Molecular devices for the spatial control of activating and inhibitory receptors help in their signal integration.
Abstract
The role of juxtaposition of activating and inhibitory receptors in signal inhibition of cytotoxic lymphocytes remains strongly debated. The challenge lies in the lack of tools that allow simultaneous spatial manipulation of signaling molecules. To circumvent this, we produced a nanoengineered multifunctional platform with molecular-scale spatial control of ligands, which was applied to elucidate KIR2DL1-mediated inhibition of NKG2D signaling—receptors of natural killer cells. This platform was conceived by bimetallic nanodot patterning with molecular-scale registry, followed by a ternary functionalization with distinct moieties. We found that a 40-nm gap between activating and inhibitory ligands provided optimal inhibitory conditions. Supported by theoretical modeling, we interpret these findings as a consequence of the size mismatch and conformational flexibility of ligands in their spatial interaction. This highly versatile approach provides an important insight into the spatial mechanism of inhibitory immune checkpoints, which is essential for the rational design of future immunotherapies.
INTRODUCTION
Cells communicate with their environment through a rich repertoire of receptors, whose signaling integration is mediated by the biochemistry of the cell-environment interface, as well as by diverse physical features, such as receptor size and spatial arrangement. In the immune synapse—the functional interface between lymphocytes and antigen-presenting cells—activating, costimulatory, and inhibitory receptors coordinate their nanoscale clustering and arrangement to regulate lymphocyte immune activity (1, 2). For example, the spatial density and organization of T cell receptors (TCRs) and their co-receptors CD3 within their nanoclusters regulate the phosphorylation of their complexes (3). Also, TCR and its linker for activation concatenate during T cell activation but form segregated clusters in resting T cells (4, 5). The inhibitory function of immune checkpoints that orchestrate the self-tolerance of the immune system, such as immunotherapeutic target programmed cell death protein 1 in T cells or KIR2DL1 in natural killer (NK) cells, is associated with their nanoscale clustering and colocalization with activating and costimulatory receptors (6, 7), yet the mechanism of these clustering and colocalization and their role in the activating-inhibitory signaling coordination are still obscure. The systematic study of how the segregation between different receptors regulates their signaling integration requires a high level of spatial control with respect to each other. However, protocols currently available for cell biologists allow controlling receptor segregation only in the direction perpendicular to the membrane, by manipulating the length of their extracellular domains. In the context of immune receptors, controlled variations in the ectodomain size of receptors were shown to affect their signaling integration in T cells and NK cells (8–11). At the same time, there has been no methodology to control the segregation of different receptors within the membrane plane, which is more relevant for the regulation of signaling cross-talk in the immune system (12). For this reason, the effect of within-membrane segregation of different receptors onto the cell function could not be systematically studied.
Receptors can be spatially guided within the cell membrane by artificial cell niches based on tunable patterns of extracellular ligands. Various concepts for such niches have evolved in the last two decades thanks to the progressing advances in nanofabrication, and they were applied to study the effect of ligand distribution on different cell functions, such as adhesion (13–15), chemokinesis (16), mechanosensing (17), differentiation (18), and immune activity (19–22). Functionalized surfaces were also the basis of mimetic cell models, which were used, in conjunction with theoretical modeling, to identify the physical foundation for the recognition of membrane-confined ligands and receptors, which mostly focused on single protein pairs (23). However, all of the state-of-the-art ligand patterns, even those including multiple ligands (19, 24), have been limited to spatially control ligands of one type only. On the other hand, simultaneous spatial control of two different ligands, i.e., ligands for immune cells, was demonstrated on the micrometer scale (25, 26). Still, molecular-scale control of multiple ligands has not been realized so far, as it requires new, paradigm-shifting fabrication and functionalization approaches with precision and complexity far beyond those exploited for the state-of-the-art ligand patterns.
In this paper, we achieved the molecular-scale spatial control of two extracellular ligands to discover how the spatial juxtaposition between two receptors—(i) NKG2D, the major activating receptor in NK cells, and (ii) KIR2DL1, an inhibitory receptor that belongs to the family of killer cell immunoglobulin-like receptors (KIRs) (27, 28)—regulates KIR2DL1-mediated inhibition of NK cell cytotoxic activity. Activating and inhibitory receptors in NK cells balance their signals to determine whether a target cell will be tolerated or attacked. In the context of NKG2D and KIR2DL1, there is emerging evidence that KIR2DL1-mediated inhibition of NKG2D signaling is closely associated with the nanoscale clustering and arrangement of the two receptors (29), yet the exact role of this arrangement in signaling cross-talk between the two receptors is still unclear. To spatially control both receptors, we realized a series of molecular-scale devices patterned with variable heterogeneous arrays of their ligands, by selectively anchoring these ligands to nanofabricated arrays of dots of two different metals. We used these arrays as artificial niches for the stimulation of NK cells. While being sized at the scale of the individual ligand-receptor complex, the anchoring dots functionalized with ligands guide the clustering of the two receptors, producing, for example, their colocalization (Fig. 1A), or controlled segregation (Fig. 1B), depending on the array geometry. We fabricated the arrays by nanoimprint lithography and double angle evaporation, which allowed us to register the anchoring dots of two different metals with molecular-scale accuracy in one lithographic step. To selectively attach the activating and inhibitory ligands to the anchoring nanodots, we developed a novel ternary chemical functionalization based on the combination (i) histidine-nitrilotriacetic acid (NTA) conjugation, (ii) biotin-avidin conjugation, and (iii) silane chemisorption (Fig. 1C), thereby forming three distinct chemical moieties onto a heterogeneous nanopattern.
Fig. 1. Platform for the molecular-scale spatial control of two extracellular ligands.
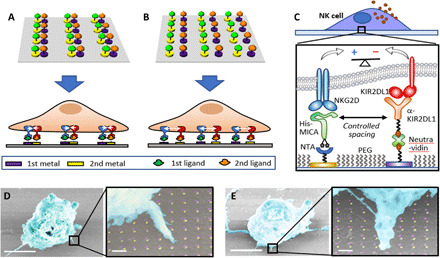
(A and B) Schematic representation of ligand patterns in which colocalized or spatially segregated ligands, respectively, control the spatial arrangement of integrating receptors (not to scale). (C) Scheme of a ligand pattern that controls activating-inhibitory balance in NK cells. PEG, polyethylene glycol. (D and E) False-colored scanning electron micrograph of NK cells stimulated in activating-inhibitory ligand array with different spacings between the ligands. Scale bars, 5 μm; scale bars in high-magnification images, 200 nm.
We stimulated NK cells on different arrays within which only one geometric parameter—the spacing between the two ligands—was systematically varied (Fig. 1, D and E). We found that NK cells sense this spacing and adjust their immune response to extremely small, molecular-scale changes: While colocalized ligands produced an insufficient inhibitory effect on the activation of NK cells, this inhibition gradually increased by inducing a nanoscale gap between the ligands and became substantial at a gap of 40 nm—the largest one within the tested range. This finding indicated the possible role of the membrane fluctuation and flexibility in the formation of proximal ligand-receptor bonds and was rationalized by theoretical modeling, by which this spacing corresponds to the optimal separation between the ligands, which stems from the limited flexibility of the receptors and the membrane, and which compensates for the size difference between the studied ligands. This strong correlation between the experimental results and the theoretical model clearly establishes a link between the spatial arrangement of activating and inhibitory receptors and their signaling integration in NK cells. Our results show that although inhibitory signaling in NK cells has been closely associated with nanoscale coclustering of activating and inhibitory ligand-receptor complexes in NK immune synapse (29–31), the colocalization between the two ligands is not a necessary condition for the inhibition in NK cells and that the inhibition is also possible for a fixed configuration of controllably distributed ligands, in which the spatial parameters, e.g., ligand spacing, can be used as a tuning knob for the inhibition efficacy. Besides the specific case study of KIR2DL1-NKG2D signaling cross-talk, our innovative nanodevice technology opens a general pathway to complex, multifunctional nanomaterials designed to model the cell environment with unprecedented precision and complexity and enables numerous molecular-scale studies of the structure and mechanism of functional biointerfaces, which have been impossible to date but are now within reach.
RESULTS
Device design and fabrication
The device surfaces were patterned with heterodimers of Au and Ti and nanodots functionalized activating and inhibitory ligands. The heterodimers were configured into orthogonal arrays with a periodicity of 200 nm in both the x and y axes. This periodicity, which is twice larger than the average membrane correlation length (32), came to minimize any possible physical interaction between the neighboring dimers and, at the same time, provided a sufficient amount of ligands to stimulate NK cells. The gap between the nanodots was the only degree of freedom in the device design, and it varied between 0 and 40 nm. Such a delicate registration between two nanofabricated metallic layers is extremely challenging to achieve with standard nanolithographic approaches, especially in a scalable manner. Here, we developed an “out-of-the-box” fabrication approach, in which we combined nanoimprint lithography with double angle evaporation. This combination allowed us to produce bimetallic nanoarrays registered with nanoscale accuracy using only one lithographic step, and with no need for alignment between different nanodots. Specifically, we first patterned a thermal nanoimprint resist on a Si substrate with periodic rectangular arrays of ~20-nm holes separated by 200 nm and transferred the imprinted pattern into a resist by angle deposition of a metallic protection mask (33), resist over-etch by plasma, two sequential metal evaporations (Au and Ti/Cr in our case), and liftoff (Fig. 2A). The resulting nanopattern consisted of bimetallic heterodimers, whose rectangular arrangement and 200-nm periodicity were determined by the initial nanoimprint pattern. The size of each nanodot was 15 to 20 nm, and the gap between the two neighboring nanodots within the dimers was determined by the evaporation angle (fig. S1). In such a way, small evaporation angles produce heterodimers of colocalized nanodots (Fig. 2B), whereas a gradual increase in the evaporation angle resulted in a gradual separation between nanodots (Fig. 2, B to E).
Fig. 2. Flowchart of the fabrication of bimetallic nanopatterns functionalized with two different biomolecules.
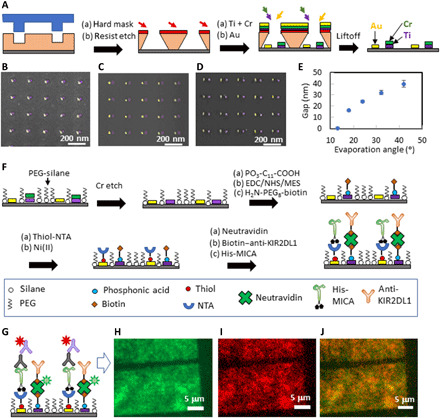
(A) Fabrication of heterogeneous metallic arrays. False-colored scanning electron micrograph of (B) colocalized nanodots, (C) nanodots separated by ~20-nm gap, and (D) nanodots separated by ~40-nm gap. (E) Segregation control via evaporation angle. (F) Functionalization scheme. (G) Scheme of the fluorescent labeling of the functionalized nanodots. (H to J) Fluorescent imaging of a functionalized array taken in the green channel, the red channel, and their merge, respectively.
The second step in the device fabrication is the biofunctionalization of the bimetallic nanoarrays with the desired ligands. The key challenge here is to achieve the absolute site selectivity with which different ligands are immobilized onto different nanodots. Not only should each of the two used ligands be specifically immobilized on the selected nanodots but also neither of these two ligands is allowed to attach to the Si background surface that surrounds the anchoring nanodots. To achieve this, as well as to prevent any nonspecific interaction between the background and the cell receptors, the Si background should be functionalized with an antifouling agent. Therefore, the surface of our nanodevice includes three distinct and discretized chemical entities—the background and two ligated nanodots. To date, state-of-the-art biomimetic patterns for controlled cellular environment consisted of up to two spatially segregated biochemical functionalities, i.e., one ligand and antifouling agent (34), or two different ligands copositioned, for instance, by binding to a bimetallic pattern (35) or DNA origami (36). Here, we developed a novel ternary functionalization process for the site-specific immobilization of three biochemical functionalities—two ligands and an antifouling agent, by combining chemical modifications of Si, Au, and TiO2 surfaces with organic silanes, thiols, and phosphonic acids, respectively (Fig. 2F). First, the Si background was passivated with silane terminated polyethylene glycol (PEG)—a commonly used antifouling agent that effectively prevents nonspecific binding interactions (37, 38). The unwanted immobilization of the silane onto oxidized Ti nanodots was prevented by covering them with a protective Cr layer during the fabrication process. This Cr layer was etched after Si passivation with PEG. The exposed oxidized Ti was then functionalized with a carboxyl-terminated phosphonic acid and subsequently modified with a biotin terminal group, while PEG-silane effectively protected Si from the nonspecific chemisorption of phosphonic acid. Then, Au was functionalized with an organic thiol terminated with NTA, which was next chelated with Ni ions to provide high affinity to the histidine conjugate. [See the Supplementary Materials and figs. S6 to S8 for polarization modulation-infrared reflection-adsorption spectroscopy (PM-IRRAS) analyses (39, 40) that confirm the chemical modification of Au and TiO2.] Last, His-tagged MICA was attached to Au nanodots via His-NTA/Ni conjugation, and biotin-labeled anti-KIR2DL1 was attached to biotin-functionalized TiO2 nanodots via a NeutrAvidin bridge. We used here a monoclonal antibody (mAb) for KIR2DL1 that was shown not only to bind KIR2DL1 receptor but also to induce KIR2DL1-mediated inhibition (41). Thus, we created here a surrogate platform with activating and inhibitory functionalities for NK cells.
To verify the functionalization efficiency and selectivity, we imaged the functionalized array by fluorescence microscopy. Specifically, we verified TiO2 functionalization by imaging NeutrAvidin labeled with Oregon Green 488. In parallel, we verified Au functionalization by immunostaining immobilized MICA with mouse anti-MICA antibody and anti-mouse Alexa Fluor 568 (Fig. 2G). Figure 2 (H and I) shows fluorescent images of the functionalized array taken separately in the green and red channels, which represent each of the immobilized bio-functionalities. Also, Fig. 2J shows the merged image of the two channels. The borders of the array areas are clearly distinguishable in all three images, as the surrounding background produces little fluorescence and therefore confirms that the ligands were immobilized specifically onto the nanodots. To verify that the efficiency and site selectivity of the functionalization process are not affected by the arrangement of nanodots, and to eliminate a scenario by which the functionalization can be influenced, for instance, by steric hindrance when two molecules are contiguous, we compared fluorescence emissions of the biofunctionalized arrays of nanodots with 0- and 40-nm spacing and found that they are identical (fig. S2, A and B). Also, we fabricated arrays of rectangular features with various orientations using sequential angle deposition of two metals, resulting in a gradually changing composition of the arrays—from pure Au to pure Ti, and functionalized the metals with the same chemistry (fig. S2, C to E). The obtained fluorescent palette changes gradually from green to red, according to the variation in the metal composition (fig. S2, F to H). Overall, these results demonstrate that our ternary process is completely site selective and insensitive to the geometry of the bimetallic nanopattern. We must note that our ternary functionalization approach was demonstrated here on a specific set of ligands. However, because NTA/Ni-His and biotin-avidin links are very commonly used bioconjugation techniques, and the variety of biomolecules that can be tagged with either biotin or histidine is practically unlimited, our ternary approach is very versatile and can be used as a general route to produce multifunctional molecular-scale ligand patterns.
In addition, we verified that our device could control ligand arrangement at the single-molecule scale. It is generally hypothesized that when biomolecules are immobilized onto lithographically patterned nanofeatures, their occupancy is determined by steric hindrance (13, 14). Recently, this hypothesis was recently confirmed experimentally using fluorescence microscopy (22, 42). In our devices, we functionalized the nanodots with MICA, whose complex with NKG2D has a footprint of 12 × 12 nm (43), and anti-KIR2DL1, whose footprint is about 7 × 19 nm (44, 45). Hence, the size of the fabricated nanodots suits the size of the immobilized molecules, so that each nanodot hosts, on average, one to two ligand-receptor complexes. To further confirm this, we immobilized KIR2DL1 and MICA onto metallic films according to our functionalization protocol and performed surface plasmon resonance analyses, which showed that the footprint of both biomolecules is about 200 nm2 each (see the Supplementary Materials and figs. S3 to S5 for details).
Activation of NK cells
With the demonstrated nanoscale control over the spatial segregation of different ligands, we could now explore how the segregation between ligands for NKG2D and KIR2DL1 receptors affects KIR2DL1-mediated inhibition of NKG2D signaling in NK cells. For this purpose, we stimulated primary NK cells on arrays of MICA and anti-KIR2DL1, which were either colocalized or separated by gaps of 20 and 40 nm. We also used three types of control samples: (i) positive control samples that contained MICA immobilized on a continuous Au film and should provide optimal conditions for the dense clustering of NKG2D and thus maximize the activation of NK cells; (ii) negative control samples that contained a continuous TiO2 film functionalized with anti-KIR2DL1, which was assumed to provide the optimal condition for the dense clustering of KIR2DL1 within the cell membrane; and (iii) neutral control samples of glass surface covered with poly-l-lysine, which enhances cell adhesion but lacks any specific functionalities for immune stimulation.
We assessed the cytotoxic activity of NK cells by monitoring the fluorescence intensity of lysosome-associated membrane protein-1 CD107a (Fig. 3A) (46, 47), which is a broadly used marker for the immune activation of T cells and NK cells (48, 49). In activated NK cells, lytic granules that contain CD107a move to the immune synapse, fuse with the cell membrane, and degranulate, thereby exposing CD107a. In our case, quantifying the expression of CD107a is the ultimate way to assess the degree of the immune activation of NK cells because the resulting biological information obtained by other methods like Western blot or flow cytometry would stem from cells activated on and outside of the ligand pattern. Figure 3 (B and C) presents representative images of NK cells stimulated on patterned and control surfaces, showing the expression of CD107a (white-colored).
Fig. 3. Impact of molecular-scale spatial control of two extracellular ligands on NK cell immune response.
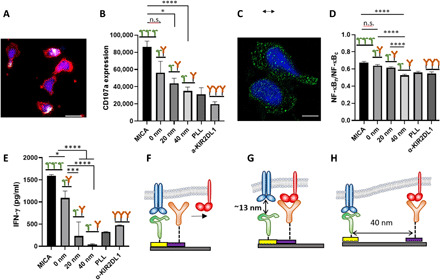
(A) NK cells stimulated on nanopattern of MICA continuous MICA film, respectively. The cells were stained with phalloidin for cytoskeleton (red), 4′,6-diamidino-2-phenylindole for nucleus (blue), and anti-CD107a (white). Scale bar, 10 μm. (B) Average amount of membrane-bound CD107a per cell on ligand patterns (n ≥ 60) compared to that of cells stimulated on control surfaces (n ≥ 60). Surfaces with continuous MICA provided the optimal condition for the activation of NK cells, whereas surfaces with poly-l-lysine and KIR2DL1 did not stimulate significant CD107a expression owing to the absence of activating ligands. Patterned arrays of MICA and anti-KIR2DL1 produced activation that depended on the gap between the ligands. (C) Nuclear factor κB (NF-κB) p65 staining (green) of activated NK cells. Scale bar, 5 μm. (D) Degree of NF-κB nuclear translocation on ligand patterns (n ≥ 60) compared to that of cells on control surfaces (n ≥ 30). (E) Interferon-γ (IFN-γ) release intensity. (F to H) Schemes of the possible mechanisms explaining the observed effect of the gap between the ligands in NK cell activation: (F) long ligand binds first, precluding binding to the colocalized shorter ligand; (G) short ligand binds first, precluding binding to the colocalized longer ligands; and (H) both ligands can bind their receptors owing to the spatial segregation. Data are representative of three experiments [means and SEM in (B), (D), and (E)]. In brackets, n represents the number of imaged regions. The results were considered to be significantly different for P < 0.05. *P < 0.05, ***P < 0.001, ****P < 0.0001. n.s., not significant.
As expected, the continuous MICA film provided the optimal conditions for the immune activation NK cells (Fig. 3B). This observation mirrors recent studies that used diverse microscale and nanoscale stimulating platforms to reveal the effect of clustering and density of activating ligands on the immune response of T cells and NK cells (20, 21, 50). It is reasonable to assume that the continuous MICA film used in our experiments provides a much higher amount of MICA ligands than the threshold amount required for the activation of NK cells; thus, the obtained average CD107a signal was relatively high. On the other hand, NK cells stimulated on poly-l-lysine and continuous anti-KIR2DL1 expressed, on average, similar amounts of CD107a, which are almost one-third of that found for the MICA surface. Specifically, low levels of CD107a expression by the cells stimulated on the anti-KIR2DL1 surface can be explained by the lack of any activating functionality rather than by the presence of anti-KIR2DL1 itself. Inhibitory signaling of KIR2DL1 suppresses NKG2D-mediated activation by blocking NKG2D clustering within the immune synapse (51). Thus, the recognition of KIR2DL1 itself is unlikely to produce any observable effect on the immune response of NK cells.
The most intriguing result, however, is the effect of the gap between anti-KIR2DL1 and MICA on the activation of NK cells. NK cells stimulated on arrays with colocalized ligands produced, on average, a CD107a signal that was slightly lower than that obtained continuous MICA, yet not significantly. This result indicates that when anti-KIR2DL1 colocalizes with MICA, it cannot produce a sufficient inhibition of the NKG2D signal. The inhibitory effect, however, progressively increased with the gap between the two ligands and became significant for the gap of 40 nm. This result demonstrates that the nanoscale architecture of activating and inhibitory ligands can encode the immune response of NK cells. Regardless of ligand spacing, ligand density is constant for all the tested activating/inhibitory conditions. Furthermore, the projected cell areas are very similar on all surfaces, suggesting that cells are exposed to the same total number of ligands of both types. Therefore, the observed pronounced effect on the inhibitory-activating balance in NK cells was obtained solely by altering the nanometric gap between the two ligands, while keeping all other compositional and spatial parameters of the cell environment constant.
To further explore the effect of the gap between the ligands on the inhibition efficiency of KIR2D1, we assessed the degree of nuclear translocation of nuclear factor κB (NF-κB), which takes part in the signaling pathway of NKG2D and is used as a marker for NK cell activation by immunofluorescence via quantification of the p65 nuclear content (52, 53) (please see experimental details in the Supplementary Materials). We stained the p65 unit of NF-κB after 18 hours of stimulation, imaged cells using confocal microscopy (Fig. 3C), and assessed the average degree of translocation (Fig. 3D) (53). We observed a similar trend to degranulation in NK cell response to the composition and configuration of the stimulating surfaces: Continuous MICA produced the highest degree of NF-κB translocation, which was not significantly different from that obtained on the arrays of colocalized activating and inhibitory ligands. However, the gradual segregation between the ligands significantly suppressed the translocation down to the level of pure anti-KIR2DL1. Last, we also checked the functional response of NK cells, by quantifying the secretion of interferon-γ (IFN-γ) by enzyme-linked immunosorbent assay (ELISA) (Fig. 3E). Notably, for ELISA, large arrays (2 × 2 mm) were fabricated and mounted in a specially designed microfluidic chamber to ensure that the secreted IFN-γ arose only from the cells on the arrays. Again, we observed a clear trend, by which cells reduce their activation with the increase in the gap between the ligands, indicating that the inhibitory function was stronger for the largest gap, thereby suppressing NKG2D-mediated activation of NK cells. Together, the observed trends in CD107a-, NF-κB–, and IFN-γ–based responses, which are consistent, clearly demonstrate that NK cells sense nanometric variation on the spatial copositioning of the activating and inhibitory ligands within the membrane plane.
The observed effect of the ligand segregation onto KIR2DL1-mediated inhibition is not obvious. We hypothesize that this effect is related to the difference in the total length of the ligand constructs (including the ligand molecule and the linker to the nanodots) and the relative flexibility of the bonds. In our case, the total length of MICA, including thiol-NTA linking molecules, is about 10 nm, while the total length of the anti-KIR2DL1 mAb including its biotin-avidin attachment chemistry on Ti is about 22 nm (see the Supplementary Materials for the estimation of the ligand length) (43, 44, 54, 55). For colocalized ligands, this length difference results in a ~12-nm mismatch between the ligands in the direction perpendicular to the membrane. Such a mismatch could prevent the simultaneous engagement of adjacent KIR2DL1 and NKG2D receptors, which could then explain how KIR2DL1 mediated inhibition is impeded (11).
More specific analysis of the effect of the gap between the ligands on the inhibition in our system must take into account that unbound receptors preferentially bind at the sites at which similarly sized ligand-receptor bonds have been already formed (56, 57). Here, two scenarios are possible for an isolated pair of colocalized activating and inhibitory receptors with a length mismatch. By the first scenario, the shorter activating ligand binds first to its receptor (Fig. 3F). This binding produces a localized proximity between the surface and the cell membrane, leaving no physical space for the binding of inhibitory receptor, which will be excluded from the site of the two colocalized ligands. By the second scenario (Fig. 3G), the longer inhibitory ligand binds first to its ligand. Then, the sequential binding of an activating receptor to the adjacent shorter activating ligand would be impeded by the large vertical gap formed by the inhibitory ligand-receptor pair. The inhibition mechanism of KIR2DL1 involves its binding to its ligand, followed by the dephosphorylation of two tyrosines within a tyrosine-based inhibitory motif, and recruitment and activation of Src homology region 2 domain-containing phosphatase-1 (SHP-1) that then dephosphorylates activating receptors and its proximal signaling molecules (58). Thus, KIR2DL1 ligation alone, without engaged activating receptors within the reach distance of SHP-1, does not produce any inhibitory effect. Because the initial binding is stochastic, both scenarios are likely to occur simultaneously, resulting in overall activating signaling. In the third scenario, at which the ligands are segregated, the above-described physical constraints for the simultaneous binding of both receptors are loosened because of the membrane flexibility (Fig. 3H). In such a case, inhibition of NKG2D signaling by its proximal KIR2DL1 is allowed.
Modeling of ligand-receptor interactions
To verify our hypothesis, we adapted the recently developed model that quantitatively captures interaction rates of ligand-receptor pairs embedded in membranes (see section S9 for details) (59). This model is particularly suitable here because no direct involvement of the cytoskeleton is expected in the formation or dissociation of KIR2DL1–anti-KIR2DL1 and NKG2D-MICA complexes. Accordingly, we mathematically represented each bond as a fluctuating flexible spring with a defined experimental rest length and stiffness. Consequently, we calculated the affinity of the long bond from its binding and dissociation rates in the presence of the shorter bond (Fig. 4, A to C), and vice versa (Fig. 4, D and E). Notably, the rates themselves are obtained from the convolution of the membrane gap-dependent distribution of positional probabilities with Bell-Dembo–like kinetics (60) (the dissociation rate is dependent on the force acting on the bond, while the binding rate reflects the energy stored in the bond). In the case of an isolated pair, consisting of one short (activating) and one long (inhibitory) bond in the membrane, these rates show a nonmonotonous behavior, and both bonds in the pair are stabilized relative to the lone bond. Furthermore, there is an optimum distance of about half of the membrane correlation length, at which each receptor has the highest binding rate and affinity for its cognate ligand. This phenomenon stems from the fact that the bonds are of different lengths and is further promoted by the difference in bond flexibilities. Membrane correlation length is of the order of 100 nm; thus, the modeled optimal spacing precisely fits the experimentally observed gap of 40 nm between the ligands (32), at which the inhibitory effect on the stimulation of NK cells was significant.
Fig. 4. Modeling the interaction of an NK cell with a substrate with immobilized ligands for KIR2DL1 and NKG2D.
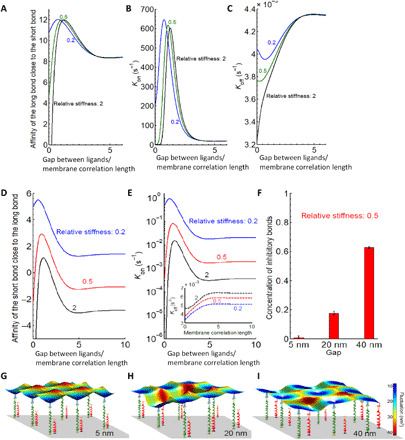
(A to C) Affinity, binding, and unbinding reaction rates of the long bond close to the short bond as a function of the gap between ligands normalized to membrane correlation length. (D and E) Affinity and binding rates of the short bond as a function of a distance (normalized to the membrane correlation length which is about 100 nm) from the existent long bond. Inset in (E): Dissociation rate of the long bond in the presence of the short bond. (F) Concentration of total number of bonds for different gap sizes, showing a clear increase in their numbers when the gap is wider. The error bars signify the variance in the total number of bonds in equilibrium and are calculated from the concentration-time curve (taking equilibrium state). (G to I) Characteristic snapshots of a small segment between the membrane and the patterned substrate are shown for a gap of (G) 5 nm, (H) 20 nm, and (I) 40 nm.
To verify whether this sensitivity to both the mechanical properties of the membrane and to length mismatch persists in geometry of 2D arrays of immobilized ligands, we performed effective Monte Carlo simulations (see section S10 for details) (59, 61). Specifically, the experimental patterns are first reproduced in our in silico model of the substrate, with respect to both the lateral distribution and the size of the ligand constructs. Next, the NK cell mimic is allowed to hover over the patterned substrate and spontaneously make and break KIR2DL1–anti-KIR2DL1 and NKG2D-MICA in the contact area with the substrate, using the same rates as discussed above. The number of formed bonds is recorded as a function of time and the system is allowed to propagate until the steady state is achieved, and the average number of formed bonds is reported (Fig. 4F). Notably, we find that similar to the two-bond case, at equilibrium, the total number of bonds increases when the gap between the nanodots on the pattern is widened within the range observed experimentally (Fig. 4G). More specifically, the number of short bonds increases with ligand spacing while the number of long bonds is barely affected (Fig. 4, G to I). Last, it should be noted that calculations here were made for anti-KIR2DL1 attached to the nanodot via a biotinylated Fc fragment, whereas immunoglobulin G has multiple residues available for chemical biotinylation (62). However, when the variability in the biotin position was considered in the calculations, it did not affect the overall trend of the bond affinity versus ligand gap, but only reduced the value of the horizontal gap at which the affinity peaks by ~0.1 to 0.2 of the membrane correlation length (see the Supplementary Materials and fig. S10).
DISCUSSION
Lymphocyte activation is controlled by numerous receptors, whose spatial arrangement with respect to each other has been hypothesized to depend on their matching sizes: Similarly sized ligand-receptor complexes come in close proximity, whereas differently sized ones remain spatially separated (63–65) This size-dependent separation is the basis of the kinetic segregation model for T cell activation, by which large molecules of tyrosine phosphatase CD45 are spatially excluded from relatively short TCR–major histocompatibility complexes (MHC) formed at the T cell–anaphase-promoting complex (APC) interface, thereby diminishing CD45-mediated dephosphorylation of TCR-MHC, and producing downstream signaling that lastly leads to the activation of T cells (66). The hypothesis that the size mismatch between TCR and CD45 causes their segregation upon TCR engagement, and the subsequent decrease in TCR-MHC dephosphorylation, was directly confirmed using several artificial model systems for T cell stimulation. For example, nanofabricated monoligand patterns were used to demonstrate that MHC ligands positioned on nanoparticles within the supported bilayer do not allow TCR association with the diffusing CD45, while MHC ligands elevated on etched nanopillars increased the spacing between the bound membranes and allowed the association of CD45 and TCR, thereby affecting the phosphorylation of the TCR, and modulating T cell immune response (19). Also, a T cell mimicking model based on membrane vesicles was used to show that large CD45 isoforms were excluded more rapidly from the TCR-MHC complex than smaller counterparts, directly indicating the crucial role of CD45 size in this exclusion (67). Furthermore, elongation of TCR ligands (8) and shortening of CD45 ectodomain (9) were both shown to suppress TCR engagement.
Similar to T cells, the immune function of NK cells is regulated by multiple receptors with different functionalities—activating, costimulatory, and inhibitory—whose delicate signaling balance determines the immune response of NK cells. Although receptor clustering in NK cells has been less studied than for T cells, it would be reasonable to presume that the association or segregation of two different receptors in NK cells is regulated by spatial exclusion: Two receptors will spatially associate only if the sizes of their complex with their cognate ligands match and segregate if the sizes are different, similarly to how TCR-MHC and CD45 in T cells do. It was previously shown that varying ectodomain size of either MICA or KIR2DL1 ligand HLA-C could greatly affect the inhibition of NK cells: Differently sized ectodomains of MICA and HLA-C produced segregation between the two ligands, resulting in a reduction of KIR2DL1 mediated inhibition, while matched ectodomain lengths, regardless of their absolute size, produced colocalization of the two ligands and thus induced an effective inhibition (11). Still, this study on NK cells, as well as previously mentioned studies on T cells, predicts that when two size-mismatched ligands are brought together, they cannot both bind their receptors simultaneously, which hampers their signal integration. However, this prediction could not be directly examined in those studies because they could not control the lateral position of the ligands within the membrane plane and, therefore, could not produce a scenario in which differently sized ligands are controllably brought together within the cell membrane. In our work, on the contrary, both the size mismatch and lateral mismatch were deterministically controlled, via the size of molecules and the spacing between the nanodots, respectively. In this way, we completely and precisely realized the scenario of bringing together two unevenly sized ligands and provided the full and exact experimental confirmation that this scenario hampers signal integration between the two receptors. Furthermore, we also showed that the constraints imposed by this scenario on signal integration can be relieved if the ligands are controllably spaced apart, allowing an effective inhibition, at least when the spacing between ligands stays within a few tens of nanometers. Last, we provided theoretical support for this demonstration based on the modeling of the membrane and receptor conformation. Thus, our work clearly confirms that the high level of proximity of inhibitory and activating ligand-receptor complexes, or tightly packed segregated clusters, could be deleterious to inhibitory signaling in the case where there is a mismatch in the size of the ligands. Our results furthermore point to the important aspect of biomechanical optimization of the membrane and the protein design in NK cells, in the context of length and stiffness.
While our model is based on functional and signaling assays of NK cells, a follow-up study should directly verify it, for example, by imaging of KIR2DL1. For this purpose, KIR2DL1 can be fused with a fluorescent protein (68), and its recruitment to ligand pairs with various spacing can be imaged by super-resolution microscopy. According to our model, the recruitment of KIR2DL1 should be inversely proportional to the spacing between the ligands and will be minimal for the ligands with zero spacing.
We also cannot exclude the idea that the spatial manipulation of two ligands could affect the recruitment of tyrosine phosphatases SHP-1 and SHP-2, which are key players in the inhibition mechanism in NK cells. SHP-1 and SHP-2 are recruited by KIR-family inhibitory receptors, and this recruitment causes tyrosine dephosphorylation of activating receptors such as 2B4, NKG2D, and their costimulatory receptors (58, 69). Whether this mechanism requires the physical recruitment of SHP-1 and SHP-2 between the activating and inhibitory receptors is unclear. If this is the case, colocalizing MICA and anti-KIR2DL1 by their immobilization onto overlapped nanodots (with a zero gap) could prevent the recruitment of SHP-1 and SHP-2 in the interstice between the two receptors, which might be needed for the effective tyrosine phosphorylation of NKG2D. It remains uncertain, however, if a ligand spacing of a few tens of nanometers, as in our most potent inhibitory system, can impede the association between NKG2D and KIR2DL1. Given that the successful regulation of NK cell activation has been observed in segregated domains, the role of the direct interactions between NKG2D and KIR2DL, and its impact on KIR2DL1 inhibition, remains an open question. To address it, our follow-up work will include fluorescence staining of SHP-1 (70). This will shed light on whether and how the effect of ligand spacing on the inhibition is related to phosphatase recruitment.
Besides the explanation based on the size consideration, an additional view can be considered, based on the scanning electron microscopy (SEM) images of NK cells stimulated on the nanoarrays (Fig. 1, D and E). It can be seen in these images that the adhesion between the cells and the surface is poor and that the contact between the cells contacts the surface through thin projections with a tip radius on the order of 40 nm called microvilli, at whose tips the activating receptors are concentrated (71). In this case, the larger link to the inhibitory receptors may interact with the preset geometry of the projection to systematically push the inhibitory receptor toward the shaft of the projection due to the longer tether and away from the activating receptor. On the basis of simple geometric considerations, this view might be even more deterministic in generating the observed outcome.
In summary, we demonstrated here that controlling the juxtaposition of activating and inhibitory ligands either on the axis perpendicular to the membrane plane or within the membrane plane, individually or in combination with each other, provides a powerful means to regulate the functional cross-talk in NK cell activation. We trace the potential origins of this behavior in the biomechanical optimization of the receptor-ligand constructs and the membrane. This exquisite control is only possible by a precise lateral fixation of ligands, which was allowed by our novel lithographic approach combined with unprecedented site specificity of ligand immobilization into three distinct functionalities. The observed effect of the gap between the two ligands stems from the size difference of ligands, but it is only the tip of the iceberg of what seems to be a complex interplay between the size of the ligands, their spatial arrangement within the membrane, and membrane fluctuations, and this interplay will be further explored in our follow-up works. In particular, to mimic physiological conditions, in which KIR2DL1 and NKG2D complexes with their ligands are of similar size (~15 nm) (72, 73), and remove the physical barrier for simultaneous formation of colocalized activating and inhibitory complexes, equally sized linker ligands should be used. For this, anti-KIR2DL1 can be replaced with a smaller HLA-C2 ligand. We should note that we used this particular type of anti-KIR2DL1 because of its high specificity to KIR2DL1 (41), whereas its HLA-C2 ligand also recognizes the activating receptor KIR2DS1 whose ectodomain is almost identical to that of KIR2DL1 (74). Notwithstanding the above, replacing anti-KIR2DL1 with HLA-C2 will be done in the follow-up experiments to study whether and how this replacement can affect the observed dependence of NK cell inhibition on the ligand arrangement. Last, the fact that the affinity of the interaction between KIR2DL1 to its antibody is likely higher than that of HLA-C2 should be also considered, and the effect of the different affinities should be examined in follow-up work. Specifically, we will extend this study to two more balanced platforms for NK cell stimulation: one combining activating and inhibitory antibodies, both with large size and high affinity, and another combining two natural ligands (MICA and HLA-C2), both with a smaller size and relatively low affinity.
Future platforms should also include variable surface stiffness because it was recently shown that NK cells are mechanosensitive and apply forces to probe the mechanical properties of their environment (75, 76). In addition to the specific case study of the inhibitory-activating signal integration in NK cells, the nanotechnological platform supported by the theoretical insights allows broad studies of the role of spatial coordination of different signaling molecules in their functional interplay. Such an interplay could be important for the activation of other immune cells, such as T cells and B cells, whose signaling mechanism is yet to be fully understood. Beyond the fundamental insights into immune cell regulation, our approach could be useful in the more applied context of immunotherapy. One example could involve systems based on dual activating and inhibitory chimeric antigen receptor (aCAR:iCAR) (77), whose efficient delivery requires the optimization of a combination of multiple ligated receptors. The work presented here establishes a reliable methodology for comprehensively addressing various systems, in which the integration of multiple signals from multiple receptors needs to be identified and optimized.
MATERIALS AND METHODS
Detailed experimental protocols are provided in the Supplementary Materials.
Preparation of the bimetallic nanodot arrays
A nanoimprint mold was prepared by electron beam lithography of hydrogen silesquioxane consisting of an orthogonal array of ~20-nm features separated by 200 nm. Thermal resist was imprinted onto silicon substrates, covered by a titanium mask using angle evaporation, and etched with oxygen plasma. Ti/Au (1 nm/5 nm) and Ti/Cr (5 nm/3 nm) were sequentially evaporated at two opposite angles to produce arrays of nanodot heterodimers after liftoff.
Ternary biofunctionalization of nanodot arrays
The Si background of the fabricated arrays was passivated by immersing the samples in a toluene solution PEG-silane and catalytic amount of acetic acid. Then, the Cr protection layer was etched in commercial Cr etchant. Oxidized Ti was biotinylated by immersing samples in a solution of carboxyl terminated alkyl phosphonic acid followed by baking and the subsequent attachment biotin terminated amino-PEG. Au was functionalized by the reaction in an ethanolic solution of nitrilotriacetic-terminated alkyl thiol, followed by Ni2+ chelation. The biotin moieties on Ti were modified with green fluorescent–labeled NeutrAvidin. The samples were then successively immersed in solutions containing biotin α-KIR2DL1 and His-MICA. For cell studies, freshly prepared samples were used as is. To confirm the selective attachment of MICA by fluorescence, the surfaces were first incubated overnight in α-MICA, followed by a red fluorescent secondary antibody. Last, surfaces were mounted and imaged with a fluorescence microscope.
Primary NK cell purification
Primary NK cells were purified from peripheral blood of healthy, adult, volunteer donors, recruited by written informed consent, as approved by the Institutional Review Board Ben-Gurion University of the Negev. The primary NK cells were used between week 1 and week 3 after isolation.
NK cell degranulation
For CD107a-mediated evaluation of degranulation, cultured NK cells were seeded onto the surfaces in interleukin-2 (IL-2) poor growth medium containing APC α-CD10a and left to adhere for 3 to 4 hours. After fixing the adherent cells with paraformaldehyde, their cytoskeleton was stained with Alexa Fluor 555 phalloidin without permeabilization to maintain the integrity of the cell membrane. Last, the nuclei were stained by mounting the samples with a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI).
NF-κB activation
To examine NF-κB p65 nuclear content, NK cells were seeded onto the surfaces in IL-2 poor growth medium for 18 to 24 hours (50 U of IL-2 compared to 300 U when cultured). Cells were fixed in paraformaldehyde and briefly permeabilized with a non-ionic surfactant and ice-cold methanol for 10 min. Cells were then blocked and incubated with anti–NF-κB p65 subunit, followed by incubation in an Alexa Fluor 647–labeled secondary antibody and mounting with DAPI-containing mounting medium.
Microscopy
Characterization of the surfaces as well as NK cell adhesion and degranulation were performed using a Nikon Ti2e epifluorescence microscope and quantified using the Fiji imaging software. For α-CD107a, we quantified the degree of degranulation, and therefore indirectly the degree of activation, which is proportional to the intensity of the APC α-CD107a signal. For NF-κB activation, surfaces were imaged with a Zeiss LSM 880 confocal microscope and quantified using the Fiji imaging software. The protocol for the quantification of NF-κB p65 nuclear content was taken from the work by others (52, 53), with modifications taken from Le Saux et al. (78). Similar to the work by others, NF-κB activation is proportional to the nuclear content of the p65 subunit NF-κB that, in our case, is equal to the fluorescence intensity ratio of the nuclear region to the total cell. To ensure consistent results in the quantification of fluorescence intensity, exposure time and magnification were set on the first imaged sample and were then kept unchanged for all the other images.
Interferon-γ secretion by NK cells
For IFN-γ detection, cultured NK cells were seeded onto the surfaces in IL-2 poor growth medium for 18 to 24 hours. To ensure that cells interacted with the nanodot arrays only, custom-made chambers were designed with wells whose contact area gave access to the nanodot array only. Following incubation, the supernatant was collected and the released IFN-γ was quantified using a standard sandwich immunoassay.
Statistics
All biological experiments were performed three times. Thirty to 60 regions were imaged and averaged for quantification. In Fig. 3, means with SEM are shown. Statistical analysis was performed by analysis of variance, and Tukey’s multiple comparison post hoc test was also performed. The results were considered to be significantly different for P < 0.05.
Acknowledgments
Funding: This work was funded by the Multidisciplinary Research Grant—The Faculty of Health Science in Ben-Gurion University of the Negev, Israel Science Foundation, Individual Grant No. 1401/15, and Israel Science Foundations: F.I.R.S.T. Individual Grant No. 2058/18. SPR and PM-IRRAS analyses were performed with the financial help of COST Action CA15126. This work has benefited from the facilities and expertise of the Biophysical and Structural Chemistry platform at IECB, CNRS UMS3033, Inserm US001, Bordeaux University. The Biacore T200 instrument was acquired with the support of the Conseil Régional d’Aquitaine, the GIS-IBiSA, and the Cellule Hôtels à Projets of the CNRS. We thank L. Minder, technical manager of the SPR Platform. A.-S.S. and L.L. thank the German Science Foundation project SM 289/8-1, AOBJ: 652939. L.L. was supported by the China Scholarship Council (CSC, File No. 201806185038). E.T. was supported by the Israel Ministry of Science and Technology, Ariane de Rothschild Women’s Doctoral scholarships program for outstanding female PhD students, and the Israel Scholarship Education Foundation (ISEF). Author contributions: E.T., G.L.S., A.P., and M.S. designed the experiments. E.T., Y.K., and V.B. fabricated the nanodot arrays. G.L.S. developed the ternary functionalization. E.T. and G.L.S. fabricated the arrays for NK cell study. M.R. fabricated and characterized bimetallic arrays with varied metal composition. G.L.S. and C.D.P. carried out the SPR study. G.L.S. and T.B. performed the PM-IRRAS analyses. A.E. produced primary NK cells. E.T., G.L.S., and O.R. did the cell experiments. E.T., G.L.S., and U.H. analyzed the experimental results. L.L. and A.-S.S. did the numerical simulations. E.T., G.L.S., A.-S.S., A.P., and M.S. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabc1640/DC1
REFERENCES AND NOTES
- 1.M. L. Dustin, Handbook of Cell Signaling, 2/e (Elsevier, 2010). [Google Scholar]
- 2.M. L. Dustin, C. T. Baldari, The Immune Synapse. Methods in Molecular Biology, vol 1584 (Humana Press, 2017). [DOI] [PubMed] [Google Scholar]
- 3.Pageon S. V., Tabarin T., Yamamoto Y., Ma Y., Bridgeman J. S., Cohnen A., Benzing C., Gao Y., Crowther M. D., Tungatt K., Dolton G., Sewell A. K., Price D. A., Acuto O., Parton R. G., Gooding J. J., Rossy J., Rossjohn J., Gaus K., Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 113, E5454–E5463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lillemeier B. F., Mörtelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T., Davis M. M., TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson D. J., Owen D. M., Rossy J., Magenau A., Wehrmann M., Gooding J. J., Gaus K., Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 12, 655–662 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Hui E., Cheung J., Zhu J., Su X., Taylor M. J., Wallweber H. A., Sasmal D. K., Huang J., Kim J. M., Mellman I., Vale R. D., T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355, 1428–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oszmiana A., Williamson D. J., Cordoba S.-P., Morgan D. J., Kennedy P. R., Stacey K., Davis D. M., The size of activating and inhibitory killer Ig-like receptor nanoclusters is controlled by the transmembrane sequence and affects signaling. Cell Rep. 15, 1957–1972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhuri K., Wiseman D., Brown M. H., Gould K., van der Merwe P. A., T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 436, 578–582 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Irles C., Symons A., Michel F., Bakker T. R., van der Merwe P. A., Acuto O., CD45 ectodomain controls interaction with GEMs and Lck activity for optimal TCR signaling. Nat. Immunol. 4, 189–197 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Brzostek J., Chai J.-G., Gebhardt F., Busch D. H., Zhao R., Anton Van Der Merwe P., Gould K. G., Ligand dimensions are important in controlling NK-cell responses. Eur. J. Immunol. 40, 2050–2059 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köhler K., Xiong S., Brzostek J., Mehrabi M., Eissmann P., Harrison A., Cordoba S., Oddos S., Miloserdov V., Gould K., Burroughs N., van der Merwe P., Davis D., Matched sizes of activating and inhibitory receptor/ligand pairs are required for optimal signal integration by human natural killer cells. PLOS ONE 5, e15374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X., Ditlev J. A., Hui E., Xing W., Banjade S., Okrut J., King D. S., Taunton J., Rosen M. K., Vale R. D., Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold M., Cavalcanti-Adam E. A., Glass R., Blümmel J., Eck W., Kantlehner M., Kessler H., Spatz J. P., Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem 5, 383–388 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Schvartzman M., Palma M., Sable J., Abramson J., Hu X., Sheetz M. P., Wind S. J., Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett. 11, 1306–1312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pi F., Dillard P., Alameddine R., Benard E., Wahl A., Ozerov I., Charrier A., Limozin L., Sengupta K., Size-tunable organic nanodot arrays: A versatile platform for manipulating and imaging cells. Nano Lett. 15, 5178–5184 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Schumann K., Lämmermann T., Bruckner M., Legler D. F., Polleux J., Spatz J. P., Schuler G., Förster R., Lutz M. B., Sorokin L., Sixt M., Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity 32, 703–713 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Oria R., Wiegand T., Escribano J., Elosegui-Artola A., Uriarte J. J., Moreno-Pulido C., Platzman I., Delcanale P., Albertazzi L., Navajas D., Trepat X., García-Aznar J. M., Cavalcanti-Adam E. A., Roca-Cusachs P., Force loading explains spatial sensing of ligands by cells. Nature 552, 219–224 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Dalby M. J., Gadegaard N., Oreffo R. O. C., Harnessing nanotopography and integrin–matrix interactions to influence stem cell fate. Nat. Mater. 13, 558–569 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Cai H., Muller J., Depoil D., Mayya V., Sheetz M. P., Dustin M. L., Wind S. J., Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat. Nanotechnol. 13, 610–617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keydar Y., Le Saux G., Pandey A., Avishay E., Bar-Hanin N., Esti T., Bhingardive V., Hadad U., Porgador A., Schvartzman M., Natural killer cells’ immune response requires a minimal nanoscale distribution of activating antigens. Nanoscale 10, 14651–14659 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Delcassian D., Depoil D., Rudnicka D., Liu M., Davis D. M., Dustin M. L., Dunlop I. E., Nanoscale ligand spacing influences receptor triggering in T cells and NK cells. Nano Lett. 13, 5608–5614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deeg J., Axmann M., Matic J., Liapis A., Depoil D., Afrose J., Curado S., Dustin M. L., Spatz J. P., T cell activation is determined by the number of presented antigens. Nano Lett. 13, 5619–5626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenz S. F., Bihr T., Schmidt D., Merkel R., Seifert U., Sengupta K., Smith A. S., Membrane fluctuations mediate lateral interaction between cadherin bonds. Nat. Phys. 13, 906–913 (2017). [Google Scholar]
- 24.Guasch J., Hoffmann M., Diemer J., Riahinezhad H., Neubauer S., Kessler H., Spatz J. P., Combining adhesive nanostructured surfaces and costimulatory signals to increase T cell activation. Nano Lett. 18, 5899–5904 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Culley F. J., Johnson M., Evans J. H., Kumar S., Crilly R., Casasbuenas J., Schnyder T., Mehrabi M., Deonarain M. P., Ushakov D. S., Braud V., Roth G., Brock R., Köhler K., Davis D. M., Natural killer cell signal integration balances synapse symmetry and migration. PLoS Biol. 7, e1000159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doh J., Irvine D. J., Immunological synapse arrays: Patterned protein surfaces that modulate immunological synapse structure formation in T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 5700–5705 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purdy A. K., Campbell K. S., Natural killer cells and cancer: Regulation by the killer cell ig-like receptors (KIR). Cancer Biol. Ther. 8, 2209–2218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretta L., Moretta A., Killer immunoglobulin-like receptors. Curr. Opin. Immunol. 16, 626–633 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Pageon S. V., Cordoba S.-P., Owen D. M., Rothery S. M., Oszmiana A., Davis D. M., Superresolution microscopy reveals nanometer-scale reorganization of inhibitory natural killer cell receptors upon activation of NKG2D. Sci. Signal. 6, ra62 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Davis D. M., Assembly of the immunological synapse for T cells and NK cells. Trends Immunol. 23, 356–363 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Faure M., Barber D. F., Takahashi S. M., Jin T., Long E. O., Spontaneous clustering and tyrosine phosphorylation of nk cell inhibitory receptor induced by ligand binding. J. Immunol. 170, 6107–6114 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Janeš J. A., Stumpf H., Schmidt D., Seifert U., Smith A. S., Statistical mechanics of an elastically pinned membrane: Static profile and correlations. Biophys. J. 116, 283–295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schvartzman M., Wind S. J., Robust pattern transfer of nanoimprinted features for sub-5-nm fabrication. Nano Lett. 9, 3629–3634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schvartzman M., Nguyen K., Palma M., Abramson J., Sable J., Hone J., Sheetz M. P., Wind S. J., Fabrication of nanoscale bioarrays for the study of cytoskeletal protein binding interactions using nanoimprint lithography. J. Vac. Sci. Technol. B Microelectron. Nanom. Struct. 27, 61–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Saux G., Edri A., Keydar Y., Hadad U., Porgador A., Schvartzman M., Spatial and chemical surface guidance of NK cell cytotoxic activity. ACS Appl. Mater. Interfaces 10, 11486–11494 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Huang D., Patel K., Perez-Garrido S., Marshall J. F., Palma M., DNA origami nanoarrays for multivalent investigations of cancer cell spreading with nanoscale spatial resolution and single-molecule control. ACS Nano 13, 728–736 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Prime K., Whitesides G., Self-assembled organic monolayers: Model systems for studying adsorption of proteins at surfaces. Science 252, 1164–1167 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Lee S. W., Laibinis P. E., Protein-resistant coatings for glass and metal oxide surfaces derived from oligo(ethylene glycol)-terminated alkytrichlorosilanes. Biomaterials 19, 1669–1675 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Desbat B., Turlet J. M., Buffeteau T., Polarization modulation FT-IR spectroscopy of surfaces and ultra-thin films: Experimental procedure and quantitative analysis. Appl. Spectrosc. 45, 380–389 (1991). [Google Scholar]
- 40.Ramin M. A., Le Bourdon G., Daugey N., Bennetau B., Vellutini L., Buffeteau T., PM-IRRAS investigation of self-assembled monolayers grafted onto SiO 2/Au substrates. Langmuir 27, 6076–6084 (2011). [DOI] [PubMed] [Google Scholar]
- 41.David G., Morvan M., Gagne K., Kerdudou N., Willem C., Devys A., Bonneville M., Folléa G., Bignon J. D., Retière C., Discrimination between the main activating and inhibitory killer cell immunoglobulin-like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology 128, 172–184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai H., Wolfenson H., Depoil D., Dustin M. L., Sheetz M. P., Wind S. J., Molecular occupancy of nanodot arrays. ACS Nano 10, 4173–4183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P., Morris D. L., Willcox B. E., Steinle A., Spies T., Strong R. K., Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2, 443–451 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Harris L. J., Skaletsky E., McPherson A., Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275, 861–872 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Harris L. J., Larson S. B., Hasel K. W., McPherson A., Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Krzewski K., Gil-Krzewska A., Nguyen V., Peruzzi G., Coligan J. E., LAMP1/CD107a is required for efficient perforin delivery to lytic granules and NK-cell cytotoxicity. Blood 121, 4672–4683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertrand F., Muller S., Roh K.-H., Laurent C., Dupre L., Valitutti S., An initial and rapid step of lytic granule secretion precedes microtubule organizing center polarization at the cytotoxic T lymphocyte/target cell synapse. Proc. Natl. Acad. Sci. U.S.A. 110, 6073–6078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aktas E., Kucuksezer U. C., Bilgic S., Erten G., Deniz G., Relationship between CD107a expression and cytotoxic activity. Cell. Immunol. 254, 149–154 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Alter G., Malenfant J. M., Altfeld M., CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 294, 15–22 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Manz B. N., Jackson B. L., Petit R. S., Dustin M. L., Groves J., T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. 108, 9089–9094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Endt J., McCann F. E., Almeida C. R., Urlaub D., Leung R., Pende D., Davis D. M., Watzl C., Inhibitory receptor signals suppress ligation-induced recruitment of NKG2D to GM1-rich membrane domains at the human NK cell immune synapse. J. Immunol. 178, 5606–5611 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Pandey R., DeStephan C. M., Madge L. A., May M. J., Orange J. S., NKp30 ligation induces rapid activation of the canonical NF-κB pathway in NK cells. J. Immunol. 179, 7385–7396 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon H. J., Choi G. E., Ryu S., Kwon S. J., Kim S. C., Booth C., Nichols K. E., Kim H. S., Stepwise phosphorylation of p65 promotes NF-kB activation and NK cell responses during target cell recognition. Nat. Commun. 7, 11686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFarland B. J., Kortemme T., Yu S. F., Baker D., Strong R. K., Symmetry recognizing asymmetry: Analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure 11, 411–422 (2003). [DOI] [PubMed] [Google Scholar]
- 55.O. Livnah, E. A. Bayer, M. Wilchek, J. L. Sussman, in Proceedings of the National Academy of Sciences of the United States of America (National Academy of Sciences, 1993), vol. 90, pp. 5076–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez-Ferreros P., Gaus K., Goyette J., Tethered Signaling in Inhibitory Immune Receptors. Front. Phys. 6, 158 (2019). [Google Scholar]
- 57.Goyette J., Nieves D. J., Ma Y., Gaus K., How does T cell receptor clustering impact on signal transduction? J. Cell Sci. 132, jcs226423 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Long E. O., Negative signaling by inhibitory receptors: The NK cell paradigm. Immunol. Rev. 224, 70–84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bihr T., Seifert U., Smith A.-S., Multiscale approaches to protein-mediated interactions between membranes—Relating microscopic and macroscopic dynamics in radially growing adhesions. New J. Phys. 17, 083016 (2015). [Google Scholar]
- 60.Dembo M., Torney D. C., Saxman K., Hammer D., The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. Biol. Sci. 234, 55–83 (1988). [DOI] [PubMed] [Google Scholar]
- 61.Bihr T., Seifert U., Smith A. S., Nucleation of ligand-receptor domains in membrane adhesion. Phys. Rev. Lett. 109, 258101 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Welch N. G., Scoble J. A., Muir B. W., Pigram P. J., Orientation and characterization of immobilized antibodies for improved immunoassays (Review). Biointerphases 12, 02D301 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Dustin M. L., Supported bilayers at the vanguard of immune cell activation studies. J. Struct. Biol. 168, 152–160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milstein O., Tseng S. Y., Starr T., Llodra J., Nans A., Liu M., Wild M. K., Van Der Merwe P. A., Stokes D. L., Reisner Y., Dustin M. L., Nanoscale increases in CD2-CD48-mediated intermembrane spacing decrease adhesion and reorganize the immunological synapse. J. Biol. Chem. 283, 34414–34422 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wild M. K., Cambiaggi A., Brown M. H., Davies E. A., Ohno H., Saito T., Van Der Merwe P. A., Dependence of T cell antigen recognition on the dimensions of an accessory receptor-ligand complex. J. Exp. Med. 190, 31–42 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis S. J., van der Merwe P. A., The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 7, 803–809 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Carbone C. B., Kern N., Fernandes R. A., Hui E., Su X., Garcia K. C., Vale R. D., In vitro reconstitution of T cell receptor-mediated segregation of the CD45 phosphatase. Proc. Natl. Acad. Sci. U.S.A. 114, E9338–E9345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis D. M., Chiu I., Fassett M., Cohen G. B., Mandelboim O., Strominger J. L., The human natural killer cell immune synapse. Proc. Natl. Acad. Sci. U.S.A. 96, 15062–15067 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matalon O., Fried S., Ben-Shmuel A., Pauker M. H., Joseph N., Keizer D., Piterburg M., Barda-Saad M., Dephosphorylation of the adaptor LAT and phospholipase C-γ by SHP-1 inhibits natural killer cell cytotoxicity. Sci. Signal. 9, ra54 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Mintz M. A., Felce J. H., Chou M. Y., Mayya V., Xu Y., Shui J. W., An J., Li Z., Marson A., Okada T., Ware C. F., Kronenberg M., Dustin M. L., Cyster J. G., The HVEM-BTLA axis restrains T cell help to germinal center B cells and functions as a cell-extrinsic suppressor in lymphomagenesis. Immunity 51, 310–323.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jung Y., Riven I., Feigelson S. W., Kartvelishvily E., Tohya K., Miyasaka M., Alon R., Haran G., Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl. Acad. Sci. 113, E5916–E5924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dushek O., Goyette J., van der Merwe P. A., Non-catalytic tyrosine-phosphorylated receptors. Immunol. Rev. 250, 258–276 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Kumar S., Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 154, 383–393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cognet C., Farnarier C., Gauthier L., Frassati C., André P., Magérus-Chatinet A., Anfossi N., Rieux-Laucat F., Vivier E., Schleinitz N., Expression of the HLA-C2-specific activating killer-cell Ig-like receptor KIR2DS1 on NK and T cells. Clin. Immunol. 135, 26–32 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Matalon O., Ben-Shmuel A., Kivelevitz J., Sabag B., Fried S., Joseph N., Noy E., Biber G., Barda-Saad M., Actin retrograde flow controls natural killer cell response by regulating the conformation state of SHP-1. EMBO J. 37, e96264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Saux G., Bar-Hanin N., Edri A., Hadad U., Porgador A., Schvartzman M., Nanoscale mechanosensing of natural killer cells is revealed by antigen-functionalized nanowires. Adv. Mater. 31, e1805954 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Ruella M., Barrett D. M., Kenderian S. S., Shestova O., Hofmann T. J., Perazzelli J., Klichinsky M., Aikawa V., Nazimuddin F., Kozlowski M., Scholler J., Lacey S. F., Melenhorst J. J., Morrissette J. J. D., Christian D. A., Hunter C. A., Kalos M., Porter D. L., June C. H., Grupp S. A., Gill S., Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. Am. Soc. Clin. Investig. 126, 3814–3826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le Saux G., Plawinski L., Parrot C., Nlate S., Servant L., Teichmann M., Buffeteau T., Durrieu M.-C., Surface bound VEGF mimicking peptide maintains endothelial cell proliferation in the absence of soluble VEGF in vitro. J. Biomed. Mater. Res. Part A. 104, 1425–1436 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabc1640/DC1


