Abstract
Exposure to ultraviolet B radiation (UV-B) stress can have serious effects on the growth and development of plants. Germin-like proteins (GLPs) may be involved in different abiotic and biotic stress responses in different plants, but little is known about the role of GLPs in UV-B stress response and acclimation in plants. In the present study, knockout of GLP 8–14 (OsGLP1) using the CRISPR/Cas9 system resulted in mutant rice (Oryza sativa L.) plants (herein called glp1) that exhibited UV-B-dependent formation of lesion mimic in leaves. Moreover, glp1 grown under solar radiation (including UV-B) showed decreased plant height and increased leaf angle, but we observed no significant differences in phenotypes between wild-type (WT) plants and glp1 grown under artificial light lacking UV-B. Fv/Fm, Y (II) and the expression of many genes, based on RNA-seq analysis, related to photosynthesis were also only reduced in glp1, but not in WT, after transfer from a growth cabinet illuminated with artificial white light lacking UV-B to growth under natural sunlight. The genes-associated with flavonoid metabolism as well as UV resistance locus 8 (OsUVR8), phytochrome interacting factor-like 15-like (OsPIF3), pyridoxal 5′-phosphate synthase subunit PDX1.2 (OsPDX1.2), deoxyribodipyrimidine photolyase (OsPHR), and deoxyribodipyrimidine photolyase family protein-like (OsPHRL) exhibited lower expression levels, while higher expression levels of mitogen-activated protein kinase 5-like (OsMPK3), mitogen-activated protein kinase 13-like (OsMPK13), and transcription factor MYB4-like (OsMYB4) were observed in glp1 than in WT after transfer from a growth cabinet illuminated with artificial white light to growth under natural sunlight. Therefore, mutations in OsGLP1 resulted in rice plants more sensitive to UV-B and reduced expression of some genes for UV-B protection, suggesting that OsGLP1 is involved in acclimation to UV-B radiation.
Germin-like protein 8–14 mutations increase rice sensitivity to UV-B and reduce expression of some genes with UV-B protective roles, suggesting the involvement of the protein in UV-B acclimation.
Introduction
UV-B light (280–315 nm and herein referred to as UV-B) is an intrinsic part of sunlight reaching the earth’s surface. Since plants use sunlight for photosynthesis, inevitably they are exposed to potentially harmful levels of UV-B under natural growth conditions. Sensing low levels of UV-B with receptor UV resistance locus 8 (UVR8) is known to trigger UVR8-mediated signaling in regulating various aspects of metabolism and development to protect plants against UV-B damage (Jenkins, 2009; Tilbrook et al., 2013). This UVR8-mediated signaling process involves two important light signaling components, namely E3 ubiquitin ligase constitutively photomorphogenic 1 (COP1) and the transcription factor elongated hypocotyl 5 (HY5). The interaction between COP1 and UVR8 activates the expression of many genes mainly associated with prevention of UV-B damage (Nawkar et al., 2013; Tilbrook et al., 2013). As a feedback regulation, repressor of UV-B photomorphogenesis 1 (RUP1) and RUP2 can disrupt the interaction between COP1 and UVR8 leading to the re-dimerization of UVR8 (Heijde and Ulm, 2013). On the other hand, RUP1/RUP2 could also be degraded following interaction between RUP1/RUP2 and COP1, leading to HY5 stability (Ren et al., 2019). Besides, the interaction of UVR8 with WRKY DNA-binding protein 36 (WRYK36) in the presence of UV-B prevents the inhibition of HY5 transcription by WRYK36. HY5 plays a role in induction of gene expression by UV-B and result in, for example, suppression of embryonic axis elongation (Yang et al., 2018). Furthermore, UVR8 seems to be able to cross talk with other signaling pathways, particularly auxin signaling, in regulating several responses. For instance, in the presence of UV-B, UVR8 inhibited the DNA-binding activities of MYB domain protein 73/77 (MYB73/MYB77) and directly repressed the transcription of their target auxin-responsive genes (Yang et al., 2020). UVR8 can also interact with methanesulfonate-suppressor (BES1) and BES1-interacting Myc-like 1 (BIM1), which are transcription factors involved in brassinosteroid signaling promoting hypocotyl elongation of Arabidopsis (Liang et al., 2018). By repressing the DNA-binding activities of these transcription factors, UVR8 can affect growth and photomorphogenesis. Moreover, BES1-RNAi transgenic plants and bim123 triple mutants were more sensitive to UV-B treatment (Liang et al., 2018). Similarly, rup1 rup2 and rup2 mutants were also more sensitive to narrowband UV-B radiation (Gruber et al., 2010), but uvr8 (Kliebenstein et al., 2002; Favory et al., 2009), cop1 (Oravecz et al., 2006), hy5 (Ulm et al., 2004; Brown et al., 2005) mutants were hypersensitive to UV-B stress.
High levels of UV-B can directly damage DNA, lipids, proteins, and photosynthesis apparatus, induce reactive oxygen species production, and affect cell integrity and vitality (Jenkins, 2009). For survival, plants need highly efficient UV-B protective mechanisms/tolerance. The response to UV-B stress involves the mitogen-activated protein kinase (MAPK) signaling cascade. UV-B stress can induce accumulation of MAP kinase phosphatase (MKP1) as well as activation of MKP1-interacting proteins MPK3 and MPK6 in Arabidopsis (González Besteiro et al., 2011; González Besteiro and Ulm, 2013). Single Arabidopsis mutants of mpk3 and mpk6 are more resistant to UV-B stress, but mkp1 mutants are hypersensitive to acute UV-B stress, which is related to sustained activities of MPK3 and MPK6 (González Besteiro et al., 2011). This pathway of MKP1-regulated stress–response is independent of the known signaling response mediated by UVR8. The role of UVR8-dependent pathway in UV-B stress is elusive, although studies have shown that the two independent pathways mediated by MKP1 and UVR8 contribute synergistically to UV-B tolerance in plants (González Besteiro et al., 2011).
Substantial knowledge of molecular, cellular, and organismal responses to UV-B has been achieved in past decades from research mainly undertaken with Arabidopsis grown under artificial light. The activities and relative importance of the UVR8-dependent and -independent pathways in plants growing under sunlight are, however, poorly understood. Since high levels of UV-B can also regulate the expression of many genes through UVR8-independent pathways, there may be other types of UV-B photoreceptors in plants (Nawkar et al., 2013; Liang et al., 2019; Tossi et al., 2019). It is, therefore, necessary to explore the functions and regulation of UVR8 in diverse plant species as well as the activities and importance of UV-B response pathways in plants growing under sunlight.
Germin-like proteins (GLPs) are the proteins that share around 30%–70% identity with germin (Bernier and Berna, 2001) and have diverse enzymatic activities, such as superoxide dismutase (SOD; Segarra et al., 2003; Gucciardo et al., 2007), oxalate oxidase (OxO; Sakamoto et al., 2015), pyrophosphatase/phosphodiesterase (AGPPase; Rodríguez-López et al., 2001; Mansilla et al., 2012) and polyphenol oxidase activity (Cheng et al., 2014). Additionally, they can also act as receptors (Swart et al., 1994; Membré et al., 2000; Gucciardo et al., 2007), protease inhibitor (Mansilla et al., 2012), and may be involved in different abiotic and biotic stress responses in different plants (Das et al., 2019). Little is, however, known about the role of GLPs in UV stress response and tolerance in plants. In a proteomic study, the abundance of a GLP in the leaves of peanut plants kept under supplementary UV-B was reported to be increased (Du et al., 2014). It is, however, not clear if a GLP might also play a role in UV radiation response in rice. In this study, new evidence was obtained in support of the participation of OsGLP1 in UV-B stress response of rice plants based on phenotypic, RNA sequencing (RNA-seq), and reverse transcription quantitative (RT-qPCR) analyses of the glp1 mutants generated using the CRISPR/Cas9 system. A rice GLP 8–14 (OsGLP1, LOC4345763) is encoded by a member of the germin-like gene family, which shares 90% amino acid identity with adenosine diphosphate glucose pyrophosphatase of barley and wheat, and 80% with the auxin-binding protein ABP20 precursor of corn (Zea may L). Besides alterations to plant architecture, lesion mimic only appeared in the leaves of glp1 mutants grown under lighting regimes that included UV-B (sunlight or artificial light supplemented with UV-B in a growth chamber). Similarly, the leaves of uvr8-1 mutants exposed to white light supplemented with UV-B for 3 d also showed necrosis (Kliebenstein et al., 2002). The results obtained in the present study support the hypothesis that OsGLP1 was involved in the acclimation of rice to UV-B.
Results
UV-B-dependent formation of lesion mimic and alterations in plant architecture in glp1 mutants
Using CRISPR/Cas9 technology, glp1 mutant rice plants were generated in the Dongjin (DJ) background. Sequence alignment and expression analysis showed that glp1-3, glp1-4, and glp1-8 mutants with different mutations in the coding sequence (CDS) of OsGLP1 were loss-of-function mutants (Supplemental Figure S1). The glp1 mutant plants displayed phenotypes that differed from those of wild-type (WT). Compared to WT, there were some brown spots on the leaf blades of glp1 grown under natural sunlight (Figure 1A). Clear trypan blue staining areas showing occurrence of cell death were observed in the leaves of glp1 mutants grown under sunlight (Supplemental Figure S2). The brown spots appeared in the leaves of glp1 after transfer from a controlled growth chamber (12-h dark at 28°C and 12 h of 600 µmol m−2 s−1 artificial light lacking UV-B at 30°C) to growth under sunlight. There were, however, significantly fewer lesions in the newly grown leaves than in old leaves (Supplemental Figure S3) and in the leaves of seedlings grown under natural sunlight after germination than in those transferred to natural sunlight from white light (Figure 2A;Supplemental Figure 3A). When glp1 was grown outdoors under a piece of glass or in a glasshouse whereby natural sunlight was filtered through, the abundance of the lesions in the leaf blades decreased remarkably compared to those of glp1 grown under 50–100 µW cm−2 UV-B in the glasshouse supplemented with extra UV-B (Supplemental Figure S4). On the other hand, lesions were not observed in the leaves of glp1 mutant plants grown in a controlled growth chamber (12-h dark, 28°C/12-h light, 30°C) under 600 µmol m−2 s−1 artificial light lacking UV-B compared to those of glp1 kept in a growth room supplemented with 50–80 µW cm−2 UV-B (Figure 1B). These results suggest that UV-B triggered the lesion development in the glp1 knockout mutants. To explore the possibility that formation of leaf lesions in the glp1 mutants could be specifically dependent on UV-B or a combination of environmental factors other than UV-B, transgenic plants grown to the 4-leaf stage in a growth chamber without supplementary UV-B were then placed at different conditions as described in Supplemental Figure S4. Interestingly, high light intensity and high temperature could not induce the appearance of lesions in the leaves of the glp1 (Supplemental Figure S4). Therefore, the formation of lesions in the leaves of glp1 appears to be dependent on UV-B.
Figure 1.
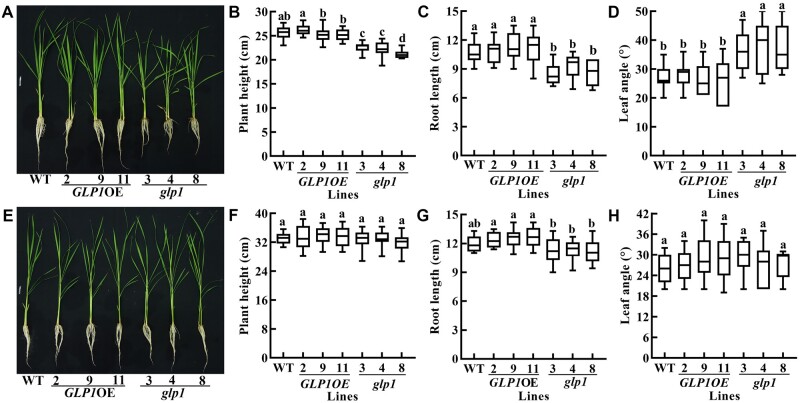
Appearance of the lesion mimic on leaves of rice glp1 mutants. A, is the second leaves from bottom to top of WT, transgenic plants overexpressing OsGLP1 (2 and 9) and glp1 mutants (3, 4, and 8) grown in a growth chamber without UV-B (12 h of 600 µmol m−2 s−1 artificial light at 30°C and 12 h of dark at 28°C) to 5-leaf stage and then exposed to natural sunlight and natural sunlight passing through a glass for 1.5 d. B, is the second leaves from bottom to top of 5-leaf stage seedlings grown in a controlled growth chamber with (left) or without (right) 50–80 µW cm−2 UV-B for 5 h, and were then grown in the chamber without UV-B for 30 h.
Figure 2.
Knockout of OsGLP1 results in dwarfness and increased leaf inclination in rice plants grown under natural sunlight. In A, B, C, and D, the phenotype, plant height, root length and leaf angle (the second leaf from bottom to top), respectively, of WT, transgenic plants overexpressing OsGLP1 (2, 9, and 11) and glp1 mutants (3, 4, and 8) grown under natural sunlight until the 5-leaf stage are shown. In E, F, G, and H, the phenotype, plant height, root length and leaf angle, respectively, of WT, transgenic plants overexpressing OsGLP1 (2, 9, and 11) and glp1 mutants (3, 4, and 8) grown in a controlled growth chamber deficient in UV-B (12-h light, 600 µmol m−2 s−1, 30°C; 12-h dark, 28°C) until the 5-leaf stage are shown. The boxplots, from top to bottom, denote maximum value, upper quartile, median, lower quartile and minimum value, and whiskers denote maximum value to minimum value of the data. The means ± sd (n ≥ 16 plants) of plant height, root length or leaf angle assigned with different letters were significantly different as determined by one-way analysis of variance (ANOVA) with post hoc Bonferroni test (P < 0.05).
Besides lesion formation, glp1 grown under natural sunlight also showed decreased plant height and root length as well as increased leaf angle at the 5-leaf stage (Figure 2, A–D). In contrast, there was no apparent difference in the phenotypes between WT and glp1 grown in a controlled growth chamber illuminated with artificial light lacking UV-B (Figure 2, E–H). Phenotypes including increased leaf angle as well as semi-dwarf plants became more prominent in glp1 grown under natural sunlight at the maturation stage (Supplemental Figure S5, A–C). There was no significant difference in stem length between WT and glp1 grown in a glasshouse, although the leaf angle of glp1 was greater than WT. It is important to note that this increase in leaf angle in glp1 grown in the glasshouse was less than when grown under natural light (Supplemental Figure S5, D–F). Taken together, these results have revealed a heretofore unrecognized role for OsGLP1 in UV-B protection and UV-B-triggered responses.
More damages on photosynthetic efficiency in glp1 than in WT plants transferred from artificial white light to growth under natural sunlight
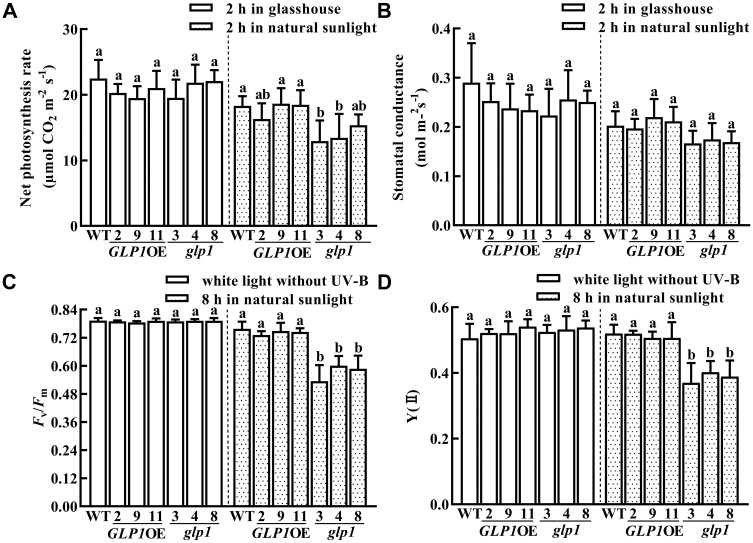
The photosynthetic organs of plants are very sensitive to UV-B. The parameters related to photosynthesis were, therefore, measured in WT, glp1, and OsGLP1OE plants. Compared to WT, the net photosynthesis rate but not the stomatal conductance in glp1 at the 6-leaf stage was decreased after transfer to growth under natural sunlight for 2 h from a controlled growth chamber deficient in UV-B, but this decrease in net photosynthesis rate in glp1 did not take place after transfer to a glasshouse (without any supplementation of UV-B) for 2 h (Figure 3, A and B). Moreover, Fv/Fm and Y (II) of glp1 displayed a significant decrease after transfer to growth under natural sunlight for 8 h. No significant difference in Fv/Fm and Y (II) was observed between WT and glp1 grown in a growth chamber under artificial light without any supplementation of UV-B (Figure 3, C and D). Taken together, these results suggest that there was more damage to photosystem in the glp1 under sunlight and are also consistent with the suggestion that glp1 may play a role in UV-B acclimation.
Figure 3.
Knockout of OsGLP1 results in lower photosynthetic efficiency in glp1 compared to those in WT plants transferred from white light to natural sunlight. Determination of net photosynthesis rate (A), stomatal conductance (B), Fv/Fm (C), and Y (II) (D) in leaves of WT, transgenic plants overexpressing OsGLP1 (2, 9, and 11) and glp1 mutants (3, 4, and 8). All the plants were grown to the 6-leaf stage in a growth chamber (conditions of the growth chamber: 600 µmol m−2 s−1 white light, 0 µW cm−2 UV-B, 12 h, 30°C; dark, 12 h, 28°C). They were then exposed to different experimental conditions as specified on the figure: under glasshouse conditions, natural sunlight (320–1080 µmol m−2 s−1, 21.1–112.9 µW cm−2 UV-B, 26°C–29°C) or white light without supplementation of UV-B. From the comparative analysis of the data obtained under a specific experimental condition, the means ± sd (n ≥ 4 plants) assigned with different letters were significantly different as determined by one-way ANOVA with the means separation post hoc Student–Newman–Keuls test (P < 0.05).
The abundance and location of OsGLP1 are unaffected by UV-B irradiation
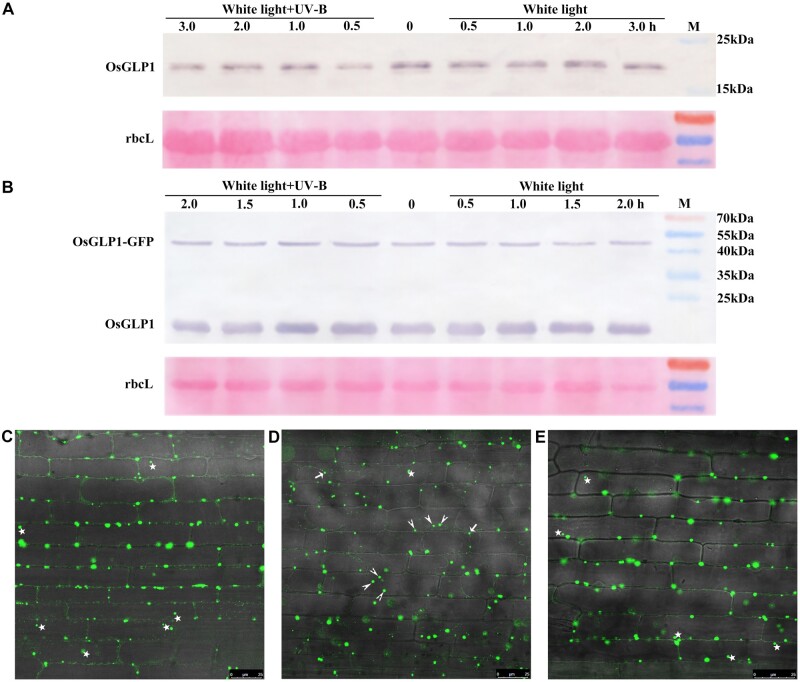
To explore the role of OsGLP1 in UV-B signal transduction, regulation of OsGLP1 by UV-B was investigated. Based on the Western blotting results (Figure 4, A and B), there was no significant difference in the levels of OsGLP1 whether WT at the 4-leaf stage or 7-d-old OsGLP1-GFPOE seedlings were exposed to 50–80 µW cm−2 of UV-B for 3 or 2 h, respectively. Similarly, the abundance of UVR8 was also unaffected by 3 µmol m−2 s−1 UV-B, but UV-B could affect its subcellular location (Kaiserli and Jenkins, 2007).
Figure 4.
Effect of UV-B on GLP1 and GLP1-GFP protein levels as well as location. Protein gel blot, with OsGLP1-His antisera, of total protein extracts from the leaves of WT (A) grown to 4-leaf stage under 600 µmol m−2 s−1 white light in a growth chamber deficient in UV-B, and GLP1-GFPOE seedlings grown in the dark on Murashige and Skoog (MS) medium for 7 d (B) before they were exposed to 600 µmol m−2 s−1 white light supplemented with or without 50–80 µW cm−2 UV-B for different times (hours). Confocal images of GFP fluorescence in leaf sheath of transgenic plants expressing GLP1-GFP from the Ubi promoter grown in the dark on MS medium for 7 d before they were exposed to white light and white light supplemented 50–80 µW cm−2 UV-B for 2 h (C and E, respectively); confocal images of GFP fluorescence in leaf sheath from GLP1-GFPOE seedlings exposed to white light for 2 h, treated with 30% sucrose about 5 min (D). Bars = 25 µm. Stars denote fluorescence in the cell, short arrows and long arrows denote fluorescence in the plasma membrane and apoplast, respectively.
When leaf sheath from 7-d-old seedlings overexpressing GLP1-GFP grown under white light lacking UV-B were examined using confocal microscopy, the fluorescence dots were found mainly in the cell margins and a few in the cytosol (Figure 4C), and some were moving during microscopic observation. There was, however, no obvious difference in the localization of the fluorescence when the seedlings overexpressing GLP1-GFP placed under white light supplemented with 50–80 µW cm−2 UV-B for 2 h during microscopic observation (Figure 4E). Plasmolysis of leaf sheaths using 30% sucrose showed some dots were in the extracellular space (Figure 4D), but fluorescence dots were mainly found in the cytosol of protoplasts isolated from WT transfected with pBI121-GLP1-GFP (Supplemental Figure S6).
OsGLP1 encodes a protein devoid of detectable SOD activity and OxO activity
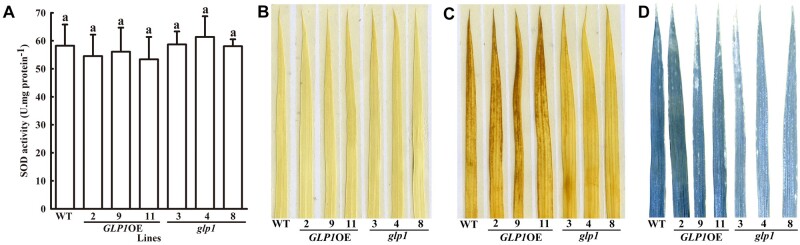
The amino acid sequence identity between OsGLP1and a GLP of wheat (TaGLPI) is 90% (Supplemental Figure S7). Both have previously been reported to have SOD activity (Segarra et al., 2003; Banerjee and Maiti, 2010). Here, using a test-tube colorimetric enzyme assay and in-gel enzyme activity staining, no difference in SOD activity was found in the crude protein extracts from WT, transgenic plants overexpressing OsGLP1 and glp1 mutants placed under natural sunlight for 8 h before SOD assay (Figure 5A;Supplemental Figure S8). In contrast, OsGLP1 abundance displayed a substantial increase in GLP1OE plants and it was knocked out in glp1 (Supplemental Figure S1). No OxO activity could be detected in leaves of WT, glp1, and GLP1OE plants using the colorimetric method. Therefore, OsGLP1 was a protein that lacked detectable activities of SOD and OxO. Moreover, 3, 3'-diaminobenzidine (DAB) staining and nitrotetrazolium blue chloride (NBT) staining showed that glp1 had slight lower contents of H2O2 and O2•−, respectively, than WT and transgenic rice plants overexpressing OsGLP1 (Figure 5, C and D).
Figure 5.
OsGLP1 encodes a protein devoid of detectable SOD activity. The levels of SOD activity (A), in situ staining for detection of H2O2 using DAB (C), O2•− using NBT (D), and staining control (water only, B) in rice leaves of WT, transgenic plants overexpressing OsGLP1 (2, 9, and 11) and glp1 mutants (3, 4, and 8) grown in a growth chamber without UV-B (600 µmol m−2 s−1 white light, 12 h, 30°C; dark, 12 h, 28°C) to the 5-leaf stage and then exposed to natural sunlight for 8 h. The means ± sd (n = 3) of SOD activity assigned with the same letter were not significantly different as determined by one-way ANOVA with post-hoc Student–Newman–Keuls test (P < 0.05).
OsGLP1 mutation causes reduced expression of some genes in UVR8 signaling pathway and increased MPK3 and MPK13 transcripts in MAPK signaling cascade
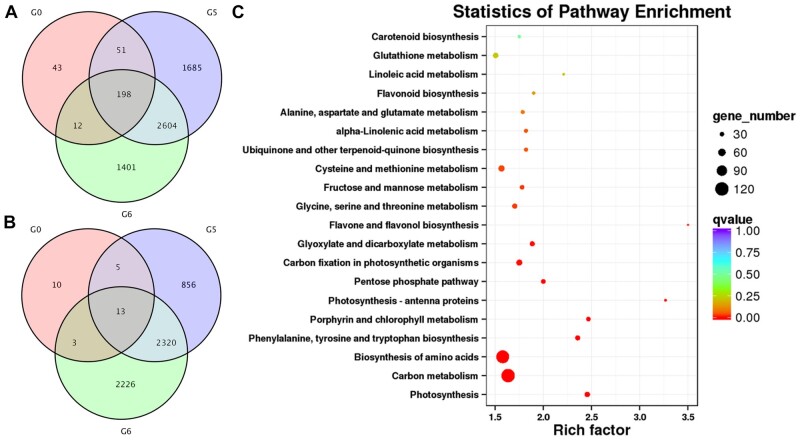
To gain insight into the molecular basis of UV-B-dependent glp1-related lesion mimic formation in rice, total RNA was extracted for RNA-seq from leaves of WT and glp1 at the 4-leaf stage transferred to growth under natural sunlight for 0 h, 4 h, and 8 h from a controlled growth chamber (12-h dark at 28°C/12 h artificial light of 600 µmol m−2 s−1 without UV-B supplementation at 30°C). It was found that 304, 4,538, and 4,215 genes were upregulated at least two-fold, while 31, 3,194, and 4,562 genes were downregulated at least two-fold in glp1 compared with WT after exposure to 0 h, 4 h, and 8 h under natural sunlight, respectively (Figure 6). The RNA-seq results suggest that transcriptional reprogramming occurred in glp1 prior to the onset of lesion formation (Figure 6, A and B; Supplemental Figure S9A and Supplemental datasets S1–8). The results of pathway enrichment analyses revealed that the genes mainly involved in photosynthesis, carbon metabolism, flavone, and flavonol biosynthesis were affected in glp1 (Figure 6C;Supplemental Figure S9B).
Figure 6.
RNA-Seq analyses revealed that many genes are regulated in the leaves of glp1 mutant transferred from white light to natural sunlight. In A and B, numbers of genes that were upregulated and downregulated in the second leaf from bottom to top of glp1 seedlings by at least two-fold compared to those in WT are shown, respectively. G0, G5, and G6 denote those in WT and glp1-4 grown in a growth chamber under 600 µmol m−2 s−1 constant white light without UV-B (12-h light, 30°C; 12-h dark, 28°C) to the 4-leaf stage and then transferred under natural sunlight for 0 h (600 µmol m−2 s−1, 0 µW cm−2 UV-B, 30°C), 4 h (1,440 µmol m−2 s−1, 174 µW cm−2 UV-B, 37°C), and 8 h (810 µmol m−2 s−1, 80.2 µW cm−2 UV-B, 35°C), respectively. Statistics of pathway enrichment analyses of the differentially regulated genes in WT and glp1-4 grown under natural sunlight for 8 h (C). Scale bar denotes the q value.
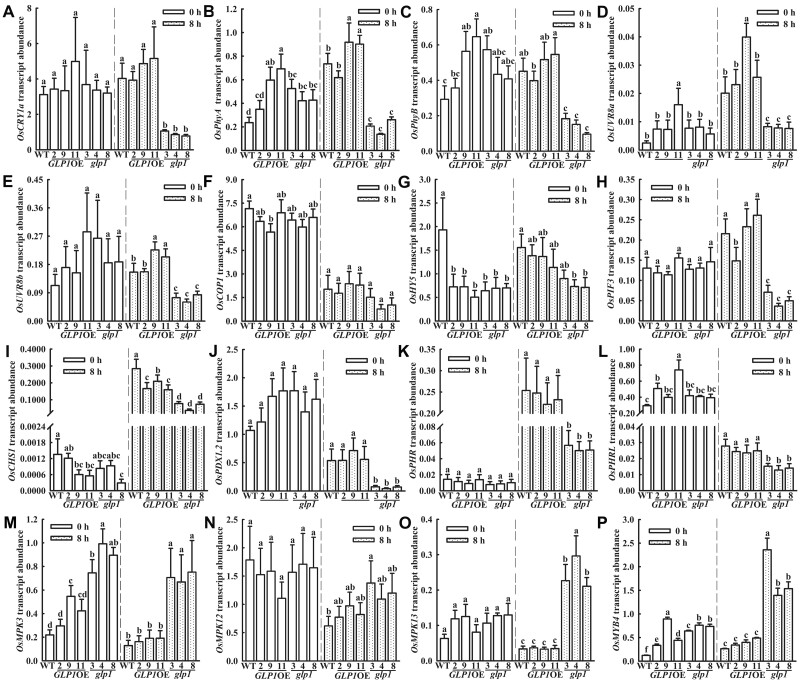
Validation using RT-qPCR of the changes in the expression of the following selected genes that seem to be associated with UV-B exposure based on the RNA-seq results was carried out (Figure 7;Supplemental Figure S10): cryptochrome 1a, (OsCRY1a), phytochrome A (OsPhyA), phytochrome B (OsPhyB), OsUVR8a, OsUVR8b, OsCOP1, OsHY5, PHYTOCHROME-INTERACTING FACTOR-LIKE 15-like (OsPIF3), chalcone synthase 1 (OsCHS1), pyridoxal 5′-phosphate synthase subunit PDX1.2 (OsPDX1.2), deoxyribodipyrimidine photolyase (OsPHR), deoxyribodipyrimidine photolyase family protein-like (OsPHRL), OsMKP1, MAPK OsMPK3, OsMPK6, OsMPK12, and OsMPK13 as well as transcription factor MYB4-like (OsMYB4). At 0 h (the time before the plants were transferred to natural sunlight), there were no significant differences in the expression of these genes except HY5, MPK3, MYB4, and PHYA between WT and glp1. Following 8 h of exposure to natural sunlight, however, these genes except COP1, MKP1, MKP12, and MPK6 were clearly differentially regulated in glp1. The vast majority of the RT-qPCR analyses are consistent with the RNA-seq results, and the expression levels of most genes were synergistically regulated by light and genotype (Supplemental Table S1). Moreover, mutating MPK3 using CRISPR-Cas9 in glp1-8 plants abated the formation of lesion (Supplemental Figure S11). These findings further gave credence to the suggestion that OsGLP1 seems to play an intriguing role in UV-B protection by involving the modulation of expression of a range of genes in UVR8-mediated and MAPK signaling cascade pathways.
Figure 7.
RT-qPCR analysis of the expression of UV-B-associated genes regulated in glp1 compared to WT. WT, transgenic plants overexpressing OsGLP1 (2, 9, and 11) and glp1 mutants (3, 4, and 8) were grown in a growth chamber under 600 µmol m−2 s−1 constant white light (12-h light, 30°C; 12-h dark, 28°C) to the 4-leaf stage, then transferred to natural sunlight for 0 h (600 μmol m−2 s−1, 0 μW cm−2 UV-B, 30°C) and 8 h (891.3 μmol m−2 s−1, 96.4 μW cm−2 UV-B, 34°C), respectively, and the second leaf from bottom to top of the seedlings were harvested separately. Expression levels of OsCRY1a (A), OsPhyA (B), OsPhyB (C), OsUVR8a (D), OsUVR8b (E), OsCOP1 (F), OsHY5 (G), OsPIF3 (H), OsCHS1 (I), OsPDX1.2 (J), OsPHR (K), OsPHRL (L), OsMPK3 (M), OsMPK12 (N), OsMPK13(O), and OsMYB4 (P) were analyzed and ACTIN was used an internal standard. The mean gene expression levels ± sd (n = 3) in the different plants exposed to natural sunlight at 0 h were compared statistically and those assigned with different letters were significantly different. The same analyses were performed with the plants exposed to natural sunlight for 8 h. All analyses were determined using one-way ANOVA with the means separation post hoc Student–Newman–Keuls test (P < 0.05).
Expression of OsGLP1 and genes associated with IAA metabolism modulated in glp1
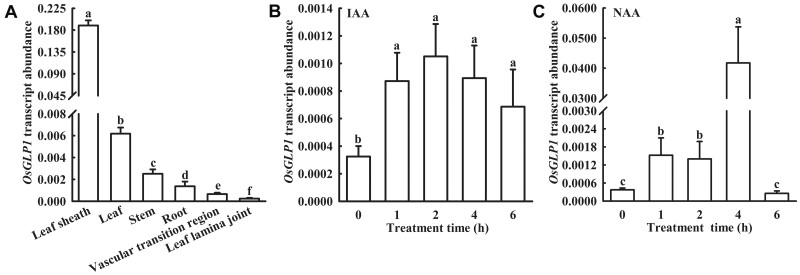
Based on BLAST search of the protein databases, many GLPs in rice can be classified into five phylogenetic subgroups (Supplemental Figure S12) referring to Carrillo et al. (2009). OsGLP1 belongs to the auxin-binding protein subgroup of GLPs, although the only member of this subgroup in peach, ABP19/20, has been shown to bind auxin (Ohmiya et al., 1998). RT-qPCR analysis showed that although OsGLP1 transcripts were detectable in various tissues, the expression was relatively stronger in the leaf sheath, leaf, and stem of WT (Figure 8A). The expression levels OsGLP1 increased rapidly in the leaves of WT following the treatment with IAA or NAA. The OsGLP1 transcript was about three-fold of that in control (treatment with Kimura B complete nutrient solution only) after the treatment with IAA or NAA for 2 h (Figure 8, B and C). Based on the results of RNA-seq, many genes related to IAA metabolism were changed, including an increase in the transcripts of probable indole-3-acetic acid-amido synthetase GH3.6 (OsGH3.6), GH3.8 (OsGH3.8), and auxin-responsive protein SAUR71 (OsSAUR71), and a reduction in the transcripts of auxin response factor 15-like (OsARF15), auxin-responsive protein OsIAA24, OsIAA21, and OsIAA13 in glp1 exposed to natural sunlight for 4 h and 8 h (Supplemental dataset S3). Moreover, in an assay of auxin-induced gene expression in plant using pDR5: GUS reporter, the GUS staining in the leaf cushions of glp1 grown under natural sunlight was much lighter than that of WT, but nearly similar to that of WT plants grown under artificial light without supplementation of UV-B in a growth chamber (Supplemental Figure S13).
Figure 8.
Analysis of OsGLP1 expression using RT-qPCR. OsGLP1 expression in various tissues of booting stage rice (A) and in leaves of rice seedlings at the 5-leaf stage treated with 10 μmol L−1 IAA (B) and 1 μmol L−1 NAA (C) at various time points was analyzed and ACTIN was used as an internal standard. The means ± sd (n = 3) of gene expression levels assigned with different letters were significantly different as determined by one-way ANOVA with Student–Newman–Keuls test (P < 0.05).
Discussion
UV-B is a trigger for the formation of leaf lesions and changes in other growth parameters in glp1
The following lines of evidence support the notion that UV-B is a trigger for the formation of lesions in glp1. At the 2-leaf stage, brown spots began to appear in the leaves of glp1 mutants transferred from a growth chamber illuminated with artificial white light deficient in UV-B to growth under natural sunlight. In contrast, there was a notable reduction in the number of lesions in the leaves of glp1 grown under natural sunlight filtered through a glasshouse. Although the light intensity and temperature in the glasshouse were comparable to outdoors, the UV-B intensity in the glasshouse was only about 20% of that in natural sunlight. Additionally, when glp1 mutants were placed in a glasshouse supplemented with UV-B, a reduction of lesions did not occur. Upon transfer to a growth chamber supplemented with 50–80 μW cm−2 UV-B, brown spots appeared in the leaves of glp1, WT and GLP1OE plants exposed to UV-B 3 h, 5 h, and 7 h, respectively. In a prior study involving downregulation of OsGLP1 using hairpin RNA silencing approach, lesions on the leaves of the transgenic rice plants, and not the untransformed control, were also apparent, but the possibility that these would be lesion mimics was not recognized (Figure 3B in Banerjee and Maiti, 2010). This was probably because the transgenic plants were assumed to be infected by blast fungi and sheath blight as the plants were grown under irrigation in a region known to be disease-prone and the transgenic plants were likely to be more susceptible to diseases compared to the untransformed control (Banerjee and Maiti, 2010). This assumption may be, however, not the sole possible explanation for the lesions on the leaves. Since the plants were grown outdoors, arguably the lesions found on the rice leaves in this prior study could also be induced by UV-B as shown in the present study. Besides, there was no corroborative evidence that the transgenic plants in the study of Banerjee and Maiti (2010) were diseased as they were not inoculated with any sheath blight pathogen or blast fungi.
Ultraviolet light has also been implicated in a prior study of the lesion mimic rice mutant generated from deletion of the heat stress transcription factor OsSpl7. When adult spl7 mutants were grown in summer under natural field conditions (30°C–35°C), there was a high density of lesions on the leaves. If ultraviolet light in solar radiation (presumably UV-B) was filtered out, the number of lesions was drastically reduced (Yamanouchi et al., 2002). Lesions were not observed on the mutant plants grown in a controlled growth chamber (26°C, artificial light), and only few lesions appeared in a high-temperature growth chamber (35°C, artificial light). It seems, therefore, that high temperature and ultraviolet light were both needed to trigger the spl7 mutants to produce a high density of lesions (Yamanouchi et al., 2002). Other environmental factors including temperature, light intensity, and sunshine duration have all been previously shown to be associated with the formation of lesions in rice leaves (Wang et al., 2005, 2015; Zhao et al., 2017). For example, brown necrotic lesions appeared in the mature leaves of antisense transgenic rice plants with reduced expression of a rice zinc finger protein (OsLSD1) grown at low temperature (21.9°C) and short daylight (11–12 h; Wang et al., 2005). In the present study, however, high light intensity and high temperature could not induce lesion formation in the leaves of glp1 mutants. UV-B seems, therefore, to be the only requisite trigger for the formation of leaf lesions in glp1. Moreover, Arabidopsis uvr8-1 mutants showed enhanced UV-B sensitivity including exhibition of necrosis in the first true leaves and cotyledons compared to WT when grown for 3 d under constant 0.2 kJ UV-B. Leaf necrosis was progressively more severe during 3 d of recovery under white light deficient in UV-B (Kliebenstein et al., 2002). In the present study, knockout of OsGLP1 may also make rice plants more sensitive to UV-B as far as induction of necrosis in the leaves and alteration of phenotype are concerned.
Plants exposed to UV-B exhibit a dwarf phenotype as UV-B can greatly inhibit hypocotyl and stem elongation (Jansen, 2002). The reduction in plant height has, therefore, become an important parameter to determine the sensitivity of plants to UV-B radiation. Additionally, UV-B can also affect the growth and development of roots (Ge et al., 2010; Yang et al., 2020). Our results showed that the phenotype of glp1 was comparable with WT and OsGLP1OE seedlings under white light deficient in UV-B (12 h of 600 μmol m−2 s−1 artificial light at 30°C and 12 h of dark at 28°C). When grown under natural sunlight, the plant height of glp1 was, however, about 80% that of WT, and the root length was also shorter than that of WT. The phenotype of OsGLP1OE seedlings was not significantly different from WT grown under natural sunlight. In contrast, transgenic rice plants with downregulated expression of OsGLP1 displayed semi-dwarf phenotype. Similarly, the cells of the stem were shorter and lower in the length: width ratio in transgenic plants compared to the control reported in a previous study (Banerjee and Maiti, 2010).
After perceiving UV-B signal, UVR8 monomerizes and interacts immediately with COP1, promoting the accumulation of HY5/HYH (Vanhaelewyn et al., 2016). HY5/HYH transcription factor could also control transcription and translation of the auxin transport proteins PIN1 and PIN3 as well as negative regulators of auxin signaling AXR2/IAA7, IAA2, and SLR/IAA14 (Vanhaelewyn et al., 2016). In the present study, RNA-seq analysis showed that the expression of Aux/IAA family members such as OsIAA2, OsIAA3, OsIAA10, OsIAA17, OsIAA24, etc. was not significantly different from WT before transfer to growth under natural sunlight, but the expression levels of these genes in the leaves of glp1 were significantly lower than that in WT at 8 h after transfer to growth under natural sunlight. In addition, the GH3 family as a class of genes encoding enzymes that catalyze the conjugation of IAA to amino acids to produce aminoacyl compounds was significantly upregulated in the leaves of glp1 (Supplemental dataset S3). The transcript of PIFs was also downregulated in glp1 mutants (Supplemental dataset S6). PIFs are direct regulators of the expression of a number of genes including genes encoding auxin biosynthesis (YUC8/YUC9) and auxin signaling (AUX/IAA; Vanhaelewyn et al., 2016). UV-B can inhibit auxin biosynthesis by triggering degradation of PIF4 and PIF5 and stabilizing DELLA proteins (Hayes et al., 2014), and also inhibit auxin responses through UVR8 (Yang et al., 2020). UV-B can alter the expression of auxin-related genes in plants and cause changes in plant morphological changes (Jansen, 2002). Under natural sunlight conditions, glp1 showed a typical auxin-deficient phenotype with dwarf and increased angles of flag leaves, as well as decreased auxin content in the leaf lamina. Similarly, OsLC1 (OsGH3-1) function-acquired mutant lc1-D also showed dwarf plant phenotype, increased leaf angle, and a decreased free auxin content in the leaf lamina (Zhao et al., 2013). It seems that the alteration in plant height of glp1 mutants kept under natural sunlight may be due to UV-B-mediated changes in the expression of auxin-related genes in glp1 mutants.
In addition, the net photosynthetic rate was also reduced in glp1 mutants after transfer to natural sunlight for 2 h, but not transfer to a glasshouse for 2 h. Fv/Fm and Y(II) in glp1 also showed a significant decrease when the plants were transferred to natural sunlight for 8 h. These two parameters were not significantly different between WT and glp1 kept under white light deficient in UV-B. Additionally, the RNA-seq analysis revealed that the expression levels of genes associated with photosynthesis and photorespiration were downregulated in glp1 after transfer to natural sunlight for 4 h and 8 h (Supplemental dataset S2). Similarly, Fv/Fm and ΦPSII were also decreased in uvr8 mutants compared to WT exposed to elevated UV-B. It is well known that some photosynthetic components are particularly susceptible to damage by UV-B, UVR8 is known to promote photosynthetic efficiency at elevated levels of UV-B (Davey et al., 2012). These results in the present study strongly support that knockout of OsGLP1 resulted in increased sensitivity to UV-B.
OsGLP1 is involved in rice acclimation to UV-B radiation: several genes with UV-B protective roles show altered expression in the glp1 mutant
Currently, UVR8 is the only known photoreceptor of UV-B. After perceiving UV-B signal, UVR8 orchestrates the expression of a range of genes, such as HY5, flavonoid synthesis genes, PHR and PDX, with vital functions in protecting plants against UV-B and enables plants to survive in sunlight. Accordingly, UV-B signal transduction in uvr8-1 has been altered as shown by a lack of UV-induced accumulation of flavonoids as well as CHS mRNA and protein which is the committing enzyme for flavonoid biosynthesis (Kliebenstein et al., 2002), therefore, uvr8-1 and hy5 mutants are highly sensitive to UV-B stress (Brown et al., 2005). In rice genome, two proteins encoded by LOC4329648 (OsUVR8a) and LOC4335903 (OsUVR8b) share 74% and 75% sequence identity with the Arabidopsis counterpart (AtUVR8). Based on RT-qPCR and RNA-seq analyses, there was no significant difference in the expression of OsUVR8a and OsUVR8b in the WT and glp1 before transfer from a growth chamber illuminated with artificial white light to outdoors with sunlight. The expression of these genes was significantly lower in the leaves of glp1 than in WT after transfer to sunlight for 8 h. Similarly, the level of CHS1 mRNA in glp1 was also lower than in WT grown under sunlight. Based on the RNA-seq analysis, the expression of genes related to flavonoid metabolism (Os10g0317900, Os10g0317950, Os10g0320100, Os10g0320201, and Os11g0530600) in the glp1 was not different from in WT before transfer to outdoors with sunlight. The expression of these five genes in the leaves of glp1 was, however, significantly downregulated compared to WT after 4 h and 8 h of transfer to natural sunlight (Supplemental dataset S1). Similarly, there is also a lack of UV-B-regulated expression of CHS in uvr8-2 (Brown et al., 2005; Cloix et al., 2012).
In contrast to the expression of CHS1, the expression level of OsMYB4 was upregulated in glp1 at 8 h after transfer to growth under natural sunlight. Plants overexpressing AtMYB4 were more sensitive to UV-B. The mutation of AtMYB4 resulted in increased levels of sinapate esters in Arabidopsis leaves and UV-B tolerance. AtMYB4 expression is down-regulated by exposure to UV-B light, suggesting that repression of its expression is an important mechanism for acclimation to UV-B in Arabidopsis (Jin et al., 2000). Moreover, the induction of the OsPHR transcript in glp1 grown under natural sunlight was substantially reduced in comparison with WT. This is consistent with a study of Arabidopsis showing that the induction of AtPHR transcript by UV-B was also substantially reduced in uvr8-2 and uvr8-6 mutants (Li et al., 2015). PHR is the repair enzyme for UV-B-induced cyclobutane pyrimidine dimers which is a principal cause of UV-B-induced growth inhibition in rice grown with supplementary UV-B (Hidema et al., 2007; Teranishi et al., 2012). An increase in PHR activity can significantly alleviate UVB-caused growth inhibition in rice and is essential for protecting cells from UV-B radiation (Li et al., 2015). In addition, the expression level of OsPDX1.2 in glp1 was also downregulated. Pyridoxine biosynthesis1 (PDX1) is a protein involved in the synthesis of pyridoxine (vitamin B6) and plays important functions in oxidative stress, photoprotection, and UV-B response (Chen and Xiong 2005; Ristilä et al., 2011). Accumulation of PDX1 under sunlight is modulated by UVR8 (Morales et al., 2013). PDX1.2, MYB4, and PHR all belong to the downstream signaling components of UVR8. Therefore, OsGLP1 might be involved in the regulation of some genes in UVR8 signaling.
UV-B stress is mediated by activating MAPK signaling pathway. MKP1, MPK3, and MPK6 may play important roles in UV-B signaling and is necessary for UV-B stress resistance in Arabidopsis (González Besteiro et al., 2011). The present RNA-seq analysis showed that the expression levels of MAPK-related genes (MPK6, MKP1, and MPK13) were similar in glp1 and WT grown under artificial white light deficient in UV-B. The expression levels of OsMKP1, OsMPK3, OsMPK6, OsMPK12, and OsMPK13 were, however, significantly higher in the leaves of glp1 than in WT grown under natural sunlight for 4 h, while only the OsMPK3 and OsMPK13 expression levels were much higher in glp1 mutant than in WT at 8 h (Supplemental dataset 1). Based on the RT-qPCR analysis, the expression levels of OsMPK3 and OsMPK13 were significantly higher in the leaves of glp1 than in WT grown under natural sunlight for 8 h. The expression level of OsMPK6 was similar in glp1 and WT. OsMPK6 shares 46% identify with AtMPK4 and has been shown to play a critical role during early embryogenesis because T-DNA insertion mutant of OsMPK6 is an embryo-lethal mutant (Yi et al., 2016). OsMPK6 might not be involved in UV-B stress response. Additionally, mutation of MPK3 in glp1 mutants could make its lesion lessened in response to solar irradiation (Supplemental Figure S11). mkp1 seedlings irradiated with broadband UV-B for 3.5 h displayed bleaching and a characteristic dark pigmentation. MKP1 can provide protection against UV-B-induced cell death by inhibiting UV-induced MPK3 and MPK6 activities (González Besteiro et al., 2011), suggesting that the lesion formation in the leaves of glp1 might be related to MAPK signaling cascade.
In a previous study, TaGLPI, which shares 90% identity with OsGLP1 is a protease inhibitor with SOD and AGPP activity (Segarra et al., 2003; Mansilla et al., 2012). The results in the present study, however, suggest that OsGLP1 is devoid of SOD activity, AGPP activity, and protease inhibitor activity based on the in-gel activity assays described by Mansilla et al. (2012). Phylogenetic analysis of 52 GLPs in plants showed that OsGLP1 belongs to the auxin-binding protein subgroup (Supplemental Figure S12) and has a conserved auxin-binding region Box A (Carrillo et al., 2009). However, only ABP19/20 in this subgroup has been shown to bind auxin (Ohmiya et al., 1998). Arabidopsis GLP4 could also bind IAA and 2, 4-D as well as its transcript could be stimulated by 10 µmol L−1 IAA (Yin et al., 2009). Like AtGLP4, the expression of OsGLP1 could also be induced by IAA and NAA under artificial white light, although it only shares 43% identity with OsGLP1. In contrast to AtGLP4, another GLP in Arabidopsis, PDGLP1, and shares 41% identity with OsGLP1 localized in the plasmodesmata and was not induced by exogenous IAA treatment. PDGLP1 could, however, interact with a non-cell-autonomous protein, NCAPP1, as well as actin, α β-1, 3-glucanase, phosphate responsive 1, and a putative ABC transporter. In addition, PDGLP1 could also interact with PDGLP2 which is found in plasmodesmata-enriched cell wall protein fraction and a regulatory component of root growth and development (Ham et al., 2012). GLPs AtGER1 and AtGER3 sharing 62% and 59% identities with OsGLP1 are also associated with extracellular matrix, but part of AtGER1 was found in soluble fraction, Membré et al. (2000) proposed that AtGER1, ATGER2, and AtGER3 could be a class of receptors involved in physiological, developmental processes and stress response. Moreover, a GLP from pea displayed receptor activity for rhicadhesin (Swart et al., 1994). Similarly, plasmolysis of leaf sheaths of GLP1-GFP overexpressing plants showed that fluorescence was found both intracellularly and extracellularly in a dot-like manner. In addition, most GLPs reported up to now are somehow associated with the extracellular matrix, more than half of the GLPs contain a Arg-Gly-Asp/Lys-Gly-Asp (RGD/KGD) tripeptide, which is also present in animal RGD-containing proteins such as fibronectin and vitronectin. In animals, these are adhesion proteins that participate in the exchange of information between the outside and the inside of the cells (Bernier and Berna, 2001). Like the above-mentioned GLPs, OsGLP1 is also associated with extracellular matrix and contains the auxin-binding region Box A, suggesting that some of the molecular mechanisms whereby OsGLP1 may be involved in UV-B response in rice.
In conclusion, new insights about UV-B signal transduction in rice acclimating to UV-B have been obtained. The results presented here suggest that OsGLP1 plays a role in upregulation of UVR8 pathway and repression of the MAPK pathway in response to UV-B. The precise mechanism underpinning these two UV-B response pathways is still elusive and warrants future investigations.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa L.) cv DJ was used for generating OsGLP1 knockout mutants and OsGLP1 overexpressing transgenic rice plants. Germinated seeds were grown in Kimura B complete nutrient solution (Yoshida et al., 1976) under different experimental conditions as described in the legend of each figure. A plant growth chamber (Percival E-41HO) was fitted with fluorescent lamps to supply artificial light. For supplementary UV-B in some experiments, a UV-B lamp (G15T8E UV-B, with a radiation spectrum of 290–310 nm and a peak at 306 nm, purchased from Sankyo Denki, Kanagawa, Japan) was used. UV intensity was measured using a 742 UV light meter (manufactured by Beijing Normal University, Beijing, China).
Construction of OsGLP1 mutants and OsGLP1-overexpression transgenic plants
OsGLP1 mutants were constructed using the CRISPR/Cas9 system. First, the vector containing a CRISPR cassette comprising a functional Cas9 under an Ubi promoter and two gRNAs with targets at different locations of OsGLP1 was constructed according to Ma et al. (2015). The vector was then introduced into Agrobacterium tumefaciens strain EHA105 for transformation of DJ rice calli using the procedure as described in Hiei et al. (1994). The mutation of OsGLP1 in transgenic plants regenerated from the transformed rice calli was genotyped by PCR amplification of the target region in DNA extracted from their leaves. The primer sequences used for vector construction and amplification of the target region are listed in Supplemental Table S2. For OsGLP1OE vector construction, the full-length OsGLP1 coding sequence was amplified using PCR with the primers GLP1-O-L and GLP1-O-R (Supplemental Table S2) and inserted into the pOx vector (provided by professor Yao-Guang Liu, College of Life Sciences, South China Agricultural University, China) containing an Ubi promoter. The OsGLP1OE vector construct was introduced into A. tumefaciens strain EHA105 for transformation of DJ rice calli according to Hiei et al. (1994). For OsGLP1-GFP construct, the full-length OsGLP1 coding sequence was inserted into pBI121-GFP for rice protoplast transformation, and OsGLP1-GFP coding sequence was inserted into pOx for constructing transgenic rice plants overexpressing OsGLP1-GFP. Fluorescence signals were observed using WLL laser and HyD detectors of Leica TCS SP8 STED 3X. The excitation/emission filters utilized for fluorescence detection were 488/501–549 nm for GFP, and the gains were 61%. The construct pBI121-GLP1-GFP was then introduced into rice protoplasts and the transfected rice protoplasts were sampled for GFP fluorescence observation using argon laser and PMT detectors of Zeiss LSM 7 DUO (Zhang et al., 2011). The excitation/emission filters utilized for fluorescence detection were 488/493–546 nm for GFP, and 488/658–735 nm for chlorophyll autofluorescence, and the gains were 700 for GFP, 790 for GLP1-GFP, and chlorophyll autofluorescence.
Total RNA extraction and RT-qPCR analysis
Total RNAs were extracted from various tissues of WT plants or OsGLP1 transgenic plants with Trizol and reverse-transcribed following the manufacturer’s instructions (Vazyme Biotech). RT-qPCR analyses were carried out using a PTC200 (BIO-RAD) PCR machine and a SYBR green probe (Bimake, China). The details of the primers used are given in Supplemental Table S3. ACTIN was used as an internal standard and was amplified with qACTIN primers.
Determination of photosynthetic parameters and chlorophyll fluorescence
WT, GLP1OE plants and glp1 mutants were grown to the 6-leaf stage in a growth chamber (600 μmol m−2 s−1 white light, 0 μW cm−2 UV-B, 12 h, 30°C; dark, 12 h, 28°C), and the plants were then placed under natural sunlight or glasshouse conditions for 2 h. The net photosynthetic rate (Pn) and stomatal conductance of the third leaf from top to bottom on each plant were determined using a Li-Cor Li-6800 portable photosynthetic apparatus (LI-COR, USA). The air velocity in the system was set to 500 μmol s−1 and a built-in light source was used with the light intensity set to 800 μmol m−2 s−1. The maximum photochemical efficiency of photosystem II (Fv/Fm) and Y (II) were determined using a DUAL-PAM700 chlorophyll fluorometer after plants were placed under natural sunlight for 8 h or white light without supplementation of UV-B light in a growth room.
SOD activity analysis and immunoblot analysis
WT, GLP1OE plants, and glp1 mutants were grown in Kimura B complete nutrient solution to the 5-leaf stage in a growth chamber (600 μmol m−2 s−1 white light, 0 μW cm−2 UV-B, 12 h, 30°C; dark, 12 h, 28°C), before they were transferred to natural sunlight for 8 h. Proteins for SOD assay were extracted using 50 mmol L−1 PBS buffer (pH 7.8) from leaves of individual transgenic lines and WT. The assay mixture contained 0.3 µmol L–1 riboflavin, 13 mmol L−1 methionine, 63 µmol L−1 nitrotetrazolium blue chloride (NBT) and the enzyme extract in a total volume of 3 mL. The reaction mixture was incubated under 60 μmol m−2 s−1 light for 14 min at 25°C before the absorbance was measured at 560 nm. One unit (U) of SOD activity was defined as the enzyme activity that brought about 50% inhibition of photo-oxidative reduction of NBT, and specific SOD activity was expressed as U/mg protein. In-gel detection of SOD activity was performed according to Gucciardo et al. (2007). The proteins from leaves were separated in 7.5% (m/v) native polyacrylamide gel, and then the gel was immersed in 20 mL of 50 mmol L−1 PBS buffer (pH 7.8) containing 0.25 mg riboflavin, 4 mg NBT and 50 μL TEMED for about 30 min under light (100 μmol m−2 s−1) until visible bands of enzyme activity could be observed. For immunoblotting analysis, proteins were separated by electrophoresis in 13% sodium dodecylsulfate polyacrylamide gel and then electrophoretically transferred to nitrocellulose paper. The OsGLP1 proteins were detected using Escherichia coli-expressed OsGLP1-His antiserum as the primary antibodies (prepared in our laboratory), and the alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin (Sigma, St Louis, USA) as the secondary antibodies. For visualization of the OsGLP1, the nitrocellulose paper was immersed in 100 mmol L−1 Tris–HCl buffer (pH 9.5) containing 100 mmol L−1 NaCl, 5 mmol L−1 MgCl2.6H2O, 0.2 mmol L−1 NBT, and 0.15 mmol L−1 5-bromo-4-chloro-3-indolyl phosphate-toluidine salt.
In situ detection of hydrogen peroxide and superoxide
Hydrogen peroxide (H2O2) accumulation was detected using 3, 3′-diaminobenzidine (DAB) staining according to Thordal-Christensen et al. (1997), and superoxide anion (O2•−) was detected using NBT staining according to May et al. (1996). Leaf blades of rice seedlings at the 5-leaf stage were cut and immersed in 0.5 mg L−1 DAB solution (pH 3.8) and 10 mmol L−1 PBS buffer (pH 7.8) containing 0.5% NBT, respectively, under white light (100 μmol m−2 s−1) until staining could be observed. Chlorophyll was then removed from the leaf blades by immersing them in industrial-grade alcohol while kept in a boiling water bath.
RNA-seq
The WT and glp1 mutants were grown to the 4-leaf stage in a growth chamber (600 μmol m−2 s−1 white light, 12 h, 0 μW cm−2 UV-B, 30°C; dark, 12 h, 28°C), and then transferred into natural sunlight. The RNA used for RNA-seq analysis was extracted using Trizol from the leaves of WT and glp1 mutants placed under natural sunlight for 0 h (8:00, 600 μmol m−2 s−1, 0 μW cm−2 UV-B, 30°C), 4 h (1440 μmol m−2 s−1, 174 μW cm−2 UV-B, 37°C), and 8 h (810 μmol m−2 s−1, 80.2 μW cm−2 UV-B, 35°C). RNA-seq libraries were constructed using the New England Biolab Next Ultra RNA Library Prep Kit for Illumina following manufacturer’s instructions. RNA-Seq libraries were sequenced on an Illumina HiSeq xTen sequencer to generate 150 bp paired-end reads. The resulting clean reads were mapped to O. sativa reference genome (IRGSP_1.0) using TopHat2 with default parameters. Differential expression analysis of glp1 and WT was performed using the DEseq and the differentially expressed genes were identified using fold change ≥2 and FDR <0.01 as significance cutoffs.
Statistical analysis
Except RNA-seq and the construction of transgenic plants, all experiments were performed two times, and the data were statistically analyzed using MS Excel for Windows. The values in the figures are means ± sd, and significant differences among various treatments were analyzed using the Spss 22 analytical software.
Accession numbers
Sequence information of major genes/proteins mentioned in this article can be found in the National Center for Biotechnology Information by the accession numbers in Supplemental dataset 9.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Molecular evaluation of glp1 mutants generated by CRISPR-Cas9 system.
Supplemental Figure S2 . Measurement of cell death in the leaves of glp1 mutants.
Supplemental Figure S3 . Appearance of the lesion mimic on leaves of the glp1 mutants at different development stages.
Supplemental Figure S4 . The effect of growth conditions on the lesion mimic on leaves of glp1 mutants.
Supplemental Figure S5 . Characterization of rice glp1 mutants grown under natural condition and in glasshouse.
Supplemental Figure S6 . Subcellular location of OsGLP1 in rice protoplast.
Supplemental Figure S7 . Alignment of amino acid sequences of OsGLP1 with its homologues.
Supplemental Figure S8 . Profiles of SOD isoforms in rice leaves.
Supplemental Figure S9 . RNA-Seq analyses.
Supplemental Figure S10 . The effect of mutation in OsGLP1 on the transcript levels of MKP1 and MPK6.
Supplemental Figure S11 . Appearance of the lesion mimic on leaves of glp1, mpk3, and glp1 mpk3 mutants.
Supplemental Figure S12 . Phylogenetic analysis of OsGLP1 homologs in rice.
Supplemental Figure S13 . Assay of IAA content using pDR5: GUS staining.
Supplemental Table S1 . Results of RT-qPCR data analyzed using 2-way analysis of variance.
Supplemental Table S2 . Primer sequences used for vector construction and amplification of the target region.
Supplemental Table S3 . Primer sequences used for RT-qPCR.
Supplemental datasets S1-9 . List of genes regulated in glp1 compared to WT.
Supplementary Material
Acknowledgments
We thank Yaoguang Liu (South China Agricultural University) for kindly providing pOX and CRISPR/Cas9 system vectors, Huili Liu and Hai Zhou (South China Agricultural University) for helpful discussions, Qinlong Zhu and Hao Wang (South China Agricultural University) for technical assistance, Menglong Zhuang and Xiaojing Zhang (South China Agricultural University) for GFP fluorescence observation.
Funding
This study was funded by Natural Science Foundation of Guangdong (2020A1515010192) and National Natural Science Foundation of China (31071345).
Conflict of interest statement. No conflict of interest is declared.
Z.H., M.T., and E.L. designed the research. Z.H., M.T., X.Y., C.L., and E.L. performed the experiments, analyzed the data. D.L., X.P., and E.L. wrote the manuscript. Z.H. and M.T. contributed equally.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: E-E. Liu (eeliu70@scau.edu.cn).
References
- Banerjee J, Maiti MK (2010) Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem Biophys Res Commun 394: 178–183 [DOI] [PubMed] [Google Scholar]
- Bernier F, Berna A (2001) Germins and germin-like proteins: Plant do-all proteins. But what do they do exactly? Plant Physiol Biochem 39: 39545–39554 [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MP, Susanti NI, Wargent JJ, Findlay JE, Quick WP, Paul ND, Jenkins GI (2012) The UV-B photoreceptor UVR8 promotes photosynthetic efficiency in Arabidopsis thaliana exposed to elevated levels of UV-B. Photosynth Res 114: 121–131 [DOI] [PubMed] [Google Scholar]
- Carrillo MGC, Goodwin PH, Leach JE, Leung H, Cruz CMV (2009) Phylogenomic relationships of rice oxalate oxidases to the cupin superfamily and their association with disease resistance QTL. Rice 2: 67–79 [Google Scholar]
- Chen H, Xiong L (2005) Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J 44: 396–408 [DOI] [PubMed] [Google Scholar]
- Cheng X, Huang X, Liu S, Tang M, Hu W, Pan S (2014) Characterization of germin-like protein with polyphenol oxidase activity from Satsuma mandarine. Biochem Biophys Res Commun 449: 313–318 [DOI] [PubMed] [Google Scholar]
- Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O’Hara A, Smith BO, Christie JM, Jenkins GI (2012) C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA 109: 16366–16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Pramanik K, Sharma R, Gantait S, Banerjee J (2019) In-silico study of biotic and abiotic stress related transcription factor binding sites in the promoter regions of rice germin-like protein genes. PLoS One 14: e0211887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZK, Li JM, Zhong ZC, Dong M (2014) A proteomic analysis of Arachis hypogaea leaf in responses to enhanced ultraviolet-B radiation. Acta Ecol Sin 34: 2589–2598 [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ et al. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 34: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Peer W, Robert S, Swarup R, Ye S, Prigge M, Cohen JD, Friml J, Murphy A, Tang D et al. (2010) Arabidopsis ROOT UVB SENSITIVE2/WEAK AUXIN RESPONSE1 is required for polar auxin transport. Plant Cell 22: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro MA, Bartels S, Albert A, Ulm R (2011) Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J 68: 727–737 [DOI] [PubMed] [Google Scholar]
- González Besteiro MA, Ulm R (2013) Phosphorylation and stabilization of Arabidopsis MAP kinase phosphatase 1 in response to UV-B stress. J Biol Chem 288: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107: 20132–20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gucciardo S, Wisniewski JP, Brewin NJ, Bornemann S (2007) A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J Exp Bot 58: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Ham B, Li G, Kang B, Zeng F, Lucasa WJ (2012) Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development. Plant Cell 24: 3630–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Velanis CN, Jenkins GI, Franklin KA (2014) UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci USA 111: 11894–11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Ulm R (2013) Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA 110: 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidema J, Taguchi T, Ono T, Teranishi M, Yamamoto K, Kumagai T (2007) Increase in CPD photolyase activity functions effectively to prevent growth inhibition caused by UVB radiation. Plant J 1: 70–79 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Jansen MAK (2002) Ultraviolet-B radiation effects on plants: induction of morphogenic responses. Physiol Plant 116: 423–429 [Google Scholar]
- Jenkins GI (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 30: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Teranishi M, Yamaguchi H, Matsushita T, Watahiki MK, Tsuge T, Li S, Hidema J (2015) UV-B-induced CPD photolyase gene expression is regulated by UVR8-dependent and -independent pathways in Arabidopsis. Plant Cell Physiol 56: 2014–2023 [DOI] [PubMed] [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y et al. (2018) UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44: 1–12 [DOI] [PubMed] [Google Scholar]
- Liang T, Yang Y, Liu H (2019) Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol 3: 1247–1252 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Mansilla AY, Segarra CI, Conde RD (2012) Structural and functional features of a wheat germin-like protein that inhibits trypsin. Plant Mol Biol Rep 30: 624–632 [Google Scholar]
- May MJ, Hammond-Kosack KE, Jones JDG (1996) Involvement of reactive oxygen species, glutathione metabolism, and lipid peroxidation in the Cf-gene-dependent defense response of tomato cotyledons induced by race-specific elicitors of Cladosporium fulvum. Plant Physiol 110: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membré N, Bernier F, Staiger D, Berna A (2000) Arabidopsis Thaliana germin-Like proteins: Common and specific features point to a variety of functions. Planta 3: 345–354 [DOI] [PubMed] [Google Scholar]
- Morales LO, Brosché M, Vainonen J,, Jenkins GI, Wargent JJ, Sipari N, Strid Å, Lindfors AV, Tegelberg R, Aphalo PJ (2013) Multiple roles for UV RESISTANCE LOCUS8 in regulating gene expression and metabolite accumulation in Arabidopsis under solar ultraviolet radiation. Plant Physiol 161: 744–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawkar GM, Maibam P, Park JH, Sahi VP, Lee SY, Kang CH (2013) UV-induced cell death in plants. Int J Mol Sci 14: 1608–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A, Tanaka Y, Kadowaki K, Hayashi T (1998) Cloning of genes encoding auxin-binding proteins (ABP19/20) from peach: significant peptide sequence similarity with germin-like proteins. Plant Cell Physiol 39: 492–499 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska MJ, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Han J, Yang P, Mao W, Liu X, Qiu L, Qian C, Liu Y, Chen Z, Ouyang X et al. (2019) Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc Natl Acad Sci USA 116: 4722–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristilä M, Strid H, Eriksson LA, Strid A, Sävenstrand H (2011) The role of the pyridoxine (vitamin B6) biosynthesis enzyme PDX1 in ultraviolet-B radiation responses in plants. Plant Physiol Biochem 49: 284–292 [DOI] [PubMed] [Google Scholar]
- Rodríguez-López M, Baroja-Fernandez E, Zandueta-Criado A, Moreno-Bruna B, Munoz FJ, Akazawa T, Pozueta-Romero J (2001) Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett 490: 44–48 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Nishimura T, Miyaki YI, Watanabe S, Takagi H, Izumi S, Shimada H (2015) In vitro and in vivo evidence for oxalate oxidase activity of a germin-like protein from azalea. Biochem Biophys Res Commun 458: 536–542 [DOI] [PubMed] [Google Scholar]
- Segarra CI, Casalongué CA, Pinedo ML, Ronchi VP, Conde RD (2003) A germin-like protein of wheat leaf apoplast inhibits serine proteases. J Exp Bot 54: 1335–1341 [DOI] [PubMed] [Google Scholar]
- Swart S, Logman TJ, Smit G, Lugtenberg BJ, Kijne JW (1994) Purification and partial characterization of a glycoprotein from pea (Pisum sativum) with receptor activity for rhicadhesin, an attachment protein of Rhizobiaceae. Plant Mol Biol 24: 171–183 [DOI] [PubMed] [Google Scholar]
- Teranishi M, Taguchi T, Ono T, Hidema J (2012) Augmentation of CPD photolyase activity in japonica and indica rice increases their UVB resistance but still leaves the difference in their sensitivities. Photochem Photobiol Sci 11: 812–820 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book 11: e0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi VE, Regalado JJ, Iannicelli J, Laino LE, Burrieza HP, Escandón AS, Pitta-Álvarez SI (2019) Beyond Arabidopsis: differential UV-B response mediated by UVR8 in diverse species. Front Plant Sci 10: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaelewyn L, Prinsen E, Van Der Straeten D, Vandenbussche F (2016) Hormone-controlled UV-B responses in plants. J Exp Bot 67: 4469–4482 [DOI] [PubMed] [Google Scholar]
- Wang J, Ye B, Yin J, Yuan C, Zhou X, Li W, He M, Wang J, Chen W, Qin P et al. (2015) Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol Biochem 97: 44–51 [DOI] [PubMed] [Google Scholar]
- Wang L, Pei Z, Tian Y, He C (2005) OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact 18: 375–384 [DOI] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K (2002) A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci USA 99: 7530–7535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, Shi N, Li X, Zhang P, Liu H (2018) UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4: 98–107 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Chen P, Liang T, Li X, Liu H (2020) UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J 39: e101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Lee YS, Lee DY, Cho MH, Jeon JS, An G (2016) OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. J Exp Bot 67: 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Han X, Xu Z, Xue H (2009) Arabidopsis GLP4 is localized to the Golgi and binds auxin in vitro. Acta Biochim Biophys Sin 41: 478–487 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. International Rice Research Institute, Manila, Philippines. [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Liu P, Li C, Wang Y, Guo L, Jiang G, Zhai W (2017) LMM5.1 and LMM5.4, two eukaryotic translation elongation factor 1A-like gene family members, negatively affect cell death and disease resistance in rice. J Genet Genomics 44: 107–118 [DOI] [PubMed] [Google Scholar]
- Zhao SQ, Xiang JJ, Xue HW (2013) Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant 6: 174–187 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.