to extend such studies to include future explorations of drug interactions and effects of membrane lipid raft stabilization relating to SARS-CoV-2 tropism in the intestine, in contrast to events in lung pneumocytes that lack B0AT1. Such tools include, for example: (1) the recent expression of B0AT1 in bacteria93; (2) HEK293 cells' coexpression of ACE2 with B0AT11 as exploited by Pfizer/BioNTech to screen their mRNA vaccine candidates against SARS-CoV-22; and (3) the recent discoveries of nimesulide94 and cinromide95 as inhibitors of B0AT1. Coexpression evidence suggests that the small intestinal BBMV SIT1 (SLC6A20), representing the IMINO transport system serving proline uptake originally described by us,17,96,97 also functionally partners with epithelial membrane ACE2.26,98 Thus, we posit that it would be beneficial to pursue the atomic structural interactions, functional relationships, and effects of targeted drugs engaging SITS1 relating to COVID-19 in the manner analogous to B0AT1 with ACE2. Furthermore, such future experimental pursuits would bear fruit relating to our in silico studies33,34,55,56 that have implicated a role for B0AT1 and SITS1 in sterically governing the role of intestinal membrane proteinase TMPRSS2 and ADAM17 as mediators of ACE2-dependent intestinal SARS-CoV-2 infection and gut inflammasome induction.
In conclusion, high-energy electron radiation inactivation analysis was used to determine that B0AT1 transport activity occurs via the [ACE2:B0AT1] heterodimer functional unit housed within the physical structure of the [ACE2:B0AT1]2 dimer-of-heterodimers quaternary complex embedded in the apical brush border membranes of small intestinal enterocytes. It is noteworthy that SARS-CoV-2 virus hijacks ACE2 as its receptor and entry point of infecting cells, and further that the small intestine is the body's site of greatest magnitude of expression of both B0AT1 and ACE2.33,34 Thus, the [ACE2:B0AT1] heterodimer functional unit is important for gut lumen activities (1) relating to pleiotropic native physiological roles in amino nitrogen metabolism of nutritive and bioactive peptides, (2) in local gut mucosa renin–angiotensin system regulating absorption of sodium and organic nutrients, and (3) as central to steering SARS-CoV-2 tropism in the GI tract with attending GI shedding of SARS-CoV-2 particles and clinical symptomology in about half of COVID-19 patients,17,25,33,34,42,55,56,81 including bacteremic inflammation of gut dysbiosis origin in COVID-19 patients.99 These findings enhance our understanding of gut pathophysiology, thereby contributing to future translational experiments designed to treat or mitigate COVID-19 variant outbreaks and/or GI symptom persistence in long-haul PASC.
Contributor Information
Bruce R Stevens, Department of Physiology and Functional Genomics, University of Florida College of Medicine, Gainesville, FL, 32610, USA; Department of Medicine, Division of Gastroenterology, University of Florida College of Medicine, Gainesville, FL, 32610, USA.
J Clive Ellory, Department of Physiology, Anatomy and Genetics, University of Oxford, Oxford, OX1 3PT, UK.
Robert L Preston, School of Biological Sciences, Illinois State University, Normal, IL, 61790, USA.
Funding
None declared
Conflict of Interest
None declared.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Yan R, Zhang Y, Li Yet al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vogel AB, Kanevsky I, Che Yet al. BNT162b vaccines are immunogenic and protect non-human primates against SARS-CoV-2. bioRxiv. 2020. doi: 10.1101/2020.12.11.421008. [Google Scholar]
- 3. Stevens BR, Ross HJ, Wright EM. Multiple transport pathways for neutral amino acids in rabbit jejunal brush border vesicles. J Membr Biol. 1982;66(3):213–225. [DOI] [PubMed] [Google Scholar]
- 4. Stevens BR, Wright SH, Hirayama BSet al. Organic and inorganic solute transport in renal and intestinal membrane vesicles preserved in liquid nitrogen. Membr Biochem. 1982;4(4):271–282. [DOI] [PubMed] [Google Scholar]
- 5. Wright EM, Gunther RD, Kaunitz JDet al. Mechanisms of Sodium Transport Across Brush Border and Basolateral Membranes. Berlin, Heidelberg: Springer; 1983:122–132. [Google Scholar]
- 6. Schell RE, Stevens BR, Wright EM. Kinetics of sodium-dependent solute transport by rabbit renal and jejunal brush-border vesicles using a fluorescent dye. J Physiol. 1983;335(1):307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stevens BR, Kaunitz JD, Wright EM. Intestinal transport of amino acids and sugars: advances using membrane vesicles. Annu Rev Physiol. 1984;46(1):417–433. [DOI] [PubMed] [Google Scholar]
- 8. Stevens B. Amino acid transport in intestine. In: Kilberg M, Haussinger D, eds. Mammalian Amino Acid Transport. New York: Plenum Press; 1992:149–163. [Google Scholar]
- 9. Stevens BR. Vertebrate intestine apical membrane mechanisms of organic nutrient transport. Am J Physiol. 1992;263(3 Pt 2):R458–R463. [DOI] [PubMed] [Google Scholar]
- 10. Souba WW, Pan M, Stevens BR. Kinetics of the sodium-dependent glutamine transporter in human intestinal cell confluent monolayers. Biochem Biophys Res Commun. 1992;188(2):746–753. [DOI] [PubMed] [Google Scholar]
- 11. Kilberg MS, Stevens BR, Novak DA. Recent advances in mammalian amino acid transport. Annu Rev Nutr. 1993;13(1):137–165. [DOI] [PubMed] [Google Scholar]
- 12. Gerencser GA, Stevens BR. Thermodynamics of symport and antiport catalyzed by cloned or native transporters. J Exp Biol. 1994;196(1):59–75. [DOI] [PubMed] [Google Scholar]
- 13. Pan M, Stevens BR. Differentiation- and protein kinase C-dependent regulation of alanine transport via system B. J Biol Chem. 1995;270(8):3582–3587. [DOI] [PubMed] [Google Scholar]
- 14. Mailliard ME, Stevens BR, Mann GE. Amino acid transport by small intestinal, hepatic, and pancreatic epithelia. Gastroenterology. 1995;108(3):888–910. [DOI] [PubMed] [Google Scholar]
- 15. Stevens BR, Preston RL. Sodium-dependent amino acid transport is preserved in lyophilized reconstituted apical membranes from intestinal epithelium. Anal Biochem. 1998;265(1):117–122. [DOI] [PubMed] [Google Scholar]
- 16. Pan M, Souba WW, Wolfgang CLet al. Posttranslational alanine trans-stimulation of zwitterionic amino acid transport systems in human intestinal Caco-2 cells. J Surg Res. 2002;104(1):63–69. [DOI] [PubMed] [Google Scholar]
- 17. Stevens BR. Amino acid transport by epithelial membranes. In: Gerencser GA, ed. Epithelial Transport Physiology. Humana Press; Totowa, New Jersey, USA: 2010:353–378. [Google Scholar]
- 18. Fairweather SJ, Bröer A, O'Mara MLet al. Intestinal peptidases form functional complexes with the neutral amino acid transporter B0AT1. Biochem J. 2012;446(1):135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munck LK, Munck BG. Amino acid transport in the small intestine. Physiol Res. 1995;44(2):335–346. [PubMed] [Google Scholar]
- 20. Broer A, Klingel K, Kowalczuk Set al. Molecular cloning of mouse amino acid transport system B0, a neutral amino acid transporter related to Hartnup disorder. J Biol Chem. 2004;279(23):24467–24476. [DOI] [PubMed] [Google Scholar]
- 21. Broer S. Apical transporters for neutral amino acids: physiology and pathophysiology. Physiology (Bethesda). 2008;23:95–103. [DOI] [PubMed] [Google Scholar]
- 22. Broer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol Rev. 2008;88(1):249–286. [DOI] [PubMed] [Google Scholar]
- 23. Talukder JR, Kekuda R, Saha Pet al. Identification and characterization of rabbit small intestinal villus cell brush border membrane Na-glutamine cotransporter. Am J Physiol Gastrointest Liver Physiol. 2008;295(1):G7–G15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Camargo SM, Singer D, Makrides Vet al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with Hartnup mutations. Gastroenterology. 2009;136(3):872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hashimoto T, Perlot T, Rehman Aet al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vuille-dit-Bille RN, Camargo SM, Emmenegger Let al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47(4):693–705. [DOI] [PubMed] [Google Scholar]
- 27. Fairweather SJ, Broer A, Subramanian Net al. Molecular basis for the interaction of the mammalian amino acid transporters B0AT1 and B0AT3 with their ancillary protein collectrin. J Biol Chem. 2015;290(40):24308–24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jando J, Camargo SMR, Herzog Bet al. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. Plos One. 2017;12(9):e0184845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hediger MA, Romero MF, Peng JBet al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 2004;447(5):465–468. [DOI] [PubMed] [Google Scholar]
- 30. Christensen HN. Distinguishing amino acid transport systems of a given cell or tissue. Methods Enzymol. 1989;173:576–616. [DOI] [PubMed] [Google Scholar]
- 31. Palacin M, Estevez R, Bertran Jet al. Molecular biology of mammalian plasma membrane amino acid transporters. Physiol Rev. 1998;78(4):969–1054. [DOI] [PubMed] [Google Scholar]
- 32. O'Mara M, Oakley A, Broer S. Mechanism and putative structure of B(0)-like neutral amino acid transporters. J Membr Biol. 2006;213(2):111–118. [DOI] [PubMed] [Google Scholar]
- 33. Sharma RK, Stevens BR, Obukhov AGet al. ACE2 (angiotensin-converting enzyme 2) in cardiopulmonary diseases: ramifications for the control of SARS-CoV-2. Hypertension. 2020;76(3):651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Obukhov AG, Stevens BR, Prasad Ret al. SARS-CoV-2 infections and ACE2: clinical outcomes linked with increased morbidity and mortality in individuals with diabetes. Diabetes. 2020;69(9):1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ponten F, Jirstrom K, Uhlen M. The human protein Atlas: a tool for pathology. J Pathol. 2008;216(4):387–393. [DOI] [PubMed] [Google Scholar]
- 36. Chen QL, Li JQ, Xiang ZDet al. Localization of cell receptor-related genes of SARS-CoV-2 in the kidney through single-cell transcriptome analysis. Kidney Diseases. 2020:6(4):258–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Human Protein Atlas. 2021Ver 20.1, 24/02/2021. Available at: http://proteinatlas.org.
- 38. Thul PJ, Akesson L, Wiking Met al. A subcellular map of the human proteome. Science. 2017;356(6340):eaal3321. [DOI] [PubMed] [Google Scholar]
- 39. Zou X, Chen K, Zou Jet al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14(2):185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wrapp D, Wang N, Corbett KSet al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H, Li HB, Lyu JRet al. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int J Infect Dis. 2020;96:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stevens BR, Goel R, Seungbum Ket al. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut. 2018;67(8):1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zang R, Gomez Castro MF, McCune BTet al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Science Immunol. 2020;5(47):eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharma A, Garcia G Jr., Wang Yet al. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep Med. 2020;1(4):100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Werion A, Belkhir L, Perrot Met al. SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020;98(5):1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lamers MM, Beumer J, van der Vaart Jet al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson EL, Alkass K, Bergmann Oet al. Genes encoding ACE2, TMPRSS2 and related proteins mediating SARS-CoV-2 viral entry are upregulated with age in human cardiomyocytes. J Mol Cell Cardiol. 2020;147:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parasa S, Desai M, Thoguluva Chandrasekar Vet al. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw Open. 2020;3(6):e2011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cholankeril G, Podboy A, Aivaliotis VIet al. High prevalence of concurrent gastrointestinal manifestations in patients with SARS-CoV-2: early experience from California. Gastroenterology. 2020;159(2):775–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020;35(5):744–748. [DOI] [PubMed] [Google Scholar]
- 51. Redd WD, Zhou JC, Hathorn KEet al. Prevalence and characteristics of gastrointestinal symptoms in patients with SARS-CoV-2 infection in the United States: a multicenter cohort study. Gastroenterology. 2020;159(2):765.e2–767.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wong MC, Huang J, Lai Cet al. Detection of SARS-CoV-2 RNA in fecal specimens of patients with confirmed COVID-19: a meta-analysis. J Infect. 2020;81(2):e31–e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Y, Chen L, Deng Qet al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92(7):833–840. [DOI] [PubMed] [Google Scholar]
- 54. Zipeto D, Palmeira JDF, Arganaraz GAet al. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front Immunol. 2020; Oct. 7, 11:576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stevens BR. TMPRSS2 and ADAM17 interactions with ACE2 complexed with SARS-CoV-2 and B0AT1 putatively in intestine, cardiomyocytes, and kidney. bioRxiv. 2020. doi: 10.1101/2020.10.31.363473. [Google Scholar]
- 56. Andring JT, McKenna R, Stevens BR. Amino acid transporter B0AT1 influence on ADAM17 interactions with SARS-CoV-2 receptor ACE2 putatively expressed in intestine, kidney, and cardiomyocytes. bioRxiv. 2020. doi: 10.1101/2020.10.30.361873. [Google Scholar]
- 57. Stevens BR, Kempner ES, Wright EM. Radiation inactivation probe of membrane-bound enzymes: gamma-glutamyltranspeptidase, aminopeptidase N, and sucrase. Anal Biochem. 1986;158(2):278–282. [DOI] [PubMed] [Google Scholar]
- 58. Stevens BR, Fernandez A, Hirayama Bet al. Intestinal brush border membrane Na+/glucose cotransporter functions in situ as a homotetramer. Proc Natl Acad Sci USA. 1990;87(4):1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kempner ES. The mathematics of radiation target analyses. Bull Math Biol. 1995;57(6):883–898. [DOI] [PubMed] [Google Scholar]
- 60. Kempner ES. Molecular size determination of enzymes by radiation inactivation. Adv Enzymol Relat Areas Mol Biol. 1988;61:107–147. [DOI] [PubMed] [Google Scholar]
- 61. Kempner ES, Miller JH, Schlegel Wet al. The functional unit of polyenzymes. Determination by radiation inactivation. J Biol Chem. 1980;255(14):6826–6831. [PubMed] [Google Scholar]
- 62. Kempner ES, Schlegel W. Size determination of enzymes by radiation inactivation. Anal Biochem. 1979;92(1):2–10. [DOI] [PubMed] [Google Scholar]
- 63. Jarvis SM, Ellory JC, Young JD. Radiation inactivation of the human erythrocyte nucleoside and glucose transporters. Biochim Biophys Acta. 1986;855(2):312–315. [DOI] [PubMed] [Google Scholar]
- 64. Dawson G, Ellory JC. Functional lysosomal hydrolase size as determined by radiation inactivation analysis. Biochem J. 1985;226(1):283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lummis SC, Sattelle DB, Ellory JC. Molecular weight estimates of insect cholinergic receptors by radiation inactivation. Neurosci Lett. 1984;44(1):7–12. [DOI] [PubMed] [Google Scholar]
- 66. Beliveau R, Demeule M, Ibnoul-Khatib Het al. Radiation-inactivation studies on brush-border-membrane vesicles. General considerations, and application to the glucose and phosphate carriers. Biochem J. 1988;252(3):807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fincham DA, Ellory JC, Young JD. Characterization of a novel variant of amino acid transport system asc in erythrocytes from Przewalski's horse (Equus przewalskii). Can J Physiol Pharmacol. 1992;70(8):1117–1127. [DOI] [PubMed] [Google Scholar]
- 68. Verkman AS, Skorecki K, Ausiello DA. Radiation inactivation of oligomeric enzyme systems: theoretical considerations. Proc Natl Acad Sci USA. 1984;81(1):150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lidzey DG, Berovic N, Chittock RSet al. A critical analysis of the use of radiation inactivation to measure the mass of protein. Radiat Res. 1995;143(2):181–186. [PubMed] [Google Scholar]
- 70. McLawhon RW, Ellory JC, Dawson G. Molecular size of opiate (enkephalin) receptors in neuroblastoma-glioma hybrid cells as determined by radiation inactivation analysis. J Biol Chem. 1983;258(4):2102–2105. [PubMed] [Google Scholar]
- 71. Stevens BR, Fernandez A, Kneer Cet al. Human intestinal brush border angiotensin-converting enzyme activity and its inhibition by antihypertensive Ramipril. Gastroenterology. 1988;94(4):942–947. [DOI] [PubMed] [Google Scholar]
- 72. Goddard TD, Huang CC, Meng ECet al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27(1):14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. [DOI] [PubMed] [Google Scholar]
- 74. The PyMOL molecular graphics system, version 2.4.0 [computer program]. 2020. [Google Scholar]
- 75. Lee J, Kim SH. PDB editor: a user-friendly Java-based Protein Data Bank file editor with a GUI. Acta Crystallogr D Biol Crystallogr. 2009;65(4):399–402. [DOI] [PubMed] [Google Scholar]
- 76. Lomize MA, Pogozheva ID, Joo Het al. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40(D1):D370–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Scalise M, Pochini L, Console Let al. The human SLC1A5 (ASCT2) amino acid transporter: from function to structure and role in cell biology. Front Cell Dev Biol. 2018;6:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Verrey F, Singer D, Ramadan Tet al. Kidney amino acid transport. Pflugers Arch. 2009;458(1):53–60. [DOI] [PubMed] [Google Scholar]
- 79. Ghosh K, Mazumder Tagore D, Anumula Ret al. Crystal structure of rat intestinal alkaline phosphatase: role of crown domain in mammalian alkaline phosphatases. J Struct Biol. 2013;184(2):182–192. [DOI] [PubMed] [Google Scholar]
- 80. Stec B, Cheltsov A, Millan JL. Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(8):866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li J, Stevens BR, Richards EMet al. SARS-CoV-2 receptor ACE2 (Angiotensin-Converting Enzyme 2) is upregulated in colonic organoids from hypertensive rats. Hypertension. 2020;76(3):e26–e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bohmer C, Broer A, Munzinger Met al. Characterization of mouse amino acid transporter B0AT1 (slc6a19). Biochem J. 2005;389(3):745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Singer D, Camargo SM, Ramadan Tet al. Defective intestinal amino acid absorption in Ace2 null mice. Am J Physiol Gastrointest Liver Physiol. 2012;303(6):G686–G695. [DOI] [PubMed] [Google Scholar]
- 84. Kleta R, Romeo E, Ristic Zet al. Mutations in SLC6A19, encoding B0AT1, cause Hartnup disorder. Nat Genet. 2004;36(9):999–1002. [DOI] [PubMed] [Google Scholar]
- 85. Kowalczuk S, Broer A, Tietze Net al. A protein complex in the brush-border membrane explains a Hartnup disorder allele. Faseb J. 2008;22(8):2880–2887. [DOI] [PubMed] [Google Scholar]
- 86. Butts M, Singh Paulraj R, Haynes Jet al. Moderate alcohol consumption inhibits sodium-dependent glutamine co-transport in rat intestinal epithelial cells in vitro and ex vivo. Nutrients. 2019;11(10):2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Arthur S, Manoharan P, Sundaram Set al. Unique regulation of enterocyte brush border membrane Na-Glutamine and Na-Alanine co-transport by peroxynitrite during chronic intestinal inflammation. Int J Mol Sci. 2019;20(6):1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sheehan SA, Hamilton KL, Retzbach EPet al. Evidence that Maackia amurensis seed lectin (MASL) exerts pleiotropic actions on oral squamous cells to inhibit SARS-CoV-2 infection and COVID-19 disease progression. Res Sq. 2020. doi:10.21203/rs.3.rs-93851/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Heurich A, Hofmann-Winkler H, Gierer Set al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88(2):1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hennighausen L, Lee HK. Activation of the SARS-CoV-2 receptor Ace2 through JAK/STAT-dependent enhancers during pregnancy. Cell Rep. 2020;32(13):108199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fignani D, Licata G, Brusco Net al. SARS-CoV-2 receptor angiotensin I-converting enzyme type 2 (ACE2) is expressed in human pancreatic beta-cells and in the human pancreas microvasculature. Front Endocrinol (Lausanne). 2020;11:596898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moughan PJ, Stevens BR, Stipanuk MH. Digestion and absorption of protein. In: Stipanuk MH, Caudill MA, eds. Biochemical, Physiological, and Molecular Aspects of Human Nutrition. W.B. Saunders Co, Philadelphia. 2018. [Google Scholar]
- 93. Galluccio M, Pantanella M, Giudice Det al. Low temperature bacterial expression of the neutral amino acid transporters SLC1A5 (ASCT2), and SLC6A19 (B0AT1). Mol Biol Rep. 2020;47(9):7283–7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Scalise M, Indiveri C. Repurposing nimesulide, a potent inhibitor of the B0AT1 subunit of the SARS-CoV-2 receptor, as a therapeutic adjuvant of COVID-19. SLAS Discov. 2020;25(10):1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yadav A, Shah N, Tiwari PKet al. Novel chemical scaffolds to inhibit the neutral amino acid transporter B0AT1 (SLC6A19), a potential target to treat metabolic diseases. Front Pharmacol. 2020;11:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Stevens BR, Wright EM. Substrate specificity of the intestinal brush-border proline/sodium (IMINO) transporter. J Membr Biol. 1985;87(1):27–34. [DOI] [PubMed] [Google Scholar]
- 97. Takanaga H, Mackenzie B, Suzuki Yet al. Identification of mammalian proline transporter SIT1 (SLC6A20) with characteristics of classical system Imino. J Biol Chem. 2005;280(10):8974–8984. [DOI] [PubMed] [Google Scholar]
- 98. Camargo SMR, Vuille-Dit-Bille RN, Meier CFet al. ACE2 and gut amino acid transport. Clin Sci (Lond). 2020;134(21):2823–2833. [DOI] [PubMed] [Google Scholar]
- 99. Prasad R, Patton MJ, Floyd JLet al. Plasma microbiome in COVID-19 subjects: an indicator of gut barrier defects and dysbiosis. bioRxiv. 2021. doi: 10.1101/2021.04.06.438634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.