Abstract
In Arabidopsis thaliana, mitochondrial-localized heat-shock cognate protein 70-1 (mtHSC70-1) plays an important role in vegetativegrowth. However, whether mtHSC70-1 affects reproductive growth remains unknown. Here, we found that the mtHSC70-1 gene was expressed in the provascular cells of the embryo proper from the early heart stage onward during embryogenesis. Phenotypic analyses of mthsc70-1 mutants revealed that mtHSC70 deficiency leads to defective embryo development and that this effect is mediated by auxin. In addition to a dwarf phenotype, the mthsc70-1 mutant displayed defects in flower morphology, anther development, and embryogenesis. At early developmental stages, the mthsc70-1 embryos exhibited abnormal cell divisions in both embryo proper and suspensor cells. From heart stage onward, they displayed an abnormal shape such as with no or very small cotyledon protrusions, had aberrant number of cotyledons, or were twisted. These embryo defects were associated with reduced or ectopic expression of auxin responsive reporter DR5rev:GFP. Consistently, the expression of auxin biosynthesis and polar auxin transport genes were markedly altered in mthsc70-1. On the other hand, mitochondrial retrograde regulation (MRR) was enhanced in mthsc70-1. Treatment of wild-type plants with an inhibitor that activates mitochondrial retrograde signaling reduced the expression level of auxin biosynthesis and polar auxin transport genes and induced phenotypes similar to those of mthsc70-1. Taken together, our data reveal that loss of function of mtHSC70-1 induces MRR, which inhibits auxin biosynthesis and polar auxin transport, leading to abnormal auxin gradients and defective embryo development.
mtHSC70-1 dysfunction induces mitochondrial retrograde regulation, which inhibits auxin biosynthesis and polar auxin transport, leading to abnormal auxin gradients and defective embryo development.
Introduction
The 70-kDa heat-shock proteins (HSP70s) are highly conserved molecular chaperones among prokaryotes and eukaryotes (Gupta and Golding, 1993). HSP70s assist protein translocation, protein folding, and quality control (Nishikawa et al., 2005; Bukau et al., 2006). In Arabidopsis thaliana (Arabidopsis), there are 18 HSP70s located in the cytosol, endoplasmic reticulum (ER), plastids, and mitochondria, respectively (Sung et al., 2001; Lin et al., 2001). Physiologically, the cytosolic HSP70s mainly participate in plant growth and development and plant responses to biotic stress such as virus infection and abiotic stresses such as high temperature, drought, and salinity (Jungkunz et al., 2011; Leng et al., 2017). Immunoglobulin-binding proteins (BiPs) are ER-localized HSP70s comprising three members in Arabidopsis, namely BiP1, BiP2, and BiP3 (Noh et al., 2003). BiP1 and BiP2 are ubiquitously expressed proteins, whereas BiP3 is expressed only under ER-stress conditions (Noh et al., 2003). BiPs are required for female and male gametophyte development and pollen tube growth (Maruyama et al., 2010, 2014). The plastid-localized cpHSC70-1 and cpHSC70-2 proteins are essential for both vegetative growth and reproduction and are required for thermotolerance during seed germination (Su and Li, 2008). There are two mitochondrial-localized HSP70s in Arabidopsis, mtHSC70-1, and mtHSC70-2 (also known as AtHscA1 and AtHscA2, respectively; Lin et al., 2001; Sung et al., 2001; Xu et al., 2009; Leaden et al., 2014; Wei et al., 2019). mtHSC70-1/AtHscA1 was demonstrated to be localized to mitochondria by co-localization of its fusion protein with a fluorescent reporter at the C-terminal with a mitotracker dye (Xu et al., 2009; Wei et al., 2019). mtHSC70-1/AtHscA1 can functionally rescue the yeast (Saccharomyces cerevisiae) Ssq1 (MtHSC70 yeast homolog) knockout mutant (Xu et al., 2009). Both mtHSC70-1 and mtHSC70-2 are demonstrated to be involved in the assembly of the mitochondrial Fe-S cluster (Xu et al., 2009; Leaden et al., 2014). Overexpression of the rice (Oryza sativa) mtHSC70 suppressed heat- and H2O2-induced cell death in rice protoplasts by maintaining mitochondrial membrane potential and reducing reactive oxygen species (ROS) generation (Qi et al., 2011). The cochaperone AtBAG5 participates in the regulation of leaf senescence through the cooperation of Ca2+ signaling and the mtHSC70 molecular chaperone system (Li et al., 2016). A recent study showed that mtHSC70-1 loss-of-function mutation in Arabidopsis led to abnormal mitochondria and decreased activity and abundance of the respiratory complex IV, and resulted in a dwarf phenotype (Wei et al., 2019). Furthermore, mtHSC70-1 protein was found to physically interact with the COX2 (cytochrome c oxidase, COX) protein of the respiratory complex IV (Zhai et al., 2020). Both ER and plastid HSP70 proteins have been shown to play important roles in reproductive growth in Arabidopsis. However, whether mitochondrial HSP70 proteins affect reproductive growth has received little attention.
Mitochondria play important roles in energy production, redox homeostasis, metabolic homeostasis, and signaling in plants (Noctor et al., 2007; Zhu et al., 2014). The mitochondrial proteome is encoded by nuclear and mitochondrial genomes, of which >90% proteins are encoded by the nuclear genome (Woodson and Chory, 2008). To produce and maintain a functional mitochondrial proteome requires the coordination of nucleus to mitochondrion (anterograde) and mitochondrion to nucleus (retrograde) signals (Woodson and Chory, 2008). Impaired mitochondrial function induces the mitochondrial retrograde regulation (MRR), during which a set of genes are consistently and reproducibly induced, such as ALTERNATIVE OXIDASE 1a (AOX1a), UDP-GLUCOSYLTRANSFERASE 74E2 (UGT74E2), ABSCISIC ACID INSENSITIVE 4 (ABI4), WRKY40, NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON 13 (ANAC013), and ANAC017 (Giraud et al., 2009; Tognetti et al., 2010; Van Aken and Whelan, 2012; De Clercq et al., 2013; Ng et al., 2013; Van Aken et al., 2013; Ivanova et al., 2014; Kerchev et al., 2014).
Increasing evidence suggests that mitochondrial retrograde signals interact with the phytohormone auxin to regulate plant growth and development and plant responses to environmental stresses. Chemicals, such as 3-(2-furyl) acrylic acid (FAA) and Antimycin A (AA), which can activate mitochondrial retrograde signaling, have been found to cause downregulation of auxin responses (Kerchev et al., 2014). On the other hand, perturbed auxin biosynthesis, polar auxin transport, or auxin signaling by genetic or pharmacological methods induce mitochondrial retrograde marker gene expression, revealing that mitochondrial stress signaling and the auxin pathway interact antagonistically to balance growth and stress responses (Ivanova et al., 2014; Kerchev et al., 2014; Zhang et al., 2015). Perturbation of mitochondrial proteome homeostasis activates a plant-specific unfolded protein response, which is mediated by plant hormones including ethylene and auxin (Wang and Auwerx, 2017). Dysfunction of the Arabidopsis mitochondria-located J-protein chaperone AtDjB1 (a homolog of the yeast DnaJ 41 KDa HSP) leads to downregulation of auxin biosynthesis genes, including YUCCA1 (YUC1), YUC2, YUC4, and YUC6, and accordingly YUC2 overexpression in the atdjb1-1 mutant rescues its dwarf phenotype (Jia et al., 2016). It has been shown that mtHSC70-1 mutation impairs mitochondria integrity and induces expression of mitochondrial retrograde marker genes (Wei et al., 2019). However, whether mtHSC70-1 dysfunction affects the auxin pathway remains to be elucidated.
In this study, we show that mtHSC70-1 mutation leads to severe embryo defects. Moreover, abnormal mthsc70-1 embryos resemble those of auxin-defective mutants. Further investigations show that indeed auxin biosynthesis, polar auxin transport, and auxin response are abnormal in the mthsc70-1 embryos. Our data reveal that the mitochondrial HSP70 protein is involved in auxin-mediated embryogenesis.
Results
Loss-of-function mutation of mtHSC70-1 results in vegetative and reproductive growth defects
To study the biological function of the mtHSC70-1 gene, transfer DNA (T-DNA)-insertion alleles were isolated for mtHSC70-1 within the second intron (SALK_081385, mthsc70-1b) and within the third exon (WiscDsLox335G07, mthsc70-1e; Figure 1A). The mthsc70-1b mutant has been reported previously (Wei et al., 2019). Reverse transcription (RT)-PCR analyses revealed that no mtHSC70-1 transcript was detectable in the homozygous mthsc70-1b and mthsc70-1e mutants (Figure 1B), indicating that these mutants are null alleles. Both mthsc70-1b and mthsc70-1e seedlings exhibited severe short-root phenotypes and their etiolated seedlings exhibited short-hypocotyl phenotypes (Figure 1, C and D; Supplemental Figure S1, A and B). Since both mthsc70-1b and mthsc70-1e were knockout mutants and their phenotypes were consistent, our subsequent studies were carried out mainly with mthsc70-1b. In order to determine the cause of the mthsc70-1 short-root phenotype, we compared the length of the root meristem zone, elongation zone, and trichoblast of mthsc70-1b with that of the wild-type (WT). The results showed that mthsc70-1b roots had a shorter meristem zone, a shorter elongation zone, and a shorter trichoblast than that of the WT (Figure 1, E and G–J), indicating that defects in cell division and cell elongation contributed to the mthsc70-1 short-root phenotype. Reduced expression of a cell-cycle marker CYCB1;1:GUS in mthsc70-1b root meristem further confirmed that root cell dividing activity was considerably decreased in the mthsc70-1 mutant compared with that of the WT (Figure 1F). The mthsc70-1b plants exhibited a smaller rosette and a shorter plant height compared to that of the WT over a period of 4 weeks (Supplemental Figure S1, C–E). The first internode of mthsc70-1b plants was also shorter than that of WT, and the stem of mthsc70-1b plants was significantly thinner than that of the WT (Supplemental Figure S2, A–C). Some phenotypes of mthsc70-1b, including short root, small rosette, and reduced plant height were consistent with previously reported data (Wei et al., 2019).
Figure 1.
Expression and phenotypic analysis of T-DNA insertion mutants of the mtHSC70-1 gene. A, Structure of the mtHSC70-1 gene with T-DNA insertion sites. Flags indicate T-DNA insertion sites; arrows indicate the positions of gene-specific primers used for PCR verification of the insertions. B, RT-PCR analysis of mtHSC70-1 gene transcription in WT Columbia-0 (Col) and mthsc70-1 mutants, the ACTIN2 gene was used as an internal control. C and D, Roots of WT, mthsc70-1b, and mthsc70-1e seedlings grown on MS medium under a 16-h light/8-h dark photoperiod for 7 d. E and G, Meristem zone and trichoblast of 5-d-old WT and mthsc70-1b seedling roots. White arrowheads indicate the root meristem boundary; orange arrowheads indicate the cell boundary. F, Expression patterns of CYCB1;1:GUS in WT and mthsc70-1b root tips. H–J, Meristem zone length (H), elongation zone length (I), and trichoblast length (J) of 5-d-old WT and mthsc70-1b seedling roots. The data are derived from three independent experiments and are presented as means ± sd (n = 30 roots per experiment). K and L, 8 DPA siliques of WT (K) and mthsc70-1b (L) plants. White arrowheads indicate unfertilized ovules; orange arrows indicate aborted seeds. M–P, Silique length (M), total seed numbers per silique (N), percentage of aborted seeds (O), and unfertilized ovules (P) of 8-DPA WT and mthsc70-1b siliques. The data are derived from three independent experiments and are presented as means ± sd (n = 10 siliques from 10 plants per experiment). Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; *P < 0.05; **P < 0.01; and ***P < 0.001). Bars = 5 mm (C), 50 μm (E–G), and 1 mm (K and L).
In addition to these vegetative growth defects, the mthsc70-1 mutants exhibited fertility defects. The mthsc70-1b and mthsc70-1e plants bore short siliques (Figure 1, K–M; Supplemental Figure S3, A and B). Flower morphology of mthsc70-1 plants was abnormal with small and shrunken petals and sepals that could not completely enclose the pistil at each developmental stage, causing the pistil to be exposed before flowering (Supplemental Figure S2, D–G). The pistil of mthsc70-1 opened flowers seemed longer than that of the WT (Supplemental Figure S2, J and K), and quantification results showed that indeed the average length of mthsc70-1b pistils was significantly longer than that of the WT (Supplemental Figure S2N). Whereas the anther filaments of mthsc70-1b opened flowers were slightly shorter than that of the WT, they were not statistically different (Supplemental Figure S2O). The length ratio of filaments/pistils in WT opened flowers was approximately 1; however, the same ratio was only 0.77 in the mthsc70-1b opened flowers (Supplemental Figure S2P). Although the mthsc70-1b pistil was longer than that of the WT, the number of ovules it contained was not significantly different from that of the WT (Supplemental Figure S2, Q–S). This result indicates that ovule density of the mthsc70-1b pistil is low. Moreover, mthsc70-1b anthers were smaller than those of the WT and contained fewer or no pollen grains (Supplemental Figure S2, L and M).
Because mthsc70-1 opened flowers had anther filaments shorter than the pistil, smaller anthers and fewer pollen grains, and produced short siliques, we wondered whether mthsc70-1 fertility was affected. To determine this, we observed the seed set of mthsc70-1 siliques. Compared to normal seed set in WT siliques (Figure 1K;Supplemental Figure S3A), there were unfertilized ovules and white or yellow wrinkled seeds in mthsc70-1b and mthsc70-1e siliques (Figure 1L;Supplemental Figure S3A). Quantification results showed that WT siliques contained approximately 55 seeds on average; however, mthsc70-1b and mthsc70-1e siliques only contained approximately 20 and 15 seeds, respectively (Figure 1N;Supplemental Figure S3C), among which approximately 16.4% in mthsc70-1b and 28.7% in mthsc70-1e were aborted (Figure 1O;Supplemental Figure S3D). Consistently, siliques from mthsc70-1b and mthsc70-1e plants had approximately 51% and 59% unfertilized ovules, respectively, compared with less than 2% in that of the WT (Figure 1P;Supplemental Figure S3E). These results suggest that mtHSC70-1 mutation affects embryogenesis and fertilization.
To investigate whether the reduced fertility in mthsc70-1 was caused by a longer pistil, fewer pollen grains, or male/female gamete defects, we performed reciprocal crosses between mthsc70-1 and WT plants. With WT pollen as the male donor used to pollinate mthsc70-1 stigmas, nearly half of the ovules in the crossed siliques were not fertilized, suggesting that ovule development in mthsc70-1 plants was defective (Supplemental Figure S4). When the pistils of WT were pollinated with mthsc70-1 pollen grains, the crossed siliques still had nearly half unfertilized ovules, suggesting that pollen development or pollen tube growth was also defective in mthsc70-1 plants (Supplemental Figure S4). Consistently, mthsc70-1 plants produced smaller anthers, and Alexander staining showed that there were three types of mthsc70-1 anthers (Supplemental Figure S5, A–D). The type-1 anthers were normal with all locules containing pollen grains (56%, n = 98; Supplemental Figure S5B). The type-2 anthers only had one or two locules containing pollen grains (32%, n = 98; Supplemental Figure S5B). The type-3 anthers had no locules containing pollen grains (12%, n = 98; Supplemental Figure S5B). Then we analyzed pollen viability in WT anthers and in the type-1 mthsc70-1b anthers from opened flowers. The results showed that only approximately 88.3% of mthsc70-1b pollen grains were normal, as compared with approximately 96.8% in the WT (Supplemental Figure S5, E–G). This result implies that pollen germination or pollen tube growth might be impaired in mthsc70-1 plants, which would also contribute to the low fertilization rate (<50%) in the WT (female) × mthsc70-1 (male) crossed silique. Interestingly, we found that when the WT plants were used as the female parent, crossed siliques showed no aborted seed (Supplemental Figure S4). In contrast, when the mthsc70-1 plants were used as the female parent, crossed siliques produced approximately 5.8% aborted seeds (Supplemental Figure S4). Although this seed abortion rate was significantly lower than that of the self-crossed siliques (Supplemental Figure S4), this finding clearly suggests that the seed-abortion phenotype of the mthsc70-1 mutant is partially due to the mother plant.
To verify whether the above described phenotypes of the mthsc70-1 mutant are caused by the lack of mtHSC70-1 function, a translational fusion between mtHSC70-1 coding sequence and the GFP reporter gene driven by the mtHSC70-1 promoter (mtHSC70-1p:mtHSC70-1-EGFP) was introduced into the mthsc70-1b mutant. All mthsc70-1b phenotypes, including short root, small rosette, reduced stature, abnormal flowers, short siliques, unfertilized ovules, and aborted seeds, were restored to WT levels in the complementation lines (Supplemental Figure S6), confirming that these phenotypes are indeed caused by mtHSC70-1 loss of function.
mtHSC70-1 mutation leads to defective embryogenesis
To determine at which developmental stage the mthsc70-1 seeds were aborted, we observed embryos from 2- to 8-d postanthesis (DPA) siliques of WT and mthsc70-1b plants. The WT embryos showed normal cell division pattern and morphology throughout the developmental process (Figure 2, A, E, I, L, and O). In contrast, the mthsc70-1b embryos displayed aberrant cell division in embryo proper or suspensor and were abnormal in morphology at various developmental stages (Figure 2, B–D, F–H, J–K, M–N, and P–R). In addition, the mthsc70-1 embryos displayed a delayed embryonic development compared to that of the WT (Figure 2S). In 2-DPA WT siliques, most embryos were at 2-, 8-, 16-, and 32-cell stages (∼28.3%, 21.4%, 25.5%, and 19.3%, respectively, n = 145; Figure 2S). However, in 2 DPA mthsc70-1b siliques, most embryos were at 1-, 2-, 8-, and 16-cell stage (∼35.4%, 32.3%, 15.6%, and 10.4%, respectively, n = 96), and approximately 4.2% embryos were abnormal (Figure 2S, and compare Figure 2, B–D with Figure 2, A). These abnormal mthsc70-1 embryos showed cell-division defects in the embryo proper (Figure 2B) or in the suspensor with some suspensor cells divided longitudinally, resulting in an enlarged multicellular structure (Figure 2, C and D). In 3-DPA WT siliques, most embryos were at 32-cell and globular stages (∼26.7% and 39.3%, respectively, n = 135; Figure 2S). However, in 3-DPA mthsc70-1b siliques, most embryos were at 8- and 16-cell stages (∼24.3% and 30.1%, respectively, n = 103), and ∼9.7% embryos were abnormal (Figure 2S). Some abnormal mthsc70-1 embryos were pear-like or olive-like rather than globular in shape due to defective cell divisions in the embryo proper (compare Figure 2, F–G with Figure 2, E) and some abnormal mthsc70-1 embryos displayed defective cell divisions in the suspensor (Figure 2H). In 4-DPA WT siliques, ∼61.6% embryos developed to heart stage (n = 219; Figure 2, I and S). However, in 4-DPA mthsc70-1b siliques, most embryos were still at globular stage (∼43%, n = 114), and ∼19.3% embryos were abnormal (Figure 2S). These abnormal mthsc70-1 embryos failed to generate cotyledonary primordia and were olive-like or ball-like rather than heart-like in shape (compare Figure 2, J and K with Figure 2, I). In 5- and 6-DPA WT siliques, most embryos developed to torpedo stage (∼70% in 5-DPA siliques, n = 217 and 75.3% in 6-DPA siliques, n = 219) and almost no embryos stayed at globular stage (Figure 2, L and S). However, in 5-DPA mthsc70-1b siliques, most embryos were at heart stage (∼42.6%;z n = 108), ∼31.5% embryos were still at globular stage, and ∼11.1% embryos were abnormal (Figure 2S). In 6-DPA mthsc70-1b siliques, the percentage of abnormal embryos was increased to ∼44.6% (n = 92; Figure 2S). These abnormal mthsc70-1 embryos either had no cotyledon protrusion (Figure 2M) or had two or three very small cotyledons (Figure 2N). In 7-DPA WT siliques, most embryos developed to bent and mature stages (∼44.3% and 47.1%, respectively, n = 140). However, in 7-DPA mthsc70-1b siliques, most embryos also developed to mature stage (∼38.5%, n = 104), whereas ∼36.5% embryos were abnormal (Figure 2S). These abnormal mthsc70-1 embryos displayed four types of morphology: (1) had no cotyledon protrusion (Figures 2M and 10; Supplemental Figure S7, G); (2) had two or three very small cotyledons (Figures 2N and 10); (3) had one or three normal sized cotyledons (Figure 2, Q and R; Supplemental Figure S7, I and K); (4) twisted in the hypocotyl or cotyledon (Figures 2P and 10; Supplemental Figure S7, C and E). These abnormal-embryo phenotypes could persist into seedling stage (Supplemental Figure S7). Seven-day-old mthsc70-1 seedlings displayed a variety of aberrations, including slender curving cotyledons (Supplemental Figure S7D), bent hypocotyl (Supplemental Figure F7F), without cotyledon (Supplemental Figure S7H), or bearing one or three cotyledons (Supplemental Figure S7, J and L) compared with the two symmetrical cotyledons in WT seedlings (Supplemental Figure S7B). These results indicate that mtHSC70-1 is involved in embryo development and pattern formation.
Figure 2.
Phenotypes of mthsc70-1b embryos at different developmental stages. Shown are DIC images of WT (A, E, I, L, O) and mthsc70-1b (B–D, F–H, J–K, M–N, P–R) embryos. A–D, Preglobular stage embryos of WT (A) and mthsc70-1b (B–D). E–H, Globular stage embryos of WT (E) and mthsc70-1b (F–H). I–K, Heart stage embryos of WT (I) and mthsc70-1b (J and K). L–N, Torpedo stage embryos of WT (L) and mthsc70-1b (M and N). O–R, Mature embryos of WT (O) and mthsc70-1b (P–R). Arrowheads indicate the cotyledons. S, Embryonic developmental stage quantification of WT and mthsc70-1b seeds from 2- to 7-DPA siliques. Note that embryos displaying an abnormal shape were counted as abnormal. Approximately 92–219 embryos were counted in each period. Bars = 50 μm (A–R).
The mtHSC70-1 gene is expressed during embryonic and postembryonic development
To explore the expression pattern of the mtHSC70-1 gene during embryo development, we used the functional mtHSC70-1p:mtHSC70-1-EGFP (mtHSC70-1-EGFP) transgenic plants in the mthsc70-1b mutant background. The mtHSC70-1-EGFP signal was first detected in the early heart embryos, which was confined to the inner cells of the embryo proper (Figure 3A). Signal intensity increased in late heart and torpedo embryos, and was specifically detected in the provascular cells (Figure 3, B and C). In bent and mature embryos, the mtHSC70-1-EGFP signal was strong in the provascular cells of the hypocotyl, whereas only very weak signal was detected in the provascular strands of the cotyledons (Figure 3, D and E). In seedling roots, mtHSC70-1-EGFP expression was strongly detected in the stele and columella root cap cells (Supplemental Figure S8). These results show that the expression patterns of mtHSC70-1 correlate with its roles in embryogenesis and postembryonic development.
Figure 3.
Dynamic expression of the mtHSC70-1 gene during embryo development. A–E, Microscope images of GFP fluorescence (green, in the left) and merged GFP and bright field images (right) of embryos at early-heart stage (A), late-heart stage (B), torpedo stage (C), bent stage (D), and mature stage (E) from the mtHSC70-1p:mtHSC70-1-EGFP transgenic plants in the mthsc70-1b mutant background. Shown are representative images of two independent experiments (n = 100 embryos from five plants per experiment). Bars = 50 μm.
Abnormal mthsc70-1 embryos are associated with impaired auxin response, reduced auxin biosynthesis, and defective polar auxin transport
Auxin is a key regulator of embryogenesis and cotyledon formation (Friml et al., 2003). Phenotypes of the mthsc70-1 embryos are similar to that of auxin-deficient mutants (Cheng et al., 2006, 2007; Robert et al., 2015). To determine whether the defective embryogenesis in the mthsc70-1 mutant was related to auxin, we crossed DR5rev:GFP (an auxin-responsive reporter; Friml et al., 2003) with the mthsc70-1b mutant and examined the GFP signals in the embryos. In WT early heart and late heart embryos, the maximum of DR5rev:GFP activity was observed in the hypophysis and the uppermost suspensor cell, and DR5rev:GFP signal was also detected in the tips of the cotyledon primordia and in the provascular cells (Figure 4, A and B). Compared with WT, the mthsc70-1 embryos displayed relatively weaker DR5rev:GFP activity in general (Figure 4, E–H). In addition, in the apical region of some of the mthsc70-1 embryos, DR5rev:GFP signal was either present throughout the apical epidermis (Figure 4E) or was completely absent (Figure 4H), indicating that no auxin activity maximum was established in these embryos to define the cotyledon primordia. Indeed, these embryos displayed no cotyledon protrusion (Figure 4, E and H). In the provascular cells of the mthsc70-1 embryos, DR5rev:GFP signal was either undetectable (Figure 4, E, F and H) or was diffuse (Figure 4G). In the basal region of the mthsc70-1 embryos, DR5rev:GFP signal was sometimes expanded into all of the suspensor cells (Figure 4F), which might be the cause of the longitudinal cell divisions in the suspensor. In WT torpedo and mature embryos, the maximum of DR5rev:GFP activity was observed in the quiescent center (QC) of the root meristem and the adjacent columella precursors and in the tips of the developing cotyledons (Figure 4, C and D). DR5rev:GFP signal was also detected in the provascular strands (Figure 4, C and D). However, in the provascular strands of the mthsc70-1 embryos, DR5rev:GFP signal was either very weak (Figure 4I) or was completely absent (Figure 4J). In the apical region of the mthsc70-1 embryos, DR5rev:GFP signal was sometimes present throughout the apical epidermis (Figure 4J). Interestingly, additional symmetric or asymmetric DR5rev:GFP signal was observed in the epidermis at the basal region of the developing hypocotyl of the mthsc70-1 embryos (Figure 4, I, K, and L). Furthermore, the asymmetric DR5rev:GFP distribution was associated with the bent-hypocotyl phenotype of the abnormal mthsc70-1 embryos (Figure 4 K). These results indicate that auxin activity and distribution are altered in the mthsc70-1 embryos.
Figure 4.
Expression patterns of DR5rev:GFP in WT and mthsc70-1b embryos. Shown are DR5rev:GFP signal distributions in WT embryos at early heart stage (A), late heart stage (B), torpedo stage (C), and mature stage (D) and DR5rev:GFP signal distributions in mthsc70-1b embryos at early heart stage (E and F), late heart stage (G and H), torpedo stage (I and J), and mature stage (K and L). The images in the left in (A–L) are GFP fluorescence in green and the images in the right are merged GFP and bright field images. The white arrow in (F) indicates abnormal DR5rev:GFP signal in the suspensor; white arrowheads in (I and L) indicate ectopic but symmetric DR5rev:GFP signal at the base of the hypocotyl; orange arrowheads in (K) indicate ectopic and asymmetric DR5rev:GFP signal at the base of the hypocotyl. Shown are representative microscope images of three independent experiments (n = 100 embryos from 15 plants per experiment). Bars = 50 μm.
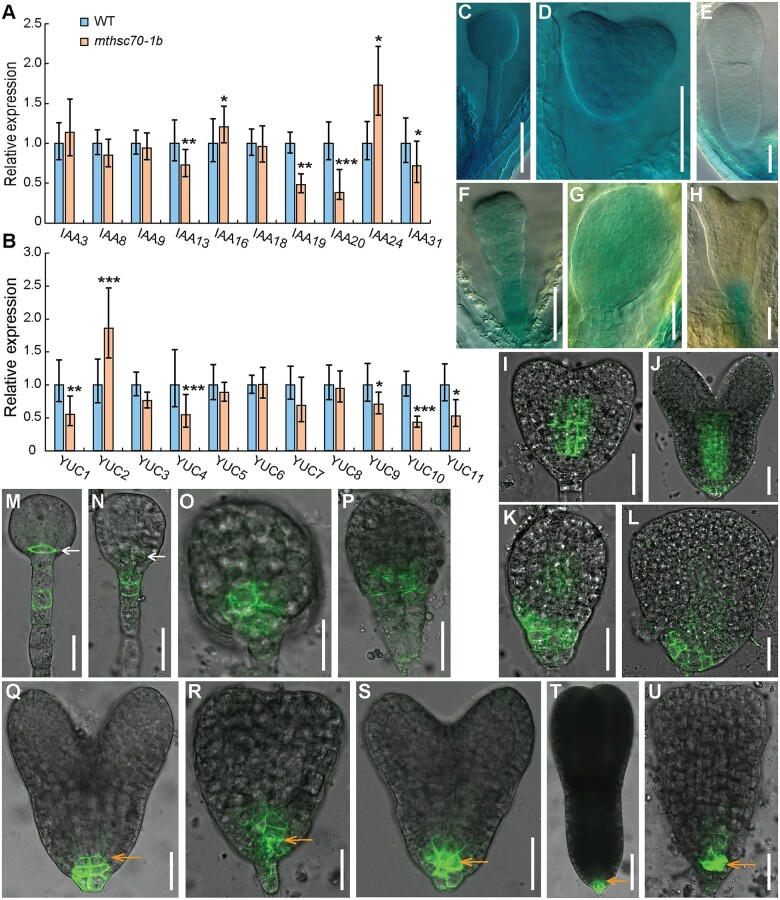
Several auxin-responsive AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) genes, including IAA3, IAA8, IAA9, IAA13, IAA16, IAA18, IAA19, IAA20, IAA24, and IAA31 that are expressed during embryo development, were selected to check their expression in 1–3-DPA siliques from WT and mthsc70-1b plants by reverse transcription quantitative PCR (RT-qPCR). The results showed that expression levels of IAA13, IAA19, IAA20, and IAA31 genes were significantly downregulated in the mthsc70-1b siliques compared with that in the WT siliques, whereas expression of IAA16 and IAA24 was upregulated (Figure 5A).
Figure 5.
Expression analyses of auxin-related genes in WT and mthsc70-1b siliques or embryos. A and B, Relative transcription levels of IAA (A) and YUCCA (YUC) (B) genes in 1–3-DPA siliques. The transcription levels were normalized to that of TAP42 INTERACTING PROTEIN OF 41 KDA (TIP41, AT4G34270). Values are means ± sd of three independent experiments. Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; *P < 0.05; **P < 0.01; and ***P < 0.001). C–H, Expression patterns of ProYUC9:GUS in WT (C–E) and mthsc70-1b (F–H) embryos at globular (C and F), heart (D and G), and torpedo (E and H) stages. Shown in (C–H) are representative images of three independent experiments (n = 100 embryos from 15 plants per experiment). I–L, AUX1-YFP localization in WT (I and J) and mthsc70-1b (K and L) embryos at early-heart (I and K) and late-heart (J and L) stages. M-U, PIN3-GFP localization in WT (M, Q, and T) and mthsc70-1b (N–P, R–S, and U) embryos at globular (M–P), heart (Q–S), and torpedo (T and U) stages. White arrows indicate the lens-shaped cell; orange arrows indicate the QC or putative QC cells. Shown are representative images of 38 embryos per genotype. Bars = 50 μm (C–H) and 25 μm (I–U).
The distribution of auxin is regulated by its local biosynthesis and polar transport, and the flavin monooxygenases YUC family proteins are key enzymes in Trp-dependent auxin biosynthesis (Zhao et al., 2001; Cheng et al., 2006, 2007). To determine whether auxin biosynthesis is altered in the mthsc70-1 mutant, YUC1–11 genes were selected to check their expression in 1–3 DPA siliques from WT and mthsc70-1b plants by RT-qPCR. The results showed that expression of YUC1, YUC4, YUC9, YUC10, and YUC11 was significantly reduced in the mthsc70-1b siliques compared with that in the WT siliques (Figure 5B). To analyze the expression patterns of the auxin biosynthesis genes in the mthsc70-1 mutant, we introduced the ProYUC9:GUS transgene (Hentrich et al., 2013) into the mthsc70-1b mutant by crossing and analyzed the expression of the GUS reporter gene in developing embryos. In WT, ProYUC9:GUS was expressed throughout the embryo at the globular and heart stages (Figure 5, C and D). Following this, its expression became weaker at the torpedo embryo stage and its expression was only detectable in the suspensor (Figure 5E). Compared with WT, mthsc70-1b displayed reduced ProYUC9:GUS expression in the globular and heart stage embryos (Figure 5, F–H). Together, these results indicate that in the mthsc70-1b embryos, the expression of auxin biosynthesis genes is reduced.
AUX1/LAX-dependent auxin influx is required for embryo patterning (Marchant et al., 1999; Robert et al., 2015). To explore whether AUX1 localization is altered in the mthsc70-1 embryos, we introduced AUX1-YFP into the mthsc70-1b mutant (Swarup et al., 2004). In WT early heart and late heart embryos, AUX1-yellow fluorescent protein (YFP) was mainly detected at the basal plasma membrane of the provascular cells (Figure 5, I and J). Weak AUX1-YFP signal was also detected at the whole plasma membrane of the hypophysis and the several epidermal cells around the hypophysis in late heart embryos (Figure 5J). In contrast, mthsc70-1b abnormal embryos displayed weaker or no AUX1-YFP signal in the provascular cells (Figure 5, K and L). In addition, AUX1-YFP signal was strongly detected at the whole plasma membrane of the epidermal cells at the basal part of the mthsc70-1b abnormal embryos (Figure 5, K and L). These results demonstrate that the expression and localization of the auxin influx carrier AUX1 are altered in the mthsc70-1b embryos.
In addition to auxin influx carriers, polar localization of PIN auxin efflux carriers is also essential for the establishment and maintenance of correct auxin gradient during early embryo development (Friml et al., 2003; Möller and Weijers, 2009). Since the mthsc70-1 embryos showed defects in the lower region, we analyzed the expression of PIN3-GFP (Friml et al., 2002, 2003). In WT globular embryos, PIN3-GFP was detected at the whole plasma membrane of the lens-shaped cell and at the basal plasma membrane of the upper suspensor cells (Figure 5M). In contrast, in mthsc70-1b globular-like embryos, PIN3-GFP signal around the lens-shaped cell was weaker (Figure 5N). Sometimes, the lens-shaped cell was not distinguishable and PIN3-GFP signal was detected in the plasma membrane of several cells at the basal part of the proembryo (Figure 5, O and P). In WT heart and torpedo embryos, PIN3-GFP signal was detected at the plasma membrane of the QC and the adjacent columella precursors (Figure 5, Q and T). In contrast, in mthsc70-1b heart and torpedo stage embryos, cells in the root pole were disorganized and PIN3-GFP signal was additionally detected in several cells above the putative QC cells (Figure 5, R, S and U). These results demonstrate that the expression pattern of the auxin efflux carrier PIN3 is altered in the mthsc70-1 embryos.
The data described above demonstrate that reduced auxin biosynthesis and abnormal auxin transport and distribution are correlated with the mthsc70-1 embryonic patterning defects. We, thus, wondered whether auxin application could rescue the mthsc70-1 embryonic patterning defects. Therefore, we performed exogenous auxin application on pistils of mthsc70-1b opened flowers and examined seed development 10 d later. Compared with the mock treatment, indole-3-acetic acid (IAA), 1-naphthaleneacetic acid (NAA), or 2,4-dichlorophenoxyacetic acid (2,4-D) application significantly reduced the seed abortion rate of mthsc70-1b siliques (Figure 6).
Figure 6.
Exogenous application of auxin partially rescues mthsc70-1b embryonic lethality. A, C, and E, 10-DPA siliques of WT and mthsc70-1b after mock or 100 μM IAA (A), 500 μM NAA (C), or 100 μM 2,4-D (E) treatment. Orange arrows indicate aborted seeds. B, D, and F, Percentage of aborted seeds in WT and mthsc70-1b siliques after mock or 100 μM IAA (B), 500 μM NAA (D), or 100 μM 2,4-D (F) treatment. For each experiment, pistils of 30 opened flowers from 15 plants of each genotype were either treated with IAA, NAA, or 2,4-D or underwent mock treatment, and aborted seeds were scored 10 d after application. Values are means ± sd of three independent experiments. Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; *P < 0.05). Bars = 1 mm.
Taken together, these data indicate that reduced auxin biosynthesis and perturbed polar auxin transport result in abnormal auxin activity in mthsc70-1 embryos, and exogenous application of auxin can partially rescue mthsc70-1 embryonic lethality.
Expression of auxin biosynthesis and polar auxin transport genes is reduced in mthsc70-1b roots, and mutant roots display lower sensitivity to auxin
The data described above suggest that reduced auxin activity and abnormal auxin distribution and transport are correlated with the mthsc70-1b embryonic patterning defects. We wondered whether these auxin-related changes also occurred in the mutant roots. Therefore, we first examined the expression pattern of DR5rev:GFP in WT and mthsc70-1b roots. Consistent with the results in the embryos, DR5rev:GFP activity was reduced in mthsc70-1 roots compared with that of the WT (Supplemental Figure S9). We then analyzed the expression patterns of auxin biosynthesis genes, including ProYUC8:GUS and ProYUC9:GUS, and polar auxin transport genes, including AUX1-YFP, PIN1-GFP, PIN2-GFP, PIN3-GFP, and PIN7-GFP (Swarup et al., 2004; Benková et al., 2003; Xu and Scheres, 2005; Friml et al., 2003), in WT and mthsc70-1b roots. The expression of ProYUC8:GUS was reduced in mthsc70-1b roots compared with that of the WT (Figure 7A). As for ProYUC9:GUS, we noticed that there were three different expression patterns in mthsc70-1b roots (Figure 7B); the main type was similar to but weaker than that of the WT (64%), whereas other types either had no GUS staining in the columella root cap cells (21%) or had no GUS staining in the elongation zone (15%). These results indicate that expression of ProYUC8:GUS and ProYUC9:GUS is downregulated in mthsc70-1b roots. Whereas the expression pattern and localization of the AUX1-YFP, PIN1-GFP, PIN2-GFP, PIN3-GFP, and PIN7-GFP auxin influx and efflux carriers were not altered in the mthsc70-1b root, their expression levels were all reduced compared with that of the WT (Figure 7, C–G). Together, these results indicate that auxin activity and expression levels of auxin biosynthesis and polar transport genes are decreased in the mthsc70-1b root.
Figure 7.
Expression patterns of auxin biosynthesis and polar transport-related genes in WT and mthsc70-1b roots. A and B, Expression patterns of ProYUC8:GUS (A) and ProYUC9:GUS (B) in WT and mthsc70-1b roots. C–G, Localization of the AUX1-YFP auxin influx carrier (C) and the PINs-GFP auxin efflux carriers, such as PIN1 (D), PIN2 (E), PIN3 (F), and PIN7 (G) in WT and mthsc70-1b roots. The images in the left panel of each genotype in (C–G) are YFP or GFP fluorescence in green and the images in the right panel are merged fluorescence and bright field images. Shown are representative images of two and three independent experiments (n = 10–20 roots per experiment). Bars = 50 μm.
To determine whether the mthsc70-1b mutant responds to auxin differently from the WT, we treated WT and mthsc70-1b seedlings with various concentrations of NAA and 2,4-D. The results showed that at higher NAA or 2,4-D concentrations, the mthsc70-1b mutant roots were less sensitive compared to WT (Supplemental Figures S10 and S11). These results might be due to reduced exogenous auxin uptake in mthsc70-1b because of the reduced expression level of auxin influx carrier AUX1, or alternatively due to lower auxin activity in mthsc70-1b.
Loss-of-function mutation of mtHSC70-1 reduces mitochondrial activity and activates MRR, which inhibits auxin biosynthesis and polar auxin transport
A recent study has shown by transmission electron microscopy that knockout of mtHSC70-1 affects mitochondrial morphology (Wei et al., 2019). We further explored mitochondrial activity in protoplasts generated from leaves of WT and mthsc70-1b plants by using the 5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) dye, which forms aggregates emitting red fluorescence in healthy mitochondria and monomers emitting green fluorescence in depolarized mitochondria (Salvioli et al., 1997). We found that mthsc70-1b protoplasts displayed higher JC-1 monomer content than the WT (Figure 8, A–C), suggesting that mthsc70-1b mitochondrial membrane potential and mitochondrial activity are compromised. To explore how the knockout of mtHSC70-1 affects mitochondrial activity, we conducted a quantitative proteomics analysis of mitochondrial protein extracts from inflorescence tissues of WT and mthsc70-1b plants. Gene ontology enrichment analysis of the differentially expressed proteins in mthsc70-1b indicated that knockout of mtHSC70-1 affects many biological processes in mitochondria, such as urea cycle, oxidative phosphorylation, ATP metabolic process, oxaloacetate metabolic process, cell redox homeostasis, respiratory electron transport, and protein folding (Figure 8D).
Figure 8.
Mitochondrial function defects in mthsc70-1. A and B, Representative histograms showing the flow cytometry analysis on 5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1)-stained protoplasts from WT (A) and mthsc70-1b (B) leaves. C, The ratio of JC-1 aggregates to monomers for WT and mthsc70-1b protoplasts. Values are means ± sd of three independent experiments. Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; *P < 0.05). D, Gene ontology enrichment analysis of differentially expressed mitochondrial proteins from inflorescence tissues between WT and mthsc70-1b plants. E, Relative transcription levels of mitochondria dysfunction stress genes in 7-d-old WT and mthsc70-1b seedlings. Values are means ± sd of three independent experiments. Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; **P < 0.01).
Since loss-of-function mutation of mtHSC70-1 lead to mitochondrial dysfunction and impaired respiration (Wei et al., 2019), and that the above results showed that the mitochondria activity is compromised in mthsc70-1b plants, we wondered whether mtHSC70-1 mutation triggers MRR. To address this, we analyzed the transcription level of MRR marker genes including AOX1a, ANAC013, and UGT74E2 (Tognetti et al., 2010; De Clercq et al., 2013) in 7-d-old WT and mthsc70-1b seedlings. The results showed that the expression of AOX1a and ANAC013 was significantly upregulated in mthsc70-1b seedlings compared to WT (Figure 8E), suggesting that the MRR was activated in the mthsc70-1b mutant.
FAA treatment activates the mitochondrial dysfunction and mitochondrial retrograde signaling pathways (Kerchev et al., 2014). To explore whether FAA treatment could induce similar phenotypes as that in the mthsc70-1 mutant, we treated WT seedlings and pistils with various concentrations of FAA. The results showed that the root lengths of WT seedlings gradually became shorter as FAA treatment concentration increased (Figure 9, A and B) and that aborted seeds were produced in WT siliques (Figure 9, C and D).
Figure 9.
FAA treatment of WT seedlings and opened flowers mimics the short-root and seed-abortion phenotypes of the mthsc70-1 mutant and downregulate expression of auxin biosynthesis and polar transport genes. A and B, Phenotypes (A) and root length (B) of WT seedlings grown on MS medium supplemented with various concentrations of FAA for 7 d. The data are derived from three independent experiments and are presented as means ± sd (n = 90 seedlings per experiment). Significant differences were analyzed using Student’s t test (one-tailed, two-sample equal variance; **P < 0.01; ***P < 0.001). C and D, Phenotypes (C) and seed abortion rates (D) of WT siliques 10 d after FAA treatment of various concentrations (n = 15 opened flowers from 15 plants per treatment). Orange arrows in (C) indicate aborted seeds. E, Effects of FAA on expression of ProYUC8:GUS, ProYUC9:GUS, and ProPIN7:GUS. Five-day-old seedlings were treated with 20 μM FAA for 1 h. Shown are representative images of two experiments (n = 20 roots per experiment). 0.1% (v/v) DMSO was used as a mock treatment. Bars = 5 mm (A), 2 mm (C), and 50 μm (E).
We further analyzed whether FAA treatment could affect the expression of auxin biosynthesis and transport genes. The results showed that FAA treatment downregulated the expression of ProYUC8:GUS, ProYUC9:GUS, and ProPIN7:GUS (Figure 9E).
Together, these results suggest that the FAA inhibitor can downregulate the transcription level of auxin biosynthesis and transport genes and induce similar phenotypes as the mthsc70-1 mutant.
Based on our studies, we propose that loss-of-function mutation of the mtHSC70-1 gene compromises mitochondria activity leading to activation of mitochondrial retrograde signaling, which inhibits the expression of auxin biosynthesis and polar transport genes, resulting in abnormal auxin gradients and ultimately affecting embryo patterning and symmetric hypocotyl elongation (Figure 10).
Figure 10.
Working model for mtHSC70-1 in embryo development. mtHSC70-1 loss-of-function mutation leads to mitochondrial dysfunction and activated mitochondrial retrograde signaling, which inhibits the expression of auxin biosynthesis and polar transport genes, resulting in abnormal auxin gradients. This ultimately affects embryo patterning and symmetric hypocotyl elongation.
Discussion
mtHSC70-1 plays important roles in embryonic and postembryonic development
In this study, we found that loss-of-function mutation of mtHSC70-1 not only leads to defects in vegetative growth but also causes defects in reproduction. The vegetative growth defects of the mthsc70-1 mutant include short root, small rosette, dwarf stature, and thin stem. Cell biological analysis revealed that the short-root phenotype of the mthsc70-1 mutant is due to defects in cell division and cell elongation. The reproductive defects of the mthsc70-1 mutant include malformed flowers, small anthers with less or no pollen grains, short siliques with low seed-setting rates, male and female gametophyte development defects, and embryo development retardation and abortion. We further demonstrated that mtHSC70-1 mutation downregulates expression of auxin biosynthesis genes and polar auxin transport genes and disrupts expression patterns of some of these genes, which results in aberrant auxin activity distribution during embryo development, thus, causing abnormal embryo patterning of the mthsc70-1 mutant. Consistent with the previous report by Wei et al. (2019), we observed no obvious phenotype in the mthsc70-2 mutant under normal growth conditions. Previous studies found that transcription levels of the mtHSC70-2 gene in various organs of Arabidopsis plants grown under normal conditions were much lower than that of the mtHSC70-1 gene except in mature seeds, dry seeds, and imbibed seeds (Sung et al., 2001; Wei et al., 2019). These findings imply that the mtHSC70-1 gene plays more important roles than the mtHSC70-2 gene in Arabidopsis development and that the mtHSC70-2 gene might have some specific roles in seed maturation, seed dormancy, and seed germination. A cross of the mthsc70-1 mutant with the mthsc70-2 mutant to generate a double mutant and subsequent phenotypic analysis of the offspring will determine whether mtHSC70-1 and mtHSC70-2 genes share redundant functions during plant development. Whereas both mtHSC70-1 and mtHSC70-2 genes were induced by cold acclimation, only the mtHSC70-2 gene was induced by heat-shock treatment (Sung et al., 2001). This finding suggests that the mtHSC70-2 gene might have a specific role in Arabidopsis responses to heat stress.
The mtHSC70-1 gene regulates embryogenesis via an auxin-related mechanism
It is known that the plant hormone auxin plays a crucial role in embryogenesis. Local auxin biosynthesis and polar auxin transport coordinate to establish the auxin maxima in developing embryos, which defines the embryonic axis and subsequent embryo pattern formation (Möller and Weijers, 2009; Mironova et al., 2017). Studies have shown that mutations in the auxin biosynthesis genes TAA1, TAR1, and TAR2 cause defects in embryo development, and the wei8 tar1 tar2 triple mutant exhibits monopteros (mp)-like (mp is an auxin signaling mutant; Hardtke and Berleth, 1998) embryo and seedling phenotypes (Stepanova et al., 2008). Similar embryo defects were observed in the yuc1 yuc4 yuc10 yuc11 quadruple mutants (Cheng et al., 2007). Dysfunction of the AUX1, LAX1, and LAX2 auxin influx proteins cause defects in apical–basal axis establishment at globular embryo stage, and the hypophysis of the aux1 lax1 lax2 triple mutant embryos starts vertical symmetric division instead of horizontal asymmetric division at heart stage, eventually forming embryos with abnormal morphology (Robert et al., 2015). Deletion of four PIN auxin efflux proteins expressed in embryos, including PIN1, PIN3, PIN4, and PIN7, also causes a defective embryonic development (Friml et al., 2003).
In this study, we found that morphology of the mthsc70-1 embryos was very similar to those of auxin-deficient mutants, especially the aux1 lax1 lax2 lax3 quadruple mutant (Robert et al., 2015). Indeed, the expression of genes related to auxin biosynthesis and polar auxin transport were altered in the mthsc70-1 mutant. In particular, AUX1-YFP and PIN3-GFP were ectopically expressed in mthsc70-1 embryos. These alterations resulted in abnormal auxin activity distribution as indicated by expression of the auxin-responsive reporter DR5rev:GFP (Figure 4). Furthermore, these abnormal auxin activity distributions were consistent with the phenotypes of mthsc70-1 embryos. For example, in those mthsc70-1 embryos without cotyledon protrusion, the DR5rev:GFP signal was either evenly distributed in the apical epidermis or was completely absent, and, therefore, no maximum auxin activity was established to define the cotyledon primordia (Figure 4, E, H, and J). However, in those mthsc70-1 embryos with a bent-hypocotyl phenotype, asymmetric DR5rev:GFP signal was observed in the epidermis at the bending region of the hypocotyl (Figure 4K). Interestingly, the expression pattern of mtHSC70-1 during embryo development was very similar to that of the AUX1 and LAX2 genes (Robert et al., 2015), suggesting that they function in the same cell file. As shown in Figure 3, the mtHSC70-1 gene is expressed in the inner cells of the early-heart embryos, and later on it is expressed in the provascular cells. These strands of cells are the location where auxin canalization occurs to direct the formation of the vasculature strands. Consistently, in mthsc70-1 embryos the DR5rev:GFP-marked vasculature strands were either interrupted or were completely absent (compare Figure 4, I and J with Figure 4, C). These results, thus, suggest very strongly that mtHSC70-1 regulates embryo development through the auxin pathway.
How a mitochondria chaperone protein like mtHSC70-1 affects the expression of nuclear-encoded auxin biosynthesis and polar transport genes is enigmatic. Mitochondria, as the main organelles that generate energy in cells, are essential for plant growth and development and environmental stress responses (Berkowitz et al., 2016). To coordinate mitochondrial function with the cellular developmental program, there are extensive signal communications between mitochondria and the nucleus, including the anterograde nucleus-to-mitochondria signals and the retrograde mitochondria-to-nucleus signals (Woodson and Chory, 2008). Since mutation of the mtHSC70-1 gene perturbs the activity and abundance of complex IV and leads to mitochondrial dysfunction (Wei et al., 2019), mitochondrial retrograde signals might be activated in the mthsc70-1 mutant. Indeed, the expression of mitochondrial retrograde signaling marker genes such as AOX1a and ANAC013 was upregulated in mthsc70-1 (Figure 8E; Wei et al., 2019). We, thus, propose that mtHSC70-1 affects the auxin pathway through MRR (Figure 10). Indeed, intimate interactions between mitochondrial retrograde signals and the auxin pathway are being revealed. Studies have found that perturbation of the mitochondrial electron transport chain negatively regulate auxin signaling, whereas in auxin polar transport mutants and auxin signaling mutants, mitochondrial retrograde signals are upregulated, suggesting a mutual regulation of mitochondrial stress and auxin signals (Ivanova et al., 2014; Kerchev et al., 2014). Mutation of the mitochondria-localized J-protein chaperone AtDjB1 perturbs the activity of complex I and leads to reduced levels of auxin (Jia et al., 2016). Interestingly, overexpression of the auxin biosynthesis gene YUC2 in the atdjb1 mutant rescues its growth defects (Jia et al., 2016). Impaired mitochondrial retrograde trafficking caused by dysfunction of a tetratricopeptide repeat domain-containing protein SSR1 also reduces auxin levels and downregulates the expression of several auxin efflux PIN proteins (Zhang et al., 2015). In contrast, mitochondrial proteostatic stress upregulates auxin signaling (Wang and Auwerx, 2017), suggesting that different types of mitochondrial perturbation can have different effects on the auxin pathway. Although ROS have been implicated as mitochondrial retrograde signals (Woodson and Chory, 2008; Wang and Auwerx, 2017), ROS accumulation is not the cause of the impaired response of auxin-deficient mutants to mitochondrial malfunction (Ivanova et al., 2014). Recent studies in mammalian cells showed that mitochondrial stress stimulates a mitochondrial protease OMA1, which cleaves a little-characterized protein DELE1 and causes accumulation of the shortened DELE1 in the cytosol, where it activates the eIF2α kinase HRI (Guo et al., 2020; Fessler et al., 2020). So far, which molecules transduce the mitochondrial retrograde signals to the cytosol in plants remains largely unknown. On the other hand, the mtHSC70-1 protein was found to localize in both mitochondria and the cytosol, implying that it might have a direct role in the cytosol (Xu et al., 2009). Interestingly, the cytosolic HSP90 protein interacts directly with the auxin receptor TRANSPORT INHIBITOR RESPONSE1/AUXIN RESPONSE F-box (TIR1/AFB) protein and increases its stability (Wang et al., 2016). Further analyses are required to confirm the cytosolic localization of the mtHSC70-1 protein and to determine whether it interacts directly or indirectly with the auxin pathway proteins.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the WT control in this study. The T-DNA insertion lines mthsc70-1b (SALK_081385; Wei et al., 2019) and mthsc70-1e (WiscDsLox335G07) were obtained from the Arabidopsis Biological Resource Center (ABRC). DR5rev:GFP, PIN3-GFP and PIN7-GFP (Friml et al., 2003), ProYUC8:GUS and ProYUC9:GUS (Hentrich et al., 2013), AUX1-YFP (Swarup et al., 2004), PIN1-GFP and ProPIN7:GUS (Benková et al., 2003), and PIN2-GFP (Xu and Scheres, 2005) were crossed with mthsc70-1b plants.
Seeds were sterilized for 5 min in 70% (v/v) ethanol and for 10 min in 1% (v/v) NaClO, then washed four times with sterile water. After incubation for 2–3 d at 4°C, the seeds were sown on Murashige and Skoog (MS) medium (pH 5.8) supplemented with 1% (w/v) sucrose and solidified by 0.8% (w/v) agar, and then cultured for germination at 22°C under a 16-h light/8-h dark photoperiod with 90–110 μmol m−2 s−1 illumination. Seven-day-old seedlings were transferred to soil and grown under the same conditions.
RNA extraction, RT-PCR, and RT-qPCR
For analyses of mtHSC70-1 transcript levels in WT and mthsc70-1 mutants, total RNA was extracted from 1- to 3 DPA siliques or 7-d-old seedlings using Trizol reagent (TransGen Biotech, Beijing, China). Next, 2 μg of total RNA was reverse transcribed into cDNA using an EasyScript First-Strand cDNA Synthesis Super-Mix kit (TransGen Biotech, Beijing, China). A 1-μL aliquot of the synthesized cDNA was used as a template for PCR analysis. The ACTIN2 gene was used as an internal control. Primers used in this experiment are listed in Supplemental Table S1.
For RT-qPCR analyses, three biological replicates of 1–3-DPA siliques or 7-d-old seedlings from WT and mthsc70-1b mutant were collected for total RNA extraction. Then 2 μg of total RNA was reverse transcribed into cDNA using an EasyScript First-Strand cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China). One microliter of each cDNA sample was mixed with 7.5 μL of SYBR Green Real-Time PCR Master Mix (DBI Bioscience, Shanghai, China) and then analyzed on a fluorescent qPCR machine (Eppendorf). The TAP42 INTERACTING PROTEIN OF 41 KDA (TIP41, AT4G34270) gene was used as an internal control. The cycle threshold (Ct) 2−ΔΔCt method was used to calculate the relative expression level. Primers used in the RT-qPCR analyses are listed in Supplemental Table S1.
Measurement of the length of root, hypocotyl, root tip meristem zone and elongation zone, and trichoblast
For root and hypocotyl length measurement, primary root and hypocotyl of 7-d-old seedlings grown on MS plates cultured vertically under a 16-h light/8-h dark photoperiod or 24-h dark were photographed using a scanner (EPSON V370), and then quantification was done using Image J software (http://rsb.info.nih.gov/ij/). To measure the length of root tip meristem zone and elongation zone and trichoblast, 5-d-old seedlings grown under a 16-h light/8-h dark photoperiod were fixed in ethanol:acetic acid (3:1, v/v) for 12 h, then were cleared with chloral hydrate:dH2O:glycerol (8:3:1, w:v:v) for 48 h. Then the roots were imaged using an Olympus BX63 (Tokyo, Japan) microscope with differential interference contrast (DIC) mode, and the measurements were done using tools from the cellSens Dimension-V1.5 software. Root meristem zone and elongation zone were determined as described by Men et al. (2008).
Observation of flower organs, siliques, and cotyledons
Opened flowers and 8-DPA siliques from 7-week-old plants and cotyledons from 7-d-old seedlings were imaged using a Leica M165FC (Leica, Wetzlar, Germany) dissection microscope. Then quantification of silique length, pistil length, and anther filaments length were done from digital images using Image J software (http://rsb.info.nih.gov/ij/).
Histology and microscopy
Pistils of opened flowers from 6-week-old plants were cleared for visualization using a clearing solution of 8:3:1 (w:v:v) chloral hydrate:dH2O:glycerol as described by Zhang et al. (2016). Then, the pistils were placed on a glass slide in a drop of clearing solution and were covered with a coverslip. Observations were performed with an Olympus BX63 (Tokyo, Japan) microscope under DIC, and the number of ovules per pistil was counted.
For embryo phenotyping, seeds of 1–8-DPA siliques from 6-week-old WT and mthsc70-1b plants were cleared and visualized as described by Song et al. (2019).
GUS (β-glucuronidase) staining and observation were carried out as previously described (Song et al., 2019).
Vector construction and plant transformation
To construct the mtHSC70-1p:mtHSC70-1-EGFP vector, the mtHSC70-1 promoter region (1,434-bp upstream sequence of the start codon) was amplified and cloned into the pGreenII0229-EGFP-TNos vector (Bao et al., 2019) between KpnI and SmaI sites. Then the mtHSC70-1 coding sequence without the stop codon was cloned into the above obtained HSC70-1p:EGFP vector between SmaI and BamHI sites. Primers used for cloning are listed in Supplemental Table S1.
The above construct was introduced into Agrobacterium tumefaciens strain C58C1 (pMP90/pJIC Sa-Rep) and transformed into ecotype Columbia-0 and mthsc70-1b plants using the floral-dip method (Clough and Bent, 1998). Transgenic plants were screened by spraying with 0.02% (v/v) Basta (Sangon Biotech, Shanghai, China) and PCR amplification.
Fluorescence detection of roots and embryos
Seven-day-old WT and mthsc70-1b seedling roots expressing DR5rev:GFP, AUX1-YFP, PIN1-GFP, PIN2-GFP, PIN3-GFP, and PIN7-GFP were imaged either using an Olympus BX63 microscope or using confocal laser scanning microscopy (Leica TCS SP5). Embryos expressing mtHSC70-1p:mtHSC70-1-EGFP, DR5rev:GFP, AUX1-YFP, or PIN3-GFP were dissected from seeds at various developmental stages and mounted in an 8% (v/v) glycerol solution. Fluorescent protein signals were detected either by an Olympus BX63 fluorescent microscope or by confocal laser scanning microscopy (Leica TCS SP5). GFP and YFP signals were detected using a 488-nm excitation filter with 500–550-nm band pass. Images were processed with Adobe Photoshop CS5 and assembled in Adobe Illustrator CS4.
Chemical reagents
IAA, NAA, and 2,4-D were purchased from Sigma-Aldrich (St. Louis, MO, USA). FAA was purchased from Yuanye Bio-technology (Shanghai, China). The JC-1 Mitochondrial Membrane Potential Assay Kit was purchased from Yeasen Biotech Co., Ltd (Shanghai, China).
Mitochondrial protein exaction and proteomic analysis
Mitochondrial proteins were extracted from 100 mg inflorescence tissues of 6-week-old WT and mthsc70-1b plants using a Mitochondria Isolation Kit for Cell Tissue (Yeasen Biotech Co., Ltd, Shanghai, China). Proteins were reduced with 10 mM dithiothreitol and alkylated with 55 mM iodoacetamide. Then, proteins were equivalented with 20 mM Tris–HCl buffer on 30-KDa ultracentrifugation tubes and digested with trypsin (modified sequencing grade; Promega, Madison, WI, USA) overnight at 37°C in ultracentrifugation tubes. The tryptic peptides were de-salted and concentrated on reverse phase C18 Stage Tips. The elution products were dried down in a vacuum centrifuge to remove solvent.
Peptides were revolved in 0.1% (v/v) formic acid (FA) and separated with EASY-n LC1000 system. Column oven was set to 60°C. Peptides were delivered to a trap column (75 μm × 2 cm, C18, 5 μm, Thermo Scientific, Waltham, MA, USA), then separated with capillary LC column (75 μm × 100 mm, C18, 3μm, Kyoto Monotech, Kyoto, Japan). The eluted gradient was 6%–28% for 48 min and 28%–95% for 4 min buffer B (0.1% [v/v] FA, 100% [v/v] ACN; flow rate, 0.6 μL/min). An Orbitrap Fusion Lumos mass spectrometer was used to analyze the eluted peptides from LC. The data were acquired with data-independent acquisition under high-sensitivity mode using the following parameters: positive mode was set. One cycle contains one full scan and 40 segments fragment scans. The full scan range is from 350 to 1300 m/z and screened at 120,000 resolution. Fragment spectra were collected at 3,000 resolution and segmented 40 MS/MS scans. Maximum injection time is 50 ms.
Raw data of DIA were analyzed by Spectronaut (version 14.3, Biognosys, Schlieren, Switzerland) with default settings. The Spectral libraries of DIA was generated using a total of six raw data with a Q value cutoff of 0.01 and a minimum of six fragment ions. The raw files were searched against the A. thaliana database downloaded from reviewed Swissprot containing 16,004 sequences. Decoy items were generated by inverse mode. The six samples were subjected to quantitative evaluation based on the MS2 area. Cross runs were normalized according to the global area.
GO enrichment analysis was performed using GENE ONTOLOGY (http://geneontology.org/).
Statistical analyses
Three independent repetitions were performed for all experiments. Statistical significance was determined using one-tailed Student’s t test. All values are presented as means ± sd. Significant differences are denoted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL database or Arabidopsis Genome Initiative database under accession numbers mtHSC70-1 (AT4G37910). T-DNA insertion lines used for mutant analyses were as follows: mthsc70-1b (SALK_081385) and mthsc70-1e (WiscDsLox335G07).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotype analysis of mthsc70-1.
Supplemental Figure S2. Inflorescence and flower morphology of WT and mthsc70-1b plants.
Supplemental Figure S3. Seed set of WT and mthsc70-1e siliques.
Supplemental Figure S4. Male and female gametophytes of mthsc70-1 plants are defective.
Supplemental Figure S5. Anther and pollen grains of WT and mthsc70-1b plants.
Supplemental Figure S6. Complementation of mthsc70-1b with mtHSC70-1p:mtHSC70-1-EGFP.
Supplemental Figure S7. Morphology of mature embryos and corresponding seedling cotyledons.
Supplemental Figure S8. Expression pattern of the mtHSC70-1 gene in the root tip.
Supplemental Figure S9. Expression patterns of DR5rev:GFP in WT and mthsc70-1b root tips.
Supplemental Figure S10. WT and mthsc70-1b seedling responses to different concentrations of NAA.
Supplemental Figure S11. WT and mthsc70-1b seedling responses to different concentrations of 2,4-D.
Supplemental Table S1. List of primers used in this research.
Supplementary Material
Acknowledgments
The authors thank Arabidopsis Biological Resource Center (ABRC) for providing mutant seeds, John Innes Centre for providing the pGREENII0229 vector. The authors thank Luhua Li for isolating the mthsc70-1b mutant. We thank Jiřı’ Friml for DR5rev:GFP, PIN1-GFP, PIN3-GFP and PIN7-GFP seeds, Stephan Pollmann for ProYUC8:GUS and ProYUC9:GUS seeds, Ben Scheres for PIN2-GFP seed, Malcolm J. Bennett for AUX1-YFP seed. The authors thank Ruming Liu and Li Jiao for technical assistance in the use of the confocal.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31870230, 32070281, and 31570247 to S.M.).
Conflict of interest statement. The authors declare no conflict of interest.
S.M. and Y.S. conceived the project. S.M. planned and designed the research. G.L. performed experiments. Z.L. and Z.Y. helped to perform FAA and auxin application on pistils. Y.L. critically read the manuscript and shared helpful discussions. S.M. and G.L. analyzed data. S.M. and G.L. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plphys/pages/general-instructions) is: Shuzhen Men (shuzhenmen@nankai.edu.cn).
References
- Bao S, Shen G, Li G, Liu Z, Arif M, Wei Q, Men S (2019) The Arabidopsis nucleoporin NUP1 is essential for megasporogenesis and early stages of pollen development. Plant Cell Rep 38:59–74 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J ( 2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602 [DOI] [PubMed] [Google Scholar]
- Berkowitz O, De Clercq I, Van Breusegem F, Whelan J (2016) Interaction between hormonal and mitochondrial signaling during growth, development and in plant defence responses. Plant Cell Environ 39:1127–1139 [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125:443–451 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20:1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25:3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT (2020) A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579:7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415:806–809 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426:147–153 [DOI] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LH, Whelan J (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150:1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X Aviles G Liu Y Tian R Unger BA Lin YHT Wiita AP Xu K Correia MA Kampmann M (2020) Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579: 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Golding GB (1993) Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol 37:573–582 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17:1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S (2013) The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74:626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, Duncan O, Lee JH, Zhang B, Van Aken O, Radomiljac JD, van der Merwe M, Yi K, et al. (2014) A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1a expression in Arabidopsis. Plant Physiol 165:1233–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Lv TT, Li MX, Wei SS, Li YY, Zhao CL, Li B (2016) The J-protein AtDjB1 is required for mitochondrial complex I activity and regulates growth and development through ROS-mediated auxin signaling. J Exp Botany 67:3481–3496 [DOI] [PubMed] [Google Scholar]
- Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, Sonnewald U (2011) AtHsp70‐15‐deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J 66:983–995 [DOI] [PubMed] [Google Scholar]
- Kerchev PI, De Clercq I, Denecker J, Mühlenbock P, Kumpf R, Nguyen L, Audenaert D, Dejonghe W, Van Breusegem F (2014) Mitochondrial perturbation negatively affects auxin signaling. Molecular Plant 7:1138–1150 [DOI] [PubMed] [Google Scholar]
- Leaden L, Busi MV, Gomez-Casati DF (2014) The mitochondrial proteins AtHscB and AtIsu1 involved in Fe–S cluster assembly interact with the Hsp70-type chaperon AtHscA2 and modulate its catalytic activity. Mitochondrion 19:375–381 [DOI] [PubMed] [Google Scholar]
- Leng L, Liang Q, Jiang J, Zhang C, Hao Y, Wang X, Su W (2017) A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. J Plant Res 130:349–363 [DOI] [PubMed] [Google Scholar]
- Li L, Xing Y, Chang D, Fang S, Cui B, Li Q, Wang X, Guo S, Yang X, Men S, et al. (2016) CaM/BAG5/Hsc70 signaling complex dynamically regulates leaf senescence. Scientific Rep 6:31889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL Wang JS Liu HC Chen RW Meyer Y Barakat A Delseny M (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18:2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Sugiyama T, Endo T, Nishikawa SI (2014) Multiple BiP genes of Arabidopsis thaliana are required for male gametogenesis and pollen competitiveness. Plant Cell Physiol 55:801–810 [DOI] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10:237. [DOI] [PubMed] [Google Scholar]
- Mironova V, Teale W, Shahriari M, Dawson J, Palme K (2017) The systems biology of auxin in developing embryos. Trends Plant Sci 22:225–235 [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harbor Perspect Biol 1: a001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25:3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Brodsky JL, Nakatsukasa K (2005) Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J Biochem 137:551–555 [DOI] [PubMed] [Google Scholar]
- Noctor G, De Paepe R, Foyer CH (2007) Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci 12:125–134 [DOI] [PubMed] [Google Scholar]
- Noh SJ, Kwon CS, Oh DH, Moon JS, Chung WI (2003) Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311:81–91 [DOI] [PubMed] [Google Scholar]
- Qi Y, Wang H, Zou Y, Liu C, Liu Y, Wang Y, Zhang W (2011) Over‐expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett 585:231–239 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J (2015) Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142:702–711 [DOI] [PubMed] [Google Scholar]
- Salvioli S, Ardizzoni A, Franceschi C, Cossarizza A (1997) JC-1, but not DiOC6(3) or rhodamine 123, is a reliable fluorescent probe to assess ΔΨ changes in intact cells: implications for studies on mitochondrial functionality during apoptosis. FEBS Lett 411:77–82 [DOI] [PubMed] [Google Scholar]
- Song J, Sun S, Ren H, Grison M, Boutté Y, Bai W, Men S (2019) The SMO1 family of sterol 4α-methyl oxidases is essential for auxin- and cytokinin-regulated embryogenesis. Plant Physiol 181:578–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191 [DOI] [PubMed] [Google Scholar]
- Su PH, Li H (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL (2001) Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol 126:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al. (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16:3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W et al. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22:2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Whelan J (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front Plant Sci 3:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162:254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M (2016) Hsp90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor f-box protein TIR1. Nat Commun 7:10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Auwerx J (2017) Systems phytohormone responses to mitochondrial proteotoxic stress. Mol Cell 68:540–551 [DOI] [PubMed] [Google Scholar]
- Wei SS, Niu WT, Zhai XT, Liang WQ, Xu M, Fan X, Lv TT, Xu WY, Bai JT, Jia N. et al. (2019) Arabidopsis mtHSC70-1 plays important roles in the establishment of COX-dependent respiration and redox homeostasis. J Exp Botany 70:5575–5590 [DOI] [PubMed] [Google Scholar]
- Woodson J D, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9:383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17:525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Lin H, Latijnhouwers M, Møller SG (2009) Dual localized AtHscB involved in iron sulfur protein biogenesis in Arabidopsis. PLoS One 4: e7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai XT, Wei SS, Liang WQ, Bai JT, Li B (2020) Arabidopsis mtHSC70-1 physically interacts with the Cox2 subunit of cytochrome c oxidase. Plant Signaling Behav 15:1714189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang C, Lin Q, Liu A, Wang T, Feng X, Liu J, Han H, Ma Y, Bonea D, et al. (2015) A tetratricopeptide repeat domain‐containing protein SSR 1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis. Plant J 83:582–599 [DOI] [PubMed] [Google Scholar]
- Zhang X, Sun S, Nie X, Boutté Y, Grison M, Li P, Kuang S, Men S (2016) Sterol methyl oxidases affect embryo development via auxin-associated mechanisms. Plant Physiol 171:468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen S, Fankhauser C, Cashman J, Cohen J, Weigel D, Chory J (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dugardeyn J, Zhang C, Mühlenbock P, Eastmond PJ, Valcke R, De Coninck B, Oden S, Karampelias M, Cammue BP, et al. (2014) The Arabidopsis thaliana RNA editing factor SLO2, which affects the mitochondrial electron transport chain, participates in multiple stress and hormone responses. Mol Plant 7:290–310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.