Abstract
Background
We investigated for change in blood stream infections (BSI) with Enterobacterales, coagulase negative staphylococci (CoNS), Streptococcus pneumoniae, and Staphylococcus aureus during the first UK wave of SARS-CoV-2 across five London hospitals.
Methods
A retrospective multicentre ecological analysis was undertaken evaluating all blood cultures taken from adults from 01 April 2017 to 30 April 2020 across five acute hospitals in London. Linear trend analysis and ARIMA models allowing for seasonality were used to look for significant variation.
Results
One hundred nineteen thousand five hundred eighty-four blood cultures were included. At the height of the UK SARS-CoV-2 first wave in April 2020, Enterobacterales bacteraemias were at an historic low across two London trusts (63/3814, 1.65%), whilst all CoNS BSI were at an historic high (173/3814, 4.25%). This differed significantly for both Enterobacterales (p = 0.013), CoNS central line associated BSIs (CLABSI) (p < 0.01) and CoNS non-CLABSI (p < 0.01), when compared with prior periods, even allowing for seasonal variation. S. pneumoniae (p = 0.631) and S. aureus (p = 0.617) BSI did not vary significant throughout the study period.
Conclusions
Significantly fewer than expected Enterobacterales BSI occurred during the UK peak of the COVID-19 pandemic; identifying potential causes, including potential unintended consequences of national self-isolation public health messaging, is essential. High rates of CoNS BSI, with evidence of increased CLABSI, but also likely contamination associated with increased use of personal protective equipment, may result in inappropriate antimicrobial use and indicates a clear area for intervention during further waves.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-021-06159-8.
Keywords: Microbiology, Blood culture, Coronavirus, SARS-CoV-2
Background
Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) began in December 2019 in China. Transmission within Europe, including the United Kingdom (UK), was confirmed by the end of February 2020. In the UK, social distancing and self-quarantine measures were subsequently implemented, aimed at slowing transmission. Furthermore, in an attempt to prevent overwhelming hospital capacity, those with high temperature and/or new continuous cough (i.e. symptoms consistent with COVID-19, but also other infections) were advised to stay home and seek advice online or via a national telephone service [1]. Elective clinical services were also reduced, with cancellation of non-urgent clinics and surgery. Extensive self-isolation practices and decreased utilisation of healthcare services has potentially impacted on direct care for both COVID-19 and non-COVID-19 clinical presentations.
Altered presentation of bloodstream infections (BSI) to healthcare is one potential indirect consequence of national public health measures. Whilst some organisms associated with BSIs have a clear communicable component (for example Streptococcus pneumoniae, Neisseria meningitidis and Staphylococcus aureus) [2–4], others are likely to arise endogenously [5]. Therefore, the incidence of communicable pathogens may decrease secondary to social distancing, or increase due to increased time spent with household contacts, while incidence of likely endogenous pathogens, such as Escherichia coli and other Enterobacterales, should be less affected. Potential changes must be considered however in the context of the seasonality for Enterobacterales (peaks seen in summer) [6, 7] and S. pneumoniae (peaks in winter). Finally, coagulase negative staphylococci (CoNS) are skin commensals and frequent contaminants of bloods cultures, although they can be implicated in BSIs for example, in the presence of intravascular catheters. Lower rates are seen with increased phlebotomy expertise/appropriate skin decontamination. International targets for contamination are less than 3% [8]. It may be hypothesised that incidence would increase with changes to working conditions, including implementation of personal protective equipment (PPE) for staff.
With NHS England data suggesting that emergency presentations to secondary care were down 29.4% in March 2020 compared to March 2019 [9], there are concerns over delayed or missed presentations with non-COVID-19 infections. To explore this, we investigated the incidence of BSIs during the initial months of the COVID-19 pandemic, focusing on Enterobacterales and CoNS, against variations in rates across the preceding 3 years. We used Enterobacterales, S. pneumoniae, and S. aureus BSI as indicator organisms for missed bacteraemia presentations and CoNS BSIs not related to a central line associated BSI (non-CLABSI) as a surrogate for contamination.
Methods
Study setting and design
A retrospective multicentre ecological analysis was undertaken evaluating all blood culture (BC) samples from adults (aged 17 years and above) over a three-year period from 01 April 2017 to 30 April 2020 across five acute hospitals in London, serving a population approximating 3 million.
A hub-and-spoke laboratory network with a centralized microbiology laboratory processes samples from multiple hospitals in accordance with UK laboratory standard operating procedures [10] with minor local variation. BCs were collected at each hospital, without pre-incubation, and transported to the centralized laboratory. They were subsequently incubated using a BACTEC system (Becton Dickinson, Franklin Lakes, NJ, USA). Organisms were identified by matrix assisted laser desorption/ionisation-time-of-flight (MALDI-TOF) mass spectroscopy (Bruker Daltonik GmbH, Bremen, Germany). Susceptibility testing was undertaken using disk diffusion using European Committee on Antimicrobial Susceptibility testing methods and interpretative criteria [11].
Data collection
Microbiological data was extracted from Sunquest Laboratory V8.3 (Tucson, AZ, USA). For the purposes of this study, Enterobacterales included Escherichia coli, Klebsiella spp., Serratia spp., Enterobacter spp., Proteus spp., Citrobacter spp., Hafnia spp., Morganella spp., and Pantoea spp. All CoNS, S.pneumoniae and S.aureus isolated from BCs were included; the seasonality of the latter two has been extensively described [12–15]. If more than one pathogen was isolated from a BC, each was recorded individually at either genus or species level. Samples with multiple Enterobacterales or CoNS were recorded as a single positive BC. Repeated positives within a 14-day period were de-duplicated except4 for CoNS related to CLABSIs. In order to determine this, blood patient notes were accessed and those with repeated BSIs with the same CoNS within a 14 day period in the presence of a central venous catheter, were classified as CLABSIs. Those where this could not be determined were not included in the statistical analysis.
Data analysis
To explore seasonal variations in BSI rates, results were classified as spring (March–May), summer (June–August), autumn (September–November) and winter (December–February). For spring 2017, only April and May data were available. March and April 2020 were considered as the period when social distancing measures were in place, encompassing the UK COVID-19 peak [16]. The same period in 2019 was analysed for comparison.
To explore changing trends in BCs results during COVID-19, two different statistical models were used; both (first) allowing for seasonality, and (second) then separately obviating any potential seasonality. The first method was a time-series analysis performed in R (R Core Team). After testing for the absence of first-order autocorrelations with the Durbin-Watson statistic, univariable autoregressive integrated moving average (ARIMA) models were fitted to data from CoNS (non-CLABSI and CLABSI), Enterobacterales, S.pneumoniae, and S.aureus from April 2017 to December 2019. These models were used to forecast estimated BSI rates for January to June 2020. To explore differences between observed and expected BSI rates during the COVID-19 peak, they were graphically compared.
The second method was a linear trend analysis of variations in observed rates of BSIs. Z-scores were calculated for Enterobacterales, CoNS (non-CLABSI and CLABSI), S.pneumoniae and S.aureus using SPSS (IBM Corp, Armonk, NY, USA). Data were checked for normality using the Shapiro-Wilk test. Where non-normal distribution was identified, data were transformed prior to analysis. Z-scores were then calculated each month by applying the standard formula z = (x-μ)/σ (x = raw score, μ = mean, σ = standard deviation). Statistical significance for z-scores were determined using R (R Core Team).
Study approval
This analysis was registered with North West London Pathology hosted by Imperial College Healthcare NHS Trust as a service evaluation (reference PAT_012) to investigate BC contamination rates. Individual consent was not indicated for this ecological level analysis reporting only aggregated data.
Results
During April 2017 to April 2020, 133,856 BCs were identified. Of those, 12,625 were from an external source (non-networked hospital), 2002 unclassifiable, and 729 not processed, leaving 119,584 for analysis. The overall number of BC requests remained stable throughout the entire period in terms of absolute numbers. From the 119,584 BCs, no pathogen/microorganism was detected in 109,144 (91.27%). Growth was demonstrated in 10,440 cultures (8.73%). Enterobacterales were the most commonly isolated pathogen (3508 cultures, 2.93% of total), followed by CoNS (2855 cultures, 2.39%). Full details of cultured microorganisms and number of tests are provided (Table 1).
Table 1.
Summary of positive and negative blood cultures collected during the period April 2017 to April 2020 across a six London hospital network
| Group | Number of blood cultures | % of total blood cultures | |
|---|---|---|---|
| Blood cultures with no growth (N; % of all blood cultures) | 109,144 | 91.27% | |
| Blood cultures with isolate identified (N; % of all blood cultures) | CoNS all | 2855 | 2.39% |
| - CLABSI | 2466 | 2.06% | |
| - Non-CLABSI | 341 | 0.29% | |
| - Unknown | 48 | 0.04% | |
| S.pneumoniae | 78 | 0.07% | |
| GNR-E | 3508 | 2.93% | |
| GNR-NE | 790 | 0.66% | |
| Other | 1975 | 1.65% | |
| S.aureus | 988 | 0.83% | |
| Yeasts | 246 | 0.21% |
Abbreviations: CoNS coagulase negative staphylococci, CLABSI Central line associated blood stream infection, Non-CLABSI Non-central line associated blood stream infection, Unknown unable to classify if CLABSI or Non-CLABSI, S.pneumoniae Streptococcus pneumoniae, GNR-E Gram Negative Rods – Enterobacterales, GNR-NE Gram Negative Rods- Non Enterobacterales, Other including Group A/B/C/G Streptococci, Gram positive rods, Actinobacillus spp., Aerococcus spp., Microbacterium spp., Micrococcus spp, S.aureus Staphylococcus aureus (both meticillin susceptible and meticillin resistant), Yeasts predominantly Candida spp.
Seasonal trends
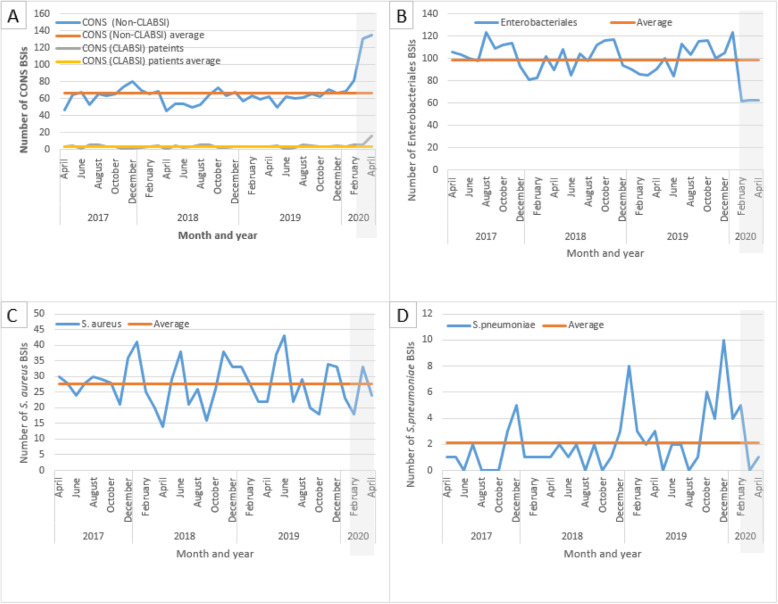
Seasonal trends were observed for S.pneumoniae and Enterobacterales. The seasonality of S.pneumoniae (preponderance towards winter months) is shown in Fig. 1d, whilst the seasonality of Enterobacterales (peaks in summer months) is shown in Fig. 1b. Across the cohort, Enterobacterales as a percentage of all BCs taken was lowest in the winter (2.49% 2017, 2.54% 2018 and 2.63% 2019), with a general trend towards sequentially rising throughout spring and into summer before peaking in autumn (3.42% 2017, 3.56% 2018 and 3.22% 2019) (Fig. 2). For all CoNS the mean percentage over the study period is 2.39%. There was little seasonal variation prior to COVID-19 (Fig. 2). For S. aureus the linear mean over the study period was 27.57, with significant variation over time, but little evidence of recursive seasonality (Fig. 1c).
Fig. 1.
Comparison of observed blood stream infection with linear trend analysis during the period April 2017 to April 2020 across a six London hospital network for a coagulase negative staphylococci, b Enterobacterales, c S.aureus, and d S.pneumoniae
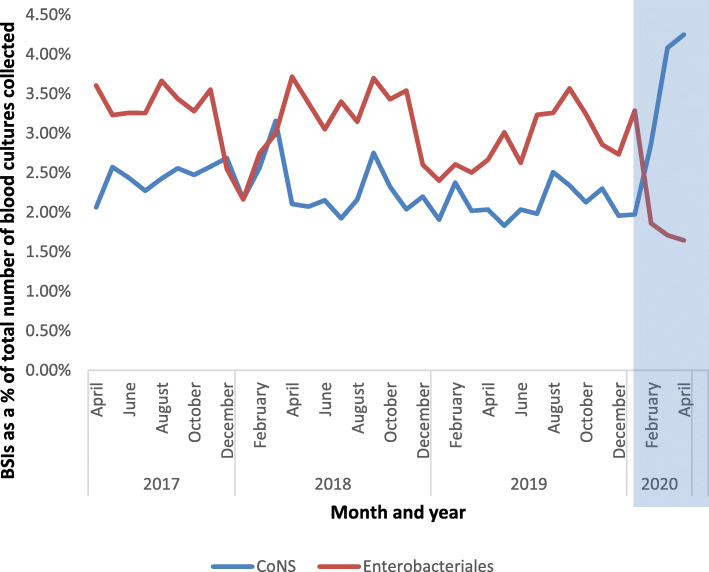
Fig. 2.
Change in blood culture isolates across six London hospitals during the COVID-19 social isolation measures in March and April 2020. Enterobacterales and CoNS (total) are shown and expressed as a percentage of the total number of blood cultures collected. For breakdown of CONS (non-CLABSI and CLABSI), please see Fig. 1
Bacteraemia variation during the UK COVID-19 peak March and April 2020
In April 2020, Enterobacterales as a percentage of all BCs taken were at their lowest during the study period at 1.65% (second lowest March 2020 at 1.71%; Fig. 2). In contrast, all CoNS, as a percentage of all BCs taken, were at their highest during the study period at 4.25% (second highest in March 2020 at 4.08%) (Fig. 2).
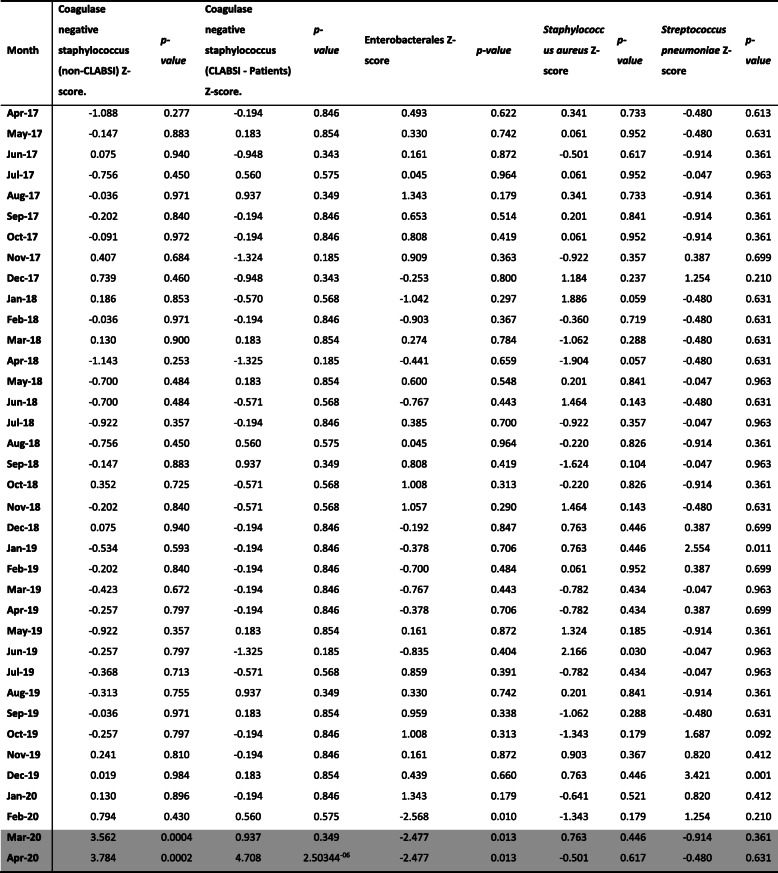
The linear trend analysis (Table 2) for Enterobacterales, CoNS, S.aureus and S.pneumoniae from April 2017 to April 2020 suggests significant differences in BSI rates, particularly during the COVID-19 period. Enterobacterales BSIs were significantly lower than the mean in February, March, and April 2020 only (Z-scores; − 2.57, − 2.48, and 2.48, respectively; p < 0.05). In contrast, CoNS (non-CLABSI) BSIs were significantly above the mean for March and April 2020 (Z-scores; 3.56, 3.78 respectively; p < 0.01). Similarly, CoNS (CLABSI patients) were significantly above the mean in April 2020 (Z-score 4.71, p < 0.01). Numbers detected did not deviate significantly from the mean from April 2017 until February and March 2020, respectively for these organisms. S.aureus BSIs did not vary significantly from mean throughout the period analysed. S.pneumoniae BSIs did not vary significantly from the mean throughout the period analysed, except for in December 2019. Before Spring 2020, the highest number of patients with a CLABSI was six per month (range 0–6), however, in April 2020 we identified 16 patients with a CLABSI. Whilst the numbers were too small to undertake statistical analysis, there was a trend towards an increased number of patients with CLABSIs in April 2020.
Table 2.
Variation in blood stream infection during the period April 2017 to April 2020 across a six London hospital network for Enterobaterales, CoNS, S.aureus and S.pneumoniae. Z-score calculated as variation from linear mean. The month of March and April 2020 during the COVID-19 social isolation measures are highlighted in grey
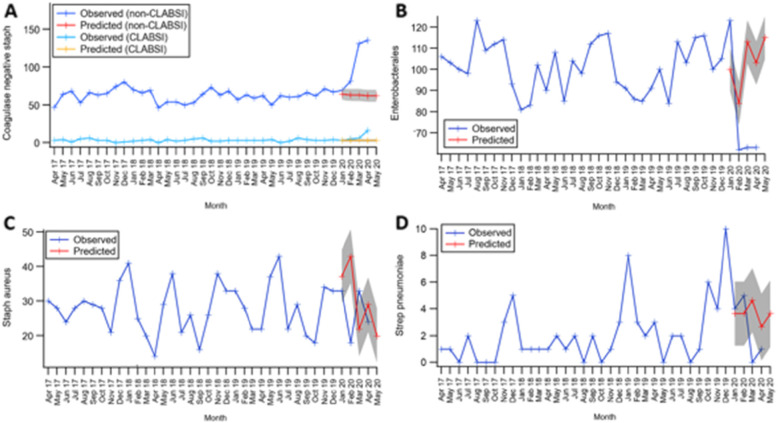
The ARIMA model, constructed to allow for any seasonality, is represented in Fig. 3a-d and demonstrates historic observed BSI (April 2017–December 2019) and then predicted versus observed BSI from January 2020 to June 2020. Once seasonality is allowed for, this also showed a significant divergence in observed versus predicted Enterobacterales and CoNS from February to April 2020. Fewer numbers of Enterobacterales BSIs and higher numbers of CoNS BSIs (non-CLABSI and patients with CLABSIs) were seen. Numbers of S.aureus and S.pneumoniae BSI remained within the limits of confidence of predicted based on historical trends.
Fig. 3.
Comparison of observed versus predicted (ARIMA model Jan-June 2020) trends in blood stream infection during the period April 2017 to April 2020 across a six London hospital network for a Coagulase negative Staphylococci, b Enterobacterales, c S.aureus, and d S.pneumoniae
Discussion
In our multi-centre BSI ecological analysis we find during the period of COVID-19 social distancing and self-isolation a significant reduction in Enterobacterales BSIs verified across two different statistical models. Similarly, there was a significant increase in non-CLABSI CoNS (presumptively contamination) BSI, whilst BSI with ostensibly communicable organisms (S.aureus and S.pneumoniae) remained stable. The observation of reduced Enterobacterales BSI immediately preceding and contemporaneous with public health measures, at a time when they would usually be increasing in incidence, requires urgent consideration [6, 7]. Given that Enterobacterales BSI could be considered non-communicable and likely endogenous in origin, incidence would be expected to remain similar allowing for seasonality. This observed reduction may reflect changes in presentation behaviour, with patients either self-isolating with ‘COVID-19 symptoms’ (e.g. fever), or avoiding healthcare due to fear of COVID-19 exposure. Certainly, emergency department attendances were lower than expected [9], in keeping with patients not presenting. Figures from England in 2018/19 show a 30-day mortality rate of 10.4 per 100,000 of the population and case fatality rate of 13.8% of cases [17] for Enterobacterales BSIs and there is concern that patients experiencing but not presenting with a BSI may have suffered fatalities in the community.
Widespread cancellation of elective surgical procedures offers an alternative explanation for the reduction in Enterobacterales BSIs. Gastrointestinal (GI) and urological procedures are a well-established source of Enterobacterales BSI [18, 19]. Furthermore, many patients presenting to hospital who fit the broad case definition for COVID-19 are rapidly started on broad-spectrum antimicrobials. As antimicrobials affect the detection rate of Enterobacterales from BCs [20], this may also be impacting our observed case rate.
The increased number of CoNS (non-CLABSI) BSIs seen during the pandemic may reflect increased rates of contamination. Contamination rates are reduced with increased phlebotomy expertise, but with staff performing procedures in unfamiliar PPE, the practicalities of venepuncture might prove more difficult. Additionally, more BCs are likely to be taken in patients with COVID-19 due to its febrile nature. This has been substantiated by Sepulveda et al. [21] who found a surge in BCs during March 2020, the majority of which were for SARS-CoV-2 positive patients, including repeated sampling. They found that CoNS accounted for 59.7% of positive BCs in these patients. Furthermore, data from Hughes et al. [22] has demonstrated infrequent confirmed secondary bacterial infection, approximately 3–6%. When planning for future COVID-19 peaks and/or other pandemics, these findings would support the need for dedicated teaching on aseptic non-touch techniques (ANTT) whilst in PPE, in an attempt to reduce contamination.
This could also potentially impact on antimicrobial stewardship and resistance [23]. Infection teams may recommend the addition of glycopeptides when Gram-positive cocci (GPC) are isolated from BCs, in the presence of a central line, whilst waiting full identification and sensitivities. To prevent unnecessary glycopeptide prescriptions, potential adverse drug events, antimicrobial resistance, and increased costs, it is important to have COVID-19 specific antimicrobial guidance reflecting the likelihood of increased BC contamination.
The absolute number of patients with CoNS CLABSI was also increased during April 2020 of the pandemic, which may reflect increased ITU capacity during this time, or alternatively, the challenges of managing patients within an ITU setting during the pandemic. For example, challenges of aseptic technique in full PPE as well as redeployment of staff from other areas to ITU and also, in patients with COVID-19, repositioning to try and improve oxygenation, increasing the risk of line displacement and/or contamination.
Our study has several limitations. We looked at total number of blood culture requests and so repeated samples from the same patients may have been included. Also, repeated positive BCs within a 14-day period were not de-duplicated for organisms other than CoNS CLABSI, which may have affected incidence. Also, the absolute number of patients with CoNS CLABSI was reported, but in order to determine if the rate of CoNS CLABSI was increased, we would have to look at ITU admission data over the same period. Furthermore, timing or setting of sampling (e.g. ED versus inpatient) was not assessed and thus whether Enterobacterales BSIs were community or hospital acquired. To gain finer resolution on the causes for the observed fall Enterobacterales BSI, it would be necessary to look at community and hospital acquired BSI related to elective GI/urological procedures over the same time frame. In addition, because this was an ecological analysis, we did not describe the epidemiology of patients including whether they had tested positive for COVID-19 or not, nor did we use a COVID-free hospital as a control.
Conclusion
During the peak of the COVID-19 pandemic in London, significantly fewer Enterobacterales BSI occurred, alongside fewer documented emergency presentations. This potentially reflects reduced presentations of Enterobacterales BSI due to patients not presenting to secondary care. We suggest more nuanced public health messaging around self-isolation for febrile illnesses to ensure patients present to healthcare where necessary. High rates of CoNS BSI during the COVID-19 peak, likely due to contamination, might reflect an unintended consequence of PPE. We suggest increased training for ANTT procedures whilst using PPE to prepare for a second CoVID-19 wave or any future pandemics. We suggest specific antimicrobial guidelines for patients with COVID-19, not only accounting for fine resolution epidemiology on the frequency of bacterial co-infection, but also reflecting caution in reacting to GPCs in BC bottles.
Supplementary Information
Acknowledgements
LSPM and TMR acknowledge support from the National Institute of Health Research (NIHR) Imperial Biomedical Research Centre (BRC) and the National Institute for Health Research Health Protection Research Unit (HPRU) in Healthcare Associated Infection and Antimicrobial Resistance at Imperial College London in partnership with Public Health England.
The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the UK Department of Health.
Abbreviations
- ANTT
Aseptic non-touch technique
- ARIMA
Autoregressive integrated moving average
- BC
Blood culture
- BSI
Blood stream infection
- CLABSI
Central line associated blood stream infection
- CoNS
Coagulase negative Staphylococci
- GI
Gastrointestinal
- GNR-E
Gram negative rods – Enterobacterales
- GNR-NE
Gram negative rods – Non Enterobacterales
- GPC
Gram-positive cocci
- MALDI-TOF
Matrix-assisted laser desorption/ionisation-time-of-flight
- Non-CLABSI
Non-Central line associated blood stream infection
- PPE
Personal protective equipment
- SARS-CoV-2
Severe Acute Respiratory Syndrome coronavirus 2
- S.pneumo
Streptococcus pneumoniae
- UK
United Kingdom
Authors’ contributions
SD, GS and LSPM designed the study methodology. SD collated the data. SD, TMR, PH, GS, SJCP, AA, SH, MG, NM and LSPM reviewed the results, SD, TMR, PH and GS lead the data analysis, with SD, TMR, PH, GS, SJCP, AA, SH, MG, NM and LSPM reviewing the resulting themes and contributing comments. SD drafted the initial manuscript with SD, TMR, PH, GS, SJCP, AA, SH, MG, NM and LSPM contributing significantly to revising this for submission. SD, TMR, PH, GS, SJCP, AA, SH, MG, NM and LSPM agreed on the final version for submission to the journal. All authors read and approved the final manuscript.
Funding
This research did not receive any grant from funding agencies in the public or commercial sectors.
Availability of data and materials
The data analysed during the current study is available from the first author (SD; sarahdenny1@nhs.net) on reasonable request, as long as this meets local ethical and research governance criteria.
Declarations
Ethics approval and consent to participate
This analysis was registered with North West London Pathology hosted by Imperial College Healthcare NHS Trust as a service evaluation (reference PAT_012) to investigate BC contamination rates. Ethics approval and individual consent was not indicated for this ecological level analysis reporting only aggregated data which was anonymised at the point of extraction. The aggregated data was analysed in accordance with the UK Secretary of State for Health and Social care notice that stipulated that patient data for COVID-19 purposed may be used for research under the Health Service Control of Patient Information Regulations (2002). Administrative permission was not required to access the raw data.
Consent for publication
Not applicable.
Competing interests
LSPM has consulted for/received speaker fees from DNAelectronics (2015–18), Dairy Crest (2017–2018), Profile Pharma (2018), Eumedica (2016–2021), Umovis Lab (2020–2021), bioMerieux (2013–2021), and Pfizer (2018–2021), and received research grants from the National Institute for Health Research (2013–2019), and CW+ Charity (2018–2021). GS is currently supported by a Clinical Academic Research Partnership awarded by the Medical Research Council. SJCP has received a research grant from the Scientific Exploration Society. NM has received speaker fees from Beyer (2016) and Pfizer (2019-2021) and received educational support from Eumedica (2016) and Baxter (2017).
All other authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.website N. Check if you or your child has coronavirus symptoms 2020 [Available from: https://www.nhs.uk/conditions/coronavirus-covid-19/symptoms/. Accessed 18 Oct 2020.
- 2.Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16(6):355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouphael NG, Stephens DS. Neisseria meningitidis: biology, microbiology, and epidemiology. Methods Mol Biol (Clifton, NJ) 2012;799:1–20. doi: 10.1007/978-1-61779-346-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knox J, Uhlemann A-C, Lowy FD. Staphylococcus aureus infections: transmission within households and the community. Trends Microbiol. 2015;23(7):437–444. doi: 10.1016/j.tim.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliveira J, Reygaert WC. Gram negative bacteria. 2019. [PubMed] [Google Scholar]
- 6.Al-Hasan MN, Lahr BD, Eckel-Passow JE, Baddour LM. Seasonal variation in escherichia coli bloodstream infection: a population-based study. Clin Microbiol Infect. 2009;15(10):947–950. doi: 10.1111/j.1469-0691.2009.02877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson J, Elgohari S, Livermore DM, Cookson B, Johnson A, Lamagni T, Chronias A, Sheridan E. Trends among pathogens reported as causing bacteraemia in England, 2004–2008. Clin Microbiol Infect. 2011;17(3):451–458. doi: 10.1111/j.1469-0691.2010.03262.x. [DOI] [PubMed] [Google Scholar]
- 8.Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19(4):788–802. doi: 10.1128/CMR.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.England N. A&E Attendances and Emergency Admissions 2020 [Available from: https://www.england.nhs.uk/statistics/statistical-work-areas/ae-waiting-times-and-activity/]. Accessed 18 Oct 2020.
- 10.England PH. UK SMI B 37: investigation of blood cultures (for organisms other than Mycobacterium species) 2019. [Google Scholar]
- 11.EUCAST. Clinical breakpoints - breakpoints and guidance 2020 [Available from: https://www.eucast.org/clinical_breakpoints/]. Accessed 18 Oct 2020.
- 12.Dowell SF, Whitney CG, Wright C, Rose CE, Jr, Schuchat A. Seasonal patterns of invasive pneumococcal disease. Emerg Infect Dis. 2003;9(5):573–579. doi: 10.3201/eid0905.020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Numminen E, Chewapreecha C, Turner C, Goldblatt D, Nosten F, Bentley SD, Turner P, Corander J. Climate induces seasonality in pneumococcal transmission. Sci Rep. 2015;5(1):11344. doi: 10.1038/srep11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberger DM, Harboe ZB, Viboud C, Krause TG, Miller M, Mølbak K, et al. Pneumococcal disease seasonality: incidence, severity and the role of influenza activity. Eur Respir J. 2014;43(3):833–841. doi: 10.1183/09031936.00056813. [DOI] [PubMed] [Google Scholar]
- 15.Leekha S, Diekema DJ, Perencevich EN. Seasonality of staphylococcal infections. Clin Microbiol Infect. 2012;18(10):927–933. doi: 10.1111/j.1469-0691.2012.03955.x. [DOI] [PubMed] [Google Scholar]
- 16.LSHTM. Peak in COVID-19 deaths occurring in English hospitals passed on 8 April 2020 [Available from: https://www.lshtm.ac.uk/newsevents/news/2020/peak-covid-19-deaths-occurring-english-hospitals-passed-8-april]. Accessed 18 Oct 2020.
- 17.England PH. Thirty-day all-cause mortality following MRSA, MSSA and Gram-negative bacteraemia and C. difficile infections 2019 [Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/850521/hcai_fatality_report_201819.pdf]. Accessed 18 Oct 2020.
- 18.Alqarni A, Kantor E, Grall N, Tanaka S, Zappella N, Godement M, Ribeiro-Parenti L, Tran-Dinh A, Montravers P. Clinical characteristics and prognosis of bacteraemia during postoperative intra-abdominal infections. Crit Care. 2018;22(1):175. doi: 10.1186/s13054-018-2099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson ES, Cookson BD. Do antimicrobials have a role in preventing septicaemia following instrumentation of the urinary tract? J Hosp Infect. 2000;45(2):85–97. doi: 10.1053/jhin.1999.0735. [DOI] [PubMed] [Google Scholar]
- 20.Scheer CS, Fuchs C, Gründling M, Vollmer M, Bast J, Bohnert JA, Zimmermann K, Hahnenkamp K, Rehberg S, Kuhn SO. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: a prospective clinical cohort study. Clin Microbiol Infect. 2019;25(3):326–331. doi: 10.1016/j.cmi.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Sepulveda J, Westblade LF, Whittier S, Satlin MJ, Greendyke WG, Aaron JG, et al. Bacteremia and Blood Culture Utilization During COVID-19 Surge in New York City. J Clin Microbiol. 2020;58:e00875-20. 10.1128/JCM.00875-20. [DOI] [PMC free article] [PubMed]
- 22.Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalised patients with COVID-19: a retrospective cohort study in a UK secondary care setting. Clin Microbiol Infect. 2020; S1198-743X(20)30369-4. doi: 10.1016/j.cmi.2020.06.025. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 23.Rawson TM, Moore LSP, Castro-Sanchez E, Charani E, Davies F, Satta G, Ellington MJ, Holmes AH. COVID-19 and the potential long-term impact on antimicrobial resistance. J Antimicrob Chemother. 2020;75(7):1681–1684. doi: 10.1093/jac/dkaa194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analysed during the current study is available from the first author (SD; sarahdenny1@nhs.net) on reasonable request, as long as this meets local ethical and research governance criteria.