Abstract
The world is facing a pandemic of Corona Virus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Adaptive immune responses are essential for SARS-CoV-2 virus clearance. Although a large body of studies have been conducted to investigate the immune mechanism in COVID-19 patients, we still lack a comprehensive understanding of the BCR repertoire in patients. In this study, we used the single-cell V(D)J sequencing to characterize the BCR repertoire across convalescent COVID-19 patients. We observed that the BCR diversity was significantly reduced in disease compared with healthy controls. And BCRs tend to skew toward different V gene segments in COVID-19 and healthy controls. The CDR3 sequences of heavy chain in clonal BCRs in patients were more convergent than that in healthy controls. In addition, we discovered increased IgG and IgA isotypes in the disease, including IgG1, IgG3 and IgA1. In all clonal BCRs, IgG isotypes had the most frequent class switch recombination events and the highest somatic hypermutation rate, especially IgG3. Moreover, we found that an IgG3 cluster from different clonal groups had the same IGHV, IGHJ and CDR3 sequences (IGHV4-4-CARLANTNQFYDSSSYLNAMDVW-IGHJ6). Overall, our study provides a comprehensive characterization of the BCR repertoire in COVID-19 patients, which contributes to the understanding of the mechanism for the immune response to SARS-CoV-2 infection.
Keywords: COVID-19, scBCR-seq, BCR repertoire, BCR bias, isotype analysis
Introduction
Corona Virus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has posed a serious threat to global health [1, 2]. Over the past year, SARS-CoV-2 has infected more than 63 000 000 people and caused more than 1 400 000 deaths worldwide [3]. Numerous studies have contributed to the diagnosis, characterization and treatment of COVID-19. Response to SARS-CoV-2 of patients differs ranging from asymptomatic to requiring intensive care. Similar to severe acute respiratory syndrome coronavirus (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) on the surface to enter cells and can be detected in multiple organs, including lungs, pharynx, brain, liver, kidneys and heart [4–6]. Studies have shown that SARS-CoV-2 suppresses the innate immune response and reduces the levels of type I and III interferons [7, 8]. It was also found that the number of lymphocytes was decreased and serum inflammatory cytokines levels were increased in the peripheral blood of patients [9, 10]. Adaptive immune responses play a central role in clearing SARS-CoV-2 infection and directly influence patients’ clinical outcomes. After entering cells through ACE2, SARS-COV-2 is mainly sensed by toll-like receptor 7 (TLR7), which exists in the endosome [11]. Activation of TLR7 results in the production of alpha interferon, TNF-alpha, and the secretion of IL-12 and IL-6. This leads to the formation of CD8+ cytotoxic T cells and, through CD4+ helper T cells, results in antigen-specific B cells formation and antibody production [12].
The diversity of B cell receptors (BCRs) can recognize and counter the invasion of multiple pathogens, and antibodies produced by B cells can provide long-term protection for bodies [13]. The initial diversity of the BCR repertoire is the result of a somatic recombination process called V(D)J recombination [14]. After encountering antigen, BCRs undergo a process of affinity maturation, in which rapid somatic hypermutation (SHM) and class switch recombination (CSR) lead to improved antigen binding [15, 16]. Multiple studies have isolated SARS-CoV-2-specific neutralizing antibodies from COVID-19 patients [17–22]. However, further exploration of the BCR repertoire in COVID-19 patients is urgently needed.
Recent studies suggest that early-stage recovery patients still maintain various immune responses in the circulation [23]. Here, we conducted a comprehensive analysis of the peripheral blood BCR repertoire in 12 early-recovery COVID-19 patients. The BCR diversity, different gene segment usage, CDR3 length distribution and the SHM and CSR of isotypes were analyzed between COVID-19 patients and healthy controls (Figure 1). Our study provides detailed insights on BCR repertoire in COVID-19, contributing to a better understanding of the humoral immune response after SARS-CoV-2 infection.
Figure 1.
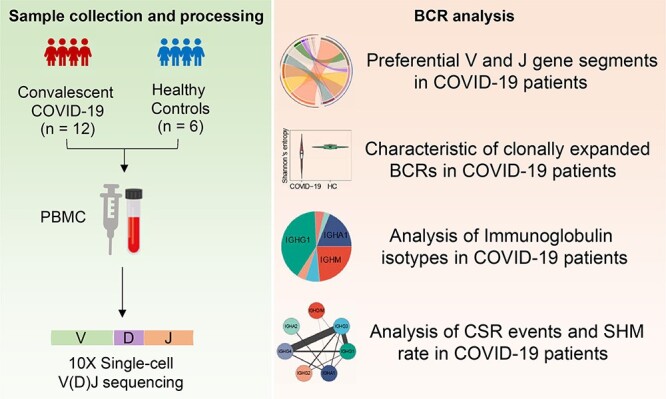
The overview of BCR repertoire characterization in COVID-19 patients.
Materials and methods
Samples
Twelve COVID-19 patients were enrolled in this study at the Harbin sixth Hospital. Fresh blood samples were collected from patients at the time of hospital discharge (Figure 1, Table S1, see Supplementary Data available online at http://bib.oxfordjournals.org/). The discharged patients must meet the following four criteria: (i) Afebrile for more than 3 days; (ii) Improved respiratory symptoms; (iii) Pulmonary imaging shows obvious absorption of inflammation; (iv) Nucleic acid tests negative for respiratory tract pathogen twice consecutively (sampling interval ≥ 24 h). Six healthy donors were enrolled as the control group, whose blood samples were collected before the COVID-19 outbreak (Table S1, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Single-cell V(D)J sequencing and data processing
Single-cell V(D)J sequencing was performed following the protocol provided by the 10X genomics Chromium Single Cell Immune Profiling Solution. The analysis pipelines in Cell Ranger (10X Genomics, version 3.1.0) were used for single-cell sequencing data processing. V(D)J sequence assembly and paired clonotype calling were performed using cellranger vdj with –reference = refdata-cellranger-vdj-GRCh38-alts-ensembl-3.1.0 for each sample.
Clonal diversity and BCR groups analysis
The clonal diversity of each sample was calculated using Shannon’s entropy [24, 25]. Entropy was calculated as follows:
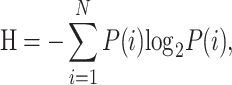 |
where N is the number of all clonotypes in each sample and P(i) represents the frequency of the ith clonotype. The higher Shannon’s entropy, the more diverse the distribution of CDR3 clones.
We conducted the BCR group analysis for each patient separately using SCOPer, which can identify B cell relationships from adaptive immune receptor repertoire sequencing data [26]. Then, we combined SCOPer results from all patients and obtained 9268 BCR groups. Groups contain at least three BCRs were defined as clonal expanded.
CDR3 motif
We first grouped BCRs according to different CDR3 lengths and then used R package ‘ggseqlogo’ to identify the motif of CDR3 with the same length [27].
SHM and CSR analysis
SHM analysis was conducted by SHazaM, which is part of the Immucantation analysis framework [28].
The Class Switch (CS) analysis took all BCRs sequences as input. For each sample, we calculated the pairwise sharing number of groups between isotypes. Then, we normalized the number of CSR by dividing the total number of groups of pairwise isotypes. Considering the real order of the CS event in the immunological process, we adjusted the CS direction of each pair of isotypes to the right order, resulting in 13 CS events in the end (IGHD/M - > IGHG1/ IGHA1/ IGHG2, IGHG3 - > IGHG1/ IGHA1/ IGHG2/ IGHG4, IGHG1 - > IGHA1/ IGHG2/ IGHG4, IGHA1 - > IGHG4/ IGHA2, IGHG2 - > IGHA2).
Statistical analysis
Fisher’s exact test was used to assess differences in usage of V and J gene segments and the proportion of Ig isotypes between COVID-19 patients and healthy controls. Wilcoxon’s signed-rank test was used to analyze differences in clonal diversity, CDR3 length and SHM rate among different groups. The significance thresholds were P < 0.05. All statistical analyses were implemented with R software (Version 3.5.1).
Results
Preferential V gene segments of BCR across COVID-19 patients
We performed single cell V(D)J sequencing of 12 COVID-19 recovery patients and 6 healthy controls (Figure 1). After filtering, a total of 19 581 B cells were obtained. On average, there were 1086 B cells per sample in COVID-19 patients and 1092 B cells per sample in healthy controls (Table S1, see Supplementary Data available online at http://bib.oxfordjournals.org/, Method). BCR is comprised of variable region and constant region, and its immensely diverse repertoire is attributed to the V(D)J recombination by assembling V, D and J gene segments into different combinations [29, 30]. We first investigated gene segment usage in the whole BCR repertoires of COVID-19 patients and healthy controls. For V gene segments in the heavy chain, IGHV3, IGHV4, IGHV1 and IGHV2 gene families were frequently used in both COVID-19 patients and healthy controls (Figure 2A), especially IGHV3 and IGHV4 family, which accounted for more than 75% of all BCRs. However, the frequencies of gene segments in each IGHV family were significantly different between COVID-19 and healthy controls (Figure 2B–E). Specifically, IGHV3-30, IGHV3-23, IGHV3-15 and IGHV3-33 of the IGHV3 family, IGHV4-4, IGHV4-31 and IGHV4-30-2 of the IGHV4 family, IGHV1-3 of the IGHV1 family and IGHV2-70 of the IGHV2 family were significantly increased in COVID-19 (Fisher’s exact test, P-value < 0.05, Odd Ratio > 1). Increased use of IGHV3-30 and IGHV3-15 has been observed in human antibodies against influenza virus, cytomegalovirus (CMV) and Ebola virus [31–33]. For V gene segments in the light chain, IGKV1 and IGKV3 were the most frequently used segments, followed by IGLV1, IGLV2, IGLV3, IGKV2 and IGKV4 (Figure S1A, see Supplementary Data available online at http://bib.oxfordjournals.org/). Similarly, gene segments in each IGKV or IGLV family were significantly different between COVID-19 and healthy controls (Figure S1B–I, see Supplementary Data available online at http://bib.oxfordjournals.org/). Analysis of IGHJ gene segments showed that the extremely preferential segment was IGHJ4, which accounted for more than 40% of all BCRs in both disease and healthy controls (Figure 2F). And the proportion of IGHJ3, IGHJ4 and IGHJ5 was significantly different between the two types of samples (Figure 2G). There was no bias in the proportion of IGL/K J gene segments within COVID-19 or healthy controls (Figure S1J, see Supplementary Data available online at http://bib.oxfordjournals.org/). But most J genes of the light chain were significantly increased or decreased in COVID-19 (Figure S1K, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Figure 2.
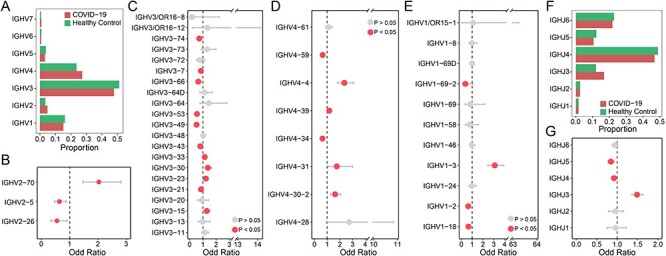
Comparison of variable (V) and joining (J) gene usage in BCR heavy chain between COVID-19 patients and healthy controls. (A) The proportion of each IGHV gene in COVID-19 patients (red) and healthy controls (green). (B–E) The differential analysis of IGHV2 (b), IGHV3(c), IGHV4 (d) and IGHV1 (e) gene family by Fisher’s exact test, the red dots represent P < 0.05 and the grey dots represent P > 0.05. (F) The same barplot as (A) for the IGHJ gene. (G) The differential analysis of IGHJ gene by Fisher’s exact test.
Characteristic of clonally expanded BCRs in COVID-19 patients
Analysis of BCR clonotypes showed that the clonal diversity of COVID-19 patients was significantly lower than healthy controls (Wilcoxon’s signed-rank test, P-value = 2.45 × 10−2, Figure 3A and Table S2, see Supplementary Data available online at http://bib.oxfordjournals.org/). Then, we clustered BCRs to identify expanded B cell clonotypes in each patient separately. We assembled a total of 9268 BCR groups from all 12 COVID-19 patients, 310 of which contain at least three BCRs, which were defined as clonal B cells. The proportion of clonal cells was much higher in COVID-19 compared with healthy controls. On average, about ~25% clonal cells in COVID-19 patients, compared with ~7% in healthy controls (Wilcoxon’s signed-rank test, P-value = 4.74 × 10−3, Figure 3B). There are no BCRs shared between COVID-19 and healthy controls. We further examined whether clonal BCRs were shared across patients and found that little identical BCRs were only present in two patients, which suggests a potentially wide range of BCRs with antivirus function. Then, we examined the usage of V and J gene segments in clonal BCRs. The most frequently used V and J gene segments were those mentioned above that were significantly higher in disease than healthy controls, such as IGHV4-39, IGHV3-23, IGHV1-3, IGHV2-70 and IGHJ3 of the heavy chain, and IGKV1D-13, IGLV1-51, IGLJ3 and IGKJ2 of the light chain (Figures 3C and S4A, see Supplementary Data available online at http://bib.oxfordjournals.org/). For the V-J pairs, the most frequently used in the heavy and light chain were IGHV4-39-IGHJ3 and IGLV1-51-IGLJ2, respectively (Figure S2, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Figure 3.
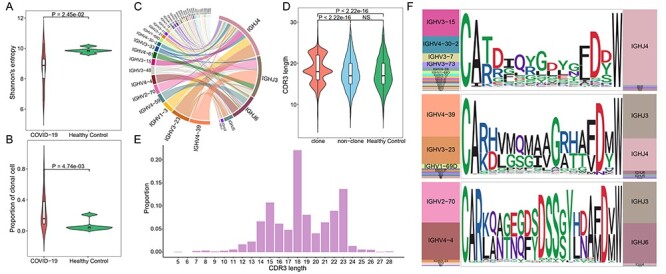
Analysis of the heavy chain of clonal expended BCR in COVID-19 patients. (A) Violin plot showing the clonal diversity of COVID-19 patients and healthy controls, statistical significance was evaluated using the Wilcoxon rank-sum test. (B) Violin plot showing the proportion of clonal B cells in COVID-19 and healthy controls, statistical significance was evaluated using the Wilcoxon rank-sum test. (C) Circos plot showing the VJ pairs in the heavy chain of clonal BCRs of disease. (D) The distribution of CDR3 length in the heavy chain, the curves of the violin plot show the density of values. Statistical significance was evaluated using the Wilcoxon rank-sum test. (E) The barplot showing the proportion of each CDR3 length in clonal BCRs of COVID-19. (F) The V and J gene usage, and CDR3 motif of clonal BCR which CDR3 length is 15aa (top), 18aa (middle) and 23aa (bottom).
The complementarity-determining region 3 (CDR3), a highly variable region in BCR, plays the most important role in specific antigen recognition in B cells [30, 34]. We next explore the characteristics of CDR3 sequences of clonal expended BCR in COVID-19. In the heavy chain, the length of CDR3 was concentrated in three lengths,15, 18 and 22 amino acids, among which 18 amino acids accounted for the largest proportion, and it was significantly longer than that in non-cloned BCRs and healthy controls (Wilcoxon’s signed-rank test, P-value < 2.22 × 10−16, Figure 3D and E). Further motif enrichment analysis showed that the amino acid sequences of CDR3s in the heavy chain were more convergent compared with that of healthy controls, especially the CDR3s with 15, 18 and 23 amino acids, suggesting high specificity of BCRs after antivirus immune (Figures 3F and S3, see Supplementary Data available online at http://bib.oxfordjournals.org/). Interestingly, the CDR3 with different lengths correspond to different V gene segments (Figure 3F). Although the average length of CDR3s in the light chain of clonal expended BCRs was significantly increased, the proportion of BCRs with different CDR3 lengths was highly consistent between disease and healthy controls. The CDR3 length of all light chains was mainly 11, 12 and 13 amino acids, and the amino acid sequence was more conserved (Figure S4B–D, see Supplementary Data available online at http://bib.oxfordjournals.org/). Altogether, our result suggested that there was a significantly clonal expansion of peripheral blood B cells and preferential usage changes of BCR repertoire in COVID-19 patients, especially in the heavy chain of BCR.
Immunoglobulin isotypes analysis of COVID-19
The constant region of the BCR heavy chain determines the type of immunoglobulin (Ig) and the effector function they can induce. There are five types of immunoglobulin in humans, including IgM, IgD, IgG, IgA and IgE. Different Ig types usually will have a different degree of antibody affinity [35, 36]. We first compared the frequency of different Ig isotypes between COVID-19 and healthy controls. IgM presents the largest proportion in all BCR repertoire (>50%), but we observed that its proportion was significantly decreased in COVID-19 patients, which is consistent with the truth of other isotypes with higher affinity arising up under anti-infective process (Fisher’s exact test, P-value < 0.05, Odd Ratio > 1, Figures 4A and S5A and B, see Supplementary Data available online at http://bib.oxfordjournals.org/). The proportions of IgG1, IgG3 and IgA1 were significantly increased in COVID-19 patients and the ratios of IgG to IgM/D and IgA to IgM/D in COVID-19 were much higher than that in healthy controls (Figure 4B). These changes indicated a strong humoral immune response to clear viral particles in the blood and respiratory mucosa [37, 38].
Figure 4.
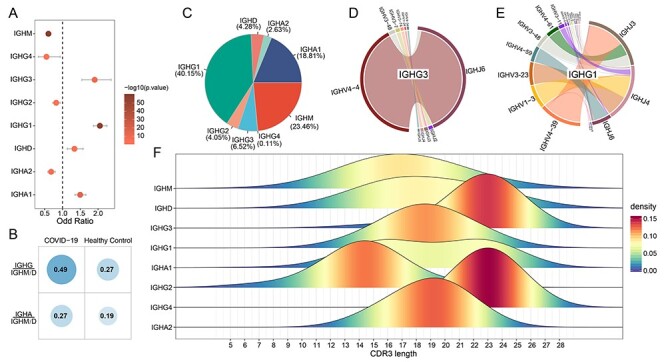
Analysis of Ig isotypes. (A) Differential analysis of Ig isotypes between COVID-19 and healthy controls by Fisher’s exact test (P < 0.05). (B) Comparison of the ratio of IgG or IgA to IgM/D between COVID-19 and healthy controls. (C) The proportion of each Ig isotype in clonal BCRs. (D) IGHV-IGHJ pairs usage of IGHG3 in clonal BCRs. (E) IGHV-IGHJ pairs usage of IGHG1 in clonal BCRs. (F) The distribution of CDR3 length of each Ig isotypes in clonal BCRs.
For clonal BCRs of COVID-19, the proportion of IgM was significantly decreased (Figures 4C and S5A, see Supplementary Data available online at http://bib.oxfordjournals.org/). And IgG1 and IgA1 were the major Ig classes, accounting for 40.15 and 18.81% of all clonal BCRs, respectively (Figure 4C). IgG-expressing B cells have higher plasma cell differentiation ability, thus increasing antibody secretion in response to viral infection [39, 40]. Statistics of the gene segments in each isotype showed that IgM and IgD utilized multiple V gene segments, while IgG and IgA showed skewing of different V gene segments (Figures 4D and E and S5C–H, see Supplementary Data available online at http://bib.oxfordjournals.org/). Specifically, IgG was primarily preferred to use IGHV4, in which IgG1 mainly used IGHV4-39, IgG2 mainly used IGHV4-30-2, IgG3 and IgG4 mainly used IGHV4-4, while IgA1 and IgA2 were primarily preferred to use IGHV2-70 and IGHV3-33, respectively, indicating respective potential local induction in serum and mucosa (Figures 4D and E and S5E–H, see Supplementary Data available online at http://bib.oxfordjournals.org/). Further analysis revealed that the length of CDR3 in each isotype was also different. The distribution of CDR3 length in IgM and IgD was consistent with that of healthy controls and non-clonal BCRs of COVID-19 (Figures 3D and 4F). However, the length of CDR3 of different isotypes in IgG and IgA tend to be concentrated in a certain length and longer than that of IgM and IgD, except for IgG2 (Figure 4F). Studies have revealed the enrichment and dominance of certain CDR3 lengths of B cells in antigen-specific populations [41, 42]. And another analysis of HIV-1+ samples showed that CDR3 lengths of IgG and IgA were longer than that in healthy donors [43]. Therefore, the enrichment and longer CDR3 with certain amino acid lengths in clonal IgG and IgA BCRs may play an important role in virus clearance.
Frequent CSR events and high SHM rate of IgG3 in COVID-19
After B cells respond to antigen, many BCRs undergo CSR to progressively refine the antibody. This process entails the DNA deletion and recombination of IgM to generate ‘downstream’ isotypes and mutation of nucleotides in the antigen-binding region (SHM), a process followed by selection and resulting in affinity maturation. We explored the SHM and CSR in COVID-19 patients compared with healthy controls. The class-switch types detectable in COVID-19 patients were increased, and most CSR only take place in clonal expanded BCRs (Figure 5A–D). Compared with other isotypes, IgG isotypes frequently undergo class switch, especially IgG3. Further analysis found that the SHM ratio of clonal BCRs was significantly higher than that of non-clonal BCRs or healthy controls (Wilcoxon’s signed-rank test, Figure 5E). Among all isotypes of clonal BCRs in COVID-19, the SHM rate of IgG was higher than that of other isotypes, and IgG3 also showed the highest SHM rate compared with other IgG isotypes (Wilcoxon’s signed-rank test, Figures 5F and S6, see Supplementary Data available online at http://bib.oxfordjournals.org/). High titers of IgG antibodies have been detected in the blood samples of discharged patients [44, 45]. Next, we focus on the IgG3 isotype due to its frequent CSR events and high SHM rate. There were 12 IgG3 associated clonal BCR groups, 7 of which had CSR events. We found that the V, J gene segment and CDR3 sequence of most IgG3 was IGHV4-4-CARLANTNQFYDSSSYLNAMDVW-IGHJ6, the CDR3 of which was a part of the motif described above (Table 1, Figure 3F). Besides, none of the CDR3 sequences of clonal IgG3 BCRs exists in healthy controls. Our results suggested that the IgG3 isotype may be a key factor in virus clearance.
Figure 5.
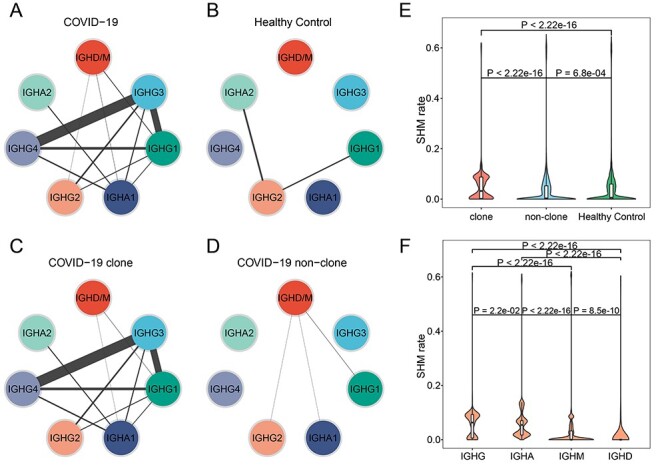
Comparison of SHM and CSR between COVID-19 and healthy controls. (A–D) CSR events of COVID-19 (A), healthy control (B), COVID-19 clonal BCR (C) and COVID-19 non-clonal BCR (D). Lines connecting two isotypes indicate the enrichment level of observing switches in the two corresponding Ig subclasses. (E) The differential analysis of SHM rate between clonal and non-clonal COVID-19 BCRs and healthy control. Statistical significance was evaluated using the Wilcoxon rank-sum test. (F) The differential analysis of SHM rate among different Ig isotypes in clonal BCRs. Statistical significance was evaluated using the Wilcoxon rank-sum test.
Table 1.
The V, J genes and CDR3H sequences of IgG3 isotype in clonal BCRs
| V | CDR3H | J | number |
|---|---|---|---|
| IGHV4-4 | CARLANTNQFYDSSSYLNAMDVW | IGHJ6 | 194 |
| IGHV3-48 | CAMFLYGSGSYYPLDVW | IGHJ6 | 7 |
| IGHV3-7 | CARRGQWLERYFALW | IGHJ2 | 7 |
| IGHV3-74 | CAREPRGYCTSISCYGGFDYW | IGHJ4 | 5 |
| IGHV3-48 | CARSMATIRVMSFGAFDIW | IGHJ3 | 5 |
| IGHV1-24 | CATDSPINWSVLDAFDIW | IGHJ3 | 3 |
| IGHV4-34 | CARSGRWLQFDHW | IGHJ5 | 3 |
Discussion
Since December 2019, COVID-19 has led to a worldwide pandemic [46]. In addition to severe respiratory pathology, COVID-19 can also cause thrombotic complications, myocardial dysfunction and arrhythmia, acute coronary syndromes, acute kidney injury, gastrointestinal symptoms, even neurologic illnesses and so on [47]. Given the serious threat COVID-19 poses to global health, a comprehensive understanding of immune responses in patients is essential for both pathological study and therapy [48]. The humoral immune responses play a central role in clearing SARS-CoV-2 infection and directly influence patients’ clinical outcomes. SARS-CoV-2 elicits a robust B cell response and virus-specific antibodies can be detected in the days following infection [46]. Analysis of BCR repertoire is critical for understanding the immune response of COVID-19 patients. In this study, we delineated the BCR repertoire in peripheral blood of convalescent COVID-19 patients.
Compared with the healthy controls, our analysis revealed higher frequent changes in the heavy chain of BCRs in disease. The BCR of disease converged on different IGHV3 and IGHV4 rearrangements, and the CDR3H was significantly longer than healthy controls and tended to converge on a few different lengths, while there was no significant difference in the light chain. These results imply that the BCR heavy chain plays a major role in clearing virus infection. Subsequent analysis of the isotype found a remarkably increased proportion of IgG and IgA in COVID-19 patients. IgA plays an important role in mucosal immunity [49]. An increase in IgA showed that it could be synthesized and widely migrated to the respiratory tract, gastrointestinal tract or other mucosal sites to perform immune functions [50]. IgG can provide long-term protection, and high titers of virus-specific IgG antibodies have been detected in the plasma of convalescent patients [51]. Besides, among all IgG isotypes, IgG3 had the most frequent SHM and CSR events, and we found an IgG3 cluster from different clonal groups which had the same IGHV, IGHJ and CDR3 sequence. These results imply that IgG3 may play an important role in fighting against SARS-CoV-2 infection.
Our study systematically characterized the BCR repertoire of COVID-19 patients. However, the data did not cover the BCR repertoire status before and during the onset of the disease, and the samples were obtained from 5 ml peripheral blood, so some clonotypes may not be included. In addition, the comparison between COVID-19 and normal infected samples was not performed because of the lack of data. Therefore, large population cohort studies and normal infected samples are urgently needed to obtain a more detailed view of the specific B cell immune response in COVID-19 patients. Taken together, our study systematically characterizes the BCR repertoire of COVID-19 patients, which provides substantial insight into exploring the immune response induced by SARS-CoV-2.
Data Availability
Data used in this study are available from the corresponding author by request.
Key Points
We performed scBCR-seq for 12 COVID-19 patients and six healthy controls.
The clonal diversity of BCR is significantly reduced in COVID-19 patients.
BCRs skew toward different V gene segments in COVID-19 and healthy controls.
IgG3 was frequently undergoing Class Switch Recombination events and the highest somatic hypermutation in COVID-19.
Supplementary Material
Xiyun Jin is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. Her research interests include bioinformatics and genomics in complex diseases.
Wenyang Zhou is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. His research interests include structural bioinformatics and single cell genomics.
Meng Luo is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. His research interests include developing algorithms and data structures for problems in the life sciences.
Pingping Wang is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. Her research interests include bioinformatics and single cell genomics in complex diseases.
Zhaochun Xu is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. His research interests include bioinformatics and machine learning.
Kexin Ma is a MS student in the School of Life Science and Technology at the Harbin Institute of Technology. Her research interests include bioinformatics and single cell sequencing.
Huimin Cao is a MS student in the School of Life Science and Technology at the Harbin Institute of Technology. Her research interests include bioinformatics and immunogenomics.
Chang Xu is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. His research interests include structural bioinformatics and machine learning.
Yan Huang is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology, China. Her research interests include bioinformatics and epigenomics of complex diseases.
Rui Cheng is a PhD student of biomedicine engineering at the Harbin Institute of Technology, China. His research interests include complex disease bioinformatics and genomics.
Lixing Xiao is a PhD student in the School of Life Science and Technology at the Harbin Institute of Technology. Her research interests include bioinformatics and immunogenomics.
Xiaoyu Lin is a PhD student of biomedicine engineering at the Harbin Institute of Technology, China. Her research interests include bioinformatics and single cell sequencing of complex diseases.
Fenglan Pang is a PhD student of biomedicine engineering at the Harbin Institute of Technology, China. Her research interests include bioinformatics and machine learning.
Yiqun Li is a postdoctoral researcher at the Harbin Institute of Technology, China. His research interests include bioinformatics and machine learning.
Huan Nie is a professor in the School of Life Science and Technology at the Harbin Institute of Technology. His research interests focus on the molecular mechanism of complex disease.
Qinghua Jiang is a professor in the School of Life Science and Technology at the Harbin Institute of Technology. His research interests cover immunoinformatics of complex diseases.
Contributor Information
Xiyun Jin, School of Life Science and Technology at the Harbin Institute of Technology, China.
Wenyang Zhou, School of Life Science and Technology at the Harbin Institute of Technology, China.
Meng Luo, School of Life Science and Technology at the Harbin Institute of Technology, China.
Pingping Wang, School of Life Science and Technology at the Harbin Institute of Technology, China.
Zhaochun Xu, School of Life Science and Technology at the Harbin Institute of Technology, China.
Kexin Ma, School of Life Science and Technology at the Harbin Institute of Technology, China.
Huimin Cao, School of Life Science and Technology at the Harbin Institute of Technology, China.
Chang Xu, School of Life Science and Technology at the Harbin Institute of Technology, China.
Yan Huang, School of Life Science and Technology at the Harbin Institute of Technology, China.
Rui Cheng, Harbin Institute of Technology, China.
Lixing Xiao, School of Life Science and Technology at the Harbin Institute of Technology, China.
Xiaoyu Lin, Harbin Institute of Technology, China.
Fenglan Pang, Harbin Institute of Technology, China.
Yiqun Li, Harbin Institute of Technology, China.
Huan Nie, School of Life Science and Technology at the Harbin Institute of Technology, China.
Qinghua Jiang, School of Life Science and Technology at the Harbin Institute of Technology, China.
Authors’ Contributions
Q.J. conceived the project. Q.J., X.J., W.Z., M.L., P.W., Z.X., K.M., H.C., C.X., Y.H., R.C., L.X., X.L., F.P., Y.L. and H.N. contributed to data analysis. Q.J. and X.J. wrote the manuscript.
Funding
National Natural Science Foundation of China (Nos. 61822108 and 62032007); Emergency Research Project for COVID-19 of Harbin Institute of Technology (No. 2020-001).
Conflict of Interest
We declare that there is no conflict of interest regarding the publication of this article.
References
- 1. Zhu N, Zhang D, Wang W, et al. . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382(8):727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Q, Guan X, Wu P, et al. . Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382(13):1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020;20(5):533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walls AC, Park YJ, Tortorici MA, et al. . Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181(2):281–292 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoffmann M, Kleine-Weber H, Schroeder S, et al. . SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271–280 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jeyanathan M, Afkhami S, Smaill F, et al. . Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol 2020;20(10):615–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. . Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020;181(5):1036–1045 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Remy KE, Mazer M, Striker DA, et al. . Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight 2020;5(17):e140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D, Hu B, Hu C, et al. . Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020;323(11):1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poulas K, Farsalinos K, Zanidis C. Activation of TLR7 and innate immunity as an efficient method against COVID-19 pandemic: imiquimod as a potential therapy. Front Immunol 2020;11:1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahmadpoor P, Rostaing L. Why the immune system fails to mount an adaptive immune response to a COVID-19 infection. Transpl Int 2020;33(7):824–5. [DOI] [PubMed] [Google Scholar]
- 13. Nielsen SCA, Boyd SD. Human adaptive immune receptor repertoire analysis-past, present, and future. Immunol Rev 2018;284(1):9–23. [DOI] [PubMed] [Google Scholar]
- 14. Soto C, Bombardi RG, Branchizio A, et al. . High frequency of shared clonotypes in human B cell receptor repertoires. Nature 2019;566(7744):398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoehn KB, Fowler A, Lunter G, et al. . The diversity and molecular evolution of B-cell receptors during infection. Mol Biol Evol 2016;33(5):1147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petrova VN, Muir L, McKay PF, et al. . Combined influence of B-cell receptor rearrangement and somatic hypermutation on B-cell class-switch fate in health and in chronic lymphocytic leukemia. Front Immunol 2018;9:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brouwer PJM, Caniels TG, van der Straten K, et al. . Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020;369(6504):643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robbiani DF, Gaebler C, Muecksch F, et al. . Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020;584(7821):437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao Y, Su B, Guo X, et al. . Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell 2020;182(1):73–84 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ju B, Zhang Q, Ge J, et al. . Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020;584(7819):115–9. [DOI] [PubMed] [Google Scholar]
- 21. Wu Y, Wang F, Shen C, et al. . A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020;368(6496):1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li F, Luo M, Zhou W, et al. . Single cell RNA and immune repertoire profiling of COVID-19 patients reveal novel neutralizing antibody. Protein Cell 2020;1–5. doi: 10.1007/s13238-020-00807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thevarajan I, Nguyen THO, Koutsakos M, et al. . Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med 2020;26(4):453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui JH, Lin KR, Yuan SH, et al. . TCR repertoire as a novel indicator for immune monitoring and prognosis assessment of patients with cervical cancer. Front Immunol 2018;9:2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schneider-Hohendorf T, Mohan H, Bien CG, et al. . CD8(+) T-cell pathogenicity in Rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing. Nat Commun 2016;7:11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nouri N, Kleinstein SH. A spectral clustering-based method for identifying clones from high-throughput B cell repertoire sequencing data. Bioinformatics 2018;34(13):i341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagih O. Ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 2017;33(22):3645–7. [DOI] [PubMed] [Google Scholar]
- 28. Gupta NT, Vander Heiden JA, Uduman M, et al. . Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 2015;31(20):3356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alt FW, Zhang Y, Meng FL, et al. . Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013;152(3):417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen H, Zhang Y, Ye AY, et al. . BCR selection and affinity maturation in Peyer's patch germinal centres. Nature 2020;582(7812):421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fu Y, Zhang Z, Sheehan J, et al. . A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat Commun 2016;7(1):12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehrhardt SA, Zehner M, Krähling V, et al. . Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat Med 2019;25(10):1589–600. [DOI] [PubMed] [Google Scholar]
- 33. Bryson S, Thomson CA, Risnes LF, et al. . Structures of preferred human IgV genes-based protective antibodies identify how conserved residues contact diverse antigens and assign source of specificity to CDR3 loop variation. J Immunol 2016;196(11):4723–30. [DOI] [PubMed] [Google Scholar]
- 34. Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000;13(1):37–45. [DOI] [PubMed] [Google Scholar]
- 35. Shi Z, Zhang Q, Yan H, et al. . More than one antibody of individual B cells revealed by single-cell immune profiling. Cell Discov 2019;5(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kerr WG, Hendershot LM, Burrows PD. Regulation of IgM and IgD expression in human B-lineage cells. J Immunol 1991;146(10):3314–21. [PubMed] [Google Scholar]
- 37. Woof JM, Kerr MA. IgA function--variations on a theme. Immunology 2004;113(2):175–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palladino G, Mozdzanowska K, Washko G, et al. . Virus-neutralizing antibodies of immunoglobulin G (IgG) but not of IgM or IgA isotypes can cure influenza virus pneumonia in SCID mice. J Virol 1995;69(4):2075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Surova E, Jumaa H. The role of BCR isotype in B-cell development and activation. Adv Immunol 2014;123:101–39. [DOI] [PubMed] [Google Scholar]
- 40. Nutt SL, Hodgkin PD, Tarlinton DM, et al. . The generation of antibody-secreting plasma cells. Nat Rev Immunol 2015;15(3):160–71. [DOI] [PubMed] [Google Scholar]
- 41. Miqueu P, Guillet M, Degauque N, et al. . Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol Immunol 2007;44(6):1057–64. [DOI] [PubMed] [Google Scholar]
- 42. Boisvert M, Zhang W, Elrod EJ, et al. . Novel E2 glycoprotein tetramer detects hepatitis C virus-specific memory B cells. J Immunol 2016;197(12):4848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waltari E, Jia M, Jiang CS, et al. . 5' Rapid amplification of cDNA ends and Illumina MiSeq reveals B cell receptor features in healthy adults, adults with chronic HIV-1 infection, cord Blood, and humanized mice. Front Immunol 2018;9:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ni L, Ye F, Cheng ML, et al. . Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 2020;52(6):971–977 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grifoni A, Weiskopf D, Ramirez SI, et al. . Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020;181(7):1489–1501 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vabret N, Britton GJ, Gruber C, et al. . Immunology of COVID-19: current state of the science. Immunity 2020;52(6):910–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta A, Madhavan MV, Sehgal K, et al. . Extrapulmonary manifestations of COVID-19. Nat Med 2020;26(7):1017–32. [DOI] [PubMed] [Google Scholar]
- 48. Su Y, Chen D, Yuan D, et al. . Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 2020;183(6):1479–1495 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen K, Magri G, Grasset EK, et al. . Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat Rev Immunol 2020;20(7):427–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Macpherson AJ, McCoy K, Johansen FE, et al. . The immune geography of IgA induction and function. Mucosal Immunol 2008;1(1):11–22. [DOI] [PubMed] [Google Scholar]
- 51. Shen C, Wang Z, Zhao F, et al. . Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA 2020;323(16):1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this study are available from the corresponding author by request.
Key Points
We performed scBCR-seq for 12 COVID-19 patients and six healthy controls.
The clonal diversity of BCR is significantly reduced in COVID-19 patients.
BCRs skew toward different V gene segments in COVID-19 and healthy controls.
IgG3 was frequently undergoing Class Switch Recombination events and the highest somatic hypermutation in COVID-19.


