SUMMARY:
Quantitative Multiplex Immunoprecipitation (QMI) uses flow cytometry for sensitive detection of changes in native protein-protein interactions at mesoscale. QMI can be performed using a small amount of biomaterial, does not require genetically engineered tags, and can be adapted for any previously defined protein interaction network.
Physical protein-protein interactions and their dynamics are the basis of a great deal of cellular physiology. While it is useful to measure changes in individual pairs of protein interactions or all interactions with one specific protein, many cellular processes or signals are modulated by interactions among multiple proteins. To address questions concerning multi-protein complexes, signaling cascades, and integration of signals, we have developed Quantitative Multiplex Immunoprecipitation (QMI), which allows quantitative assessment of fold changes in protein interactions based on relative fluorescence measurements. In QMI, protein complexes from lysate are immunoprecipitated onto microspheres, then probed with a second antibody. Flow cytometry measures the changes in protein-protein interactions between samples. Different classes of microspheres contain distinct ratios of fluorescent dyes, which allows each class to be conjugated to a unique immunoprecipitation antibody, combined for simultaneous co-immunoprecipitation, and distinguished by flow cytometry. QMI does not require genetic tagging and can be done with minimal biomaterial compared to other immunoprecipitation methods. The initial set-up of a QMI assay involves screening a panel of antibodies and empirically determining an appropriate lysis buffer. The subsequent reagent preparation includes covalently coupling immunoprecipitation antibodies to microspheres and biotinylating probe antibodies so they can be labeled by a streptavidin-conjugated fluorophore. To run the assay, lysate is immunoprecipitated onto beads overnight, then the beads are divided and incubated with different probe antibodies and a fluorophore label, and read by flow cytometry. Two statistical tests are performed to generate high-confidence “hits,” which can then be visualized using either a heatmap or a node-edge diagram. QMI can be adapted for any defined group of interacting proteins, and has thus far been used to characterize signaling networks in T cells and neuronal glutamate synapses. Results have led to new hypothesis generation with potential diagnostic and therapeutic applications.
Keywords: IP-FCM, proteomics, protein interaction, signaling
INTRODUCTION:
Cellular processes are largely controlled by changes in protein-protein interactions.1 These changes are often thought of in terms of linear signaling pathways that switch between steady states based on inputs, but they actually function as integrated networks made up of dynamic multiprotein complexes in which frequency, duration, and amplitude of signals all change in response to one another.2,3 In the case of G proteins, for example, different receptors often have the ability to activate the same G protein, and one receptor can also activate more than one type of G protein.4,5 In order for the relatively small number of G protein classes to specifically modulate a vast array of cellular functions such as synaptic transmission, hormone regulation, and cell migration, cells must both integrate and differentiate these signals.3,4 Evidence has shown that this signal specificity, for G proteins as well as others, is primarily derived on the basis of delicately balanced protein-protein interactions and their temporal dynamics.1–6 Signaling ‘pathways’ comprised of dynamic protein complexes have multiple inputs, outputs, and feedback loops, so a single perturbation has the opportunity to alter the overall homeostatic balance of a cell’s physiology.3,6 Rather than changing in isolation, elements of a signaling pathway can interact with other pathways to initiate or modify additional cellular processes.6,7 Similarly, regulators act on pathways at different points, and even a single pathway element likely has multiple regulators.3,4,8 It is now widely agreed that signaling should be examined from a network perspective in order to better understand how the integration of multiple inputs controls discrete cellular functions in health and disease.6–12
Techniques such as fluorescence resonance energy transfer (FRET) and co-immunoprecipitation (co-IP) followed by western blotting are useful for analyzing changes in individual protein interactions, but due to technological limitations are currently unable to be scaled up sufficiently to investigate network-level changes in protein complex composition. While co-IP with subsequent mass spectrometry is a highly effective method for determining a protein’s interaction partners, quantification of protein interactions from small samples, such as those from primary patients or rodent models, still presents a substantial technological challenge. Quantitative Multiplex Immunoprecipitation (QMI) was developed to gather medium-throughput, quantitative data about fold changes in dynamic protein interaction networks using the small amount of starting material available in these types of samples.
QMI is an antibody-based assay in which cell lysate is incubated with a panel of immunoprecipitation antibodies that are covalently coupled to magnetic beads containing distinct ratios of fluorescent dyes. Having specific antibodies coupled to distinct bead classes allows for simultaneous co-immunoprecipitation of multiple target proteins from the same lysate. Following immunoprecipitation, beads are incubated with a second, fluorophore-conjugated probe antibody (or biotinylated antibody in conjunction with fluorophore-conjugated streptavidin). Fold changes in the co-associations between the proteins recognized by each bead-probe pair, or PiSCES (proteins in shared complexes detected by exposed surface epitopes), are then detected by flow cytometry and can be quantitatively compared between different sample conditions.13 The sensitivity of QMI depends on the protein concentration of the lysate relative to the number of beads used for immunoprecipitation, and achieving a resolution to detect 10% fold changes requires only a small amount of starting material compared to other co-IP methods.13,14 For example, primary T cells isolated from a 4 mm skin biopsy, synaptosomes from a 2 mm coronal section of mouse prefrontal cortex, cortical slices from a single hemisphere of adult mouse brain, and 3 × 106 cultured primary neurons have each been used with up to 20×20 matrices of IP and probe targets.13,15,16 This sensitivity makes QMI useful for analysis of cells or tissue with limited availability, such as clinical samples. Additionally, proteins can be analyzed in their native states without any genetically engineered tags that could interfere with physiological function.
QMI can be adapted for any protein interaction network, and to date has been developed to analyze the T cell antigen receptor (TCR) signalosome and a subset of proteins at glutamatergic synapses in neurons.13,16,17,18 In T Cells, QMI was first used to identify stimulation-induced changes in PiSCES, then to distinguish autoimmune patients from a control group, detect endogenous autoimmune signaling, and finally to generate a hypothesis involving an unbalanced disease-associated subnetwork of interactions.13 More recently the same QMI panel was used to determine that thymocyte selection is determined by a quantitative rather than qualitative differences in TCR-associated protein signaling.18 In the neuronal synapse system, QMI was used to describe input-specific rearrangement of the protein interaction network for distinct types of signals in a manner which supports newly emerging models of synaptic plasticity.16 Additionally, this synaptic QMI panel was used to identify differences in seven mouse models of autism, cluster the models into subgroups based on their PiSCES biosignatures, and accurately hypothesize a shared molecular deficit that was previously unrecognized in one member of a cluster.15 This has implications for potential therapeutics that could be tested for that specific model, and a similar approach could be used to screen for other models that might respond to a given treatment. Ultimately, this information is useful for both diagnostics and treatment, in addition to basic science.
Any previously defined protein interaction network can be studied using QMI to gain insight into complex signal transduction and network dysfunction in disease. QMI is particularly useful to those who have limited biomaterial and want to study protein networks in their native state without genetic engineering. Once a network’s behavior is defined, QMI has the potential to be used as a tool for both diagnostics and drug screening, in health and disease.
PROTOCOL:
1. Assay Design
1.1. Candidate Antibody Preparation
-
1.1.1.
For each protein of interest, choose three to five antibodies to screen. When possible, use monoclonal antibodies that recognize different epitopes. Also include one non-specific control antibody.
-
1.1.2.
Carrier proteins and buffers with free amine groups (such as Tris) will react with COOH groups and quench the subsequent bead coupling and biotinylation reactions. Ensure that all antibodies are purified (no carrier proteins) and in a buffer free of primary amines (i.e. no Tris). To remove Tris, perform buffer exchange by adding the antibody to a 30 kDa spin filter, spinning down to its minimum volume, adding 500 μL of PBS, and repeating 3 times. To remove carrier proteins, perform Melon Gel purification according to the Thermo Scientific protocol.
-
1.1.3.
Couple each antibody to CML beads as described by Davis and Schrum19. To conserve the antibody, scale down bead coupling reactions by up to 1/5 (i.e. 3.6 × 106 beads with 10 uL of 0.2–1 mg/mL antibody). Estimate bead numbers using a hemocytometer (typically ~108/ml) and store at 4 °C.
-
1.1.4.
Biotinylate a portion of each antibody (see section 2.2 below). Store at 4 °C.
-
1.1.5.
Confirm effective bead coupling and accurate counting by staining 1×105 beads with a PE-conjugated antibody reactive to the species in which the antibody was raised and reading on a flow cytometer.
-
1.1.6.
Confirm antibody biotinylation by dot blot using streptadivin-HRP.
-
1.1.7.
Once the lab has generated reagents that are known to be effective, use those reagents as positive controls in confirmation reactions 1.1.5 and 1.1.6.
1.2. Antibody Screening by IP-FCM (Immunoprecipitation Detected by Flow Cytometry)
-
1.2.1.
Calculate your IP volume. If you are screening X probe antibodies, each IP will use (X+1) * 10ul * 1.1; X+1 to account for the required IgG probe control, and 1.1 to account for 10% pipetting error and ensure sufficient lysate in subsequent steps. For example, in a 3×3 antibody screen, each IP should use 44ul. Remember to include an IgG IP control (Figure 2).
-
1.2.2.
Calculate your bead number. If you are screening X probe antibodies, use (X+1) * 5×104 beads. Ideally, 5,000 beads per well will result in >2,000 beads per well being read by the flow cytometer. For example, in a 3×3 antibody screen, each IP should use 20,000 beads, which is approximately 0.66 ul of bead stock.
-
1.2.3.
Incubate the volume of lysate from 1.2.1 with the volume of each bead to be screened from 1.3.2, overnight at 4 °C with rotation to prevent beads from settling. Typically, incubations are performed in the first column of a 96-well plate, and capped with PCR tube strip caps.
-
1.2.4.
Spin down beads at 3200 x g for 1 min and remove lysate by a single, rapid flicking of the plate over the sink. A tiny white pellet should be visible at the bottom of each well.
-
1.2.5.
Resuspend beads in a volume of FlyP buffer calculated as (X+1) * 1.1* 20ul, similar to 1.2.1. For a 3×3 screen, resuspend in 88ul. FlyP buffer is 100 mM NaCl, 50 mM Tris pH 7.4, 1% BSA, 0.01% NaN3.
-
1.2.6.
Distribute each IP across (X+1) wells of a 96-well PCR plate, where X is the number of probe antibodies being screened, using 20 μL/well (Figure 2).
-
1.2.7.
Wash 2 additional times using 200 μL FlyP buffer per well. Spin the plate as in 1.3.4 and flick the plate to remove wash buffer after each wash. Pellets will be extremely small, but should be visible after each wash.
-
1.2.8.
For each biotinylated antibody to be screened, calculate the total volume as (Y+1) * 1.1 * 50ul, where Y is the number of IP antibodies being screened. Dilute antibody in this volume of FlyP buffer, typically starting with 1:200 dilution of a 0.5mg/ml stock.
-
1.2.9.
Distribute each diluted antibody down each column of the 96 well plate, and ensure beads are resuspended.
-
1.2.10.
Incubate at 4°C for 1 hour, either with rotation or pipetting at 15 min intervals to ensure beads remain in suspension.
-
1.2.11.
Wash 3x in 200 μL FlyP buffer.
-
1.2.12.
Resuspend all beads in 50 μL of 1:200 Streptavidin-PE in FlyP buffer.
-
1.2.13.
Incubate at 4°C in the dark for 30 min.
-
1.2.14.
Wash 3x in 200 μL FlyP buffer.
-
1.2.15.
Resuspend in 200 μL of FlyP buffer, then run on a flow cytometer.
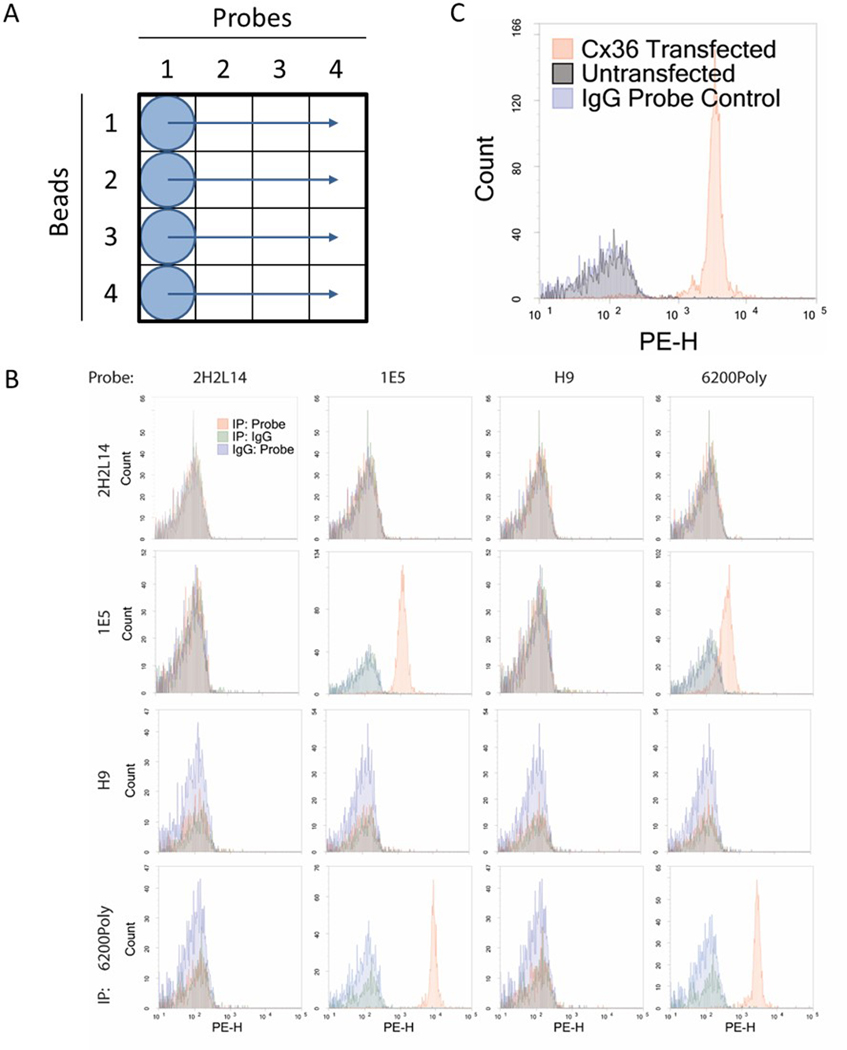
Figure 2. Cx36 antibody screening using IP-FCM.
(a) IP-FCM was performed on mouse brain lysate using a 4×4 panel of Connexin 36 (Cx36) beads and probes. Lysate was immunoprecipitated with each bead in a separate row of the plate. After washes, each bead was distributed across its row so that one probe antibody can be added per column. (b) Most antibody combinations show no signal (red) over IgG background (green, blue). The IE-5H5 IP with the 6200Poly probe shows some positive signal. The IE-5H5 bead/probe and UW bead/probe pairs each show acceptable signal, but it is not ideal to use the same antibody for both bead and probe. The 6200Poly bead with the IE-5H5 probe gives the strongest signal and was chosen to use in the multiplex assay pending specificity confirmation. (c) IP-FCM using the pair of Cx36 antibodies selected from screening was performed on the lysate of 293T cells transfected with Cx36 and non-transfected controls. There is clear signal from the Cx36-transfected cells, but the non-transfected cells are indistinguishable from IgG bead/probe controls.
1.3. Choosing Antibodies to Include in Assay
-
1.3.1.
Gate on size using FSC-H vs SSC-H, and eliminate doublets using FSC-H vs FSC-A.
-
1.3.2.
Generate histograms of PE fluorescence intensity and overlay both IgG controls (IgG bead-test probe, test bead-IgG probe) onto tested pairs (Figure 2).
-
1.3.3.
Look for a bead-probe pair that gives clear signal over noise. If there are no acceptable options, repeat screen with additional antibodies (Figure 2).
1.4. Confirmation of Antibodies
-
1.4.1.
To ensure antibody specificity for the intended targets, use a lysate sample in which the target has been knocked out; for example, a knockout mouse or an RNAi cell line. Alternatively, use lysate from a target-negative cell line in which the target protein has been artificially expressed.
-
1.4.2.
Perform IP-FCM as described in 1.2, modifying to fit the experiment.
1.5. Detergent Selection
-
1.5.1.
Detergents are critical in co-IP experiments, and different variations should be empirically tested to ensure that the assay has maximum likelihood of detecting changes. To start, choose a relatively small panel of interactions (4–8) that are known to change in a given condition and/or are of particular interest to your study.
-
1.5.2 Using the non-fluorescent, antibody-conjugated CML beads made for initial screens, perform IP-FCM as described in 1.2 using varied lysis buffer detergent conditions.
Detergent screens can be performed with detergent as the only variable, or with different cell conditions for each detergent. Always use IgG controls for both beads and probes.
-
1.5.3 Based on the MFIs from the screen, choose a detergent that optimizes the signal for the pairs or changes that are believed to be the most important to detect.
It is likely that some compromises will need to be made.
2. Multiplex Reagent Preparation
2.1. Magnetic Bead Coupling
-
2.1.1.
Using the bead region map, select bead regions to use in a pattern that minimizes risk of cross-talk. Beads typically smear up and to the right, so avoid bead regions that are diagonally next to each other. We typically use beads from every other column of the bead diagram shown on the Luminex website (https://www.luminexcorp.com/magplex-microspheres/).
-
2.1.2.
Prepare carrier-free antibody at 0.1 mg/mL in PBS (as in 1.1.2) in 250 μL. Keep on ice for later use.
-
2.1.3.
Vortex Luminex beads extensively, then aliquot 250 μL into an amber microcentrifuge tube (to protect beads from photobleaching).
-
2.1.4.
Magnetically separate beads for 60 sec and remove supernatant.
-
2.1.5.
Add 250 μL MES buffer (50 mM MES pH 6.0, 1mM EDTA), vortex, magnetically separate 60 sec, and remove supernatant. Repeat and resuspend beads in 200 μL MES buffer.
-
2.1.6.
Add 40 μL MES to a 2 mg single use tube of Sulfo-NHS to make a 50 mg/mL stock.
-
2.1.7.
Add 25 μL freshly made Sulfo-NHS to beads. Vortex.
-
2.1.8.
Add 25 μL of 50mg/mL freshly dissolved EDAC in MES buffer. Vortex.
-
2.1.9.
Cover and shake on a vortexer with a tube-holding attachment for 20 min at room temp, 1000 rpm.
-
2.1.10.
Magnetically separate 60 sec and remove supernatant.
-
2.1.11.
Resuspend in 500 μL PBS, vortex, magnetically separate 60 sec and remove supernatant. Repeat.
-
2.1.12.
Resuspend in 250 μL antibody solution from 2.1.2. Vortex.
-
2.1.13.
Incubate 2 hrs at room temp with shaking on a vortexer at 1000 rpm.
-
2.1.14.
Magnetically separate 60 sec, remove the antibody solution and reserve for troubleshooting.
-
2.1.15.
Add 500 μL PBS to beads, wash, vortex, magnetically separate 60 sec, and remove supernatant.
-
2.1.16.
Add 750 μL Blocking/Storage (B/S) buffer (1% BSA in PBS pH 7.4, 0.01% NaN3). Cover and incubate 30min at room temp, 1000rpm.
-
2.1.17.
Magnetically separate 60 sec and remove supernatant. Resuspend in 100 μL B/S buffer.
-
2.1.18.
Store at 4 ˚C.
-
2.1.19.
Validate bead coupling by staining ~0.25 μL of coupled beads with a fluorescent anti-host species secondary and reading on a flow cytometer, as in 1.1.5.
2.2. Biotinylation
-
2.2.1.
Ensure that antibodies are in PBS with no carrier protein.
-
2.2.2.
Calculate the total ug of antibody to be biotinylated (100–200 ug recommended for use in multiplex, 25–50 ug recommended for screening).
-
2.2.3.
Prepare fresh 10 mM sulfo-NHS-biotin (can be done by adding 224 μL ddH2O to a 1 mg no-weigh tube).
-
2.2.4.
Add 1 μL of 10 mM sulfo-NHS-biotin per 25 ug of antibody, vortex or pipette up and down to mix.
-
2.2.5.
Incubate at room temp for 1 hr.
-
2.2.6.
Incubate at 4 °C for 1 hr.
-
2.2.7.
Use a 30 kDa spin filter to remove unbound biotin and stop the reaction. Add 500ul of PBS and spin the column until the minimum volume is reached. Add 500ul additional PBS and repeat for 3 total buffer exchanges.
-
2.2.8.
Estimate concentration by measuring absorbance on a Nanodrop and bring antibody concentration to 0.5 mg/mL.
3. Quantitative Multiplex Immunoprecipitation
3.1. Plate Layout
-
3.1.1.
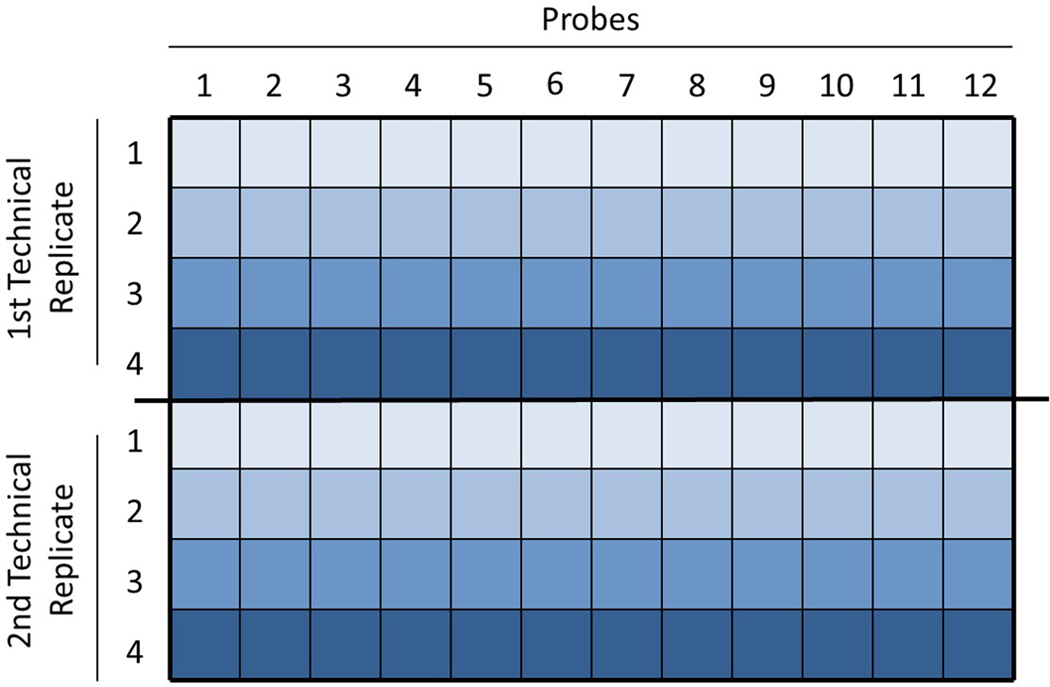
The assay is performed using 96-well plates and works best with 2–4 sample conditions. Appropriate controls (i.e. stimulated v. unstimulated cells) must always be run on the same plate in order to detect changes between conditions. Generally, each sample should be distributed horizontally across the plate, and each column should be used for a different probe antibody. A set of technical replicates for each probe should be run immediately after the first set. See Figure 3 for an example.
Figure 3. Example plate layout.
A 4-condition multiplex is set up in a 96-well plate. Samples are loaded in consecutive rows, and technical replicates are loaded in the same order in the following rows. One probe is used per column.
3.2. Sample preparation & Immunoprecipitation (Day 1)
-
3.2.1.
Lyse tissue or cells in appropriate detergent with protease and phosphatase inhibitors and incubate on ice for 15 minutes. Take care to keep the lysate cold at all times.
-
3.2.2.
Spin down at 4 °C for 15 min at 16,000 x g to remove membranes and debris, keep supernatant as lysate.
-
3.2.3.
Perform a BCA Assay or similar to determine protein concentrations, then normalize protein concentration between samples. If using cells, begin with an equal number of cells per condition and normalization is optional.
-
3.2.4.
Prepare a master bead mix that contains ~250 beads of each class per well in the assay. Bead numbers should be adjusted based on data analysis so that in future assays an average of 110 beads of each class will be read per well. Example calculation: New bead volume = [(Run Average) / 110] * (previous bead volume). Bead volumes should be adjusted in this way about every 8 runs or as needed. Typically, we use 3–4ul of beads (prepared as above) for a 2 plate experiment.
-
3.2.5.
Wash the bead mix 2x in FlyP buffer with magnetic separation, then resuspend in FlyP buffer. For resuspension, use 10 μL per sample per plate.
-
3.2.6.
After thoroughly vortexing the bead mix, aliquot 10 μL into microcentrifuge tubes (one tube per sample per plate). Add equal volumes of lysate (with normalized concentrations) to each tube for immunoprecipitation. A separate immunoprecipitation should be done for each plate. Place tubes on a rotator at 4 °C overnight for immunoprecipitation, covered to keep out light.
3.3. Running the Assay (Day 2)
-
3.3.1.
Start with the IPs for Plate #1. Use a magnetic bead rack to remove lysate from beads, and reserve lysate for future analysis. Wash beads 2x in 500 μL ice cold FlyP buffer. The IPs should always be kept on ice or at 4 °C.
-
3.3.2.
Calculate resuspension volume as (number of probes) × (2 technical replicates) × (25ul per well) × (1.1 for pipetting error). Resuspend IPs in calculated volume of ice-cold FlyP buffer.
-
3.3.3.
After thoroughly resuspending beads by gentle pipetting, distribute 25 μL per well across a flat-bottomed 96 well plate, on ice.
-
3.3.4.
In a different 96-well plate, dilute biotinylated probe antibodies to 2X working concentration (typically 1:100) in FlyP buffer so that their order matches the columns on the plate layout (see figure 3).
-
3.3.5.
Use a multichannel pipette to distribute 25 μL of each probe antibody dilution into the bead-containing assay plate.
-
3.3.6.
Shake on a horizontal plate shaker to mix and resuspend beads, then incubate at 4 °C for 1 hour, shaking at 450 rpm in the dark.
-
3.3.7.
Wash 3x with FlyP buffer on a magnetic plate washer at 4 °C.
-
3.3.8.
Resuspend beads in 50 μL of 1:200 Streptavidin-PE.
-
3.3.9.
Shake to mix and resuspend beads, then incubate at 4 °C for 30 min, shaking at 450 rpm in the dark.
-
3.3.10.
Wash 3x with FlyP buffer on a magnetic plate washer at 4 °C.
-
3.3.11.
Resuspend in 125 μL FlyP buffer.
-
3.3.12.
Shake for 1 min at 900 rpm to thoroughly resuspend beads.
-
3.3.13.
Run on refrigerated flow cytometer at high RP1 target with a stop condition of 1000 beads per region (greatly overshooting the number that should be in any individual well) and sample volume of 80 μL.
-
3.3.14.
Pause the run half way through and resuspend the beads to prevent settling
-
3.3.15.
Export files in .xls and .xml formats.
-
3.3.16.
Repeat the process for the remaining plates.
4. Data Analysis
The ANC code was designed to compare two conditions from N = 4 experiments, each with 2 technical replicates for each condition. For example, cells are stimulated four independent times, QMI is run on four different days on control (unstimulated) and stimulated cells, with technical replicates as above, and data analysis proceeds as described below.
4.1. Adaptive Non-parametric with Adjustable Alpha Cutoff. ANC
-
4.1.1
Open MATLAB and set the active directory to a folder containing the ANC program components and the .xml files exported from the Bioplex.
-
4.1.2
Fill in the “ANC input” file to reflect the details of your experimental design.
-
4.1.3
Run the program. The ANC “hits” are output as a .csv file.
4.2. Weighted Correlation Network Analysis
-
4.2.1
Use Microsoft Excel and the .xls file exported in 3.3.15 to calculate the average Median Fluorescent Intensity (MFI) of the technical replicates for each IP_bead combination for each sample.
-
4.2.2
Create a spreadsheet with each interaction IPi_ProbeJ on a different row, and each sample in a different column. The top row of each column should contain the name of each sample, for example “WildtypeControl1”. Save the file as a .csv and name it “input”.
-
4.2.3
Create a second spreadsheet. Starting with row 2, paste-transpose the column titles from “input” into column 1. Starting with column 2, title each column as an experimental “trait”, for example, genotype or treatment with a chemical stimulation. Note that the traits must be coded numerically. One of the titles must be “experiment”, that indicates which samples were run on the same plate and allow for batch effect correction.
-
4.2.4
Open R studio and set the working directory to a folder containing the “input” and “traits” files.
-
4.2.5
Run the R commands as indicated in the commented command file. The WCNA modules significantly correlated with each experimental trait are output as a graphic file, and the correlation of each interaction IPi_ProbeJ with each module is output as a .csv file.
4.3. Positive ‘Hits’ & Visualization
-
4.3.1
For each interaction in the “3/4 hits” list in the ANC output file, identify if that interaction is also a “CNA hit” by checking the CNA output file. We define CNA hits as interactions with module membership (MM) > 0.7 and p < 0.05 for membership in a module that was identified as significantly correlated with the experimental variable of interest. Create a new column that indicates if each ANC hit is also a CNA hit.
-
4.3.2
Calculate the average log2 fold change value for each ANC∩CNA hit by averaging the values given in the ANC output spreadsheet. The numbers following each significant interaction signify the p values for each experiment, then the fold change values for each experiment. Convert values to log2 fold change before averaging. For interactions that were significant in only 3/4 replicates, delete the outlier value.
-
4.3.3
Make a spreadsheet with each ANC∩CNA hit listed as an IP in one column, a probe in the second column, and the fold change value in the third column. Use this spreadsheet to create a node-edge diagram in Cytoscape by importing the file as a network.
REPRESENTATIVE RESULTS:
Antibody Screening.
A compatible bead-probe pair from the initial antibody screening should clearly show signal over noise when comparing MFI histograms (Figure 2b). When confirming specificity of a pair, a negative control lysate from a knockout animal or cell line without the target protein should have an MFI similar to the IgG controls (Figure 2c).
Bead Coupling.
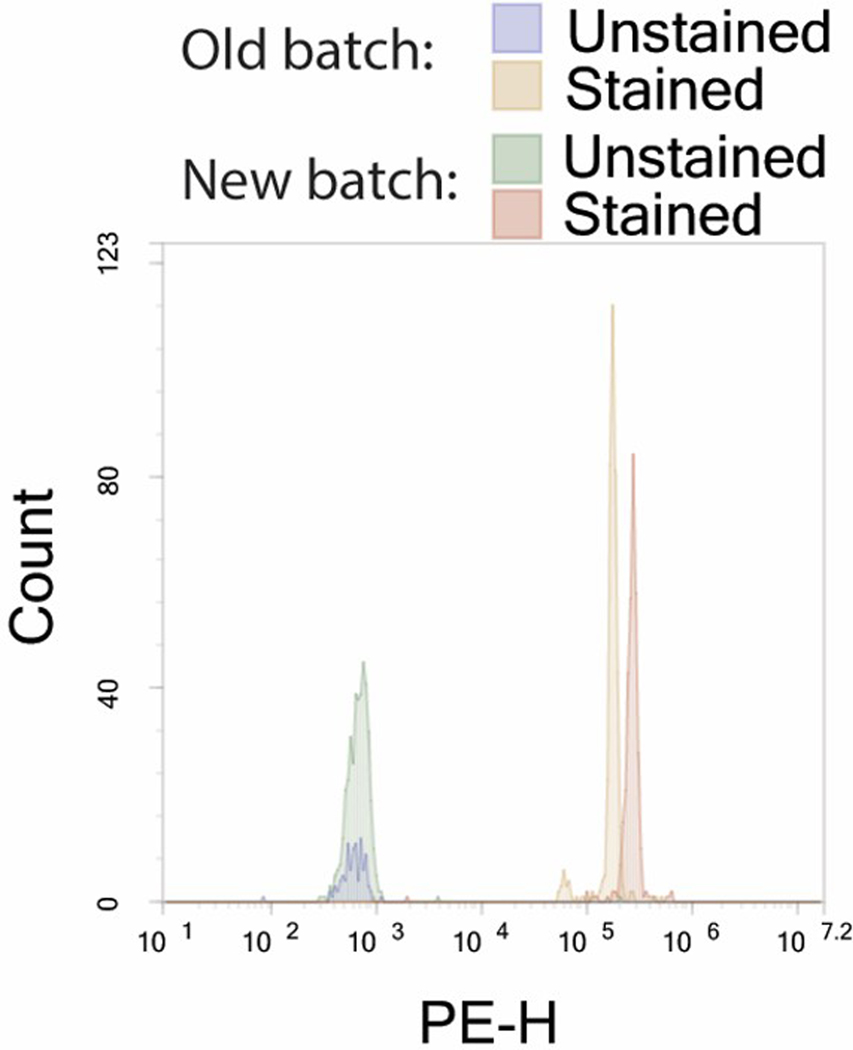
A typical bead coupling quality control reaction will give an MFI 3–4 logs above background when stained with a secondary antibody conjugated to a fluorophore with a brightness index between 3 and 5 (such as PE or FITC). Figure 4 shows a typical quality control reaction comparing the conjugation of a new bead compared to the older batch being replaced.
Figure 4. A typical quality control reaction comparing the conjugation of a new bead compared to the older batch being replaced.
The bead gives an MFI 3–4 logs above background, and the new batch has an MFI similar to that of the old batch.
Data Analysis.
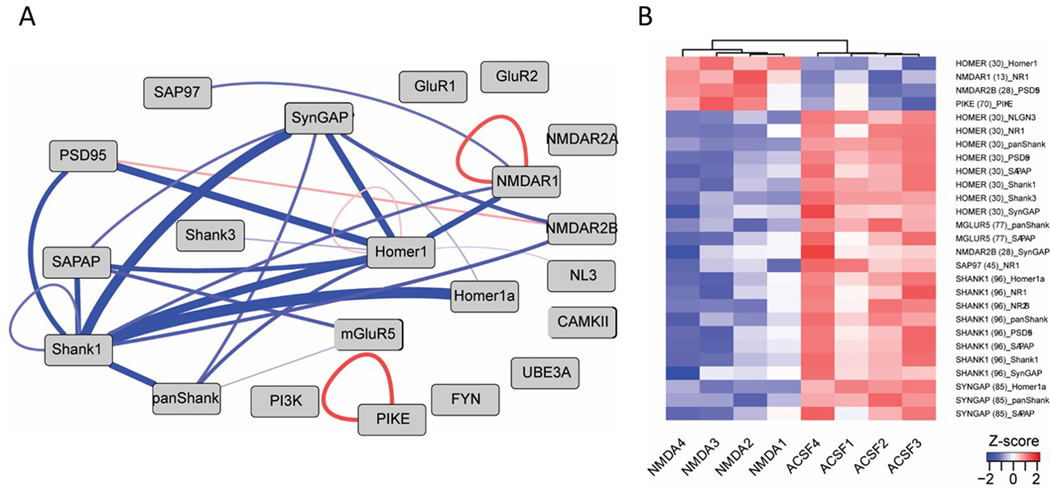
After a QMI assay is run, ANC analysis is performed and will call “hits” based on interactions with significant fold changes between conditions in >70% of runs. CNA analysis then identifies modules that change together with a given variable. Interactions that appear in both ANC hits and the relevant CNA module are ANC∩CNA hits, which have a high level of confidence and are reported as significant changes. ANC∩CAN hits can then be visualized using a node-edge diagram or a heatmap (Figure 5).
Figure 5. ANC U CNA hits in an NMDA stimulation experiment.
Visualizations of the results of a QMI experiment comparing NMDA stimulated neurons to aCSF (unstimulated) controls. Only protein-protein interactions that were statistically significant by both ANC and CNA data pipelines are shown in the node-edge diagram (a) made in Cytoscape and the heatmap (b) made in R.
DISCUSSION:
QMI requires careful pipetting and tracking of sample and antibody well locations. Carefully labeling the assay plates is useful, as is making a detailed template of well locations on paper, which is then saved for data analysis. The importance of keeping the beads and lysate cold at all times, including in the Bioplex bead reader, cannot be overstated. PPIs will rapidly dissociate at room temperature, and our early attempts at using an unmodified, room temperature Bioplex 200 ended with the identification of many temperature-labile interactions, but not those that changed with our intended stimulation.
QMI is an antibody-based assay, so the initial selection of antibodies is critical. Monoclonal or recombinant antibodies should be used whenever possible to reduce variability in results. Polyclonals show lot-to-lot variation, but peptide-based polyclonals to a short epitope seem to be relatively stable over time in our experience. Drift can be minimized by buying large batches of antibodies; this also allows for custom-orders of carrier-free antibodies which precludes the need to purify antibodies using Melon Gel and spin columns, and the associated antibody loss.
It is also important to note that, because detecting a signal is reliant upon available epitopes, the lack of a signal does not necessarily indicate the lack of an interaction, a limitation that is common with other protein interaction methodologies.13 Further, when we do detect a signal, it is impossible to unambiguously state weather the protein interaction is direct (A interacts with B) or indirect (A interacts with X and Y, which then interact with B).
It is critical in QMI to select an appropriate lysis buffer. Too weak of a detergent can leave membranes intact and hold together proteins that aren’t in complex, while too strong of a detergent can destroy protein-protein interactions (PPIs). Additional factors such as the presence of calcium or its chelators can dramatically affect PPIs and should be carefully considered before screening antibodies to include in a QMI panel. For IP-western experiments, lysis conditions are usually optimized for each PPI on a case-by-case basis, but the best conditions for detecting a single PPI often don’t translate to other PPIs in the same protein network. This is in essence a chicken-and-egg dilemma in that a lysis buffer is needed to screen antibody candidates, but the panel of antibodies is also needed to screen for an appropriate lysis buffer. While not a perfect solution, we suggest selecting a small panel of beads and probes that are of particular interest and/or have known associations or dissociations in response to a stimulus, and testing their behavior under different lysis conditions on white (non-fluorescent) CML beads. An ideal detergent should allow for both reliable detection of PPIs and recapitulation of known physiological PPI behavior (association/dissociation) with a given stimulus. If there is any concern that a detergent does not fully solubilize membranes, a negative control antibody can be added that would only give signal if two proteins were linked by membrane.20
Previous work using QMI in neurons and T cells has both carefully confirmed previous findings in order to increase confidence in the validity of the new results, and generated new hypotheses that led to discoveries about signal transduction and disease pathways. In the future, QMI can be adapted to other protein interaction networks and expanded up to 500 proteins with the current microsphere classes available. We believe that using QMI to study the ways in which networks of multi-protein complexes change in response to stimuli to control cellular processes will yield important insights into both health and disease.
Supplementary Material
Figure 1. Overview of Quantitative Multiplex Immunoprecipitation.
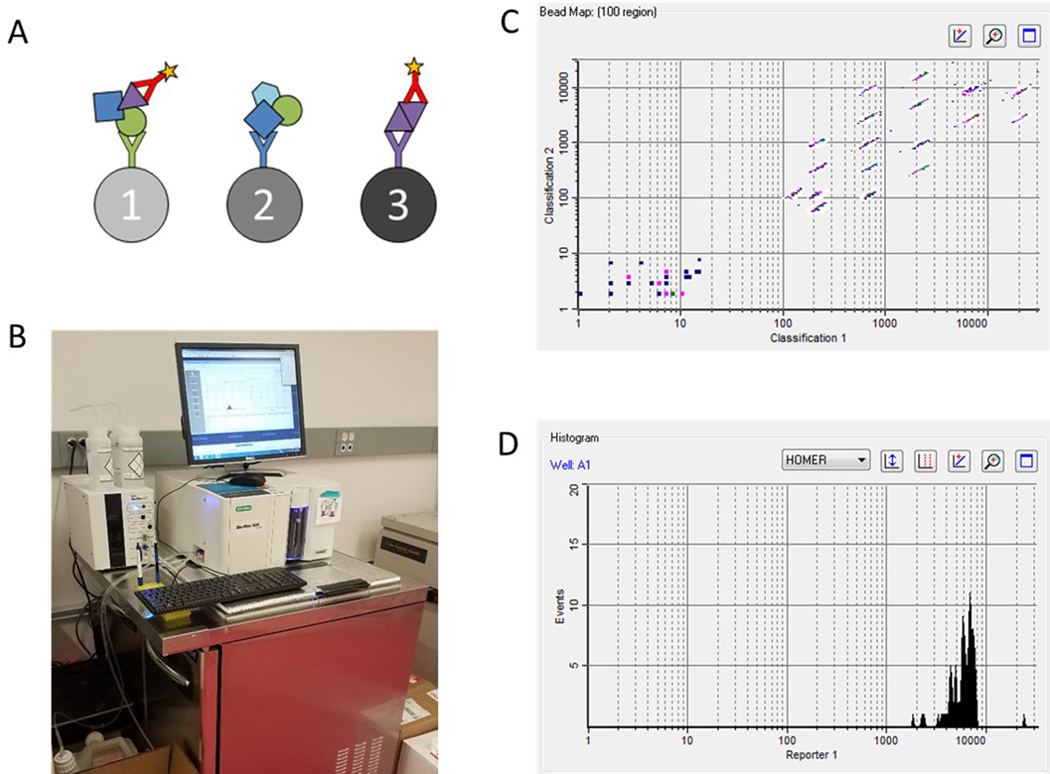
(a) Protein complexes are immunoprecipitated by antibody-coupled microspheres overnight, then co-immunoprecipitated proteins are labeled by a probe antibody and a fluorophore. (b) Microspheres are run through a refrigerated flow cytometer to quantify relative amounts of proteins occurring in shared complexes. (c) The flow cytometer separates microspheres by class and creates (d) fluorescence histograms from each bead region.
Table of Materials: Materials.
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description | ||
|---|---|---|---|---|---|
| MagPlex® Microspheres | Luminex | MC12xxx-01 | xxx is the 3 digit bead region | ||
| Sulfo NHS | Thermo Scientific | A39269 | |||
| CML beads | Invitrogen | C37481 | |||
| EDTA | Sigma | E6758 | |||
| BSA | Sigma | ||||
| Tris | Fisher Scientific | BP152 | |||
| MES | Sigma | M3671 | |||
| 96-well plates | Bio Rad | 171025001 | |||
| Microplate film, non-sterile | USA Scientific | 2920–0000 | |||
| Melon™ Gel IgG Spin Purification Kit | Thermo Scientific | 45206 | |||
| EZ-Link™ Sulfo-NHS-Biotin | Thermo Scientific | A39256 | |||
| Bioplex 200 System with HTF | Bio Rad | 171000205 | modiefied to keep partially refrigerated | ||
| Bio-Plex Pro™ Wash Station | Bio Rad | 30034376 | |||
ACKNOWLEDGMENTS:
The authors wish to acknowledge Tessa Davis for important contributions to QMI assay development, and current and former members of the Smith and Schrum labs for technical guidance and intellectual input. This work was funded by NIMH grants R01 MH113545 and R00 MH 102244.
Footnotes
DISCLOSURES: The authors have no conflicts of interest to disclose.
REFERENCES:
- 1.Pawson T. & Nash P Protein-protein interactions define specificity in signal transduction. Genes Dev. 14 (9), 1027–1047 (2000). [PubMed] [Google Scholar]
- 2.Bugaj LJ et al. Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science. 361 (6405), doi: 10.1126/science.aao3048, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr Opin Cell Biol. 19 (2), 112–116, doi: 10.1016/j.ceb.2007.02.013, (2007). [DOI] [PubMed] [Google Scholar]
- 4.Hamm HE The many faces of G protein signaling. J Biol Chem. 273 (2), 669–672 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Lohse MJ & Hofmann KP Spatial and Temporal Aspects of Signaling by G-Protein-Coupled Receptors. Mol Pharmacol. 88 (3), 572–578, doi: 10.1124/mol.115.100248, (2015). [DOI] [PubMed] [Google Scholar]
- 6.Eltschinger S. & Loewith R. TOR Complexes and the Maintenance of Cellular Homeostasis. Trends Cell Biol. 26 (2), 148–159, doi: 10.1016/j.tcb.2015.10.003, (2016). [DOI] [PubMed] [Google Scholar]
- 7.Ron D. & Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 8 (7), 519–529, doi: 10.1038/nrm2199, (2007). [DOI] [PubMed] [Google Scholar]
- 8.Bogdan AR, Miyazawa M, Hashimoto K. & Tsuji Y. Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem Sci. 41 (3), 274–286, doi: 10.1016/j.tibs.2015.11.012, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papin JA, Hunter T, Palsson BO & Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 6 (2), 99–111, doi: 10.1038/nrm1570, (2005). [DOI] [PubMed] [Google Scholar]
- 10.Komarova NL, Zou X, Nie Q. & Bardwell L. A theoretical framework for specificity in cell signaling. Mol Syst Biol. 1 2005.0023, doi: 10.1038/msb4100031, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrum AG & Gil D. Robustness and Specificity in Signal Transduction via Physiologic Protein Interaction Networks. Clin Exp Pharmacol. 2 (3), S3.001, doi: 10.4172/2161-1459.S3-001, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Las Rivas J. & Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 6 (6), e1000807, doi: 10.1371/journal.pcbi.1000807, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith SE et al. Multiplex matrix network analysis of protein complexes in the human TCR signalosome. Sci Signal. 9 (439), rs7, doi: 10.1126/scisignal.aad7279, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrum AG et al. High-sensitivity detection and quantitative analysis of native protein-protein interactions and multiprotein complexes by flow cytometry. Sci STKE. 2007 (389), pl2, doi: 10.1126/stke.3892007pl2, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown EA et al. Clustering the autisms using glutamate synapse protein interaction networks from cortical and hippocampal tissue of seven mouse models. Molecular Autism. 9 (1), 48, doi:doi: 10.1186/s13229-018-0229-1, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lautz JD, Brown EA, Williams VanSchoiack AA & Smith SEP Synaptic activity induces input-specific rearrangements in a targeted synaptic protein interaction network. J Neurochem. 146 (5), 540–559, doi: 10.1111/jnc.14466, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith SE et al. Signalling protein complexes isolated from primary human skin-resident T cells can be analysed by Multiplex IP-FCM. Exp Dermatol. 23 (4), 272–273, doi: 10.1111/exd.12362, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neier SC et al. The early proximal αβ TCR signalosome specifies thymic selection outcome through a quantitative protein interaction network. Sci Immunol. 4 (32), doi: 10.1126/sciimmunol.aal2201, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis TR & Schrum AG IP-FCM: immunoprecipitation detected by flow cytometry. J Vis Exp. (46), doi: 10.3791/2066, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrum AG, Gil D, Turka LA & Palmer E. Physical and functional bivalency observed among TCR/CD3 complexes isolated from primary T cells. J Immunol. 187 (2), 870–878, doi: 10.4049/jimmunol.1100538, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.