Abstract
Background:
Clostridium perfringens causes necrotic enteritis (NE) and is considered a major economic burden in the broiler industry and a significant foodborne pathogen, worldwide.
Aims:
Clostridium perfringens isolated from NE affected broiler chickens was aimed to characterize and the presence of β-lactamase and quinolone resistant genes were also investigated in the isolates.
Methods:
A total of 224 intestinal and caecal specimens were collected from NE affected broiler chickens and cultured to isolate C. perfringens. The toxicogenic characterization of C. perfringens was appraised using polymerase chain reaction (PCR) and antibiotic susceptibility testing (disc diffusion method). The selected C. perfringens isolates were characterized for β-lactamase and quinolone encoding genes by PCR analysis.
Results:
All isolates were cultured positive for C. perfringens and the toxin-encoding genes of C. perfringens (α-, β-, β2-, ε-, ι-, and enterotoxin) were also identified. About 65.6% of isolates had a multi-drug resistant (MDR) profile but none of these isolates were resistant or susceptible to all screened antibiotics. A subset of isolates, 160 and 98 were analyzed for β-lactamase and quinolone genes, respectively, and recognized blaTEM, blaSHV, and blaOXA in 64 (40%; CI: 32.35-48.03%; P<0.001) isolates, and qnrB and qnrS in 28 (28.57%; CI: 19.90-38.58%; P<0.001) isolates except qnrA.
Conclusion:
Therefore, the isolates of C. perfringens were toxicogenic and carried β-lactamase, and quinolone resistance genes. Nowadays, the rational use of antibiotics and safe production of broiler chickens are the major concern to save public health.
Key Words: Broiler, β-lactamase, Clostridium perfringens, Antibiotic resistance, Quinolone
Introduction
Necrotic enteritis (NE) is a major interest for the broiler industry due to it being a major economic burden. Necrotic enteritis infection can reduce body weight by 12.0% and increase feed conversion ratio (FCR) by 10.9% compared to healthy birds. As a result, NE causes a loss of 878.19$ to 1480.52$ per flock in the USA and USD 2 to 6 billion worldwide per annum (Skinner et al., 2010 ▶; Wade and Keyburn, 2015 ▶). Broiler chickens need maximum utilization of feeds by maximum digestion, absorption, and metabolism of nutrients. Necrotic enteritis disrupts the intestinal mucosa lining of chicken that decreased nutrient absorption resulting in reduced feed efficiency or FCR which is the principal parameter of good nutrition (Yang et al., 2019 ▶). The major cause of NE is Clostridium perfringens, a Gram-positive, spore-forming, rod shaped, anaerobic bacteria that is ubiquitously present in the environment and normal gut flora (Miller et al., 2010 ▶; Freedman et al., 2015 ▶). It could become pathogenic when the gut microbiome is changed from the dynamic balance of the gut environment by stress, starvation, sudden change of feed, the therapeutic use of antibiotics or anthelmintic resulting in NE in poultry, enteritis in ruminants, non-ruminants, and humans (Prescott, 2016 ▶).
Clostridium perfringens has significant importance in public health as it can cause foodborne illness and has been reported as the second most common bacterial foodborne illness in the USA (Scallan et al., 2011 ▶).
The concerning point of C. perfringens is the ability to produce potent exotoxins which are responsible for developing specific enterotoxemia in animals and humans. It can encode for up to 17 exotoxins (Freedman et al., 2015 ▶) and based on the production of four major lethal exotoxins (α: alpha, β: beta, ε: epsilon, and ι: toxins), C. perfringens is classified into five classes from type A to type E which are responsible for specific diseases (Yoo et al., 1997 ▶). Both type A and B can produce α toxin, in addition to β toxin, that is highly necrotizing and lethal and responsible for severe intestinal necrosis (Cooper and Songer, 2010 ▶). The β toxin is encoded by plasmid-mediated cpb gene (Gkiourtzidis et al., 2001 ▶). Type B and D both can produce ε-toxin that is encoded by plasmid-mediated etx gene and responsible for NE after cleavage by proteolytic enzyme (Chen et al., 2011 ▶). The type E can produce α-toxin and ι-toxin encoded by cpa and iap gene, responsible for gas gangrene, intestinal disorder, and diarrhea (Park et al., 2015 ▶). Enteric bacteria are able to acquire and exchange genetic materials via transposons, plasmids or through chromosomal exchange or mutation in DNA (Gyles and Boerlin, 2014 ▶). Several investigations revealed that C. perfringens is intrinsically resistant to several antimicrobial agents commonly used against Gram-positive bacterial infection and perform a role as multi-drug resistant (MDR) pathogens (Lee, 2016 ▶). This study also identified an MDR characteristic of C. perfringens as phenotypical and the responsible antibiotic resistant genes. The emergence and rapid spread of MDR C. perfringens poses a serious therapeutic challenge for simple foodborne illness by effective antimicrobial therapy, due to the scarcity of newer antimicrobial agents. Antibiotic resistance is an alarming issue now due to an emerging public health crisis and one of the top health challenges of the 21st century (Marshall and Levy, 2011 ▶; Ali et al., 2019 ▶). It is assumed that the antibiotic use in feed (e.g. bacitracin, tetracycline, avoparcin, vancomycin, virginiamycin erythromycin, tylosin, avilamycin, lincomycin, bambermycins, carbadox, etc) mainly focused on C. perfringens control as well as growth promoter called antibiotic growth promoters (AGPs) (Arnold et al., 2004 ▶; Marshall and Levy, 2011 ▶; Landers et al., 2012 ▶). Therefore, bacteria become resistant by adapting against repeated use of antibiotics with feeds and escape the mode of action of antibiotics by developing a resistance gene in their plasmid or chromosome (Diarra et al., 2007 ▶; Nhung et al., 2017 ▶).
There is a need to explore and elucidate the safe alternatives of antibiotics to control NE and to better understand the toxicogenic properties of C. perfringens isolated from NE affected broiler chickens with their antimicrobial resistance phenotypes and genotypes. There is a need to explore and elucidate the safe alternatives of antibiotics to control NE. In this context, the aim of the present work was to gain insights into the toxicogenic properties, antibiotic resistance patterns, presence of β-lactamase and quinolone encoded genes in C. perfringens isolated from broiler chickens with NE.
Materials and Methods
Sample collection and processing
A total of 224 clinically NE diagnosed Cobb500 broiler chickens between 7 and 30 days old came in Nourish Customer Service Laboratory, Cumilla and Rajshahi from 2017 to 2018. Chickens were inspected during the postmortem examination and were confirmed by observing standard NE lesions as previously described (Immerseel et al., 2004 ▶). Part of the small intestine and caecum were collected (2.5 cm of each) into a 10 ml tube containing 5.0 ml sterile phosphate buffered saline (PBS). The samples of intestine and caecum were homogenized by sterile pestle and mortar then clarified by centrifugation at 2147 × g for 10 min. The supernatant was collected and used as an inoculum.
Bacterial isolation
An inoculum of 1 ml sample was diluted into 9 ml of brain heart infusion (BHI) broth (Oxoid, UK) then incubated for 24 h at 37°C in an anaerobic vented jar with an appropriate amount of anaerobic gas generating from packs (Mitsubishi Gas Chemical America Inc., New York). Subsequently, 100 µL of pre-enriched BHI broth was inoculated into plates of tryptose sulfite cycloserine (TSC) agar base (Oxoid, UK) enriched with 5% egg yolk and supplemented with β D-cycloserine (Oxoid, UK). The plates were sealed tightly, incubated for 24 h at 37°C upsides down in an anaerobic condition. The black colonies on plates were examined for typical colonies of C. perfringens (Mwangi et al., 2018 ▶). Finally, the pure isolates were confirmed by the biochemical (lactose fermentation) and Gram-staining test.
Polymerase chain reaction (PCR) test for C. perfringens toxin genes screening
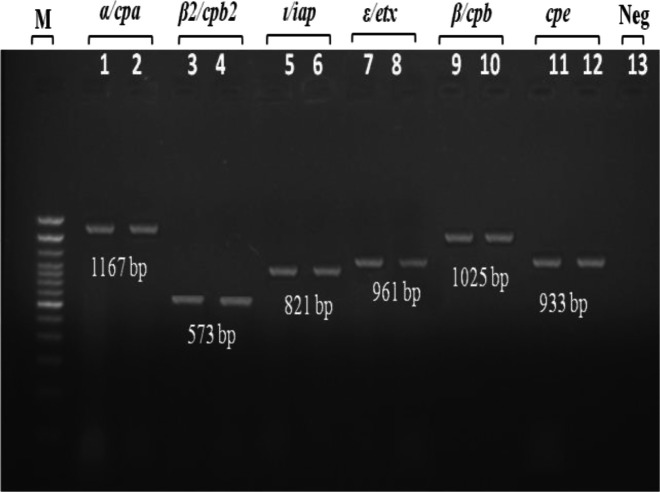
The DNA extraction of isolated C. perfringens was done by using the direct lysis method as described by Kim et al. (2011) ▶. Screening the presence of six major toxin genes (α/cpa, β/cpb, B2/cpb2, ε/etx, ι/ipa, and enterotoxin/cpe) by using six pairs of primers shown in Table 1. Total 25 µL of PCR amplification reaction mix were prepared using GoTaq® Green Master Mix (2×) (Promega, Madison, WI), 0.4 mM of each oligonucleotide primers, 6 µL of template DNA, and dH2O. The PCR assay was performed in a thermal cycler (MiniAmp Plus Thermal Cycler, Applied Biosystems, USA) with amplification conditions as following: denaturation for 30 s at 94°C; primer annealing for 30 s at the respective temperature as 46°C for cpa gene, etx gene and ipa gene, 39°C for cpb gene, 48°C for cpb2 gene, and 52°C for cpe gene; finally the chain extension for 30 s at 72°C. Then PCR products were separated by electrophoresis through 1% agarose gel stained with 0.5 μg/ml ethidium bromide at 95 V for 40 min. Then the amplification fragments were visualized under ultraviolet light at 254 nm wavelength (UVP, Inc., Upland, CA) (Fig. 1).
Table 1.
Oligonucleotide primer sequences used for C. perfringens toxin genes cpa (α-toxin), cpb (β-toxin), cpb2 (β2-toxin), cpe (enterotoxin), etx (ε-toxin), and iap (ι-toxin)
| Toxin/gene | Primer | Oligonucleotide sequence | Fragment length (bp) | References |
|---|---|---|---|---|
| α/cpa | CPALPHATOX1-F | AAGATTTGTAAGGCGCTT | 1167 bp | |
| CPALPHATOX1-R | ATTTCCTGAAATCCACTC | |||
| β/cpb | CPBETATOX1-F | AGGAGGTTTTTTTATGAAG | 1025 bp | |
| CPBETATOX1-R | TCTAAATAGCTGTTACTTTGTG | |||
| β2/cpb2 | P319BETA2 | GAAAGGTAATGGAGAATTATCTTAATGC | 573 bp | |
| P320BETA2 | GCAGAATCAGGATTTTGACCATATACC | |||
| ε/etx | CPETOXIN1-F | AAGTTTAGCAATCGCATC | 961 bp | |
| CPETOXIN1-R | TATTCCTGGTGCCTTAATT | |||
| Enterotoxin/cpe | CPENT1-F | TAACAATTTAAATCCAATGG | 933 bp | |
| CPENT1-R | ATTGAATAAGGGTAATTTCC | |||
| ι/iap | CPIOTA1-F | AATGCCATATCAAAAAATAA | 821 bp | |
| CPIOTA1-R | TTAGCAAATGCACTCATATT |
Fig. 1.
Detection of seven toxin genes of C. perfringens by PCR. Lane M: Marker (DNA ladder, 100 bp), Lanes 1-2: α/cpa, Lanes 3-4: β2/cpb2, Lanes 5-6: ι/ipa, Lanes 7-8: ε/etx, Lanes 9-10: β/cpb, Lanes 11-12: cpe, and Lane 13: Negative control
Antimicrobial susceptibility assay
Antibiotic susceptibility test was carried out for all 224 isolates by disc diffusion method on Muller Hinton agar (MHA) (Oxoid, UK) according to Clinical and Laboratory Standards Institute guideline (CLSI, 2012 ▶). The antibiotic discs were selected based on available antibiotics used in poultry and human medicine in Bangladesh including β-lactam (amoxicillin/clavulanic acid 30 µg; penicillin G 10 µg), quinolones (ciprofloxacin 5 µg; norfloxacin 5 μg), macrolides (erythromycin 15 µg), sulfonamides (co-trimoxazole 25 μg), aminoglycosides (gentamycin 30 µg; streptomycin 10 µg; amikacin 30 µg), cephalosporin (ceftriaxone 5 μg), and tetracyclines (tetracycline 30 µg) (Oxoid, UK).
The 0.5 McFarland standard inoculum of pure colonies was streaked homogenously on MHA using a sterile cotton swab and incubated at 37°C for 12 h. Then inhibition zone was measured and categorized as susceptible, resistant, and intermediate according to CLSI (2012) ▶ guideline.
Antibiotic resistance gene identification
The isolates identified as resistant against amoxicillin/clavulanic acid and penicillin G were considered for β-lactamase encoding genes (blaTEM, blaSHV, blaOXA) screening. Quinolone resistant encoding genes (qnrA, qnrB, qnrS) were screened for those isolates containing phenotypic resistance against ciprofloxacin and norfloxacin. Total of 5 µL DNA was subjected to each multiplex PCR in a 25 µL reaction mixture containing 1 × PCR buffer (0.025U/µL Taq DNA polymerase, reaction buffer, 2 mM MgCl2, 0.2 mM of each dNTP (dATP, dCTP, dGTP, and dTTP) (Thermo Scientific PCR Master Mix 2×) and a variable concentration of specific primers (Table 2). β-lactamase genes amplification was carried out as follows: initial denaturation at 94°C for 10 min, 30 cycles of 94°C for 40 s, 60°C for 40 s, and 72°C for 1 min and, a final
Table 2.
PCR primer sequences for β-lactamase and quinolone resistance genes detection
| Target gene | Primer | Sequence (5´-3´) | Amplicon size (bp) | References |
|---|---|---|---|---|
| bla TEM | MultiTSO-T_F | CATTTCCGTGTCGCCCTTATTC | 800 | |
| MultiTSO-T_R | CGTTCATCCATAGTTGCCTGAC | |||
| bla SHV | MultiTSO-S_F | AGCCGCTTGAGCAAATTAAAC | 713 | |
| MultiTSO-S_R | ACCCGCAGATAAATCACCAC | |||
| bla OXA | MultiTSO-O_F | GGCACCAGATTCAACTTTCAAG | 564 | |
| MultiTSO-O_R | GACCCCAAGTTTCCTGTAAGTG | |||
| qnrA | QnrAm-F | AGAGGATTTCTCACGCCAGG | 580 | |
| QnrAm-R | TGCCAGGCACAGATCTTGAC | |||
| qnrB | QnrBm-F | GGMATHGAAATTCGCCACTG | 264 | |
| QnrBm-R | TTTGCYGYYCGCCAGTCGAA | |||
| qnrS | QnrSm-F | GCAAGTTCATTGAACAGGGT | 428 | |
| QnrSm-R | TCTAAACCGTCGAGTTCGGCG |
PCR: Polymerase chain reaction
elongation step at 72°C for 7 min. Quinolone resistant genes (qnrA, qnrB, and qnrS) amplification was carried out at 10 min at 95°C and 35 cycles of amplification consisting of 1 min at 95°C, 1 min at 54°C and 1 min at 72°C and 10 min at 72°C for the final extension.
Statistical analysis
The data from sample collection and laboratory results were recorded as coding into Microsoft Excel spreadsheet 2010 (Microsoft Corporation, WA, USA) and analyzed by exporting to STATA version 25.0 (Stata Corp. College Station, TX, USA) (IBM Corp, 2017 ▶). Descriptive cross tabulation statistics and frequency statistics were used for prevalence data analysis. The Chi-square test (χ2 value) was used for calculating statistical significance level. The confidence levels at 95% (95% CI) were calculated and probability value of P≤0.05 was considered for statistical significance level.
Results
Isolation and identification of C. perfringens
Out of 224 clinically suspected NE samples, C. perfringens were identified in 100% samples by the conventional cultural characteristics method. Finally, all isolates were confirmed for C. perfringens by identifying presence of six toxin genes including cpa, cpb, cpb2, etx, ipa, and cpe among which cpa gene was present in all isolates.
Toxinotyping of isolated C. perfringens
The toxin typing of C. perfringens were done by PCR assay in which 100% (n=224) isolates were positive for cpa, while individually only 32.59% (73/224; CI: 26.50-39.15%) isolates carried cpa gene. 8.04% (18/224; CI: 4.83-12.40%) of isolates were identified positive for both cpa and cpe genes; moreover, 24.01% (54/224; CI: 18.66-30.25%) isolates were cpa+ and cpb2+ classified as C. perfringens type A. Type C was identified 11.61% in which cpa+, cpb+ and cpb2+ genes were 7.59% (17/224; CI: 4.48-11.87%), and cpa+, cpb+, cpb2+ and cpe genes were 4.02% (9/224; CI: 1.85-7.49%). Type D was identified 18.64% in which cpa+ and etx+ genes were 7.59% (17/224; CI: 4.48-11.87%), and cpa+, cpb2+ and etx+ genes were found 12.05% (27/224; CI: 8.10-17.05%). Furthermore, type E was recognized 4.02% (9/224; CI: 1.85-7.49%) (with cpa+ and iap genes) (Table 3). Type B however was not found in any isolates. This results were shown statistically significant relation (χ2 = 154; P<0.001).
Table 3.
Distribution of the different C. perfringens types and their toxin genes isolated from intestine and caecal samples of necrotic enteritis in broiler chicken
| C. perfringens types | C. perfringens toxin genes | Number of isolates (%) | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|
| A | cpa | 73 (32.59) | 26.50-39.15 | 154 | 0.001 |
| A | cpa, cpe | 18 (8.04) | 4.83-12.40 | ||
| C | cpa, cpb, cpb2 | 17 (7.59) | 4.48-11.87 | ||
| C | cpa, cpb, cpb2, cpe | 9 (4.02) | 1.85-7.49 | ||
| D | cpa, etx | 17 (7.59) | 4.48-11.87 | ||
| D | cpa, cpb2, etx | 27 (12.05) | 8.10-17.05 | ||
| A | cpa, cpb2 | 54 (24.11) | 18.66-30.25 | ||
| E | cpa, iap | 9 (4.02) | 1.85-7.49 | ||
| Total: 224 | |||||
Antimicrobial susceptibility test
Among the isolates, about 147 (65.62%) were found resistant to three or more antibiotic classes termed as MDR. While none of them were resistant or susceptible to all tested antibiotics. Among the tested isolates, most resistance observed against co-trimoxazole 96.42% (216/224) and streptomycin 92.85% (208/224), followed by tetracycline 75% (168/224), gentamicin 70.98% (159/224), penicillin G 68.75% (154/224), norfloxacin 39.73% (89/224); erythromycin 34.82% (78/224); amoxicillin/clavulanic acid 30% (67/224); ciprofloxacin 17.85% (40/224); and ceftriaxone 2.67% (6/224) (Table 4). However, the highest susceptible antibiotics were shown in amikacin 97.76% (219/224), ceftriaxone 96.42% (216/224), and ciprofloxacin 71.87% (161/224) in comparison with the other antibiotics. But no significant correlation (P=0.85) was found among antibiotics.
Table 4.
Antibiotic susceptibility patterns of C. perfringens isolates (n=224) recovered from broiler chickens affected by necrotic enteritis
| Antibiotics | Susceptible % (n) | Resistant % (n) | Intermediate % (n) | Non-susceptibility (resistant+intermdiate) % (n) |
|---|---|---|---|---|
| Amoxicillin/clavulanic acid | 50% (112) | 30% (67) | 20% (45) | 54.46% (122) |
| Penicillin G | 5.8% (13) | 68.75% (154) | 25.44% (57) | 94.19% (211) |
| Ciprofloxacin | 71.87% (161) | 17.85% (40) | 10.26% (23) | 28.12% (63) |
| Norfloxacin | 44.64% (100) | 39.73% (89) | 15.62% (35) | 55.35% (124) |
| Erythromycin | 46.87% (105) | 34.82% (78) | 18.3% (41) | 53.12% (119) |
| Co-trimoxazole | 3.57% (8) | 96.42% (216) | 0 | 96.93% (216) |
| Ceftriaxone | 96.42% (216) | 2.67% (6) | 0.9% (2) | 3.57% (8) |
| Amikacin | 97.76% (219) | 0 | 2.23% (5) | 0.89% (2) |
| Gentamycin | 16.96% (38) | 70.98% (159) | 12% (27) | 83.03% (186) |
| Streptomycin | 7.14% (16) | 92.85% (208) | 0 | 92.85% (208) |
| Tetracycline | 15.17% (34) | 75% (168) | 9.82% (22) | 84.82% (190) |
| P=0.85 | ||||
Identification of β - lactamase and quinolone encoded genes
A total of 160 isolates was shown resistant against amoxicillin/clavulanic acid and penicillin G that was considered screening for three β-lactamase genes including blaTEM, blaSHV and blaOXA. The β-lactamase encoding genes were identified in 64 (n=160; 40%; CI: 32.35-48.03%) isolates and the prevalence was found 13.75% (22/160; CI: 8.82-20.07%), 10.62% (17/160; CI: 6.31-16.47%), and 15.63% (25/160; CI: 10.37-22.20%) for blaTEM, blaSHV and blaOXA, respectively. Contrarily, total of 98 isolates was identified as resistant against ciprofloxacin and norfloxacin encoding genes. Whereas, quinolone encoding genes were identified in 28 (n=98; 40%; CI: 19.90-38.58%) isolates including 18.37% (18/98; CI: 11.26-27.47%) qnrB gene and 10.20% (10/98; CI: 5.00-17.97%) qnrS gene, but no qnrA gene was found. The results of both β-lactamase and quinolone encoding genes prevalence were shown statistically significant (P<0.001) relation (Table 5).
Table 5.
Antibiotic resistance gene prevalence of C. perfringens isolates (β-lactamase resistant isolates= 160; quinolones resistant isolates= 98) recovered from broiler chickens affected by necrotic enteritis
| Resistant genes | Number of isolates (%) | 95% CI | χ2 value | P-value | |
|---|---|---|---|---|---|
| β-lactamase | bla TEM | 22 (13.75) | 8.82-20.07 | 54.6 | 0.001 |
| bla SHV | 17 (10.62) | 6.31-16.47 | |||
| bla OXA | 25 (15.63) | 10.37-22.20 | |||
| Sub-total | 64/160 (40.00) | 32.35-48.03 | |||
| Quinolone | qnrA | 0 | - | 35.03 | 0.001 |
| qnrB | 18 (18.37) | 11.26-27.47 | |||
| qnrS | 10 (10.20) | 5.00-17.97 | |||
| Sub-total | 28/98 (28.57) | 19.90-38.58 | |||
Discussion
The broiler industries all over the world are facing significant challenges with NE caused by C. perfringens. Clostridium perfringens can directly affect the feed digestion and nutrient absorption as well as affecting feed conversion ratios (FCR) (Mwangi et al., 2018 ▶). The standard clinical signs of NE like roughened intestinal surfaces, low absorption, and retardation of growth were the sampling criteria of this study (Cooper and Songer, 2010 ▶). Clostridium perfringens type A is the most prevalent type of this bacterium in poultry that was found about 64.73% (145/224), herein (Yoo et al., 1997 ▶). Besides, type A has public health importance and is responsible for gas gangrene and food poisoning in humans (Uzal et al., 2014 ▶). Enterotoxigenic C. perfringens is the major pathogen of foodborne diseases to humans where the main sources of this organism are poultry and meat products (Cooper et al., 2013 ▶). It was estimated that about one million people are affected by C. perfringens per year and it has become the second highest foodborne pathogen in the USA (Scallan et al., 2011 ▶).
Clostridium perfringens is also called a commensal bacterium found in the gastrointestinal tract without showing pathogenicity except with high infective dose and stress (Miller et al., 2010 ▶; Cooper et al., 2013 ▶; Mwangi et al., 2018 ▶). Toxins are the major virulence factor of C. perfringens that plays a significant role in gut pathogenicity and public health concerns (Cooper et al., 2013 ▶). This study demonstrated that the isolated C. perfringens were capable to produce six types of toxins which should be seriously noticed in poultry enteric health and public health. The cpb2 gene represents the β2 toxin the most lethal enterotoxin for poultry found in a maximum number of isolates (Gkiourtzidis et al., 2001 ▶). There is a lack of in-depth studies on C. perfringens and to the best of our knowledge, C. perfringens toxin typing is absence in Bangladesh although the prevalence of NE is demonstrated at 8% in broiler chicken (Miah et al., 2011 ▶).
Antimicrobial resistance is the burning issue in Bangladesh as well as globally (Ahmed et al., 2019 ▶). Antibiotic used in animal feeds as a growth promoter is also an issue in Bangladesh including the subcontinent. Since 2010 the government of Bangladesh made a decision to completely ban any antibiotics as growth promoter in animal feeds through the legislation of Fish and Animal Feed Act (2010) ▶. Using anticoccidial drugs and antibiotics in animal feeds to control coccidiosis and NE were the prime focus of animal feed manufacturers (M’Sadeq et al., 2015 ▶). The present study revealed that 100% of the C. perfringens isolates were resistant to at least one antibiotic and there was no single isolate susceptible to all tested antibiotics. It was also found that the isolates were resistant to amoxycillin/clavulanic acid, ciprofloxacin, and norfloxacin. The findings showed alarming signs to the veterinary and public health aspects due to the number and variation of human antibiotics used in veterinary practices. It was encouraging to Bangladesh that in May 2019 Bangladesh government prohibited using of human antibiotics to veterinary practice and at July 2019 High Court of Bangladesh gave a rule that only registered veterinarians have the right to prescribe antibiotics to animals (The Daily Star, 2019 ▶). The most antibiotic classes used in poultry feeds include glycolipids (bambermycin), ionophores (salinomycin), polypeptides (bacitracin), and β-lactams (penicillin) as
AGP (Singer et al., 2019 ▶). Although these antibiotics improve feed efficiency and increase growth, they modify intestinal flora and create a selective pressure to develop antibiotic resistance (Collier et al., 2003 ▶). Therefore, several European countries banned or restricted the use of AGP in the feed (Casewell et al., 2003 ▶). The β-lactam antibiotic like penicillin had been used in poultry feeds for growth promotion (Collier et al., 2003 ▶). We found β-lactamase encoding genes like blaTEM, blaSHV, and blaOXA in C. perfringens which indicated a major public health concern because they can be shed into the environment and share across other microflora resulting in superbugs in the future (Burki, 2018 ▶). Prevalence of extended spectrum beta-lactamase (ESBL) producing isolates was demonstrated in 37.8% isolates from Bangladesh and 42.3% isolates from India (Abrar et al., 2019 ▶).
The fluoroquinolone-resistant antibiotics can carry several classes of plasmid-mediated quinolone resistance genes like qnrB, qnrC, qnrD, qnrS, and qnrVC that can reduce the susceptibility to fluoroquinolone (Garoff et al., 2017 ▶). We found qnrB and qnrS genes in C. perfringens isolates that supported the evidence of the presence of qnrB and qnrS genes in C. perfringens isolated from poultry (Poirel et al., 2012 ▶).
Toxigenic C. perfringens plays an important role to develop NE in broiler chickens. Six different types of toxigenic encoding genes were identified from 224 C. perfringens isolates of NE diagnosed in broiler chickens. The C. perfringens isolates have developed resistance to more than one antibiotic. The irrespective use of β-lactamase and quinolone in the poultry ration or therapeutics can cause hazards for human health through intestinal microflora imbalance, antimicrobial resistance as well as impaired therapeutic efficacy. Clostridium perfringens is an important public health pathogen and monitoring should be taken into account the prevention and control from poultry feeds and products. Any ignorance can cause severe foodborne health hazards. Therefore, rationing use of any antimicrobials and strict legislation and enforcement to antimicrobial use in food animals along with maintaining withdrawal period is mandatory to combat antimicrobial resistance.
Acknowledgements
The authors received no specific funding for this work. The Nourish Poultry Diseases Diagnostic Laboratory, Nourish Poultry and Hatchery Ltd. were supported by their laboratory facilities during the research. We gratefully acknowledge the excellent laboratory support of M. S. Hossain and M. A. Sagor and review support of D. Smith and R. Begum.
Conflict of interest
None of the authors have any conflict of interest to declare.
References
- Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of blaCTX-M, blaTEM, blaSHV and blaOXA genes in extended-spectrum-β-lactamase-producing clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrob. Resist. Infect. Control. 2019;8:1–10. doi: 10.1186/s13756-019-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed I, Rabbi MB, Sultana S. Antibiotic resistance in Bangladesh: A systematic review. Int. J. Infect. Dis. 2019;80:54–61. doi: 10.1016/j.ijid.2018.12.017. [DOI] [PubMed] [Google Scholar]
- Ali MZ, Islam E, Giasuddin M. Outbreak investigation, molecular detection, and characterization of foot and mouth disease virus in the Southern part of Bangladesh. J. Adv. Vet. Anim. Res. 2019;6:346–354. doi: 10.5455/javar.2019.f353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S, Gassner B, Giger T, Zwahlen R. Banning antimicrobial growth promoters in feedstuffs does not result in increased therapeutic use of antibiotics in medicated feed in pig farming. Pharmacoepidemiol. Drug Saf. 2004;13:323–331. doi: 10.1002/pds.874. [DOI] [PubMed] [Google Scholar]
- Burki TK. Superbugs: An arms race against bacteria. Lancet Respir. Med. 2018;6:668. doi: 10.1016/S2213-2600(18)30271-6. [DOI] [PubMed] [Google Scholar]
- Casewell M, Friis C, Marco E, McMullin P, Phillips I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. Antimicrob. Agents Chemother. 2003;52:159–161. doi: 10.1093/jac/dkg313. [DOI] [PubMed] [Google Scholar]
- Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007a;60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- Cattoir V, Weill FX, Poirel L, Fabre L, Soussy CJ, Nordmann P. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 2007b;59:751–754. doi: 10.1093/jac/dkl547. [DOI] [PubMed] [Google Scholar]
- Chen J, Rood JI, McClane BA. Epsilon-toxin production by Clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR two-component regulatory system. 2011;MBio. 2:e00275–00311. doi: 10.1128/mBio.00275-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- CLSI (Clinical and Laboratory Standards Institute); Wayne, PACLSICLSI Document M11-A8 2012, authors. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard. 8th Edn; 2012. [Google Scholar]
- Collier CT, Van DKJD, Deplancke B, Anderson DB, Gaskins HR. Effects of tylosin on bacterial mucolysis, Clostridium perfringens colonization, and intestinal barrier function in a chick model of necrotic enteritis. Antimicrob. Agents Chemother. 2003;47:3311–3317. doi: 10.1128/AAC.47.10.3311-3317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KK, Songer JG. Virulence of Clostridium perfringens in an experimental model of poultry necrotic enteritis. Vet. Microbiol. 2010;142:323–328. doi: 10.1016/j.vetmic.2009.09.065. [DOI] [PubMed] [Google Scholar]
- Cooper KK, Songer JG, Uzal FA. Diagnosing clostridial enteric disease in poultry. J. Vet. Diagn. Invest. 2013;25:314–327. doi: 10.1177/1040638713483468. [DOI] [PubMed] [Google Scholar]
- Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- Diarra MS, Silversides FG, Diarrassouba F, Pritchard J, Masson L, Brousseau R, Bonnet C, Delaquis P, Bach S, Skura BJ, Topp E. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolates. Appl. Environ. Microbiol. 2007;73:6566–6576. doi: 10.1128/AEM.01086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish and Animal Feed Act. [2010]. Available at: http://dls.portal.gov.bd/sites/default/files/files/dls.portal.gov.bd/page/b4ea0518_7cf7_43e5_b0b8_756b831991d4/1.%20Fish%20feed%20and%20Animal%20Feed%20Act%20-2010.pdf.
- Freedman JC, Theoret JR, Wisniewski JA, Uzal FA, Rood JI, McClane BA. Clostridium perfringens type A-E toxin plasmids. Res. Microbiol. 2015;166:264–279. doi: 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff L, Yadav K, Hughes D. Increased expression of Qnr is sufficient to confer clinical resistance to ciprofloxacin in Escherichia coli. J. Antimicrob. Chemother. 2017;73:348–352. doi: 10.1093/jac/dkx375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkiourtzidis K, Frey J, Bourtzi-Hatzopoulou E, Iliadis N, Sarris K. PCR detection and prevalence of α-, β-, β2-, ε-, ι- and enterotoxin genes in Clostridium perfringens isolated from lambs with Clostridial dysentery. Vet. Microbiol. 2001;82:39–43. doi: 10.1016/s0378-1135(01)00327-3. [DOI] [PubMed] [Google Scholar]
- Gyles C, Boerlin P. Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet. Pathol. 2014;51:328–340. doi: 10.1177/0300985813511131. [DOI] [PubMed] [Google Scholar]
- IBM Corp. Released. 2017. [IBM SPSS Statistics for Windows, Version 25.0, Armonk, NY: IBM Corp]. Available at: https://www.ibm.com/support/pages/how-cite-ibm-spss-statistics-or-earlier-versions-spss.
- Immerseel FV, Buck JD, Pasmans F, Huyghebaert G, Haesebrouck F, Ducatelle R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 2004;33:537–549. doi: 10.1080/03079450400013162. [DOI] [PubMed] [Google Scholar]
- Kim JB, Kim JM, Cho SH, Oh HS, Choi NJ, Oh DH. Toxin genes profiles and toxin production ability of Bacillus cereus isolated from clinical and food samples. J. Food Sci. 2011;76:25–29. doi: 10.1111/j.1750-3841.2010.01958.x. [DOI] [PubMed] [Google Scholar]
- Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 2012;127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ. Antimicrobial resistance and molecular characterization of Clostridium perfringens isolated from chicken. J. Prev. Vet. Med. 2016;40:71–79. [Google Scholar]
- Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 2011;24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miah MS, Asaduzzaman M, Sufian MA, Hossain MM. Isolation of Clostridium perfringens, causal agents of necrotic enteritis in chickens. J. Bangladesh Agril. Univ. 2011;9:97–102. [Google Scholar]
- Miller RW, Skinner J, Sulakvelidze A, Mathis GF, Hofacre CL. Bacteriophage therapy for control of necrotic enteritis of broiler chickens experimentally infected with Clostridium perfringens. Avian Dis. 2010;54:33–40. doi: 10.1637/8953-060509-Reg.1. [DOI] [PubMed] [Google Scholar]
- M’Sadeq SA, Wu S, Swick RA, Choct M. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim. Nutr. 2015;1:1–11. doi: 10.1016/j.aninu.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi S, Timmons J, Fitz-Coy S, Parveen S. Characterization of Clostridium perfringens recovered from broiler chicken affected by necrotic enteritis. Poult. Sci. 2018;98:128–135. doi: 10.3382/ps/pey332. [DOI] [PubMed] [Google Scholar]
- Nhung NT, Chansiripornchai N, Carrique-Mas JJ. Antimicrobial resistance in bacterial poultry pathogens: a review. Front. Vet. Sci. 2017;10:126. doi: 10.3389/fvets.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim S, Oh JY, Kim HR, Jang I, Lee HS, Kwon YK. Characterization of Clostridium perfringens isolates obtained from 2010 to 2012 from chickens with necrotic enteritis in Korea. Poult. Sci. 2015;94:1158–1164. doi: 10.3382/ps/pev037. [DOI] [PubMed] [Google Scholar]
- Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 2012;3:24. doi: 10.3389/fmicb.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott JF. Brief description of animal pathogenic Clostridia. In: Uzal FA, editor. ption of animal pathogenic Clostridia. In: Uzal, FA; Songer, JG and Prescott, JF (Eds.), 1st Edn. Iowa, John Wiley and Sons: Ames; 2016. pp. 13–19. [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RS, Porter LJ, Thomson DU, Gage M, Beaudoin A, Wishnie JK. Potential impacts on animal health and welfare of raising animals without antibiotics. BioRxiv. 2019:600965.. doi: 10.3389/fvets.2019.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JT, Bauer S, Young V, Pauling G, Wilson J. An economic analysis of the impact of subclinical (mild) necrotic enteritis in broiler chickens. Avian Dis. 2010;54:1237–1240. doi: 10.1637/9399-052110-Reg.1. [DOI] [PubMed] [Google Scholar]
- The Daily Star. 2019. [Prescription from vet must for giving antibiotics to cows: HC]. Available at: https://www.thedailystar.net/city/without-prescription-no-antibiotics-to-cows-high-court-1772242.
- Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, Adams V, Moore RJ, Rood JI, McClane BA. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014;9:361–377. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, B, Keyburn, A. The cost of necrotic enteritis is huge. World Poultry News 2015. 31: 5. 2015. [Accessed Oct.16.2019]. Available at: https://www.poultryworld.net/Meat/Articles/2015/10/The-true-cost-of-necrotic-enteritis-2699819W.
- Yang WY, Lee YJ, Lu HY, Branton SL, Chou CH, Wang C. The netB-positive Clostridium perfringens in the experimental induction of necrotic enteritis with or without predisposing factors. Poult. Sci. 2019;98:5297–5306. doi: 10.3382/ps/pez311. [DOI] [PubMed] [Google Scholar]
- Yoo HS, Lee SU, Park KY, Park YH. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J. Clin. Microbiol. 1997;35:228–232. doi: 10.1128/jcm.35.1.228-232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]