Abstract
The phragmoplast separates daughter cells during cytokinesis by constructing the cell plate, which depends on interaction between cytoskeleton and membrane compartments. Proteins responsible for these interactions remain unknown, but formins can link cytoskeleton with membranes and several members of formin protein family localize to the cell plate. Progress in functional characterization of formins in cytokinesis is hindered by functional redundancies within the large formin gene family. We addressed this limitation by employing Small Molecular Inhibitor of Formin Homology 2 (SMIFH2), a small-molecule inhibitor of formins. Treatment of tobacco (Nicotiana tabacum) tissue culture cells with SMIFH2 perturbed localization of actin at the cell plate; slowed down both microtubule polymerization and phragmoplast expansion; diminished association of dynamin-related proteins with the cell plate independently of actin and microtubules; and caused cell plate swelling. Another impact of SMIFH2 was shortening of the END BINDING1b (EB1b) and EB1c comets on the growing microtubule plus ends in N. tabacum tissue culture cells and Arabidopsis thaliana cotyledon epidermis cells. The shape of the EB1 comets in the SMIFH2-treated cells resembled that of the knockdown mutant of plant Xenopus Microtubule-Associated protein of 215 kDa (XMAP215) homolog MICROTUBULE ORGANIZATION 1/GEMINI 1 (MOR1/GEM1). This outcome suggests that formins promote elongation of tubulin flares on the growing plus ends. Formins AtFH1 (A. thaliana Formin Homology 1) and AtFH8 can also interact with EB1. Besides cytokinesis, formins function in the mitotic spindle assembly and metaphase to anaphase transition. Our data suggest that during cytokinesis formins function in: (1) promoting microtubule polymerization; (2) nucleating F-actin at the cell plate; (3) retaining dynamin-related proteins at the cell plate; and (4) remodeling of the cell plate membrane.
Formins regulate phragmoplast expansion, microtubule turnover rate, actin nucleation, and cell plate membrane remodeling during cytokinesis.
Introduction
Plant cells divide by constructing a partition called the cell plate between two newly forming daughter cells (Bajer, 1968; Pickett-Heaps and Northcote, 1966). The cell plate is composed of membrane and cell wall components assembled by plant-specific cytokinetic machinery known as the phragmoplast (Becker, 1938). The phragmoplast originates from the remnants of spindle microtubules during late anaphase and is positioned by signals originating from the preprophase band of microtubules (Segui-Simarro et al., 2004). Along with microtubules, the phragmoplast consists of actin microfilaments, vesicle transporters, membrane compartments, and regulatory proteins (Boruc and Van Damme, 2015; Lipka et al., 2015).
Initially, the phragmoplast appears as a disk of two opposing sets of microtubules positioned between the two daughter nuclei (Asada et al., 1991; Wasteneys, 2002). Cells are commonly wider than the initial phragmoplast diameter, requiring the phragmoplast to expand centrifugally toward the parental cell walls in order to complete formation of the cell plate (Wasteneys, 2002). Phragmoplast function and expansion rely on dynamic lateral and axial structural asymmetries (Supplemental Figure S1; Smertenko et al., 2018). Phragmoplast lateral asymmetry includes microtubule nucleation and polymerization in the leading zone and microtubule depolymerization in the lagging zone (Segui-Simarro et al., 2007; Murata et al., 2013). Leading-zone polymerization coincides with new cell plate formation (Lipka et al., 2015; Smertenko et al., 2017). Between the leading and lagging zones, there is a transition zone where microtubules are less susceptible to the microtubule-destabilizing drugs (Murata et al., 2013). This stability is thought to be due to intimate associations of microtubule plus-end proteins with the cell plate assembly matrix (Austin et al., 2005).
Other processes contributing to phragmoplast lateral asymmetry include delivery of vesicles to the midzone in the leading zone, fusion of vesicles in the transition zone, and establishment of a continuous membranous network in the lagging zone (Samuels et al., 1995; Segui-Simarro et al., 2007). Microtubule depolymerization in the lagging zone occurs only after the cell plate reaches the tubular network stage (Samuels et al., 1995). In fact, inhibition of cell plate assembly prevents microtubule depolymerization, suggesting that crosstalk between the nascent cell plate and cytoskeletal elements dictates phragmoplast expansion (Yasuhara, 2005; Yasuhara and Shibaoka, 2000). Phragmoplast axial asymmetry includes faster microtubule dynamics in the midzone where the majority of microtubule plus-ends reside, and slower dynamics in the distal zone where microtubule minus ends reside (Smertenko et al., 2011). Reduction of microtubule depolymerization by treating plant cells with the tubulin-binding drug taxol or by knocking out the microtubule severing factor katanin disrupts phragmoplast expansion (Yasuhara et al., 1993; Panteris et al., 2011; Smertenko et al., 2011; Komis et al., 2017), suggesting microtubule dynamicity is important to phragmoplast reorganization and/or function.
It remains unclear what governs phragmoplast asymmetries. A class of proteins having the requisite characteristics that may be centrally important are formins. Formins have been shown to localize on phragmoplast microtubules (AtFH14; Li et al., 2010) and to the cell plate (AtFH1, AtFH5 and AtFH8) in Arabidopsis thaliana (Ingouff et al., 2005; Oulehlova et al., 2019; Xue et al., 2011) and Physcomitrium patens (van Gisbergen et al., 2020; Wu and Bezanilla, 2014). Formins bind actin filaments in vitro and promote actin nucleation and polymerization through their evolutionarily conserved formin homology (FH1FH2) domain (Cheung and Wu, 2004; Yi et al., 2005; Kovar, 2006; Cheung et al., 2010). However, regulation of actin may not be the sole function of formins in the phragmoplast. The FH2 domain of formins can also bind microtubules and promote their stabilization (Palazzo et al., 2001; Bartolini et al., 2008; Cheng et al., 2011; Gaillard et al., 2011). In animal cells, formins align microtubules with F-actin and mediate a number of processes during interphase including cellular organization, motility, and polarity (Breitsprecher and Goode, 2013). The A. thaliana type II plant formins, AtFH14 and AtFH16, bind to both microtubules and F-actin and potentially crosslink and coordinate organization of both (Li et al., 2010; Wang et al., 2013). The rice type II formin, FH5, also facilitates organization of both microtubules and F-actin (Yang et al., 2011; Zhang et al., 2011).
Type I plant formins are defined by the presence of an N-terminal signal peptide and a membrane-anchoring transmembrane domain (Deeks et al., 2002). The A. thaliana type I formins AtFH4, AtFH6, and AtFH1 have been shown to localize to the plasma membrane (Cheung and Wu, 2004; Favery et al., 2004; Cheung et al., 2010; Deeks et al., 2010; Martiniere et al., 2011). AtFH1 also contains an N-terminal extracellular domain that anchors the protein to the cell wall (Martiniere et al., 2011). Mutations in AtFH1 affect dynamic properties of both microtubule and actin cytoskeletal systems (Rosero et al., 2013). Taking into account all available knowledge, we hypothesize that formins govern phragmoplast expansion and cell plate synthesis.
Determining the roles played by formins in plants has been complicated by the fact that formins constitute large gene families with overlapping expression patterns and functional redundancies. The A. thaliana genome contains 21 formins, with 11 being of type I, and 10 of type II (Deeks et al., 2002). Thus far, genetic approaches have demonstrated the importance of a few of these formins in cell expansion (Vidali et al., 2009; Yang et al., 2011; Zhang et al., 2011) and polarized growth (Cheung and Wu, 2004; Vidali et al., 2009; Ye et al., 2009; Cheung et al., 2010; van Gisbergen et al., 2012). With respect to cell division, mutations in AtFH5 result in delayed cellularization of endosperm and the appearance of multi-nucleate cells (Ingouff et al., 2005). Knockout of AtFH14 causes abnormal phragmoplast organization and multi-nucleate microspores (Li et al., 2010). However, formin functions during cytokinesis in somatic cells remain poorly understood.
Here, we circumvent the problem of formin functional redundancies by employing a Small Molecular Inhibitor of Formin Homology 2 domains (SMIFH2), which inhibits the ability of formin to promote actin polymerization and reduces binding of formins to microtubules (Rizvi et al., 2009). Although SMIFH2 has been used for analysis of formins in various processes of animal cells (Isogai et al., 2015), its application to plant systems has been limited to the study of formin functions in interphase cells (Rosero et al., 2013; Cao et al., 2016; Rosero et al., 2016). For example, application of SMIFH2 to A. thaliana seedlings was found to phenocopy the root growth, cell expansion, and cytoskeletal structural defects of A. thaliana Atfh1 mutants (Rosero et al., 2013). Here, we show that during phragmoplast expansion, formins regulate actin polymerization, microtubule dynamics and facilitate cell plate membrane remodeling.
Results
SMIFH2 inhibits formin-dependent actin polymerization in vitro and suppresses cell proliferation
To establish that SMIFH2 directly inhibits function of plant formins, we performed in vitro actin polymerization assays with the highly conserved FH1FH2 protein domain from the A. thaliana formin AtFH1 (Michelot et al., 2006). Under our in vitro conditions, 40 µM SMIFH2 incubated with the FH1FH2 domain and pyrene-iodoacetamide-labeled G-actin substantially reduced the rate of FH1FH2 domain-mediated actin polymerization. SMIFH2 inhibition of F-actin polymerization was much less pronounced at a concentration of 10 µM (Supplemental Figure S2A). Cao et al. (2016) found that 50 µM SMIFH2 dramatically inhibited in vitro actin polymerization mediated by the minimal FH2 nucleation domain from AtFH1, a result consistent with our observations.
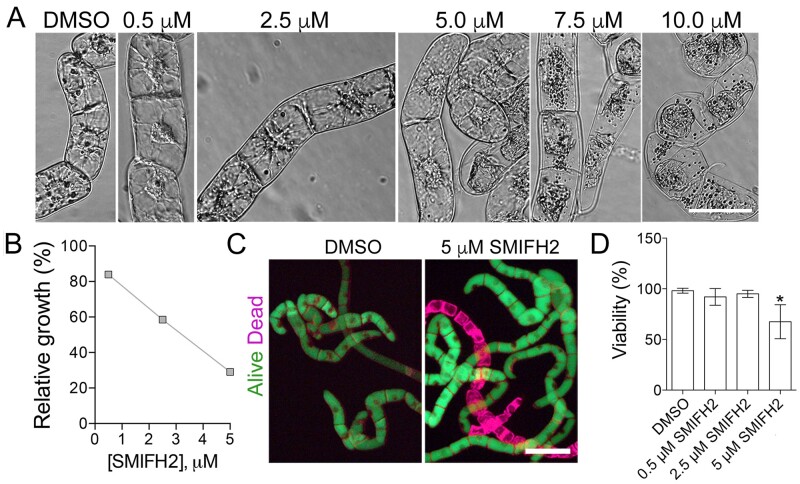
To determine what concentrations of SMIFH2 were necessary to impact plant cell proliferation and morphology, we exposed Nicotiana tabacum BY-2 cells to 0.5 µM, 2.5 µM, 5 µM, 7.5 µM, and 10 µM SMIFH2. We found that material of the vessels affects the experimental outcomes. All cells were found dead following 5 d treatment with 7.5 or 10 µM SMIFH2 in glass vessels, whereas no discernible effects on viability were observed in plastic vessels (data not shown). The second factor was drug dosage per cell; using higher cell density resulted in much weaker effect. The reproducibility was achieved by keeping consistent the age and the volume of the liquid cell culture used for each treatment.
After 5 d of growth, concentrations 7.5 and 10 µM caused protoplast contraction in all cells (Figure 1A). Concentrations of 0.5 µM, 2.5 µM, and 5 µM caused no apparent morphological defects, but the cell growth was significantly inhibited relatively to the control treated with dimethyl sulfoxide (DMSO; Figure 1B). Requirement of a higher SMIFH2 concentration for suppressing formins ability to promote actin polymerization in vitro than for inhibiting cell proliferation indicates other activities of formins besides regulation of F-actin.
Figure 1.
SMIFH2 inhibits BY-2 cell growth. A, Morphology of BY-2 cells after 5-d treatment with different concentrations of SMIFH2. Scale bar, 50 µm. B, Impact of SMIFH2 on cell growth rate. Relative fresh weight of BY-2 cells was measured after 5-d treatment with SMIFH2. Cultures treated with carrier (DMSO) were taken as 100%. C, Representative images of BY-2 cells stained with fluorescein diacetate (FDA; green, alive) and FM4-64 (purple, dead) from cultures treated for 5 d with DMSO or SMIFH2. Scale bar, 100 µm. D, Viability of BY-2 cells after treatment with DMSO or SMIFH2 for 5 d. Error bars in B and D show standard deviation of three independent replicates containing three technical repeats (n = 3). Asterisks denote samples that are significantly different from the DMSO-treated control (t test P < 0.05).
Reduction of cell proliferation can occur due to cell death, longer cell cycle, or both. To examine which of these are affected by SMIFH2, we measured cell mortality following treatment with 0.5 µM, 2.5 µM, and 5 µM of MSIFH2 for 5 d (Figure 1, C and D), representing the same treatments as in Figure 1, A and B. Concentrations of 7.5 µM and 10 µM of SMIFH2 were not included in this experiment as both induced protoplast collapse indicating cell death (Figure 1A). The cell death was measured using fluorescein diacetate (FDA), which fluorescently labels only live cells, and FM4-64, which labels both live and dead cells (Minina et al., 2013). Representative fluorescent images of cells treated for 5 d with the mock solution (DMSO) or with 5 µM SMIFH2 are shown in Figure 1C. In these assays, SMIFH2 at concentrations of 0.5 µM and 2.5 µM did not impact cell viability, whereas 5 µM SMIFH2 caused 25% cell death (Figure 1D). As treatment with 5 µM SMIFH2 inhibited cell growth by over 60%, reduction of cell proliferation appears to be the major effect of SMIFH2.
SMIFH2 disruption of specific cell cycle stages suggests formins are essential for cytokinesis
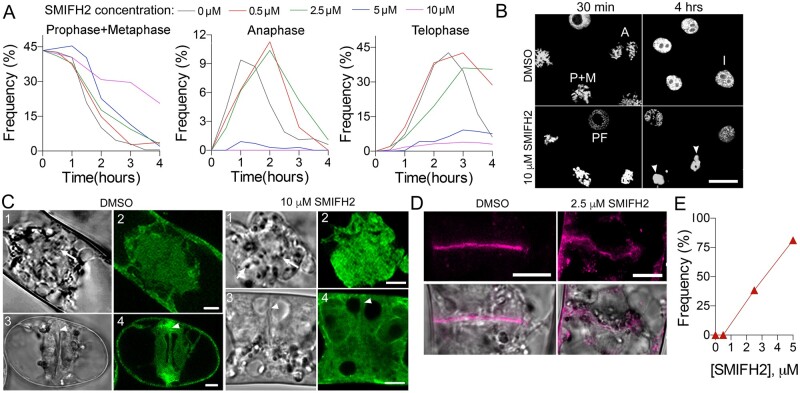
Reduction of cell proliferation in response to SMIFH2 treatment is most likely caused by the M-phase delay. To test this hypothesis, we arrested BY-2 cells in metaphase by sequential treatment with an inhibitor of DNA synthesis aphidicolin followed by the inhibitor of microtubule polymerization propyzamide (Asada et al., 1991). The metaphase block was released by washing out propyzamide. Immediately after the propyzamide washout, we added DMSO (mock treatment), 0.5 µM, 2.5 µM, 5 µM, or 10 µM SMIFH2. Cell cultures were then fixed at different time points, stained with DAPI (4′,6-diamidino-2-phenylindole) for DNA, and the frequencies of cells at prophase+metaphase (condensed chromosomes), anaphase (separating sister chromatids), and telophase (decondensing sister chromatids) were scored (Figure 2A).
Figure 2.
Impact of SMIFH2 on cell division. A, Frequency of cells in prophase plus metaphase, anaphase, or telophase at different concentrations of SMIFH2 following propyzamide washout. At least 300 cells were scored for each time point and the treatment. B, Representative images of DNA staining in cells taken at 30 min or 4 h after treatment with DMSO or 10 µM SMIFH2 in the experiment presented in panel A. Scale bar, 25 µm. A, anaphase; I, interphase; P+M, prophase and metaphase, PF, prophase. Arrowheads indicate aggregated condensed chromosomes in cells treated with SMIFH2 for 4 h. C, GFP-Lifeact labels G- and F-actin in metaphase (parts 1 and 2) or telophase (parts 3 and 4) cells after treatment with DMSO or 10 µM SMIFH2 for 3 h. Parts 1 and 2 show metaphase cells, and 3 and 4 show telophase cells. Cells were imaged using confocal microscope. Scale bars, 5 µm. Arrows denote condensed chromosomes. Arrowheads denote the midzone. D, Representative images of cell plates following treatment with DMSO or SMIFH2 (2.5 µM) for 3 h. Confocal images of BY-2 cells expressing SCAMP2-mCherry. Scale bar, 10 µm. E, The frequency of abnormal cell plates (shown in D) among cytokinetic cells treated with SMIFH2. Thirty cells were counted per each treatment.
Altogether, SMIFH2 exhibited four major effects on the mitotic progression. First, delayed metaphase exits at all concentrations with the most prominent effect at 10 µM. In fact, chromosomes remained condensed in 20% of cells after 4 h treatment with 10 µM SMIFH2 relative to 0.2% DMSO in the control (Figure 2, A and B). Second, the entry into anaphase was delayed by 0.5 and 2.5 µM, reduced by 5 µM, and almost completely inhibited by 10 µM SMIFH2. Third, the telophase entry was inhibited by 5 and 10 µM SMIFH2. Fourth, the exit from telophase was delayed by all SMIFH2 concentrations.
Based on the above results and the fact that SMIFH2 disrupts formin-dependent F-actin nucleation and polymerization, one would expect SMIFH2 treatments to cause abnormal F-actin densities and organization in cellular structures formed during the cell cycle M-phase, such as the spindle and phragmoplast. To visualize this, we employed a BY-2 cell line expressing the F-actin marker Green Fluorescent Protein (GFP)-Lifeact (Smertenko et al., 2010). In mock-treated cells (DMSO), G-actin labeling was detected in the mitotic spindle area with reduced signal in the cell division zone (Figure 2C;Supplemental Figure S2B). During cytokinesis, a strong GFP-Lifeact labeling of F-actin was visible in the phragmoplast midzone in addition to G-actin labeling in the cytoplasm (Figure 2C, DMSO treatment). Treatment with 10 µM SMIFH2 that inhibited metaphase exit and telophase entrance for 3 h abolished the actin-depleted zone during metaphase, and caused loss of GFP-Lifeact signal at the midzone.
Treatment with 5-µM SMIFH2 reduced the width of the GFP-Lifeact-GFP signal in the midzone compared to mock-treated (DMSO) cells (Supplemental Figure S3, A andB). We quantified the signal intensity differences using the dimensionless sensitivity technique. This method has high sensitivity to variability of integral characteristics due to its determination of the differential curve slope (Smertenko et al., 2005). The dimensionless sensitivity technique analysis demonstrated a much steeper signal decay curve in SMIFH2-treated cells that could be described by one function, y∼x3. In mock-treated cells, the decay of signals was less steep, and the distributions could be described by two functions, y∼x3/2 and y∼x2 (Supplemental Figure S3C).
Formins could delay transition between mitotic phases by disrupting microtubule organization. To test this hypothesis, we immunolabeled microtubules and co-stained chromosomal DNA in fixed BY-2 cells that had been incubated for 3 h in different concentrations of SMIFH2. The overall organization of the mitotic spindle and the phragmoplast was not disrupted by SMIFH2 (Supplemental Figure S3D).
Another reason for the delay in telophase exit could be abnormal cell plate assembly. We explored this possibility using a BY-2 cell line expressing the cell plate marker SECRETORY CARRIER MEMBRANE PROTEIN 2 tagged with mCherry (SCAMP2-mCherry; Toyooka et al., 2009). Treatment with 0.5 µM SMIFH2 for 3 h did not affect the cell plate organization, but higher concentrations caused cell plate deformation and bulging at the leading edge (Figure 2, D and E).
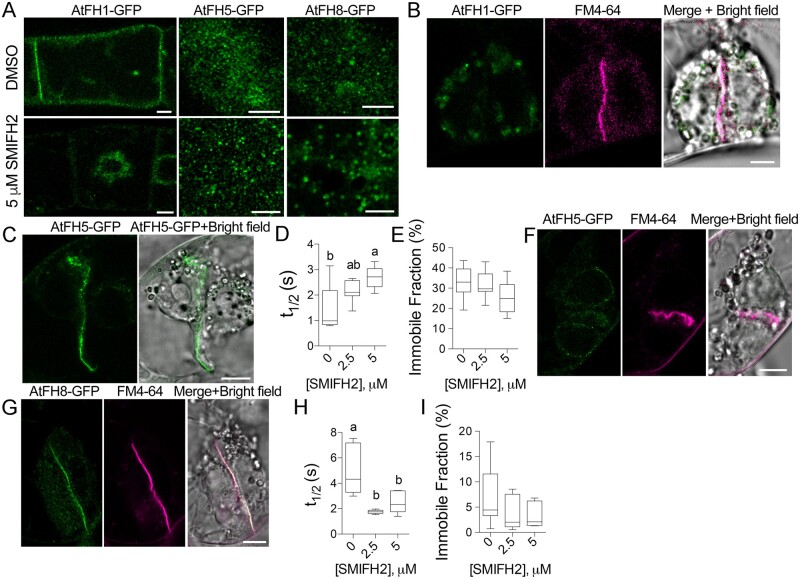
To test the impact of SMIFH2 on localization and dynamics of formins, we created BY-2 cell lines stably expressing C-terminal GFP fusions of three class I formins AtFH1, AtFH5, and AtFH8. During interphase, AtFH1 localized to the plasma membrane and cytoplasmic vesicles, AtFH5 formed puncta and localized in vesicles, and AtFH8 localized to the puncta, cytoplasmic vesicles, and the nuclear envelope (Figure 3A;Supplemental Figure S4). Treatment with 5 µM SMIFH2 resulted in the loss of AtFH1 signal from the plasma membrane and re-localization of the signal to the vicinity of the nucleus, but the AtFH5 and AtFH8 puncta persisted (Figure 3A).
Figure 3.
Localization and dynamics of formins in the cells treated with SMIFH2. A, A single optical section through the center of an interphase cells expressing AtFH1-GFP, and maximal projection of several optical sections through the plasma membrane of cells expressing AtFH5-GFP or AtFH8-GFP following treatment with DMSO or 5 µM SMIFH2 for 3 h. Scale bars, 5 µm. B, AtFH1-GFP does not localize to the cell plate after treatment with 2.5 µM SMIFH2 for 3 h. Scale bar, 5 µm. C, A representative image of AtFH5-GFP in the cell plate following treatment with 5 µM SMIFH2. Scale bar, 10 µm. D and E, Turnover rate (t1/2) and the immobile fraction of AtFH5-GFP on the cell plate in cells treated with DMSO, 2.5 µM SMIFH2, or 5 µM SMIFH2 for 3 h. Different letters denote significant difference between the datasets according to one-way ANOVA. At least six phragmoplasts were analyzed per each dataset. F and G, Representative images of cell plates without (F) or with (G) AtFH8-GFP signal following treatment with 5 µM SMIFH2 for 3 h. Scale bars, 10 µm. H and I, Turnover rate (t1/2) and the immobile fraction of AtFH8-GFP on the cell plate in cells treated with DMSO, 2.5 µM SMIFH2, or 5 µM SMIFH2 for 3 h. Different letters denote significant difference between the datasets according to one-way ANOVA. At least six phragmoplasts were analyzed per each dataset.
During telophase, all three formins localize to the cell plate (Supplemental Figure S4). Treatment with 2.5 µM or 5 µM SMIFH2 abrogated cell plate localization of AtFH1 (Figure 3B). In cells treated with 0.5 µM SMIFH2, 83% of the cell plates retained AtFH1-GFP signal. AtFH5-GFP remained at the cell plates at 0.5, 2.5, or 5 µM SMIFH2 even with the misshaped cell plates (Figure 3C). The turnover of AtFH5-GFP at the cell plate was slower (Figure 3D), whereas the immobile fraction was not significantly affected by the SMIFH2 treatment (Figure 3E). AtFH8-GFP signal was diminished in all misshapen cell plates (Figure 3F) and in the normal cell plates, AtFH8-GFP turnover was faster (Figure 3, G and H), whereas the immobile fraction was not affected (Figure 3I). In summary, SMIFH2 perturbs both localization and turnover of formins at the cell plate.
SMIFH2 disrupts microtubule dynamics in the phragmoplast
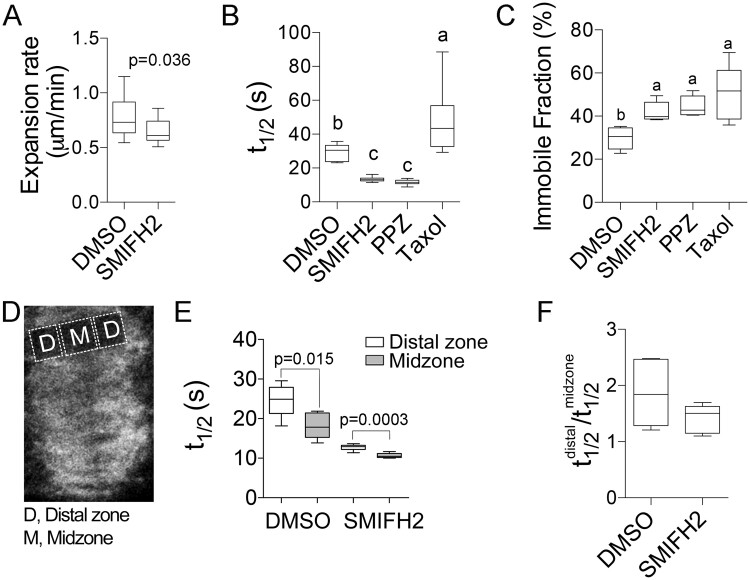
Next, we examined the impact of SMIFH2 on microtubule dynamics in the phragmoplast. These experiments were performed using 2.5 µM SMIFH2 as this concentration delayed cell cycle and affected cell plate synthesis, but did not impact cell viability. By performing time-lapse image analyses on BY-2 cells expressing GFP-NtTUA1, we determined that 2.5 µM SMIFH2 treatment for 3 h significantly reduced the rate of phragmoplast expansion (Figure 4A). Analysis of microtubule dynamics using fluorescence recovery after photobleaching (FRAP) revealed faster microtubule turnover in SMIFH2-treated cells (Figure 4B;Supplemental Figure S5). In addition, immobile fractions of tubulin were found to be greater in SMIFH2-treated cells relatively to control (Figure 4C). The effect of SMIFH2 was similar to that of microtubule polymerization inhibitor propyzamide that also increased the microtubule turnover and increased the immobile fraction (Figure 4, B and C). Treatment with an inhibitor of microtubule depolymerization taxol, reduced the turnover of tubulin, but also increased the immobile fraction (Figure 4, B and C).
Figure 4.
Effects of SMIFH2 on phragmoplast microtubule dynamics in BY-2 cells. A, Phragmoplast expansion rate. B and C, Phragmoplast microtubule turnover (t1/2; B) and tubulin immobile fraction (C) in cells treated with SMIFH2, 1 µM propyzamide (PPZ), or 5 µM taxol. D and E, Schematic showing location of the phragmoplast distal zone and midzone regions for measuring microtubule turnover in cells expressing GFP-NtTUA1 (D). Microtubule turnover in different phragmoplast zones (E). F, Ratio of t1/2 in distal zone to midzone. The concentration of SMIFH2 in all experiments was 2.5 µM and treatment time was 3 h. Whiskers denote minimal to maximal range of values. P-values were calculated using Mann–Whitney U test. Different letters denote statistically different values determined by one-way ANOVA. At least five cells were measured for each treatment.
Phragmoplast expansion depends on axial asymmetry, which can be quantified as the ratio of microtubule turnover in the distal zone versus the midzone (Figure 4D; Smertenko et al., 2011). While the rates of tubulin turnover in these zones as measured by FRAP were found to be significantly faster in cell treated with 2.5 µM SMIFH2 compared to mock-treated cells (Figure 4E), the ratio of tubulin turnover in the distal zone versus midzone was not significantly different (Figure 4F). Thus, formins play an essential role in promoting microtubule polymerization in the phragmoplast but not in the phragmoplast asymmetry.
Impact of SMIFH2 on microtubule polymerization
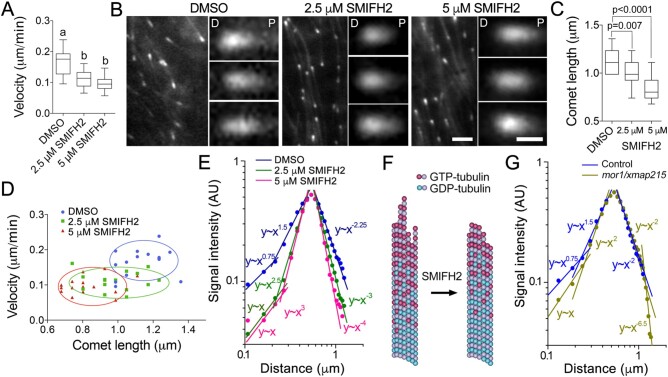
As SMIFH2 disrupted microtubule turnover in the phragmoplast, we explored how SMIFH2 affected microtubule dynamics. Tracking individual microtubules in the phragmoplast of living plant cells is challenging due to the high background of free tubulin and limited resolution of current microscopy methods (Vyplelová et al., 2018). To examine the effect of SMIFH2 on individual microtubules, we tracked microtubule plus-ends in BY-2 cells during interphase using mCherry fusion of A. thaliana EB1b, mCherry-AtEB1b (Eisinger et al., 2012). We found that treatments with 2.5 µM or 5 µM SMIFH2 for 3 h significantly decreased microtubule polymerization rates (Figure 5A). However, SMIFH2 did not impact on tubulin polymerization in vitro (Supplemental Figure S2C).
Figure 5.
SMIFH2 inhibits microtubule polymerization and alters EB1 plus-end localization. A, Microtubule polymerization rates in interphase BY-2 cells treated with DMSO (mock) or SMIFH2. Cells were treated with SMIFH2 for 3 h in all experiments shown in this figure. B, Confocal microscopic images of mCherry-AtEB1b in interphase BY-2 cells. The distal or proximal comet ends are denoted by letter D and P respectively. Scale bar are 2 µm or 0.5 µm on zoomed images. C, Average mCherry-AtEB1b comet lengths in cells treated with DMSO, 2.5 or 5 µM SMIFH2. D, Relationships between microtubule polymerization rates and mCherry-AtEB1b comet lengths in cells treated with DMSO and SMIFH2. Each data point represents measurement of one comet. E, Impact of SMIFH2 on mCherry-AtEB1b comet shape. X-axis in E and G shows absolute distance from the distal to the proximal ends of the EB1 comet, y-axis shows normalized fluorescence signal intensity. F, A model of SMIFH2 activity. SMIFH2 reduces the length of tubulin protofilament “flares” and increases β-tubulin GTP hydrolysis rate resulting in less GTP-tubulin in the microtubule lattice to which EB1 binds. G, Analysis of ProEB1::EB1b-GFP comet shapes in Arabidopsis mor1-1 mutant dark-grown hypocotyl epidermis cells compared to wild-type. Whiskers in A and C denote minimal to maximal range of values. Statistically different average values are denoted by different letters as determined by one-way ANOVA test. P-value was calculated using unpaired t test (n = 15).
AtEB1b comets appeared shorter compared to those in the mock-treated control cells (Figure 5B). Measuring the comets revealed that treatments with both 2.5 µM and 5 µM SMIFH2 reduced AtEB1b comet lengths compared to the mock-treated control (P = 0.007 and P < 0.0001; Figure 5C). Reduction of comet size could be a consequence of slower microtubule polymerization, so we tested correlation between the comet size and microtubule growth rate. The analysis showed no correlation in cells treated with DMSO (Supplemental Figure S6A). However, slower microtubule polymerization in cells treated with 5 µM SMIFH2 correlated with reduced comet size (Figure 5D).
In order to further assess roles of formins in processes at the growing microtubule tips, we compared the profiles of mCherry-AtEB1b fluorescence signal intensity in control and SMIFH2-treated cells using the dimensionless sensitivity technique. The signal declined at a greater rate on both sides of the brightest central point of each comet in cells treated with either 2.5 µM or 5 µM SMIFH2 (Figure 5E;Supplemental Figure S6, B–D). For example, in the case of 5 µM SMIFH2, the distal end slope changed from y∼x1.5 in control to y∼x3, and the proximal end slope changed from y∼x−2.25 to y∼x−4 (Figure 5E).
EB1 proteins exhibit stronger affinity to the GTP-bound form of tubulin in the microtubule lattice (Maurer et al., 2011). Steeper signal decline on the distal (polymerizing) side suggests shorter tubulin protofilament flares (Figure 5F), whereas steeper signal decline on the proximal side suggests faster GTP hydrolysis. Conversion of tubulin from GTP-bound to GDP-bound form causes dissociation of EB1 from the microtubule lattice on the proximal side of the comet (Bieling et al., 2007; Maurer et al., 2011).
To test the accuracy of our analytical method, we took advantage of the temperature-sensitive mor1-1 mutant with genetically disrupted plant XMAP215 homolog MOR1/GEM1 (Whittington et al., 2001). Animal XMAP215 promotes formation of flared protofilaments at the growing microtubule tips, but does not have an apparent effect on tubulin GTP hydrolysis rate (Kerssemakers et al., 2006). Inactivation of plant MOR1/GEM1 at the restrictive temperature is expected to reduce the size of the flares leading to steeper distal slope of EB1-comets, whereas the proximal slope determined by the GTP hydrolysis rate should not be affected. EB1b-GFP comets in mor1-1 plants transformed with ProEB1::EB1b-GFP construct and grown under restrictive temperature were reported to be shorter (Dixit et al., 2006; Kawamura and Wasteneys, 2008). Analysis of AtEB1b-GFP comets in epidermal cells within the dark-grown cotyledon of mutant and wild-type plants under restrictive temperature (29°C) revealed that only the distal part of the comet exhibited steeper signal decline in mor1-1 relative to the control, whereas the proximal slope was not affected (Figure 5G;Supplemental Figure S6E). Such outcome is consistent with prediction that MOR1/MAP215 reduces the size of protofilament flares, but does not affect tubulin GTPase activity. This outcome shows: (1) the ability of our technique to quantify meaningful changes in the shapes of EB1 comets and (2) formins play a role in the processes affecting both the distal and proximal sides of EB1 comets.
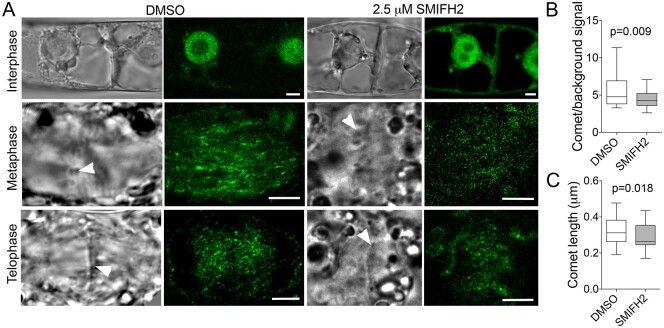
To test the impact of SMIFH2 on AtEB1 comet size during cell division, we analyzed AtEB1c-GFP driven by EB1c native promoter (Novák et al., 2015) in BY-2 cells treated with DMSO or SMIFH2 for 3 h. In A. thaliana, this GFP-tagged AtEB1c localizes to the nucleus during interphase and becomes associated with the growing microtubule ends only after nuclear envelope breakdown (Komaki et al., 2010; Novák et al., 2015). We found that AtEB1c-GFP localized the same way in BY-2 cells (Figure 6A). Although the localization pattern remained unaffected in SMIFH2-treated cells, the intensity of AtEB1c-GFP comets relative to the background signal was significantly lower in the phragmoplasts of SMIFH2-treated cells compared to mock-treated control (Figure 6B). Furthermore, the comets were shorter (Figure 6C).
Figure 6.
Impact of SMIFH2 on AtEB1c-GFP comet size and intensity during cell division. A, Confocal microscopic images (right) showing SMIFH2 does not affect nuclear localization of AtEB1c-GFP in BY-2 cells during interphase, but results in less intensive labeling of comets during both metaphase and telophase. Scale bars, 5 µm. Arrowheads in bright-field images (left) denote position of chromosomes during metaphase and cell plate during telophase. B, The ratio of signal intensity of AtEB1c-GFP comets to the background signal in phragmoplasts of BY-2 cells treated with DMSO or 2.5 µM SMIFH2. P-value was calculated using unpaired t test (n = 31). Whiskers in B and C denote minimal to maximal range of values. C, Size of AtEB1c-GFP comets in the phragmoplasts of BY-2 cells treated with DMSO or 2.5 µM SMIFH2. P-value was calculated using unpaired t test (n = 31). Cells were treated with SMIFH2 for 3 h in all experiments.
Physical and genetic interaction between EB1 and formins
It has been shown that mammalian formin mDia1 and EB1 physically interact (Wen et al., 2004). To examine interaction between plant formins and EB1s, we expressed and purified GST-tagged full-length AtEB1a, AtEB1b, and AtEB1c proteins from A. thaliana, along with 6xHistidine-tagged FH1FH2 protein domains from the A. thaliana formins AtFH1 and AtFH8, and performed in vitro pull-down assays. Individually, all three A. thaliana EB1 isoforms could pull down FH1FH2 domain of AtFH1 and AtFH8 (Supplemental Figure S7A).
The outcomes of the pull-down experiments were verified using Bimolecular Fluorescence Complementation (BiFC) in Nicotiana benthamiana leaf pavement cells. Considering similar primary structure, functional redundancy, and microtubule plus-end binding during cytokinesis of EB1a, EB1b, and EB1c (Bisgrove et al., 2008; Komaki et al., 2010), the likelihood of all EB1 proteins binding formins is high. We selected EB1c for the BiFC experiments for two reasons: (1) nuclear localization of EB1c minimizes chances of unspecific interactions (Komaki et al., 2010; Novák et al., 2015) and (2) this design allows testing which partner controls localization of the complex. Two outcomes are possible: EB1c recruits formins to the nucleus, or formins can recruit EB1c to their locations.
Co-expression of EB1c with AtFH1 resulted in reconstitution of Yellow Fluorescent Protein (YFP) fluorescence on the plasma membrane; and co-expression with AtFH8 resulted in reconstitution of YFP fluorescence on filaments and the plasma membrane (Supplemental Figure S7, B andC). Appearance of the fluorescence signal on the plasma membrane indicates close contact between EB1c and AtFH1 or AtFH8. Treatment of leaves expressing EB1c and AtFH8 with a strong inhibitor of microtubule polymerization amiprophos methyl abolished the signal on filaments, but retained the signal on the plasma membrane (Supplemental Figure S7D). This outcome is consistent with EB1c being targeted to the plasma membrane by AtFH1 or AtFH8. However, the YFP signal was not reconstituted in the formin foci. It means EB1c-binding regions of formins in these structures are inaccessible. As a negative control, fluorescence was not detected when YFP fragments were in the opposite orientation (Supplemental Figure S7, E andF). It means reconstitution of YFP fluorescence was not the consequence of unspecific aggregation of EB1 and formins but requires interaction between the molecules in the head-to-head orientation. When expressed individually, AtFH1 localized to the plasma membrane and AtFH8 localized to microtubules (Supplemental Figure S7, G–J).
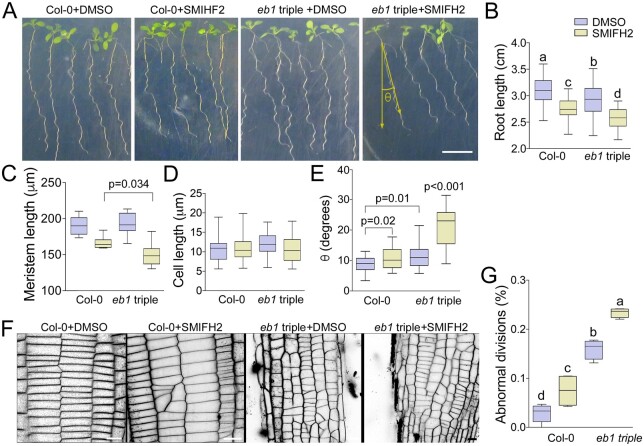
Next, we tested genetic interaction between formins and EB1s using the A. thaliana Ateb1a Ateb1b Ateb1c triple mutant (Galva et al., 2014). When grown on vertically oriented 1% agar media plates, roots of this mutant grew shorter than wild-type (Figure 7, A and B), indicating EB1 proteins promote cell division and/or cell expansion. When 10 µM SMIFH2 was added into the media, both wild-type and the Ateb1 triple mutant roots grew significantly shorter than when grown on the control media, a result consistent with both formins and EB1s contributing to root growth (Figure 7B).
Figure 7.
Formins and EB1s cooperate in regulating root development. A, Representative images f Arabidopsis wild-type Col-0 and Ateb1a Ateb1b Ateb1c triple mutant roots grown on vertically oriented agar media supplemented with DMSO or SMIFH2 (viewed through the agar). Scale bar, 1 cm. B–E, Measurements of root lengths (B), root apical meristem lengths (C), epidermal cell lengths (D), and deviation of root growth trajectories from the gravity vector, n > 21 (E). F, Impact of SMIFH2 on cell division patterns in wild type and Ateb1 triple mutant roots, visualized with FM4-64 staining and confocal microscopy. Scale bar, 20 µm. G, Frequencies of abnormal cell divisions in wild type and Ateb1 triple mutant roots after treatment with DMSO (mock) or SMIFH2. Roots were treated with 10 µM SMIFH2 in all experiments, n = 6. Whiskers denote minimal to maximal range of values. Statistically different average values are denoted by different letters as determined by one-way ANOVA test. P-values were calculated using unpaired t test.
Then, we measured root apical meristem lengths using light microscopy and found reductions in both wild-type and the Ateb1 triple mutant seedlings when grown in the presence of 10 µM SMIFH2 (Figure 7C). This result is consistent with SMIFH2 slowing down cell division as it was observed in tobacco BY-2 cells (Figure 4A). Importantly, SMIFH2 reduced the average meristem size of Ateb1 triple mutant roots significantly more than of wild-type Columbia-0 (Col-0) roots. No discernible differences in meristem length were observed between the root meristems of Col-0 and Ateb1 triple mutant seedlings grown on control plates. The lengths of root cells were similar in Col-0 and Ateb1 triple mutant seedlings grown in the presence of SMIFH2 (Figure 7D), indicating that under our experimental conditions inhibition of root growth by SMIFH2 was mostly due to inhibition of cell proliferation.
As A. thaliana root tips traverse vertically oriented or tilted hard-agar surfaces, they form sinusoidal wave patterns known as root waving. Galva et al. (2014) reported that A. thaliana Ateb1b mutant roots wave similarly to wild-type, but have an overall trajectory of growth that deviates up to 10° to the right of wild-type when viewed through the back of the agar plate (to the left when viewed from above the agar surface). Similarly, we found that Ateb1 triple mutant roots formed normal sinusoidal wave patterns of growth, but had overall growth trajectories skewed up to 10° to the right relative to the wild-type (Figure 7, A and E). When Col-0 and Ateb1 triple mutant seedlings were grown on agar media containing SMIFH2, trajectories of mutant roots deviated twice as far to the right when compared with that of wild-type (Figure 7, A and E). Right root skewing has been associated with reduced microtubule stability and dynamics (Ishida et al., 2007). Hence, our findings suggest formins work together with EB1 to control microtubule stability/dynamics and the overall direction of root growth in relation to the gravity vector.
We also tested cooperation between formins and EB1s in cell division. Treatment of Col-0 roots with 10 µM SMIFH2 caused formation of disoriented cross-walls between cells (Figure 7, F and G). The frequency of the disoriented cross-walls in untreated Ateb1 triple mutant roots was higher than in the wild type when grown on control media. Growing the Ateb1 triple mutant on media supplemented with SMIFH2 further increased cell plate orientation defects (Figure 7G), however, to a lesser extent when compared with well-known cytokinetic mutants (Muller et al., 2002, 2004; Eleftheriou et al., 2005; Kawamura and Wasteneys, 2008; Beck et al., 2011; Lipka et al., 2014; Steiner et al., 2016).
Formins contribute to cell plate membrane remodeling
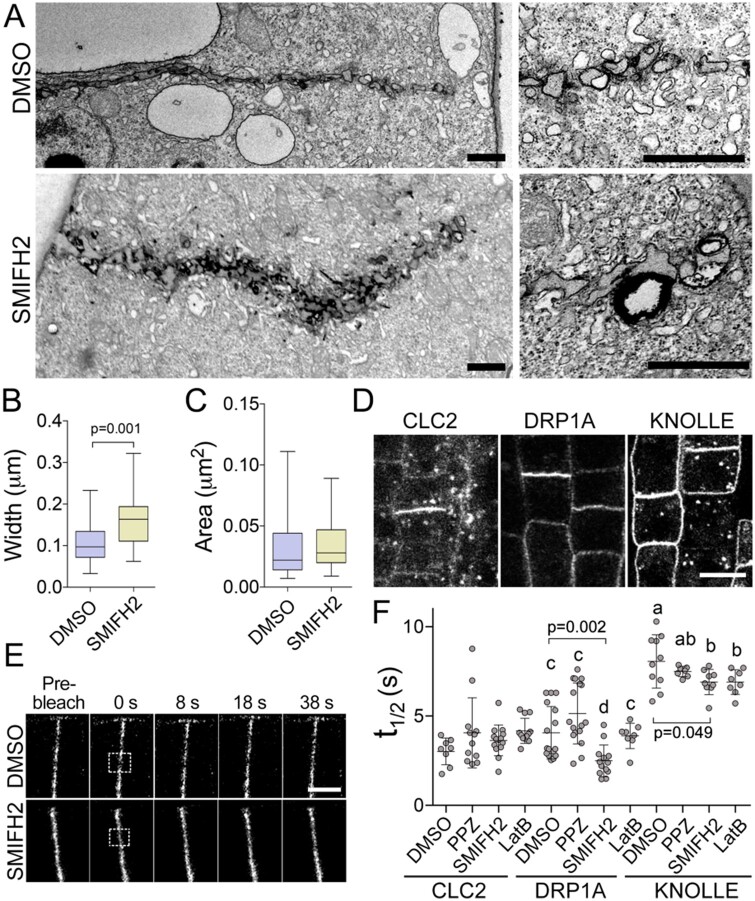
To characterize the cell plate defects in more detail, we fixed and sectioned BY-2 cells treated with 2.5 µM SMIFH2 for 3 h and performed transmission electron microscopy. Image analysis revealed cell plates were irregularly shaped and were significantly thicker than those in mock-treated cells (on average, 171 ± 59 nm versus 107 ± 49 nm; t test P = 0.001; Figure 8, A and B). The mean area of individual cytokinetic vesicles at the cell plate leading edge was not significantly different (35 ± 23 nm2 in SMIFH2-treated cells versus 31 ± 21 nm2 in control; t test P = 0.08; Figure 8C). These data indicate impairment of the cell plate remodeling by SMIFH2.
Figure 8.
SMIFH2 inhibits cell plate membrane remodeling. A, Electron microscopic images of cell plate morphologies in BY-2 cells after treatment with 2.5 µM SMIFH2 for 3 h. Scale bar, 1 µm. B, Statistical analysis of cell plate morphology using electron micrographs. P-value was calculated using unpaired t test (n = 28). C, Area of the individual vesicles at the growing edge of the cell plate in BY-2 cells treated with DMSO or 2.5 µM SMIFH2 (n = 28). Whiskers denote minimal to maximal range of values. D, Confocal microscopy images showing CLC2-GFP, DRP1A-GFP, and KNOLLE-GFP localization to the cell plates of Arabidopsis root epidermal cells. Scale bar, 5 µm. E, Representative confocal time-lapse images of photobleaching experiments in Arabidopsis root apical meristem cells expressing DRP1A-GFP after treatment with DMSO or 25 µM SMIFH2. The photobleached areas are indicated by the rectangles. Scale bar, 5 µm. F, Turnover of CLC2-GFP, DRP1A-GFP, and KNOLLE-GFP in root epidermis cell after treatment with 0.5% (v/v) DMSO, 10 µM SMIFH2, 5 µM propyzamide (PPZ), or 5 µM latruncnulin B (LatB) for 6 h. Significantly different average values are denoted by different letters as determined by one-way ANOVA test.
Abnormal cell plate remodeling could be the consequence of membrane recycling defects. To determine which processes were impaired by SMIFH2 treatment, we used FRAP to measure turnover (t1/2) of the exocytosis component syntaxin KNOLLE fused to GFP, the endocytosis component clathrin light chain (CLC2) fused to GFP, and the endocytosis and membrane remodeling factor dynamin-related protein 1A (DRP1A) fused to GFP (Figure 8, D and E). While t1/2 values of CLC2-GFP were unaffected by 10 µM SMIFH2 treatment for 6 h, the turnover of DRP1A-GFP and KNOLLE-GFP were faster (Figure 8F), indicating shorter dwell time of both proteins in the cell plate. To examine whether changes in the turnover were associated with defects in microtubules or actin dynamics, we measured the impact of propyzamide and latrunculin B on the turnover of the above fusion proteins. CLC2-GFP and DRP1A-GFP turnovers were not affected by either treatment. These results suggest that SMIFH2 affects turnover of DRP1A independently of microtubules or actin. Slight reduction of the turnover values of KNOLLE-GFP in cells treated by SMIFH2 could be mimicked by the treatment with latrunculin B, but not by treatment with propyzamide.
Discussion
Formins, EB1s, and microtubule dynamics
Plant formins are known to regulate microtubule dynamics. Knockout of class I formin AtFH1 causes reduced microtubule stability and greater frequency of depolymerization events (Rosero et al., 2013). In our experiments, SMIFH2 caused reduction of microtubule polymerization during interphase. Another phenotype was inhibition or delay of exit from metaphase and telophase, both are microtubule-dependent processes. This regulation can occur through direct binding to microtubule lattice. Such hypothesis is supported by several observations. Arabidopsis thaliana type II formins AtFH14 and AtFH16 bind microtubules in vivo as well as bind, bundle, and stabilize microtubules in vitro (Li et al., 2010; Wang et al., 2013). In our experiments, AtFH8 binds microtubules in N. benthamiana leaf pavement cells.
Formins can also regulate microtubule dynamics through interaction with EB1. It has been shown that animal FH2 binds microtubule plus-tip protein EB1 (Bartolini et al., 2008; Gaillard et al., 2011). Interaction of formins with EB1 contributed to stabilization of microtubules in cooperation with another EB1 interacting partner kinesin Kif4 (Morris et al., 2014). AtFH1 and AtFH8 interact with all three A. thaliana EB1 isoforms in vitro and target AtEB1c to their localization sites in vivo. AtEB1c was re-directed from the nucleus, where it normally localizes during interphase, to the plasma membrane by AtFH1 and to microtubules by AtFH8. This fact explains why formins do not exhibit microtubule plus-end tracking properties. Instead, association with formins could modulate EB1 affinity to microtubule lattice and facilitate interaction of microtubule growing tips with other structures, e.g. plasma membrane or the cell plate. Analysis of A. thaliana wild-type and Ateb1 triple mutant seedlings growth on vertical agar plates with or without SMIFH2 showed that formins cooperate with EB1s in promoting root growth and controlling the root apical meristem size.
One of the SMIFH2 phenotypes was reduction of AtEB1b or AtEB1c comet size during interphase or mitosis, respectively. EB1 proteins interact stronger with GTP-tubulin than with GDP-tubulin lattice (Maurer et al., 2011). Consequently, the shape and size of the comets informs on the association of tubulin dimers with growing microtubule tips and on the tubulin GTPase activity. We fitted exponential curves into the GFP-EB1 signal decay values on the proximal and distal ends of the comet to determine the role of formins in both processes. The EB1 fluorescence signal decay curves were steeper on both slopes of mCherry-AtEB1b comets in SMIFH2-treated cells relative to DMSO-treated controls. Faster decay of the proximal signal suggests formins either reduced tubulin GTPase activity or increased affinity of EB1 to microtubules.
The distal slope reflects the size of tubulin protofilament flares that form as a consequence of the stochastic nature of tubulin addition to the microtubule end (Kerssemakers et al., 2006; Akhmanova and Steinmetz, 2008). According to electron microscopy analyses, the presence of flares corresponds with microtubule polymerization in vitro (Mandelkow et al., 1991) and in vivo (Hoog et al., 2011). We found that AtEB1b-GFP signal decay in cells of the temperature-sensitive mor1-1 mutant was faster on the distal slope, but not affected on the proximal slope. This outcome is consistent with the function of MOR1 in promoting addition of tubulin on the growing microtubule ends (Kawamura and Wasteneys, 2008) and supports conclusion that formins facilitate microtubule polymerization by stabilizing the tubulin protofilament flares on the growing microtubule end. Slower microtubule polymerization rates and shorter EB1-comets in SMIFH2-treated cells support the above scenario.
Regulation of EB1 comet size by proteins that promote microtubule polymerization has been observed in other systems. In Neurospora crassa, the length of MTB-3 (EB1 homolog) comets was independent of microtubule polymerization rates, but was affected by microtubule binding proteins (Mourino-Perez et al., 2013). In neurons, knockdown of Tau reduced the size of EB1 comets (Sayas et al., 2015). SMIFH2 treatment of EB2-depleted human cells also diminished EB1 association with the microtubule lattice (Goldspink et al., 2013). Hence, regulation of the growing microtubule tips by formins appears to be evolutionarily conserved.
Interaction of formins with microtubules and their localization to other structures appears to be dependent on the cell cycle regulation. We found AtFH8 binds microtubules in N. benthamiana leaf pavement cells but not in proliferating BY-2 cells. Also, FH8-GFP did not label microtubules in the A. thaliana hypocotyl epidermal cells and in the root apical meristem (Xue et al., 2011; Diao et al., 2018). However, AtFH8 localized to the nuclear envelope in BY-2 cells and in cells of root apical meristem (Xue et al., 2011). These observations suggest cell cycle and tissue-specific regulation of FH8 localization to microtubules, nuclear envelope, and the plasma membrane. It is also possible that species-specific features could affect localization of formins.
Formins regulate cytoskeletal dynamics in the phragmoplast
Our work shows multiple roles of plant formins in cytokinesis as illustrated in the working model in Figure 9. Plant formins can regulate actin dynamics during cytokinesis. AtFH1 binds plasma membrane in the pollen tubes through its N-terminus and induces F-actin polymerization from the plasma membrane (Cheung and Wu, 2004). AtFH5 nucleates F-actin and localizes to the plasma membrane region at the growing pollen tube tips (Ingouff et al., 2005; Cheung et al., 2010). Several facts indicate that formins function in nucleating F-actin in the phragmoplast: AtFH1, AtFH5, and tFH8 localized to the BY-2 cell plate; F-actin disappeared from the cell plate region following treatment with high concentrations of SMIFH2; and width of the actin zone at the cell plate was reduced by treatment with lower concentration of SMIFH2.
Figure 9.
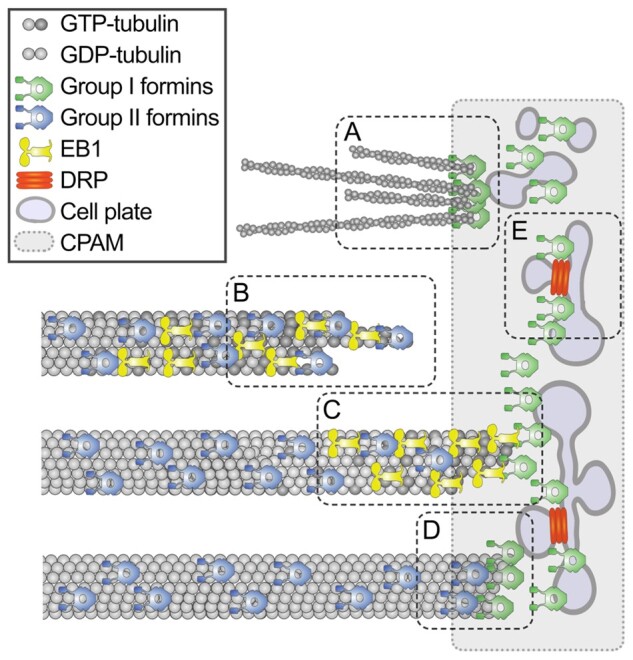
Model of formin localization and functions during cytokinesis. During cytokinesis group I formins localize on the cell plate membrane and in the cell plate assembly matrix; group II formins localize on microtubules. The first function of formins is nucleating F-actin by the cell plate (A). The second function is promoting microtubule elongation by stabilizing tubulin protofilament flares on the growing plus-tips (B). When microtubule tip approaches the cell plate, formins dock the microtubule tip to the cell plate assembly matrix (CPAM) through interaction with EB1 proteins (C). Formins localized within CPAM and on the cell plate stabilizes microtubule plus-ends (D). Formins can also contribute to the cell plate membrane recycling by facilitating recruitment of DRPs to the cell plate through yet unknown mechanism (E). Only one-half of the phragmoplast is shown in the model.
F-actin contributes to the phragmoplast expansion rate, cell plate morphology, and cell plate orientation (Baluska et al., 2001; Sano et al., 2005; Higaki et al., 2008; Zhang et al., 2009; Kojo et al., 2013; Wu and Bezanilla, 2014). However, much stronger cytokinetic defects were observed when actin polymerization inhibitors were applied before anaphase than during cytokinesis (Sano et al., 2005). Inhibition of formins in BY-2 cells in our work slowed down phragmoplast expansion, but in contrast with the actin polymerization inhibitors, SMIFH2 also perturbed the cell plate synthesis. This fact points out additional roles of formins in the phragmoplast.
The second function of formins in the phragmoplast is promoting microtubule polymerization (Figure 9). SMIFH2 treatment caused faster microtubule turnover and greater fraction of nondynamic microtubules (the immobile fraction). Treatment with a low concentration of an inhibitor of microtubule polymerization propyzamide also resulted in faster microtubule turnover and greater fraction of nondynamic microtubule. Treatment with an inhibitor of microtubule depolymerization taxol reduced the turnover rate, but also resulted in greater immobile fraction. A slower microtubule growth rate in cells treated with SMIFH2 or propyzamide would cause slower depletion of free tubulin. Consequently, concentration of free tubulin will be higher leading to: (1) more frequent microtubule nucleation and faster turnover and (2) reduced microtubule depolymerization and greater fraction of stable microtubules.
The greater immobile fraction can also be a consequence of the cell plate assembly defects. Electron tomography analysis discerned that some microtubules terminate proximal to the assembling cell plate (Otegui et al., 2001; Segui-Simarro et al., 2004). These microtubules have blunt ends, which are characteristic of stable microtubules (Austin et al., 2005), suggesting interactions with the cell plate can stabilize microtubules. The lifetime of some microtubules in tobacco BY-2 cell phragmoplasts was found to be longer than 3 min (Murata et al., 2013), while an average microtubule turnover time in the phragmoplast is in the range of 40–60 s (Hush et al., 1994; Smertenko et al., 2011). Formins at the cell plate can capture the growing microtubule ends through interaction with EB1 thus preventing their depolymerization (Figure 9).
Phragmoplast microtubules detach from the cell plate and depolymerize when the cell plate assembly reaches tubular network stage (Samuels et al., 1995). It was hypothesized that both microtubule detachment and depolymerization are governed by a cell plate quality control mechanism (Smertenko et al., 2018). Consequently, cell plate assembly defects would attenuate microtubule depolymerization leading to a greater fraction of stable microtubules.
Formins function in cell plate remodeling
The fourth function of formins in cytokinesis is cell plate remodeling (Figure 9). Gradual transition of the cell plate from initial membrane fusion to continuous sheet depends on a balance between exocytosis and endocytosis (Samuels et al., 1995; Otegui et al., 2001). Growth of pollen tube also relies on the balance between exocytosis and endocytosis. Formins play an essential role in this process. AtFH5 binds to the plasma membrane region at the growing pollen tube tips where exocytosis takes place (Cheung et al., 2010). Over-expression of AtFH1 induced ballooning of the pollen tip and excessive invagination that resemble exocytic or endocytic vesicles (Cheung and Wu, 2004).
The role of individual formins in the exocytosis and endocytosis at the cell plate appears to be redundant. No discernible cytokinetic phenotype was reported in atfh1 mutant (Rosero et al., 2016) and knockout mutant atfh5 shows a delay in the endosperm cellularization but no cell plate assembly defects (Ingouff et al., 2005). We found that SMIFH2 treatment phenocopies the abnormally thick cell plates reported in the DRP1a knockout (ADL1) in A. thaliana (Kang et al., 2003). The cytokinetic vesicles are shaped at the cell plate by dynamin-related proteins (Segui-Simarro et al., 2007; McMichael and Bednarek, 2013). Faster turnover of DRP1a at the cell plate in SMIFH2-treated cells suggests that formins facilitate recruitment of DRP1a to the cell plate. Treatment with propyzamide or latrunculin B could not phenocopy this effect indicating that formins govern membrane remodeling independently of their role in regulating dynamics of actin filaments and microtubules.
The diversity of formin functions is consistent with their localization to both phragmoplast microtubules and the cell plate (Ingouff et al., 2005; Li et al., 2010; Xue et al., 2011; Wu and Bezanilla, 2014). A similar phenomenon was reported in other organisms. For example, Saccharomyces cerevisiae formin Bni1p partners with myosin heavy chain Myo1p in constructing the actomyosin contractile ring, whereas a second formin Bnr1p cooperates with the septum assembly protein Hof1p (Vallen et al., 2000). Interaction between Myo1p and Bni1p is essential for recruiting a key cytokinetic protein, myosin light chain Mlc1, to the cytokinesis site (Feng et al., 2015).
Formins govern the vesicle trafficking to the cell plate and rely on the vesicle trafficking for the membrane localization. During interphase, AtFH1 localizes to early and late endosomes (Oulehlova et al., 2019). Localization of AtFH1 was affected by treatments with inhibitors of vesicular trafficking such as wortmannin and brefeldin A (Oulehlova et al., 2019). Treatment with SMIFH2 caused slower turnover of AtFH5 and faster turnover or loss of AtFH8 on the cell plate. It means individual members of formin family could be regulated by specific mechanisms of trafficking and retention at the cell plate. Faster turnover of FH8 in response to SMIFH2 treatment indicates that microtubules and F-actin contribute to the retention of FH8 at the cell plate. Slower turnover of FH5 suggests that it could be recycled with the cell plate membrane. Bigger cell plate compartments along with faster DRP1a turnover suggest defects in the endocytosis. FH5 turnover could be facilitated by the endocytosis. The retention of formins at the cell plate could also be governed by interaction of formin extracellular domain with specific polysaccharides in the cell plate lumen. For example, AtFH1 and AtFH8 co-localize with the callose in the cell wall, whereas localization of FH5-GFP did not correlate with the callose staining (Diao et al., 2018; Oulehlova et al., 2019).
In conclusion, our work identifies four major functions of formins during cytokinesis (Figure 9). Some of these functions, such as reduction of phragmoplast expansion rate and abnormal shape of the cell plate are likely to be associated with the defect in F-actin, whereas slower microtubule growth and abnormal EB1-comet shape are indicative of microtubule-related functions. Reduced retention of DRP1A is likely to be a consequence of a yet unknown role of formins in regulating membrane remodeling. It is likely that several members of the formin protein family perform each of these functions. The next important step would be refining and strengthening of the model by assigning specific functions to individual formin members.
Materials and methods
BY-2 cell culture and cell cycle synchronization
Tobacco (N. tabacum) BY-2 cells were grown in Murashige and Skoog (MS) medium supplemented with 200 mg L−1 KH2PO4, 3% (w/v) sucrose and 0.2 mg L−1 2,4-dichlorophenoxyacetic acid (2.4-D), pH 5.8, at 25°C in the dark (Nagata et al., 1981). Cells were sub-cultured every 7 d by transferring 1 mL of old culture into 60 mL of fresh medium. For the cell cycle synchronization experiments, 6 mL of 7-d-old BY-2 culture were inoculated into 60 mL of fresh medium containing 3-µM aphidicolin, and incubation for 24 h under normal growth conditions. Then, cells were washed intensively in several changes of 30% (w/v) sucrose solution (total 1.5 L) in sintered glass funnel to remove aphidicolin and incubated in 60 mL of fresh medium for 4–6 h. Propyzamide was added to the final concentration of 5 µM. Accumulation of the metaphase cells was monitored every hour starting from 4 h by staining cell aliquots in DAPI solution (Smertenko et al., 2016). Once metaphase index plateaued (∼5–7 h), the propyzamide was washed out with 30% sucrose as described above. The cells were incubated in 60 mL of fresh medium.
Drug treatments
Stock solutions of SMIFH2 (40 mM), propyzamide (10 mM), and taxol (20 mM) were prepared in DMSO and stored at −80°C. Aliquots of SMIFH2 were allowed through one freeze–thaw cycle. To measure the impact of SMIFH2 on cell growth, 1 mL of 7-d-old BY-2 cell suspension was seeded into 60 mL of medium in a 250 mL flask containing the desired concentration of SMIFH2. The control flask was supplemented by 0.0125% (v/v) of DMSO that is equivalent concentration in the 5 µM SMIFH2. On days 3 and 5, the fresh weight of cells was measured in 10 mL of medium. The inhibition of growth was calculated relatively to the fresh weight in 10 mL of cells taken from the control flask. For short treatments of BY-2 cells, 1 mL of 3-d-old culture was transferred to 30 mL BY-2 medium in 150 mL glass flask and grown at 25°C for 36 h, after which the culture was supplemented with the drug and incubated as specified in the text. For taking live-cell imaging, 1 mL of cell culture was transferred in a small petri dish (35-mm diameter) with 20-mm diameter coverslip at the base. Alternatively, cells were fixed and stained as described below. For treating roots, 2-d-old seedlings were transferred onto 1% (w/v) agar plus half-strength MS medium supplemented with 10 µM SMIFH2.
Testing cell viability
Evan’s Blue solution at 1% (w/v) was prepared in BY-2 tissue culture medium and mixed at a ratio of 1:1 with the cells, incubated for 5 min, then washed several times in fresh BY-2 tissue culture medium. FM4-64 and FDA staining were performed as described (Minina et al, 2013). Cells were imaged using a Leica DMI4000 fluorescence microscope.
Arabidopsis thaliana lines and growth conditions
All A. thaliana transgenic and mutant lines used in this study (GFP-KNOLLE, GFP-DRP1a, GFP-CLC2 (clathrin light chain), GFP-EB1a, Ateb1a Ateb1b Ateb1c) were derived from Columbia ecotype, so plants of this ecotype were used as wild-type controls in all experiments. Generation of the Ateb1a Ateb1b Ateb1c triple mutant is described by Galva et al. (2014). Seeds stratification was performed at 4°C for 48 h and plants were grown at 22°C with 16-h light at 100–150 mE/m2. For seedling treatment experiments, surface-sterilized seeds were sown on 1% (w/v) agar plus half-strength MS medium plates and stratified for 2 d at 4°C. Germination was performed in growth chambers on horizontal plates at 22°C with 16/8-h light/dark cycle for 7 d. Then seedlings were transferred onto and grown on vertically oriented 1% (v/w) agar plus half-strength MS plates containing either drug or DMSO under the condition detailed above.
Cloning and constructs
Total RNA was isolated from Col-0 leaves or BY-2 cells (RNeasy mini kit, Qiagen). cDNA was synthesized using oligo-dT primer. Tobacco α-tubulin NtTUA1 (GB accession number NM_001325628.2) and Arabidopsis full-length AtFH1, AtFH5,and AtFH8 coding sequences were amplified by PCR with primers listed in Supplemental Table S1. The PCR products were cloned into Gateway entry vector pDONR207 (Invitrogen), and confirmed by sequencing. The coding sequences of the formins were cloned into the binary destination vector pMDC83 (Curtis and Grossniklaus, 2003) and NtTUA1 into pUBCGFP.
In addition, we used the previously published constructs for transformation of BY-2 cells: GFP-Lifeact (Smertenko et al., 2010), mCherry-AtEB1b (Eisinger et al., 2012), pEB1c::EB1c:GFP (Novák et al., 2015), and mCherry-SCAMP2 (Toyooka et al., 2009).
Transient expression and bimolecular fluorescence complementation
Full-length CDS of AtFH1 and AtFH8 were amplified from A. thaliana Col-0 leaf cDNA via PCR using corresponding primers (Supplemental Table S1). The PCR products were cloned into pDONR207 vector using Gateway technology (Invitrogen, ThermoFisher Scientific, USA). Full-length CDS of EB1c was cloned into pDONR221 vector as previously described (Novák et al., 2015) and introduced into binary vectors as follows: to fuse the gene N-terminally to N-terminal or C-terminal portion of enhanced yellow fluorescent protein (EYFP) using pSITE-nEYFP-N1 (for AtFH1 and AtFH8) and pSITE-cEYFP-N1 (for EB1c), respectively; to fuse the gene C-terminally to C-terminal portion of EYFP using pSITE-cEYFP-C1 (for EB1c)(Martin et al., 2009). The constructs were transformed into Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures were grown as described previously (Schmidt and Smertenko, 2019) except that the final OD600 of bacteria solution for infiltration was adjusted to 0.6. Cells containing EB1c in pSITE-cEYFP-N1 were mixed with cells harboring either AtFH1 or AtFH8 in pSITE-nEYFP-N1 and p19 at the ratio 1:1:0.5. For the negative controls, cells harboring EB1c in pSITE-cEYFP-C1 were mixed with cells harboring either AtFH1 or AtFH8 in pSITE-nEYFP-N1 and p19 at the ratio 1:1:0.5. N. benthamiana plants were infiltrated as described in Schmidt and Smertenko (2019). Leaves were imaged after 2 d of infiltration.
Plant transformation
Destination vectors were transformed into A. tumefaciens strains GV3101 or LBA4404. Arabidopsis thaliana ecotype Columbia (Col-0) was transformed by floral dip (Clough and Bent, 1998). T0 seeds were surface-sterilized, stratified as above and grown in half-strength MS agar medium containing 1% (w/v) agar hygromycin B (40 mg/L) or kanamycin (50 mg/L).
For BY-2 cell transformation, 100 µL of overnight Agrobacterium culture was added to 5 mL of 3-d-old BY-2 cell culture and co-cultured at 25°C for 3 d. Then, BY-2 cells were collected, washed three times using BY-2 medium and transferred to plates containing solidified MS medium supplemented with 0.7% (w/v) agar, 100 mg/mL of carbenicillin, and 40 mL/L of hygromycin B or 100 mg/mL of kanamycin. After 3–4 weeks’ growth, calli were examined under a fluorescent dissecting microscope, and positive clones transferred to plates with solidified medium or flasks with 20 mL of liquid medium. Transient expression in N. benthamiana leaf pavement cells was performed as described earlier (Smertenko et al., 2008). The images were acquired 3–5 d after the infiltration.
Fluorescence microscopy
Fixation and staining of cells was performed as described (Smertenko et al., 2004). Rat monoclonal anti α-tubulin clone YOL-34 was used as primary at dilution 1:400. Anti-rat Alexa-Fluor 488 conjugate was used as secondary antibody at dilution 1:400. For the cell membrane staining, Arabidopsis roots were incubated in 1-µM solution of FM4-64 for 10 min and imaged using a Leica SP8 laser confocal scanning microscope. Arabidopsis hypocotyl cells expressing AtEB1c-GFP were imaged using a Leica TIRF microscope equipped with iLas targeted laser illuminator (Roper Scientific). The exposure time was set at 0.1 s.
Live cell imaging was performed using Leica SP8X confocal microscope equipped with hybrid detectors that were used in BrightR mode. GFP was excited with 488 nm line of argon laser line for exciting GFP and mCherry was excited with 561 nm solid state laser. The emitted light was collected between 495 and 550 nm for GFP and 570 and 650 nm for mCherry. The laser intensity varied between 1% and 10% and gain was between 75 and 450. Fluorescence recovery after photobleaching experiments was performed using a Leica SP8 laser confocal scanning microscope using a previously established protocol (Chang et al., 2005). At least five biological replicates were performed per each experimental condition.
Electron microscopy
Cells were fixed overnight at 4°C in 2% (v/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), post fixed overnight at 4°C in 2% (w/v) OsO4 with 0.8% (w/v) potassium ferrocyanide in 0.05 M cacodylate buffer, and then, after a standard ethanol/acetone dehydration procedure, embedded in Spurr’s epoxy resin. Ultra-thin sections (70 nm) were made on a Reichert Ultracut R ultramicrotome (Reichert-Jung GmbH, Heidelberg, Germany) and stained for transmission electron microscopy with 4% (w/v) uranyl acetate followed by 2% (w/v) lead citrate. Zeiss Libra 120 transmission electron microscope (Oberkochen, Germany) equipped with a Dual Speed 2K-On-axis CCD-Camera (Moorenweis, Germany) was used for observation and photography.
Image analysis and cell cycle quantification
All image measurements and image analyses were performed using Fiji. The intensity of GFP-Lifeact was measured along the line perpendicular to the cell plate orientation. For the analysis of cell cycle, cells were treated with the inhibitors as specified above. Then cells were fixed and stained with DAPI for counting mitotic index, or immunolabeled with anti-tubulin according to the published procedure (Smertenko et al., 2004) and then stained with DAPI for counting specific stages of the M-phase. Sufficient number of cells was counted to accumulate at least 100 cells at M-phase. On average, 1,500 cells were counted per each biological repeat. EB1 comets were measured in three roots, 4–5 comets per root.
Protein expression and purification
The FH1FH2 domains of AtFH1 and AtFH8 were amplified by PCR from total cDNA generated from Arabidopsis plant tissue and cloned into the pET-30a Gateway vector. AtFH1 and AtFH8 proteins were expressed and purified using previously established procedures (Ingouff et al, 2005; Michelot et al., 2006; Xue et al., 2011).
Generation of the pGEM-T Easy GST-AtEB1b expression construct along with the protocols for expressing/purifying the protein and performing GST pull-out assays can be found in Galva et al. (2014). pGEM-T Easy GST-AtEB1a and GST-AtEB1c constructs were generated the same way, but with gene-specific primers.
Actin and microtubule polymerization assays
G-actin was purified from chicken muscle acetone powder using the procedure published in (Ly et al., 2016). Pyrene-iodoacetamide-labeled G-actin was prepared as described (Kouyama and Mihashi, 1981; Cooper et al., 1983), and the protein was stored in liquid nitrogen. Before experiments, pyrene-actin was thawed at 37°C and centrifuged at 50,000 r.p.m. (Beckman Optima MAX-XP Tabletop Ultracentrifuge, MLA-80 Rotor), 4°C, for 20 min to remove aggregates. Actin polymerization was detected as the change in pyrene-actin fluorescence (Kouyama and Mihashi, 1981) using a Photon Technology International fluorometer (Edison, NJ; excitation, 366 nm, and emission, 387 nm, with a 2 nm slit). To measure actin nucleation, 1.5 µM G-actin was mixed with 0.15 µM pyrene-labeled G-actin, FH1 (12.5, 50, and 100 nM) and SMIFH2 (10 and 40 µM). The reaction was initiated by addition of polymerization buffer (100 mM KCl, 2 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0). Spontaneous actin nucleation in the absence of FH1 and SMIFH2 was measured as a control.
Turbidimetric assay of microtubule polymerization was performed using previously published procedure using 30-µM tubulin (Smertenko et al., 2004). SMIFH2 was diluted to final concentration 10 µM in the microtubule polymerization buffer (50 mM PIPES, pH = 6.9, 1 mM EGTA, 1 mM MgCl2) and then added to the tubulin polymerization reaction at the desired concentration.
Statistical analysis
Datasets were analyzed using Student’s t test or Analysis of Variance (ANOVA) using Prism Software. Densitometric profiles of actin localization in the phragmoplast midzone or EB1 comets were generated using “Plot profile” function of the ImageJ (Schindelin et al., 2012). EB1-comet profiles were normalized by maximal value and analyzed with a dimensionless sensitivity method using a MatLab software package as described before (Smertenko et al., 2005). This approach fitted experimental data in to the exponential equation:
where x is argument, α is the dimensionless differential slope determined using the following equation:
Densitometric profiles of Lifeact in the midzone were normalized by the maximal signal value and aligned so that the maximum value located on the y-axes. Curves were then averaged using the ORIGIN 7.5 software package and analysed as above.
Accession numbers
AtFH1 (AT3G25500), AtFH5 (AT5G54650), AT1G70140 (AtFH8), EB1a (AT3G47690), EB1b (AT5G62500), EB1c (AT5G67270), MOR1/GEM1 (AT2G35630), KNOLLE (AT1G08560), DRP1A (AT5G42080), CLC2 (AT2G40060)
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phragmoplast asymmetries.
Supplemental Figure S2. In vitro and in vivo effects of SMIFH2.
Supplemental Figure S3. Effects of SMIFH2 on F-actin and microtubules in phragmoplasts of BY-2 cells.
Supplemental Figure S4. Localization of AtFH1-GFP, AtFH5-GFP, and AtFH8-GFP in interphase and telophase BY-2 cells.
Supplemental Figure S5. Representative images of phragmoplasts in FRAP experiments.
Supplemental Figure S6. Impact of SMIFH2 treatment on the mCherry-EB1b comet shape in vivo.
Supplemental Figure S7. EB1c interact with formins in vivo and in vitro.
Supplemental Table S1. Sequence of primers used in this study.
Supplementary Material
Acknowledgments
The authors are grateful to Viktor Kirik (Illinois State University) for the mCherry-AtEB1b construct; Chris Staiger (Purdue University) for the pET30a AtFH1 and AtFH8 FH1FH2 domain expression constructs; Ken Matsuoka (Kyushu University) and Kiminori Toyooka (RIKEN) for the mCherry-SCAMP2 construct; Jordy Chan (John Innes Centre) for the GFP-AtEB1c construct; Panagiotis Moschou (Swedish Agricultural University) for seeds of Arabidopsis GFP-CLC2, GFP-KNOLLE, and GFP-DRP1a lines; Jeoffrey Wasteneys (University of British Columbia) for seeds of Arabidopsis EB1b-GFP in mor1 background; Despoina Kaloriti for assistance in genotyping Ateb1 mutant plants; Core Facility Center “Cell and Molecular Technologies in Plant Science” of Komarov Botanical Institute and Franceschi Microscopy and Imaging Center of Washington State University for use of their facilities.
Funding
This research was supported by Washington State University startup grant, USDA NIFA Hatch grant #1015621, and NSF-CAREER award #1751204 to A.S.; by ERDF project “Plants as a tool for sustainable global development” No. CZ.02.1.01/0.0/0.0/16_019/0000827 to J.Š.; by grant #0524355 from the National Science Foundation to J.C.S.; and by Washington State University startup grant to A.S.K.
Conflict of interest statement. The authors declare no conflict of interests.
Conceptualization, A.S., J.C.S., J.Š.; Investigation, L.Z., D.F., N.K., N.M., C.G., T.S., A.S.; Formal Analysis, E.M., P.S.; Resources, D.N., A.K., J.C.S.; Writing—Original Draft, L.Z., A.S., A.S.K.; Writing—Review & Editing, L.Z., D.F., N.K., N.M., C.G., A.S., P.S., A.S.K., J.Š.; Funding Acquisition, A.S., J.C.S., J. Š., A.S.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Andrei Smertenko (andrei.smertenko@wsu.edu)
References
- Akhmanova A, Steinmetz MO (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9: 309–322 [DOI] [PubMed] [Google Scholar]
- Asada T, Sonobe S, Shibaoka H (1991) Microtubule translocation in the cytokinetic apparatus of cultured tobacco cells. Nature 350: 238–241 [Google Scholar]
- Austin JR II, Segui-Simarro JM, Staehelin LA (2005) Quantitative analysis of changes in spatial distribution and plus-end geometry of microtubules involved in plant-cell cytokinesis. J Cell Sci 118: 3895–3903 [DOI] [PubMed] [Google Scholar]
- Bajer A (1968) Fine structure studies on phragmoplast and cell plate formation. Chromosoma 24: 383–417 [Google Scholar]
- Baluska F, Jasik J, Edelmann HG, Salajova T, Volkmann D (2001) Latrunculin B-induced plant dwarfism: Plant cell elongation is F-actin-dependent. Dev Biol 231: 113–124 [DOI] [PubMed] [Google Scholar]
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG (2008) The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol 181: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Komis G, Ziemann A, Menzel D, Šamaj J (2011) Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol 189: 1069–1083 [DOI] [PubMed] [Google Scholar]
- Becker WA (1938) Recent investigations in vivo on the division of plant cells. Bot Rev 4: 446–472 [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T (2007) Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450: 1100–1105 [DOI] [PubMed] [Google Scholar]
- Bisgrove SR, Lee YR, Liu B, Peters NT, Kropf DL (2008) The microtubule plus-end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis. Plant Cell 20: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van Damme D (2015) Endomembrane trafficking overarching cell plate formation. Curr Opin Plant Biol 28: 92–98 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL (2013) Formins at a glance. J Cell Sci 126: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Henty-Ridilla JL, Blanchoin L, Staiger CJ (2016) Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol 170: 220–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Smertenko AP, Igarashi H, Dixon DP, Hussey PJ (2005) Dynamic interaction of NtMAP65-1a with microtubules in vivo. J Cell Sci 118: 3195–3201 [DOI] [PubMed] [Google Scholar]
- Cheng LN, Zhang JY, Ahmed S, Rozier L, Yu HQ, Deng HT, Mao YH (2011) Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell 20: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Niroomand S, Zou YJ, Wu HM (2010) A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc Natl Acad Sci U S A 107: 16390–16395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2004) Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooper JA, Walker SB, Pollard TD (1983) Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil 4: 253–262 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Fendrych M, Smertenko A, Bell KS, Oparka K, Cvrckova F, Žárský V, Hussey PJ (2010) The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J Cell Sci 123: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ, Davies B (2002) Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci 7: 492–498 [DOI] [PubMed] [Google Scholar]
- Diao M, Ren SL, Wang QN, Qian LC, Shen JF, Liu YL, Huang SJ (2018) Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. Elife 7: e36316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Chang E, Cyr R (2006) Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol Biol Cell 17: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger WR, Kirik V, Lewis C, Ehrhardt DW, Briggs WR (2012) Quantitative changes in microtubule distribution correlate with guard cell function in Arabidopsis. Mol Plant 5: 716–725 [DOI] [PubMed] [Google Scholar]
- Eleftheriou EP, Baskin TI, Hepler PK (2005) Aberrant cell plate formation in the Arabidopsis thaliana microtubule organization 1 mutant. Plant Cell Physiol 46: 671–675 [DOI] [PubMed] [Google Scholar]
- Favery B, Chelysheva LA, Lebris M, Jammes F, Marmagne A, de Almeida-Engler J, Lecomte P, Vaury C, Arkowitz RA, Abad P (2004) Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Okada S, Cai G, Zhou B, Bi E (2015) MyosinII heavy chain and formin mediate the targeting of myosin essential light chain to the division site before and during cytokinesis. Mol Biol Cell 26: 1211–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN (2011) Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell 22: 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galva C, Kirik V, Lindeboom JJ, Kaloriti D, Rancour DM, Hussey PJ, Bednarek SY, Ehrhardt DW, Sedbrook JC (2014) The microtubule plus-end tracking proteins SPR1 and EB1b interact to maintain polar cell elongation and directional organ growth in Arabidopsis. Plant Cell 26: 4409–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink DA, Gadsby JR, Bellett G, Keynton J, Tyrrell BJ, Lund EK, Powell PP, Thomas P, Mogensen MM (2013) The microtubule end-binding protein EB2 is a central regulator of microtubule reorganisation in apico-basal epithelial differentiation. J Cell Sci 126: 4000–4014 [DOI] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Hasezawa S (2008) Quantitative analysis of changes in actin microfilament contribution to cell plate development in plant cytokinesis. BMC Plant Biol 8: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog JL, Huisman SM, Sebo-Lemke Z, Sandblad L, McIntosh JR, Antony C, Brunner D (2011) Electron tomography reveals a flared morphology on growing microtubule ends. J Cell Sci 124: 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hush JM, Wadsworth P, Callaham DA, Hepler PK (1994) Quantification of microtubule dynamics in living plant-cells using fluorescence redistribution after photobleaching. J Cell Sci 107: 775–784 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Fitz Gerald JN, Guerin C, Robert H, Sorensen MB, Van Damme D, Geelen D, Blanchoin L, Berger F (2005) Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat Cell Biol 7: 374–380 [DOI] [PubMed] [Google Scholar]
- Ishida T, Thitamadee S, Hashimoto T (2007) Twisted growth and organization of cortical microtubules. J Plant Res 120: 61–70 [DOI] [PubMed] [Google Scholar]
- Isogai T, van der Kammen R, Innocenti M (2015) SMIFH2 has effects on Formins and p53 that perturb the cell cytoskeleton. Sci Rep 5: 9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY (2003) Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15: 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E, Wasteneys GO (2008) MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J Cell Sci 121: 4114–4123 [DOI] [PubMed] [Google Scholar]
- Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M (2006) Assembly dynamics of microtubules at molecular resolution. Nature 442: 709–712 [DOI] [PubMed] [Google Scholar]
- Kojo KH, Higaki T, Kutsuna N, Yoshida Y, Yasuhara H, Hasezawa S (2013) Roles of cortical actin microfilament patterning in division plane orientation in plants. Plant Cell Physiol 54: 1491–1503 [DOI] [PubMed] [Google Scholar]
- Komaki S, Abe T, Coutuer S, Inze D, Russinova E, Hashimoto T (2010) Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J Cell Sci 123: 451–459 [DOI] [PubMed] [Google Scholar]
- Komis G, Luptovčiak I, Ovečka M, Samakovli D, Šamajová O, Šamaj J (2017) Katanin effects on dynamics of cortical microtubules and mitotic arrays in Arabidopsis thaliana revealed by advanced live-cell imaging. Front Plant Sci 8: 866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K (1981) Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem 114: 33–38 [PubMed] [Google Scholar]
- Kovar DR (2006) Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol 18: 11–17 [DOI] [PubMed] [Google Scholar]
- Li Y, Shen Y, Cai C, Zhong C, Zhu L, Yuan M, Ren H (2010) The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22: 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E, Gadeyne A, Stockle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Muller S (2014) The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26: 2617–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E, Herrmann A, Mueller S (2015) Mechanisms of plant cell division. Wiley Interdiscip Rev Dev Biol 4: 391–405 [DOI] [PubMed] [Google Scholar]
- Ly T, Moroz N, Pappas CT, Novak SM, Tolkatchev D, Wooldridge D, Mayfield RM, Helms G, Gregorio CC, Kostyukova AS (2016) The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2's function. Mol Biol Cell 27: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA (1991) Microtubule dynamics and microtubule caps - a time-resolved cryoelectron microscopy study. J Cell Biol 114: 977–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Martiniere A, Gayral P, Hawes C, Runions J (2011) Building bridges: formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J 66: 354–365 [DOI] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T (2011) GTP gamma S microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs). Proc Natl Acad Sci U S A 108: 3988–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael CM, Bednarek SY (2013) Cytoskeletal and membrane dynamics during higher plant cytokinesis. New Phytol 197: 1039–1057 [DOI] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L (2006) A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol 16: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, et al. (2013) Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol 203: 917–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Nader GPF, Ramalingam N, Bartolini F, Gundersen GG (2014) Kif4 interacts with EB1 and stabilizes microtubules downstream of rho-mDia in migrating fibroblasts. PLos One 9: e91568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourino-Perez RR, Linacre-Rojas LP, Roman-Gavilanes AI, Lew TK, Callejas-Negrete OA, Roberson RW, Freitag M (2013) MTB-3, a microtubule plus-end tracking protein (plus TIP) of neurospora crassa. PLoS One 8: e70655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Fuchs E, Ovecka M, Wysocka-Diller J, Benfey PN, Hauser MT (2002) Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol 130: 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, Machida Y, Hasebe M (2013) Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun 4: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Okada K, Takebe I, Matsui C (1981) Delivery of tobacco mosaic-virus RNA into plant protoplasts mediated by reverse-phase evaporation vesicles (liposomes). Mol Gen Genet 184: 161–165 [Google Scholar]
- Novák D, Kuchařová A, Ovečka M, Komis G, Šamaj J (2015) Developmental nuclear localization and quantification of GFP-tagged EB1c in Arabidopsis root using light-sheet microscopy. Front Plant Sci 6: 1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA (2001) Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13: 2033–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulehlova D, Kollarova E, Cifrova P, Pejchar P, Žárský V, Cvrckova F (2019) Arabidopsis class I formin FH1 relocates between membrane compartments during root cell ontogeny and associates with plasmodesmata. Plant Cell Physiol 60: 1855–1870 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG (2001) mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol 3: 723–729 [DOI] [PubMed] [Google Scholar]
- Panteris E, Adamakis IDS, Voulgari G, Papadopoulou G (2011) A role for katanin in plant cell division: microtubule organization in dividing root cells of fra2 and lue1 Arabidopsis thaliana mutants. Cytoskeleton 68: 401–413 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD, Northcote DH (1966) Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J Cell Sci 1: 109–120 [DOI] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui JY, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR (2009) Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol 16: 1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosero A, Oulehlova D, Stillerova L, Schiebertova P, Grunt M, Žárský V, Cvrckova F (2016) Arabidopsis FH1 formin affects cotyledon pavement cell shape by modulating cytoskeleton dynamics. Plant Cell Physiol 57: 488–504 [DOI] [PubMed] [Google Scholar]
- Rosero A, Žárský V, Cvrckova F (2013) AtFH1 formin mutation affects actin filament and microtubule dynamics in Arabidopsis thaliana. J Exp Bot 64: 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Giddings TH, Staehelin LA (1995) Cytokinesis in tobacco BY-2 and root-tip cells - a new model of cell plate formation in higher-plants. J Cell Biol 130: 1345–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Higaki T, Oda Y, Hayashi T, Hasezawa S (2005) Appearance of actin microfilament ‘twin peaks’ in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP-fimbrin. Plant J 44: 595–605 [DOI] [PubMed] [Google Scholar]
- Sayas CL, Tortosa E, Bollati F, Ramirez-Rios S, Arnal I, Avila J (2015) Tau regulates the localization and function of End-binding proteins 1 and 3 in developing neuronal cells. J Neurochem 133: 653–667 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Smertenko A (2019) Identification and characterization of the land-plant-specific microtubule nucleation factor MACET4. J Cell Sci 132: jcs232819. [DOI] [PubMed] [Google Scholar]
- Segui-Simarro JM, Austin JR II, White EA, Staehelin LA (2004) Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell 16: 836–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui-Simarro JM, Otegui MS, Austin JR II, Staehelin LA (2007) Plant cytokinesis - insights gained from electron tomography studies. InVerma DPS, Hong Z., eds, Cell Division Control in Plants. Berlin Heidelberg: Springer-Verlag, pp 251–287 [Google Scholar]
- Smertenko A, Assaad F, Baluska F, Bezanilla M, Buschmann H, Drakakaki G, Hauser MT, Janson M, Mineyuki Y, Moore I, et al. (2017) Plant cytokinesis: terminology for structures and processes. Trends Cell Biol 27: 885–894 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Hewitt SL, Jacques CN, Kacprzyk R, Liu Y, Marcec MJ, Moyo L, Ogden A, Oung HM, Schmidt S, et al. (2018) Phragmoplast microtubule dynamics - a game of zones. J Cell Sci 131: jcs203331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Moschou P, Zhang L, Fahy D, Bozhkov P (2016) Characterization of cytokinetic mutants using small fluorescent probes. Methods Mol Biol 1370: 199–208 [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser MT, Hussey PJ (2004) The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16: 2035–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Deeks MJ, Hussey PJ (2010) Strategies of actin reorganisation in plant cells. J Cell Sci 123: 3019–3028 [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Kaloriti D, Chang HY, Fiserova J, Opatrny Z, Hussey PJ (2008) The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20: 3346–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Piette B, Hussey PJ (2011) The origin of phragmoplast asymmetry. Curr Biol 21: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Smertenko P, Fenenko L, Brehmer L, Schrader S (2005) Differential approach to the study of integral characteristics in polymer films. Adv Colloid Interface Sci 116: 255–261 [DOI] [PubMed] [Google Scholar]
- Steiner A, Muller L, Rybak K, Vodermaier V, Facher E, Thellmann M, Ravikumar R, Wanner G, Hauser MT, Assaad FF (2016) The membrane-associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis. Mol Plant 9: 528–540 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Goto Y, Asatsuma S, Koizumi M, Mitsui T, Matsuoka K (2009) A mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant cell exterior. Plant Cell 21: 1212–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E (2000) Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell 11: 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen P, Wu SZ, Cheng XH, Pattavina KA, Bezanilla M (2020) In vivo analysis of formin dynamics in the moss P. patens reveals functional class diversification. J Cell Sci 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen PA, Li M, Wu SZ, Bezanilla M (2012) Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol 198: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PAC, Guerin C, Franco P, Li M, Burkart GM, Augustine RC, Blanchoin L, Bezanilla M (2009) Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci U S A 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyplelová P, Ovečka M, Komis G, Šamaj J (2018) Advanced microscopy methods for bioimaging of mitotic microtubules in plants. Methods Cell Biol 145: 129–158 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Wu J, Meng L, Ren H (2013) AtFH16, [corrected] an Arabidopsis type II formin, binds and bundles both microfilaments and microtubules, and preferentially binds to microtubules. J Integr Plant Biol 55: 1002–1015 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO (2002) Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci 115: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol 6: 820–830 [DOI] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Wu SZ, Bezanilla M (2014) Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. Elife 3: e03498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue XH, Guo CQ, Du F, Lu QL, Zhang CM, Ren HY (2011) AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol Plant 4: 264–278 [DOI] [PubMed] [Google Scholar]
- Yang W, Ren S, Zhang X, Gao M, Ye S, Qi Y, Zheng Y, Wang J, Zeng L, Li Q, et al. (2011) BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara H (2005) Caffeine inhibits callose deposition in the cell plate and the depolymerization of microtubules in the central region of the phragmoplast. Plant Cell Physiol 46: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Yasuhara H, Shibaoka H (2000) Inhibition of cell-plate formation by brefeldin A inhibited the depolymerization of microtubules in the central region of the phragmoplast. Plant Cell Physiol 41: 300–310 [DOI] [PubMed] [Google Scholar]
- Yasuhara H, Sonobe S, Shibaoka H (1993) Effects of taxol on the development of the cell plate and of the phragmoplast in tobacco BY-2 cells. Plant Cell Physiol 34: 21–29 [Google Scholar]
- Ye JR, Zheng YY, Yan A, Chen NZ, Wang ZK, Huang SJ, Yang ZB (2009) Arabidopsis Formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Guo C, Chen D, Zhao B, Yang B, Ren H (2005) Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol 138: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Baluska F, Menzel D, Ren H (2009) Dynamics and roles of phragmoplast microfilaments in cell plate formation during cytokinesis of tobacco BY-2 cells. Chin Sci Bull 54: 2051–2061 [Google Scholar]
- Zhang Z, Zhang Y, Tan H, Wang Y, Li G, Liang W, Yuan Z, Hu J, Ren H, Zhang D (2011) RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23: 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.