Abstract
Neurodegenerative proteinopathies are characterized by progressive cell loss that is preceded by the mislocalization and aberrant accumulation of proteins prone to aggregation. Despite their different physiological functions, disease-related proteins like tau, α-synuclein, TAR DNA binding protein-43, fused in sarcoma and mutant huntingtin, all share low complexity regions that can mediate their liquid-liquid phase transitions. The proteins’ phase transitions can range from native monomers to soluble oligomers, liquid droplets and further to irreversible, often-mislocalized aggregates that characterize the stages and severity of neurodegenerative diseases.
Recent advances into the underlying pathogenic mechanisms have associated mislocalization and aberrant accumulation of disease-related proteins with defective nucleocytoplasmic transport and its mediators called karyopherins. These studies identify karyopherin abnormalities in amyotrophic lateral sclerosis, frontotemporal dementia, Alzheimer’s disease, and synucleinopathies including Parkinson’s disease and dementia with Lewy bodies, that range from altered expression levels to the subcellular mislocalization and aggregation of karyopherin α and β proteins.
The reported findings reveal that in addition to their classical function in nuclear import and export, karyopherins can also act as chaperones by shielding aggregation-prone proteins against misfolding, accumulation and irreversible phase-transition into insoluble aggregates. Karyopherin abnormalities can, therefore, be both the cause and consequence of protein mislocalization and aggregate formation in degenerative proteinopathies. The resulting vicious feedback cycle of karyopherin pathology and proteinopathy identifies karyopherin abnormalities as a common denominator of onset and progression of neurodegenerative disease.
Pharmacological targeting of karyopherins, already in clinical trials as therapeutic intervention targeting cancers such as glioblastoma and viral infections like COVID-19, may therefore represent a promising new avenue for disease-modifying treatments in neurodegenerative proteinopathies.
Keywords: karyopherin, nucleocytoplasmic transport, phase transition, protein aggregation, neurodegeneration
Pasha et al. review the evidence that karyopherins – which mediate nucleocytoplasmic transport – can shield aggregation-prone proteins against misfolding and irreversible phase transition into insoluble aggregates. Abnormalities in karyopherins could be a novel pathogenic mechanism underlying neurodegeneration.
Introduction
Neurodegeneration is the selective and progressive loss of neurons, eventually leading to cognitive and behavioural deficits for which currently no cure or effective treatments are available.1 Neurodegenerative diseases are distinguished by clinical appearance and pathology, such as movement disorders like Parkinson’s disease, Huntington’s disease and amyotrophic lateral sclerosis (ALS), and dementias like Alzheimer’s disease, frontotemporal dementia (FTD) and dementia with Lewy bodies (DLB). Despite their clinical heterogeneity, the majority of neurodegenerative diseases are characterized by proteinaceous inclusions that are often mislocalized and accumulate in the extracellular milieu or intracellular compartments of affected cells. The disease-related proteins are frequently post-translationally modified (phosphorylated, ubiquitinated, acetylated and cleaved into fragments), which is often related to their transition from native soluble monomers into oligomers, protofibrils and the ultimate deposition of β-sheet fibrils and insoluble aggregates.2 The quantity and distribution of these aggregates correlates with the stage and severity of the diseases,3,4 and different proteins aggregate in different neurodegenerative diseases. For example, in Alzheimer’s disease, proteins aggregate as amyloid plaques comprising amyloid-β fragments of aberrantly cleaved amyloid precursor protein; in Parkinson’s disease and DLB as Lewy bodies comprising aberrant forms of α-synuclein (α-syn); in Alzheimer’s disease, FTD, Parkinson’s disease and ALS as tangles comprising phosphorylated tau; and in ALS, FTD, Alzheimer’s disease and Parkinson’s disease as insoluble aggregates containing TAR DNA binding protein 43 (TDP-43), with some TDP-43-negative cases of ALS and FTD as insoluble aggregates of fused in sarcoma (FUS); and in Huntington’s disease as mutant huntingtin (mHTT) with aberrant polyglutamine (polyQ) extension.
The disease-related proteins differ significantly in function and, until recently, it remained elusive how such functionally unrelated proteins can lead to comparable pathogenic alterations and degenerative cell death. However, structure-function analyses revealed the majority of these proteins harbour unstructured, intrinsically disordered amino acid sequences called low-complexity regions that have recently been linked with unprecedented insights into the pathogenic mechanisms underlying neurodegenerative diseases.5-11 These studies showed that low-complexity regions of disease-related proteins enable their aberrant phase transition into liquids and the subsequent mislocalization and accumulation as fibrillary aggregates.12–14 Further mechanistic insights revealed that nuclear transport receptors, termed karyopherins, and their abnormalities are associated with protein mislocalization and aggregation.15,16
These findings provide evidence that in addition to nucleocytoplasmic transport, karyopherins can also function as chaperones to maintain their protein cargos in a soluble non-aggregated state, thereby reducing aberrant phase transition and protein accumulation.17–20 For example, in the case of ALS and FTD, it has been shown that the underlying pathogenesis involves defective nucleocytoplasmic transport across the nuclear envelope,21–24 mediated by karyopherins known to shuttle proteins across the nuclear membrane.25 These findings suggest a dual role of karyopherins as a molecular switch between protein aggregation states and as a gatekeeper of protein localization, deregulation of which can trigger onset and progression of disease. Here we review recent evidence and current understanding of karyopherin function, their role in onset and progression of disease, and discuss their potential as therapeutic targets in neurodegenerative proteinopathies.
Karyopherin protein families
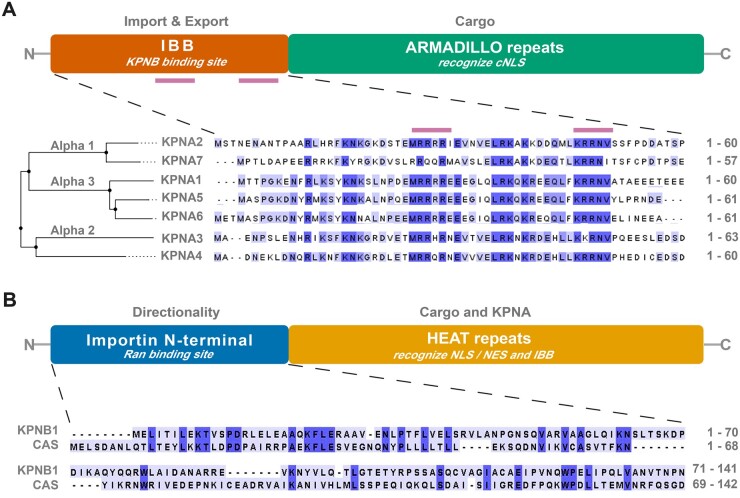
Karyopherins are classified into karyopherin-α (KPNA) and karyopherin-β (KPNB) families (Fig. 1), with each member recognizing its own set of cargo proteins or RNAs.26 KPNA orthologues are grouped into subfamilies, α1, α2 and α3 (Fig. 1A andTable 1) based on differences in primary amino acid sequences.27 The human genome encodes seven KPNA orthologues with a common structure consisting of 10 helical armadillo (ARM) repeats that recognize the nuclear localization signal (NLS) of their specific cargos; a short C-terminal region that functions as a specific binding site for the export factor chromosome segregation 1 like (CSE1L, also known as cellular apoptosis susceptibility, CAS); and an N-terminal importin beta binding (IBB) domain for KPNB1 binding.27 Evidence from cancer research reveals that these subfamilies have distinct cargo specificities. For instance, members of the α2 subfamily KPNA3 and KPNA4 show particular affinity for NF-κB and the nuclear regulator of chromosome condensation 1 (RCC1), whilst KPNA1 of the α3 subfamily specifically binds to signal transducer and activator of transcription 1 and 2 (STAT1, STAT2).28 Furthermore, the N-terminal IBB domain of KPNAs (Fig. 1A) also controls the accessibility of the NLS binding site by mimicking an NLS structure and folding back to occupy the ARM repeats when KPNAs are not bound to their cargos.29,30 Thus, only cargos with high affinity are bound to KPNAs (Table 1). Cargos with a relatively lower affinity bind to the ARM repeats of KPNAs upon formation of KPNA-KPNB1 heterodimer, which frees the NLS binding site.30
Figure 1.
Functional architecture of karyopherins. (A) KPNA family proteins consist of two main functional domains: an importin beta (KPNB) binding (IBB) domain for nuclear import and export and armadillo repeats for the recognition of canonical NLS (cNLS) of cargo proteins. The IBB consists of cNLS binding boxes (bars in purple), major (‘KRR’) and minor (‘RRRR’ or ‘RRQR’ or ‘RRHR’) binding sites, which in the absence of KPNB occupy the canonical NLS-binding surface of armadillo repeats. This prevents import of ‘unloaded’ karyopherin complexes in the nucleus. KPNA family proteins are subdivided into three subfamilies, α1, α2 and α3, based on differences in amino acid sequence in their IBB domain and cNLS binding sites. See Table 1 for details on all KPNA family members. (B) KPNB family proteins comprise an N-terminal (20–120 amino acids) importin domain responsible for binding with Ran and for directed protein translocation across the nuclear envelope, and HEAT repeats that are distinguishable by their flexibility and recognition/binding of cargo-proteins via the NLS or the nuclear export signal and binding to KPNA. Shown are two functionally important representatives of the KPNB family, KPNB1 and CAS (see Table 2 for details on all KPNB family members). Protein sequences were obtained from UniProt (see Tables 1 and 2 for ID numbers); Clustal Omega was used for sequence alignment. CAS = cellular apoptosis susceptibility protein; NES = nuclear export signal.
Table 1.
KPNA proteins and their essential cargos
| Gene/Protein name | NCBI ID | Subfamily | Synonyms |
Cargos | References | |||
|---|---|---|---|---|---|---|---|---|
| Importin | SRP | NPI/QIP | Other | |||||
| KPNA1 | 3836 | α3 | IPOA-5 | SRP1β | NPI-1 | RCH2 | TDP-43 NF-κB (p50/p65; c-Rel; p52; RelB) | Pumroy and Cingolani,27 Lee et al.,50 Nishimura et al.54 |
| KPNA2 | 3838 | α1 | IPOA-1 | SRP1α | NPI-3 | RCH1 | TDP-43 NF-κB (p65) | Pumroy and Cingolani,27 Nishimura et al.54 |
| KPNA3 | 3839 | α2 | IPOA-4 | SRP4, hSRP1, SRP1γ | QIP2 | n/a | TDP-43a NF-κB (p50/p65; p52) ataxin-3 α1ACT | Pumroy and Cingolani,27 Nishimura et al.,54 Khsoravi et al.,208 Sowa et al.,209 Tsou et al.,210 Fagerlund et al.211 |
| KPNA4 | 3840 | α2 | IPOA-3 | SRP3 | QIP1 | MGC12217 | TDP-43a NF-κB (p50/p65; p52) | Pumroy and Cingolani,27 Nishimura et al.54 Khosravi et al.,208 Fagerlund et al.211 |
| KPNA5 | 3841 | α3 | IPOA-6 | SRP6 | n/a | n/a | TDP-43 NF-κB (p50/p65; c-Rel; p52; RelB) | Pumroy and Cingolani,27 Nishimura et al.54 |
| KPNA6 | 23633 | α3 | IPOA-7 | n/a | NPI-2 | MGC17918 | TDP-43 NF-κB (c-Rel; RelB) | Pumroy and Cingolani.,27 Nishimura et al.54 |
| KPNA7 | 402569 | α1 | IPOA-8 | n/a | n/a | n/a | n/a | n/a |
IPOA = importin alpha; MGC = Mammalian Gene Collection; n/a = not applicable; NF-κB = nuclear factor-κB; NPI = nucleoprotein interactor; QIP = Importin alpha Q; RCH = RAG cohort protein; SRP = signal recognition particle.
Whilst TDP-43 acts as a cargo for KPNA1–6, KPNA3 and 4 show the strongest interactions.
KPNB proteins (Fig. 1B and Table 2) play an essential role in nucleocytoplasmic transport and, as a secondary consequence downstream of their role in nuclear transport, also affect the regulation of gene expression, the immune response and signal transduction,31 as well as viral pathogenesis and tumorigenesis.32,33 Members of the KPNB family share an importin N-terminal domain responsible for RanGTP/GDP binding that drives active nucleocytoplasmic transport (Fig. 2). KPNBs are characterized by multiple tandem helical repeats termed HEAT repeats located in the C-terminal region, each composed of two antiparallel helices A and B arranged into ring-like structures.34,35 The HEAT repeats are responsible for binding to KPNA and to cargos.36 In humans, the KPNB family contains 18 members (Table 2), which can be divided into three groups based on their major role in nucleocytoplasmic transport as either exportins, importins or for bidirectional transport. For example, KPNBs such as exportin 4 (XPO4) or exportin 5 (XPO5) mediate nuclear export of cargos,37–41 whereas KPNB1 or TNPO1 and TNPO2 (also called KPNB2 and KPNB2B, respectively) mediate nuclear import (see Table 2 for synonyms and cargos). Despite significant differences in protein structure, localization and function of the majority of disease-associated proteins, their nucleocytoplasmic shuttling is mediated by karyopherins.
Table 2.
KPNB proteins and their essential cargos
| Gene/Protein |
Cargos | References | |||
|---|---|---|---|---|---|
| Name | NCBI ID | Synonyms | Full name | ||
| XPO1 | 7514 | EXP1 | Exportin-1 | TDP-43?; rRNA; snRNA; mRNA | Archbold et al.,205 Lu et al.212 |
| CRM1 | Chromosomal maintenance 1 | ||||
| CAS | 1434 | XPO2 | Exportin-2 | Importin α; TDP-43? | Kutay et al.,37 Nishimura et al.54 |
| CAS | Cellular apoptosis susceptibility | ||||
| CSE1L | Chromosome segregation 1-like | ||||
| XPOT | 11260 | XPO3 | Exportin-3 | tRNA; Scyl1 | Kutay et al.,38 Chafe and Mangroo213 |
| XPO5 | 57510 | EXP5 | Exportin-5 | Scyl1 miRNA precursors | Chafe and Mangroo,213 Wang et al.214 |
| RANBP21 | Ran-binding protein 21 | ||||
| XPO6 | 23214 | EXP6 | Exportin-6 | Actin | Stueven et al.215 |
| RANBP20 | Ran-binding protein 20 | ||||
| XPO7 | 23039 | EXP7 | Exportin-7 | p50RhoGAP; TDP-43; Tubulin; Histones | Archbold et al.,205 Mingot et al.,216 Aksu et al.217 |
| RANBP16 | Ran-binding protein 16 | ||||
| KPNB1 | 3837 | IPO1 | Importin-1 | HTT; RPL23A; RPS7; RPL5; SNAI1 PRKCI | Desmond et al.68 Jäkel and Görlich218 UniProtKB-Q14974 |
| IPO90 | Importin-90 | ||||
| IPOB | Importin-beta | ||||
| IMB1 | Importin beta 1 subunit | ||||
| KPNB1 | Karyopherin-beta 1 | ||||
| PTAC97 | Pore targeting complex 97 kDa subunit | ||||
| NTF97 | Nuclear factor p97 | ||||
| TNPO1 | 3842 | TRN | Transportin 1 | HTT; FUS; TAF15; EWS; hnRNP-a1/a2; RPL23A; RPS7; RPL5; ADAR/ADAR1 (isoform 1 and 5) | Desmond et al.,68 Jäkel and Görlich,218 Neumann et al.,219 Barraud et al.220 |
| IPO2 | Importin beta-2 | ||||
| KPNB2 | Karyopherin-beta 2 | ||||
| MIP | M9 region interaction protein (MIP) | ||||
| TNPO2 | 30000 | TRN2 | Transportin-2 | FUS; hnRNP A1/H/M | Güttinger et al.221 |
| IPO3 | Importin 3 | ||||
| KPNB2B | Karyopherin β2-2B | ||||
| TNPO3 | 23534 | TRN3 | Transportin-3 | SFRS1; SFRS2 | Kataoka et al.222 |
| TRNSR | Transportin-SR | ||||
| IPO12 | Importin-12 | ||||
| LGMD1F | Limb girdle muscular dystrophy 1 F | ||||
| IPO4 | 79711 | IMP4 | Importin-4 | RPS3A | Jäkel et al.223 |
| IMP4b | Importin-4b | ||||
| RANBP4 | Ran-binding protein 4 | ||||
| IPO5 | 3843 | IMP5 | Importin-5 | RPL23A RPS7; RPL5 | Jäkel and Görlich218 |
| IMB3 | Importin subunit β-3 | ||||
| KPNB3 | Karyopherin β-3 | ||||
| RANBP5 | Ran-binding protein 5 | ||||
| PSE1 | Protein Secretion Enhancer 1 | ||||
| IPO7 | 10527 | IMP7 | Importin-7 | RPL23A RPS7; RPL5 | Jäkel and Görlich218 |
| RANBP7 | Ran-binding protein 7 | ||||
| IPO8 | 10526 | IMP8 | Importin-8 | SRP19 | Dean et al.224 |
| RANBP8 | Ran-binding protein 8 | ||||
| IPO9 | 55705 | IMP9 | Importin-9 | Actin; RPS7 RPL18A RPL6 | Jäkel et al.223 UniProtKB—Q96P70 |
| RANBP9 | Ran-binding protein 9 | ||||
| KIAA1192 | Identified by Kazusa Institute | ||||
| HSPC273 | Homo sapiens HSPC273 mRNA, partial cds | ||||
| IPO11 | 51194 | IMP11 | Importin-11 | UbcM2 | Plafker and Macara225 |
| RANBP11 | Ran-binding protein 11 | ||||
| XPO4 | 64328 | EXP4 | Exportin-4 | n/a | n/a |
| IPO13 | 9670 | IMP13 | Importin-13 | RBM8A SUMO1; UBC9; EIF1A | Mingot et al.226 |
| KAP13 | Karyopherin 13 | ||||
| RANBP13 | Ran binding protein 13 | ||||
| KIAA0724 | Identified by Kazusa Institute | ||||
| LGL2 | Late gestation lung 2 | ||||
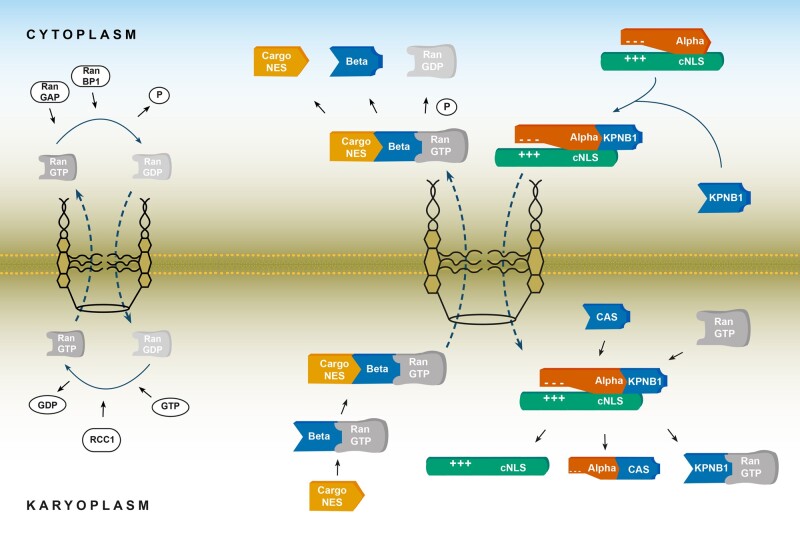
Figure 2.
Nucleocytoplasmic transport and chaperone function of karyopherins. Left: Schematic diagram showing the RanGTP/GDP cycle through the nuclear pore complex. In the cytoplasm, RanGAP1, together with RanBP, hydrolyses RanGTP to maintain high cytoplasmic concentrations of RanGDP. In the nucleus, RCC1 (nuclear RanGEF) facilitates GTP-GDP exchange, causing high concentrations of RanGTP. These regulators maintain higher concentrations of RanGDP in the cytoplasm whilst preserving high levels of RanGTP in the karyoplasm, leading to a gradient required for energy-dependent nucleocytoplasmic transport. Right: Schematic diagram of the classical nuclear import and export pathway. During nuclear import, KPNB1 binds to KPNA, which itself is bound to the classical NLS (cNLS) of cargo, forming a trimeric complex. KPNA also exerts chaperone function to cargo-cNLS by shielding basic residues from hydrophobic/ionic interactions, which maintains a cargo protein in its native soluble state. KPNB1 carries the complex through the nuclear pore complex, where RanGTP binds, causing a conformational change in the bound importin (note, some KPNBs shown as beta bind directly to cargo forming a dimeric complex which directly translocates to the nucleus). This results in a trimeric complex of KPNA, nuclear export factor CAS, and RanGTP, and a dimeric complex consisting of KPNB1 and RanGTP. Both complexes then translocate back to the cytoplasm where their respective RanGTPs are hydrolysed to bind to the next cargo. During export, Exportin/KPNB (beta) bound to RanGTP binds to the cargo-NES of the cargo in the nucleoplasm. This complex is exported through the nuclear pore complex into the cytoplasm where RanGTP is hydrolysed, which triggers cargo release. CAS = cellular apoptosis susceptibility protein; RAN = Ras-related nuclear; RCC1 = regulator of chromosome condensation 1; Ran-GAP = Ran GTPase activating protein; RanBP1 = Ran binding protein 1; RanGEF = Ran guanine exchange factor.
Karyopherins in nucleocytoplasmic transport
The nucleus of cells provides a protective environment through tightly regulated protein import and export.42 While smaller proteins as well as ions, salts and nucleotides can enter the nucleus via passive diffusion,34 proteins larger than 40–65 kDa require binding to nuclear transport receptors to traverse the nuclear pore complex.26 This process (Fig. 2) is regulated by three key elements: the nuclear pore complex; nuclear transport factors of the karyopherin families; and a Ras-related nuclear (RAN) protein gradient.43 The nuclear pore complex consists of three rings: the nucleoplasmic ring, the middle inner ring, and the cytoplasmic ring that form an aqueous channel spanning the nuclear envelope.44 At the molecular level, it consists of an assembly of ∼30 nucleoporins classified into six groups based on their localization.45 The nucleoporins are also grouped by their molecular function into those that form the nuclear pore complex scaffold, the transmembrane nucleoporins and the phenylalanine and glycine (FG) containing nucleoporins—where phase separation occurs. In the absence of stable nucleoporin complexes, the nucleus would be pore-free with a continuous nuclear envelope.43 Ran, a small GTPase protein, drives the direction of cargo transport, depending on its bound nucleotide state. The nuclear regulator RCC1 functions as a guanine nucleotide exchange factor and maintains RanGTP levels by facilitating GDP-GTP exchange. In the cytoplasm, Ran GTPase activating protein (Ran-GAP) and Ran binding protein 1 (RanBP1) catalyse the hydrolysis of GTP to GDP. Cooperation of these two systems forms a Ran gradient, which guides directionality of nucleopcytoplasmic transport (Fig. 2) and facilitates the rapid and selective cargo transport into and out of the nucleus.26
Karyopherins are central to nucleocytoplasmic trafficking as they recognize the nuclear localization signal or nuclear export signal (NES) of cargo proteins.31 In the ‘classical’ nuclear import pathway,46 cargo proteins such as TDP-43 contain a ‘classical’ NLS, which KPNAs bind to mediate nuclear import. Classical NLSs are either monopartite or bipartite sequences. Whilst monopartite sequences contain a single cluster of basic amino acids (Lys/Arg) as exemplified by the SV40 large T antigen NLS 126-PKKKRRV-132,47 bipartite sequences are made up of two clusters of basic residues separated by an unconserved linker region (e.g. nucleoplasmin NLS 155-KRPAATKKAGQAKKKK-170).48
During classical nuclear import (Fig. 2), the cargo-KPNA bipartite complex binds to KPNB1 creating a trimeric complex (cargo-KPNA-KPNB1) that interacts with the FG motifs of the nucleoporins in the central channel, and subsequently, driven by the RanGTP/GDP gradient, this complex translocates to the nucleus.42,49 In the nucleus, the trimeric complex starts to dissociate once RanGTP binds to KPNB1, with the physical separation of KPNB1 and KPNA. Subsequent dissociation of the KPNA-cargo complex is slow and can be catalysed by CSE1L/CAS that also mediates export of KPNA. The KPNA, RanGTP and CSE1L/CAS proteins are then recycled back to the cytoplasm (Fig. 2). In addition to the classical NLS-dependent pathway, other import pathways involve non-classical NLSs, which unlike classical NLSs, bind directly to KPNBs without the need for interaction with KPNA.34 One such example is the proline-tyrosine NLS (PY-NLS) motif of FUS, which is directly imported via TNPO1 and 2 (KPNB2 and KPNB2B).7,50,51 Although FUS and TDP-43 have putative nuclear export signal sequences, it should be noted that TDP-43 and FUS could also be exported through the nucleus via passive diffusion.52,53 Bioinformatics analysis indicates that the nuclear export signals in TDP-43 and FUS are non-functional and both proteins can be exported independent of the export receptors XPO1 and XPO5.52
Karyopherin abnormalities in neurodegenerative diseases
Karyopherin abnormalities have been implicated in ALS and FTD with TDP-43 pathology,15–20,54 in Alzheimer’s disease and FTD with tau pathology,55–60 in synucleinopathies,61–66 and in cell and animal models of Huntington’s disease,67–70 suggesting karyopherin-associated pathology as a common denominator in the aetiology of neurodegenerative proteinopathies. In the following sections we review how karyopherin abnormalities manifest in these neurodegenerative diseases, which is summarized in Table 3.
Table 3.
Karyopherin abnormalities in neurodegenerative proteinopathies
| Karyopherin | Pathogenesis | References |
|---|---|---|
|
ALS/FTD-TDP | ||
| KPNA2 | Reduced levels in FTD-TDP frontal cortex; accumulating TDP-43 causes cytoplasmic mislocalization in a Drosophila model of C9ALS/FTD. | Chou et al.,15 Solomon et al.,16 Nishimura et al.54 |
| KPNA4 | Accumulating TDP-43 causes cytoplasmic mislocalization in a Drosophila model of C9ALS/FTD; depletion of KPNA directly contributes to impaired nuclear import and cytoplasmic accumulation of TDP-43. | Chou et al.,15 Solomon et al.,16 Park et al.103 |
| XPO1 | In ALS models, XPO1 inhibition showed neuroprotective effects against C9orf72-related disease. | Zhang et al.21 |
| CAS | Reduced levels in FTD-TDP frontal cortex; knockdown of CAS dysregulates the import of TDP-43. | Nishimura et al.54 |
| KPNB1 | Reduced levels in spinal cords of patients with ALS; irregular and disrupted nuclear staining in sporadic ALS with TDP-43; depletion of KPNB1 directly contributes to impaired nuclear import and cytoplasmic accumulation of TDP-43; ALS-related mutations in FUS reduce its sensitivity to the chaperone activity of KPNB1, ultimately leading to increased phase separation. | Solomon et al.,16 Hofweber et al.,17 Yamashita et al.,78 Aizawa et al.,80 Park et al.103 |
| TNPO 1 | ALS related mutations in FUS reduce its sensitivity to the chaperone activity of TNPO1, ultimately leading to increased phase separation. | Hofweber et al.17 |
|
| ||
|
Alzheimer’s disease/FTD-Tau | ||
| KPNA2 | Accumulation and aggregation of KPNA2 is found in neurofibrillary tangles and Hirano bodies of hippocampal CA1 neurons of Alzheimer’s disease patients. | Lee et al.,56 Carter57 |
| KPNA3 | Abnormally upregulated levels of KPNA3 found in cDNA microarray studies of Alzheimer’s disease human brains. | Wang et al.55 |
| KPNA6 | Upregulated KPNA6 identified in association with small non-coding RNAs. | Roy et al.58 |
| KPNB1 | Found within cytoplasmic granules in hippocampal neurons in Alzheimer’s disease cases and co-localizes with hyperphosphorylated tau. | Nuovo et al.,59 Sheffield and Mirra107 |
| KPNB2 | Found in cytoplasmic granules in hippocampal neurons and in tangle-bearing cells of Alzheimer’s disease cases. | Sheffield and Mirra107 |
| XPO1 | Tau‐induced nuclear envelope invaginations sequester XPO1 in a Drosophila model of tauopathy. | Cornelison et al.108 |
|
| ||
|
Synucleinopathies | ||
| KPNA2 | Targeted knockdown linked to nuclear aggregation of α-syn; substrate of LRRK2/PARK8. | Ma et al.,62 Han et al.64 |
| KPNA3 | α-Syn mediated cytotoxicity involves interaction with KPNA3 | Büttner et al.137 |
| KPNA6 | Substrate of LRRK2/PARK8 | Han et al.64 |
| KPNA7 | Lewy body formation triggers alterations in the expression level of KPNA7. | Ma et al.62 |
| XPO1 | FBXO7/PARK15 was found mislocalized together with α-syn in Lewy bodies, Lewy neurites and cytoplasmic inclusions in glial cells in both Parkinson’s disease and MSA cases; Lewy body formation triggers the alterations in the expression level. | Ma et al.,62 Zhao et al.142 |
| KPNB1 | Alterations in the expression level of Parkinson’s disease patients with a triplication in the SNCA locus encoding α-syn. | George et al.,65 Devine et al.135 |
| KPNB2 | Mutant DJ-1/PARK7 was shown to interact with KPNB2 in an oxidative stress-dependent manner leading to its mislocalization. | Björkblom et al.63 |
| KPNB3 | α-Syn-mediated cytotoxicity lead to upregulation of KPNB3. | Zhou et al.61 |
| KPNA1 | Target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Langfelder et al.159 |
|
| ||
|
Huntington’s disease | ||
| KPNA2 | mHTT-transfected mouse neurons cause aggregation of mHTT and KPNA2/KPNA4. Some KPNA2/4 aggregates were associated with mHTT aggregates; target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Woerner et al.,70 Langfelder et al.159 |
| KPNA3 | Target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Langfelder et al.159 |
| KPNA4 | mHTT-transfected mouse neurons cause aggregation of mHTT and KPNA2/KPNA4. Some KPNA2/4 aggregates were associated with mHTT aggregates; target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Woerner et al.,70 Langfelder et al.159 |
| KPNA6 | Binding partner of mHTT in mouse brain expressing mHTT expanded with 97 polyQ; target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Shirasaki et al.,158 Langfelder et al.159 |
| KPNB1 | Binding partner of mHTT in mouse brain expressing mHTT expanded with 97 polyQ; Target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Shirasaki et al.,158 Langfelder et al.159 |
| KPNB2 | Target of mHTT in genomics and proteomics study of transgenic mice expressing mHTT. | Langfelder et al.159 |
| XPO7 | Binding partner of mHTT in mouse brain expressing mHTT expanded with 97 polyQ. | Shirasaki et al.158 |
| CAS | Binding partner of mHTT in mouse brain expressing mHTT expanded with 97 polyQ. | Shirasaki et al.158 |
MSA = multiple system atrophy.
Karyopherin abnormalities in amyotrophic lateral sclerosis and frontotemporal dementia with TDP-43 pathology
ALS and FTD are part of a common disease spectrum with overlapping clinical, pathological and genetic features.71,72 ALS is a debilitating and ultimately fatal disease, characterized by the progressive loss of upper and lower motor neurons, which inevitably leads to degeneration of corticospinal tracts and muscle wasting.73,74 In contrast, FTD is characterized by progressive neuronal atrophy, which occurs in the frontal and temporal lobes with the resulting loss of cortical tissue leading to personality and behavioural changes, and/or a gradual impairment of language skills.72,75 A common pathological feature found in both diseases are abnormally aggregated cytoplasmic inclusions of TDP-4376,77 that characterize 97% of ALS cases and ∼45% of patients with FTD,71 which are therefore termed FTD-TDP in contrast to FTD cases with tau pathology termed FTD-Tau.
Reduced protein levels of KPNA2 and CAS are observed in FTD-TDP frontal cortex54 and reduced KPNB1 levels are found in the spinal cord of patients with ALS.78 Immunohistochemical analysis of nucleoporins in ALS revealed an irregular, fractured nuclear pore complex,79 possibly preventing the nuclear import of TDP-43. In addition, irregular, disrupted nuclear staining for NUP62 or KPNB1 has been reported in sporadic ALS with TDP-43 mislocalization, providing further evidence for disruption of the nuclear pore complex in TDP-43 pathologies.80 Moreover, cell culture experiments revealed that KPNB1 and CAS knockdown dysregulates the classical nuclear import of TDP-43, leading to its cytoplasmic accumulation comparable to disease pathology.54
Corresponding phenotypes were also observed in the most common genetic form of ALS and FTD caused by an intronic hexanucleotide repeat expansion (GGGGCC) in the C9orf72 gene, termed C9ALS/FTD.21,22,81–84 Several pathogenic mechanisms have been related to the C9ORF72 mutation, including haploinsufficiency85,86 and homozygous loss of C9orf72 function,87 as well as toxic gain-of-function of the repeat RNA,83,88–90 and the accumulation of dipeptide repeat proteins (DPRs) that are translated from G4C2 mRNA templates through repeat-associated non-ATG translation (RAN translation).89,91 However, the exact contribution of each of these mechanisms to onset and progression of disease in C9ALS/FTD is still inconclusive.92 Functional studies in C9ALS/FTD cell and animal models identified defective nucleocytoplasmic transport and nuclear pore complex deficits as potential pathogenic mechanisms underlying neurodegeneration.21–24,93,94 These findings are supported by rescue experiments in yeast and Drosophila models of C9ALS/FTD, which identified importins, exportins, Ran-GTP cycle regulators and nuclear pore components as modifiers of G4C2 RNA and DPR-mediated neurodegeneration.21,23,24 Moreover, a recent study, using a permeabilized cell assay, showed that arginine-rich DPRs bind KPNB1, ultimately leading to dose- and length-dependent disruption of nuclear import.95 Of note, this study also revealed that transport disruption was neither due to sequestration of nucleocytoplasmic transport components, nor to blockade of the FG nucleoporin-mediated permeability barrier, but rather due to perturbed KPNB-related cargo-binding.95
Comparable nucleocytoplasmic transport and nuclear pore complex deficits were also reported in models of TDP-43 proteinopathy.15,16 These studies provide experimental evidence that accumulating cytosolic TDP-43 causes cytoplasmic mislocalization of KPNAs in Drosophila models of C9ALS/FTD, a phenotype that is also observed in post-mortem frontal cortex tissue of C9FTD patients and sporadic cases of FTD-TDP.15,16 Cytosolic TDP-43 functions in the stabilization, translation and transport of mRNA.96 However, imbalanced cytoplasmic accumulation of TDP-43 can induce its nuclear depletion and aggregate formation,97 cellular phenotypes that are both related to onset and progression of disease.98 Nuclear depletion of TDP-43, through decreased expression or aggregate formation, has also been shown to lead to reduced levels of RanBP1, suggesting that TDP-43 mislocalization may be both cause and consequence of errors in nucleocytoplasmic transport.98 However, the initiating causes leading to cytoplasmic accumulation of TDP-43 remain elusive.
Interaction studies have identified direct protein-protein interactions between TDP-43 and KPNA254 and KPNA4,15 in addition to TDP-43 targeting the pre-mRNA of KPNA4 and, to a lesser extent, that of KPNA2.99 Furthermore, cytoplasmic TDP-43 droplets that are not associated with stress granules have been shown to sequester KPNA4 and NUP62 and induce mislocalization of RanGap1, Ran, and NUP107.20 These findings support the feedback loop model proposed by Solomon et al.,16 whereby cytoplasmic, non-aggregated droplets of TDP-43 recruit karyopherins, which in turn causes nucleocytoplasmic transport deficits. Of note, KPNAs also bind arginine-rich DPRs,93 the most toxic DPR species observed in models of C9ALS/FTD,100,101 which in turn impairs KPNA/KPNB-mediated nuclear import, including cargo transport of TDP-43.102 Moreover, cell culture studies revealed that DPRs can be sequestered into cytoplasmic inclusions by β-sheet protein aggregates, including fragments of TDP-43.70 Furthermore, experiments in cell culture54 and in Drosophila103 revealed the depletion of KPNB1 and KPNAs directly contributes to impaired nuclear import and cytoplasmic accumulation of TDP-43. These data suggest a vicious cycle of TDP-43 and karyopherin abnormalities16 and in turn impaired nucleocytoplasmic transport and a defective nuclear pore complex, that trigger onset and progression of disease in ALS and FTD-TDP.
Karyopherin abnormalities in Alzheimer’s disease and frontotemporal dementia with tau pathology
Alzheimer’s disease is the most common cause of dementia, accounting for up to 75% of all dementia cases.104 Patients suffer from insidiously progressive disturbances in memory and other cognitive capabilities such as dysfunction in language, visuospatial deficits and impaired executive function.105 Alzheimer’s disease-related proteinopathy is characterized by neurofibrillary tangles enriched in hyperphosphorylated tau, and amyloid plaques comprising amyloid-β fragments of aberrantly cleaved amyloid precursor protein.4,106 Karyopherin abnormalities in Alzheimer’s disease have been repeatedly reported, yet how they contribute to disease formation and/or progression is only starting to emerge.
Accumulated and aggregated KPNA2/Importin-α1 has been identified in neurofibrillary tangles57 and in Hirano bodies in hippocampal CA1 neurons of Alzheimer’s disease patient cases.56 cDNA microarray studies using Alzheimer’s disease patient brain samples identified abnormally upregulated levels of KPNA3,55 while KPNA6 upregulation was identified in association with small non-coding RNAs in Alzheimer’s disease-affected brain.58 In addition to KPNA abnormalities, several members of the KPNB family have been found to be mislocalized, accumulated or aggregated in Alzheimer’s disease patient brain. KPNB1 and KPNB2 were found in cytoplasmic granules in hippocampal neurons, while KPNB2 accumulation was also detected in tangle-bearing cells of Alzheimer’s disease cases.107 Expression of KPNB1 and XPO5/exportin-5 were both found to be upregulated in Alzheimer’s disease cases and both proteins co-localized with hyperphosphorylated tau.59 These data suggest that karyopherin abnormalities may play a direct role in tau pathology related to Alzheimer’s disease and possibly also FTD-Tau.
In support of this hypothesis, experiments using a Drosophila model of tauopathy revealed that tau-induced nuclear envelope invaginations sequestered the KPNB family member XPO1/exportin-1.108 Another study using Alzheimer’s disease patient brain material together with transgenic mouse models of FTD-Tau identified a direct link between tau pathology and nucleocytoplasmic transport deficits.60 Using co-immunoprecipitation from human Alzheimer’s disease brain tissue, these studies demonstrated direct binding between tau and NUP98 and that tau interacts with nuclear pore proteins enriched in phenylalanine-glycine (FG) repeat domains. In addition, these FG-motif nucleoporins were shown to mislocalize from and in turn disrupt the nuclear membrane in primary cortical neurons derived from transgenic mouse models of FTD-Tau.60 As a likely consequence of these nuclear pore complex deficits, alterations in active protein import and export were observed including decreased expression of Ran required for energy-dependent nucleocytoplasmic shuttling.60 This reduction in Ran expression and reduced nucleocytoplasmic transport has also been observed in an earlier study using Alzheimer’s disease brain samples and neuroblastoma cells,109 suggesting that nucleocytoplasmic transport deficits may directly contribute to tau pathology and disease progression. This is further supported by in vitro experiments, which showed that NUP98 triggers tau aggregation and accelerates tau fibrilization seen in Alzheimer’s disease and tauopathy brains.60
Despite these functional insights into tau phenotypes and nucleocytoplasmic transport deficits, it remains to be established whether the karyopherin abnormalities observed in Alzheimer’s disease patient brain55–59,107 are a cause or a consequence of tau pathology. A possible pathogenic mechanism might be related to the functional link between nuclear pore proteins enriched in FG-motifs like NUP98 and karyopherins of the beta family. These FG-motif nucleoporins and KPNBs are known to interact with each other at the nuclear membrane110,111 and their functional interactions are essential to allow repeated Ran-dependent cycles of nucleocytoplasmic shuttling.112 These data suggest that alterations in levels or localization of FG nucleoporins, Ran and/or KPNBs could trigger tau pathology—and vice versa—and lead to the cellular and molecular phenotypes already observed in transgenic mouse models of FTD-Tau.60
Karyopherin abnormalities in synucleinopathies
Synucleinopathies are a group of neurodegenerative disorders that include Parkinson’s disease, Parkinson’s disease with dementia (PDD), DLB and multiple system atrophy (MSA).113,114 Their common pathological feature is the accumulation and aggregation of α-syn in Lewy bodies found in neurons and glial cells, and in Lewy neurites within neuronal processes.66,115–117 Native α-syn is found in presynaptic terminals of neurons where it acts as a chaperone for neurotransmitter release.118 Its initial discovery in Torpedo identified α-syn not only in synaptic terminals but also in neuronal nuclei,119 suggesting a physiological function in both synapse and the nucleus. Indeed, recent experiments using transgenic mice and human cell culture revealed translocation of α-syn to the nucleus where it modulates DNA repair,120 raising the question how α-syn can shuttle into the nucleus.
α-Syn does not possess a nuclear localization signal and due to its small size of 14 kDa,121 may enter the nucleus via passive diffusion. Recent findings, however, identified three intermediates that interact with α-syn and together translocate into the nucleus: by direct binding of α-syn to the retinoic acid receptor and its calcium-dependent nuclear import122; by sumoylation of α-syn and its subsequent binding to KPNA6, which mediates transport into the nucleus123; and by interaction with TRIM28124 and its karyopherin-alpha mediated nuclear import.125 Dysfunction of either pathway has been directly implicated in α-syn pathology in transgenic mice and in cell culture experiments expressing human mutant forms of α-syn, as well as in post-mortem brain tissue of DLB cases.120,122,126 These findings could explain the pathological phenotypes observed in synucleinopathies in which α-syn has been detected in aberrant forms in the nucleus120,127,128 and in presynaptic terminals129–132 where it can spread across synaptically connected brain regions.133,134
Consistent with these findings, karyopherin abnormalities have been associated with synucleinopathies. Alterations in KPNB1 expression were identified using microarray analysis of tissue material derived from Parkinson’s disease patients with a triplication in the α-syn encoding SNCA gene.65,135 Lewy body formation triggered by prefibrillary α-syn was found to alter the expression levels of KPNA7 and XPO1/exportin-1.66 The Parkinson’s disease-related α-syn mutation G51D was shown to cause nuclear aggregation,136 while nuclear aggregation of α-syn was shown to be mitigated by targeted knockdown of KPNA2.62 Cell culture studies revealed that α-syn-mediated cytotoxicity caused upregulation of KPNB361 and involved interaction with KPNA3.137 In addition to these phenotypes directly related to α-syn accumulation, several other studies reported karyopherin abnormalities in cell and animal models of familial forms of Parkinson’s disease that are characterized by α-syn pathology. KPNA2 and KPNA6 have been identified as substrates of LRRK2/PARK864 whose mutations cause one of the most common forms of Parkinson’s disease.138,139 Mutant DJ-1/PARK7140 was shown to interact with KPNB2 in an oxidative stress-dependent manner leading to its mislocalization.63 FBXO7/PARK15, which requires XPO1 for its normal localization141 was found mislocalized together with α-syn in Lewy bodies, Lewy neurites and cytoplasmic inclusions in glial cells in both Parkinson’s disease and MSA cases.142
Despite this multitude of phenotypes, it should be noted that unlike in ALS, FTD and Alzheimer’s disease cases, conspicuous karyopherin pathology has not been reported in any of the synucleinopathies to date. It therefore seems likely that the detected phenotypes described above are a consequence rather than a cause contributing to the cellular and molecular mechanisms underlying synucleinopathies. However, this situation may change with future work specifically focusing on karyopherin abnormalities and their potential role in the onset and progression of α-syn pathology.
Karyopherin abnormalities in cell and animal models of Huntington’s disease
Huntington’s disease is an autosomal dominant disorder characterized by neurodegeneration and gliosis in the striatum and cortex,143 leading to progressive loss of motor function, psychiatric symptoms, and cognitive impairment.144 Huntington’s disease is caused by a CAG triplet repeat expansion encoding for a polyglutamine (polyQ) tract in the N-terminus of huntingtin (HTT).145 Expansions of >36–39 glutamine residues trigger disease, and longer polyQ stretches above this threshold cause early-onset Huntington’s disease.146,147 HTT is a large (∼350 kDa) protein that shuttles between the cytoplasm and nucleus148; it contains a XPO1/CRM1-dependent nuclear export signal at its carboxyl-terminus149,150 and its nuclear import occurs via its PY-NLS by the TNPO/KPNB2 pathway.68,151 In mHTT, the polyQ domain is flanked by the first 17 amino acids at the N-terminus (Nt17) and by a poly-P stretch at its C-terminus. Mutant HTT is more prone to proteolysis, resulting in smaller fragments of the N-terminus containing the polyQ expansion of exon 1.152 Nt17 plays an important role in HTT aggregation and toxicity, in addition to acting as another nuclear export signal via interactions with XPO1/CRM1 in an α-helical and RanGTP-dependent manner.149
The subcellular localization and actual function of wild-type HTT is still debated153,154; however, there is consensus that polyQ expanded mHTT causes gain-of-function proteinopathy in both the nucleus and the cytoplasm.144 Consistent with this hypothesis, both intranuclear and cytoplasmic aggregates have been observed in Huntington’s disease patient brains,155,156 while cell culture experiments using human embryonic kidney 293 T cells, neuroblastoma cells and mouse primary neurons transfected with fragments of mHTT revealed its nuclear and cytoplasmic aggregation.70 Moreover, in the striatum and cortex of Huntington’s disease mouse models, mHTT formed both nuclear and cytoplasmic aggregates, which resulted in the accumulation of mRNA in the nucleus and an overall reduction in mRNA levels.70,157 These cellular and molecular phenotypes may point towards a role of nucleocytoplasmic transport deficits in Huntington’s disease.
Indeed, polyQ and mHTT pathology has been related to nuclear pore complex deficits and defective nucleocytoplasmic transport, including karyopherin abnormalities.67,69,150,158–161 Transfection of mouse primary neurons with mHTT expanded with 96 polyQ fragments not only caused nuclear and cytoplasmic aggregation of mHTT; it also caused aggregation of KPNA2 and KPNA4.70 Some of these cytoplasmic KPNA2/4 aggregates were found associated with mHTT aggregates, suggesting that Huntington’s disease-related polyQ pathology directly causes karyopherin abnormalities. Consistent with this hypothesis, affinity-purification mass spectrometry using mouse brain tissue expressing mHTT expanded with 97 polyQ, identified the karyopherins KPNA6, XPO7, CAS and KPNB1 as binding partners of mHTT.158 Several of these karyopherins were independently confirmed as targets of mHTT in a genomics and proteomics study using transgenic mice expressing human mHTT exon 1 carrying disease-related CAG repeats.159 These experiments identified altered expression levels of the KPNA proteins KPNA1, KPNA2 and KPNA3 together with KPNA4 and KPNA6, as well as altered expression levels of KPNB1 and KPNB2.159 Despite this experimental evidence in cell and animal models of Huntington’s disease, conspicuous karyopherin pathology has not been reported to date in brains of patient cases.
Interestingly, however, it has been shown that Huntington’s disease-related CAG repeat expansion RNAs can also function as a template for repeat-associated non-AT (RAN) translation. The resulting homo-polymeric RAN translation products include poly-Ala, poly-Ser, poly-Leu, and poly-Cys peptides that have been detected in brains of Huntington’s disease patients, especially in regions of microglial activation and degenerative apoptotic cell loss, as well as throughout the degenerating cerebellar layers of juvenile-onset Huntington’s disease cases.162 These data indicate that the aetiology of Huntington’s disease may share similarities with C9ALS/FTD, both in its RAN translation-derived peptide accumulation161,163 and in nuclear transport deficits.70,161 However, further work is required to establish whether karyopherin pathology characterizes patient brain cases and whether these phenotypes are mere consequences of, or directly contribute to mHTT, polyQ and RAN-derived peptide pathology that have been detected in cell and animal models of Huntington’s disease.
Phase transition and protein aggregation of karyopherin cargos
Despite their functional heterogeneity and disease-specificity, TDP-43, tau, α-syn and FUS, as well as mutant huntingtin with polyQ, all harbour low complexity regions rendering them intrinsically disordered proteins.9,164–167 Low complexity regions are devoid of charged amino acids but contain polar ones (Q, N, S, G) with interspersed aromatic residues (F, Y), making these chains highly flexible and interactive.168 Because of their similarity to infectious yeast proteins, low complexity regions are also called prion-like domains.5 Recent functional studies provide compelling evidence that prion-like low complexity regions make intrinsically disordered proteins vulnerable to changes in the surrounding milieu, which can alter their physical properties, eventually leading to their accumulation in pathological aggregates.13 These changes in aggregate state are similar to phase separations of mixed solutions, ranging from liquid-liquid demixing into liquid droplets, to phase transitions into gel-like structures (hydrogels) that over time, can mature into solid fibrillary aggregates such as amyloid-like fibres.169,170 These phase transitions, especially the initiating phase separation of soluble proteins with low complexity regions into droplets and hydrogels, tend to accumulate in membraneless compartments such as P granules, paraspeckles, nucleoli and stress granules and are very sensitive to changes in temperature, salt, pH or molecular concentration.169,170 These environmental alterations characterize protein accumulation and cellular stress observed in neurodegenerative diseases.12–14
Recent studies showed that tau,166,171–174 mHTT-polyQ,165 TDP-43,9,175–178 FUS7,17–19,179,180 and α-syn167,181 can undergo liquid-liquid phase separation. In the case of polyQ pathology, it was shown that huntingtin exon 1 aggregates are present in liquid-like as well as solid-like forms.165 The polyQ and proline-rich regions of exon 1 facilitate liquid-like formation, which can convert into solid-like assemblies when polyQ length expands beyond the threshold for Huntington’s disease.165 Similar to polyQ-related phase separation, recent studies showed that phosphorylated tau, recombinant mutant aggregate prone tau, and soluble phospho-tau isolated from human Alzheimer’s disease brain can all undergo phase separation.166,171–174 These findings are consistent with previous studies, in which TIA1, a major component of stress granules, promoted tau misfolding and aggregation by sequestering tau into stress granules, whilst TIA1 knockout inhibited tau mediated pathology and toxicity.182
Karyopherin-mediated phase transition in neurodegeneration
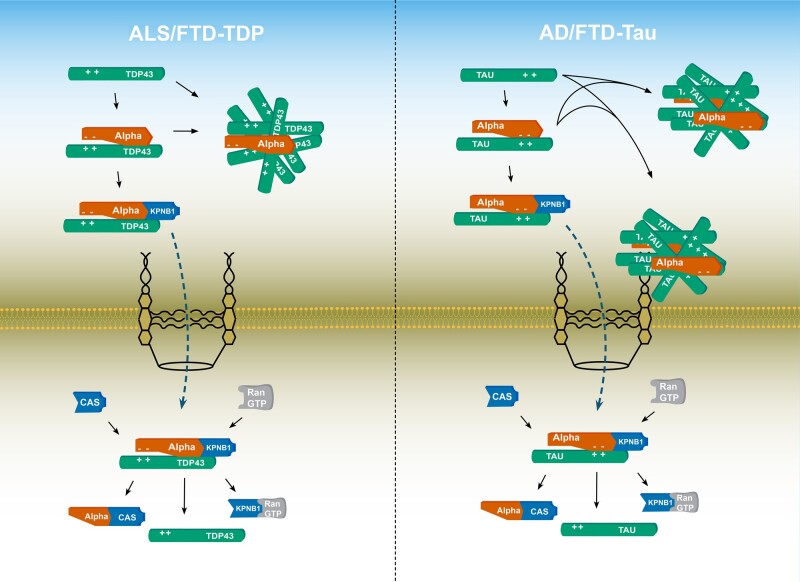
Previously known for their role in nucleocytoplasmic transport, karyopherins are now also recognized for their important functions as chaperones and molecular switches in the pathogenesis of neurodegenerative proteinopathies (Fig. 3).13,14,183 In the case of TDP-43 and FUS, recent studies established that this process is mediated by karyopherins acting as molecular chaperones in phase separation.17–20 These data suggest that karyopherin abnormalities are not only a common denominator in neurodegenerative proteinopathies but misregulation of karyopherins can also trigger onset and progression of disease. In line with this notion, in vitro studies demonstrated that TDP-43, which assembled into protein-rich droplets via liquid-liquid phase separation, could fibrillate over time.8,94 Liquid-liquid phase separation of TDP-43 is crucially dependent on the C-terminal low complexity region,175 especially its α-helical structure9,178,184 and three tryptophan residues within it, W334, W385 and W412.185 Using hydrogels, Guo and colleagues19 showed that KPNA and KPNB1 interact with the canonical nuclear localization signal of TDP-43 (Fig. 3), the binding of which reverses and prevents its aberrant phase transition.19,102 ALS-related mutations in the low complexity region of TDP-43 accelerate the transition of liquid-like droplets to pathologically fibrillized states,8,186 whereas overexpression of KPNA and KPNB1 resulted in reversal and prevention of TDP-43 fibrillization.19 Moreover, studies in sporadic and familial forms of ALS and FTD revealed that disease-initiating cytoplasmic accumulation of TDP-43 is associated with a vicious cycle of KPNA dysfunction that is accompanied by depletion of KPNB1 and a dysfunctional nuclear pore complex.15,16,20,93
Figure 3.
Karyopherins abnormalities in neurodegenerative proteinopathies. Left: In ALS/FTD-TDP, cytoplasmic accumulation of TDP-43 and its aggregates not associated with stress granules have been shown to sequester KPNAs. Right: In Alzheimer’s disease and FTD-Tau, pathological tau has been shown to sequester KPNAs (Table 3). AD = Alzheimer’s disease; CAS = cellular apoptosis susceptibility.
Comparable phenotypes have been observed in FUS-related ALS. TNPO1/KPNB2 was shown to inhibit liquid-liquid phase separation of FUS18 and to reverse and prevent the fibrillization of FUS and other RNA binding proteins by interacting with the PY-NLS domain.19 Using NMR spectroscopy and small angle X-ray scattering techniques, Yoshizawa et al.18 showed that a combination of strong and weak binding between TNPO1/KPNB2 and FUS inhibits phase separation. The interactions between TNPO1/KPNB2 and FUS competed with FUS-FUS interactions, thereby disrupting phase separation.18 Moreover, in vitro studies revealed that TNPO1/KPNB2 suppresses FUS phase separation and stress granule association by interacting with its arginine residues in its PY-NLS and RGG domains.17 These studies also showed that ALS-related mutations in FUS reduce its sensitivity to the chaperone activity of TNPO1/KPNB2 both in cells and in vitro, thereby leading to a model in which FUS mutations both enhance phase separation and elude the chaperone activity of TNPO1/KPNB2.17–19 Arginine residues appear to be critical to aberrant phase separation and karyopherin-mediated chaperoning as overexpression of arginine-containing DPRs derived from RAN translated, C9ALS/FTD-related G4C2 repeat RNA, can induce the assembly of stress granules, triggering the co-localization of Ran and karyopherins into these membraneless organelles.187 The resulting decrease in karyopherin availability impairs karyopherin-mediated cargo transport and chaperone function, thereby increasing cytoplasmic accumulation and phase transitions, which ultimately leads to disease-related fibrillization and aggregate formation.
Corresponding molecular and cellular phenotypes have been observed in Alzheimer’s disease, FTD-Tau and synucleinopathies. Liquid-liquid phase separation of tau protein was shown to initiate tau aggregation166,174,188,189 that was significantly enhanced by FTD-tau associated mutations including P301L-tau linked to inherited tauopathy.173 Tau positive neurofibrillary tangles and Hirano bodies were found enriched in KPNA2/Importin-α1 aggregates,56,57 while KPNB1 and XPO5 were found co-localized with hyperphosphorylated tau in affected neurons.59 α-Syn has been shown to undergo spontaneous liquid-liquid phase separation, which was accelerated by disease-related mutations A53T, E46K and S129E and formation of fibrils167 that could mature into Lewy body-like assemblies.181 Of note, Lewy body formation related to synucleinopathies has been associated with altered KPNA7 and XPO1/exportin-1 expression,66 while inhibition of KPNA2 impaired the pathological accumulation of α-syn in the nucleus.62 Similarly, triggered by its disease-related polyQ tracts, mHTT exon 1 was shown to form liquid-like assemblies that could convert to solid fibrillar structures.165
These findings suggest that karyopherin abnormalities are interlinked with alterations of aggregation-prone proteins that can phase separate into disease-related intracellular inclusions and aggregates. In line with these observations, work on nucleocytoplasmic transport receptors, including KPNB3, showed that karyopherins can act as molecular chaperones, shielding basic domains of positively charged proteins and preventing their aggregation.102,187 Together, these data indicate that both karyopherin abnormalities and affected karyopherin-cargo binding promote phase transition, which accelerates protein mislocalization and aggregate formation characteristic of neurodegenerative proteinopathies. As a result of these new findings, it is becoming increasingly evident that karyopherins can act as chaperones comparable to molecular disaggregases14,190,191 that protect aggregation-prone proteins against misfolding, accumulation and irreversible phase-transition into insoluble aggregates. It is therefore reasonable to consider karyopherins as therapeutic targets for the treatment of neurodegenerative diseases for which currently no cure or effective treatments are available.
Targeting karyopherin abnormalities in degenerative proteinopathies
Current treatments, such as riluzole for ALS192 or donepezil, rivastigmine, galantamine and memantine for Alzheimer’s disease,193 offer only short-term amelioration of clinical symptoms but do not arrest progressive cell loss. Similarly, Parkinson’s disease-related treatment with levodopa offers symptomatic improvements but it does not arrest synapse loss and subsequent neurodegeneration.118 However, there is time between when a patient is first diagnosed and when severe mental and physical decline occurs, indicating the existence of a potential therapeutic window during which surviving, unaffected neurons could be protected against their progressive dysfunction leading to further cognitive decline and degenerative cell loss. As biomarkers for early diagnosis and effective treatments become available, the development of therapies to halt progressive cell death becomes an increasingly hopeful prospect. Karyopherins have already been described as diagnostic and prognostic biomarkers in cancer and viral infections, where they are usually upregulated.194–196 As a result, several compounds have been developed to target nuclear import and export receptors197 and phase III clinical trials are under way targeting the KPNA/KPNB1 interface highjacked by viruses causing Dengue fever and COVID-19, respectively.196
In neurodegenerative diseases, karyopherins are generally downregulated and mislocalized, which together with impaired nucleocytoplasmic transport and defective chaperone function, leads to aberrant phase transition and the subsequent accumulation and aggregation of affected proteins.14,191 Recent studies demonstrate that targeting karyopherins can reverse aberrant phase transition and extract aggregated proteins from stress granules.17–20,102 Thus, targeting karyopherins, especially their chaperone function, provides a potential novel therapeutic avenue to shield and maintain vulnerable proteins in their native and soluble state, thereby protecting them against phase transition, mislocalization and aberrant aggregate formation, ultimately protecting affected neurons against cytotoxicity and degenerative loss.
The role of compounds inhibiting nuclear transport of specific cargo substrates has been reported for their use as anticancer and antiviral agents, with more than 20 nuclear transport inhibitors being identified,196–198 including mifepristone, ivermectin and verdinexor.199–202 Inhibitors of XPO1/CRM1 like verdinexor, commonly also known as selective inhibitors of nuclear export (SINE) compounds, have been developed via structure-based drug design.198 Previous studies investigating SINE compounds focused on their use as anticancer and antiviral agents197; however, they are also candidates for the development of therapeutic strategies to target neurodegeneration.203 For example, XPO1 inhibition in preclinical models of inflammatory demyelination and axonal damage resulted in attenuation of disease progression.203 Comparably, in ALS models, XPO1 inhibition showed neuroprotective effects against C9orf72-related disease phenotypes caused by G4C2 hexanucleotide repeat expansion.21 Targeted manipulation of the nuclear import receptor karyopherin-β2 was able to reverse the defects caused by mutant FUS.19,204 Pharmacological treatment with verdinexor and KPT-276, two XPO1 inhibitor, reduced cell death by >50% caused by selective overexpression of C-terminal fragments of human TDP-43 or its ALS-related mutant Q331K in mouse cortical neurons.15 A parallel study showed that SINE compounds targeting XPO1 could prolong, at least to some extent, neuronal survival of rodent primary neurons and mitigate motor symptoms in an in vivo rat model expressing wild-type and mutant forms of TDP-43.205 This study revealed that none of the SINE compounds could enhance nuclear TDP-43 levels, while depletion of XPO1 or other exportins had little effect on TDP-43 localization,205 consistent with recent studies suggesting that nuclear egress of TDP-43 does not require XPO1-mediated nuclear export.52,53
These findings suggest that pharmacological targeting of karyopherin function might represent a new strategy for disease-modifying treatments of neurodegenerative proteinopathies. However, given the different topology of disease-related protein accumulation—nuclear and/or cytoplasmic—a more balanced application will be required that considers not only nuclear transport inhibitors but also their targeted gain-of-function, which will require careful assessment and calibration. Karyopherins modulate an essential cellular process, energy-dependent nucleocytoplasmic transport of cargo proteins required for cellular homeostasis. Any interference or manipulation of this process may therefore cause unwanted side effects affecting the localization and function of cargo proteins essential for neuronal and glial function. It will be of great interest to await and analyse the outcome of currently ongoing clinical trials targeting the KPNA/KPNB1 interface as a strategy to combat several types of cancer such as leukaemia and glioblastoma, as well as viral infections including Dengue fever and COVID-19.196,197,206 If proven to be clinically safe, some of these compounds might be suitable for repurposing as a novel therapeutic strategy for the targeted treatment of neurodegenerative proteinopathies.
Conclusions and future directions
Nucleocytoplasmic transport is a fundamental process that moves macromolecules across the nuclear membrane, thereby allocating proteins to their principal destination of activity, which is an essential requirement for proper cell function and viability. Key players in this process are karyopherins that mediate the transport of cargo proteins in and out of the nucleus. The mislocalization of proteins and their subsequent accumulation in the nucleus or cytoplasm are characteristic features, and likely the direct cause of several neurodegenerative proteinopathies. Why these proteins mislocalize and accumulate in the first place, has remained elusive and the lack of insights into the underlying pathogenic mechanisms constitutes a major obstacle for the development of efficient therapeutic strategies to go beyond current symptomatic treatments.
Despite their differences in function, ranging from nucleic acid processing by TDP-43 to tau-mediated microtubule stabilization and chaperone activity of α-synuclein in synaptic terminals, all of these proteins harbour low complexity regions that make them intrinsically vulnerable to phase transitions of their aggregation state. Phase transitions can be initiated by changes in cellular acidity, temperature and alterations in pH or salt but also by protein mutations, all of which have been associated with neurodegeneration. However, in vivo determination of these pathomechanisms, their detection in patient tissue and a direct role in disease formation and/or progression remain to be shown, and although first generation preclinical biomarkers of liquid-liquid phase separation are becoming available, their use and efficiency remains to be established.207
Nucleocytoplasmic transport deficits and in particular karyopherin abnormalities are now recognized as a major player, and possibly a trigger, in mediating the mislocalization and accumulation of disease-related proteins in degenerative proteinopathies, especially in ALS and FTD. Furthermore, karyopherins can also act as chaperones by shielding proteins from irreversible phase transitions, thereby retaining their targets in a soluble form and thus devoid of disease-related aggregation comparable to disaggregase activity. It is currently unclear and remains to be elucidated, how these chaperone functions are exerted, whether they include specific amino acids/protein interaction domains and how these interactions are accomplished. Thus, an exciting area of future research will be to determine how karyopherins exert their chaperone and disaggregase activity. Work in this area will lead to both a better understanding of how neurodegenerative diseases develop and may uncover novel treatment opportunities.
Pharmacological targeting of karyopherins represents a promising new strategy for therapeutic intervention in neurodegenerative diseases. It remains a major task for an ever-growing elderly population to identify therapies that can stop or at least ameliorate degenerative cell loss. Even after decades of intense research, the destiny of a patient remains the same: when they enter the hospital with disease-specific clinical symptoms, a significant number of neurons are already lost, while remaining functional ones are doomed to degenerate over time due to the lack of efficient treatments. Several compounds targeting karyopherins are already in clinical trials to treat specific forms of cancer and viral infections, with an emphasis on the karyopherin-α/karyopherin-β1 interface. A number of neurodegenerative diseases are characterized by intracellular proteinaceous inclusions and aggregates for which nucleocytoplasmic transport-modifying compounds may yield promising outcomes for an effective treatment of these devastating illnesses. Targeting nuclear transport may therefore be an advantageous strategy for halting and perhaps reversing the progression of neurodegenerative proteinopathies.
Acknowledgements
We thank D. Solomon, R.B. Parsons and M. Atwal for comments on the manuscript.
Funding
This work was supported by a Bolashak International scholarship (Sharipov/Aug28/2019) of the government of Kazakhstan to D.S.; Slovenian Research Agency grants (P4-0127, J3-9263, J3-8201, J7-9399 and N3-0141) and CRP-ICGEB research grant (CRP/SVN19-03) to B.R.; the Hungarian Brain Research Program (2017-1.2.1-NKP-2017-00002) and NKFIH-SNN-132999 grant to T.H.; and an Alzheimer's Research UK—King's College London Network grant to F.H. and T.H. F.H. acknowledges funding from the UK Medical Research Council (G0701498; MR/L010666/1); the UK Biotechnology and Biological Sciences Research Council (BB/N001230/1); Alzheimer's Research UK (Hirth/ARUK/2012); and the UK MND Association (Hirth/Oct07/6233; Hirth/Mar12/6085; Hirth/Oct13/6202; Hirth/Nov15/914-793; Hirth/Oct16/890-792).
Competing interests
The authors report no competing interests.
Glossary
- ALS
amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
- KPNA/B
karyopherin-α/β
- mHTT
mutant huntingtin
- NLS
nuclear localization signal
- TDP-43
TAR DNA binding protein 43
References
- 1.World Health Organization. Global action plan on the public health response to dementia 2017–2025. World Health Organization; 2017. Accessed 30 September 2021. https://www.who.int/publications/i/item/global-action-plan-on-the-public-health-response-to-dementia-2017-2025 [Google Scholar]
- 2. Moreno-Gonzalez I, Soto C.. Misfolded protein aggregates: Mechanisms, structures and potential for disease transmission. Semin Cell Dev Biol. 2011;22(5):482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braak H, Tredici K, Del, Rüb U, de Vos RA, Jansen Steur EN, Braak E.. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24(2):197–211. [DOI] [PubMed] [Google Scholar]
- 4. Jucker M, Walker LC.. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alberti S, Halfmann R, King O, Kapila A, Lindquist S.. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han TW, Kato M, Xie S, et al. Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell. 2012;149(4):768–779. [DOI] [PubMed] [Google Scholar]
- 7. Patel A, Lee HO, Jawerth L, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–1077. [DOI] [PubMed] [Google Scholar]
- 8. Molliex A, Temirov J, Lee J, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conicella AE, Zerze GH, Mittal J, Fawzi NL.. ALS mutations disrupt phase separation mediated by-helical structure in the TDP-43 low complexity C-terminal domain. Structure. 2016;24(9):1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monahan Z, Ryan VH, Janke AM, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36(20):2951–2967. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murray T, Kato M, Lin Y, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171(3):615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aguzzi A, Altmeyer M.. Phase separation: Linking cellular compartmentalization to disease. Trends Cell Biol. 2016;26(7):547–558. [DOI] [PubMed] [Google Scholar]
- 13. Boeynaems S, Alberti S, Fawzi NL, et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018;28(6):420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo L, Fare CM, Shorter J.. Therapeutic dissolution of aberrant phases by nuclear-import receptors. Trends Cell Biol. 2019;29(4):308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chou CC, Zhang Y, Umoh ME, et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci. 2018;21(2):228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Solomon DA, Stepto A, Au WH, et al. feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-a mediates C9orf72-related neurodegeneration. Brain. 2018;141(10):2908–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hofweber M, Hutten S, Bourgeois B, et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173(3):706–713. [DOI] [PubMed] [Google Scholar]
- 18. Yoshizawa T, Ali R, Jiou J, et al. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell. 2018;173(3):693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo L, Kim HJ, Wang H, et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173(3):677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gasset-Rosa F, Lu S, Yu H, et al. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 2019;102(2):339–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang K, Donnelly CJ, Haeusler AR, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freibaum BD, Lu Y, Lopez-Gonzalez R, et al. GGGGCC repeat expansion in C0ORF72 compromises nucleocytoplasmic transport. Nature. 2015;525(7567):129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jovičič A, Mertens J, Boeynaems S, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18(9):1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boeynaems S, Bogaert E, Michiels E, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Görlich D, Kutay U.. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev. Biol. 1999;15:607–660. [DOI] [PubMed] [Google Scholar]
- 26. Cautain B, Hill R, de Pedro N, Link W.. Components and regulation of nuclear transport processes. FEBS J. 2015;282(3):445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pumroy RA, Cingolani G.. Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem J. 2015;466(1):13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Köhler M, Speck C, Christiansen M, et al. Evidence for distinct substrate specificities of importin-α family members in nuclear protein import. Mol Cell Biol. 1999;19(11):7782–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herold A, Truant R, Wiegand H, Cullen BR.. Determination of the functional domain organization of the importin alpha nuclear import factor. J Cell Biol. 1998;143(2):309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6(4):388–397. [DOI] [PubMed] [Google Scholar]
- 31. Soniat M, Chook YM.. Nuclear localization signals for four distinct karyopherin-β nuclear import systems. Biochem J. 2015;468(3):353–362. [DOI] [PubMed] [Google Scholar]
- 32. Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM.. Viral subversion of nucleocytoplasmic trafficking. Traffic. 2014;15(2):127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mor A, White MA, Fontoura BM.. Nuclear trafficking in health and disease. Curr Opin Cell Biol. 2014;28:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chook Y, Blobel G.. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11(6):703–715. [DOI] [PubMed] [Google Scholar]
- 35. Quan Y, Ji ZL, Wang X, Tartakoff AM, Tao T.. Evolutionary and transcriptional analysis of karyopherin beta superfamily proteins. Mol Cell Proteomics. 2008;7(7):1254–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Di Francesco L, Verrico A, Asteriti IA, et al. Visualization of human karyopherin beta-1/importin beta-1 interactions with protein partners in mitotic cells by co-immunoprecipitation and proximity ligation assays. Sci Rep. 2018;8:1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D.. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90(6):1061–1071. [DOI] [PubMed] [Google Scholar]
- 38. Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA specific nuclear export receptor. Mol Cell. 1998;1(3):359–369. [DOI] [PubMed] [Google Scholar]
- 39. Lipowsky G, Bischoff FR, Schwarzmaier P, et al. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000;19(16):4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yi R, Qin Y, Macara IG, Cullen BR.. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kurisaki A, Kurisaki K, Kowanetz M, et al. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell. 2006;26(4):1318–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112(4):441–451. [DOI] [PubMed] [Google Scholar]
- 43. Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O.. Structure and gating of the nuclear pore complex. Nat Commun. 2015;6:7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. von Appen A, Kosinski J, Sparks L, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526(7571):140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoelz A, Glavy JS, Beck M.. Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat Struct Mol Biol. 2016;23(7):624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chook YM, Suel KE.. Nuclear import by karyopherin-betas: Recognition and inhibition. Biochim Biophys Acta. 2011;1813(9):1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dingwall C, Robbins J, Dilworth SM, Roberts B, Richardson¢ WD.. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J Cell Biol. 1988;107(3):841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kalderon D, Roberts BL, Richardson WD, Smith AE.. A short amino acid sequence able to specify nuclear location. Cell. 1984;39(3):499–509. [DOI] [PubMed] [Google Scholar]
- 49. Bednenko J, Cingolani G, Gerace L.. Importin b contains a COOH-terminal nucleoporin binding region important for nuclear transport. J Cell Biol. 2003;162(3):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM.. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126(3):543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vance C, Scotter EL, Nishimura AL, et al. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet. 2013;22(13):2676–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ederle H, Funk C, Abou-Ajram C, et al. Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci Rep. 2018;8(1):7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pinarbasi ES, Cağatay T, Yee H, et al. Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci Rep. 2018;8(1):7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nishimura AL, Zupunski V, Troakes C, et al. Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain. 2010;133(Pt 6):1763–1771. [DOI] [PubMed] [Google Scholar]
- 55. Wang G, Zhang Y, Chen B, Cheng J.. Preliminary studies on Alzheimer's disease using cDNA microarrays. Mech Ageing Dev. 2003;124(1):115–124. [DOI] [PubMed] [Google Scholar]
- 56. Lee H, Ueda M, Miyamoto Y, et al. Aberrant localization of importin alpha1 in hippocampal neurons in Alzheimer disease. Brain Res. 2006;1124(1):1–4. [DOI] [PubMed] [Google Scholar]
- 57. Carter CJ. Alzheimer's disease plaques and tangles: cemeteries of a pyrrhic victory of the immune defence network against herpes simplex infection at the expense of complement and inflammation-mediated neuronal destruction. Neurochem Int. 2011;58(3):301–320. [DOI] [PubMed] [Google Scholar]
- 58. Roy J, Sarkar A, Parida S, Ghosh Z, Mallick B.. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer's disease and their probable role in pathogenesis. Mol Biosyst. 2017;13(3):565–576. [DOI] [PubMed] [Google Scholar]
- 59. Nuovo G, Amann V, Williams J, et al. Increased expression of importin-β, exportin-5 and nuclear transportable proteins in Alzheimer’s disease aids anatomic pathologists in its diagnosis. Ann Diagn Pathol. 2018;32:10–16. [DOI] [PubMed] [Google Scholar]
- 60. Eftekharzadeh B, Daigle JG, Kapinos LE, et al. Tau protein disrupts nucleocytoplasmic transport in Alzheimer's disease. Neuron. 2018;99(5):925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou Y, Gu G, Goodlett DR, et al. Analysis of alpha-synuclein-associated proteins by quantitative proteomics. J Biol Chem. 2004;279(37):39155–39164. [DOI] [PubMed] [Google Scholar]
- 62. Ma KL, Song LK, Yuan YH, et al. The nuclear accumulation of alpha-synuclein is mediated by importin alpha and promotes neurotoxicity by accelerating the cell cycle. Neuropharmacology. 2014;82:132–142. [DOI] [PubMed] [Google Scholar]
- 63. Björkblom B, Maple-Grødem J, Puno MR, Odell M, Larsen JP, Møller SG.. Reactive oxygen species-mediated DJ-1 monomerization modulates intracellular trafficking involving karyopherin β2. Mol Cell Biol. 2014;34(16):3024–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han KA, Shin WH, Jung S, et al. Leucine-rich repeat kinase 2 exacerbates neuronal cytotoxicity through phosphorylation of histone deacetylase 3 and histone deacetylation. Hum Mol Genet. 2017;26(1):1–18. [DOI] [PubMed] [Google Scholar]
- 65. George G, Singh S, Lokappa SB, Varkey J.. Gene co-expression network analysis for identifying genetic markers in Parkinson's disease - a three-way comparative approach. Genomics. 2019;111(4):819–830. [DOI] [PubMed] [Google Scholar]
- 66. Mahul-Mellier AL, Burtscher J, Maharjan N, et al. The process of Lewy body formation, rather than simply α-synuclein fibrillization, is one of the major drivers of neurodegeneration. Proc Natl Acad Sci U S A. 2020;117(9):4971–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chan WM, Tsoi H, Wu CC, et al. Expanded polyglutamine domain possesses nuclear export activity which modulates subcellular localization and toxicity of polyQ disease protein via exportin-1. Hum Mol Genet. 2011;20(9):1738–1750. [DOI] [PubMed] [Google Scholar]
- 68. Desmond CR, Atwal RS, Xia J, Truant R.. Identification of a karyopherin β1/β2 proline-tyrosine nuclear localization signal in huntingtin protein. J Biol Chem. 2012;287(47):39626–39633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rué L, López-Soop G, Gelpi E, Martínez-Vicente M, Alberch J, Pérez-Navarro E.. Brain region- and age-dependent dysregulation of p62 and NBR1 in a mouse model of Huntington's disease. Neurobiol Dis. 2013;52:219–228. [DOI] [PubMed] [Google Scholar]
- 70. Woerner AC, Frottin F, Hornburg D, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351(6269):173–176. [DOI] [PubMed] [Google Scholar]
- 71. Ling SC, Polymenidou M, Cleveland DW.. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hortobágyi T, Cairns N.. Amyotrophic lateral sclerosis and non-tau frontotemporal lobar degeneration. In: Kovacs GG,, Alafuzoff I, eds. Handbook of clinical neurology (3rd series). Elsevier; 2017;145:369–381. [DOI] [PubMed] [Google Scholar]
- 73. Strong MJ, Hortobágyi T, Okamoto K, Kato S.. Amyotrophic lateral sclerosis, Primary lateral sclerosis and Spinal muscular atrophy. In: Dickson DW, Weller RO, eds. Neurodegeneration: The molecular pathology of dementia and movement disorders. 2nd ed. Wiley-Blackwell; 2011:418–433. [Google Scholar]
- 74. Taylor JP, Brown RH Jr, Cleveland DW.. Decoding ALS: From genes to mechanism. Nature. 2016;539(7628):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Deleon J, Miller BL.. Frontotemporal dementia. Handb Clin Neurol. 2018;148:409–430. [DOI] [PubMed] [Google Scholar]
- 76. Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351(3):602–611. [DOI] [PubMed] [Google Scholar]
- 77. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- 78. Yamashita T, Aizawa H, Teramoto S, Akamatsu M, Kwak S.. Calpain-dependent disruption of nucleo-cytoplasmic transport in ALS motor neurons. Sci Rep. 2017;7:39994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kinoshita Y, Ito H, Hirano A, et al. Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2009;68(11):1184–1192. [DOI] [PubMed] [Google Scholar]
- 80. Aizawa H, Yamashita T, Kato H, Kimura T, Kwak S.. Impaired nucleoporins are present in sporadic Amyotrophic Lateral Sclerosis motor neurons that exhibit mislocalization of the 43-kDa TAR DNA-Binding Protein. J Clin Neurol. 2019;15(1):62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Al-Sarraj S, King A, Troakes C, et al. P62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122(6):691–702. [DOI] [PubMed] [Google Scholar]
- 82. Murray ME, DeJesus-Hernandez M, Rutherford NJ, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122(6):673–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dejesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-Linked FTD and ALS. Neuron. 2011;72(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Renton AE, Majounie E, Waite A, et al. ; ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Waite AJ, Bäumer D, East S, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35(7):1779.e5-1779–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shi Y, Lin S, Staats KA, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24(3):313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fratta P, Poulter M, Lashley T, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126(3):401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lee YB, Chen HJ, Peres JN, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zu T, Liu Y, Bañez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110(51):E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cooper-Knock J, Walsh MJ, Higginbottom A, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137(7):2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mori K, Weng S-M, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. [DOI] [PubMed] [Google Scholar]
- 92. Braems E, Swinnen B, Van Den Bosch L.. Van Den Bosch, L. C9orf72 loss-of-function: a trivial, stand-alone or additive mechanism in C9 ALS/FTD? Acta Neuropathol. 2020;140(5):625–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lee KH, Zhang P, Kim HJ, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167(3):774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Boeynaems S, Bogaert E, Kovacs D, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65(6):1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hayes LR, Duan L, Bowen K, Kalab P, Rothstein JD.. C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. Elife. 2020;9:e51685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ederle H, Dormann D.. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 2017;591(11):1489–1507. [DOI] [PubMed] [Google Scholar]
- 97. Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, M-Y Lee V.. Disturbance of nuclear and cytoplasmic TAR DNA-binding Protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem. 2008;283(19):13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tziortzouda P, Van Den Bosch L, Hirth F.. Triad of TDP43 control in neurodegeneration: autoregulation, localization and aggregation. Nat Rev Neurosci. 2021;22(4):197–208. [DOI] [PubMed] [Google Scholar]
- 99. Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mizielinska S, Grönke S, Niccoli T, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345(6201):1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wen X, Tan W, Westergard T, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84(6):1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hutten S, Usluer S, Bourgeois B, et al. Nuclear Import Receptors Directly Bind to Arginine-Rich Dipeptide Repeat Proteins and Suppress Their Pathological Interactions. Cell Rep. 2020;33(12):108538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Park JH, Chung CG, Park SS, et al. Cytosolic calcium regulates cytoplasmic accumulation of TDP-43 through Calpain-A and Importin alpha3. Elife. 2020;9:e60132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Qui C, Kivipelto M, von Strauss E.. Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci. 2009;11:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang J, Gu BJ, Masters CL, Wang Y-J.. A systemic view of Alzheimer disease-insights from amyloid-β metabolism beyond the brain. Nat Rev Neurol. 2017;13(10):612–623. [DOI] [PubMed] [Google Scholar]
- 106. Hyman BT, Phelps CH, Beach TG, et al. National Insitute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimer’s Dement. 2012;8(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sheffield LG, Mirra SS.. The distribution of karyopherin-β proteins in Alzheimer’s disease. FASEB J. 2008;22:707.6. [Google Scholar]
- 108. Cornelison GL, Levy SA, Jenson T, Frost B.. Tau-induced nuclear envelope invagination causes a toxic accumulation of mRNA in Drosophila. Aging Cell. 2019;18(1):e12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mastroeni D, Chouliaras L, Grover A, et al. Reduced RAN expression and disrupted transport between cytoplasm and nucleus; a key event in Alzheimer's disease pathophysiology. PLoS One. 2013;8(1):e53349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bayliss R, Littlewood T, Stewart M.. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102(1):99–108. [DOI] [PubMed] [Google Scholar]
- 111. Fontoura BM, Blobel G, Yaseen NR.. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin beta 2-mediated nuclear import. J Biol Chem. 2000;275(40):31289–31296. [DOI] [PubMed] [Google Scholar]
- 112. Wagner RS, Kapinos LE, Marshall NJ, Stewart M, Lim RYH.. Promiscuous binding of Karyopherinβ1 modulates FG nucleoporin barrier function and expedites NTF2 transport kinetics. Biophys J. 2015;108(4):918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Goedert M, Spillantini MG, Del Tredici K, Braak H.. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9(1):13–24. [DOI] [PubMed] [Google Scholar]
- 114. Goedert M, Jakes R, Spillantini MG.. The synucleinopathies: Twenty years on. J Parkinsons Dis. 2017;7(s1):S51–S69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Spillantini MG, Schmidt ML, Lee VM-Y, Trojanowski JQ, Jakes R, Goedert M. αSynuclein in Lewy bodies. Nature. 1997;388(6645):839–840. [DOI] [PubMed] [Google Scholar]
- 116. Baba M, Nakajo S, Tu PH, et al. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol. 1998;152(4):879–884. [PMC free article] [PubMed] [Google Scholar]
- 117. Takeda A, Mallory M, Sundsmo M, Honer W, Hansen L, Masliah E.. Abnormal accumulation of NACP/alpha-synuclein in neurodegenerative disorders. Am J Pathol. 1998;152(2):367–372. [PMC free article] [PubMed] [Google Scholar]
- 118. Bridi J, Hirth F.. Mechanisms of a-synuclein induced synaptopathy in Parkinson's disease. Front Neurosci. 2018;12:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Maroteaux L, Campanelli JT, Scheller RH.. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8(8):2804–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Schaser AJ, Osterberg VR, Dent SE, et al. Alpha-synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci Rep. 2019;9(1):10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ueda K, Fukushima H, Masliah E, et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(23):11282–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Davidi D, Schechter M, Elhadi SA, Matatov A, Nathanson L, Sharon R.. α-Synuclein translocates to the nucleus to activate retinoic-acid-dependent gene transcription. iScience. 2020;23(3):100910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ryu S, Baek I, Liew H.. Sumoylated α-synuclein translocates into the nucleus by karyopherin α6. Mol Cell Toxicol. 2019;15(1):103–109. [Google Scholar]
- 124. Rousseaux MW, de Haro M, Lasagna-Reeves CA, et al. TRIM28 regulates the nuclear accumulation and toxicity of both alpha-synuclein and tau. Elife. 2016;5:e19809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Moriyama T, Sangel P, Yamaguchi H, et al. Identification and characterization of a nuclear localization signal of TRIM28 that overlaps with the HP1 box. Biochem Biophys Res Commun. 2015;462(3):201–207. [DOI] [PubMed] [Google Scholar]
- 126. Rousseaux MW, Revelli J-P, Vázquez-Vélez GE, et al. Depleting Trim28 in adult mice is well tolerated and reduces levels of α-synuclein and tau. Elife. 2018;7:e36768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Nishie M, Mori F, Yoshimoto M, Takahashi H, Wakabayashi K.. A quantitative investigation of neuronal cytoplasmic and intranuclear inclusions in the pontine and inferior olivary nuclei in multiple system atrophy. Neuropathol Appl Neurobiol. 2004;30(5):546–554. [DOI] [PubMed] [Google Scholar]
- 128. Pinho R, Paiva I, Jercic KG, et al. Nuclear localization and phosphorylation modulate pathological effects of alpha-synuclein. Hum Mol Genet. 2019;28(1):31–50. [DOI] [PubMed] [Google Scholar]
- 129. Kramer ML, Schulz-Schaeffer WJ.. Presynaptic alpha-synuclein aggregates, not Lewy bodies, cause neurodegeneration in dementia with Lewy bodies. J Neurosci. 2007;27(6):1405–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Colom-Cadena M, Pegueroles J, Herrmann AG, et al. Synaptic phosphorylated α-synuclein in dementia with Lewy bodies. Brain. 2017;140(12):3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bereczki E, Francis PT, Howlett D, et al. Synaptic proteins predict cognitive decline in Alzheimer`s disease and Lewy body dementia. Alzheimer’s Dement. 2016;12(11):1149–1158. [DOI] [PubMed] [Google Scholar]
- 132. Bereczki E, Branca RM, Francis PT, et al. Synaptic markers of cognitive decline in neurodegenerative diseases: A proteomic approach. Brain. 2018;141(2):582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Braak H, Rub U, Gai WP, Del Tredici K.. Idiopathic Parkinson’s disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J Neural Transm (Vienna). 2003;110(5):517–536. [DOI] [PubMed] [Google Scholar]
- 134. Morgan SA, Lavenir I, Fan J, et al. Goedert, M. alpha-Synuclein filaments from transgenic mouse and human synucleinopathy-containing brains are major seed-competent species. J Biol Chem. 2020;295(19):6652–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Devine MJ, Ryten M, Vodicka P, et al. Parkinson's disease induced pluripotent stem cells with triplication of the α-synuclein locus. Nat Commun. 2011;2:440- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Fares MB, Ait-Bouziad N, Dikiy I, et al. The novel Parkinson's disease linked mutation G51D attenuates in vitro aggregation and membrane binding of α-synuclein, and enhances its secretion and nuclear localization in cells. Hum Mol Genet. 2014;23(17):4491–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Büttner S, Habernig L, Broeskamp F, et al. Endonuclease G mediates α-synuclein cytotoxicity during Parkinson's disease. EMBO J. 2013;32(23):3041–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Paisán-Ruíz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44(4):595–600. [DOI] [PubMed] [Google Scholar]
- 139. Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. [DOI] [PubMed] [Google Scholar]
- 140. Bonifati V, Rizzu P, van Baren MJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–259. [DOI] [PubMed] [Google Scholar]
- 141. Nelson DE, Laman H.. A Competitive binding mechanism between Skp1 and exportin 1 (CRM1) controls the localization of a subset of F-box proteins. J Biol Chem. 2011;286(22):19804–19815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhao T, Severijnen LA, van der Weiden M, et al. FBXO7 immunoreactivity in α-synuclein-containing inclusions in Parkinson disease and multiple system atrophy. J Neuropathol Exp Neurol. 2013;72(6):482–488. [DOI] [PubMed] [Google Scholar]
- 143. McColgan P, Tabrizi SJ.. Huntington's disease: A clinical review. Eur J Neurol. 2018;25(1):24–34. [DOI] [PubMed] [Google Scholar]
- 144. Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Prim. 2015;1:15005. [DOI] [PubMed] [Google Scholar]
- 145. Finkbeiner S. Huntington's disease. Cold Spring Harb Perspect Biol. 2011;3(6):a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ashizawa T, Wong LJ, Richards CS, Caskey CT, Jankovic J.. CAG repeat size and clinical presentation in Huntington's disease. Neurology. 1994;44(6):1137–1143. [DOI] [PubMed] [Google Scholar]
- 147. Kieburtz K, MacDonald M, Shih C, et al. Trinucleotide repeat length and progression of illness in Huntington's disease. J Med Genet. 1994;31(11):872–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Truant R, Atwal RS, Burtnik A.. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington’s disease. Prog Neurobiol. 2007;83(4):211–227. [DOI] [PubMed] [Google Scholar]
- 149. Maiuri T, Woloshansky T, Xia J, Truant R.. Truant, R. The huntingtin N17 domain is a multifunctional CRM1 and ran-dependent nuclear and cilial export signal. Hum Mol Genet. 2013;22(7):1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zheng Z, Li A, Holmes BB, Marasa JC, Diamond MI.. An N-terminal nuclear export signal regulates trafficking and aggregation of huntingtin (Htt) protein exon 1. J Biol Chem. 2013;288(9):6063–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Twyffels L, Gueydan C, Kruys V.. Transportin-1 and Transportin-2: Protein nuclear import and beyond. FEBS Lett. 2014;588(10):1857–1868. [DOI] [PubMed] [Google Scholar]
- 152. Kim M, Roh JK, Yoon BW, et al. Huntingtin is degraded to small fragments by calpain after ischemic injury. Exp Neurol. 2003;183(1):109–115. [DOI] [PubMed] [Google Scholar]
- 153. Hughes A, Jones L.. Huntingtin localisation studies - a technical review. PLoS Curr. 2011;3:RRN1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Saudou F, Humbert S.. The Biology of Huntingtin. Neuron. 2016;89(5):910–926. [DOI] [PubMed] [Google Scholar]
- 155. DiFiglia M, Sapp E, Chase KO, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. [DOI] [PubMed] [Google Scholar]
- 156. Wellington CL, Ellerby LM, Hackam AS, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273(15):9158–9167. [DOI] [PubMed] [Google Scholar]
- 157. Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. [DOI] [PubMed] [Google Scholar]
- 158. Shirasaki DI, Greiner ER, Al-Ramahi I, et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron. 2012;75(1):41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Langfelder P, Cantle JP, Chatzopoulou D, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19(4):623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, et al. Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron. 2017;94(1):48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Grima JC, Daigle JG, Arbez N, et al. Mutant huntingtin disrupts the nuclear pore complex. Neuron. 2017;94(1):93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Bañez-Coronel M, Ayhan F, Tarabochia AD, et al. RAN translation in Huntington Disease. Neuron. 2015;88(4):667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Yamada SB, Gendron TF, Niccoli T, et al. RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat Neurosci. 2019;22(9):1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Bogaert E, Boeynaems S, Kato M, et al. Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 2018;24(3):529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Peskett TR, Rau F, O'Driscoll J, Patani R, Lowe AR, Saibil HR.. A liquid to solid phase transition underlying pathological Huntingtin exon1 aggregation. Mol Cell. 2018;70(4):588–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wegmann S, Eftekharzadeh B, Tepper K, et al. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 2018;37(7):e98049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ray S, Singh N, Kumar R, et al. α-synuclein aggregation nucleates through liquid-liquid phase separation. Nat Chem. 2020;12(8):705–716. [DOI] [PubMed] [Google Scholar]
- 168. Cascarina M, Elder R, Ross D.. Atypical structural tendencies among low-complexity domains in the Protein Data Bank proteome. PLoS Comput Biol. 2020;16(1):e1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Alberti S, Hyman AA.. Are aberrant phase transitions a driver of cellular aging? Bioessays. 2016;38(10):959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Shin Y, Brangwynne CP.. Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357):eaaf4382. [DOI] [PubMed] [Google Scholar]
- 171. Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M.. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Boyko S, Surewicz K, Surewicz WK.. Regulatory mechanisms of tau protein fibrillation under the conditions of liquid-liquid phase separation. Proc Natl Acad Sci U S A. 2020;117(50):31882–31890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kanaan NM, Hamel C, Grabinski T, Combs B.. Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat Commun. 2020;11(1):2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ash PEA, Lei S, Shattuck J, et al. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc Natl Acad Sci U S A. 2021;118(9):e2014188118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Babinchak WM, Haider R, Dumm BK, et al. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J Biol Chem. 2019;294(16):6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. French RL, Grese ZR, Aligireddy H, et al. Detection of TAR DNA-binding protein 43 (TDP-43) oligomers as initial intermediate species during aggregate formation. J Biol Chem. 2019;294(17):6696–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Watanabe S, Inami H, Oiwa K, et al. Aggresome formation and liquid-liquid phase separation independently induce cytoplasmic aggregation of TAR DNA-binding protein 43. Cell Death Dis. 2020;11(10):909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Conicella AE, Dignon GL, Zerze GH, et al. TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proc Natl Acad Sci U S A. 2020;117(11):5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Murthy AC, Dignon GL, Kan Y, et al. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol. 2019;26(7):637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Dasmeh P, Wagner A.. Natural selection on the phase-separation properties of FUS during 160 my of mammalian evolution. Mol Biol Evol. 2021;38(3):940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Hardenberg MC, Sinnige T, Casford S, et al. Observation of an α-synuclein liquid droplet state and its maturation into Lewy body-like assemblies. J Mol Cell Biol. 2021;mjaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Vanderweyde T, Yu H, Varnum M, et al. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J Neurosci. 2012;32(24):8270–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Springhower CE, Rosen MK, Chook YM.. Karyopherins and condensates. Curr Opin Cell Biol. 2020;64:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Schmidt HB, Barreau A, Rohatgi R.. Phase separation-deficient TDP43 remains functional in splicing. Nat Commun. 2019;10(1):4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Li H-R, Chiang W-C, Chou P-C, Wang W-J, Huang J.. TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. J Biol Chem. 2018;293(16):6090–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Lin Y, Protter DSW, Rosen MK, Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60(2):208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhang K, Daigle JG, Cunningham KM, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173(4):958–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zhang X, Vigers M, McCarty J, et al. The proline-rich domain promotes Tau liquid-liquid phase separation in cells. J Cell Biol. 2020;219:e202006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Lin Y, Fichou Y, Longhini AP, et al. Liquid-liquid phase separation of tau driven by hydrophobic interaction facilitates fibrillization of tau. J Mol Biol. 2021;433(2):166731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Shorter J. Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev. 2017;44:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Darling AL, Shorter J.. Combating deleterious phase transitions in neurodegenerative disease. Biochim Biophys Acta Mol Cell Res. 2021;1868(5):118984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Dorst J, Ludolph AC, Huebers A.. Disease-modifying and symptomatic treatment of amyotrophic lateral sclerosis. Ther Adv Neurol Disord. 2018;11:1756285617734734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Long JM, Holtzman DM.. Alzheimer Disease: An update on pathobiology and treatment strategies. Cell. 2019;179(2):312–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Stelma T, Chi A, Van Der Watt PJ, Verrico A, Lavia P, Leaner VD.. Critical review targeting nuclear transporters in cancer: diagnostic, prognostic and therapeutic potential. IUBMB Life. 2016;68(4):268–280. [DOI] [PubMed] [Google Scholar]
- 195. Çağatay T, Chook YM.. Karyopherins in cancer. Curr Opin Cell Biol. 2018;52:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Martin AJ, Jans DA.. Antivirals that target the host IMPα/β1-virus interface. Biochem Soc Trans. 2021;49(1):281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Jans DA, Martin AJ, Wagstaff KM.. Inhibitors of nuclear transport. Curr Opin Cell Biol. 2019;58:50–60. [DOI] [PubMed] [Google Scholar]
- 198. Kosyna FK, Depping R.. Controlling the gatekeeper: Therapeutic targeting of nuclear transport. Cells. 2018;7(11):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA.. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443(3):851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Tay MY, Fraser JE, Chan WK, et al. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir Res. 2013;99(3):301–306. [DOI] [PubMed] [Google Scholar]
- 201. Panchal M, Rawat K, Kumar G, et al. Plasmodium falciparum signal recognition particle components and anti-parasitic effect of ivermectin in blocking nucleo-cytoplasmic shuttling of SRP. Cell Death Dis. 2014;5:e994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Widman DG, Gornisiewicz S, Shacham S, Tamir S.. In vitro toxicity and efficacy of verdinexor, an exportin 1 inhibitor, on opportunistic viruses affecting immunocompromised individuals. PLoS One. 2018;13(10):e0200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Haines JD, Herbin O, De La Hera B, et al. Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. Nat Neurosci. 2015;18(4):511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Niaki AG, Sarkar J, Cai X, et al. Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol Cell. 2020;77(1):82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Archbold HC, Jackson KL, Arora A, et al. TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci Rep. 2018;8(1):4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Wang AY, Liu H.. The past, present, and future of CRM1/XPO1 inhibitors. Stem Cell Investig. 2019;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Quiroz FG, Fiore VF, Levorse J, et al. Liquid-liquid phase separation drives skin barrier formation. Science. 2020;367(6483):eaax9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Khosravi B, Hartmann H, May S, et al. Cytoplasmic poly-GA aggregates impair nuclear import of TDP-43 in C9orf72 ALS/FTLD. Hum Mol Genet. 2017;26(4):790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Sowa AS, Martin E, Martins IM, et al. Karyopherin α-3 is a key protein in the pathogenesis of spinocerebellar ataxia type 3 controlling the nuclear localization of ataxin-3. Proc Natl Acad Sci U S A. 2018;115(11):E2624–E2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Tsou WL, Ouyang M, Hosking RR, et al. The deubiquitinase ataxin-3 requires Rad23 and DnaJ-1 for its neuroprotective role in Drosophila melanogaster. Neurobiol Dis. 2015;82:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Fagerlund R, Kinnunen L, Köhler M, Julkunen I, Melén K.. NF-κB is transported into the nucleus by importin α3 and importin α4. J Biol Chem. 2005;280(16):15942–15951. [DOI] [PubMed] [Google Scholar]
- 212. Lu C, Figueroa JA, Liu Z, et al. Nuclear export as a novel therapeutic target: The CRM1 connection. Curr Cancer Drug Targets. 2015;15(7):575–592. [DOI] [PubMed] [Google Scholar]
- 213. Chafe SC, Mangroo D.. Scyl1 facilitates nuclear tRNA export in mammalian cells by acting ath nuclear pore complex. Mol Biol Cell. 2010;21(14):2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Wang J, Lee JE, Riemondy K, et al. XPO5 promotes primary miRNA processing independently of RanGTP. Nat Commun. 2020;11(1):1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Stueven T, Hartmann E, Goerlich D.. Exportin 6: A novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Mingot JM, Bohnsack MT, Jakle U, Görlich D.. Exportin 7 defines a novel general nuclear export pathway. EMBO J. 2004;23(16):3227–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Aksu M, Pleiner T, Karaca S, et al. Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J Cell Biol. 2018;217(7):2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Jäkel S, Görlich D.. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Neumann M, Bentmann E, Dormann D, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134(Pt 9):2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Barraud P, Banerjee S, Mohamed WI, Jantsch MF, Allain FH.. A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1. Proc Natl Acad Sci U S A. 2014;111(18):E1852-E1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Güttinger S, Mühlhäusser P, Koller-Eichhorn R, Brennecke J, Kutay U.. Transportin2 functions as importin and mediates nuclear import of HuR. Proc Natl Acad Sci U S A. 2004;101(9):2918–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Kataoka N, Bachorik JL, Dreyfuss G.. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145(6):1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Jäkel S, Mingot J-M, Schwarzmaier P, Hartmann E, Görlich D.. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21(3):377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Dean KA, von Ahsen O, Görlich D, Fried HM.. Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J Cell Sci. 2001;114(19):3479–3485. [DOI] [PubMed] [Google Scholar]
- 225. Plafker SM, Macara IG.. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 2000;19(20):5502–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Mingot JM, Kostka S, Kraft R, Hartmann E, Görlich D.. Importin 13: A novel mediator of nuclear import and export. EMBO J. 2001;20(14):3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]