Polθ reverse transcribes RNA by undergoing a significant conformational change and promotes RNA-templated DNA repair.
Abstract
Genome-embedded ribonucleotides arrest replicative DNA polymerases (Pols) and cause DNA breaks. Whether mammalian DNA repair Pols efficiently use template ribonucleotides and promote RNA-templated DNA repair synthesis remains unknown. We find that human Polθ reverse transcribes RNA, similar to retroviral reverse transcriptases (RTs). Polθ exhibits a significantly higher velocity and fidelity of deoxyribonucleotide incorporation on RNA versus DNA. The 3.2-Å crystal structure of Polθ on a DNA/RNA primer-template with bound deoxyribonucleotide reveals that the enzyme undergoes a major structural transformation within the thumb subdomain to accommodate A-form DNA/RNA and forms multiple hydrogen bonds with template ribose 2′-hydroxyl groups like retroviral RTs. Last, we find that Polθ promotes RNA-templated DNA repair in mammalian cells. These findings suggest that Polθ was selected to accommodate template ribonucleotides during DNA repair.
INTRODUCTION
Polymerase θ (Polθ) is a unique DNA polymerase-helicase fusion protein in higher eukaryotes whose A-family polymerase domain evolved from Pol I enzymes (Fig. 1A) (1, 2). However, contrary to most Pol I enzymes, Polθ is highly error-prone and promiscuous (3–6), performs translesion synthesis (TLS) opposite DNA lesions (3, 7, 8), and facilitates microhomology-mediated end-joining (MMEJ) of double-strand breaks (DSBs) by extending partially base-paired 3′ single-stranded DNA (ssDNA) overhangs at DSB repair junctions (5, 9–12). Polθ is not expressed in most tissues but is highly expressed in many cancer cells, which corresponds to a poor clinical outcome (13, 14). Furthermore, Polθ confers resistance to genotoxic cancer therapies and promotes the survival of cells deficient in DNA damage response pathways (11, 13–16). Thus, Polθ represents a promising cancer drug target.
Fig. 1. Polθ exhibits reverse transcriptase activity.
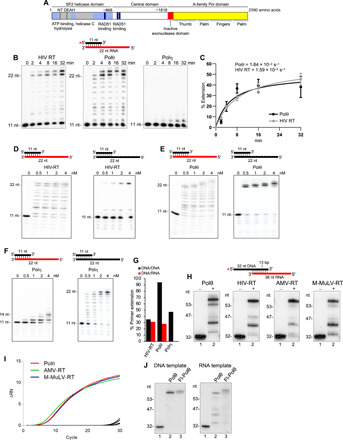
(A) Schematic of full-length Polθ. (B) Denaturing gels showing a time course of DNA/RNA primer-template extension by the indicated polymerases. (C) Plot showing relative rate of DNA/RNA extension by Polθ and HIV RT. Data represent mean ± SD; n = 3. (D to F) Denaturing gels showing DNA/RNA (left) and DNA/DNA (right) extension by the indicated polymerases. (G) Bar plot showing percent extension of DNA/RNA (red) and DNA/DNA (black) by the indicated polymerases (4 nM) [data from (D) to (F)]. (H) Denaturing gels showing DNA/RNA extension by the indicated polymerases. (I) Quantitative PCR chromatogram showing cDNA synthesis by the indicated polymerases. (J) Denaturing gels showing DNA/DNA (left) and DNA/RNA (right) extension by Polθ and Fl-Polθ.
Intriguingly, Polθ has an inactive proofreading domain due to acquired mutations (Fig. 1A) (2). Inactivating the 3′-5′ proofreading function of closely related A-family bacterial Pol I Klenow fragment (KF) enables this polymerase to reverse transcribe RNA like retroviral reverse transcriptases (RTs), which lack proofreading activity (fig. S1A) (17, 18). Because Polθ is highly error-prone and promiscuous and contains an inactive proofreading domain, we hypothesized that it has RNA-dependent DNA synthesis activity. Given that ribonucleotides are the most frequently occurring nucleotide lesion in genomic DNA that arrest replicative Pols and cause DNA breaks (19, 20), we also envisaged that Polθ would tolerate template ribonucleotides during its DNA repair activities and thus promote RNA-templated DNA repair synthesis (RNA-DNA repair). Although RNA-DNA repair mechanisms have been demonstrated in genetically engineered yeast cells (21, 22), they remain obscure in mammalian cells.
RESULTS
Polθ exhibits RNA-dependent DNA synthesis activity
We tested whether the polymerase domain of Polθ (herein referred to as Polθ) reverse transcribes RNA like HIV RT using a DNA primer annealed to a RNA template (DNA/RNA). Polθ exhibits a similar rate of RT activity as HIV RT under identical conditions using substoichiometric amounts of enzyme relative to template (Fig. 1, B and C). Previous studies indicated that human Polη has RT activity at high micromolar concentrations (23). We find that Polη fails to perform reverse transcription beyond 3 nucleotides (nt) using conditions identical to those of Polθ and HIV RT at multiple concentrations (Fig. 1, B and C; compare Fig. 1F with Fig. 1, D and E; Fig. 1G). However, at significantly higher concentrations, Polη can further extend the DNA/RNA (figs. S1, B to E). Controls show that Polη is active on a DNA/DNA template like Polθ and HIV RT (compare Fig. 1F with Fig. 1, D and E; Fig. 1G). Overall, Polη exhibits increased stalling on RNA and requires higher enzyme concentration relative to Polθ for reverse transcription (figs. S1, B to E). Polθ RNA-dependent DNA synthesis activity is observed under various conditions and on different template constructs (figs. S1, F and G), and sequences (fig. S1H). Despite its robust activity on RNA, Polθ strongly discriminates against incorporating ribonucleotides (fig. S1I). Complementary DNA (cDNA) sequencing confirms Polθ’s RNA-dependent DNA synthesis activity (fig. S2A) and reveals nucleotide misincorporations and indels, which is consistent with its low-fidelity DNA synthesis activity (fig. S2A) (3). HIV RT and other retrovirus RTs are also highly error-prone, demonstrating a shared characteristic between Polθ and retroviral RTs (18, 24–28). In addition to HIV RT, Polθ activity on RNA is nearly identical to RTs encoded by Moloney murine leukemia virus (M-MuLV) and avian myeloblastosis virus (AMV) (Fig. 1H). Polθ exhibits pausing events nearly identical to those of retrovirus RTs, which is consistent with pausing tendencies for RTs (Fig. 1H) (29, 30). Polθ cDNA synthesis on synthetic RNA is similar to M-MuLV and AMV RTs (Fig. 1I and fig. S2B), and Polθ also promotes cDNA synthesis of purified Escherichia coli 16S ribosomal RNA similar to M-MuLV and AMV RTs (fig. S2C).
The efficient RT activity of Polθ appears to be unique among human Pols. Eight other human Pols, representative of at least two enzymes from each polymerase family in humans (A, B, X, and Y), fail to reverse transcribe DNA beyond 2 to 3 nt under conditions identical to those of Polθ (Fig. 1F and fig. S3). Y-family polymerase κ (Polκ) exhibits limited RT activity under various conditions similar to Polη (Fig. S3H). Replicative Pols δ and ε degrade the DNA primer on RNA due to exonuclease activity (fig. S3, F and G). All Pols are active on DNA as expected (Fig. 1F and fig. S3). Full-length Polθ (Fl-Polθ) containing an N-terminal superfamily 2 and disordered central domain (Fig. 1A) (1) also has RT activity, suggesting that the endogenous protein performs RNA-DNA repair in cells (Fig. 1J). Because recombinant human Polθ and Fl-Polθ were purified from different organisms (E. coli and Saccharomyces cerevisiae, respectively) and by different methods (1, 9), the observed RT activity is not due to a protein contaminant. Consistent with this, human recombinant Polθ purified from S. cerevisiae also has robust RT activity (fig. S4A) (31).
Polθ exhibits higher velocity and fidelity of deoxyribonucleotide incorporation on RNA
We tested the relative velocity of Polθ deoxyribonucleoside monophosphate (dNMP) incorporation on RNA versus DNA. Single deoxyribonucleoside triphosphates (dNTPs) were incubated with Polθ during a time course on DNA/RNA versus DNA/DNA with identical sequence (Fig. 2A). Polθ exhibits a significantly higher velocity of incorporating deoxycytidine monophosphate (dCMP), deoxythymidine monophosphate (dTMP), and deoxyadenosine monophosphate (dAMP) on RNA (Fig. 2, B to D, and fig. S4B). The velocity of deoxyguanosine monophosphate (dGMP) incorporation was similar on RNA and DNA (Fig. 2E). Fl-Polθ also exhibits higher rates of dCMP, dTMP, and dAMP incorporation on RNA (fig. S5). Polθ exhibits a twofold higher affinity for DNA/RNA (fig. S4C), which may contribute to the higher velocity of nucleotide incorporation.
Fig. 2. Polθ exhibits a higher fidelity and velocity of deoxyribonucleotide incorporation on RNA.
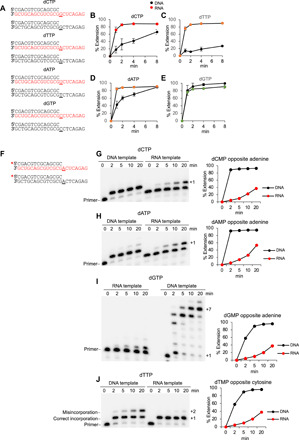
(A) Schematic of DNA/DNA and DNA/RNA templates used for the indicated dNTPs. Underlined base codes for the incoming nucleotide. Red, RNA; black, DNA. (B to E) Plots showing relative velocity of the indicated deoxyribonucleotide incorporation by Polθ on DNA/RNA and DNA/DNA primer-templates. Red, DNA/RNA; black, DNA/DNA. Data represent mean ± SD; n = 3. (F) Schematic of DNA/RNA and DNA/DNA primer-templates. Underlined base codes for the correct incoming nucleotide. Red, RNA; black, DNA. (G to J) Denaturing gels showing a time course of Polθ primer extension on the indicated DNA/RNA and DNA/DNA primer-templates in the presence of the indicated deoxyribonucleotide (left). Plots showing percent extension over time (right). Red, DNA/RNA; black, DNA/DNA. dTMP.thymidine monophosphate.
To assess the relative fidelity of dNMP incorporation by Polθ on RNA versus DNA, we measured the relative velocity of nucleotide misincorporation. Remarkably, Polθ is significantly more accurate on RNA versus DNA as demonstrated by its severely limited ability to misincorporate nucleotides on RNA versus DNA (Fig. 2, F to J). For example, in Fig. 2 (G and H), Polθ fully misincorporates dCMP and dAMP, respectively, on DNA in less than 2 min. Yet, on RNA, Polθ is unable to fully misincorporate dCMP or dAMP even after 20 min (Fig. 2, G and H). Polθ also fails to effectively misincorporate dTMP on RNA but efficiently misincorporates dTMP on DNA (Fig. 2J). Remarkably, Polθ rapidly misincorporates several consecutive dGMPs on DNA yet fails to fully misincorporate a single dGMP on RNA even after 20 min (Fig. 2I). Consecutive dGMP misincorporation events on DNA suggest that Polθ more easily misaligns the DNA/DNA primer-template (fig. S6A). The higher fidelity of Polθ on RNA versus DNA is also observed in a different sequence context (fig. S6, B to F). Hence, despite the enzyme’s overall higher rate of correct dNMP incorporation on RNA, it exhibits a substantially slower rate of misincorporation on RNA. These data suggest that Polθ evolved to be more accurate on RNA similar to HIV RT (32).
Ternary structure of Polθ on a DNA/RNA primer-template
The higher fidelity of Polθ on RNA suggests that it binds the DNA/RNA and/or active site deoxyribonucleotide:ribonucleotide base pair in a distinct conformation. To investigate the molecular basis of Polθ RT activity, we solved a 3.2-Å crystal structure of Polθ on DNA/RNA with incoming 2′,3′-dideoxyguanosine triphosphate (ddGTP) (Fig. 3). The construct used for x-ray crystallography (PolθΔL) was engineered to achieve higher E. coli expression by replacing five small disordered loops, which were not resolved in previous Polθ:DNA/DNA structures (2), with short glycine-serine inserts (Fig. 3A and fig. S7, A to C). The endogenous insert 2 loop promotes Polθ MMEJ of DNA/DNA with 3′ ssDNA overhangs containing microhomology (9), and inserts 2 and 3 contribute to TLS (2, 7). WT (wild type) and PolθΔL exhibit similar DNA/DNA and DNA/RNA primer extension activities, demonstrating that the disordered loops do not substantially contribute to reverse transcription and canonical DNA/DNA extension (Fig. 3B and fig. S7D). In contrast to prior Polθ:DNA/DNA:ddNTP structures that were captured in the closed conformation (2), the PolθΔL:DNA/RNA:ddGTP complex is in the open form, whereby the O helix of the fingers subdomain is rotated outward by 42° and the bound ddGTP is solvent-exposed (Fig. 3, C and D, and fig. S8a). This demonstrates that Polθ shares the induced-fit nucleotide incorporation mechanism with related A-family polymerases (2, 33).
Fig. 3. Ternary structure of Polθ on a DNA/RNA primer-template.
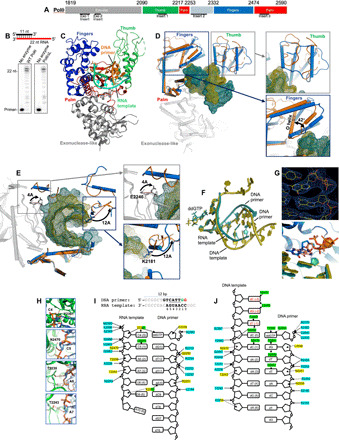
(A) Polθ polymerase. (B) DNA/RNA extension by Polθ and PolθΔL. (C) Structure of Polθ:DNA/RNA:ddGTP. (D) Superposition of Polθ:DNA/RNA (marine) and Polθ:DNA/DNA (orange, 4x0q). The fingers and thumb subdomains undergo reconfiguration. (E) Superposition of Polθ:DNA/RNA (marine) and Polθ:DNA/DNA (orange, 4x0q) highlighting a 12-Å shift of K2181 (blue box; thumb) and a 4.4-Å shift of E2246 (gray box; palm). (F) Superposition of nucleic acids and ddGTP from Polθ:DNA/RNA:ddGTP and Polθ:DNA/DNA:ddGTP structures. (G) Top: Electron density of ddGTP and 3′ primer terminus in Polθ:DNA/RNA structure. Bottom: Zoomed-in image of the superposition of active sites, illustrating a different conformation of ddGTP in the Polθ:DNA/RNA (blue) and Polθ:DNA/DNA (salmon) complexes. (H) Interactions between ribose 2′-hydroxyl groups of the RNA template and residues in the Polθ:DNA/RNA structure. Red dashed lines, hydrogen bonds. (I) DNA/RNA used for cocrystallization with Polθ and ddGTP (top). Strong electron density is present for four base pairs [nucleotides located at positions 2 to 5 (underlined) of the DNA/RNA] and two base pairs resulting from an incorporated ddGMP (2’,3’ dideoxyguanosine monophosphate) (green; position 1) and a bound unincorporated ddGTP (red; position 0) in the active site (top). Interactions between Polθ and nucleic acids in Polθ:DNA/RNA:ddGTP (bottom). Interactions between residues and phosphate backbone, sugar oxygen, or nucleobase are shown in blue, yellow, and green, respectively. Hydrogen bonds between Polθ and ribose 2′-hydroxyl groups are indicated (boxed residues). (J) Interactions between Polθ and nucleic acids in Polθ:DNA/DNA:ddGTP (4x0q). Color scheme identical to (I).
Unexpectedly, the thumb subdomain undergoes a major reconfiguration. Fifty-seven percent of thumb subdomain residues refold from α helices to loops (Fig. 3D and fig. S7C, thumb domain), which may be necessary to accommodate the thicker A-form DNA/RNA. A loop shift in the palm subdomain involving E2246 is also observed on the opposite side of the DNA/RNA, suggesting a specific ribose 2′-hydroxyl interaction with its main-chain carbonyl likely mediated through a water molecule (Fig. 3E, gray). Superposition of the DNA/RNA from our structure onto DNA/DNA from the prior Polθ:DNA/DNA:ddGTP structure reveals that the DNA/RNA has a shorter distance between neighboring base pairs near the 3′ primer terminus (Fig. 3F and fig. S8B). The general features of the DNA/RNA are A-form–like, whereas the DNA/DNA is B-form–like upstream from the active site (Fig. 3F and fig. S8b). Structural irregularities (weakly paired bases and mismatches) are observed near the upstream portion of the DNA/RNA (Fig. 3I). The incoming ddGTP and complementary cytidine on the RNA template also show a significant shift relative to the Polθ:DNA/DNA:ddGTP complex (Fig. 3G).
Polθ accommodation of DNA/RNA also involves many RNA template interactions (compare Fig. 3, I and J). Multiple hydrogen bonds with ribose 2′-hydroxyl groups along the RNA template are observed (Fig. 3, H and I). Polθ, HIV RT, and M-MuLV RT form a similar carbonyl hydrogen bond with the 2′-hydroxyl group of the active site template ribose, suggesting a conserved mechanism of active site RNA template binding (fig. S9, C and D) (34, 35). This interaction and additional Polθ:2′-hydroxyl ribose hydrogen bonds along the RNA template may suppress Polθ template misalignment errors and thus potentially contribute to its higher fidelity on RNA. Overall, the formation of multiple specific Polθ:RNA interactions combined with the major thumb subdomain reconfiguration and palm subdomain loop shift reveal how the polymerase becomes active on a DNA/RNA hybrid.
Polθ promotes RNA-templated DNA repair
To test whether Polθ promotes RNA-templated DNA repair synthesis (RNA-DNA repair) in a biological setting, we developed a cellular green fluorescent protein (GFP) reporter assay that can simultaneously quantitate Polθ MMEJ and RNA-DNA repair (Fig. 4B). Polθ is essential for MMEJ of DSBs, such as those caused by ionizing radiation (1, 9–11), and performs TLS (7, 31). Polθ therefore regularly uses aberrant template bases during its DNA repair activities. Polθ promotes MMEJ by using microhomology [≥2 base pairs (bp)] between 3′ ssDNA overhangs generated by 5′-3′ resection of DSBs and then extends the partially base-paired overhangs (Fig. 4A). The MMEJ GFP reporter assay is described as follows. Left and right DNA constructs respectively encoding the upstream and downstream portion of a GFP expression vector with 6 bp of overlapping sequence (microhomology) were conjugated with streptavidin at 5′ DNA termini opposite from the microhomology tract to suppress 5′-3′ exonuclease activity at these ends (Fig. 4B). Introduction of 5 or 10 ribonucleoside monophosphates (NMPs) into the nontemplate transcription strand immediately upstream from the microhomology tract within the left construct enables analysis of RNA-templated DNA repair synthesis during MMEJ (Fig. 4, B and C). In the case of the left construct with five NMPs, base mutations within the transcription template strand directly opposite the RNA tract were added to engineer a stop codon in the GFP coding sequence (Fig. 4C, middle). Cotransfection of the right construct and left construct with or without NMP tracts into mouse induced pluripotent stem cells (iPSCs) activates GFP, demonstrating the capacity of MMEJ to use deoxyribose and ribose nucleobases as templates for DNA repair synthesis (Fig. 4D). Sequencing confirms MMEJ of the left and right DNA constructs in cells as well as error-prone repair (Fig. 4E and fig. S10). Cotransfection of the right construct and left construct without NMPs results in significantly higher %GFP in Polq+/+ versus Polq−/− iPSCs that were previously characterized (Fig. 4F, left) (10). This demonstrates that Polθ promotes MMEJ as expected. Cotransfection of the right construct with the left construct containing 5 or 10 NMPs also results in significantly higher %GFP in Polq+/+ iPSCs, demonstrating that Polθ promotes RNA-DNA repair (Fig. 4F, middle and right). In the case of the left construct containing the stop codon opposite the 5-NMP tract (Fig. 4C, middle), RT activity is essential for generating a transcription template strand lacking the stop codon. Hence, these data confirm that Polθ promotes RNA-dependent DNA synthesis in cells.
Fig. 4. Polθ promotes RNA-templated DNA repair synthesis in cells.
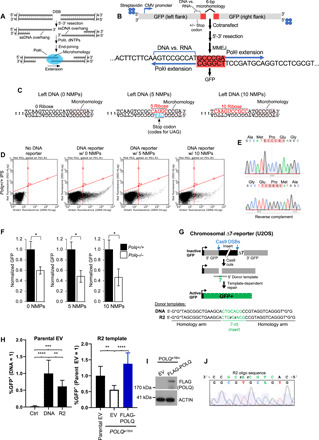
(A) Schematic of MMEJ. (B) Schematic of GFP MMEJ reporter assay. (C) Sequences of downstream end of left DNA GFP constructs with and without NMPs. (D) GFP FACS plots following no transfection (left) and cotransfection of the indicated right DNA constructs in Polq+/+ iPSCs. (E) Sequencing chromatograms showing MMEJ of the left and right constructs. Red, microhomology. (F) Bar plots showing normalized GFP cells following cotransfection of the indicated reporter constructs. Data pooled from three independent experiments performed at least in duplicate. ± SEM. *Statistical significance from paired t test: P = 0.04, 0 NMPs; P = 0.025, 5 NMPs; P = 0.04, 10 NMPs. (G) ∆7-GFP reporter integrated in U2OS cells for measuring CRISPR-Cas9 donor template [DNA (*phosphorothioate linkages) or DNA with two RNA bases (R2)]–mediated genome engineering. Ribonucleotides, rCrA; Green text, knock-in donor sequence. (H) Shown are frequencies of GFP+ cells (±SD) for the oligonucleotides shown, cotransfected with FLAG-POLQ, or empty vector (EV), normalized to transfection efficiency and to the DNA template (=1, left) and the R2 template in the parental cell line (=1, right). **P ≤ 0.0043, ***P = 0.0001, and ****P < 0.0001, one-way analysis of variance with Tukey’s (left) and Dunnett’s (right) posttests. (I) Immunoblot analysis of FLAG-POLQ and actin control. (J) Sequence of an amplification product from GFP+ cells isolated from POLQe16m cells with the ∆7 reporter assay using the R2 donor template and POLQ expression vector.
We further investigated Polθ RNA-DNA repair in cells using a previously published GFP knock-in reporter assay (∆7-reporter) that is chromosomally integrated in human U2OS cells (Fig. 4G) (36). This assay has a GFP expression cassette disrupted by an insert sequence, along with a deletion of 7 bp of GFP sequence (∆7). An oligonucleotide donor with these 7 nt flanked by 16-nt homology arms on each side can template restoration of active GFP+ (36). This event is induced via CRISPR-Cas9 to generate two DSBs that excise the insert. We compared the frequency of such repair from the positive control DNA donor-template versus a variant (R2) donor-template that contains two ribonucleotides within the 7 nt missing from the ∆7-reporter (Fig. 4G, bottom), finding only a modest decrease with the R2 template versus DNA (Fig. 4H, left). To examine the influence of Polθ on the R2-templated event, we used a previously described POLQ-polymerase deficient cell line (POLQe16m) with this reporter (36), which we compared both to the parental cell line and to the mutant cells with expression of FLAG-POLQ (Fig. 4I). From either comparison, we found that Polθ promotes a significant increase in the frequency of GFP+ cells using the R2 donor template, which further confirms its ability to reverse transcribe template ribonucleotides during DNA repair in a biological setting (Fig. 4H, right).
DISCUSSION
Our study unexpectedly reveals that Polθ reverse transcribes RNA and undergoes a significant structural transformation to accommodate a DNA/RNA template. The structural transformation of Polθ’s thumb subdomain is likely needed to maintain productive interactions on DNA/RNA, which adopts a significantly different conformation relative to DNA/DNA in the Polθ complex (Fig. 3F and fig. S11B). In contrast, structurally characterized retroviral RTs, such as HIV-RT, do not exhibit structural refolding of their thumb subdomain when acting on DNA/RNA (fig. S11A). The marked structural-functional switch within the thumb subdomain observed in Polθ has not been previously observed in other DNA polymerases or retroviral RTs and therefore may be unique to Polθ, which is an unusually promiscuous enzyme that is capable of acting on a variety of different templates including DNA/DNA, DNA/RNA, ssDNA, partial ssDNA, and single-stranded RNA (1, 5, 9, 37). Together, these structural studies reveal that Polθ has an extraordinary degree of structural plasticity that enables it to efficiently transcribe template ribonucleotides and accommodate a full RNA-DNA hybrid within its active site. Although future studies will be required to fully elucidate the physiological relevance of Polθ RT activity, our findings demonstrate that Polθ accommodates template ribonucleotides in an active configuration and promotes RNA-DNA repair, which may contribute to cellular tolerance of genome-embedded ribonucleotides.
MATERIALS AND METHODS
Primer-template extension assays
Relative velocity of RT activity (Fig. 1B)
Polθ (0.5 nM), HIV RT, and Polη (catalytic core, residues 1 to 514) were incubated with 10 nM radiolabeled DNA/RNA template (RP559/RP493R) for the indicated times in buffer A [25 mM tris-HCl (pH 7.8), 10 mM MgCl2, 0.01% (v/v) NP-40, 1 mM dithiothreitol (DTT), bovine serum albumin (BSA; 0.1 mg/ml), and 10% (v/v) glycerol] with 100 μM dNTPs at 37°C. Percent extension was determined by dividing the intensity of the extended product by the intensity of the sum of extended and unextended products for each lane. All primer-template reactions were terminated with 25 mM EDTA and 45% (v/v) formamide then resolved in urea denaturing polyacrylamide gels and visualized by PhosphorImager. The rate of RT activity was determined from the slope of the linear portion of the plot representing steady-state conditions.
Comparison of polymerase activities on DNA/DNA and DNA/RNA (Fig. 1, D to G)
The primer extension assays on DNA/RNA (RP559/RP493R) and DNA/DNA (RP559/RP493D) templates were performed using the conditions in Fig. 1B with the following changes. The indicated concentrations of the indicated polymerases were used, and the reactions were performed for 32 min.
Relative RT activity (Fig. 1H)
The indicated polymerases were incubated with 10 nM radiolabeled DNA/RNA template (SM98/SM44R) for 20 min in the presence of 10 μM dNTPs at 37°C. Polθ and HIV RT reactions were performed in 25 mM tris-HCl (pH 8.0), 10 mM KCl, 10 mM MgCl2, 0.01% (v/v) NP-40, 1 mM DTT, BSA (0.1 mg/ml), and 10% (v/v) glycerol. AMV RT (20 units, New England Biolabs)–containing reactions were performed in buffer [50 mM tris-acetate (pH 8.3), 75 mM potassium acetate, 8 mM magnesium acetate, and 10 mM DTT] and contained BSA (0.1 mg/ml). M-MuLV RT (400 units, New England Biolabs)–containing reactions were performed in buffer [50 mM tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 10 mM DTT] and contained BSA (0.01 mg/ml).
Comparison of RT and DNA-dependent DNA synthesis activities by truncated and full-length Polθ (Fig. 1J)
One hundred nanomolar of the indicated polymerases were incubated with 10 nM radiolabeled DNA/RNA (SM98/SM44R) or DNA/DNA (SM98/SM44) for 45 min with 50 μM dNTPs and 25 mM tris-HCl (pH 7.8), 2 mM MgCl2, 4 mM KCl, 6 mM NaCl, 0.01% (v/v) NP-40, 1 mM DTT, BSA (0.1 mg/ml), 10% (v/v) glycerol, and 750 μM adenosine triphosphate (ATP) at 37°C.
Relative velocity of single dNMP incorporation on radiolabeled DNA/RNA (Fig. 2, A to E)
Polθ (2 nM) was incubated with 100 nM of the indicated radiolabeled DNA/RNA and DNA/DNA templates for the indicated times with 300 μM of the indicated dNTP in buffer [25 mM tris-HCl (pH 8.0), 10 mM MgCl2, 4 mM KCl, 6 mM NaCl, 0.01% NP-40, 1 mM DTT, BSA (0.01 mg/ml), and 10% (v/v) glycerol] at 37°C. Percent extension was determined as described above.
Relative velocity of nucleotide (dNMP) misincorporation (Fig. 2, F to J)
Polθ (20 nM) was incubated with 100 nM of the indicated radiolabeled DNA/RNA and DNA/DNA templates and 300 μM of the indicated dNTP for the indicated times in buffer [25 mM tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM KCl, 0.01% (v/v) NP-40, 1 mM DTT, BSA (0.1 mg/ml), and 10% (v/v) glycerol]. Percent extension was determined as described above. All oligonucleotides were radiolabeled using T4 polynucleotide kinase (New England Biolabs) and 32P-γ-ATP (PerkinElmer) in recommended buffer for 37°C for at least 1 hour.
MMEJ cellular assay
Some of the methods used here are similar to those in a previously published article (38). iPSCs (2 × 105) were transfected in suspension with 0.25 μg each of the indicated left- and right-flanking DNA GFP constructs using Lipofectamine 2000 (Invitrogen). As a negative control, similar volume of buffer that was used in experimental wells was used for transfection in control wells. As a positive control to measure transfection efficiency, a wild-type linear DNA GFP expression construct was transfected simultaneously. GFP-positive cell frequencies were measured 3 days after transfection by flow cytometry using GUAVA easyCyte 5-HT (Luminex Corp.) in independent replicates and corrected for transfection efficiency and background events. Data are represented as the mean and SEM of three independent experiments, with at least duplicates per experiment. Statistical analysis was carried out by paired t test.
Preparation of GFP MMEJ reporter constructs
Some of the methods used here are similar to those in a previously published article (38). Polymerase chain reaction (PCR) preparation followed recommended conditions for the Phusion High-Fidelity DNA Polymerase (New England Biolabs M0530) using 10 ng of pCMV-GFP plasmid as template in 1× Phusion HF Buffer. PCR for the left-flank DNA was performed with primers RP500B and RP501. PCR for the right-flank DNA was performed with primers RP502 and RP503B. Following PCR, left- or right-flank DNA products were pooled together and digested with Dpn I (New England Biolabs) in 1× CutSmart buffer and then purified via Qiagen QIAquick PCR Purification Kit. PCR was then conjugated to streptavidin using PCR (110 ng/μl) and streptavidin (0.8 μg/μl) in 10 mM tris-HCl (pH 7.5) and 100 mM NaCl at 37°C for 1 hour. PCR for the left-flank DNA with five consecutive ribonucleotides was performed with primers RP500B and RP501. PCR was purified and then digested with Dpn I (New England Biolabs) and Sap I in 1× CutSmart buffer. PCR was purified again and then ligated to double-stranded DNA composed of annealed oligonucleotides RP550R-P and RP530a [oligos were annealed in the presence of a ribonuclease (RNase) inhibitor] for 16 hours at 16°C with T4 DNA Ligase (New England Biolabs) in 1× T4 DNA Ligase Buffer. Ligated PCR was purified and then conjugated to streptavidin as described above. PCR for the left-flank DNA with 10 consecutive ribonucleotides was prepared by the same methodology with ligation to the double-stranded DNA composed of oligos RP546P and RP530. Streptavidin conjugation and DNA amplification steps were confirmed in agarose gels stained with ethidium bromide.
CRISPR-Cas9 knock-in GFP reporter assay
The U2OS parental and POLQe16m cell lines with the ∆7-reporter and CAS9/single guide RNA (sgRNA) plasmids to target the DSBs in this reporter, DNA oligonucleotide template, and control oligonucleotide (LUC) were previously described (36). The R2 oligonucleotide has the same sequence as the DNA oligonucleotides, but with two RNA bases in the 7 nt missing from the ∆7-reporter (IDT). The cell lines were seeded at 1 × 105 on a 12-well dish and transfected the following day, and % GFP was analyzed 3 days after transfection using a CyAN-ADP (DAKO) cytometer and normalized to transfection efficiency, as previously described (36). Transfections for the reporter assay contained 400 ng each CAS9/sgRNA plasmid, 10 pmol oligonucleotide, and 100 ng of either pCAGGS-BSKX (empty vector) or FLAG-POLQ expression vector (38). Transfections for transfection efficiency contained 400 ng of pCAGGS-NZE-GFP (GFP expression vector), 500 ng of empty vector (EV), and 10 pmol control oligonucleotide (36). Transfections were performed with 4 μl of Lipofectamine 2000 (Thermo Fisher Scientific) in 0.2 ml of Optimem (Thermo Fisher Scientific) and incubated with cells in 1 ml of antibiotic-free medium for 4 hours. Immunoblotting analysis for FLAG-POLQ involved extraction with ELB [250 mM NaCl, 5 mM EDTA, 50 mM Hepes, 0.1% (v/v) Ipegal, and Roche protease inhibitor] with sonication (Qsonica, Q800R) and using antibodies for FLAG (Sigma-Aldrich, A8592) or ACTIN (Sigma-Aldrich, A2066). Sequence analysis of reporter assay with R2 oligo: Cells were transfected as for the reporter assay with the R2 oligonucleotide template in the POLQe16m cells with the POLQ expression vector, GFP+ cells were isolated by fluorescence-activated cell sorting (FACS) (Becton Dickinson Aria Sorter), and the GFP repair product was amplified with CMVFWDFRT5 5′ CGCAAATGGGCGGTAGGCGTG and BGHREVFRT5 5′ TAGAAGGCACAGTCGAGG and sequenced with the CMVFWDFRT5 primer.
cDNA synthesis
cDNA synthesis reactions were performed by the indicated polymerase in the presence of 100 μM dNTPs and optimal buffer for each enzyme: Polθ [25 mM tris-HCl (pH 8.0), 10 mM KCl, 10 mM MgCl2, 0.01% (v/v) NP-40, 1 mM DTT, BSA (0.1 mg/ml), and 10% (v/v) glycerol]; AMV RT [50 mM tris-acetate (pH 8.3), 75 mM potassium acetate, 8 mM magnesium acetate, 10 mM DTT, and BSA (0.1 mg/ml)]; and M-MuLV RT [50 mM tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 10 mM DTT]. Reactions with synthetic DNA/RNA contained 10 ng of template. Polθ (100 nM), AMV-RT (20 units), or M-MuLV RT (400 units) were incubated at 37°C (Polθ) or 42°C (AMV-RT and M-MuLV RT) for 1 hour. Enzymes were then heat-inactivated at 85°C for 20 min. cDNA (0.1875 ng) was used for real-time PCR (Power SYBR Green Master Mix, Thermo Fisher Scientific) using primers SM246 and SM247.
Proteins
Polθ, Fl-Polθ, and Polδ were purified as previously described (1, 9). Pols β and λ were provided by S. Wilson. Polκ was purchased from Enzymax LLC. HIV RT RT52A optimized for x-ray crystallography was provided by Dr. E. Arnold. Polη was provided by Dr. S. Arora. Pols ε and α were provided by Dr. M. O’Donnell. PolγExo- was provided by Dr. W. Copeland. M-MuLV, AMV RTs, and KF Pols were purchased from New England Biolabs.
Protein purification for x-ray crystallography
The gene encoding Polθ (residues 1819 to 2590) was codon-optimized and cloned into the pSUMO vector to generate a sumo fusion that carries an N-terminal 6×His tag and a PreScission protease cleavage site. Polθ-expressing E. coli Rosetta(DE3)pLysS cells were cultured at 37°C in LB medium until optical density at 600 nm (OD600) reached 0.3, the growth temperature was then lowered to 16°C, and E. coli cells were further cultured. Protein expression was induced by the addition of 0.1 mM isopropyl β-d-thiogalactopyranoside when OD600 reached 0.7 to 0.9. E. coli cells were cultured at 16°C overnight and harvested by centrifugation. Cell pellet was resuspended in buffer L [50 mM Hepes (pH 8.0), 500 mM NaCl, 0.005% (v/v) Igepal CA 630, and 0.5 mM TCEP]; lysed by sonication or French Press in the presence of DNase I (20 μg/ml), RNase A (30 μg/ml), 5 mM MgCl2, 2 mM CaCl2, and 1 mM phenylmethylsulfonyl fluoride; and then centrifuged at 12,000g for 45 min. 6×His sumo fusion was captured by Ni-NTA agarose gravity-flow chromatography followed by a series of washes by buffer W [50 mM Hepes (pH 8.0), 500 mM NaCl, 0.005% (v/v) Igepal CA 630, 0.5 mM TCEP, and 10 mM imidazole]. Five milliliters of buffer L and Precission Protease was added to cleave Polθ from the 6×His tag by staying overnight at 4°C. The cleaved Polθ was eluted another two times by 5 ml of buffer L. The eluted protein was purified to homogeneity using a HiTrap Heparin column (GE Healthcare Life Sciences). The protein was concentrated to 5 mg/ml in a buffer of ammonium acetate (150 mM), KCl (150 mM), tris-HCl buffer (pH 8.0) (40 mM), TCEP (2.5 mM), and glycerol (1% v/v).
Crystallization and structure determination
The crystallization condition was identified by wide matrix screening. Sitting-drop crystallization screening plates were set at 18°C using ARI Crystal Gryphon Robot (ARI) and crystallization screening solutions (Qiagen and Hampton Research). Reacting Polθ (2.5 mg/ml) with a DNA/RNA hybrid (DNA primer: 5′-GCGGCTGTCATT and RNA template: 5′-CGUCCAAUGACAGCCGC) in the presence of ddGTP (1 mM), sucrose monolaurate (300 μM), MgCl2 (1 mM), and spermine tetrahydrochloride (20 mM) prepared the sample for sitting drop vapor diffusion over a 50-μl reservoir containing 20% (w/v) ethanol by mixing 0.3 μl of the reservoir solution with an equal part of the reaction solution. Crystals of approximate final dimensions 20 μm by 20 μm by 20 μm grew over the next 2 weeks. Cryoprotection was achieved by looping the crystals into mother solution with additional 25% (v/v) glycerol before flash cooling into liquid nitrogen. Diffraction data were collected at beamline 23ID-D of the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility at the Advanced Photon Source. A complete dataset for Polθ was collected, indexed, integrated, and scaled by the autoprocessing package in APS server (GMCAproc). The Phaser-MR program in the PHENIX package was used for molecular replacement, and COOT was used for model rebuilding and Phenix for simulated annealing and refinement. The structure of Polθ was determined by molecular replacement (MR) using Polθ [Protein Data Bank (PDB): 4x0q] as a search model. The full PDB structure of 4x0q could not yield a solution. An MR solution was only obtained by removing some of the loops and deleting the DNA/DNA duplex from the model. The initial phases of the MR protein model were improved by cyclic model building and refinement, which allow us to slowly build in the missing loops, refolded and reoriented finger and thumb domains, and the DNA/RNA residues on the basis of improved electron density maps. A final model with very good statistics for this resolution range of 3.2 Å has been achieved (table S1).
Nucleic acids
The following nucleic acids were purchased from Integrated DNA Technologies (listed as 5′-3′ polarity):
RP559 CGACGTCGCAG
RP493R rUrUrUrUrUrUrUrCrGrCrGrCrUrGrCrGrArCrGrUrCrG
RP493R TTTTTTTCGCGCTGCGACGTCG
RP500B/5Biosg/GCTAGCCAGTCAGTGGGCCCGC
RP501 TCGGGCATGGCGGACTTGAAGAAGTCG
RP502 GCCCGAAGGCTACGTCCAGGAGCGCACC
RP503B/5Biosg/TGATTACGCCAAGTTAATTAAGGACGTCCTCCTGCTGG
RP550R-P/5Phos/TCTTCAAGTCCGCrCrArUrGrCCCGA
RP530a TCGGGCTAGGCGGACTTGA
RP546P/5Phos/TCTTrCrArArGrUrCrCrGrCrCATGCCCGA
RP530 TCGGGCATGGCGGACTTGA
RP563 CCACAAGTTCAGCGTGTCCGGCGAGG
RP566 CCTTGATGCCGTTCTTCTGCTTGTCGGCC
RP564 CGAGGGCGATGCCACCTACGGC
RP565 GTGGCTGTTGTAGTTGTACTCCAGCTTGTGCC
RP560 AGAACCTGTTGAA
RP468R rArCrArGrUrUrUrUrUrUrUrUrGrCrUrUrUrUrGrUrUrCrArArCrArGrGrUrUrCrU
SM98 ATTATGCGGCCGTGTACAACGGGTGTCGGGGC
SM44R GCUCUGAUGCCGCAUAGUUAAGCCAGCCCCGACACCCG
SM44 GCTCTGATGCCGCATAGTTAAGCCAGCCCCGACACCCG
SM246 ATTATGCGGCCGTGTACAAC
SM247 GCTCTGATGCCGCATAGTTAAG
SM249 TTCGACTTGCATGTGTTAGG
SM248 AATTGAAGAGTTTGATCATGGC
SM270FAM/56-FAM/CGACGTCGCAGCGC
SM271 GAGACTCGGCGCTGCGACGTCG
SM271R GAGACUCGGCGCUGCGACGUCG
SM106 ATTATGCGGCCGTGTAC
SM107 GCTCTGATGCCGCATAG
RP561R2 G*G*TAGCGGCTGAAGCACTGrCrACGCCGTAGGTCAGGGT*G*G.
RP561 G*G*TAGCGGCTGAAGCACTGCACGCCGTAGGTCAGGGT*G*G.
Acknowledgments
We are grateful to Dr. E. Arnold (Rutgers University) for providing recombinant HIV RT. We are grateful to Dr. A. Sfeir (Sloan-Kettering) for providing isogenic Polq+/+ and Polq−/− iPSCs and for discussions and input on the studies. We thank Dr. S. Arora (Fox Chase Cancer Center) for providing recombinant human Polη. We thank Dr. S. Wilson (NIH) for providing Polβ. We thank Dr. W. Copeland (NIH) for providing PolγExo−. We thank Dr. M. O’Donnell (Rockefeller University) for providing recombinant Polε and Dr. L. Prakash (University of Texas) for providing human recombinant Polθ purified from S. cerevisiae. We acknowledge the use of synchrotron 23ID beamline of APS at Argonne National Laboratory. Last, we are grateful to Dr. S. Li for assistance in the structural determination process. Funding: This research was supported by NIH grants 1R01GM130889-01 and 1R01GM137124-01 to R.T.P., and R01CA197506 and R01CA240392 to J.M.S. This research was also supported in part by a Tower Cancer Research Foundation grant to X.S.C. Author contributions: G.C. Performed cell biology assays and edited the manuscript. J.Z. Performed protein purification, X-ray crystallization studies, and solved the ternary structure of Polq:DNA/RNA:ddGTP. S.M. co-conceived the idea for the study and performed protein purification and biochemical assays. T.R., T.H., N.B., T.T., L.A.S., J.M., and Z.S. performed biochemical assays. F.W.L. performed cell biology assays. T.K. performed protein purification and biochemical assays. J.H. performed DNA sequencing assays. E.K. performed protein purification. A.B. performed cell biology assays. J.M.S. guided and interpreted cell biology assays and contributed to manuscript writing. X.S.C. guided the X-ray crystallization studies, interpreted the data, and contributed to manuscript writing. R.T.P. wrote and edited the manuscript, and co-conceived the idea for the study. Competing interests: Richard Pomerantz is a co-founder and chief scientific officer for Recombination Therapeutics, LLC. The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabf1771/DC1
REFERENCES AND NOTES
- 1.Black S. J., Ozdemir A. Y., Kashkina E., Kent T., Rusanov T., Ristic D., Shin Y., Suma A., Hoang T., Chandramouly G., Siddique L. A., Borisonnik N., Sullivan-Reed K., Mallon J. S., Skorski T., Carnevale V., Murakami K. S., Wyman C., Pomerantz R. T., Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 10, 4423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahn K. E., Averill A. M., Aller P., Wood R. D., Doublie S., Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 22, 304–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arana M. E., Seki M., Wood R. D., Rogozin I. B., Kunkel T. A., Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36, 3847–3856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent T., Rusanov T. D., Hoang T. M., Velema W. A., Krueger A. T., Copeland W. C., Kool E. T., Pomerantz R. T., DNA polymerase theta specializes in incorporating synthetic expanded-size (xDNA) nucleotides. Nucleic Acids Res. 44, 9381–9392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent T., Mateos-Gomez P. A., Sfeir A., Pomerantz R. T., Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5, e13740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki M., Wood R. D., DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair 7, 119–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogg M., Seki M., Wood R. D., Doublie S., Wallace S. S., Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J. Mol. Biol. 405, 642–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon J. H., Roy Choudhury J., Park J., Prakash S., Prakash L., A role for DNA polymerase theta in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J. Biol. Chem. 289, 13177–13185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent T., Chandramouly G., McDevitt S. M., Ozdemir A. Y., Pomerantz R. T., Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateos-Gomez P. A., Gong F., Nair N., Miller K. M., Lazzerini-Denchi E., Sfeir A., Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yousefzadeh M. J., Wyatt D. W., Takata K., Mu Y., Hensley S. C., Tomida J., Bylund G. O., Doublie S., Johansson E., Ramsden D. A., McBride K. M., Wood R. D., Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLOS Genet. 10, e1004654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Vidal A., Guitton-Sert L., Cadoret J. C., Drac M., Schwob E., Baldacci G., Cazaux C., Hoffmann J. S., A role for DNA polymerase theta in the timing of DNA replication. Nat. Commun. 5, 4285 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Higgins G. S., Boulton S. J., Beyond PARP—POLθ as an anticancer target. Science 359, 1217–1218 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wood R. D., Doublie S., DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair 44, 22–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceccaldi R., Liu J. C., Amunugama R., Hajdu I., Primack B., Petalcorin M. I., O’Connor K. W., Konstantinopoulos P. A., Elledge S. J., Boulton S. J., Yusufzai T., D’Andrea A. D., Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng W., Simpson D. A., Carvajal-Garcia J., Price B. A., Kumar R. J., Mose L. E., Wood R. D., Rashid N., Purvis J. E., Parker J. S., Ramsden D. A., Gupta G. P., Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 10, 4286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi C., Shen X., Niu S., Ma C., Innate reverse transcriptase activity of DNA polymerase for isothermal RNA direct detection. J. Am. Chem. Soc. 137, 13804–13806 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Menendez-Arias L., Sebastian-Martin A., Alvarez M., Viral reverse transcriptases. Virus Res. 234, 153–176 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Williams J. S., Lujan S. A., Kunkel T. A., Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 17, 350–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams J. S., Kunkel T. A., Ribonucleotides in DNA: Origins, repair, and consequences. DNA Repair 19, 27–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazina O. M., Keskin H., Hanamshet K., Storici F., Mazin A. V., Rad52 inverse strand exchange drives RNA-templated DNA double-strand break repair. Mol. Cell 67, 19–29.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meers C., Keskin H., Banyai G., Mazina O., Yang T., Gombolay A. L., Mukherjee K., Kaparos E. I., Newnam G., Mazin A., Storici F., Genetic characterization of three distinct mechanisms supporting RNA-driven DNA repair and modification reveals major role of DNA polymerase zeta. Mol. Cell 79, 1037–1050.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Y., Egli M., Guengerich F. P., Human DNA polymerase eta accommodates RNA for strand extension. J. Biol. Chem. 292, 18044–18051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H., Goodman M. F., Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J. Biol. Chem. 267, 10888–10896 (1992). [PubMed] [Google Scholar]
- 25.Creighton S., Huang M. M., Cai H., Arnheim N., Goodman M. F., Base mispair extension kinetics. Binding of avian myeloblastosis reverse transcriptase to matched and mismatched base pair termini. J. Biol. Chem. 267, 2633–2639 (1992). [PubMed] [Google Scholar]
- 26.Sebastian-Martin A., Barrioluengo V., Menendez-Arias L., Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transcriptases. Sci. Rep. 8, 627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakhanashvili M., Hizi A., Fidelity of the RNA-dependent DNA synthesis exhibited by the reverse transcriptases of human immunodeficiency virus types 1 and 2 and of murine leukemia virus: Mispair extension frequencies. Biochemistry 31, 9393–9398 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Ji J. P., Loeb L. A., Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31, 954–958 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Klarmann G. J., Schauber C. A., Preston B. D., Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 268, 9793–9802 (1993). [PubMed] [Google Scholar]
- 30.Gao L., Balakrishnan M., Roques B. P., Bambara R. A., Insights into the multiple roles of pausing in HIV-1 reverse transcriptase-promoted strand transfers. J. Biol. Chem. 282, 6222–6231 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Yoon J. H., McArthur M. J., Park J., Basu D., Wakamiya M., Prakash L., Prakash S., Error-prone replication through UV lesions by DNA Polymerase θ protects against skin cancers. Cell 176, 1295–1309.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyer J. C., Bebenek K., Kunkel T. A., Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. U.S.A. 89, 6919–6923 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Korolev S., Waksman G., Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 17, 7514–7525 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowak E., Potrzebowski W., Konarev P. V., Rausch J. W., Bona M. K., Svergun D. I., Bujnicki J. M., Le Grice S. F., Nowotny M., Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid. Nucleic Acids Res. 41, 3874–3887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarafianos S. G., Das K., Tantillo C., Clark A. D. Jr., Ding J., Whitcomb J. M., Boyer P. L., Hughes S. H., Arnold E., Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20, 1449–1461 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelso A. A., Lopezcolorado F. W., Bhargava R., Stark J. M., Distinct roles of RAD52 and POLQ in chromosomal break repair and replication stress response. PLOS Genet. 15, e1008319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas C., Rusanov T., Hoang T., Augustin T., Kent T., Gaspar I., Pomerantz R. T., One-step enzymatic modification of RNA 3′ termini using polymerase theta. Nucleic Acids Res. 47, 3272–3283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandramouly G., Liao S., Rusanov T., Borisonnik N., Calbert M. L., Kent T., Sullivan-Reed K., Vekariya U., Kashkina E., Skorski T., Yan H., Pomerantz R. T., Polθ promotes the repair of 5′-DNA-protein crosslinks by microhomology-mediated end-joining. Cell Rep. 34, 108820 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zelensky A. N., Schimmel J., Kool H., Kanaar R., Tijsterman M., Inactivation of Polθ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 8, 66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabf1771/DC1


