Abstract
Novel coronavirus disease 2019 (COVID-19) is an emerging, rapidly evolving crisis, and the ability to predict prognosis for individual COVID-19 patient is important for guiding treatment. Laboratory examinations were repeatedly measured during hospitalization for COVID-19 patients, which provide the possibility for the individualized early prediction of prognosis. However, previous studies mainly focused on risk prediction based on laboratory measurements at one time point, ignoring disease progression and changes of biomarkers over time. By using historical regression trees (HTREEs), a novel machine learning method, and joint modeling technique, we modeled the longitudinal trajectories of laboratory biomarkers and made dynamically predictions on individual prognosis for 1997 COVID-19 patients. In the discovery phase, based on 358 COVID-19 patients admitted between 10 January and 18 February 2020 from Tongji Hospital, HTREE model identified a set of important variables including 14 prognostic biomarkers. With the trajectories of those biomarkers through 5-day, 10-day and 15-day, the joint model had a good performance in discriminating the survived and deceased COVID-19 patients (mean AUCs of 88.81, 84.81 and 85.62% for the discovery set). The predictive model was successfully validated in two independent datasets (mean AUCs of 87.61, 87.55 and 87.03% for validation the first dataset including 112 patients, 94.97, 95.78 and 94.63% for the second validation dataset including 1527 patients, respectively). In conclusion, our study identified important biomarkers associated with the prognosis of COVID-19 patients, characterized the time-to-event process and obtained dynamic predictions at the individual level.
Keywords: COVID-19, longitudinal data, dynamic risk prediction, time-to-event
Introduction
The coronavirus disease 2019 (COVID-19), which emerged in December 2019, has become a major worldwide public health problem. As of 1 April 1 2021 severe acute respiratory syndrome coronavirus 2, or SARS-CoV-2, has rapidly spread to approximately 218 countries, causing 130 163 234 cases and 2 839 661 deaths [1].
Because of the high mortality rate of the COVID-19, it is important to identify prognostic factors and develop predictive models to estimate survival probability and better individualize treatments, especially for severe or critically ill COVID-19 patients. Demographic and clinical features have predictive power and studies have also demonstrated the significance of laboratory measurements, such as lactate dehydrogenase (LDH), hypersensitive c-reactive protein (Hs-CRP), aspartate aminotransferase (AST), creatine and d-dimer, in discriminating between COVID-19 patients who survived or deceased [2].
Clinical and laboratory biomarkers are usually measured at either regular or irregular intervals in COVID-19 patients during hospitalization [3]. Previous studies mainly focused on risk prediction based on laboratory measurements at a single time point (at baseline or the last measure before death or discharge from the hospital), ignoring the time course of biomarkers varying over time [3–5]. However, disease progression is a dynamic process. Static prediction based on a single time point may not provide sufficient information on how an individual patient’s risk dynamically will update over time and how the risk is influenced by time-varying predictors [6]. Thus, it is attractive to make dynamic predictions by fully utilized information from longitudinal data measured at multiple time points, which may be beneficial for making timely and effective individual treatment recommendations [7, 8]. As an example, it is recognized that psychopathology is highly dynamic. Static prediction based on single baseline assessments may fail to produce accuracy and replicability predictions, whereas analytical methods built on the dynamic nature of psychopathology may be more powerful for predicting which individual may change from one clinical state to another and when this change will happen [9].
In this study, we aimed to develop a dynamic risk prediction model for COVID-19 prognosis based on the information of 1997 patients from Hubei Province, China, applying a random forest-based machine learning method and a joint modeling technique. The predictive model was firstly developed based on a publicly available dataset of 375 patients from Wuhan. In addition, our study characterized the time-dependent effect of laboratory biomarkers on prognosis. The model was further validated in two independent cohorts, including a cohort of 112 patients from Huangshi, Hubei and a cohort of 1527 patients from Wuhan Huoshenshan Hospital.
Methods
Data sources
For the discovery set, data of 375 COVID-19 patients from Tongji hospital collected between10 January and 18 February 2020 were obtained from a recently published literature [4]. Details about recruitment and inclusion/exclusion procedures have been described before [4]. Briefly, the dataset includes 197 general, 27 severe, and 151 critically ill COVID-19 patients. For the 375 patients, the age distribution (mean ± standard deviation) was 58.83 ± 16.46 years, 59.7% were male and 201 (53.6%) recovered from COVID-19 and were discharged from hospital. Period of follow-up was defined as the duration from hospital admission to death or discharge, and the maximal follow-up time was 35 days.
We validated the predictive model generated from the discovery stage in two independent cohorts. The first dataset includes 112 severe or critically ill COVID-19 patients recruited from three hospitals (Huangshi Central Hospital, Huangshi Hospital of Traditional Chinese Medicine and Daye People’s Hospital) during 21 January–6 March 2020. All three hospitals are located in Huangshi City, Hubei Province, China. Detailed demographics and clinical characteristics, including initial symptoms, comorbidities and disease severity, were recorded at admission. Laboratory measurements, such as routine blood tests, lymphocyte subsets and inflammatory biomarkers, were obtained at admission and during hospitalization. More details about the study have been described previously [10]. The second validation set includes laboratory test results of 1527 severe or critically ill COVID-19 patients recruited between 4 February and 30 March 2020 from Wuhan Huoshenshan Hospital in China. Data on the clinical characteristics and laboratory findings of all patients were extracted from the hospital electronic medical records, and more information can be found in a previously published article [11].
The discovery dataset used in our study had been published and is publicly available, which was approved by the Tongji Hospital Ethics Committee [4]. As for the first validation dataset, the ethics committee of the hospitals (Huangshi Central Hospital, Huangshi Hospital of Traditional Chinese Medicine, and Daye People’s Hospital) waived the written informed consent from patients with COVID-19, and all the procedures being performed were part of the routine care [10]. Written informed consent of Wuhan Huoshenshan Hospital was obtained from each patient, and the study was approved by the Medical Ethical Committee of Wuhan Huoshenshan Hospital and the Ethical Committee of Nanjing Medical University [11].
Statistical analysis
Descriptive statistics were obtained for all study variables. Continuous variables were summarized as means (± standard deviations), and the characteristics of patients who survived or died were compared using t-test or Mann–Whitney U test for continuous variables depending on the data distributions. Categorized variables were described by frequency (n) and proportion (%) and compared using chi-square test. As longitudinal biomarker measurements were only taken intermittently during hospitalization, missing values are unavoidable. Missing laboratory values were imputed using the “Mice” package in R [12]. All concentrations of laboratory measures were loge transformed for further statistical analysis.
Model development
Development of the predictive model consisted of two main stages: (i) feature selection and (ii) dynamic characterization of the longitudinal process and time-to-event outcome. Each process was externally verified, and the analysis workflow is shown in Figure 1.
Figure 1.
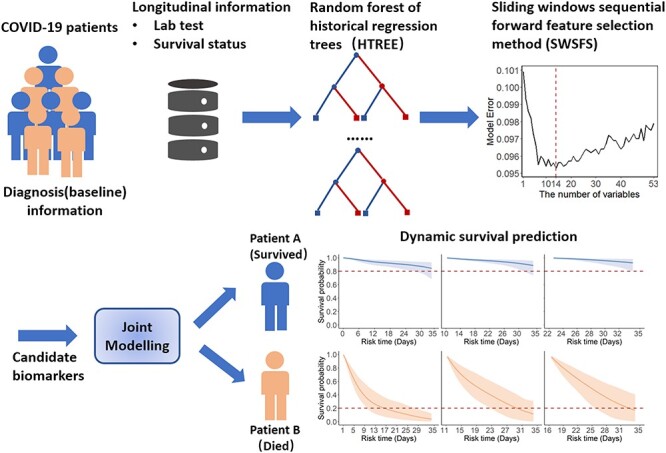
Strategy for incorporating longitudinal measurements to make dynamically predictions on individual prognosis for COVID-19 patients. First, baseline and laboratory tests information during hospitalization were used in HTREEs, and the SWSFS was used to determine the number of important biomarkers, identifying a set of important prognostic biomarkers. Then, joint modeling was applied to characterize the disease process and obtain dynamic predictions at the individual level in COVID-19 patients.
Feature selection
We applied historical regression trees (HTREEs) to screen for important biomarkers in COVID-19 patients by summarizing variable importance with longitudinal measurements using the R package “htree”. HTREE is an extension of the standard random forest model appropriate for longitudinal data and a procedure for non-parametric estimates of how the response depends on all prior observations as well as that of any time-varying predictor variables. To evaluate stability of the model and reliability of the biomarkers, the prediction forest was evaluated by 3-fold cross-validation and was further assessed in two external validation datasets based on the candidate biomarkers.
In the discovery phase, all clinical characteristics during follow-up, including demographics, were evaluated in HTREE model. A sliding windows sequential forward feature selection method (SWSFS) was used to determine the number of “important” biomarkers. Briefly, the importance of each variable was evaluated by the average mean squared error which was first obtained by a HTREE analysis calculated via the varimp_hrf function. Variables were then included one by one to the HTREE model in the order of importance. We plotted the marginalized error, which measures the performance of each model consisting of different numbers of variables to identify a feature set with the lowest error rate. Finally, prognostic biomarkers and covariates in the selected feature set were used as HTREE input to construct a prediction forest model, which was further validated in two external validation datasets.
Joint model construction
We applied the joint modeling technique to fully use the repeated laboratory measurements and time-to-event data. Joint modeling is a method that can simultaneously analyze the longitudinally measured laboratory measurements and survival outcome [13]. It consists of two linked sub-models: a mixed model specifying the time-course of biomarkers, and a survival model using the Cox proportional hazards model. For each of the prognostic biomarkers in the selected feature set, we developed a generalized linear mixed effect sub-model. The estimation of regression parameters within the multivariate joint model framework was performed using the R package “JMbayes” [13], which fits joint model under a Bayesian approach using the Markov Chain Monte Carlo algorithms [14]. For the survival sub-model, a Cox proportional hazard model with a penalized-spline-approximated baseline risk function was used [15] (Details about joint model can be found in the Supplementary Methods).
With the joint model, we were able to estimate the subject-specific marker trajectories and predict conditional survival probabilities for subject i, providing a set of longitudinal measurements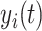 [13]. The conditional probability of surviving to time u given that the patient has survived up to time t (u > t) was
[13]. The conditional probability of surviving to time u given that the patient has survived up to time t (u > t) was
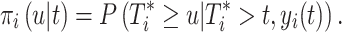 |
An estimate of 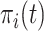 was computed by resampling from posterior distributions produced using the survfitJM function, assuming that the patient was event-free up to the time point of the last measurements [13, 14]. We calculated 5-day, 10-day and 15-day time-dependent areas under the receiver-operator characteristic curve (AUC) to evaluate the predictive accuracy of the joint model in both discovery and external validation datasets, as the 5th, 10th and 15th days after admission are critical time points at which the physician can take action to improve survival chances of the COVID-19 patients.
was computed by resampling from posterior distributions produced using the survfitJM function, assuming that the patient was event-free up to the time point of the last measurements [13, 14]. We calculated 5-day, 10-day and 15-day time-dependent areas under the receiver-operator characteristic curve (AUC) to evaluate the predictive accuracy of the joint model in both discovery and external validation datasets, as the 5th, 10th and 15th days after admission are critical time points at which the physician can take action to improve survival chances of the COVID-19 patients.
To investigate the potential nonlinear trend of biomarker effects varying over time, we allowed a time-varying coefficient to link longitudinal and survival processes using P-splines for the logarithm of the baseline hazard, with the idea to include interactions of biomarkers with an appropriate pre-defined time function (see Supplementary Methods) [16]. For the case of splines, baseline hazard was defined using P-splines with seven internal knots placed at equally spaced percentiles of the observed survival time t.
Statistical analyses were performed using R version 4.0.2 (The R Foundation of Statistical Computing). P ≤ 0.05 was considered statistically significant unless otherwise specified.
Results
Characteristics of the study population
In the discovery phase, we used a previously described cohort that, after exclusion of 17 patients lacking more than 80% laboratory test data (see Supplementary Table 1), consisted of 358 COVID-19 patients enrolled from Tongji Hospital in Wuhan, Hubei Province, China [4]. The mean age of all 358 patients was 58.84 (16.51) years, and 210 (58.66%) were male. The median follow-up time since admission was 10 days, and 148 (41.34%) patients died in hospital. A comparison of baseline laboratory measures of patients who survived and those who died is presented in Table 1. In the validation phase, the first dataset consists of 112 patients from three hospitals in Huangshi City, Hubei Province, China. The mean age of these patients was 60.99 (14.87) years, and 73 (65.18%) were male. The median follow-up time since admission was 11 days, and 31 (27.68%) died (see Supplementary Table 2). For the second validation set including hospital records of 1527 severe or critically ill COVID-19 patients from Wuhan Huoshenshan Hospital in China, the mean age of these patients was 61.81 (14.13) years, and 775 (50.75%) were male. The median follow-up time since admission was 15 days, and 57 (3.73%) died (see Supplementary Table 3).
Table 1.
Demographics, baseline clinical laboratory test and mortality outcomes collected from medical records in the discovery dataset
| Total (n = 358) | Survived (n = 195) | Dead (n = 163) | P-value | |
|---|---|---|---|---|
| Age | 58.84 ± 16.51 | 50.39 ± 15.00 | 68.94 ± 11.94 | <0.0001b |
| Gender, n (%) | <0.0001c | |||
| Male | 210 (58.66) | 92 (43.80) | 118 (56.19) | |
| Female | 148 (41.34) | 103 (69.59) | 45 (30.40) | |
| Median follow-up (days) | 10 | 14 | 6 | <0.0001b |
| Laboratory tests (baseline) | ||||
| LDH, U/L | 440.24 ± 308.77 | 270.23 ± 102.86 | 649.66 ± 347.17 | <0.0001b |
| WBC, 10^9/L | 9.66 ± 14.19 | 7.75 ± 16.64 | 12 ± 10.03 | <0.0001b |
| NEU, 10^9/L | 6.34 ± 5.18 | 3.53 ± 2.17 | 9.78 ± 5.69 | <0.0001b |
| Hs-CRP, mg/L | 76.32 ± 75.49 | 35.23 ± 43.45 | 128.31 ± 75.46 | <0.0001b |
| MPV, fL | 10.86 ± 0.94 | 10.63 ± 0.87 | 11.16 ± 0.94 | <0.0001a |
| Lymphocyte, % | 17.17 ± 12.65 | 24.85 ± 11.25 | 7.71 ± 6.28 | <0.0001b |
| Monocyte, % | 7.01 ± 4.27 | 8.56 ± 3.52 | 5.11 ± 4.36 | <0.0001b |
| Procalcitonin, ng/ml | 0.7 ± 3.36 | 0.09 ± 0.3 | 1.39 ± 4.85 | <0.0001b |
| Creatinine, umol/L | 98 ± 126.81 | 90.02 ± 152.6 | 107.83 ± 84.41 | <0.0001b |
| PT, S | 15.19 ± 5.65 | 13.82 ± 0.91 | 16.87 ± 8.09 | <0.0001b |
| RDW, % | 12.81 ± 1.61 | 12.33 ± 0.99 | 13.38 ± 1.97 | <0.0001b |
| Urea, nmol/L | 7.53 ± 6.77 | 4.88 ± 4.2 | 10.8 ± 7.84 | <0.0001b |
| Glucose, mmol/L | 8.30 ± 4.26 | 6.91 ± 2.93 | 9.94 ± 4.95 | <0.0001b |
| AST, U/L | 42.63 ± 61.57 | 28.08 ± 19.82 | 60.56 ± 86.14 | <0.0001b |
Note: Continuous variables are presented as mean ± standard deviations; categorical variables are presented as frequency and proportion n (%).
a P-value was derived from Student’s t-test.
b P-value was derived from rank-sum test.
c P-value was derived from χ2 test.
Feature selection
In the discovery dataset, two covariates (age and gender) and 53 laboratory test markers (see Supplementary Figure 1) with sufficient numbers of replicates and examined in at least 80% of COVID-19 patients were included in the HTREE model. SWSFS identified 14 top biomarkers: LDH, white blood cell (WBC) counts, neutrophil (NEU), mean platelet volume (MPV), creatinine, lymphocyte (%), Hs-CRP, prothrombin time (PT), red blood cell distribution width (RDW), urea, AST, glucose, monocytes (%) and procalcitonin (see Figure 1 and Supplementary Figure 2A). Three-fold cross-validation using an internal validation dataset further verified the importance of the candidate prognostic biomarkers. All 14 selected biomarkers were ranked in the top 25, and 7 were in the top 10 in the internal validation dataset, as shown in Supplementary Figure 2B. The mean AUC of this model was 98.32% [95% confidence interval (CI): 0.98–0.99] in the internal training dataset, and 96.49% (95% CI: 0.93–1.00) in the internal test dataset. Further, the predictive forest model was validated in two external validation datasets. The AUCs in the external validation datasets from Huangshi and Wuhan Huoshenshan Hospital reached 99.76% (95% CI: 0.99–1.00) and 97.63% (95% CI: 0.97–0.98), respectively (Figure 2A and B), showing the stability of the HTREE model and reliability of the predictive value of the selected biomarkers.
Figure 2.
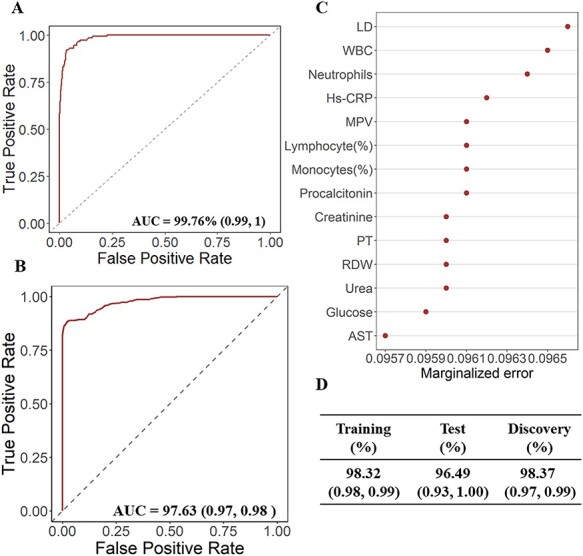
Performances of HTREE model and importance of 14 selected biomarkers. (A) Area under the receiver-operator characteristic curve (AUC) of the HTREE model including the 14 candidate biomarkers in the first validation dataset from Huangshi City. (B) Area under the receiver-operator characteristic curve (AUC) of the HTREE model including the 14 candidate biomarkers in the second validation dataset from Wuhan Huoshenshan Hospital. (C) Importance of the 14 selected biomarkers in the final model based on the discovery dataset. (D) Area under the receiver-operator characteristic curve (AUC) of the HTREE model including the 14 candidate biomarkers in discovery dataset and internal training and test sets using 3-fold cross-validation.
Joint model construction and assessment
With the linear mixed effect model in the joint modeling procedure, individualized prediction was made for longitudinal covariates using mean posterior fixed and random effects. Each participant would have predicted longitudinal measurements of biomarkers from hospital admission up to death or censoring. With the predicted biomarkers, we further made dynamic predictions on the risk of death for each individual since hospitalization. For example, the event-free probability curve of Patient A (survived) showed no apparent changes, whereas Patient B (died at the 32th day) showed considerable decline in event-free probability, indicating that Patient B had a higher risk of death, thus deserving frequent monitoring and close watching (see Figure 3). Dynamic prediction results for Patient B also indicated that long-term risk of death was higher in early visits but declined in later visits, which is in agreement with experiences obtained from medical practice. Predictive performance of the multivariate joint model is presented in Table 2. In particular, the time-dependent AUC is presented assuming a different prediction interval (5, 10 or 15 days), starting at a specific day (the 5th, 10th and 15th days after admission, respectively). The mean 5-day AUC was 87.61% (range: 83.43%–93.67%), mean 10-day AUC was 87.55% (range: 85.52%–88.95%) and mean 15-day AUC was 87.03% (range: 86.70%–87.29%) in the first validation dataset from Huangshi City. Meanwhile, the mean 5-day, 10-day and 15-AUC were 94.97% (range: 91.00%–97.86%), 95.78% (range: 93.58%–97.02%) and 94.63% (range: 93.48%–95.43%) in the second validation dataset from Wuhan Huoshenshan Hospital.
Figure 3.
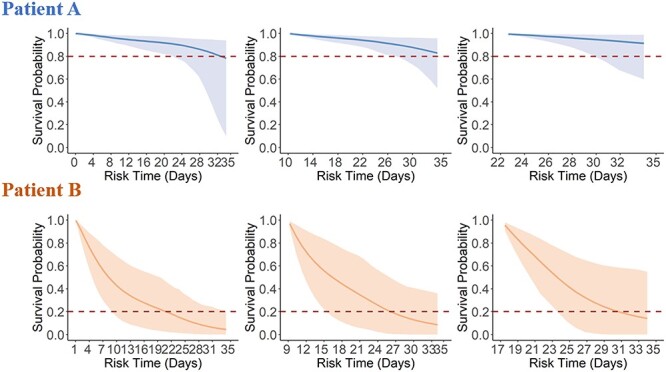
Dynamic predictions on the risk of death for Patient A and Patient B since hospitalization in the discovery dataset. Patient A had a total of seven laboratory test time windows and was discharged at the 35th day. The three graphs show survival probabilities of patient A at the follow-up visit 1th (day 0), visit 5th (day 10), and visit 7th (day 22). Patient B had a total of nine laboratory test time windows and was dead at the 32th days. The graphs show survival probabilities of patient B at follow-up visit 1th (day 1), visit 5th (day 11) and visit 8th (day 16).
Table 2.
Joint model results showing the 5-day, 10-day and 15-day areas under the receiver-operator characteristic curve (AUC) by different start follow-up time
| Start day | At-risk | 5-day AUC (%) | 10-day AUC (%) | 15-day AUC (%) |
|---|---|---|---|---|
| Discovery | ||||
| 5 | 258 | 94.87 | 95.36 | 92.82 |
| 10 | 171 | 93.37 | 89.29 | 84.57 |
| 15 | 97 | 78.20 | 69.78 | 79.47 |
| Mean | 88.81 | 84.81 | 85.62 | |
| Validationa | ||||
| 5 | 92 | 83.43 | 85.52 | 87.11 |
| 10 | 80 | 85.73 | 88.19 | 87.29 |
| 15 | 49 | 93.67 | 88.95 | 86.70 |
| Mean | 87.61 | 87.55 | 87.03 | |
| Validationb | ||||
| 5 | 1431 | 91.00 | 93.58 | 93.48 |
| 10 | 1035 | 96.03 | 96.73 | 95.43 |
| 15 | 746 | 97.86 | 97.02 | 94.99 |
| Mean | 94.97 | 95.78 | 94.63 |
Note: AUC: the areas under the receiver-operator characteristic curve.
Validation a: the first validation dataset from Huangshi City.
Validation b: the second validation dataset from Wuhan Huoshenshan Hospital.
At-risk: the number of COVID-19 patients who were still at hospital.
The predicted longitudinal measurements of patient A and patient B were characterized separately using smoothed curves (see Supplementary Figures 3). Biomarkers, excluding lymphocyte and monocytes, of Patient B were higher than those of Patient A. By fitting the changing trajectories of biomarkers over time for all patients, we found that patients who died and those who survived showed distinct patterns (see Figure 4 for the discovery dataset, Supplementary Figures 4 and 6 for the validation datasets).
Figure 4.
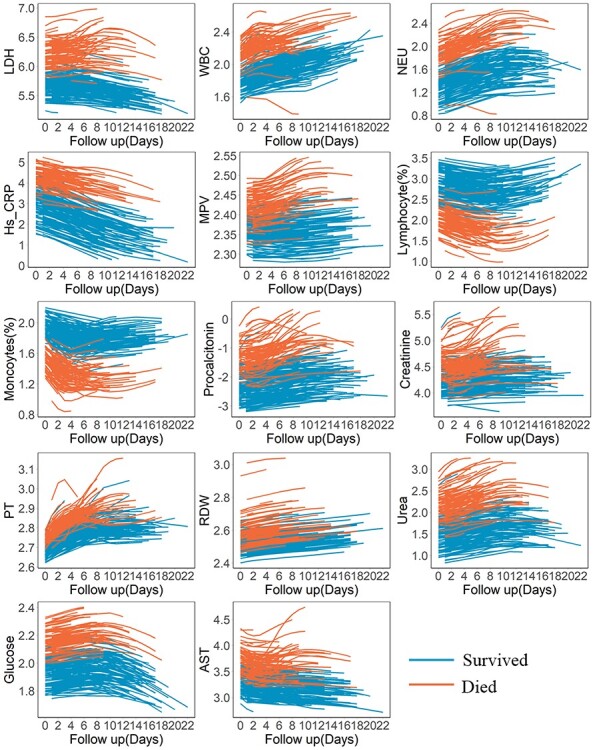
Fitting trajectory patterns of longitudinal biomarkers for patients who survived or deceased in the discovery dataset. Lines represent averaged trajectories of patients who survived (blue) or deceased (red) during hospitalization using natural cubic splines with two degrees of freedom.
Figure 5 shows how the effects of prognostic biomarkers on the event vary over time in the discovery dataset (see Supplementary Figures 5 and 7 for the validation datasets). The risk effects of NEU, Hs-CRP, MPV and urea gradually increased over time, whereas the effects of procalcitonin, PT and RDW declined. We observed relatively constant effects of LDH, WBC, creatinine, glucose and AST. Only lymphocyte and monocyte counts showed protective effects.
Figure 5.
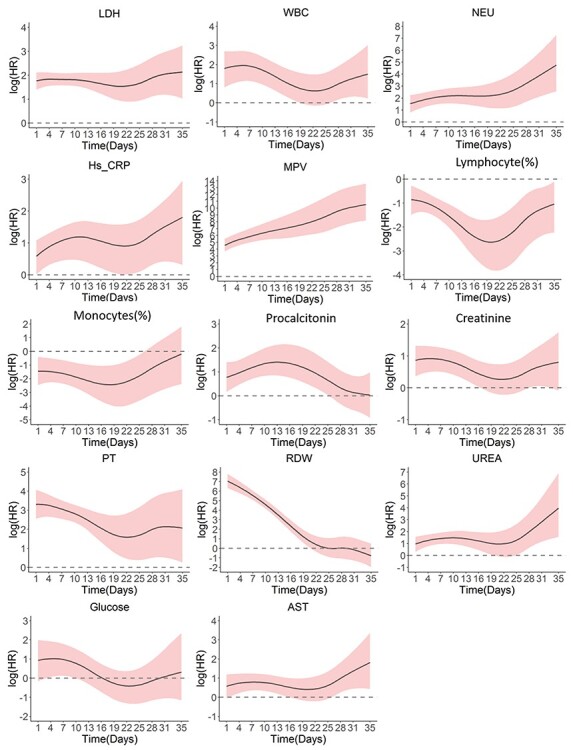
Time-varying effects of the biomarkers in the discovery dataset.
Web-based predictive tool
We developed an online tool to facilitate the application of our predictive model in practice (http://218.2.247.110:19040/COVID-19/Prediction). By inputting the values of prognostic factors of a COVID-19 patient, the tool would produce the conditional probability of death at specific time points.
Discussion
Laboratory tests during the hospitalization of COVID-19 patients are usually repeated at regular or irregular intervals. It is of the clinicians’ interest to make predictions of future outcomes based on all available information known up to a certain point of time. However, there is little research on dynamically prediction of COVID-19 prognosis based on repeated measurements and trajectories of laboratory biomarkers. In our study, we used the HTREE, an extension of random forests, to screen for biomarkers to predict the outcome of COVID-19 patients, thus boosting efficiency by incorporating information of longitudinal biomarker profiles [17]. The accuracy of our model was clinically satisfactory (98.37% of AUC in the discovery set, 99.76 and 97.63% of AUCs in two external validation datasets). We also used multivariate joint model to characterize the time-to-event process and obtain dynamic survival predictions at the individual level, providing useful and practical information to support individualized decisions for the treatment on COVID-19 patients. Also, time-varying associations between survival status and longitudinal biomarkers may be helpful for choosing timely treatment strategy that could improve prognosis.
We identified 14 important biomarkers among 53 clinical features, of which 12 were risk factors associated with the mortality of COVID-19 patients. LDH had the highest importance for predicting COVID-19 patient survival, consistent with previous studies [4, 18]. Decreased LDH level was related to the elimination of viral messenger RNA and correlated with shorter hospital stay, indicating better COVID-19 prognosis [19].
Like LDH, most of the remaining identified biomarkers (WBC, NEU, Hs-CRP, MPV, procalcitonin, creatinine, urea, AST, PT, RDW and glucose levels) are associated with poor COVID-19 prognosis. Leukocytosis and neutrophilia are hallmarks of acute infection [18, 20–22]. Hs-CRP and procalcitonin are commonly used inflammatory markers in the clinic, the increase of which may indicate poor prognosis in COVID-19 patients [23, 24]. It has been reported that high MPV level, related to thrombotic events in COVID-19, is an independent risk factor for disease progression [25, 26]. Further, concentrations of creatinine, urea and AST, related to myocardial injury, were markedly higher in COVID-19 patients who died than in those patients who survived [23, 27, 28]. Elevated procalcitonin and PT levels were reported to indicate the degree of severity for COVID-19 cases [29, 30]. RDW is a readily available laboratory measure reflecting the extent of anisocytosis, which is a proposed part of the routine panel of laboratory tests offered for monitoring COVID-19 patients [31, 32]. Glucose level is an independent risk factor for severe or critical COVID-19, and well-controlled blood glucose correlates with improved prognosis [33]. On the other hand, monocytes and lymphocyte, which play central roles in the maintenance of immune system function of humans and in protecting the body from virus infections, showed protective effects in our research [34, 35]. In this study, the numbers of monocytes and lymphocyte of the patients who survived were significantly higher than those who died. Moreover, lower lymphocyte levels, platelet counts, higher blood urea and neutrophils have been reported to be associated with neurological disorders which has been a public concern for COVID-19 survivors [36–38].
All beneficial and risk biomarkers identified in this study have been previously reported to be associated with the prognosis of COVID-19 patients. Our study extends the findings to show how the time-varying patterns of biomarkers can dynamically predict individual outcome, as well as inform timely and precise treatment recommendations. As an example, the clinician may need to pay attention to LDH changes during the whole course of treatment for its constant and strong risk effect, whereas it may be significant to prevent high level of NEU in the late stage of treatment. Our results provide the possibility for clinicians to adapt appropriate treatment timely by monitoring the change of easily available biomarkers [39].
The analytical strategy we applied in this study, which is a combination of machine learning method and joint modeling technique, provides insights on the comprehensive analysis on clinical data arising from personalized medicine and real-world circumstances, in particular longitudinal measurements provided by electronic health records [40]. Fully usage of longitudinal measurements can inform the probability of an outcome of interest occurring at a future time [41]. Specially, joint modeling is able to account for measurement errors of the biomarkers and model biomarkers’ trajectories over time. This strategy also enables the identification of potential interactions between nonlinear independent effects and time.
Our study has several strengths. Firstly, our study includes 1997 COVID-19 patients, which is one of the largest studies on COVID-19 prognosis. Secondly, we used an advanced machine learning technique, HTREEs, to identify important biomarkers. This method makes full use of longitudinal biomarkers and inherits the advantages of random forest algorithm, ensuring good stability and accuracy for further prediction [42, 43]. We also adopted multivariate joint model to characterize the time-to-event process, obtain dynamic predictions at the individual level and describe time-varying associations between the longitudinal biomarkers and the event. This facilitates the identification of critical time points and can alert the clinician when to apply patient-tailored therapies [44]. The performance of our final predictive model was also externally validated, achieving satisfactory AUC results in two independent datasets. Furthermore, we developed an online tool for clinicians to facilitate the application of our model.
However, our study also has several limitations. First, some patients had missing values for laboratory tests. Second, the mechanism of time-varying dynamic effects requires further investigation from a clinical perspective. Third, the prediction model of this study was trained and validated using Chinese population. Therefore, caution should be exercised when generalizing out findings to other race populations.
In conclusion, our study identified important biomarkers for early prediction on the outcome of COVID-19 patients by using a novel strategy suitable for dynamic risk prediction incorporating longitudinal laboratory measurements. The well-performing predictive model, with its accompanying online web application, is of great importance for the clinician to identify patients under high risk of death, as well as to characterize how the effects of biomarkers vary over time.
Key Points
Longitudinal data could provide more information on disease progression and the possibility of the dynamic prediction on probabilities of survival over time in COVID-19 patients.
Fourteen important biomarkers, including lactate dehydrogenase, white blood cell counts, neutrophil, mean platelet volume, creatinine, lymphocyte (%), hypersensitive c-reactive protein, prothrombin time, red blood cell distribution width, urea, aspartate aminotransferase, glucose, monocytes (%) and procalcitonin, were identified to be associated with the mortality of COVID-19 patients.
This research characterized the time-to-event process, obtained dynamic predictions at the individual level and demonstrated the time-varying associations between the candidate longitudinal biomarkers and the mortality in COVID-19 patients.
Our study provides a novel strategy suitable for dynamic risk prediction incorporating repeated measurements, which is very practical for clinical research.
Supplementary Material
Xin Chen is a PhD candidate at School of Public Health, Nanjing Medical University. Her research focuses on heterogeneous treatment effect for clinical research.
Wei Gao is a Professor at Sir Run Run Hospital, Nanjing Medical University. His research focuses on cardiovascular diseases.
Jie Li is a Master’s student at Department of Bioinformatics, Nanjing Medical University. His research focuses on cancer genomics.
Dongfang You is a PhD candidate at School of Public Health, Nanjing Medical University. Her research focuses on individual treatment effect for clinical research and random forest study.
Zhaolei Yu is a Master’s candidate at School of Public Health, Nanjing Medical University. His research focuses on simulating clinical trials using clinical big data.
Mingzhi Zhang is a Master’s candidate at School of Public Health, Nanjing Medical University. His research focuses on high order interaction effects using random forest.
Fang Shao is a Lecturer at School of Public Health, Nanjing Medical University. His research focuses on survival analysis, nonparametric regression model and statistical computation.
Yongyue Wei is an Associate Professor at School of Public Health, Nanjing Medical University. His research focuses on the theory and method of causal inference and disease prognosis.
Ruyang Zhang is an Associate Professor at School of Public Health, Nanjing Medical University. His research focuses on theoretical methods and clinical application of biomedical high dimensional data statistics.
Theis Lange is Professor at Department of Public Health, Faculty of Health and Medical Sciences, University of Copenhagen. His research focuses on the theory and method of causal inference.
Qianghu Wang is a Professor at Department of Bioinformatics, Nanjing Medical University. His research mainly focuses on bioinformatics and oncology genomics research.
Feng Chen is a Professor at School of Public Health, Nanjing Medical University, whose lab is interested in the statistical theories and methods of independent data, biomedical high-dimensional data and clinical research.
Xiang Lu is a Professor at Sir Run Run Hospital, Nanjing Medical University. His research focuses on hospital management and evidence-based medicine.
Yang Zhao is a Professor at School of Public Health, Nanjing Medical University, whose lab is interested in methods for integration and analysis of complex biomedical data, causal inference, statistical methods in clinical trials, real world research and biomedical big data and clinical trial data management.
Contributor Information
Xin Chen, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Wei Gao, Department of Geriatrics, Sir Run Run Hospital, Nanjing Medical University, 109 Longmian Avenue, Nanjing, 211166, China.
Jie Li, Research Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing, 211166, Jiangsu, China; Department of Bioinformatics, Nanjing Medical University, 101 Longmian Avenue, Jiangning District, Nanjing, 211166, Jiangsu, China; Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center for Personalized Cancer Medicine, Nanjing Medical University, Nanjing, 211166, Jiangsu, China.
Dongfang You, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Zhaolei Yu, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Mingzhi Zhang, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Fang Shao, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Yongyue Wei, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; China International Cooperation Center for Environment and Human Health, Center for Global Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; The Center of Biomedical Big Data and the Laboratory of Biomedical Big Data, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Ruyang Zhang, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; China International Cooperation Center for Environment and Human Health, Center for Global Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; The Center of Biomedical Big Data and the Laboratory of Biomedical Big Data, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Theis Lange, Section of Biostatistics, Department of Public Health, Faculty of Health and Medical Sciences, University of Copenhagen, Øster Farimagsgade 5, 1353, Copenhagen, Denmark.
Qianghu Wang, Research Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing, 211166, Jiangsu, China; Department of Bioinformatics, Nanjing Medical University, 101 Longmian Avenue, Jiangning District, Nanjing, 211166, Jiangsu, China; Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center for Personalized Cancer Medicine, Nanjing Medical University, Nanjing, 211166, Jiangsu, China.
Feng Chen, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center for Personalized Cancer Medicine, Nanjing Medical University, Nanjing, 211166, Jiangsu, China; China International Cooperation Center for Environment and Human Health, Center for Global Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; The Center of Biomedical Big Data and the Laboratory of Biomedical Big Data, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Xiang Lu, Department of Geriatrics, Sir Run Run Hospital, Nanjing Medical University, 109 Longmian Avenue, Nanjing, 211166, China.
Yang Zhao, Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center for Personalized Cancer Medicine, Nanjing Medical University, Nanjing, 211166, Jiangsu, China; China International Cooperation Center for Environment and Human Health, Center for Global Health, Nanjing Medical University, Nanjing, Jiangsu, China, 211166; The Center of Biomedical Big Data and the Laboratory of Biomedical Big Data, Nanjing Medical University, Nanjing, Jiangsu, China, 211166.
Author contributions
X.C., W.G., D.Y., F.C., X.L. and Y.Z. contributed to the study design. X.C., W.G., J. L., Y.D., Z.Y., M.Z., Q.W. and F.S. contributed to data collection. X.C., W.G. and D.Y. performed statistical analyses and interpretation and drafted the manuscript. Z.Y. developed the online application. J.L., M.Z., F.S., Y.W., R.Z., T.L., Q.W., F.C., X.L. and Y.Z. revised the manuscript. All authors approved the final version. Financial support and study supervision were provided by Y.Z., F.C., Y.W., W.G. and X.L..
Data Availability
In discovery phase, data are available from the published literature, which has been published online [4]. The relevant validation datasets are available from the corresponding author upon reasonable request.
Consent for publication
All authors have reviewed the manuscript and approved the final draft for publication.
Funding
This study was supported by the National Natural Science Foundation of China (81872709 to Y.Z., 82041024 to F.C., 81973142 to Y.W., 81970217 to W.G. and 81770440 and 81970218 to X.L.).
References
- 1. COVID-19 Coronavirus-Update . https://virusncov.com/ (1 April 2021, date last accessed).
- 2. Qiu H, Wu J, Hong L, et al. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis 2020;20:689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gao Y, Cai G-Y, Fang W, et al. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat Commun 2020;11:5033–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan L, Zhang H-T, Goncalves J, et al. An interpretable mortality prediction model for COVID-19 patients. Natre Mach Intell 2020;2:283–8. [Google Scholar]
- 5. Hu C, Liu Z, Jiang Y, et al. Early prediction of mortality risk among patients with severe COVID-19, using machine learning. Int J Epidemiol 2021;49:1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerig G, Fishbaugh J, Sadeghi N. Longitudinal modeling of appearance and shape and its potential for clinical use. Med Image Anal 2016;33:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maziarz M, Heagerty P, Cai T, et al. On longitudinal prediction with time-to-event outcome: comparison of modeling options. Biometrics 2017;73:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Os J. The dynamics of subthreshold psychopathology: implications for diagnosis and treatment. Am J Psychiatry 2013;170:695–8. [DOI] [PubMed] [Google Scholar]
- 9. Nelson B, McGorry PD, Wichers M, et al. Moving from static to dynamic models of the onset of mental disorder: a review. JAMA Psychiat 2017;74:528–34. [DOI] [PubMed] [Google Scholar]
- 10. He J, Wei Y, Chen J, et al. Dynamic trajectory of platelet-related indicators and survival of severe COVID-19 patients. Crit Care 2020;24:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li K, Huang B, Wu M, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun 2020;11:6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Z. Multiple imputation with multivariate imputation by chained equation (MICE) package. Ann Transl Med 2016;4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rizopoulos D. The R package JMbayes for fitting joint models for longitudinal and time-to-event data using MCMC. J Stat Softw 2016;72:1–46. [Google Scholar]
- 14. Rizopoulos D. Dynamic predictions and prospective accuracy in joint models for longitudinal and time-to-event data. Biometrics 2011;67:819–29. [DOI] [PubMed] [Google Scholar]
- 15. Simon N, Friedman J, Hastie T, et al. Regularization paths for cox's proportional hazards model via coordinate descent. J Stat Softw 2011;39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andrinopoulou ER, Eilers PHC, Takkenberg JJM, et al. Improved dynamic predictions from joint models of longitudinal and survival data with time-varying effects using P-splines. Biometrics 2018;74:685–93. [DOI] [PubMed] [Google Scholar]
- 17. Zhao L, Murray S, Mariani LH, et al. Incorporating longitudinal biomarkers for dynamic risk prediction in the era of big data: A pseudo-observation approach. Stat Med 2020;39:3685–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol 2020;146:110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J, Pan Y, Li G, et al. Distinguishing between COVID-19 and influenza during the early stages by measurement of peripheral blood parameters. J Med Virol 2021;93:1029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med 2020;217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J, Jiang M, Chen X, et al. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol 2020;108:17–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaur SP, Gupta V. COVID-19 vaccine: a comprehensive status report. Virus Res 2020;288:198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song Y, Gao P, Ran T, et al. High inflammatory burden: a potential cause of myocardial injury in critically Ill patients with COVID-19. Front Cardiovasc Med 2020;7:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu S, Du Z, Shen S, et al. Identification and validation of a novel clinical signature to predict the prognosis in confirmed COVID-19 patients. Clin Infect Dis 2020;71:3154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barrett TJ, Lee AH, Xia Y, et al. Platelet and vascular biomarkers associate with thrombosis and death in coronavirus disease. Circ Res 2020;127:945–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhong Q, Peng J. Mean platelet volume/platelet count ratio predicts severe pneumonia of COVID-19. J Clin Lab Anal 2021;35:e23607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol 2020;5:802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 2020;368:m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terpos E, Ntanasis-Stathopoulos I, Elalamy I, et al. Hematological findings and complications of COVID-19. Am J Hematol 2020;95:834–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henry BM, Benoit JL, Benoit S, et al. Red blood cell distribution width (RDW) Predicts COVID-19 severity: a prospective, observational study from the cincinnati SARS-CoV-2 emergency department cohort. Diagnostics (Basel) 2020;10:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goyal H, Lippi G, Gjymishka A, et al. Prognostic significance of red blood cell distribution width in gastrointestinal disorders. World J Gastroenterol 2017;23:4879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu L, She ZG, Cheng X, et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab 2020;31:1068–1077 e1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol 2020;20:355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang F, Nie J, Wang H, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis 2020;221:1762–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mao L, Jin H, Wang M, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 2020;77:683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mazza MG, De Lorenzo R, Conte C, et al. Anxiety and depression in COVID-19 survivors: role of inflammatory and clinical predictors. Brain Behav Immun 2020;89:594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taquet M, Geddes JR, Husain M, et al. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 2021;8:416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schumacher M, Hieke S, Ihorst G, et al. Dynamic prediction: a challenge for biostatisticians, but greatly needed by patients, physicians and the public. Biom J 2020;62:822–35. [DOI] [PubMed] [Google Scholar]
- 40. Bica I, Alaa AM, Lambert C, et al. From real-world patient data to individualized treatment effects using machine learning: current and future methods to address underlying challenges. Clin Pharmacol Ther 2021;109:87–100. [DOI] [PubMed] [Google Scholar]
- 41. Schachterle SE, Hurley S, Liu Q, et al. An implementation and visualization of the tree-based scan statistic for safety event monitoring in longitudinal electronic health data. Drug Saf 2019;42:727–41. [DOI] [PubMed] [Google Scholar]
- 42. Zhang H, Singer BH. Recursive Partioning and Applications. New York: Springer, 2010. [Google Scholar]
- 43. Yan J, Fine J. Estimating equations for association structures. Stat Med 2004;23:859–74 discussion 875-857, 879-880. [DOI] [PubMed] [Google Scholar]
- 44. Studerus E, Beck K, Fusar-Poli P, et al. Development and validation of a dynamic risk prediction model to forecast psychosis onset in patients at clinical high risk. Schizophr Bull 2020;46:252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In discovery phase, data are available from the published literature, which has been published online [4]. The relevant validation datasets are available from the corresponding author upon reasonable request.


