Abstract
In many fruiting plant species, flower abscission is induced by low light stress. Here, we elucidated how signaling mediated by the peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) controls low light-induced flower drop in tomato (Solanum lycopersicum). We analyzed the expression patterns of an IDA-Like gene (SlIDL6) during low light-induced flower abscission, and used tandem mass spectrometry to identify and characterize the mature SlIDL6 peptide. Tomato knockout lines were created to investigate the in vivo function of SlIDL6. In addition, yeast one-hybrid assays were used to investigate the binding of the SlWRKY17 transcription factor to the SlIDL6 promoter, and silencing of SlWRKY17 expression delayed low light-induced flower abscission. SlIDL6 was specifically expressed in the abscission zone and at high levels during low light-induced abscission and ethylene treatment. SlIDL6 knockout lines showed delayed low light-induced flower drop, and the application of SlIDL6 peptide accelerated abscission. Overexpression of SlIDL6 rescued the ida mutant phenotype in Arabidopsis (Arabidopsis thaliana), suggesting functional conservation between species. SlIDL6-mediated abscission was via an ethylene-independent pathway. We report a SlWRKY17-SlIDL6 regulatory module that functions in low light promoted abscission by increasing the expression of enzymes involved in cell wall remodeling and disassembly.
A SlWRKY17- SlIDL6 regulatory module that functions in low light promoted abscission by increasing the expression of enzymes involved in cell wall remodeling and disassembly.
Introduction
Plants have the capacity to shed their vegetative and reproductive organs in response to developmental signals and environmental stimuli (Addicott, 1982). Although this abscission process has many adaptive benefits, in an agricultural context, premature abscission can cause a major reduction in crop productivity, as exemplified by fruit set failure. Thus, there is considerable interest in understanding the mechanism underlying organ abscission and its regulation.
Abscission is a highly regulated process involving structural, biochemical, and molecular changes in a specialized tissue of the organ called the abscission zone (AZ; Sexton and Roberts, 1982; Estornell et al., 2013). These changes include protein degradation, cell wall remodeling, dissolution of the middle lamellar, loss of cell membrane permeability, and programmed cell death (Cho et al., 2008; Meir et al., 2010; Bar-Dror et al., 2011).
Multiple phytohormones are known to play roles in triggering and modulating abscission, and ethylene is a well-documented accelerator of leaf, flower, and fruit abscission. Accordingly, enhancing ethylene production and promoting ethylene signaling by elevating the expression of ethylene response genes, including transcription factor (TF) genes, have been shown to promote abscission (Butenko et al., 2006; Wilson et al., 2011). In tomato (Solanum lycopersicum), transcriptome analysis of the AZ pedicel indicated that ethylene biosynthesis and related signal transduction, including the expression of bZIP and WRKY TF family members, is upregulated during abscission, and pretreatment with the ethylene inhibitor 1-methylcyclopropene (1-MCP) suppressed this upregulation (Meir et al., 2010). Furthermore, WRKY genes were also identified as abscission-specific transcriptional regulators of Arabidopsis (Arabidopsis thaliana) flower abscission and soybean (Glycine max) leaf abscission (Nath et al., 2007; Niederhuth et al., 2013). However, while WRKY TFs are known to integrate ethylene responses during senescence, their role in abscission is not well understood (Robatzek and Somssich, 2001; Koyama, 2014).
In addition to ethylene, a small peptide, INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), has been identified as an essential signaling element in the induction of Arabidopsis floral organ abscission (Butenko et al., 2003; Stenvik et al., 2008). This IDA signal functions during phases 3 to 4 of A. thaliana abscission, which are the last steps, and it has also been shown that the ida mutant was completely blocked during flower abscission (Butenko et al., 2003). Before cleavage, IDA-LIKE (IDL) and IDA peptides are usually smaller than 100 amino acids and consist of an N-terminal export signal, a variable region, and a C-terminal extension with a PIP motif. Overexpression of AtIDL1-5 compensated for the abscission defect in the ida mutant (Stenvik et al., 2008). Using both the AtIDA promoter to drive the litchi (Litchi chinensis) LcIDL1 gene and the constitutive 35S promoter to drive the Citrus IDA3 gene resulted in rescue of the abscission-deficient phenotype of the ida mutant, suggesting functional conservation of IDA peptides between species (Estornell et al., 2015; Ying et al., 2016). Five tomato IDA homologs have also been shown to have specific expression patterns during pedicel abscission (Tucker and Yang, 2012). In vitro analysis indicated that the mature A. thaliana IDA peptide is 14 amino acids long after cleavage between a Lys and a Gly residue by the subtilase (SBT) family proteases SBT4.12, SBT4.13, or SBT5.2 (Schardon et al., 2016). The leucine-rich repeat (LRR) receptor-like kinases (RLK) HAESA (HAE) and HAESA-LIKE2 (HSL2) are thought to act as IDA receptors. Indeed, the hae hsl2 double mutant has been reported to have a similar phenotype to the ida mutant (Cho et al., 2008; Stenvik et al., 2008). Other studies have indicated that the hydroxylated IDA peptide binds to HAE and HSL2, and a structural model of the ligand and receptor has been proposed (Santiago et al., 2016). In A. thaliana, the KNOTTED-LIKE FROM ARABIDOPSIS THALIANA (KNAT) TFs and cell wall modifying proteins have been identified as downstream components in the IDA-HAE/HSL2 pathway (Shi et al., 2011).
An essential step in abscission is the loss of cell–cell adhesion, which is associated with degradation of the pectin-rich middle lamella. Consistent with this idea, disruption of the expression of an abscission-related pectinase, polygalacturonase, in A. thaliana has been shown to significantly delay floral organ drop (González-Carranza et al., 2007). In tomato, abscission-related polygalacturonases (TAPG 1, -2, -3, and -4) are specifically expressed in the flower pedicel AZ during abscission, and their silencing significantly delayed leaf abscission (Jiang et al., 2008). Furthermore, endo-β-1,4-glucanases Cel1 and Cel2, which have been associated with cell wall modification, are expressed in ripening fruit and abscising flowers, and anti-sense based silencing of Cell has been shown to delay tomato flower abscission (Gonzalez-Bosch et al., 1997; Lashbrook et al., 1998). In A. thaliana, IDA signaling has been reported to be independent from ethylene-triggered abscission, although its transcript levels are lower in etr1-1 lines (Butenko et al., 2006). However, the role of IDA signaling in tomato abscission and its relationship with ethylene is still unclear.
Drought and low light stress can trigger flower, flower bud, leaf, and fruit abscission in many plant species. In pepper (Capsicum annuum), 80% shade for 6 d was shown to significantly promote the abscission of flower buds and positively corresponded with ethylene levels (Wien et al., 1996). However, application of the synthetic auxin 1-naphthaleneacetic acid (NAA) effectively prevented low light-induced flower bud abscission. Furthermore, an eight-fold increase in ethylene production was observed in dark-induced lily (Lilium candidum) flower buds during abscission, and application of the ethylene inhibitor silver thiosulfate abolished dark-induced abscission (Van Meeteren and De Proft, 1982). The expression of IDA and IDL genes is also induced by biotic and abiotic stresses (Vie et al., 2015; Patharkar and Walker, 2016). Although IDA-HAE/HSL2 is known to be involved in drought-induced leaf abscission, it is not known whether IDA signaling also mediates low light stress-induced abscission.
Recently, the phytosulfokine (PSK:Tyr[SO3H]-Ile-Tyr[SO3H]-Thr-Gln) signal has been reported to function in drought-induced tomato flower abscission by promoting the expression of the abscission-related polygalacturonase TAPG4 in an auxin- and ethylene-independent manner (Reichardt et al., 2020). In tomato, PSK is perceived by membrane-bound PSK receptors (PSKR1 and PSKR2), which triggers downstream signaling (Zhang et al., 2018). Thus, PSK and IDA are both involved in abscission and have been associated with cell wall remodeling, but act through different receptors, and their relationship and relative contributions to the abscission process are unclear.
In this study, we investigated the regulatory mechanism underlying low light-induced flower abscission in tomato, one of the most economically important vegetable crops. Blossom drop is a common phenotype in tomatoes and peppers suffering from dark and low light stress. We report that a low light-induced tomato IDA-like gene, SlIDL6, is involved in abscission.
Results
Expression of SlIDL6 is induced in the AZ following low light-induced abscission
We identified 11 IDA-like genes, including the previously identified SlIDL1-5 (Tucker and Yang, 2012), using a TBLASTN search (https://blast.ncbi.nlm.nih.gov/) with AtIDA and AtIDL sequences from the tomato genome sequence (Supplemental Figure S1). We named them SlIDA and SlIDL1-10 (Supplemental Table S2). To establish the relationship between SlIDA family genes and low light-induced abscission, the expression patterns of SlIDA/SlIDL genes were evaluated in a transcriptome database of low-light-treated pedicels. This showed a significant upregulation of SlIDL6 by the treatment compared with the control conditions (Supplemental Table S3). A RT-qPCR analysis further revealed that SlIDL6 was specifically expressed in the AZ with a low expression in the distal and proximal regions (Figure 1A), and expression was induced in the AZ under low light stress (Figure 1B). SlIDL6 expression was significantly increased in the AZ after flower removal induced abscission (Figure 1C). Pedicels treated with ethylene or an ethylene inhibitor (1-MCP) showed elevated or suppressed SlIDL6 expression, respectively (Figure 1D). By in situ hybridization, we observed that SlIDL6 was specifically expressed in the AZ and a particularly strong signal was present in the vascular tissue, while no signal was found in the distal or proximal regions, which was consistent with the RT-qPCR results (Figure 1, E–G).
Figure 1.
SlIDL6 expression analysis in flower pedicels. A, The distal, proximal, and abscission zones (AZ) of flower pedicels were used to study SlIDL6 expression by quantitative real-time PCR. B, SlIDL6 expression levels in the pedicel AZ under low light treatment compared with the control (normal light), and low light-induced SlIDL6 expression with 1-methylcyclopropene (1-MCP) treatment. C, The effect of flower removal on SlIDL6 expression patterns in the wild-type pedicel AZ. D, The effect of 10 μL·L−1 ethylene and 2 μL·L−1 1-MCP on SlIDL6 expression in the pedicel AZ. E–G, Expression analysis of SlIDL6 at different development stages of pedicel abscission by in situ hybridization. Signal in the pedicel AZ at 8 h (E) or 16 h (F) after flower removal, and in a negative control stained with the positive probe (G). Scale bar = 100 μm. H, LC–MS/MS revealing 12 amino acid mature SlIDL6 peptide sequences in the tomato pedicel AZ. Different letters indicate significant differences (Student’s t test, P < 0.05). The results are the means of three replicates ± sd. P, pith; VB, vascular bundle.
As further evidence that the SlIDL6 peptide is involved in abscission, the mature 12-mer SlIDL6 peptide was detected by liquid chromatography mass spectrometry (LC–MS/MS) in culture exudates from the pedicel AZ (Figure 1H; Supplemental Table S4).
Flower drop is enhanced under low light stress in a SlIDL6 dependent manner
We generated SlIDL6 knockout lines by CRISPR/Cas9 to investigate the possible role of SlIDL6 signaling during low light stress-induced flower abscission. Eight independent knockout transgenic lines were obtained through A. tumefaciens-mediated transformation (Supplemental Figure S2), five of which were genome deleted. Three independent Cas9-free T2 plants carrying mutations were screened and identified (CR-). In addition, off-target analysis suggested that no off-target activity occurred in the CRslidl6 lines (Table S5). Delayed flower abscission was observed in the knockout lines under low light stress, and the flower drop was 8.93% ± 0.82% compared with 26%.3 ± 2.28% in the wild-type (Figure 2, A and B). We further analyzed the effect of CRslidl6 on flower removal abscission in explants. The wild-type plants showed 50% abscission at 16 h and 100% at 40 h, while the CRslidl6 knockout plants had significantly delayed abscission, with pedicels that showed only 50% abscission at 40 h (Figure 2C). To investigate the effect of the SlIDL6 peptide on pedicel abscission, wild-type and CRslidl6 pedicels were incubated in a medium containing various concentrations of the 12 aa synthetic SlIDL6 peptide. A SlIDL6 peptide of 1-μΜ had little effect on wild-type abscission, but the 5- and 10-μM SlIDL6 peptides significantly enhanced abscission. Notably, 5- and 10-μM SlIDL6 peptides had similar effects on abscission, suggesting that the amount of active peptide was saturated at 5-μM concentration. Furthermore, 5-μM SlIDL6 peptide also complemented the CRslidl6 abscission defect (Figure 2D), indicating that tomato flower abscission is regulated by SlIDL6.
Figure 2.
Effects of knocking out SlIDL6 on the kinetics of flower pedicel abscission. A, Inflorescence phenotypes of CRslidl6 (knockout, KO) and the wild-type under low light stress. Wild-type shows premature abscission in the pedicel AZ (arrowhead). B, Low light-induced flower abscission in AZ (less than 0.5 mm) of CRslidl6 and wild-type lines after fruit set, and the percentage of flowers per inflorescence. The results are the means of three replicates ± sd, with at least 18 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). C, The effect of flower removal on CRslidl6 and wild-type pedicel (about 4 cm) abscission. The results are the means of three replicates ± sd, with at least 18 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). D, The effect of different peptide concentrations on CRslidl6 and wild-type pedicel abscission. The black, blue, purple, and yellow solid curves correspond to the abscission of wild-type lines after 0, 1, 5, and 10 μm peptide treatment. The gray, blue, purple, and yellow dotted curves correspond to the abscission of CRslidl6 lines after 0, 1, 5, and 10 μm peptide treatment. The results are the means of three replicates ± sd, with at least 18 samples per replicate.
35S::SlIDL6 can rescue the ida2 abscission defect
To test whether SlIDL6 has a similar role to AtIDA in control of floral organ abscission, we generated a construct where SlIDL6 expression was driven by the cauliflower mosaic virus 35S promoter and transformed into A. thaliana ida2 mutant plants. Five out of 20 plants were shown to have been transformed, and three lines with high expression were chosen for further experiments (Supplemental Figure S3A). The ida2 mutants had a flower organ abscission defect phenotype at anthesis, while the ida2 35S::SlIDL6 transgenic plants showed wild-type phenotype (Supplemental Figure S3B). Furthermore, measurement of the force of petal attachment by petal break strength (pBS) also revealed that overexpression of SlIDL6 was sufficient to complement the abscission defect in ida2 (Supplemental Figure S4).
SlIDL6 induced abscission via a partially ethylene-independent manner
To understand whether SlIDL6 regulates abscission through ethylene signaling, CRslidl6 and wild-type abscission assays were performed following ethylene treatment. In contrast to the wild-type, abscission was observed to be significantly delayed, but not entirely blocked in the CRslidl6 lines, while application of 5-µM SlIDL6 peptide accelerated abscission (Figure 3A). We also used the ethylene inhibitor 1-MCP to pretreat the wild-type explants before incubating them with 5-µM SlIDL6 peptide, while the controls were not treated. Abscission was significantly accelerated in peptide-treated pedicels compared with the control (Figure 3B). Furthermore, application of 5-µM SlIDL6 peptide to the tomato ethylene response mutant, Neverripe (Nr), led to faster abscission (Figure 3C). However, it is notable, 5-µM SlIDL6 could not restore the abscission of Nr- and 1-MCP-treated plants to levels of the wild-type control. These results indicate that SlIDL6 can induce abscission via both ethylene-dependent and -independent pathway.
Figure 3.

Kinetics of pedicel abscission is mediated by SlIDL6, which functions downstream from ethylene. A, Pedicel explant abscission assay for SlIDL6 knockout (KO) and wild-type plants with or without 5-μM peptide treatment and 10-μL·L−1 ethylene. B, Pedicel explant abscission assay for 1-methylcyclopropene (1-MCP) pretreated wild-type pedicel explants with, or without, 5-μM peptide. C, Pedicel explant abscission assay for Never ripe (Nr) mutant explants with, or without, 5-μM peptide. Different letters indicate significant differences (Student’s t test, P < 0.05). The results are the means of three replicates ± sd, with at least 18 samples per replicate.
Delayed abscission caused by knocking out SlIDL6 is associated with cell wall degradation
We observed that the expression levels of genes encoding cell wall degrading proteins associated with abscission, including TAPG1 (Solyc02g067630.2), TAPG4 (Solyc12g096750), and CEL2 (Solyc09g010210.2), were downregulated in CRslidl6 lines compared with the wild-type and upregulated by the peptide treatment (Figure 4, A–C). In addition, PTAPG4::GUS staining intensity was enhanced by the SlIDL6 peptide treatment, and a stronger GUS signal was observed after the 5-µm peptide treatment than after the 1-µm treatment (Figure 4D). These results indicate that the SlIDL6 signal may modulate TAPG1, TAPG4, and CEL2 expression.
Figure 4.
The expressions of genes encoding proteins involved in cell wall remodeling are affected by SlIDL6. RT-qPCR analysis of TAPG1 (A), TAPG4 (B), and CEL2 (C) expressions in CRslidl6, wild-type, and SlIDL6 peptide-treated plants. Different letters indicate significant differences (Student’s t test, P < 0.05). The results are the means of three replicates ± sd. D, β-glucosidase (GUS) staining revealed that the TAPG4::GUS signal is induced by different concentrations of SlIDL6 peptide 16 h after flower removal. Scale bar = 100 µm.
SlIDL6 and PSK regulate abscission via different pathways
A transcript analysis showed that both SlPSKR1 (Solyc01g008140) and SlPSKR2 (Solyc07g063000) were expressed during tomato pedicel abscission (Figure 5A). We also observed that SlPSKR2 was significantly upregulated under low light stress (Supplemental Table S3; Figure 5B). Silencing SlPSKR1 and SlPSKR2 expression to inhibit the PSK signal significantly suppressed low light-induced abscission (Figure 5C). Furthermore, knockdown of SlPSKR1 and SlPSKR2 expression in the CRslidl6 lines further suppressed low light-induced abscission (Figure 5D), and similar results were observed after flower removal-induced abscission (Figure 5E). To further elucidate the relationship between SlIDL6 and PSK signals during abscission, SlIDL6, PSK, and 5 or 12 mer control peptides were used to treat tomato pedicel explants. Co-treatment with SlIDL6 and PSK led to significantly accelerated abscission compared with the single SlIDL6 or PSK peptide treatments, whereas application of SlIDL6 and the 5 aa control peptide, or PSK and the 12 aa control peptide, did not result in any differences compared with the single SlIDL6 or PSK peptide treatments. These results indicate that SlIDL6 and PSK act synergistically during abscission (Figure 5F).
Figure 5.
Effect of PSK and SlIDL6 peptide treatments on tomato flower pedicel abscission. A, SlPSKR1 and SlPSKR2 expression in the pedicel abscission zone. B, SlPSKR2 expression in the pedicel abscission zone after 7 d of low light treatment compared with the control grown under normal light. The results are the means of three replicates ± sd, with at least 20 samples per replicate (Student’s t test, P < 0.05). C, Effect of SlPSKR1 and SlPSKR2 on low light-induced abscission. The results are the means of three replicates ± sd, with at least 30 samples per replicate (Student’s t test, P < 0.05). D, Effects of low light stress on pedicel abscission in pTRV0 control plants, CRslidl6 and SlPSKR1- and SlPSKR2-silenced pedicels of wild-type and SlPSKR1-, and SlPSKR2-silenced pedicels of CRslidl6 plants. The results are the means of three replicates ± SD, with at least 30 samples per replicate (Student’s t test, P < 0.05). E, The effect of flower removal on pedicel abscission in pTRV0 control plants, CRslidl6 and SlPSKR1-, and SlPSKR2-silenced pedicels of wild-type, and SlPSKR1- and SlPSKR2-silenced pedicels from CRslidl6 plants. The results are the means of three replicates ± sd, with at least 30 samples per replicate (Student’s t test, P < 0.05). F, Percentage abscised wild-type flower explants after exposure to 5-μM PSK peptide, 5-μM SlIDL6 peptide and both 5-μM PSK peptide and 5-μM SlIDL6 peptide, both 5-μM PSK peptide and 5-μM 12AA, and both 5-μM 5AA peptide and 5-μM SlIDL6 peptide compared with the control flowers. Different letters indicate significant differences between treatments (Student’s t test, P < 0.05). The results are the means of three replicates ± sd, with at least 30 samples per replicate.
SlIDL6 is directly regulated by SlWRKY17
To address the regulation of SlIDL6 expression, a 1,284-bp promoter fragment upstream from its coding sequence was chosen. A search of the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) suggested that the promoter included two ABRE (ACGTG located at −795 and −618) binding sites and one WRKY (GTCAGC located at −121) binding site (Figure 6A). A search of the low-light-treated tomato pedicel transcriptome data revealed a member of the WRKY II b subfamily, WRKY17, to be differentially expressed under low light conditions compared with wild-type light levels (Supplemental Table S3).
Figure 6.
SlWRKY17 promotes SlIDL6 transcription. A, The SlIDL6 promoter fragment containing two ABRE (ACGTG located at −795 and −618) and one WRKY (GTCAGC located at −121) binding sites. wt, intact W-box element; mt, mutated W-box element. B, Yeast one-hybrid analysis revealing that SlWRKY17 binds to the SlIDL6 promoter fragment (Pro SlIDL6) containing the W-box motif (−121 bp). AbA (Aureobasidin A) was used as a screening marker. Rec-P53 and the P53-promoter were chosen as positive controls while the empty vector and the SlIDL6 promoter were used as negative controls. C, Electrophoretic mobility shift assay (EMSA) indicating that SlWRKY17 binds to the W-box motif of the SlIDL6 promoter. The biotin-labeled fragment of the SlIDL6 promoter containing the W-box motif (wt), a W-box motif with two mutated nucleotides (mt), and with a nonlabeled competitive probe (100-fold that of the biotin-labeled probe). His-tagged SlWRKY17 was used in the analysis. D, β-glucosidase (GUS) activity analysis showing that SlWRKY17 activates the SlIDL6 promoter. The SlWRKY17 overexpression vector combined with the reporter vector containing the wild-type SlIDL6 promoter, or a mutated promoter sequence, were co-infiltrated into wild N. benthamiana leaves to analyze the regulation of GUS activity. The transfection experiments were performed using three replicates. Values represent means ± sd. Different letters indicate significant differences between treatments (Student’s t test, P < 0.05).
We investigated the binding of SlWRKY17 to the SlIDL6 promoter using a yeast one-hybrid assay and found that it bound and triggered the expression of the LacZ reporter gene (Figure 6B). To further confirm this interaction, we performed an electrophoretic mobility shift assay (EMSA) analysis using the full-length SlWRKY17 protein and fragments of biotin-labeled SlIDL6 promoter containing the W-box motif as well as a competitor probe containing three mutated nucleotides. We observed that SlWRKY17 was bound to the W-box motif of SlIDL6 (Figure 6C). Regulation of the SlIDL6 promoter by SlWRKY17 was investigated using a GUS transactivation assay in Nicotiana benthamiana leaves. An increase in the GUS activity was observed upon coexpression of 35S::WRKY17 with the wild-type SlIDL6 promoter::reporter construct, but not for the mutated SlIDL6 promoter (Figure 6D). The results indicated that SlWRKY17 significantly promoted the activity of the SlIDL6 promoter.
Knockdown of SlWRKY17 delays pedicel abscission
We evaluated the expression of SlWRKY17 in flower AZs after the low light treatments and observed elevated expressions in both cases, which we found to be blocked by treatment with 1-MCP (Figure 7A). Flower removal also induced SlWRKY17 expression, which reached a peak at 24 h after removal before decreasing (Figure 7B). Ethylene treatment induced SlWRKY17 expression, while 1-MCP treatment suppressed its expression (Figure 7C).
Figure 7.
Silencing of SlWRKY17 causes delayed flower abscission. A, SlWRKY17 expression in the pedicel abscission zone (AZ) after 7 d of low light treatment compared with the control (normal light), and the effect of 1-methylcyclopropene (1-MCP) treatment on low light-induced SlWRKY17 expression. B, Effect of flower removal on the SlWRKY17 expression pattern in wild-type pedicel AZ. C, Effect of 10-μL·L−1 ethylene and 2-μL·L−1 1-MCP on SlWRKY17 expression in the pedicel AZ. D, Expression of SlWRKY17 and SlIDL6 in SlWRKY17-silenced and TRV control plants. E, Effect of low light stress on pedicel abscission in the TRV control and SlWRKY17-silenced plants. F, Effect of flower removal on SlWRKY17-silenced and wild-type pedicel abscission. Different letters indicate significant differences between treatments (Student’s t test, P < 0.05). Results are the means of three replicates ± sd.
Virus-induced gene silencing (VIGS) of SlWRKY17 resulted in a significant downregulation of SlWRKY17 but no downregulation of SlWRKY6 (Solyc02g080890.2) or SlWRKY16 (Solyc02g032950), while SlIDL6 expression was lower in the pedicels of silenced tomato plants than in the control (Figure 7D; Supplemental Figure S5). Downregulation of SlWRKY17 also decreased low light- and ethylene-induced SlIDL6 expression (Supplemental Figure S6). The SlWRKY17-silenced flowers showed lower flower drop rates under low light stress and delayed pedicel abscission after flower removal (Figure 7, E and F).
Discussion
SlIDL6 is involved in low light-induced tomato flower abscission
Blossom drop is observed in many species following biotic or abiotic stress (Addicott, 1982; Taylor and Whitelaw, 2001; Tranbarger et al., 2017) and the IDA peptide is thought to play a conserved and central role in this process as well as in aspects of development (Shi et al., 2019). The expression of IDA and IDA-like genes is known to increase during abscission, and most have been shown to function in mediating organ abscission by controlling cell separation. In a previous study, ProAtIDA::GUS staining was not observed until the position 4 flower stage in Arabidopsis, but a strong signal in the floral AZ from positions 5 to 8 was seen, and an abscission-related expression pattern was observed. The overexpression of each of the genes AtIDL1 to -5 was found to rescue the ida abscission defect (Butenko et al., 2003; Stenvik et al., 2008). Here, we found that SlIDA, SlIDL6, and SlIDL7 were upregulated during pedicel abscission and that SlIDL6 was induced by low light stress. We chose SlIDL6 for further studies as it had the highest expression during pedicel abscission and low light stress; however, we cannot rule out the possibility that SlIDA and SlIDL7 also contribute to tomato blossom drop (Supplemental Figure S7), which will be addressed in future studies.
The IDA peptide interacts with HAE and HSL2 to form a ligand-receptor module that triggers an abscission signal (Stenvik et al., 2008; Tang et al., 2017). A sequence alignment showed that SlIDL6 does not have a PIPP sequence, but does contain a conserved core SGPS domain with a proline that may be post-translationally modified to form hydroxyproline. Notably, the presence of hydroxyproline residues was shown to increase the activity of CLAVATA3 (CLV3) and PLANT PEPTIDE CONTAINING SULFATED TYROSINE 1 (PSY1; Kondo et al., 2006). A positive effect of proline hydroxylation on peptide activity has also been reported for IDA (Butenko et al., 2014; Supplemental Figure S1). Furthermore, our results showed that SlIDL6 plays a similar role to AtIDA in mediating floral abscission since its expression rescued the ida2 mutant. In summary, our results show that the canonical IDA-HAE/HSL2 abscission signaling found in A. thaliana, Citrus, and litchi is conserved in tomato (Aalen et al., 2013; Estornell et al., 2015; Ying et al., 2016).
SlIDL6 expression required ethylene but SlIDL6-induced abscission was partially ethylene independent
Ethylene is a well-known accelerator of flower abscission (Botton and Ruperti, 2019). In A. thaliana, the ida mutant showed a normal ethylene triple-response and senescence but not abscission following ethylene treatment, indicating that ida is not defective in ethylene signaling and that IDA signaling mediates organ abscission through an ethylene-independent pathway (Butenko et al., 2006). Recent studies have indicated that ethylene is required for mediating IDA expression and that wounding also induces IDA expression, although whether wounding directly induces IDA expression or acts via the induction of ethylene production is not clear (Meir et al., 2019). A previous study indicated that flower removal instantly depletes auxin levels and promotes AZ ethylene sensitivity (Meir et al., 2010). Our results showed that flower removal significantly induced SlIDL6 expression after 4 h compared with flowers that remained attached to the explants, and that there was increased expression of AZ in the pedicel explants as abscission progressed. SlIDL6 expression levels decreased after 1-MCP treatment and expression in the Nr ethylene receptor mutant was depressed, indicating that SlIDL6 expression is induced by ethylene. Low light-induced flower bud and flower abscission is influenced by ethylene action (Van Meeteren and De Proft, 1982; Wien et al., 1996); however, SlIDL6 upregulation under low light stress is most likely ethylene dependent because 1-MCP suppressed low light stress-induced SlIDL6 expression. We conclude that ethylene is partially dependent on SlIDL6 when mediating abscission since abscission in CRslidl6 plants was significantly delayed, but not entirely blocked, following ethylene treatment. Furthermore, partially restored abscission in 1-MCP treated and Nr mutant explants after SlIDL6 peptide treatment, indicated that SlIDL6 could induce tomato pedicel abscission through a partially ethylene-independent pathway. As CLV3 and IDA peptides both carry hydroxyprolines, could be further modified by glycosylation, which resulting in enhancing peptide signaling (Ohyama et al., 2009). The FASCIATED INFLORESCENCE (FIN), which encodes a hydroxyproline O-arabinosyltransferase (HPAT). HPATs are enzymes that catalyze the transfer of l-arabinose to the hydroxyl group of hydroxyproline residues, and the mutant phenotype can be rescued by application tri-arabinosylated CLV3 peptides (Ogawa-Ohnishi et al., 2013; Shinohara and Matsubayashi, 2013). It is also reported the hydroxyprolination of IDA affects the affinity of the peptide for the HSL2 receptor (Butenko et al., 2014). We proposed that ethylene might regulate post-translational enzyme activities, which involved in hydroxyprolinated and glycosylated SlIDL6 peptide, to mediate abscission.
In A. thaliana, IDA is thought to act as a master regulator governing the final cell separation stages in the abscission process (Butenko et al., 2006; Stenvik et al., 2008; Aalen et al., 2013). RNA-seq and microarray analyses have revealed that xyloglucan endotransglycosylase cell wall modification associated genes are significantly downregulated in both the hae hsl2 and the ida mutant (Niederhuth et al., 2013; Reichardt et al., 2020). Similar to A. thaliana, knocking out SlIDL6 suppressed TAPG1, TAPG4, and CEL2 expression, but applying SlIDL6 peptide had the opposite effect. It is highly acceptable that ethylene triggers IDA signaling, which in turn promotes the expression of cell wall hydrolases that break down polysaccharides in the cell wall and middle lamella, leading to abscission.
SlIDL6 and PSK signaling may regulate abscission in different ways
PSK signaling has been reported to be involved in drought-induced tomato flower abscission (Reichardt et al., 2020). PSK acts through an auxin- and ethylene-independent pathway to induce TAPG4 expression and accelerate abscission. In tomato, the PSK peptide is perceived by both PSKR1 and PSKR2 (Zhang et al., 2018), and low light stress also induces the expression of SlPSKR2, suggesting a role in low light-induced abscission. Silencing PSKR1 and PSKR2 in tomato flowers resulted in delayed abscission under low light stress, indicating that PSK signaling is also involved in low light-induced flower abscission. We investigated the relationship between SlIDL6 and PSK signaling because the SlIDL6 peptide treatment also induced TAPG1 and TAPG4 expression. PSK induced pedicel abscission, while the additional application of SlIDL6 has a synergistic effect on inducing abscission. Silencing PSKR1 and PSKR2 in the CRslidl6 lines further delayed abscission compared with the CRslidl6 lines. Given that SlIDL6 and PSK are both predominantly expressed in later abscission stages and are perceived by different receptors, we conclude that SlIDL6 and PSK signaling regulate abscission in different ways. Interestingly, AtIDA signaling has also been reported to be involved in drought-triggered leaf abscission, as the expression of the HAESA signaling receptor is induced in the leaf AZ under limited water conditions (Patharkar and Walker, 2016). In N. benthamiana, IDA-like genes also show induced expression under water stress (Ventimilla et al., 2020). During drought-induced tomato flower drop, SlIDL1-5 is expressed at very low levels (Reichardt et al., 2020). However, the role of SlIDL6-11 and SlIDA in drought-induced abscission still needs to be elucidated. It may be that plants use multiple complex peptide-signaling systems to better control abscission and to coordinate the many associated cellular processes.
WRKY17 acts upstream of SlIDL6 to trigger flower abscission
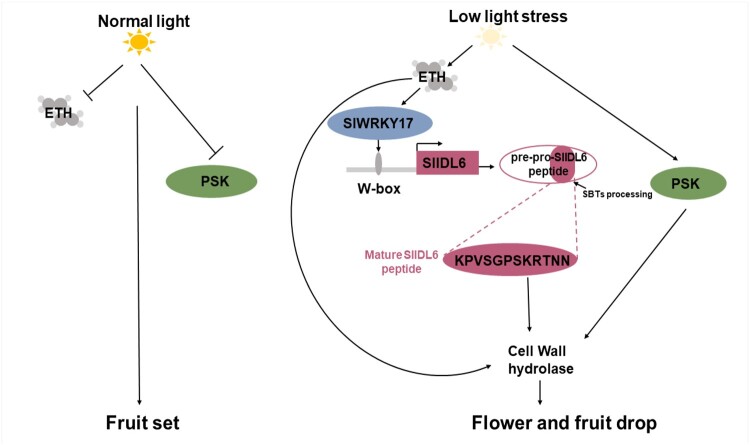
Based on their sequence, WRKY TFs have been grouped into six different classes (Zhang and Wang, 2005; Liang and Jiang, 2017). WRKY II b TFs have been shown to be involved in light signaling, stress responses, hormone signaling, leaf senescence, and abscission (Cheong et al., 2002; Meir et al., 2010; Koyama, 2014; Liang and Jiang, 2017). In this study, SlWRKY17 was found to act as a positive regulator of SlIDL6 by directly binding to the W-box elements of its promoter. SlWRKY17 is also upregulated during low light-induced tomato flower abscission and ethylene treatment, and therefore probably plays a role in low light- and ethylene-induced abscission. The results of the EMSA, yeast one-hybrid assay, and GUS activity analyses were consistent with a regulatory SlWRKY17-SlIDL6 module function in flower pedicel abscission. In tomato, SlWRKY17 is upregulated by ethylene and is involved in ethylene signaling by mediating fruit ripening through its interaction with RIPENING INHIBITOR (SlRIN), ETHYLENE RESPONSE FACTOR 2b (SlERF2b), and ETHYLENE RESPONSE FACTOR 7 (SlERF7), and silencing SlWRKY17 caused reduced red fruit coloration (Wang et al., 2017). Furthermore, SlWRKY17 knockdown suppressed low light stress- and ethylene-induced SlIDL6 expression and delayed abscission during low light stress and ethylene treatment, which is consistent with the phenotypes of the CRslidl6 lines. We propose that low-light stress enhances ethylene signaling to trigger the SlWRKY17-SlIDL6 regulatory module, which induces flower pedicel abscission (Figure 8).
Figure 8.
Model of SlIDL6 signaling during low light-induced tomato pedicel abscission. Normal light guarantees fruit set by inhibiting ethylene signaling and PSK expression. Low light stress enhances ethylene signaling, which actives SlWRKY17 to induce SlIDL6 expression in the AZ. The mature SlIDL6 peptide is secreted and acts as a signal to promote expression of cell wall hydrolases, leading to flower drop. ETH, ethylene; PSK, phytosulfokine; SBT, subtilisin-like proteinase; W-box, WRKY binding site.
Materials and methods
Plant materials
Transgenic tomato plants were generated using the wild-type Solanum lycopersicum cv Ailsa Craig (AC), and TAPG4::GUS seeds were provided by Prof. Mark L. Tucker (Hong et al., 2000). The wild-type, transgenic, and tomato ethylene response mutant, Nr seeds were washed in 50% (v/v) bleach for 3 min, rinsed, and placed onto paper soaked in water in culture dishes for 3 d at 28°C. Then the seedlings were grown in soil for seven weeks in a greenhouse under standard greenhouse conditions (25°C under 16 h of light, followed by 8 h of dark at 15°C).
Tomato plants were subjected to low light conditions in the greenhouse by covering them with shading nets. The photosynthetically active radiation levels under normal and shade conditions were 600 and 60 μmol, respectively. For the ethylene treatments, tomato flower explants were incubated in 0.9% agar in a 40 × 25 × 20 cm3 glass container, which was injected with ethylene to a final concentration of 20 μL L−1. In the 1-MCP pretreatments, plants were incubated with 1-MCP in a sealed 200 L chamber at a final concentration of 0.4 μL L−1 at 22°C for 8 h. The cumulative abscission rates of the tomato pedicels were determined as previously described (Wang et al., 2005).
Plasmid construction and plant transformation
The SlIDL6 (Solyc06g050140) was overexpressed by amplifying the full-length SlIDL6 cDNA using gene-specific primers. The full-length fragment was cloned into the pENTR/D-TOPO vector (Invitrogen) and then cloned into the binary pB7YWG2 vector using BP and LR clonase (Invitrogen), creating the 35S::SlIDL6 plasmid. To make deletions in the SlIDL6 coding sequence and produce knockout lines, a construct (CRslidl6) was designed that contained two single-guide RNAs (sgRNAs) and the Cas9 endonuclease gene. Two single-guide RNAs (sgRNA1 and sgRNA2; Supplemental Figure S2), were designed to target the SlIDL6 gene, using the CRISPR-P tool (http://cbi.hzau.edu.cn/crispr). The U6-26-SlIDL6-gRNA cassettes were cloned into the CRISPR/Cas9 binary vector, pCBC-DT1T2_tomatoU6, to generate pCBC-DT1T2_tomatoU6- SlIDL6.
The CRslidl6 and 35S::SlIDL6 vectors were introduced into Agrobacterium tumefaciens LBA4404 cells by electroporation and then transformed into wild-type tomato by leaf disc cocultivation (Wang et al., 2018), or into the A. thaliana ida2 mutant using the floral dip technique. Positive CRslidl6 lines were identified by sequencing PCR products amplified from the targeted region. Cas9-free T2 plants carrying mutations were screened and identified. Positive A. thaliana 35S::SlIDL6 lines were obtained by spraying them with a 250-mg L−1 Basta solution two to three times and confirmed by PCR. All primers used are listed in Supplemental Table S1.
Virus-induced gene silencing
A 325 bp SlWRKY17 (Solyc07g051840) specific fragment (from 809 to 1,133 bp) was amplified by PCR and purified using the TaKaRa PCR fragment recovery kit before being cloned into the pMD-18T T-vector and verified by sequencing. The EcoRI restriction enzyme site was then used to insert the fragment into the tobacco rattle virus pTRV2 vector. The pTRV1 and pTRV2-SlWRKY17 vectors were individually transformed into A. tumefaciens strain GV3101. A mix of the two A. tumefaciens cultures were used to infiltrate the tomato leaves.
Phylogenetic analysis
A BLASTp search was performed with the conserved amino acid sequences from AtIDA and AtIDL1-8 against the tomato genomic database (https://www.solgenomics.net/). Phylogenetic analysis of SlIDA amino acid sequences (SlIDA [Solyc05g010000.1]; SlIDL1 [CP023762.1]; SlIDL2 [CP023760.1]; SlIDL3 [Solyc07g044890.1]; SlIDL4 [Solyc05g007040.1]; SlIDL6 [Solyc06g050140.1]; and SlIDL7 [Solyc09g005780.1]) with AtIDAs amino acid sequences was performed using the MEGA5 software with the neighbor-joining method (Tamura et al., 2011).
In situ hybridization
The different abscission stages of the wild-type pedicel explants were sampled and immediately fixed in phosphate-buffered saline solution containing 4% (w/v) paraformaldehyde. After submersion in an ethanol gradient for dehydration, the samples were embedded in Paraplast Plus (Leica) and sectioned into 10 μm slices using a microtome (Leica). In situ hybridization analysis of SlIDL6 was performed as previously described (Wang et al., 2018). The DIG Oligonucleotide 3ʹ-End Labeling Kit (Roche) was used to generate antisense and sense RNA probes. Probes that contained the unique SlIDL6 region were 76 bp long (from 174 to 249 bp).
RNA extraction and RT-qPCR analysis
TRIzol reagent (Invitrogen) was used to extract RNA from different tomato pedicel AZ samples. The RNA was treated with recombinant DNase I (TaKaRa) according to the manufacturer’s instructions. After removing the DNA, 2-μg RNA (20-μL reactions containing 4-μL 5X PrimeScript RT Master Mix) was used to synthesize first-strand cDNA with an oligo (dT) primer and reverse transcriptase (TaKaRa). RT-qPCR was performed in 20-μL reactions containing 1 μL of cDNA, 200 nM of each primer and SuperReal PreMix Plus (Tiangen) on an Applied Biosystems 7500 instrument (Applied Biosystems). The mRNA and SlIDL6 were detected using amplification reactions and conditions that had been set according to the manufacturer’s protocols for SYBR SuperReal PreMix plus. Actin (NCBI: NM_001330119.1) was used as a control for mRNA. The program of RT-qPCR was 95°C 30 s, 95°C 5 s (40 cycles), 60°C 34 s (40 cycles). The RT-qPCR experiments were performed on three biological samples with three technical replicates. All primers used are listed in Supplemental Table S1.
Peptide treatment
Wild-type tomato flower pedicel explants were obtained at anthesis rinsed briefly in 70% ethanol and distilled water, and then incubated on 0.9% agar plates containing 0, 1, 5, or 10 μM concentrations of the SlIDL6 peptide or 0, 1, and 5 μM of the sulfated PSK peptide, or both, with or without 5AA(TNGTK) and 12AA(PFLGVYYHKNNK) control peptide substitutes. Chromatography water was used as a control. The plates were placed under a glass cover in a growth chamber at 25°C for 8 h in the dark and under 16 h of light. Data were collected and analyzed from three independent treatments. The peptides were synthesized by the Shenggong Company (China), the SlIDL6 had a purity of > 90%, and the PSK had a purity of >80%.
pBS assay
The force needed to remove the petals from 2, 4, and 6 positions on the inflorescence was quantified using pBS, as previously described (Stenvik et al., 2008). Data were collected and analyzed from at least 15 plants at each position.
Electrophoretic mobility shift assay
The conserved domains of SlWRKY17 (631 amino acids, aa) were cloned and inserted into the pEASY-E1 vector (Transgene) containing a His tag sequence and then transformed into Escherichia coli BL21 cells (Transgene). The transformed cells were cultured in LB medium containing 50 μg mL−1 of ampicillin at 37°C for OD600 = 0.5, and a final concentration of 0.2-mM isopropyl β-d-1-thiogalactopyranoside (IPTG, TaKaRa) was used to induce protein expression after incubation for 16 h at 23°C. Purification of the recombinant fusion proteins was performed. The 3′ biotin end-labeled DNA probes were prepared by the Shenggong Company (China). A Light Shift Chemi Luminescent EMSA Kit (Thermo Scientific) was used to perform the EMSA assay. Nonmutated or mutated probes (20 fmol final concentration) were added to a solution of 2% glycerol, 50-mM KCl, and 10-mM EDTA buffer, with or without SlWRKY17 protein, and incubated for 10 min at 23°C.
LC–MS/MS analysis
After removal of the flower, 0, 16, and 32 h pedicel AZs were collected and about 600 AZs were pooled respectively. The peptides were extracted by homogenization in 10 mL of phenol saturated with 50-mM Tris–HCl (pH 8.0) and precipitated with acetone. Size exclusion chromatography and reversed-phase chromatography were used to concentrate the samples. The Huada Protein Research Center (Beijing, China) performed the LC–MS/MS analysis, and Proteome Discoverer 2.1 software (Thermo Fisher Scientific) was used to analyze the data by searching against SlIDL6 peptide databases, which contained 1,140 sequences, ranging in length from 10 to 22 amino acids to ensure that all possible peptides were identified. The scan selected two to eight charged state ions in positive mode with a minimum and maximum precursor mass of 350 and 5,000 Da, respectively. The threshold values of the precursor and fragment mass high-energy collision dissociation used for the search were 10 ppm and 0.08 Da, respectively.
Yeast one-hybrid assay
The full-length SlWRKY17 sequence was ligated into the pGADT7 vector and the SlIDL6 promoter fragment was cloned into the pAbAi vector. The MatchmakerTM Gold Yeast One-Hybrid Library Screening System kit was used to conduct a yeast one-hybrid assay, according to the manufacturer’s instructions.
β-glucuronidase activity assay
The effector construct was obtained by cloning the coding sequence (CDS) of SlWRKY17 (1–957 bp) into the binary pRI101 vector using the HindIII and EcoRI enzymes. To generate reporter constructs, the SlIDL6 promoter sequence (1,284 bp) was cloned into the binary pBI101 vector, which contains the GUS reporter gene. The reporter and effector vectors were transformed into A. tumefaciens EHA105 and co-infiltrated into N. benthamiana leaves. Infected leaves were incubated in a growth chamber for 3 d. Then the proteins were extracted. A BCA Protein Assay Kit (Beyuntian) was used to determine protein concentration, and GUS activity was determined from three biological replicates and leaves obtained at time zero were used as a control.
Accession numbers
Sequence data from this article can be found in the Genome Database for Tomato (https://www.solgenomics.net/) or GenBank/EMBL libraries under accession numbers SlIDA (Solyc05g010000.1); SlIDL1 (CP023762.1); SlIDL2 (CP023760.1); SlIDL3 (Solyc07g044890.1); SlIDL4 (Solyc05g007040.1); SlIDL6 (Solyc06g050140.1); and SlIDL7 (Solyc09g005780.1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1. Primer sequences.
Supplemental Table S2. SlIDA/SlIDLs DNA and protein sequences.
Supplemental Table S3. IDL6, PSKR2 and WRKY17 expression in tomato flower pedicels AZ under low light.
Supplemental Table S4. Determination of mature peptides.
Supplemental Table S5. Potential off-target sites in CRISPR mutants.
Supplemental Figure S1. IDA/IDL protein sequence analysis using Arabidopsis thaliana and tomato sequences.
Supplemental Figure S2. Sequences of the eight CRslidl6 lines.
Supplemental Figure S3. Overexpression of SlIDL6 in floral AZ of the Arabidopsis thaliana ida2 mutant.
Supplemental Figure S4. Petal break strength in ida2 and ida2 35S::SlIDL6 plants.
Supplemental Figure S5. Expressions of the SlWRKY6 and SlWRKY16 genes in SlWRKY17-silenced and TRV control plants.
Supplemental Figure S6. Silencing of SlWRKY17 causes decreased SlIDL6 expression.
Supplemental Figure S7. RT-qPCR analysis of SlIDLs expression.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2018YFD1000800), the National Natural Science Foundation of China (grant numbers 31991184, 31572167, 31672197, U1708232, and 31861143045), and the Liaoning revitalization talents program (2018050). We acknowledge Prof. Mark L. Tucker (Soybean and Alfalfa Research Laboratory, United States Department of Agriculture-Agricultural Research Service) for donating TAPG4::GUS transgenic tomato seeds and PlantScribe (www.plantscribe.com) for editing this manuscript.
Conflicts of interest: The authors declare no conflicts of interest.
Supplementary Material
T.X. and T.L. conceived the original screening and research plans; T.X. supervised the experiments; T.X., C.-L.S., and C.-Z.J. designed the research; R.L., Y.M., X.W., L.C., and M.Q. performed the experiments; C.-L.S., T.X. and R.L. analyzed the data and wrote the manuscript with contributions from all the authors; T.X. agrees to serve as the author responsible for contact and communication.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Tianlai Li (tianlaili@126.com) and Tao Xu (syauxutao@syau.edu.cn).
References
- Aalen RB, Wildhagen M, Stø IM, Butenko MA (2013) IDA: a peptide ligand regulating cell separation processes in Arabidopsis. J Exp Bot 64: 5253–5261 [DOI] [PubMed] [Google Scholar]
- Addicott FT (1982) Abscission. Berkeley, CA, University of California Press [Google Scholar]
- Bar-Dror T, Dermastia M, Kladnik A, Žnidarič MT, Novak MP, Meir S, Burd S, Philosoph-Hadas S, Ori N, Sonego L (2011) Programmed cell death occurs asymmetrically during abscission in tomato. Plant Cell 23: 4146–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botton A, Ruperti B (2019) The yes and no of the ethylene involvement in abscission. Plants 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik G-E, Amundsen SS, Mandal A, Aalen RB (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Stenvik G-E, Alm V, Sæther B, Patterson SE, Aalen RB (2006) Ethylene-dependent and-independent pathways controlling floral abscission are revealed to converge using promoter::reporter gene constructs in the ida abscission mutant. J Exp Bot 57: 3627–3637 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M,, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G (2014) Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26: 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn T-L, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell LH, Agustí J, Merelo P, Talón M, Tadeo FR (2013) Elucidating mechanisms underlying organ abscission. Plant Sci 199: 48–60 [DOI] [PubMed] [Google Scholar]
- Estornell LH, Wildhagen M, Pérez-Amador MA, Talón M, Tadeo FR, Butenko MA (2015) The IDA peptide controls abscission in Arabidopsis and Citrus. Front Plant Sci 6: 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Bosch C, del Campillo E, Bennett AB (1997) Immunodetection and characterization of tomato endo-[beta]-1, 4-glucanase cel1 protein in flower abscission zones. Plant Physiol 114: 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Elliott KA, Roberts JA (2007) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58: 3719–3730 [DOI] [PubMed] [Google Scholar]
- Jiang C-Z, Lu F, Imsabai W, Meir S, Reid MS (2008) Silencing polygalacturonase expression inhibits tomato petiole abscission. J Exp Bot 59: 973–979 [DOI] [PubMed] [Google Scholar]
- Hong S-B, Sexton R, Tucker ML (2000) Analysis of gene promoters for two tomato polygalacturonases expressed in abscission zones and the stigma. Plant Physiol 123: 869–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Koyama T (2014) The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front Plant Sci 5: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Giovannoni JJ, Hall BD, Fischer RL, Bennett AB (1998) Transgenic analysis of tomato endo‐β‐1, 4‐glucanase gene function. Role of cel1 in floral abscission. Plant J 13: 303–310 [Google Scholar]
- Liang MH, Jiang JG (2017) Analysis of carotenogenic genes promoters and WRKY transcription factors in response to salt stress in Dunaliella bardawil. Sci Rep 7: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Riov J, Tucker ML, Patterson SE, Roberts JA (2019) Re-evaluation of the ethylene-dependent and-independent pathways in the regulation of floral and organ abscission. J Exp Bot 70: 1461–1467 [DOI] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KV, Burd S, Ophir R, Kochanek B, Reid MS, Jiang C-Z, Lers A (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154: 1929–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath P, Sane AP, Trivedi PK, Sane VA, Asif MH (2007) Role of transcription factors in regulating ripening, senescence and organ abscission in plants. Stewart Postharvest Rev 3: 1–14 [Google Scholar]
- Niederhuth CE, Patharkar OR, Walker JC (2013) Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genom 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol 9: 726–730 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Patharkar OR, Walker JC (2016) Core mechanisms regulating developmentally timed and environmentally triggered abscission. Plant Physiol 172: 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt S, Piepho H-P, Stintzi A, Schaller A (2020) Peptide signaling for drought-induced tomato flower drop. Science 367: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence‐and defence‐related processes. Plant J 28: 123–133 [DOI] [PubMed] [Google Scholar]
- Santiago J, Brandt B, Wildhagen M, Hohmann U, Hothorn LA, Butenko MA, Hothorn M (2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A (2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Sexton R, Roberts JA (1982) Cell biology of abscission. Annu Rev Plant Physiol 33: 133–162 [Google Scholar]
- Shi C-L, Stenvik G-E, Vie AK, Bones AM, Pautot V, Proveniers M,, Aalen RB, Butenko MA (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C-L, Alling RM, Hammerstad M, Aalen RB (2019) Control of organ abscission and other cell separation processes by evolutionary conserved peptide signaling. Plants 8: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y (2013) Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol 54: 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G-E, Tandstad NM, Guo Y, Shi C-L, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Whitelaw CA (2001) Signals in abscission. New Phytol 151: 323–340 [Google Scholar]
- Tranbarger TJ, Tucker ML, Roberts JA, Meir S (2017) Plant organ abscission: from models to crops. Front Plant Sci 8: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ML, Yang R (2012) IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. AoB Plants doi: 10.1093/aobpla/pls035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meeteren U, De Proft M (1982) Inhibition of flower bud abscission and ethylene evolution by light and silver thiosulphate in Lilium. Physiol Plant 56: 236–240 [Google Scholar]
- Ventimilla D, Domingo C, González-Ibeas D, Talon M, Tadeo FR (2020) Differential expression of IDA (INFLORESCENCE DEFICIENT IN ABSCISSION)-like genes in Nicotiana benthamiana during corolla abscission, stem growth and water stress. BMC Plant Biol 20: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vie AK, Najafi J, Liu B, Winge P, Butenko MA, Hornslien KS, Kumpf R, Aalen RB, Bones AM, Brembu T (2015) The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. J Exp Bot 66: 5351–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang X-l, Wang L, Tian Y, Jia N, Chen S, Shi N-b, Huang X, Zhou C, Yu Y (2017) Regulation of ethylene-responsive SlWRKY s involved in color change during tomato fruit ripening. Scient Rep 7: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li T, Meng H, Sun X (2005) Optimal and spatial analysis of hormones, degrading enzymes and isozyme profiles in tomato pedicel explants during ethylene-induced abscission. Plant Growth Regul 46: 97–107 [Google Scholar]
- Wang Y, Zou W, Xiao Y, Cheng L, Liu Y, Gao S, Shi Z, Jiang Y, Qi M, Xu T (2018) MicroRNA1917 targets CTR4 splice variants to regulate ethylene responses in tomato. J Exp Bot 69: 1011–1025 [DOI] [PubMed] [Google Scholar]
- Wien H, Turner A, Nyankanga R (1996) Low light stress influences lower abscission and yield of six bell pepper cultivars. HortScience 31: 586 [Google Scholar]
- Wilson ZA, Song J, Taylor B, Yang C (2011) The final split: the regulation of anther dehiscence. J Exp Bot 62: 1633–1649 [DOI] [PubMed] [Google Scholar]
- Ying P, Li C, Liu X, Xia R, Zhao M, Li J (2016) Identification and molecular characterization of an IDA-like gene from litchi, LcIDL1, whose ectopic expression promotes floral organ abscission in Arabidopsis. Sci Rep 6: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu Z, Lei C, Zheng C, Wang J, Shao S, Li X, Xia X, Cai X, Zhou J (2018) A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 30: 652–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.