Abstract
Apurinic/apyrimidinic endonuclease 1 (APE1) is an essential multifunctional protein in mammals that plays critical roles in DNA repair and redox signaling within the cell. Impaired APE1 function or dysregulation is associated with disease susceptibility and poor cancer prognosis. Orchestrated regulatory mechanisms are crucial to ensure its function in a specific subcellular location at specific time. Here, we report arginine methylation as a post-translational modification (PTM) that regulates APE1 translocation to mitochondria in HeLa and HEK-293 cells. Protein arginine methyl-transferase 1 (PRMT1) was shown to methylate APE1 in vitro. Site-directed mutagenesis identified R301 as the major methylation site. We confirmed that APE1 is methylated in cells and that the R301K mutation significantly reduces its methylation. Baseline mitochondrial APE1 levels were low under standard culture conditions, but they could be induced by oxidative agents. Methylation-deficient APE1 showed reduced mitochondrial translocation. Methylation affected the interaction of APE1 with Tom20, translocase of the outer mitochondrial membrane. Methylation-deficient APE1 resulted in increased mitochondrial DNA damage and increased cytochrome c release after stimuli. These data suggest that methylation of APE1 promotes its mitochondrial translocation and protects cells from oxidative damage. This work describes a novel PTM regulation model of APE1 subcellular distribution through arginine methylation.
Keywords: APE1, Post-translational modification, Arginine methylation, PRMT1, Mitochondria
1. Introduction
Genetic material is constantly challenged by internal and external stressors [1]. To maintain genome stability, elements of the DNA repair system are required to function in a temporally and spatially coordinated manner to repair the DNA lesions [2]. Defective DNA repair can potentially lead to DNA mutations, which are widely considered as a driving force for cancer development and aging. DNA base lesions due to reactive oxygen species (ROS) produced from normal metabolic byproducts and deamination are main types of endogenous DNA damage [3]. Meanwhile, DNA base damage may also result from alkylation agents. Base excision repair (BER) corrects small lesions that do not significantly change DNA structure [4]; it is known as short-patch repair when only one nucleotide is replaced and long-patch repair, which is processed by a different group of proteins, when two to ten nucleotides are replaced [5,6]. In both cases, the damaged base is recognized and removed by a DNA glycosylase, leaving an abasic site (AP site), which is further processed by APE1, a critical enzyme that cleaves at AP sites [7–9]. APE1 specifically recognizes AP sites and uses its endonuclease activity to break the 5’-phosphodiester bond, thus generating a 3’-hydroxyl for subsequent gap-filling and ligation [10]. Souza-Pinto and colleagues have reported that BER is the major DNA repair system in mitochondria [11,12] and mitochondria are much more susceptible to oxidative damage compared to nuclei, due to their proximity to ROS, their lack of histones, and their limited choices of DNA repair pathways. Oxidative damage in mitochondria is implicated in the high rate of metabolism in cells/tissues in cancer, cardiovascular diseases, and neurodegenerative diseases [13–16].
APE1 is of interest, because it is a multi-functional protein, exhibiting both nuclease and transcriptional regulatory activities. Four nuclease activities have been reported for APE1, including AP endonuclease activity (3’-repair diesterase), 3’−5’ exonuclease activity, RNA cleavage activity, and nucleotide incision repair activity [17]. APE1 exerts its transcriptional activities through redox regulation of transcriptional factors like AP-1 and NF-κB or by trans-acting modulation of transcriptional factors, like negative Ca2+ response elements (nCaRE) [18,19]. Delicate and combinatorial regulatory mechanisms ensure that APE1 executes its activities correctly in response to environmental and physiological cues.
Here, we applied the concept of the PTM “barcode,” proposed by Benayoun and Veitia, to describe the regulation of APE1 [20]. According to this concept, combinations of PTMs control APE1 activities in different processes. This theory is supported by the currently known functional roles of APE1 PTMs. APE1 undergoes a wide range of PTMs from acetylation to ubiquitination, proteolytic cleavage, phosphorylation, nitrosylation, and glutathionylation. It has been suggested that PTMs may regulate the enzyme activities of APE1 and other aspects of its function, such as protein-protein interactions [17]. For instance, APE1 phosphorylation at residue T233 by the Cdk5 complex leads to decreased nuclease activity and increased vulnerability of cells to DNA damage [21]. K6/K7 lysine acetylation of APE1 by p300/SIRT1 decreases its nuclease activity and promotes the trans-acting modulation of transcriptional factors by APE1 [22,23]. Further, K24/K25/K27/K31/K32/35 residues of APE1 are acetylated [24,25] and ubiquitinated [26–28], presumably in a competing manner. Acetylation of APE1 stabilizes its binding to mRNAs and promotes its nuclear accumulation, whereas ubiquitination of APE1 has been reported to promote its nuclear exclusion and regulate its protein levels. Nitrosylation of APE1 at the C93/C310 residues regulates its intracellular localization [29]. There is PTM cross-talk between phosphorylation at T233 and monoubiquitination at K24/K25/K27 of APE1 [28]. Phosphorylation of APE1 by PKC and glutathionylation by glutathione are increased by oxidative stress [30,31]. The fact that APE1 harbors different modifications across different domains and there is cross-talk between the modifications, ensuring combinatorial regulation, provides strong evidence for PTM-barcoded regulation. Identification of novel PTMs and their regulatory roles in cells would give us a better understanding of the finetuned responses to stimuli, not restricted to APE1. A high-throughput screening study suggested that APE1 has the potential to be mono-methylated at R156 [32].
Arginine methylation is catalyzed by a group of enzymes, termed protein arginine methyltransferases (PRTMs) that utilize S-adenosyl methionine (SAM) as a methyl donor [33–35]. PRMT1, PRMT3, PRMT4, PRMT6 and PRMT8 are classified as type I PRMTs and catalyze the formation of asymmetrical di-methylation arginine (ADMA) [33,36,37], whereas PRMT5 and PRMT9 are classified as type II PRMTs and catalyze the formation of symmetrical di-methylation arginine (SDMA) [33,38,39]. PRMT7 is the only type III enzyme that catalyzes the formation of mono-methylation arginine by far [40,41]. PRMTs have diverse substrates among histone and non-histone proteins, suggesting that methylation may be part of the proposed PTM barcode, in layers of complex functional regulation mechanisms. Our previous work on FEN1 (Flap Endonuclease 1) showed that arginine methylation suppresses the phosphorylation of nearby residues and facilities PCNA binding [42]. In addition, there are other evidence that arginine methylation is implicated in signal transduction [43], DNA damage response, and DNA repair pathways [44,45].
In this study, we focused on the identification of arginine methylation sites in APE1 and characterization of its cellular functions complementing the APE1 PTM diagram. We show that APE1 is methylated at the R301 residue by PRMT1. Methylation of APE1 enhanced its interaction with the mitochondrial protein, Tom20, and stimulated the mitochondrial translocation of APE1 in cells exposed to oxidative agents. Methylation-deficient APE1 had reduced capacity for mitochondrial BER (mtBER) and increased susceptibility to oxidative agents. Our work contributes to a better understanding of how PTM regulates the subcellular distribution of APE1.
2. Materials and methods
2.1. Cell lines and materials
HeLa cells (ATCC, CCL-2) were used for APE1 arginine methylation detection in cells. HEK-293 (ATCC, CRL-1573) cells were used to construct APE1 site-specific knock-in (KI) cell lines.
Anti-APE1 (ab194), anti-PRMT1 (ab12189), anti-GAPDH (ab9485), anti-COX IV (ab16056), anti-TOM20 (ab56783), anti-Mia40 (ab87033), anti-mono methyl arginine and dimethyl arginine (ab412), and anti-β tubulin (ab6160) antibodies were from Abcam (Cambridge, UK); the anti-Histone H3 antibody was from Cell Signaling Technology (Danvers, MA, USA; 4499); anti-β-actin (GTX11003) anti-GFP antibodies were from GeneTex (Irvine, CA, USA; 26673); and anti-asymmetric dimethyl-arginine (ASYM24, 07-414), anti-symmetric dimethyl-arginine (SYM10, 07-412), and anti-mitochondrial HSP70 antibodies were from Upstate (Lake Placid, NY, USA; H1830-91A). M2-beads (M8823) and menadione were purchased from Sigma (Cas No. 58-27-5), while the 30% H2O2 solution was purchased from Sangon (Shanghai, China; Cas No. 7722-84-1).
2.2. Plasmids and recombinant proteins
Human APE1 sequence was PCR amplified from HEK293 cell cDNA. Primers are listed in Table S2. His-tagged APE1 was cloned to pET-14b plasmid. APE1 mutant plasmid were PCR generated using wild type pET-14b-His-APE1 as template with TianGen Fast HiFidelity PCR Kit (KP202) following standard protocol. Mutagenesis primers were indicated in Table S3. PCR products were digested with DpnI(NEB, R0176S) in 37 °C for 1 h. Digested products were transformed in DH5α strain. Clones were picked and subjected for sequencing to screen for mutated clones and to ensure APE1 sequence fidelity.
APE1 recombinant proteins were purified in BL21 strain using His-Tagged Protein Purification Kit (Soluble Protein) (CWBio, China, CW0894). Recombinant Histone H2A was purchased from Active Motif (31490).
PRMT1 (10350–50), PRMT4 (10750–50), and PRMT6 (10752–50) proteins were purchased from Cayman, Ann Arbor, Michigan. Tom20 protein (PRO-1471) was obtained from ProSpec, United States.
2.3. In vitro methyl incorporation assay
Methyl incorporation assays were performed, as described by Bikkavilli et al. [46], with minor modifications. Recombinant proteins (His-tagged APE1 and APE1 mutants) were purified using a GE-Ni Sepharose Fast Flow Kit, following a standard protocol (http://www.gelifesciences.com/handbooks, Affinity Chromatography Vol. 2: Tagged Proteins, Chapter 2). Purified proteins were incubated for 2 h at 25 °C with 1 μg of recombinant PRMT1 in 30 μl of methylation buffer (DTT 0.5 mM, EDTA 4 mM, PMSF 1 mM, Tris-Cl 20 mM, pH = 8.0), supplemented with 2 μl of S-adenosyl-l-methyl-3H methionine (3H-SAM, 0.03 mCi/mL, PerkinElmer, hot methylation for autoradiography) or SAM (NEB, #B9003S. It was prepared by mix of 0.005 M H2SO4 and 10% ETOH followed with filtration. cold methylation for mass spectrometry). Reactions were stopped by the addition of 2× SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE, followed by autoradiography or MS assay to identify methylation sites.
2.4. Cellular methylation assay
Cellular methylation assays were conducted as reported by Shen’s lab [42] and Li’s lab [47] with modifications.
HeLa cells were seeded to reach subconfluent (~50%) in the next day. Cells were chased with or without MS023 (30 nM) for 24 h before use. MS023 is typeⅠPRMTs inhibitor with distinguishable IC50s in vitro and in cells [48]. At the concentration of 30 nM, MS023 showed predominantly PRMT1 inhibition effect in cells. PRMTs inhibitor treatment would increase the methyl acceptable proteins in cells. When removed, proteins would show hypermethylated pattern in loading. Cycloheximide and chloramphenicol were used as protein synthesis inhibitor to prevent newly synthesized protein. Methyl group were added to methyl acceptable proteins accumulated during MS023 chasing. Immunoprecipitation using APE1 antibody was used to enrich APE1 to detect its methylation status.
Cells were cultured in 10 cm dishes and washed twice with 1×PBS. Replace medium with medium A (DMEM without L-methionine (Sigma), supplemented with 0.584 g/L L-glutamine (Gibico), 100 μg/ml streptomycin, 100 U/ml penicillin, and 10% FBS) for 30 min. Cycloheximide (100 μg/ml) and 40 μg/ml chloramphenicol were added to prevent protein synthesis. L-[methyl-3H] methionine (Perkin Elmer) (10 Ci/ml) were added to start the labeling. 4 h later, cells were lysed and subjected to immunoprecipitation of APE1 using anti-APE1 antibody. The precipitates or lysates were separated on 12% SDS-PAGE. Proteins were transferred onto PVDF membrane and subjected to autoradiography or western blot for APE1 loading control.
2.5. Generation of APE1 knock-in cell lines using CRISPR-Cas9 system
The PX459 plasmid from Feng Zhang’s lab was used to generate APE1 KI cell lines [49]. Three pairs of sgRNA sets were designed and optimized [50,51]. The oligonucleotides used in these experiments are listed in Supplementary Table S1. The single-stranded oligonucleotide (ssODN), 5′-TTGATTACTTTTTGTTGTCCCACTCTCTGTTACCTGCATTGTGACAGCAAGATCAAGTCCAAGGCCCTCGGCAGTGATCACTGTCCTATCACCCTAGGATCCTACCTAG-3′ (R301K mutation) was synthesized as the donor strand. SgRNA-plasmids were premixed (1:1:1) and cotransfected into HEK-293 cells with the donor, using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA, 1 μg: 0.1 nmol). At 24 h after transfection, cells were screened using puromycin for 72 h. After 48 h of recovery, cells were seeded in 10 cm dishes (200 cells/dish). Sixty clones were successfully picked for sequencing validation. Two R301K KI clones and eight partial knock-out clones were identified. HEK-293-R301K cell lines were used in APE1 mitochondrial localization experiments and cell survival assays after H2O2 treatment.
2.6. Subcellular fractionation
Microsomal, cytoplasmic, mitochondrial, and nuclear fractions were prepared using a mitochondria isolation kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Alternatively, we followed a published protocol to fractionate the cell lysates into nuclear, cytoplasmic, and mitochondrial fractions [52] with small modification. Briefly, HeLa cells were washed twice with 1×PBS and resuspended in 500 μl buffer A (10 mM Hepes, 10 mM KCl, 0.1 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 5 mM PMSF, pH = 7.9). Nuclei were collected after incubation for 10 min by centrifugation for 10 min at 500 g at 4 °C. The supernatant was used as cytoplasmic fraction in Fig. 2. The pellet was resuspended and incubated with 200 μl of buffer B (10 mM Hepes, 100 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 5 mM PMSF, pH = 7.9) for 20 min on ice. The supernatant was collected after centrifugation at 10,000 g at 4 °C for 20 min.
Fig. 2. Oxidative stress induces APE1 methylation and mitochondrial translocation.
In all co-IP assays, the loading percentage (to whole cell lysate) is 1% for input and 12.5% for IP samples. HeLa cells were treated with 100 μM H2O2 (a) or 10 μM menadione (b) for 2 h. An anti-arginine specific antibody was used to detect methylated APE1 (MeAPE1). (a) Immunoprecipitation of HeLa lysates with (right panel) or without (left panel) H2O2 treatment, using an anti-APE1 antibody. (b) Relative MeAPE1 levels compared to input showed significant elevation after H2O2 treatment. (p = 0.0126). (c) Immunoprecipitation of HeLa lysates with (right) or without (left) menadione treatment, using an anti-APE1 antibody. (d) Relative MeAPE1 levels compared to input showed significant increase after menadione treatment. (p = 0.024). (e) HeLa cells were treated with 100 μM H2O2 or 60 mM NAC (N-Acetyl-L-cysteine) for 2 h. Immunoprecipitation of HeLa lysates of untreated cells, H2O2 treated cells and NAC treated cells, using an anti-APE1 antibody. Anti-arginine specific antibody (ASYM24) was used to detect methylated APE1 (meAPE1) and anti-Tom 20 antibody was used to detect co-IPed Tom20. (f) bar chart for (e). Relative MeAPE1 levels showed that H2O2 treatment significantly increased compared to untreated group (p < 0.001). NAC treated group showed significant decrease comparing to untreated group (p < 0.001). (g) Subcellular fractionation of HeLa cells and immunoprecipitation of N (nucleus), C (cytosol), and M (mitochondria) fractions. The cytoplasmic protein extracts used for IP contained mitochondria from the nuclei-cytoplasm fractionation. Mitochondria was isolated in an independent experiment to achieve equivalent APE1 reading in western blot. GAPDH, cytosol control; mtHSP70 (mitochondrial HSP70), mitochondria control; Lamin A/C, nucleus control. (h) Bar chart for mitochondrial relative meAPE1 level in (g). p < 0.001.
For mitochondrial fractions, cells were washed once with 1 ml of grinding buffer (sucrose 250 mM, EDTA 2 mM, BSA 1 mg/ml, pH 7.4) and collected by centrifugation for 5 min at 800 g at 4 °C. The pellet was resuspended in 1 ml grinding buffer and lysed with 20 G needle/1 ml syringe by repetitive pipetting. The lysate was examined under microscope to ensure sufficient cell lysis (~60%). Discard the pellet after centrifugation at 800g at 4 °C for 12 min. The supernatant was immediately centrifuged for 20 min at 8500 g 4 °C. The pellet contains mitochondrial fraction and was washed with grinding buffer twice and resuspended with buffer S (sucrose 150 mM, KCl 40 mM, Tris-HCl 25 mM, BSA 1 mg/ml, pH 7.4) for storage or Western/IP cell lysis buffer (Beyotime Technology, P0013) for IP.
2.7. Pull-down assays and co-immunoprecipitation assays
Pull-down assays were performed using protein A/G beads purchased from Beyotime Technology (Jiangsu, China; Cat. P2012). We followed the standard protocol provided by the manufacturer with minor adjustments. Specifically, 2 μg antibody was used in each reaction. Protein A/G beads were immediately added to the protein-antibody solution and incubated at 4 °C overnight. Beads were washed with 1× PBS (diluted from 10× PBS, ST476, Beyotime Technology) at least 3 times before western blotting (WB) analysis.
2.8. APE1 endonuclease activity assay
The AP endonuclease activity of APE1 was determined as previously described with minor adjustments, using isotope [53] and fluorescence [54] labeling. 2 ng purified APE1 WT or mutant protein and 1 pmol annealed substrates were used in each reaction. Specifically, for isotope-labeled endonuclease assay [53], enzymatic reactions were carried out in a final volume of 20 μl. The protein were mixed with AP buffer to a final concentration of 50 mM Hepes, 50 mM KCl, 10 mM MgCl2, 1 μg/ml BSA, 0.05% (w/v) Triton X-100, pH = 7.5. 32P-labeled double stranded abasic DNA substrate (Table S2) obtained by (γ−32P) dATP phospho-labeling and subsequent annealing reaction. Reactions were started by incubating at 37 °C. 20min later, reactions were halted by the addition of 2×Stop Solution (96% (v/v) formamide, 10 mM EDTA, Xylene cyanol and bromophenol blue). Samples were separated onto 20% denaturing polyacrylamide gel and analyzed by autoradiography. The endonuclease activity was quantified by a gray scale ratio of cleaved products and loading substrates.
For fluorescence-based nuclease assay [54], the protein was incubated in a buffer comprising of 50 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 50 mM NaCl and 2 mM dithiothreitol (DTT) at 37 °C for 10 min. Reactions were initiated by addition of annealed FAM-Quencher labeled substrates (Table S2). Fluorescence readings were taken continuously by Tecan Infinite F200 Pro using ELISA program (495 nm Excitation, a 530 nm Emission, 30 s detection interval) for 25 min incubation at 37 °C. We used data when the relative fluorescent unit (RFU) and incubation time was linear regression to calculate APE1 endonuclease activity efficiency. We compared the slope of APE1-WT, R301K, R301F purified proteins in Fig. 4d.
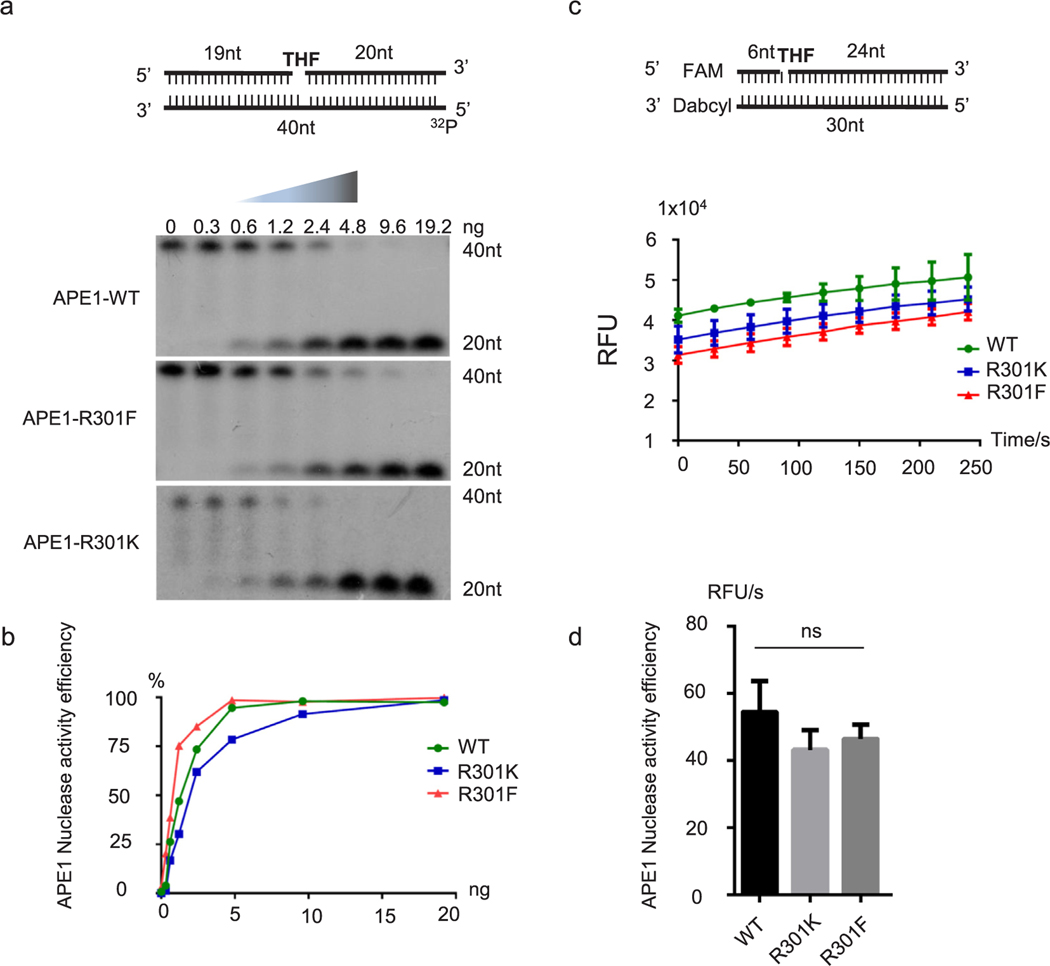
Fig. 4. Mutations at R301 do not affect nuclease activity of APE1.
Purified APE1, APE1-R301K, and APE1-R301F proteins were used in in vitro nuclease activity assays. (a) AP nuclease activity was assessed using the indicated substrate, with a 5’−32P label. Gradient concentrations were used and APE1 cutting efficiency was analyzed (b). A fluorescently labeled substrate was used in (c). Relative fluorescence units (RFU) were detected by Tecan Infinite F200 Pro. APE1 nuclease activity efficiency was quantified in (d).
2.9. Immunofluorescence and confocal assay
Immunofluorescence and confocal microscopy were conducted as reported [55] with adjustments. HEK293 cells and HEK293-APE1-R301K KI cells were used in this assay. The pCDNA6.1-APE1-R301F plasmid was transfected into R301K KI cells and labeled R301F group. Empty vectors were transfected into HEK293 cells and R301K KI cells, labeled APE1-WT and APE1-R301K group, respectively. Cells were grown on glass coverslips for 24 h to the desired confluency. The cells were treated with 200 μM H2O2 for 1 h and allowed cells to recover for 2 h before fixation. The cells were gently rinsed with 1×PBS and fixed with paraformaldehyde (3.7%) for 20 min at room temperature. After blocking with 3% fetal bovine serum for 1 h, primary antibodies against APE1, COX Ⅳ were incubated at 4 °C overnight and Alexa 488/568 (Invitrogen) secondary antibodies were used to detect. Cells were mounted in anti-fade solution with DAPI and examined under Zeiss LMS 880 using Airyscan confocal channel with 40×oil objective. Images were collected and processed using Zeiss blue software and sized in Adobe Photoshop CS5.0. The co-localization signal (green + red) beyond nucleus area was determined with Adobe Photoshop by measuring the fluorescence normalized to the number of cell count.
2.10. Analysis of mtDNA damage by quantitative PCR (qPCR)
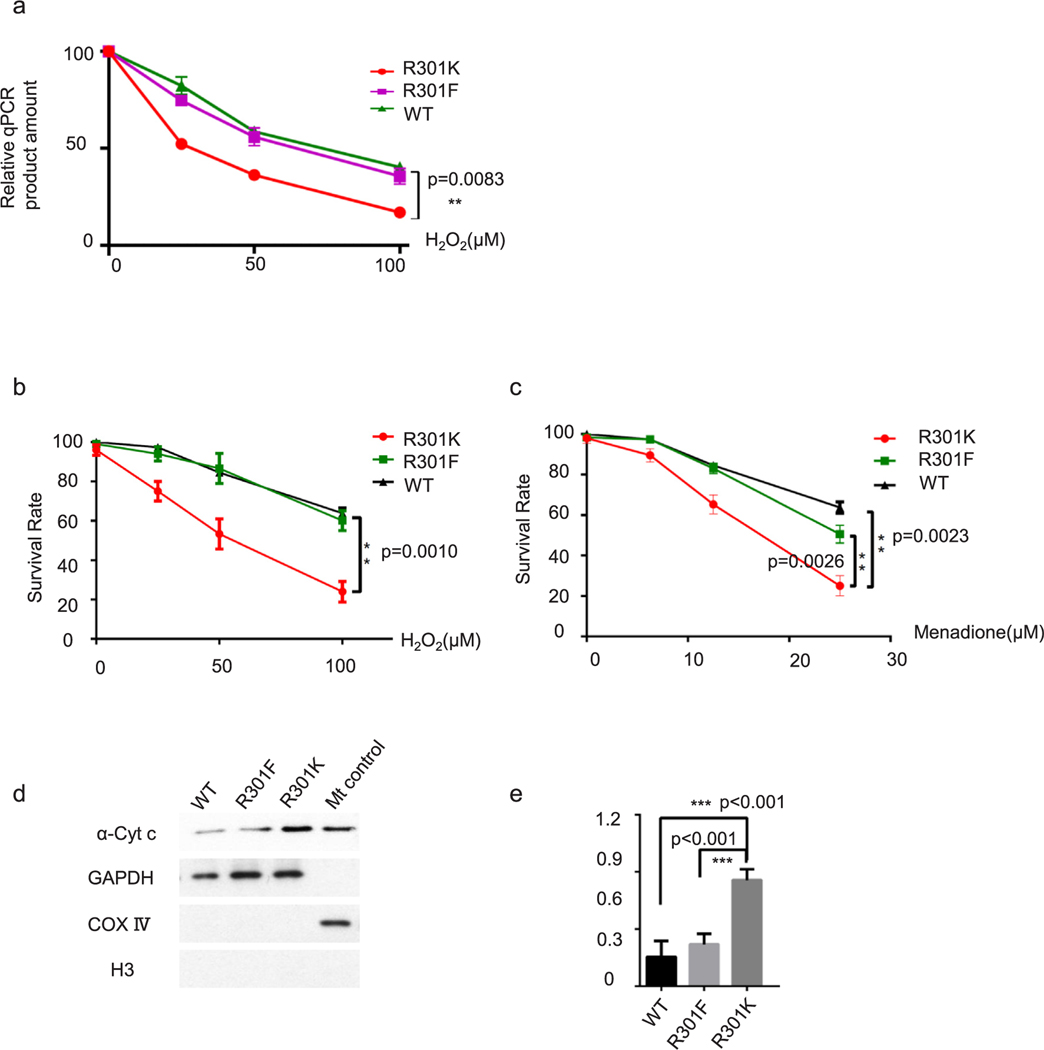
mtDNA damage was analyzed by qPCR, as described previously [56,57,70]. Many kinds of DNA lesions can slow down or block the progression of DNA polymerase. When equal amounts of DNA are qPCR amplified under identical conditions, DNA with more damage will get less PCR products. PCR conditions for β-globin (13.5 kb), large mtDNA (8.9 kb), small mtDNA and β-actin are Tm 64 °C 27 cycles, Tm 64 °C 19 cycles, Tm 60 °C 18 cycles, Tm 62 °C 18 cycles, respectively, to keep the PCR in the exponential phase.
Cells were transfected with APE1-WT, APE1-R301K, or APE1-R301F plasmids using Lipofectamine 2000. At 48 h after transfection, the cells were treated with H2O2 or menadione for 2 h. The cells were then washed three times and incubated for an additional 6 h. DNA was isolated using a Qiagen DNA Isolation Kit. PCR was performed using the following primers: mtDNA (8.9 kb), 5′-TCTAAGCCTCCTTATTCGAGCCGA-3′ (sense) and 5′-TTTCATCATGCGGAGATGTTGGATGG-3′ (antisense); mtDNA (221 bp), 5′-CCCCACAAACCCCATTACTAAACCCA-3′ (sense); β-globin, 5′-CGAGTAAGAGACCATTGTGGCAG-3′ (sense) and 5′-GCACTGGCTTAGGAGTTGGACT-3′ (antisense); β-actin, 5′-ACATCCGCAAAGACCTGTAC-3′ (sense) and 5′-TGATCTTCATTGTGCTGGGTG-3′ (antisense). Template DNA and PCR products were quantified using a Quant-iT™ PicoGreen™ dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA, P7589). We used kit provided DNA standards mixed with PicoGreen reagent (1:200, final concentration) to generate a standard curve (0–200 ng). Incubate diluted DNA template or PCR product with PicoGreen for 10 min at room temperature in the dark and read the fluorescence using Tecan Infinite F200 Pro using ELISA program (495 nm Excitation, a 530 nm Emission, shake for 20 s).
2.11. MTT cell survival assay
Cells were transfected with pCDNA6.1-APE1-WT, APE1-R301K, or APE1-R301F plasmids using Lipofectamine 2000. After 24 h, the cells were treated with H2O2 or menadione at various concentrations for 2 h. The cells were then allowed to recover for 8 h and incubated with 2.0 μg/mL MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazoliumbromide) for 1 h, followed by lysis in 20% SDS and 50% DMSO in 1× PBS buffer. The absorbance of samples was measured at 570 nm. MTT reduction in treated samples was normalized to MTT reduction in non-treated control samples.
2.12. Statistical analysis
We used student’s t-test for all comparisons between untreated and treated groups. We put three replicates of three independent data in Graphpad software. Data passed D’Agostino-Pearson omnibus normality test and F test. All data are presented as the mean ± SD from three independent experiments. P-values <0.05 were considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, n.s. = not significant).
3. Results
3.1. PRMT1 methylates APE1 in vitro
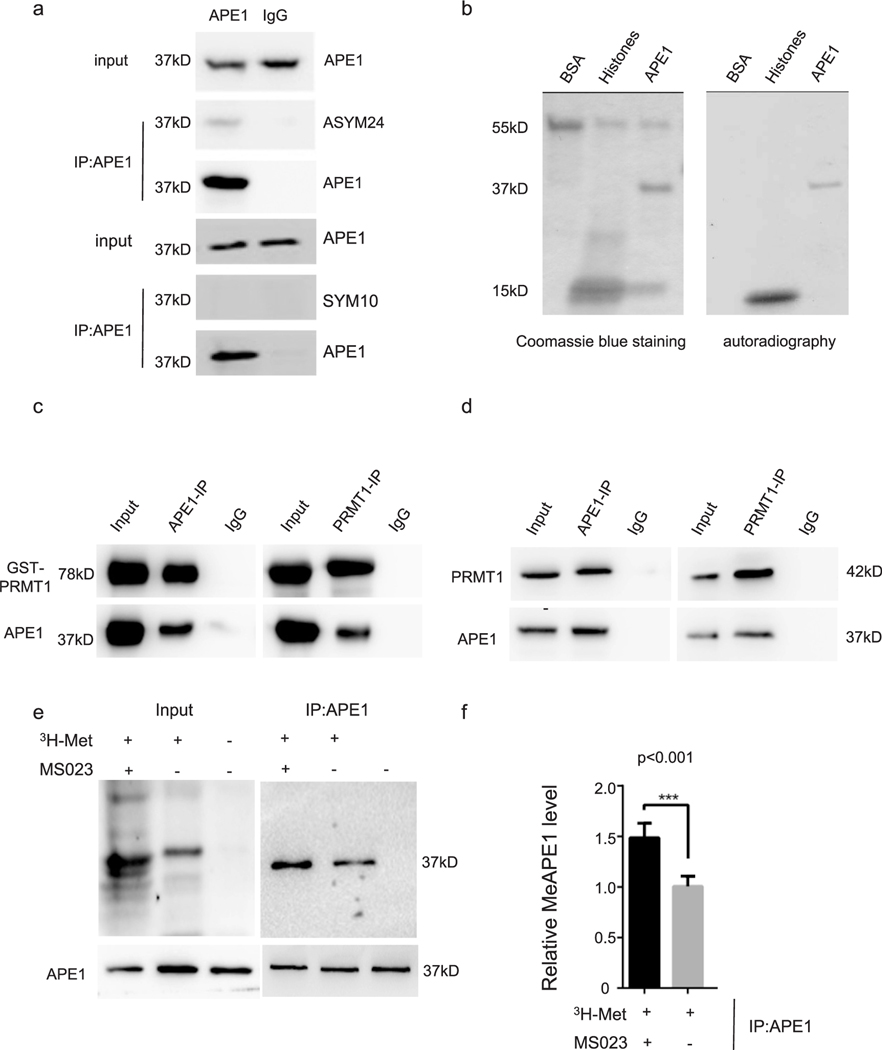
To determine whether APE1 is methylated at arginine residues, we immunoprecipitated endogenous APE1 from HeLa cells and immunoprecipitated samples were analyzed by western blotting for APE1 methylation, using pan-ADMA (ASYM24) and pan-SDMA (SYM10) antibodies. The asymmetric arginine methylation antibody detected a distinct band with the same molecular weight as endogenous APE1 in samples immunoprecipitated using the APE1 antibody, but not in those immunoprecipitated using control IgG (Fig. 1a upper panel). There was no detectable band when using the symmetric arginine methylation antibody (Fig. 1a lower panel).
Fig. 1. PRMT1 methylates APE1 in vitro.
In all co-IP assays, the loading percentage (to whole cell lysate) is 1% for input and 12.5% for IP samples. (a) Immunoprecipitates extracted from HeLa cells using an anti-APE1 antibody were detected using arginine methylation-specific antibodies. (b) In vitro methyl incorporation assay. Proteins were resolved by SDS-PAGE, stained with Coomassie blue (left panel), dried, and analyzed by autoradiography (right panel). BSA, negative control; Histone, positive control; (c) Co-immunoprecipitation assay of purified recombinant APE1 and PRMT1 proteins. Left panel, anti-APE1 antibody; right panel, anti-PRMT1 antibody. Anti-APE1 and anti-PRMT1 antibodies were used for western blotting. (d) Co-immunoprecipitation assay of APE1 and PRMT1 proteins in HeLa cell lysates. Left panel, anti-APE1 antibody; right panel, anti-PRMT1 antibody. Anti-APE1 and anti-PRMT1 antibodies were used for western blotting. (e) 3H labeled-methyl incorporation assay for HeLa cells with or without MS023 treatment, respectively. (f) Bar chart for (e) showing relative methylated APE1 comparing to cells without MS023 treatment (p < 0.001).
To determine which PRMT catalyzed APE1 arginine methylation, we used commercially available asymmetric arginine methyl transferases (PRMT1, PRMT4, and PRMT6; Cayman, Ann Arbor, Michigan; 10350–50, 10750–50, and 10752–50, respectively) to perform in vitro methyl incorporation assays for purified APE1 protein [46]. Methylated APE1 was detected in the PRMT1-mediated reaction (Fig. 1b), but not in the PRMT4-or PRMT6-mediated reactions (data not shown). As expected, recombinant histone H2A (Active Motif), which was used as a positive control, was methylated by PRMT1, but BSA, s a negative control, was not (Fig. 1b). To confirm that APE1 directly interacted with PRMT1, purified APE1 (0.5 μg) and GST-PRMT1 (0.5 μg) were mixed and co-immunoprecipitated (co-IPed) using an anti-APE1 (2 μg) or anti-PRMT1 antibody (2 μg), respectively. The western blotting analysis showed that APE1 and PRMT1 co-precipitated with each other in vitro (Fig. 1c).
We performed co-IP assay using HeLa cell lysate to investigate APE1 and PRMT1 interaction in cells. Co-IP of endogenous APE1 and PRMT1 from HeLa cell lysates showed that APE1 co-immunoprecipitated with PRMT1 (Fig. 1d). In Fig. 1e, cellular methylation assay was conducted as described in Materials and Methods section. At the concentration of 30 nM, PRMT inhibitor MS023 showed predominantly PRMT1 inhibition effect in cells. PRMTs inhibitor treatment would increase the methyl acceptable proteins in cells. When removed, proteins would show hypermethylated pattern in loading. IPed samples (Fig. 1e, right panel) showed that 3H-labeled APE1 was detected in cells and accumulated after MS023 chase for 24 h (Fig. 1f, p < 0.001). These data suggested that PRMT1 interacted with and methylated APE1.
3.2. Oxidative stress induces APE1 methylation and mitochondrial translocation
We observed a low level of APE1 arginine methylation in HeLa cells under normal culture conditions (Fig. 1a). Because APE1 is a critical enzyme involved in BER and redox regulation, we hypothesized that oxidative stress induces the arginine methylation of APE1. To test this hypothesis, we treated HeLa cells with the oxidative agents H2O2 and menadione. H2O2 is a common oxidative damage agent and menadione is a mitochondrial-specific damage agent, inducing ROS through futile redox cycling [74]. We observed that H2O2 (Fig. 2a, b) and menadione (Fig. 2c, d) treatment elevated APE1 arginine methylation levels in HeLa cells. We treated HeLa cells with 100 μM H2O2 or 60 mM NAC (N-Acetyl-L-cysteine) an anti-oxidant to evaluate APE1 methylated level (Fig. 2e). H2O2 treated cells showed increased meAPE1 compared to untreated cells (Fig. 2f, p < 0.001). NAC treated cells showed significantly less meAPE1 comparing to untreated cells (Fig. 2f, p < 0.001). This suggest that oxidative stress triggered by both exogenous and endogenous oxidants contribute to APE1 methylation.
APE1 is predominantly localized in the nucleus, but also has been detected in the cytoplasm and mitochondria. Considering the proportion of methylated APE1 and the fact that both H2O2 and menadione treatment cause mitochondrial damage, we assessed the correlation between APE1 methylation status and its subcellular localization (Fig. 2g, h). Methylated APE1 was detected in both the cytoplasm and mitochondria, but predominantly in the mitochondria. Surprisingly, no methylated APE1 was detected in the nucleus. Comparing the ratio of methylated APE1 to total APE1 for H2O2-treated cells and non-treated cells, methylated APE1 was shown to increase after treatment (Fig. 2b, d). These results suggested the possible association between APE1 arginine methylation and its mitochondrial translocation.
3.3. Identification and validation of methylation sites
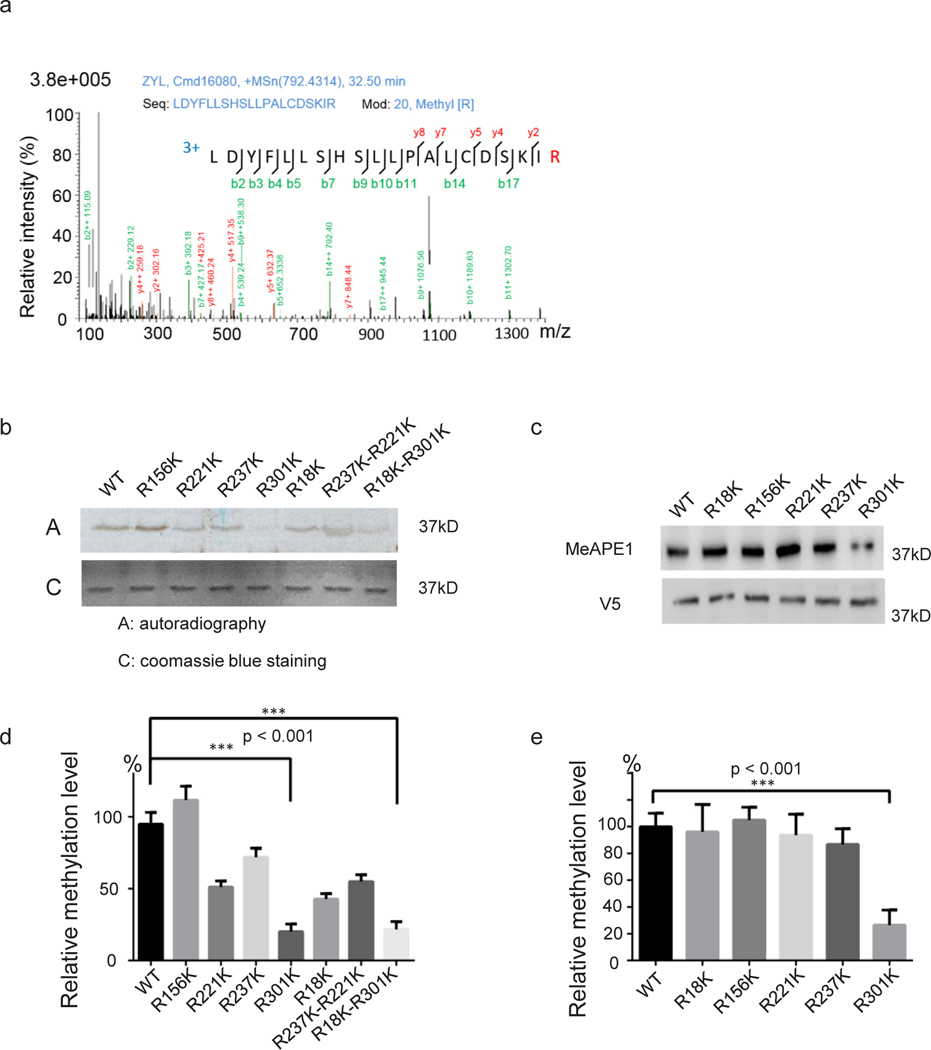
To identify which arginine residues were methylated, we conducted APE1 in vitro methylation reaction as stated in “Method and materials” section using PRMT1 and SAM (cold reaction). Methylation products were subjected to SDS-PAGE analysis and stained with coomassie brilliant blue R250. After destaining, APE1 band at 37 kDa was cut out and subjected to ESI-LC-MS/MS analysis [42,58]. R18, R156, R221, R237, and R301 residues were identified as potential methylation sites (Table S1, and Supplementary Fig. S1).
Next, we performed a mutagenesis study, constructing a series of individual or combined mutants at these arginine residues to confirm that they are indeed arginine methylation sites. In vitro methyl incorporation assays (hot reactions) were performed to determine if APE1 mutations cause methylation defects. We found that R301K led to a significant reduction in methylation levels (Fig. 3b). However, the R18K, R156, R221, and R237K mutations and the combined R221K-R237K mutation caused minimal reduction in methylation levels (Fig. 3b and Fig. 3d). This suggests that the R301 residue is the primary methylation site of APE1. To determine if the R301 residue is the main methylation site in cells, we transfected HeLa cells with His-V5 tagged APE1 plasmids expressing His-V5-tagged wild-type (WT) or R18K, R156, R221, and R237K, R310K APE1. Consistent with in vitro methyl incorporation data, His-V5 tagged APE1 R301K had significantly less methylation than WT APE1 (Fig. 3c and Fig. 3e). These data suggest R301 is the predominant methylation site for APE1.
Fig. 3. Identification and validation of APE1 arginine methylation site.
(a) Mass spectrometry profile of methylated APE1-R301 site. The mass spectrometry profiles of other detected arginine methylation candidates are shown in Supplementary Fig. S1. (b) Mutation of the methylation sites individually or in combination, resulted in reduced PRMT1 methylation of APE1 in vitro. APE1 proteins (wild type and mutants) were methylated in vitro using 3H-SAM as the methyl donor, separated on SDS-PAGE, and detected using Coomassie blue staining (bottom panel) or autoradiography (top panel). Quantitative results are shown in (d). (c) V5-His tagged wild-type APE1 or APE1 mutants harboring mutations at five arginine methylation sites were transfected into HeLa cells. Immunoprecipitates of HeLa cell lysates using an anti-V5 antibody were detected using an arginine methylation-specific and anti-V5 antibodies. Bar chart (e) shows relative methylation levels compared to wild-type APE1.
3.4. Mutations at R301 do not affect APE1 nuclease activity
Arginine methylation modification tends to make the arginine residue more hydrophobic for the bulky methyl group created steric hindrance. Alternatively, methylation can enhance van der Waals or London forces interaction. Phenylalanine(F) mutation introduced hydrophobicity has been used to partially mimic methylated arginine residues [42,59–62]. Since F cannot provide the positive charge that methyl-arginine has, R to F mutation mimicking arginine methylation was tested functionally in comparison with wild type and methylation deficiency cells (Figs. 5–7).
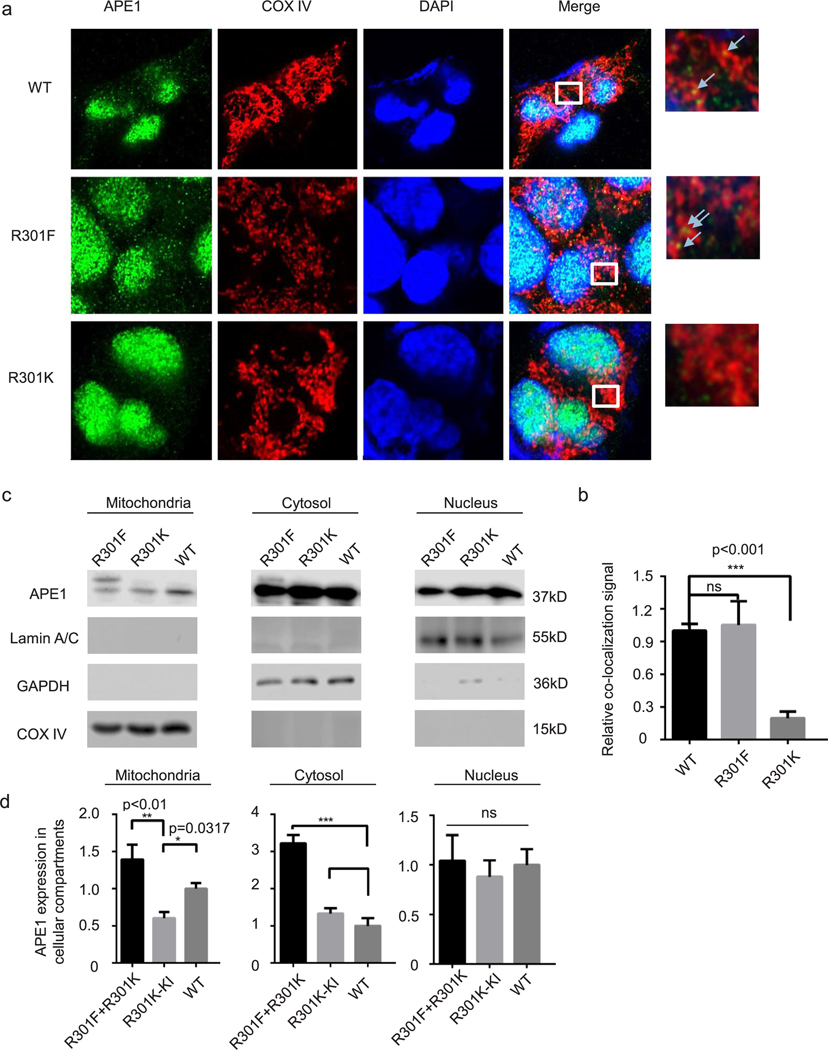
Fig. 5. APE1 methylation deficiency decreases mitochondrial translocation.
HEK-293 and APE1-R301K knock-in cell lines were used in this experiment and sequencing verification data are shown in Supplementary Fig. 2. An R301F-APE1 (methylation mimic) plasmid was transfected into the APE1-R301K knock-in cell line. Empty vector was transfected into HEK-293 and R301K knock-in cells. (a) Immunofluorescence assays were performed to assess the co-localization of APE1 proteins and mitochondria. The anti-APE1 antibody stained green, COX IV stained mitochondria, and DAPI stained nuclei. Merged yellow dots outside of the nucleus indicate APE1 localized in the mitochondria and were counted for quantification (b) (n = 20 cells). (c) Western blotting analysis of subcellular fractions obtained using a Qiagen Mitochondrial Isolation Kit. Lamin A/C, nuclear control; GAPDH, cytosolic control, COX IV, mitochondrial control. The relative APE1 level was determined for each cellular compartment (d).
Fig. 7. Methylation deficient mutation sensitizes cells to oxidative treatment.
(a) A mitochondrial DNA integrity assay was performed using qPCR. PCR products were quantified using a Quant-iT™ PicoGreen™ dsDNA Assay Kit. Survival assay for HEK-293-R301K cells transfected with wild-type APE1-expressing, R301F APE1-expressing, and vector plasmids after gradient H2O2 (b) and menadione treatment (c). (d) A cytochrome c release assay was performed to detect mitochondrial impairment after H2O2 (50 μM) treatment for 2 h. GAPDH, cytosolic marker; COX IV, mitochondrial marker; H3, nucleus marker. (e) Bar chart for cytochrome c release assay. R301K cells showed significantly more cytochrome c in cytosolic fraction comparing to WT (p < 0.001) or R301F (p < 0.001) mutation, respectively.
To investigate the role of R301 methylation of APE1, we tested whether wild type, unmethylatable mimetic R to K or methylation mimetic R to F affected the endonuclease activity, redox activity, or other biological functions of APE1. Methylation status had no obvious effects on APE1 redox activity, as predicted, since the R301 residue is in the nuclease domain. Surprisingly, there was no significant difference in APE1 endonuclease activity between WT APE1 and either the R-to-K or the R-to-F mutant (Fig. 4). As shown in Fig. 4a, 32P-labeled substrates with tetrahydrofuran (THF) were used to mimic AP sites. If APE1 nuclease activity was intact, there would be a 20-nt fragment cleaved from the substrate. There was no significant difference in the cleavage between WT and R301K or between WT and R301F APE1 (Fig. 4a–4d). Fig. 4c illustrates nuclease assays that were performed using fluorescently labeled substrates. Detailed methods are defined in Methods and Materials section. Consistent with the isotope-labeled method, there was no significant difference in nuclease activity between mutant and WT APE1 (Fig. 4d). In light of the recent available APE1 crystal structure from Whitaker et al. [63], we compared APE1 R301 methylation form with wild type in the context of endonuclease or exonuclease substrates binding. Methylation of R301 didn’t affect the binding active site structure. This is consistent with our in vitro endonuclease assays.
Since there was no significant difference found in the in vitro biochemical assays, we investigated indirect effects at the cellular level caused by potential protein-protein interactions, neutralized positive charge, or even interplay with other PTMs. Li et al. reported that an R-to-A mutation at R301 (and K299) in the APE1 C-terminal peptide diminishes mitochondrial targeting potential, possibly due to the loss of positively charged status [52]. Despite the fact that a partial peptide was used in their research, instead of the full-length APE1 protein, the authors failed to investigate APE1 mitochondrial translocation when the positive charge was maintained using an R-to-K mutation. Though positive charges may play an important role in this process, we propose that methylation and its associated protein-protein interactions may be also critical.
3.5. APE1 methylation deficient mutation decreases mitochondrial translocation
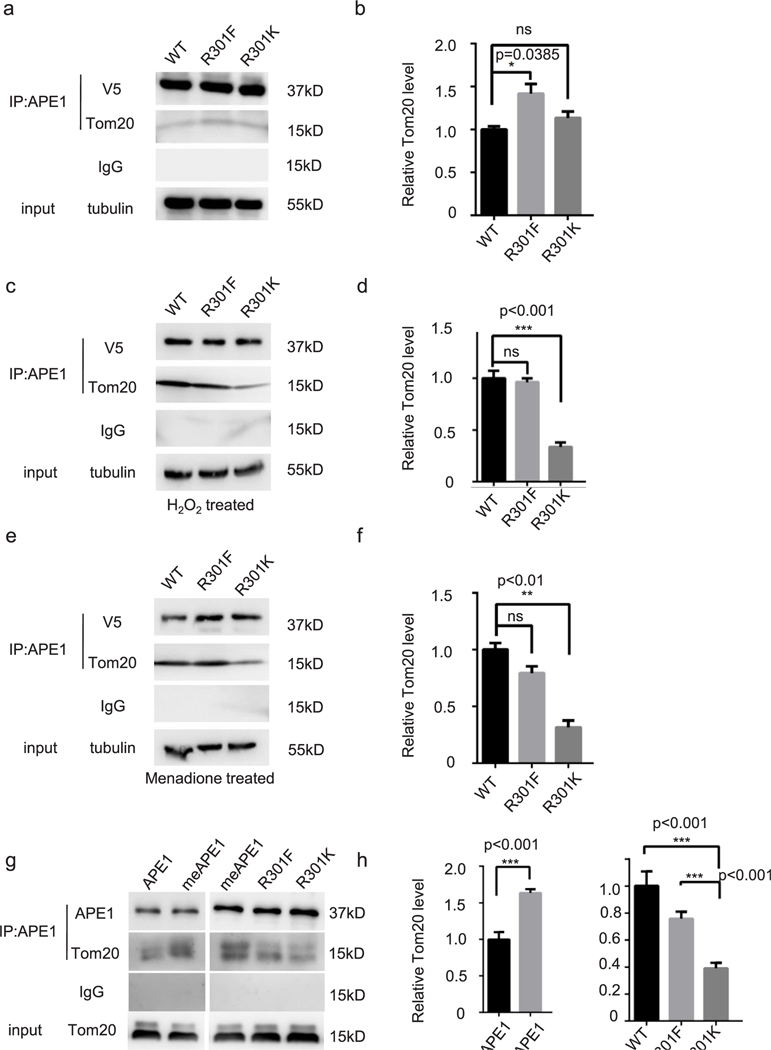
Next, we determined the effects of APE1 methylation on mitochondrial localization, since the R301 residue was previously found to be important for APE1 mitochondrial targeting [52]. To address this issue, we constructed HEK-293-R301K (arginine methylation deficient, but provide positive charge like arginine and methyl-arginine) KI cell line using the CRISPR/Cas9 system. We transfected R301F plasmid into R30K-KI cell line. Control plasmid are transfected into HEK-293-R301K-KI cells and WT-HEK-293 cells. As shown in Fig. 5a, we used anti-APE1 and anti-COX IV antibodies to identify APE1 localization and mitochondria, respectively. The relative localization of the arginine methylation mimic APE1 and R301K APE1 to the mitochondria were significantly different (Fig. 5b). We further investigated the mitochondrial localization of WT, R301F, and R301K APE1 using cellular fractionation and WB. In the mitochondrial fraction, we observed significantly less APE1 in methylation defect mutation group comparing to wild type group and transfected R to F APE1 mutant compensates it (Fig. 5c and Fig 5d). Intriguingly, there was significantly more R301F APE1 than WT or R301K APE1 in the cytoplasmic fraction (Fig. 5c and Fig. 5d). Cellular fractionation experiments did not detect any differences in the amount of WT, R301F, or R301K APE1 proteins in the nucleus (Fig. 5c and Fig. 5d).
Next, we aimed to define how APE1 methylation stimulates its mitochondrial localization. It has been reported that the mitochondrial outer membrane translocase, Tom20 and the mitochondrial inner assembly protein, Mia40 are involved in APE1 mitochondrial translocation [52,53]. Thus, we tested if the methylation of APE1 enhances its interaction with Tom20. HEK-293 cells were transfected with a plasmid encoding V5-tagged APE1 (WT, R301F, or R301K). We conducted co-IP experiments to determine if Tom20 was pulled down with V5-tagged APE1. We observed only a small amount of Tom20 that was co-pulled down with WT or R301K APE1 in cells under normal culture conditions (Fig. 6a). The amount of Tom20 that pulled down with R301F-APE1 was significantly greater than the amount pulled down with WT or R301K APE1 (Fig. 6b). This result supported our previous conclusion that cells maintain a low level of methylated APE1 in normal culture conditions. In cells treated with H2O2 (Fig. 6c, d) or menadione (Fig. 6e, f), a considerable amount of Tom20 was pulled down with WT and R301F APE1, but not with R301K APE1. This suggests that oxidative stress induced APE1 R301 methylation and the interaction of APE1 with Tom20.
Fig. 6. Methylation affectes the APE1-Tom20 interaction in HEK-293 cells and in vitro.
In all co-IP assays, the loading percentage (to whole cell lysate) is 1% for input and 12.5% for IP samples. Immunoprecipitation using an anti-V5 antibody in HEK-293-R301K cells transfected with V5-tagged wild type APE1-, R301F APE1-, and R301K APE1-expressing plasmids. Cells were maintained under standard culture conditions (a) or treated with H2O2 (200 μM for 1 h) (c)/menadione (10 μM for 1 h (e) to increase APE1 arginine methylation. Bar chart for relative pulled-down Tom20 protein in normal cells (b), H2O2-treated cells (d), and menadione-treated cells (f). Co-IP assay for APE1 and Tom20 purified proteins using anti-APE1 antibody (g). APE1 was methylated through in vitro methyl incorporation assay (g, second and third lane). APE1 samples in the first lane, fourth lane and fifth lane omitted SAM during in vitro methyl incorporation assay. (h) Bar chart for relative pulled down Tom20 in co-IP assays.
We conducted co-IP assay for purified APE1 proteins and Tom20 protein (ProSpec, cat. PRO1471) to address whether methylation status affects their interaction. In Fig. 6g left panel, we did APE1 in vitro methyl incorporation assay before incubating APE1 and Tom20 with anti-APE1 antibody. In the sample loaded on the first lane, SAM was omitted from the reaction to get unmethylated sample while for the second lane sample we used methylated APE1. The amount of IPed Tom20 of methylated APE1 is significantly more than unmethylated APE1 (Fig. 6g, h). In the right panel, APE1 wild type, R301F and R301K mutant protein were used in methyl incorporation assay and then subjected to co-IP assay with Tom20 protein. Methylated APE1 and R301F mutant showed more interaction with Tom20 than R301K mutant (Fig. 6g, h).
Taken together these data demonstrated that methylated APE1 preferentially interacts with Tom20 protein.
3.6. Methylation deficient mutation sensitized cells to oxidative treatment
Since APE1 is a critical enzyme that participates in BER, it is expected that mtDNA integrity would be compromised when APE1 mitochondrial translocation is perturbed, especially under oxidative stress challenge. MtDNA damage may cause mutations in mitochondrial-derived proteins, which would affect electronic chain reactions or membrane potential. These effects, in turn, would further impair mtDNA integrity, forming a vicious cycle.
To test this hypothesis, we measured the frequency of mtDNA damages using qPCR [57]. We harvested HEK-293-APE1-R301K cells and HEK-293-APE1-R301K cells transfected with R301F APE1 or wild type APE1, after challenging the cells with different concentrations of H2O2 or menadione. After DNA extraction, qPCR was performed to evaluate mtDNA integrity, with low PCR efficiency associated with increased DNA damage. Quantification of PCR products using Pico Green (Thermo Fisher Scientific, Waltham, MA, USA) indicated damage to mtDNA. As shown in Fig. 7a, cells expressing R301K mutant APE1 had significantly more mtDNA damage after H2O2 treatment than cells expressing R301F mutant or WT APE1 (Fig. 7a). As shown in Fig. 7b and c, cells expressing APE1 R301K exhibited significantly lower survival than cells expressing WT APE1, when treated with H2O2 or menadione. The APE1 R301F mutation rescued (Fig. 7b), either fully or partially, cells from oxidative damage.
The APE1 R301K mutation increased the frequency of mtDNA damages and impaired the mitochondrial membrane potential, which is catastrophic for cells with respect to sustaining metabolism. An apoptotic stimulus triggers cytochrome c release from the mitochondrial inter membrane space to the cytosol. A cytochrome c release assay was used to detect mitochondria-related apoptosis and assess membrane potential integrity. Mitochondrial-free cytosolic fraction was obtained during mitochondria hypertonic purification process. As shown in Fig. 7d, cells expressing R301K mutant APE1 had more cytochrome c in the cytosolic compartment compared to cells expressing WT APE1. The expression of R301F APE1 partially counteracted this effect.
These data suggest that R301F and WT APE1 partially rescued cellular sensitivity to oxidative damage caused by arginine methylation deficiency in HEK293 cells.
4. Discussion
APE1 has been detected in the cytoplasm and in mitochondria, but it is predominantly located in the nucleus where DNA repair and transcriptional regulation activities take place [10]. Abnormal subcellular translocation patterns of APE1 are associated with disease susceptibility and poor prognosis in cancer [24,64,65]. Therefore, it is of interest to gain a better understanding of the mechanisms that regulate APE1 localization. In this study, we identified arginine methylation as a novel PTM of APE1 and demonstrated its role in regulating the mitochondrial translocation of APE1. Since BER is the main repair system in mitochondria, mtAPE1 levels are essential for the maintenance of mitochondrial genome stability, especially under oxidative stress [66]. Understanding how methylation regulates APE1 will help to complete our understanding of the post-translational regulation network of APE1, which may further be exploited for targeted manipulation in both research and translational applications.
4.1. PRMT1 methylates APE1 at R301
Methylation at arginine residues is an indispensable part of PTM. Its emerging role in coordinating non-histone proteins involved in DNA replication and repair suggests that arginine methylation fits in the general PTM network of DNA damage response (DDR) pathways [42,55,62]. PRMT1, the major arginine methyltransferase in mammalian cells, generates approximately 85% of all methylated arginine residues and catalyzes both mono- and di-methylated (asymmetric) arginine modifications [67]. The currently known substrates involved in DDR are MRE11, 53BP1, BRCA1, and Pol β; the first three proteins are required for double-stranded break (DSB) repair and Pol β is required for BER. Our work provides evidence that APE1 can now be regarded as a BER substrate of PRMT1, especially in mtBER.
As mentioned in the “Introduction” section, R156 was reported to be mono-methylated in a high-throughput methylation modification screening study [32]. Although it was one of our five candidate methylation sites, we ruled out R156 as a predominant site through in vitro and cellular methylation experiments (Fig. 3). PRMT1 directly interacted with APE1 and catalyzed asymmetric methylation at R301 (Fig. 1, Fig. 3). Unlike conventional PRMT1 substrates harboring the RG/RGG motif, APE1 arginine methylation at the sequence PALCDSKIRSKALG supported the hypothesis that PRMT1 selectively recognizes amino acid sequences beyond the RG/RGG paradigm [68].
Overexpression or siRNA-mediated downregulation of PRMT1 did not significantly affect APE1 methylation levels and chemical inhibition of PRMT1 using MS023 showed similar effects (data not shown). Methylated APE1 appeared to be at a baseline level under all three conditions. After oxidative treatment, APE1 arginine methylation and mitochondrial translocation significantly increased (Fig. 2). This suggests that APE1 arginine methylation acts as a stress-response strategy.
4.2. APE1 arginine methylation deficiency decreases its mitochondrial translocation
Although the PTM regulation mechanism underlying the subcellular distribution of APE1 is not conclusive, several studies have shed light on this subject. Vascotto et al. reported that the redox state of C65 could control APE1 translocation into the mitochondrial inner membrane space, by interacting with Mia40 [53]. A deletion analysis also showed that residues 64–80 likely harbor a nuclear exclusion signal (NES) and a hidden “MTS” at the C-terminal 69 amino acids, as previously described in the “Introduction” section. One study showed that S-nitrosylation of C93 and C310 residues after nitric oxidant treatment induces nuclear export [29]. Arginine residues are present at nucleic acid-binding domains and have been considered to have the potential to alter ionic and hydrogen bonds [69]. Methylation may increase the hydrophobicity of arginine [34].
The R301 residue is within the nuclease domain of APE1, but our data showed that methylation of this residue did not significantly affect the nuclease activity of APE1 (Fig. 4). However, APE1 methylation level was elevated under conditions of oxidative stress (Fig. 2) and we showed that arginine methylation promoted the mitochondrial translocation of APE1 (Fig. 5). However, R301K KI cells did not show an absolute abolishment of mitochondrial APE1 localization. The R301F mutant and wild type APE1 showed normal nuclear localization and increased mitochondrial translocation comparing to R301K mutant. This suggest R301F partially mimics arginine methylation as to APE1 mitochondrial distribution. There appeared to be a basal level of mitochondrial APE1, with an unknown source. Nuclear exclusion and mitochondrial targeting could both be implicated in this case. Taken together, there may be cooperating partners, an interacting protein, or “PTM crosstalk” involved in facilitating APE1 nuclear export and subsequent mitochondrial targeting. Further study of the co-existence and crosstalk of APE1 PTMs is warranted.
Further investigation revealed that the methylation of APE1 at R301 affected its interaction with the mitochondrial outer membrane translocase, Tom20 (Fig. 6). Consistent with the observation that methylated APE1 was maintained at a baseline level under normal conditions and oxidative treatment elevated methylated APE1 levels (Fig. 2), the APE1-Tom20 interaction showed a similar pattern (Fig. 2e, Fig. 6). Our work corroborates the body of data showing that PRMT1 affected protein-protein interactions and protein subcellular distribution [33,34].
4.3. mtAPE1 and cell sensitivity to oxidative stress
mtAPE1 has become an emerging focus in recent studies since mtDNA is more vulnerable than nuclear DNA (nDNA) to oxidative stress-induced damage [70]. Predominantly localized in the nuclei in most cells, mitochondrial APE1 levels are crucial for the repair of mtDNA. Although APE1 is up-regulated in both the nucleus and mitochondria, overexpression studies of nuclear/mitochondrial-targeted proteins showed that the latter had reduced sensitivity to H2O2. This may be due to the fact that nuclear APE1 is overcompensated in the nucleus. It has been reported that mtAPE1 suppresses phorbol 12-myristate 13-acetate (PMA)-induced mitochondrial dysfunction more effectively than general overexpression of APE1 [16]. The authors reported that APE1 levels in mitochondrial fractions were elevated within 1 h of PMA (an activator of protein kinase C [PKC]) treatment, and showed increased mitochondrial translocation. APE1 phosphorylation by PKC may be implicated in processes upstream of arginine methylation. Another possibility is that PKC-induced, mitochondrial p66shc phosphorylation-mediated ROS production triggers subsequent responses. This supports our proposal that there may be crosstalk among APE1 PTMs.
Accumulation of mtDNA mutations may be a critical factor in eliciting persistent mitochondrial defects and consequently, contributing to cancer initiation, progression, and metastasis [71]. However, increased mtDNA mutations can form a viscous cycle by perturbing the electron transport chain reaction, thus generating more ROS. APE1 function is tightly regulated within the mitochondrial compartment. Xie et al. reported that the anti-apoptotic molecule, oncoprotein Bcl2, suppresses mtDNA repair by directly interacting with mtAPE1 and subsequently inhibiting its endonuclease activities [70]. More than 90% of Bcl2 is localized in the mitochondria and this mitochondrial localization is thought to be required for the suppression of apoptosis [72,73]. However, it is not known whether this process promotes cancer progression. Methylation-deficient mutants of APE1 display reduced mitochondrial targeting, resulting in the accumulation of mtDNA damage, increased cytochrome c release, and increased sensitivity to oxidative damage agents (Fig. 7).
A recent study has shown that overexpression of GADD45a sensitizes radioresistant cervical cancer by decreasing the cytoplasmic distribution of APE1. Li et al. suggested that GADD45a inhibits the production of nitric oxide (NO), a nitrosylation signal of APE1 that serves as a nuclear export stimulator. There has been extensive research on the correlation between cytoplasmic levels of APE1 and cancer progression and survival. However, the mechanisms responsible for the abnormal phenotype, with an emphasis on APE1 mitochondrial localization, have not been investigated in detail due to limited number of samples, relatively low detection resolution, and inability to perform high-throughput screening. This is an urgent matter to resolve through the development of cutting-edge techniques, such as proximity ligation assays and high-resolution Airyscan confocal microscopy.
In summary, APE1 has always been an intriguing target for novel cancer therapeutic strategies as an effective adjuvant, due to its essential role in DNA repair and its distinctive expression pattern in cancerous tissues. However, the clinical translation of such therapies has been difficult, since current reports regarding the subcellular translocation of APE1 are scarce and inconsistent. Here, we suggested a PTM regulation model that complements the known APE1 PTM mechanism, indicating that arginine methylation of APE1 mediates its mitochondrial translocation to protect cells from oxidative damage. Our findings open new translational perspectives for cancer treatment, based on the use of antibodies against arginine-methylated APE1 as adjuvants to DNA-damaging chemotherapeutic agents. However, there are several aspects that warrant further investigation, particularly the potential correlation between the mitochondrial translocation of APE1 and cancer progression/survival. The details of APE1 PTM-barcoded regulation, with emphasis on crosstalk among the PTMs, require in-depth investigation.
Supplementary Material
Acknowledgement
This work was supported by National Natural Science Foundation of China (81872284), National Natural Science Foundation of China (31701179), and the Natural Science Foundation of Jiangsu Province (Grant No. BE2018714), grants awarded to Zhigang Guo. This work was supported by National Cancer Institute/National Institutes of Health (NCI/NIH) (R01CA085344) grant awarded to Binghui Shen. The National Natural Science Foundation of China (81601539) and Nanjing Medical Science & Technique Development Foundation (QRX17057) grants awarded to Dan Mu.
We thank professor Grigory Dianov for critically reading the manuscript.
Abbreviations:
- APE1
Apurinic/apyrimidinic Endonuclease 1
- PTM
Post-Translational Modification
- PRMT1
Protein Arginine Methyl-Transferase 1
- BER
Base Excision Repair
- ROS
Reactive Oxygen Species
- ADMA
Asymmetrical Di-Methylation Arginine
- SDMA
Symmetrical Di-Methylation Arginine
- Tom20
Translocase of the Outer-mitochondrial Membrane
- THF
tetrahydrofuran
- F
phenyalanine
Footnotes
Declaration of competing interest
We declare no financial or other relationships that may lead to a conflict of interest in this study.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.freeradbiomed.2020.06.027.
References
- [1].Lindahl T, Instability and decay of the primary structure of DNA, Nature 362 (1993) 709–715. [DOI] [PubMed] [Google Scholar]
- [2].Almeida KH, Sobol RW, A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification, DNA Repair 6 (2007) 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tubbs A, Nussenzweig A, Endogenous DNA damage as a source of genomic instability in cancer, Cell 168 (2017) 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Krokan HE, Bjoras M, Base excision repair, Cold Spring Harb. Perspect. Biol 5 (2013) a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].David SS, O’Shea VL, Kundu S, Base-excision repair of oxidative DNA damage, Nature 447 (2007) 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wilson DM 3rd, Bohr VA, The mechanics of base excision repair, and its relationship to aging and disease, DNA Repair 6 (2007) 544–559. [DOI] [PubMed] [Google Scholar]
- [7].Demple B, Sung JS, Molecular and biological roles of Ape1 protein in mammalian base excision repair, DNA Repair 4 (2005) 1442–1449. [DOI] [PubMed] [Google Scholar]
- [8].Hazra TK, Hill JW, Izumi T, Mitra S, Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions, Prog. Nucleic Acid Res 68 (2001) 193–205. [DOI] [PubMed] [Google Scholar]
- [9].Huffman JL, Sundheim O, Tainer JA, DNA base damage recognition and removal: new twists and grooves, Mutat. Res 577 (2005) 55–76. [DOI] [PubMed] [Google Scholar]
- [10].Tell G, Damante G, Caldwell D, Kelley MR, The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxidants Redox Signal. 7 (2005) 367–384. [DOI] [PubMed] [Google Scholar]
- [11].Hashiguchi K, Bohr VA, de Souza-Pinto NC, Oxidative stress and mitochondrial DNA repair: implications for NRTIs induced DNA damage, Mitochondrion 4 (2004) 215–222. [DOI] [PubMed] [Google Scholar]
- [12].Maynard S, de Souza-Pinto NC, Scheibye-Knudsen M, Bohr VA, Mitochondrial base excision repair assays, Methods 51 (2010) 416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Abbotts R, Madhusudan S, Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer, Canc. Treat Rev 36 (2010) 425–435. [DOI] [PubMed] [Google Scholar]
- [14].Bosshard M, Markkanen E, van Loon B, Base excision repair in physiology and pathology of the central nervous system, Int. J. Mol. Sci 13 (2012) 16172–16222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wilson DM, McNeill DR, Base excision repair and the central nervous system, Neuroscience 145 (2007) 1187–1200. [DOI] [PubMed] [Google Scholar]
- [16].Joo HK, Lee YR, Park MS, Choi S, Park K, Lee SK, Kim CS, Park JB, Jeon BH, Mitochondrial APE1/Ref-1 suppressed protein kinase C-induced mitochondrial dysfunction in mouse endothelial cells, Mitochondrion 17 (2014) 42–49. [DOI] [PubMed] [Google Scholar]
- [17].Li M, Wilson DM 3rd, Human apurinic/apyrimidinic endonuclease 1, Antioxidants Redox Signal. 20 (2014) 678–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kelley MR, Georgiadis MM, Fishel ML, APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1, Curr. Mol. Pharmacol 5 (2012) 36–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E, A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium, J. Biol. Chem 269 (1994) 27855–27862. [PubMed] [Google Scholar]
- [20].Benayoun BA, Veitia RA, A post-translational modification code for transcription factors: sorting through a sea of signals, Trends Cell Biol. 19 (2009) 189–197. [DOI] [PubMed] [Google Scholar]
- [21].Huang E, Qu D, Zhang Y, Venderova K, Haque ME, Rousseaux MW, Slack RS, Woulfe JM, Park DS, The role of Cdk5-mediated apurinic/apyrimidinic endonuclease 1 phosphorylation in neuronal death, Nat. Cell Biol 12 (2010) 563–571. [DOI] [PubMed] [Google Scholar]
- [22].Sengupta S, Mantha AK, Mitra S, Bhakat KK, Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1, Oncogene 30 (2011) 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung SB, Kim CS, Irani K, SIRT1 deacetylates APE1 and regulates cellular base excision repair, Nucleic Acids Res. 38 (2010) 832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fantini D, Vascotto C, Marasco D, D’Ambrosio C, Romanello M, Vitagliano L, Pedone C, Poletto M, Cesaratto L, Quadrifoglio F, et al. , Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions, Nucleic Acids Res. 38 (2010) 8239–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lirussi L, Antoniali G, Vascotto C, D’Ambrosio C, Poletto M, Romanello M, Marasco D, Leone M, Quadrifoglio F, Bhakat KK, et al. , Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells, Mol. Biol. Cell 23 (2012) 4079–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Busso CS, Iwakuma T, Izumi T, Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway, Oncogene 28 (2009) 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Busso CS, Wedgeworth CM, Izumi T, Ubiquitination of human AP-endonuclease 1 (APE1) enhanced by T233E substitution and by CDK5, Nucleic Acids Res. 39 (2011) 8017–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N, Tasaki T, Kessler BM, Parsons JL, Kwon YT, et al. , Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability, Nucleic Acids Res. 40 (2012) 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qu J, Liu GH, Huang B, Chen C, Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310, Nucleic Acids Res. 35 (2007) 2522–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kim YJ, Kim D, Illuzzi JL, Delaplane S, Su D, Bernier M, Gross ML, Georgiadis MM, Wilson DM, S-glutathionylation of cysteine 99 in the APE1 protein impairs abasic endonuclease activity, J. Mol. Biol 414 (2011) 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yacoub A, Kelley MR, Deutsch WA, The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation, Canc. Res 57 (1997) 5457–5459. [PubMed] [Google Scholar]
- [32].Onwuli DO, Rigau-Roca L, Cawthorne C, Beltran-Alvarez P, Mapping arginine methylation in the human body and cardiac disease, Proteonomics Clin. Appl 11 (2017). [DOI] [PubMed] [Google Scholar]
- [33].Bedford MT, Clarke SG, Protein arginine methylation in mammals: who, what, and why, Mol. Cell 33 (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bedford MT, Richard S, Arginine methylation an emerging regulator of protein function, Mol. Cell 18 (2005) 263–272. [DOI] [PubMed] [Google Scholar]
- [35].Clarke SG, Protein methylation at the surface and buried deep: thinking outside the histone box, Trends Biochem. Sci 38 (2013) 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bedford MT, Clarke SG, Protein arginine methylation in mammals: who, what, and why, Mol. Cell 33 (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Peng C, Wong CC, The story of protein arginine methylation: characterization, regulation, and function, Expet Rev. Proteonomics 14 (2017) 157–170. [DOI] [PubMed] [Google Scholar]
- [38].Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, Gong W, Xu RM, Structural insights into protein arginine symmetric dimethylation by PRMT5, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 20538–20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, et al. , PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145, Nat. Commun 6 (2015) 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jain K, Clarke SG, PRMT7 as a unique member of the protein arginine methyltransferase family: a review, Arch. Biochem. Biophys 665 (2019) 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Boisvert FM, Cote J, Boulanger MC, Richard S, A proteomic analysis of arginine-methylated protein complexes, Mol. Cell. Proteomics : MCP 2 (2003) 1319–1330. [DOI] [PubMed] [Google Scholar]
- [42].Guo ZG, Zheng L, Xu H, Dai HF, Zhou MA, Pascua MR, Chen QM, Shen BH, Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding, Nat. Chem. Biol 6 (2010) 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weber S, Bauer UM, Arginine methylation in interferon signaling: new light on an old story (vol 8, pg 1464, 2009), Cell Cycle 10 (2011) 2400–2400. [PubMed] [Google Scholar]
- [44].Du W, Amarachintha S, Erden O, Wilson A, Pang QS, The Fanconi anemia pathway controls oncogenic response in hematopoietic stem and progenitor cells by regulating PRMT5-mediated p53 arginine methylation, Oncotarget 7 (2016) 60005–60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB, Arginine methylation regulates the p53 response, Nat. Cell Biol 10 (2008) 1431–U1122. [DOI] [PubMed] [Google Scholar]
- [46].Bikkavilli RK, Avasarala S, Van Scoyk M, Karuppusamy Rathinam MK, Tauler J, Borowicz S, Winn RA, In vitro methylation assay to study protein arginine methylation, JoVE: JoVE (2014) e51997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen DH, Wu KT, Hung CJ, Hsieh M, Li C, Effects of adenosine dialdehyde treatment on in vitro and in vivo stable protein methylation in HeLa cells, J. Biochem 136 (2004) 371–376. [DOI] [PubMed] [Google Scholar]
- [48].Eram MS, Shen Y, Szewczyk M, Wu H, Senisterra G, Li F, Butler KV, Kaniskan HU, Speed BA, Dela Sena C, et al. , A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases, ACS Chem. Biol 11 (2016) 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system, Nat. Protoc 8 (2013) 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang YL, Ge XL, Yang FY, Zhang LP, Zheng JY, Tan XF, Jin ZB, Qu J, Gu F, Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells, Sci. Rep 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Heigwer F, Kerr G, Boutros M, E-CRISP: fast CRISPR target site identification, Nat. Methods 11 (2014) 122–124. [DOI] [PubMed] [Google Scholar]
- [52].Li M, Zhong Z, Zhu J, Xiang D, Dai N, Cao X, Qing Y, Yang Z, Xie J, Li Z, et al. , Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1, J. Biol. Chem 285 (2010) 14871–14881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Barchiesi A, Wasilewski M, Chacinska A, Tell G, Vascotto C, Mitochondrial translocation of APE1 relies on the MIA pathway, Nucleic Acids Res. 43 (2015) 5451–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJE, et al. , Isolation of a small molecule inhibitor of DNA base excision repair, Nucleic Acids Res. 33 (2005) 4711–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rehman I, Basu SM, Das SK, Bhattacharjee S, Ghosh A, Pommier Y, Das BB, PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes, Nucleic Acids Res. 46 (2018) 5601–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Santos JH, Meyer JN, Mandavilli BS, Van Houten B, Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells, Methods Mol. Biol 314 (2006) 183–199. [DOI] [PubMed] [Google Scholar]
- [57].Furda A, Santos JH, Meyer JN, Van Houten B, Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells, Methods Mol. Biol 1105 (2014) 419–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Domon B, Aebersold R, Mass spectrometry and protein analysis, Science 312 (2006) 212–217. [DOI] [PubMed] [Google Scholar]
- [59].Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S, Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains, J. Biol. Chem 275 (2000) 16030–16036. [DOI] [PubMed] [Google Scholar]
- [60].Mostaqul Huq MD, Gupta P, Tsai NP, White R, Parker MG, Wei LN, Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation, EMBO J. 25 (2006) 5094–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Abeywardana T, Oh M, Jiang L, Yang Y, Kong M, Song J, Yang Y, CARM1 suppresses de novo serine synthesis by promoting PKM2 activity, J. Biol. Chem 293 (2018) 15290–15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Huang L, Wang Z, Narayanan N, Yang Y, Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization, Nucleic Acids Res. 46 (2018) 3061–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Whitaker AM, Flynn TS, Freudenthal BD, Molecular snapshots of APE1 proofreading mismatches and removing DNA damage, Nat. Commun 9 (2018) 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Huamani J, McMahan CA, Herbert DC, Reddick R, McCarrey JR, MacInnes MI, Chen DJ, Walter CA, Spontaneous mutagenesis is enhanced in Apex heterozygous mice, Mol. Cell Biol 24 (2004) 8145–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, Zhang YS, APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells, Lung Canc. 66 (2009) 298–304. [DOI] [PubMed] [Google Scholar]
- [66].Li MX, Wang D, Zhong ZY, Xiang DB, Li ZP, Xie JY, Yang ZZ, Jin F, Qing Y, Targeting truncated APE1 in mitochondria enhances cell survival after oxidative stress, Free Radic. Biol. Med 45 (2008) 592–601. [DOI] [PubMed] [Google Scholar]
- [67].Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR, PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells, J. Biol. Chem 275 (2000) 7723–7730. [DOI] [PubMed] [Google Scholar]
- [68].Nicholson TB, Chen T, Richard S, The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation, Pharmacol. Res 60 (2009) 466–474. [DOI] [PubMed] [Google Scholar]
- [69].Godin KS, Varani G, How arginine-rich domains coordinate mRNA maturation events, RNA Biol. 4 (2007) 69–75. [DOI] [PubMed] [Google Scholar]
- [70].Xie M, Doetsch PW, Deng X, Bcl2 inhibition of mitochondrial DNA repair, BMC Canc. 15 (2015) 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J, ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis, Science 320 (2008) 661–664. [DOI] [PubMed] [Google Scholar]
- [72].Deng XM, Gao FQ, Flagg T, Anderson J, May WS, Bcl2’s flexible loop domain regulates p53 binding and survival, Mol. Cell Biol 26 (2006) 4421–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW, Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types, EMBO J. 15 (1996) 4130–4141. [PMC free article] [PubMed] [Google Scholar]
- [74].Guthrie HD, Welch GR, Determination of intracellular reactive oxygen species and high mitochondrial membrane potential in Percoll-treated viable boar sperm using fluorescence-activated flow cytometry, 84 (8) (2006) 2089–2100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.