A plant virus-based delivery system is a useful tool for gene and microRNA function studies in cucurbits.
Abstract
Cucurbits are economically important crops worldwide. The genomic data of many cucurbits are now available. However, functional analyses of cucurbit genes and noncoding RNAs have been impeded because genetic transformation is difficult for many cucurbitaceous plants. Here, we developed a set of tobacco ringspot virus (TRSV)-based vectors for gene and microRNA (miRNA) function studies in cucurbits. A TRSV-based expression vector could simultaneously express GREEN FLUORESCENT PROTEIN (GFP) and heterologous viral suppressors of RNA silencing in TRSV-infected plants, while a TRSV-based gene silencing vector could knock down endogenous genes exemplified by PHYTOENE DESATURASE (PDS) in Cucumis melo, Citrullus lanatus, Cucumis sativus, and Nicotiana benthamiana plants. We also developed a TRSV-based miRNA silencing vector to dissect the functions of endogenous miRNAs. Four representative miRNAs, namely, miR159, miR166, miR172, and miR319, from different cucurbits were inserted into the TRSV vector using a short tandem target mimic strategy and induced characteristic phenotypes in TRSV-miRNA-infected plants. This TRSV-based vector system will facilitate functional genomic studies in cucurbits.
Introduction
Cucurbits, including cucumber (Cucumis sativus), melon (Cucumis melo), watermelon (Citrullus lanatus), and zucchini (Cucurbita pepo), are economically important vegetables globally. Although the genomic sequencing of many cucurbits has been completed (Huang et al., 2009; Garcia-Mas et al., 2012; Guo et al., 2013; Shin et al., 2019; Wu et al., 2019; Wu et al., 2020), the functions of most miRNA or noncoding RNA have not yet been characterized. Some cucurbitaceous plants are highly recalcitrant to genetic transformation, while in others it takes a long time to obtain transgenic plants, which impedes functional genomic studies in cucurbits.
A virus-based gene delivery system is an attractive, quick, and promising approach for the transient gain- and loss-of-function studies in plants recalcitrant to genetic transformation (Zhang and Ghabrial, 2006; Seo et al., 2016; Ding et al., 2018). For the overexpression of a gene in vivo, plant virus-based vectors can elevate the amount of protein in a short time (Gleba et al., 2007; Baltes et al., 2014). For the knockdown of plant genes, plant virus-induced gene silencing has been widely employed as a fast and convenient tool in plant biology studies (Liu et al., 2002; Robertson, 2004; Bachan and Dinesh-Kumar, 2012; Senthil-Kumar and Mysore, 2014). Recently, some plant RNA or DNA viruses, including barley stripe mosaic virus, Chinese wheat mosaic virus, cotton leaf crumple virus, cucumber mosaic virus (CMV), potato virus X (PVX), and tobacco rattle virus, have been engineered into virus-based miRNA silencing (VbMS) vectors to inhibit the miRNA function in maize (Zea mays), wheat (Triticum aestivum), cotton (Gossypium hirsutum), Arabidopsis (Arabidopsis thaliana), potato (Solanum tuberosum), and Nicotiana benthamiana (Du et al., 2014; Gu et al., 2014; Sha et al., 2014; Jiao et al., 2015; Liao et al., 2015; Jian et al., 2017; Yang et al., 2018; Zhao et al., 2020). However, to the best of our knowledge, there has been no virus-based miRNA silencing tool developed for cucurbits up to now.
Two reciprocal reverse genetic strategies have been used to dissect the function of a specific miRNA. For gain-of-function studies, the miRNA activity can be enhanced through transgenically overexpressing miRNA (Sainsbury et al., 2010; Zhang et al., 2013; Yang et al., 2015). For loss-of-function studies, miRNA activity can be blocked by expressing a miRNA-resistant target that contains uncleavable mutations in the miRNA-binding sequence (Javier et al., 2003; Franco-Zorrilla et al., 2007). Some alternative approaches, including miRNA target mimicry (Franco-Zorrilla et al., 2007), short tandem target mimic (STTM; Yan et al., 2012), transcriptional gene silencing of miRNA gene promoters (Vaistij et al., 2010), and artificial miRNA-directed silencing of miRNA precursors (Eamens et al., 2011), have also been applied in the function investigation of miRNA. The above approaches all require time-consuming and expensive transformation. Furthermore, transgenic approaches are not applicable to all cucurbit species. Therefore, it is necessary to develop a fast and efficient approach to investigate the function of cucurbit miRNAs.
Tobacco ringspot virus (TRSV; genus Nepovirus, family Secoviridae) can infect plants including cucurbits, soybean, blueberry, and N. benthamiana (Zellner et al., 2011; Abdalla et al., 2012; Shakiba et al., 2012; Zhao et al., 2015). The genome of TRSV consists of two single-stranded RNAs. RNA1 contains an open reading frame encoding a polyprotein precursor, which is then cleaved into five mature proteins designated as32K, helicase, genome-linked protein, protease, and RNA-dependent RNA polymerase. The RNA2 encodes a polyprotein that can be cleaved into three mature proteins, namely, 2a, movement protein (MP), and coat protein (CP).
In this study, we first engineered TRSV into a gene expression vector, which can simultaneously express two proteins in virus-infected plants, and a gene-silencing vector to knock down the expression of the PDS gene in four kinds of plants. Then, we engineered TRSV into a VbMS vector to investigate the biological roles of four representative miRNAs in different cucurbitaceous crops.
Results
Engineering of TRSV as a gene expression vector
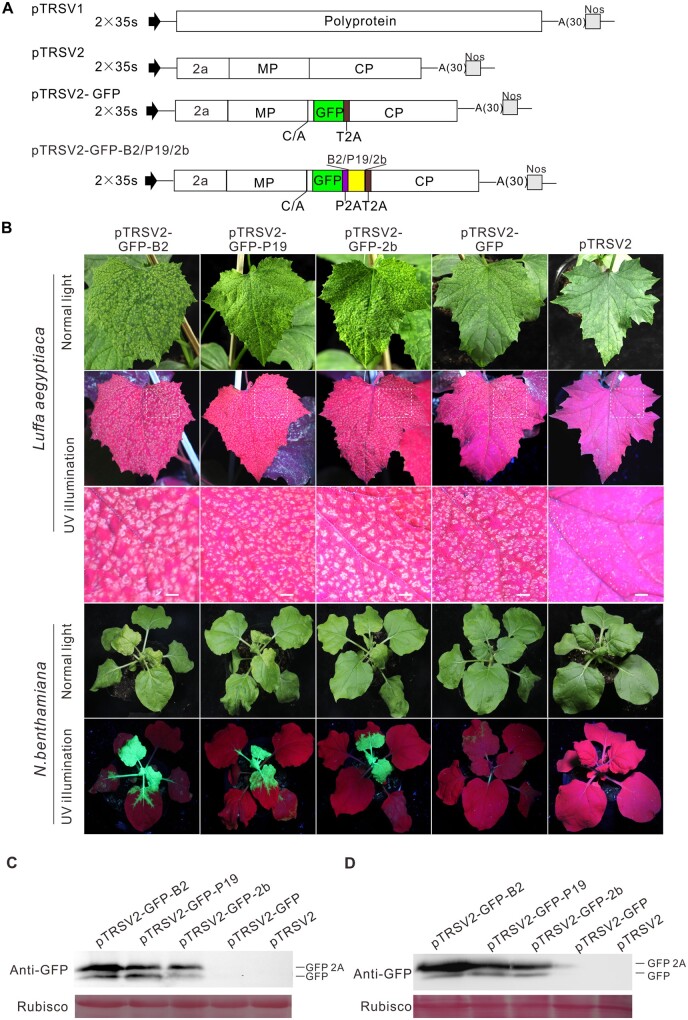
To determine whether TRSV can be used to express foreign genes, we cloned its genomic RNA1 and RNA2 into the binary vector pCB301 to generate plasmids pTRSV1 and pTRSV2, respectively. Then, the sequences coding GREEN FLUORESCENT PROTEIN (GFP) and thosea asigna virus 2A (T2A) catalytic peptide were inserted after the coding sequence for the MP/CP cleavage site in pTRSV2, producing pTRSV2-GFP (Figure 1A). The Agrobacterium tumefaciens cells harboring pTRSV1 and pTRSV2-GFP were infiltrated into leaves of Luffa aegyptiaca and N. benthamiana plants. Green fluorescence appeared in the noninoculated upper (i.e. systemic) leaves at 7 d post agroinfiltration (dpai), but became weak and disappeared in 10–12 d along with viral symptom recovery (Figure 1B). To overcome this disadvantage, we tested whether the co-expression of heterologous viral suppressors of RNA silencing (VSRs) could increase the foreign gene’s expression level. The coding sequences of three VSRs, namely, the flock house virus (FHV) B2, tomato bushy stunt virus (TBSV) P19, and CMV 2b, were individually inserted into plasmid pTRSV2-GFP through T2A and porcine teschovirus-1 2A (P2A) in tandem, and the resulting plasmids were designated as pTRSV2-GFP-B2, pTRSV2-GFP-P19, and pTRSV2-GFP-2b, respectively (Figure 1A). Upon co-agroinfiltration of each construct with pTRSV1 into L. aegyptiaca and N. benthamiana plants, much brighter green fluorescence appeared at 12 dpai (Figure 1B), indicating that the expression of heterologous VSRs could significantly increase GFP expression. Western blot analysis of total proteins extracted from virus-infected L. aegyptiaca and N. benthamiana leaves using a GFP antibody showed that two bands corresponding to GFP-2A and GFP, respectively, were detected in TRSV-GFP-VSRs-infected plants, less protein was detected in TRSV-GFP-infected plants, and no protein was detected in the mock-inoculated plants (Figure 1, C and D). These results showed that the newly modified TRSV-based vector can significantly increase the expression levels of foreign proteins.
Figure 1.
TRSV-based gene expression in L. aegyptiaca and N. benthamiana plants. A, Schemes of TRSV infectious clone and derivatives. The full-length sequences of RNA1 and RNA2 were inserted between a duplicated cauliflower mosaic virus 35S promoter (2x35S) and the NOS terminator (Nos). To construct pTRSV2-GFP, the GFP gene and the sequence encoding thosea asigna virus 2A (T2A) were inserted after the coding sequence for the MOVEMENT PROTEIN/COAT PROTEIN (MP/CP) cleavage site (C/A). Heterologous VSRs and GFP were simultaneously expressed and separated by porcine teschovirus-1 2A (P2A) and T2A in TRSV-based dual genes expression vectors. The release of GFP relied on 2A mediated self-cleavage and polyprotein cleavage. B, Effects of heterologous VSRs on GFP expression using TRSV-based expression vectors in plants at 12 dpai. Symptoms and fluorescence signals of L. aegyptiaca and N. benthamiana infected with TRSV-GFP-B2, -GFP-P19, -GFP-2b, -GFP, and the control pTRSV2. Scale bars= 1 cm in the second row. The third row shows a magnification of dotted rectangle regions in the second row. C and D, Effects of heterologous VSRs on GFP expression in L. aegyptiaca and N. benthamiana leaves as determined with Western blotting
Development of TRSV as a virus-induced gene silencing vector
Virus-induced gene silencing (VIGS) is widely used to investigate the function of genes (Liu et al., 2002; Senthil-Kumar and Mysore, 2014; Choi et al., 2019). Because TRSV only caused mild symptoms, we hypothesized that TRSV could be modified as a VIGS vector. To test this point, we inserted a 300-bp fragment of C. melo PHYTOENE DESATURASE (PDS) into pTRSV2 to generate pTRSV2-CmPDS (Figure 2A). The plasmids pTRSV2-CmPDS and pTRSV1 were first agroinfiltrated into N. benthamiana plants. After 7 dpai, the extracts of systemically infected leaves were used as inoculum for C. melo, C. lanatus, and C. sativus plants. Nearly all of the systematically infected leaves became completely white at 18 dpi (Figure 2B). In contrast, the control plants inoculated with TRSV showed no photo-bleaching phenotype. The results of three independent experiments showed that the silencing efficiency could reach 75.0%–100.0% in C. melo and C. lanatus, and 33.3%–41.7% in C. sativus (Supplemental Table S1). The VIGS experiments were also performed in N. benthamiana plants. The systemic leaves and even the flowers of TRSV2-NbPDS-infected N. benthamiana plants showed a photo-bleaching phenotype (Figure 2B). The silencing efficiency was 100.0% in three independent experiments (Supplemental Table S1). We also used TRSV to silence the expression of eukaryotic initiation factor 4E (eIF4E) and eukaryotic initiation factor (iso) 4E [eIF(iso)4E] in melon. The results of reverse transcription quantitative polymerase chain reaction (RT-qPCR) showed that the mRNA expression levels of eIF4E and eIF(iso)4E were significantly decreased (Supplemental Figure S1). Taken together, these results indicated that TRSV can be utilized as an effective VIGS tool in different plants.
Figure 2.
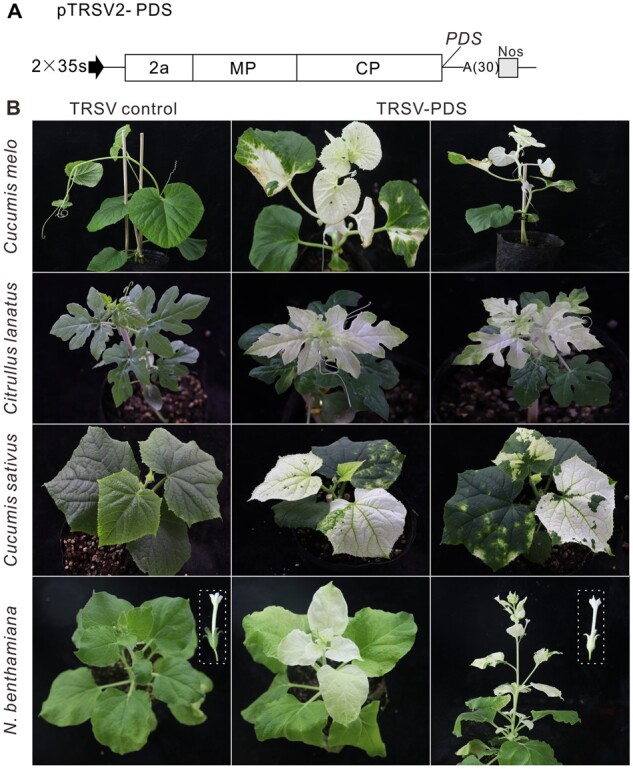
Engineering TRSV as a VIGS vector. A, Schematic representation of pTRSV2. The Sna BI restriction enzyme site was inserted into pTRSV2 downstream of the CP stop codon. The PCR products of the respective PDS genes from cucurbits and N. benthamiana genome were digested with Sna BI and cloned into pTRSV2. B, Phenotypes of PDS-silenced C. melo, C. lanatus, C. sativus, and N. benthamiana plants at 18 dpai. The boxes indicate the flowers of TRSV control and TRSV-PDS-infected N. benthamiana plants. The experiment was independently repeated three times
TRSV-based silencing of miR166 reduced leaf polarity
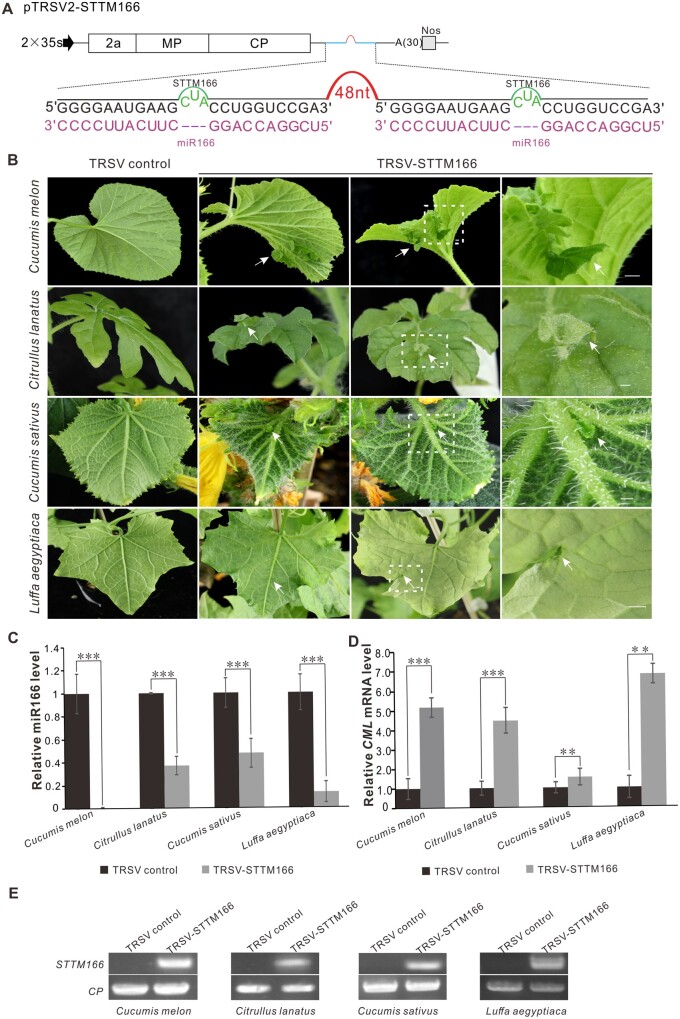
To test whether TRSV can be used as a VbMS vector, the well-characterized miR166 was cloned into pTRSV2 using an STTM strategy (Figure 3A; Yan et al., 2012; Sha et al., 2014; Peng et al., 2018; Zhao et al., 2020). By 23 dpi, TRSV2-STTM166-infected C. melo, C. lanatus, C. sativus, and L. aegyptiaca plants showed a strong silencing phenotype in which an ectopic leaf outgrew from the vein in the underside of the leaves. The abnormal bilateral symmetric leaves with a double-layered lamina were separated by the midvein due to the alteration of adaxial–abaxial regulation in C. melo (Figure 3B). The results of RT-qPCR detection showed that the miR166 level was significantly reduced while the target mRNA level increased in all of the plants infected with TRSV2-STTM166 (Figure 3, C and D). RT-PCR detection confirmed that STTM166 was expressed in the TRSV-STTM166-infected plants (Figure 3E). The silencing efficiency was 62.5%–87.5% in C. melo and L. aegyptiaca plants and 12.5%–37.5% in C. sativus and C. lanatus plants. A similar phenotype was observed in the model plant N. benhamiana, in which the silencing efficiency could reach 88.0%–100.0% (Supplemental Figure S2 and Supplemental Table S2). Taken together, these results show that TRSV-based VbMS using the STTM strategy can effectively suppress the miRNA function in cucurbits and N. benthamiana plants.
Figure 3.
TRSV-based silencing of miR166 in different cucurbits plants. A, Schematic representation of VbMS of mi166. “–-” represents no nucleotide in this position. The STTM166 sequence was introduced into pTRSV2 downstream of the CP stop codon. The 48 nucleotides stem-loop spacer contributes to STTM stability and efficiency. B, Leaf phenotypes of plants inoculated with TRSV-STTM166 at 23 dpi. The arrows indicate the ectopic leaf outgrowing from the vein. Scale bars = 1 cm in the third column. The fourth column shows a magnification of dotted rectangle regions in the third column. C, Stem-loop RT-qPCR detection of miR166 level in TRSV- and TRSV-STTM166-infected cucurbits plants. D, Relative expression level of miR166 target gene in various cucurbits plants inoculated with TRSV or TRSV-STTM166. In (C) and (D), the biological experiment was repeated three times. Error bars represent ± SD. Statistical significance between treatments was determined using paired Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. E, RT-PCR detection confirmed the TRSV infection as indicated by the presence of CP transcripts in all plants and the expression of STTM166
TRSV-based miR159 silencing caused pleiotropic developmental defects in C. melo plants
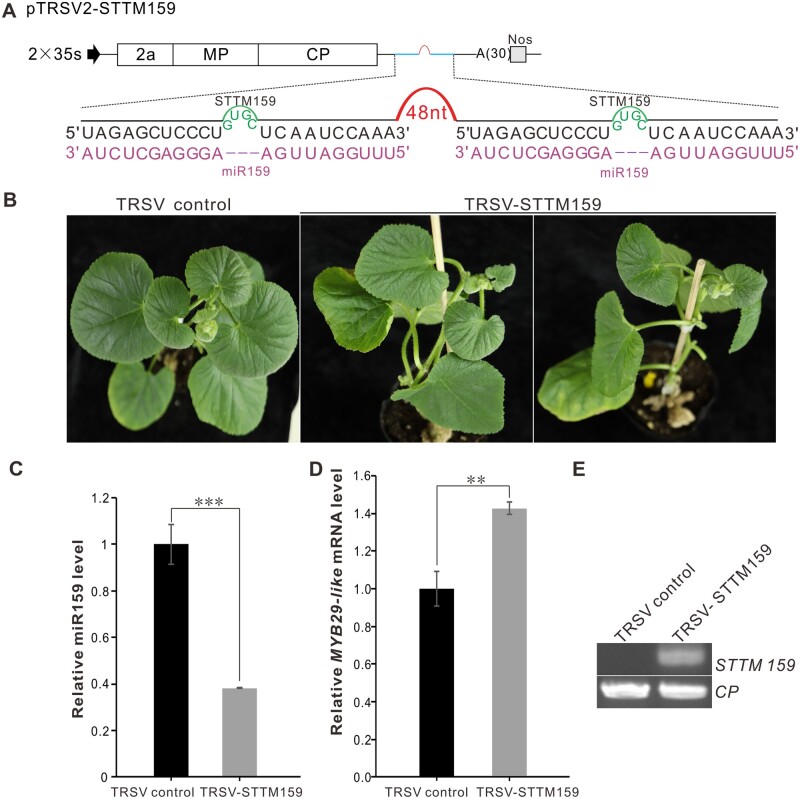
MiR159, which targets GAMYB or GAMYB-like genes, is one of the most ancient and abundant small RNAs in most land plants (Fahlgren et al., 2007; Jeong et al., 2011; Kozomara et al., 2019; Millar et al., 2019). However, the function of miR159 in cucurbits has remained largely unknown (Liang et al., 2019; Millar et al., 2019). To investigate the miR159-GAMYB regulatory circuit in C. melo, we modified TRSV to express STTM159 (Figure 4A). The TRSV-STTM159-infected plants were stunted and the leaves were smaller than the control plants inoculated with TRSV (Figure 4B). Compared with control plants, the miR159 level in TRSV-STTM159-infected plants was significantly decreased while the mRNA level of the target gene MYB29-like was significantly increased (Figure 4, C and D). RT-PCR detection demonstrated that the STTM159 was expressed in C. melo plants (Figure 4E). The silencing efficiency was 37.5%–50.0% in C. melo plants. A similar phenotype was observed in N. benthamiana plants, in which the silencing efficiency was 25.0%–31.3% (Supplemental Figure S3 and Supplemental Table S3). These results indicate that TRSV can effectively block miR159 activity in plants.
Figure 4.
Silencing of miR159 in C. melo using TRSV-based VbMS vector. A, Schematic representation of VbMS of miR159. The STTM159 sequence was cloned into pTRSV2 after the CP stop codon. “–-” represents no nucleotides in this position. B, Leaf phenotypes of plants inoculated with TSRV and TRSV-STTM159 at 25 dpi. C, Stem-loop RT-qPCR detection of miR159 level in TRSV- and TRSV-STTM159-infected plants. D, Relative expression levels of miR159 target gene MYBL29-like in the TRSV- and TRSV-STTM159-infected plants. In (C) and (D), the results were presented as means ± SD from three independent biological replicates. Statistical significance between treatments was determined using paired Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. E, RT-PCR detection confirmed the infection of TRSV and the expression of STTM159
TRSV-based VbMS of miR319 caused leaf morphogenesis in C. lanatus plants
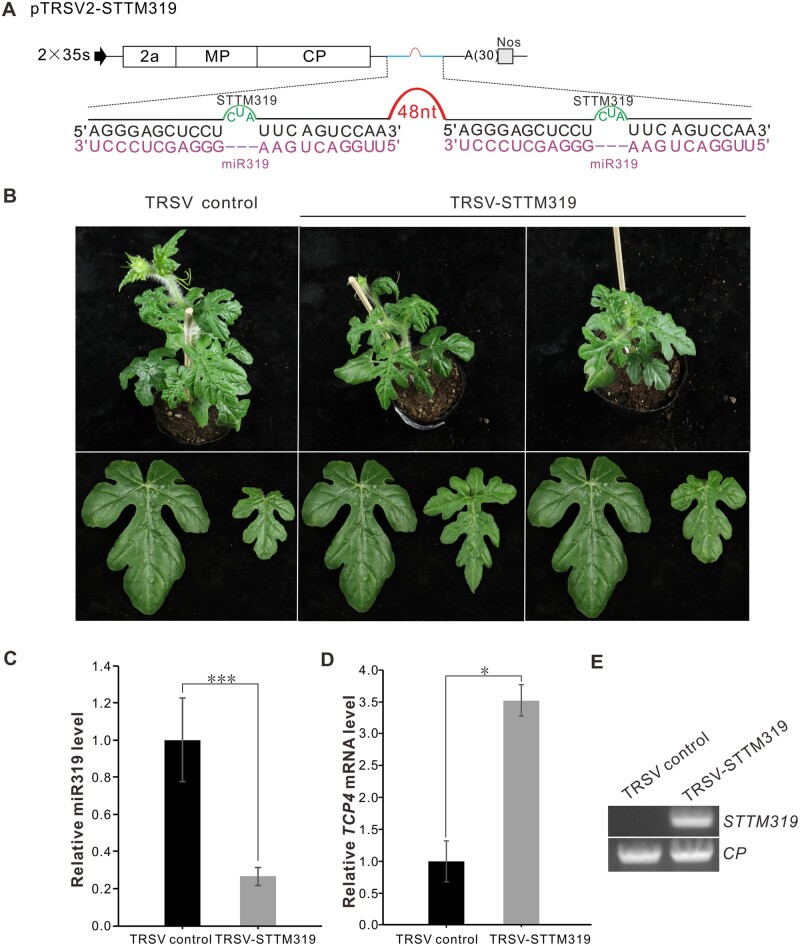
MiR319, a conserved miRNA family that participates in leaf development, controls the activity of TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATING CELL NUCLEAR ANTIGEN BINDING FACTOR (TCP) genes (Ori et al., 2007; Koyama et al., 2017). To test whether miR319 could suppress the expression of the TCP4 gene in C. lanatus, we modified TRSV to express STTM319 and inoculated it into watermelon plants (Figure 5A). At 30 dpi, the TRSV-STTM319-infected C. lanatus leaves were smaller than the control plants (Figure 5B). The miR319 level was lower and the expression level of the target gene TCP4 was higher in TRSV-STTM319-infected C. lanatus leaves than in the control plants (Figure 5, C and D). RT-PCR detection showed that STTM319 was expressed in TRSV-STTM319-infected C. lanatus plants (Figure 5E). The silencing efficiency of miR319 was 43.8%–50.0% (Supplemental Table S4). These results indicate that VbMS of miR319 has an impact on leaf development in C. lanatus plants.
Figure 5.
TRSV-based VbMS of miR319 in C. lanatus. A, Schematic representation of VbMS of mi319. B, Leaf phenotypes of plants inoculated with TRSV-STTM319 at 30 dpai. C, Stem-loop RT-qPCR analysis of miR319 expression level of TRSV- or TRSV-STTM319-infected plants. D, Detection of the relative expression level of miR319 target gene TCP4. In C and D, error bars represent the ± SD of three independent experiments. Statistical significance between treatments was determined using paired Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. E, RT-PCR detection confirmed the expression of STTM319 in TRSV-STTM319-infected plants
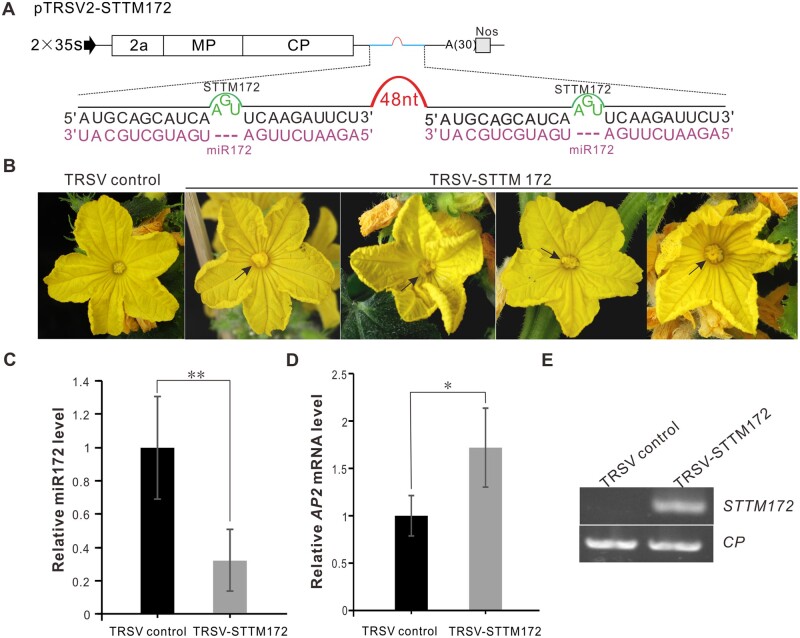
TRSV-based VbMS of miR172 caused flower development defects in C. sativus plants
Arabidopsis miR172 is a crucial regulator of flower organ identity and flower stem cell proliferation (Chen, 2004). Over-expression of a miR172-resistant version of (APETALA2) AP2 elevated AP2 protein levels and led to an indeterminate floral meristem producing supernumerary stamens in Arabidopsis (Zhao et al., 2007). To test whether TRSV-based VbMS of STTM172 could inhibit miR172 activity, we amplified STTM172 and cloned it into pTRSV2 to generate pTRSV-STTM172 (Figure 6A). Extra petals arose from inner floral meristem in TRSV-STTM172-infected cucumber plants while the control plants grew normally, suggesting that TRSV-based VbMS of miR172 suppressed the function of miR172 (Figure 6B). The miR172 level was lower and the mRNA level of the target gene C. sativus APETALA2 (CsAP2) was higher in TRSV-STTM172-infected cucumber plants than in the control plants (Figure 6, C and D). RT-PCR detection showed that STTM172 was expressed in TRSV-STTM172-infected cucumber plants (Figure 6E). The silencing efficiency of miR172 in C. sativus plants was 25.0%–37.5%. We also inoculated N. benthamiana plants with TRSV-STTM172. Flowers in plants inoculated with TRSV-STTM172 displayed shortened petals that could not enclose the interior stamens and carpels. In certain extreme cases, some calyces even turned into petals, causing abnormal flower phenotypes (Supplemental Figure S4B). The level of miR172 in TRSV-STTM172-infected N. benthamiana plants was reduced, but the expression level of target gene NbAP2-LIKE 1 (NbAP2L1) was increased (Supplemental Figure S4, C and D). The silencing efficiency of miR172 in N. benthamiana plants could reach 81.3%–100.0% (Supplemental Table S5). All of these results suggest that TRSV-based VbMS of miR172 using the STTM approach effectively inhibits miR172 function.
Figure 6.
Suppression of miR172 by TRSV-STTM172 in C. sativus. A, Schematic representation of TRSV-STTM172. B, The flowers of TRSV- or TRSV-STTM172-infected cucumber plants were photographed at 45 dpi. Arrows indicate the extra petals arising from inner floral meristem. C, Detection of miR172 expression level in TRSV- or TRSV-STTM172-infected plants through stem-loop RT-qPCR. D, The RT-qPCR analysis of relative expression level of miR172 target RAP2. In (C) and (D), the results presented were repeated three times. Error bars represent ± SD. Statistical significance between treatments was determined using paired Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. E, RT-PCR detection confirmed the expression of miR172 in TRSV-STTM172-infected plants
Discussion
Some cucurbit species are recalcitrant to genetic transformation. For example, the efficiency in C. melo ranges from 0.7% to 7.0% (Fang and Grumet, 1990; Guis et al., 2000; Akasaka-Kennedy et al., 2004; Çürük et al., 2005; Rhimi et al., 2007; Chovelon et al., 2011; Choi et al., 2012; Ren et al., 2012). VIGS provides an alternative strategy to study the function of plant genes. However, very few plant virus-based silencing systems work well in cucurbits (Zhao et al., 2016; Liu et al., 2020). Recently, a cucumber green mottle mosaic virus-based VIGS vector was developed, but it only induced a silencing phenotype in leaf veins (Liu et al., 2020). TRSV has displayed potential in knocking down the expression of genes in different crop plants (Zhao et al., 2016). Here, we presented a set of TRSV-based vectors with high efficiency in silencing genes or miRNAs in cucurbits, which will help to validate the function of candidate genes involved in agronomic traits. The efficiency of gene silencing was above 80% in C. melo and C. lanatus, while it was about 35% in C. sativus (Supplemental Table S1). This may be due to the infectivity of TRSV in C. sativus being weaker than that in C. melo and C. lanatus plants. In addition to serving as a reverse genetic tool, VIGS is also a forward genetics tool to perform high-throughput genetic screens and functional genomics (Bachan and Dinesh-Kumar, 2012). The TRSV-based VIGS vectors developed in this study can induce a silencing phenotype within 18 d in four important cucurbitaceous plants (Figure 2B). Therefore, they have great potential for rapid and high-throughput forward genetic screens in cucurbits.
Except for engineering TRSV as VIGS vectors, we developed its function in miRNA silencing. This is the first VbMS vector for the function investigation of cucurbit miRNAs. It is noteworthy that the efficiency of most TRSV-based miRNA silencing is high in both model and crop plants. In a previous study, only 20.0%–30.0% of N. benthamiana plants expressing target mimics against miR165/166 showed the expected phenotype (Sha et al., 2014). However, all of the TRSV-miR165/166-infected N. benthamiana plants showed defective symptoms in leaves (Supplemental Figure S2 and Supplemental Table S2). Furthermore, the silencing efficiency of miRNA was 75.0%–87.6% in L. aegyptiaca plants and 68.8%–75.0% in C. melo. In most cases, the silencing efficiency of miRNAs in N. benthamiana plants was higher than that in cucurbitaceous plants. Only miRNA159 was an exception. In a previous study, no visible phenotype was observed in TRV-miR159-infected N. benthamiana plants, but all of the PVX-miR159-infected N. benthamiana plants displayed the expected phenotype (Sha et al., 2014; Zhao et al., 2016). Therefore, the development of VbMS systems provides potential solutions for the functional study of some specific miRNAs.
Plant virus-based vectors can be utilized for transient gain-of-function studies, especially in recalcitrant species for genetic transformation (Choi et al., 2019). TRSV can be used to express GFP in N. benthamiana plants. However, the GFP fluorescence becomes nearly invisible in newly emerging leaves after 15 dpi (Zhao et al., 2016). The same problem occurred in our study with both N. benthamiana and L. aegyptiaca plants (Figure 2A). To overcome this drawback, we engineered TRSV into a dual gene expression vector that simultaneously expressed GFP and heterologous VSR separated through two different 2A peptides in tandem. Here, we tested three different VSRs and found that the GFP accumulation in TRSV-GFP-B2-infected plants was slightly higher than that in TRSV-GFP-p19 or TRSV-GFP-2b-infected plants (Figure 1, C and D), which indicated that FHV B2 is more effective for TRSV-based protein expression in L. aegyptiaca and N. benthamiana. The 2A peptides lead to relatively high levels of downstream protein expression compared with other strategies for multigene co-expression, and they are small in size, thus bearing a lower risk of interfering with the function of co-expressed genes (Donnelly et al., 2001; Liu et al., 2017). Two 2A peptides are different in the sequence, which avoids the loss of foreign genes caused by viral RdRP leap. Different viral 2A peptides are available in the GenBank database and may be used for multiple gene expression of TRSV-based vectors. In addition, we also used TRSV to express Cas9 from Staphylococcus aureus (SaCas9, 3,162 bp) in N. benthamiana plants (Supplemental Figure S5). The molecular weight of SaCas9 was 124 kDa. The gene-expression capacity of TRSV indicated the possibility of extending the usage of TRSV-based vectors. In recent studies, two plant virus-based vectors were modified for heritable gene editing (Ellison et al., 2020; Ma et al., 2020). It will be interesting to investigate whether TRSV can be used for gene editing in the future.
Taken together, we have shown the great potential of TRSV-based vectors in dissecting the biological roles of genes and miRNA. The versatility of this set of vectors will significantly facilitate functional genomic studies in cucurbitaceous plants.
Materials and methods
Plasmid construction
To construct a full-length infectious clone, the genomic fragments of TRSV RNA1 and RNA2 were obtained by one-step amplification and cloned into the binary vector pCB301 downstream of a double cauliflower mosaic virus (CaMV) 35S promoter to generate pTRSV1 and pTRSV2, respectively. The pTRSV2 vector was further modified to express foreign genes and knockdown endogenous genes or miRNAs.
To improve the foreign gene expression level, the codons for N-terminal 19 amino acids of CP with some nucleotide substitutions for degenerate codons were duplicated and added to the 5′ of the GFP and SaCas9 genes (Zhao et al., 2016). The sequence encoding TaV 2A (5′AGAGCAGAAGGGAGAGGAAGCTTGCTAACCTGTGGAGACGTTGAGGAAAATCCAGGGCCA-3′) was also added to the 3′ of GFP and SaCas9 genes to guarantee the efficient cleavage of GFP. The Arabidopsis (A. thaliana) codon-optimized SaCas9 gene was synthesized as the sequence published previously (Kaya et al., 2016). To construct pTRSV2-GFP-B2, pTRSV2-GFP-P19, and pTRSV-GFP-2b, sequences encoding heterologous VSR and P2A (5′GGCTCGGGCCAGTGTACTAATTATGCTCTCTTGAAATTGGCTGGAGATGTTGAGAGCAACCCAGGTCCC3′) were inserted between the GFP-T2A and CP region in the pTRSV2-GFP vector through PCR using specific primers as described in Supplemental Table S6.
The Sna BI restriction enzyme sequences downstream of the CP translational stop codon were introduced into TRSV RNA2 through one-step site-directed mutation (Liu and Naismith, 2008). The CuPDS gene and NbPDS gene were RT-PCR amplified separately from the total RNAs from C. melo and N. benthamiana leaves using specific primers and inserted into pTRSV2 (Supplemental Table S6). A 48-bp oligonucleotide was utilized as a linker to construct STTM miRNA. The full sequence STTM miRNA was amplified and cloned into pTRSV2 (Yan et al., 2012). All of the constructs were confirmed by DNA sequencing before further use. Primers are listed in Supplemental Table S6.
Plant growth and inoculation
Cucumis melo (cultivar “Huoyingua”), C. lanatus (cultivar “Fufengshijilong”), C. sativus (cultivar “Nairewangzhongwang”), L. aegyptiaca (cultivar “Changfengfiangsigua”), and N. benthamiana plants were grown at 22°C in a greenhouse under a 16 h light/8 h dark cycle with 60.0% humidity. Plasmids were individually transformed into A. tumefaciens GV3101 cells using a freeze-thaw method (Hofgen and Willmitzer, 1988). The A. tumefaciens cultures were grown overnight in Luria–Bertani liquid medium with appropriate antibiotics at 28°C and individually re-suspended in an induction buffer containing 10-mM MgCl2, 10-mM MES, and 150-μM acetosyringone as described earlier (Geng et al., 2015). Separate cultures of A. tumefaciens (OD600= 1.0) harboring pTRSV1, pTRSV2, or pBinP19 were mixed in a 1:1:1 ratio and infiltrated into N. benthamiana plants. The inoculated plants were grown and observed until the phenotype appeared. The N. benthamiana leaves agroinfiltrated with pTRSV2-CuPDS or pTRSV2-miRNA were harvested at 6 dpi for rub-inoculation.
Protein extractions and western blotting
For total protein extraction, plant leaf tissue infected with TRSV was ground in liquid nitrogen followed by suspension in a protein extraction buffer. Samples were boiled for 10 min before centrifugation for 2 min at 10,000g at 4°C. The supernatant was kept for SDS-PAGE. Proteins were then transferred into a nitrocellulose membrane in tris-glycine transfer buffer (25-mM Tris base, 192-mM glycine, and 3.5-mM SDS) at 100 V for 60 min. The membrane was then blocked in 5% (w/v) nonfat milk in Tris Buffered Saline with Tween-20 (TBST, 20-mM Tris base, 150-mM NaCl, 0.05% (v/v) Tween-20, pH = 7.5) with gentle agitation for 60 min at room temperature or 4°C overnight. The GFP-specific antibody was added to the nonfat milk at the appropriate dilution. Membranes were washed with TBST three times for 10 min each wash. Secondary goat anti-rabbit IgG conjugated with horseradish peroxidase (Sigma-Aldrich) was incubated for 1 h at a working dilution. The nitrocellulose membranes were then washed briefly with TBS (20-mM Tris base, 150-mM NaCl, pH = 7.5) after two washes in TBST for 10 min each and then photographed. GFP imaging was observed under long-wavelength UV light, and photographs were taken using a Canon 80D camera.
RNA extraction and RT-PCR
Total RNAs were extracted from plant leaves using TransZol reagent (TransGen Biotech, ET101-01). The resulting total RNA samples were treated with a gDNA wiper enzyme (Vazyme, R233-01-AB) to remove plant genomic DNA. Stem-loop RT-PCR was performed to detect the miRNA expression level. Total RNA of 1–5 μg was reverse transcribed using a specific reverse primer and miRNA 1st Strand cDNA Synthesis Kit (Vazyme, MR101-01). Then, PCR was performed using miRNA Universal SYBR qPCR Master Mix (Vazyme, MQ101-01). To detect the target gene level of inoculated plants, 1 μg total RNA (per sample) was reverse transcribed using a HiScriptII Q RT SuperMix kit supplemented with random primers (Vazyme, R222-01-AB). RT-qPCR was conducted using a ChamQ SYBR qPCR Master Mix (Vazyme, Q311-02) on a thermocycler (LightCycler® 96, Roche). The specific detection primers are listed in the Supplemental Table S6. The CmACTIN gene was used as an internal control for the detection of miRNA and target genes. The N. benthamiana eIF4A and EF1α were used as internal controls for the analysis of miRNA and target genes.
Accession numbers
The sequence data of this paper can be obtained from GenBank and the Sol Genomic Network under the following accession numbers: LC376026.1 (CMV 2b), AJ288942.1 (TBSV P19), X77156.1 (FHV B2), EU165355.1 (NbPDS), XM_011654729 (CsPDS), NM_001297480.1 (CmeIF4E), XM_008462461.2 [CmeIF(ISO)4E], XM_004137620.3 (CsCML42), XM_004137168.3 (CsTCP4), XM_031885023.1 (CsAP2), Niben101Scf01383g07027.1 (NbMYBL1), and CK287095 (NbAP2L1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. TRSV-based silencing of eIF4E and eIF(iso)4E in C. melo plants.
Supplemental Figure S2. TRSV-based silencing of miR165/166 in N. benthamiana plants.
Supplemental Figure S3. TRSV-based knockdown of miR159 in N. benthamiana plants.
Supplemental Figure S4 . TRSV-based VbMS of miR172 in N. benthamiana plants.
Supplemental Figure S5. The expression of SaCas9 gene using TRSV in N. benthamiana plants.
Supplemental Table S1. Silencing efficiency of PDS genes.
Supplemental Table S2. Silencing efficiency of miR166.
Supplemental Table S3. Silencing efficiency of miR159.
Supplemental Table S4. Silencing efficiency of miR319.
Supplemental Table S5. Silencing efficiency of miR172.
Supplemental Table S6. Primers used in this study.
Funding
This study was supported by the National Natural Science Foundation of China (NSFC; 31801704, 31720103912) and the “Taishan Scholar” Construction Project (TS201712023).
Conflict of interest statement. The authors declare no conflict of interest.
Supplementary Material
C.G. and X.-D.L. conceived the research plan and designed the experiments. L.F. and X.-Y.W performed the experiments and prepare the Figures and Tables. C.G., X.-D.L., and L.F. co-wrote and edited the article. All authors analyzed the results.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Chao Geng (chaogeng@sdau.edu.cn) and Xiang-Dong Li (xdongli@sdau.edu.cn).
References
- Abdalla OA, Bruton BD, Fish WW, Ali A (2012) First confirmed report of Tobacco ringspot virus in cucurbits crops in Oklahoma. Plant Dis 96: 1705. [DOI] [PubMed] [Google Scholar]
- Akasaka-Kennedy Y, Tomita KO, Ezura HJPS (2004) Efficient plant regeneration and Agrobacterium-mediated transformation via somatic embryogenesis in melon (Cucumis melo L.). Plant Sci 166: 763–769 [Google Scholar]
- Bachan S, Dinesh-Kumar SP (2012) Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods Mol Biol 894: 83–92 [DOI] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B, Kwon SJ, Kim MH, Choe S, Kwak HR, Kim MK, Jung C, Seo JK (2019) A plant virus-based vector system for gene function studies in pepper. Plant Physiol 181: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Shin JS, Chung YS, Hyung N-I (2012) An efficient selection and regeneration protocol for Agrobacterium-mediated transformation of oriental melon (Cucumis melo L. var. makuwa). Plant Cell Tiss Organ Cult 110: 133–140 [Google Scholar]
- Chovelon V, Restier V, Giovinazzo N, Dogimont C, Aarrouf J (2011) Histological study of organogenesis in Cucumis melo L. after genetic transformation: why is it difficult to obtain transgenic plants? Plant Cell Rep 30: 2001–2011 [DOI] [PubMed] [Google Scholar]
- Çürük S, Çetiner S, Elman C, Xia X, Wang Y, Yeheskel A, Zilberstein L, Perl-Treves R, Watad AA, Gaba V (2005) Transformation of recalcitrant melon (Cucumis melo L.) cultivars is facilitated by wounding with carborundum. Eng Life Sci 5: 169–177 [Google Scholar]
- Ding XS, Mannas SW, Bishop BA, Rao X, Lecoultre M, Kwon S, Nelson RS (2018) An improved Brome mosaic virus silencing vector: greater insert stability and more extensive VIGS. Plant Physiol 176: 496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MLL, Hughes LE, Luke G, Mendoza H, Dam Et, Gani D, Ryan MD (2001) The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Mol Virol 82: 1027–1041 [DOI] [PubMed] [Google Scholar]
- Du Z, Chen A, Chen W, Westwood JH, Baulcombe DC, Carr JP (2014) Using a viral vector to reveal the role of microRNA159 in disease symptom induction by a severe strain of Cucumber mosaic virus. Plant Physiol 164: 1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Agius C, Smith NA, Waterhouse PM, Wang MB (2011) Efficient silencing of endogenous microRNAs using artificial microRNAs in Arabidopsis thaliana. Mol Plant 4: 157–170 [DOI] [PubMed] [Google Scholar]
- Ellison EE, Nagalakshmi U, Gamo ME, Huang PJ, Dinesh-Kumar S, Voytas DF (2020) Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants 6: 620–624 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Grumet R (1990) Agrobacterium tumefaciens mediated transformation and regeneration of muskmelon plants. Plant Cell Rep 9: 160–164 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, Gonzalez VM, Henaff E, Camara F, Cozzuto L, Lowy E, et al. (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA 109: 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C, Cong QQ, Li XD, Mou AL, Gao R, Liu JL, Tian YP (2015) DEVELOPMENTALLY REGULATED PLASMA MEMBRANE PROTEIN of Nicotiana benthamiana contributes to potyvirus movement and transports to plasmodesmata via the early secretory pathway and the actomyosin system. Plant Physiol 167: 394–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba Y, Klimyuk V, Marillonnet S (2007) Viral vectors for the expression of proteins in plants. Curr Opin Biotechnol 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Gu Z, Huang C, Li F, Zhou X (2014) A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol J 12: 638–649 [DOI] [PubMed] [Google Scholar]
- Guis M, Amor MB, Latché A, Pech JC, Roustan JPJSH (2000) A reliable system for the transformation of cantaloupe charentais melon (Cucumis melo L. var. cantalupensis) leading to a majority of diploid regenerants. Sci Hortic 84: 91–99 [Google Scholar]
- Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, et al. (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45: 51–58 [DOI] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16 9877–9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Javier F., Palatnik EA, Wu Xuelin (2003) Control of leaf morphogenesis by micoRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian C, Han R, Chi Q, Wang S, Ma M, Liu X, Zhao H (2017) Virus-based microRNA silencing and overexpressing in common wheat (Triticum aestivum L.). Front Plant Sci 8: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Wang Y, Selvaraj JN, Xing F, Liu Y (2015) Barley stripe mosaic virus (BSMV) induced microRNA silencing in common wheat (Triticum aestivum L.). PLoS One 10: e0126621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Mikami M, Endo A, Endo M, Toki S (2016) Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Scientific Rep 6: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175: 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47: D155–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Liu H, Hao J, Li J, Luo L (2019) Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Arch Virol 164: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Tu Y, Carr JP, Du Z (2015) An improved cucumber mosaic virus-based vector for efficient decoying of plant microRNAs. Sci Rep 5: 13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Liang Z, Aranda MA, Hong N, Liu L, Kang B, Gu Q (2020) A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, Vaseghi HR, Qian L, Liu J (2017) Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep 7: 2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat Plants 6: 773–779 [DOI] [PubMed] [Google Scholar]
- Millar AA, Lohe A, Wong G (2019) Biology and function of miR159 in plants. Plants (Basel) 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Peng T, Qiao M, Liu H, Teotia S, Zhang Z, Zhao Y, Wang B, Zhao D, Shi L, Zhang C, et al. (2018) A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol Plant 11: 1400–1417 [DOI] [PubMed] [Google Scholar]
- Ren Y, Bang H, Curtis IS, Gould J, Patil BS, Crosby KMJPCT, Culture O (2012) Agrobacterium-mediated transformation and shoot regeneration in elite breeding lines of western shipper cantaloupe and honeydew melons (Cucumis melo L.). Plant Cell Tiss Organ Cult 108: 147–158 [Google Scholar]
- Rhimi A, Hernould M, Boussaid M (2007) Agrobacterium mediated transformation of Tunisian Cucumis melo cv maazoun. Afr J Biotechnol 6 [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Canizares MC, Lomonossoff GP (2010) Cowpea mosaic virus: the plant virus-based biotechnology workhorse. Annu Rev Phytopathol 48: 437–455 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2014) Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc 9: 1549–1562 [DOI] [PubMed] [Google Scholar]
- Seo JK, Choi HS, Kim KH (2016) Engineering of soybean mosaic virus as a versatile tool for studying protein-protein interactions in soybean. Sci Rep 6: 22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha A, Zhao J, Yin K, Tang Y, Wang Y, Wei X, Hong Y, Liu Y (2014) Virus-based microRNA silencing in plants. Plant Physiol 164: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba E, Chen PY, Gergerich R, Li SX, Dombek D, Shi AN, Brye K (2012) Reactions of mid-southern U.S. soybean cultivars to Bean pod mottle virus and Tobacco ringspot virus. Crop Sci 52: 1980–1989 [Google Scholar]
- Shin AY, Koo N, Kim S, Sim YM, Choi D, Kim YM, Kwon SY (2019) Draft genome sequences of two oriental melons, Cucumis melo L. var. makuwa. Sci Data 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Elias L, George GL, Jones L (2010) Suppression of microRNA accumulation via RNA interference in Arabidopsis thaliana. Plant Mol Biol 73: 391–397 [DOI] [PubMed] [Google Scholar]
- Wu H, Zhao G, Gong H, Li J, Luo C, He X, Luo S, Zheng X, Liu X, Guo J, Chen J, Luo J (2020) A high-quality sponge gourd (Luffa cylindrica) genome. Hortic Res 7: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang X, Reddy U, Sun H, Bao K, Gao L, Mao L, Patel T, Ortiz C, Abburi VL, et al. (2019) Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the U.S. National Plant Germplasm System watermelon collection. Plant Biotechnol J 17: 2246–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang TY, Liao QS, He L, Li J, Zhang HM, Chen X, Li J, Yang J, Li JB, Chen JP (2018) Chinese wheat mosaic virus-induced gene silencing in monocots and dicots at low temperature. Front Plant Sci 9: 1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Mu X, Liu C, Cai J, Shi K, Zhu W, Yang Q (2015) Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol 57: 1078–1088 [DOI] [PubMed] [Google Scholar]
- Zellner W, Frantz J, Leisner S (2011) Silicon delays Tobacco ringspot virus systemic symptoms in Nicotiana tabacum. J Plant Physiol 168: 1866–1869 [DOI] [PubMed] [Google Scholar]
- Zhang C, Ghabrial SA (2006) Development of Bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344: 401–411 [DOI] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, et al. (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31: 848–852 [DOI] [PubMed] [Google Scholar]
- Zhao F, Hwang US, Lim S, Yoo RH, Igori D, Lee SH, Lim HS, Moon JS (2015) Complete genome sequence and construction of infectious full-length cDNA clones of tobacco ringspot Nepovirus, a viral pathogen causing bud blight in soybean. Virus Genes 51: 163–166 [DOI] [PubMed] [Google Scholar]
- Zhao F, Lim S, Igori D, Yoo RH, Kwon SY, Moon JS (2016) Development of tobacco ringspot virus-based vectors for foreign gene expression and virus-induced gene silencing in a variety of plants. Virology 492: 166–178 [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu Q, Hu P, Jia Q, Liu N, Yin K, Cheng Y, Yan F, Chen J, Liu Y (2016) An efficient Potato virus X-based microRNA silencing in Nicotiana benthamiana. Sci Rep 6: 20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Rios CG, Song J (2020) Potato virus X-based microRNA silencing (VbMS) in Potato. J Vis Exp 159: e61067. [DOI] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X (2007. ) miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J 51: 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.