Abstract
Fluoride is everywhere in the environment, yet it is toxic to living things. How biological organisms detoxify fluoride has been unknown until recently. Fluoride-specific ion transporters in both prokaryotes (Fluoride channel; Fluc) and fungi (Fluoride Exporter; FEX) efficiently export fluoride to the extracellular environment. FEX homologs have been identified throughout the plant kingdom. Understanding the function of FEX in a multicellular organism will reveal valuable knowledge about reducing toxic effects caused by fluoride. Here, we demonstrate the conserved role of plant FEX (FLUORIDE EXPORTER) in conferring fluoride tolerance. Plant FEX facilitates the efflux of toxic fluoride ions from yeast cells and is required for fluoride tolerance in plants. A CRISPR/Cas9-generated mutation in Arabidopsis thaliana FEX renders the plant vulnerable to low concentrations (100-µM) of fluoride at every stage of development. Pollen is particularly affected, failing to develop even at extremely low levels of fluoride in the growth medium. The action of the FEX membrane transport protein is the major fluoride defense mechanism in plants.
Plants rely on a fluoride transporter to tolerate toxic fluoride ions.
Introduction
Fluorine, the smallest and most electronegative of the halogens, is abundant in the environment as the ion fluoride (F−). Fluoride is naturally released into the biosphere by weathering of fluoride-containing minerals and from volcanoes and marine aerosols (Symonds et al., 1988; Weinstein and Davison, 2004). Natural concentrations in fresh water can vary from <26 to 260-µM (Wang and Cheng, 2001). Concentrations of fluoride in soil also vary widely depending on mineral deposits, acidity, and local environmental factors. Additional fluoride is released into the environment by human activities, including coal burning; aluminum smelting; and the manufacture of ceramics, bricks, and glass (Weinstein and Davison, 2004). One substantial and widespread source of fluoride contamination is phosphate fertilizer. The wet production of phosphate fertilizers from phosphate rock (mostly fluorapatite) releases toxic byproducts HF and SiF4 (Weinstein and Davison, 2004). However, the fertilizers produced still retain a sizeable amount of fluoride, which is then applied to soil, directly increasing fluoride levels (Oelschläger, 1971; McLaughlin et al., 1996; Cronin et al., 2000). A recent survey of fertilizers revealed that seven out of eight tested contained high levels of fluoride (average ∼4.7 mM; Ramteke et al., 2018). The added fluoride stays in the top layer of soil and accumulates in plants (Anbuvel et al., 2014). Thus, the application of phosphate fertilizers increases fluoride stress in crop plants and introduces additional fluoride into food and feed products. Despite the occurrence of bioavailable fluoride compounds in water, air, and soil, we know little about how plants avoid fluoride toxicity.
Plant species vary in fluoride uptake, accumulation, and tolerance. In vascular plants, fluoride, if airborne, can enter through the stomatal pores or, if present in water or soil, can move through the transpiration stream. Fluoride concentrates in leaf tips, which can reach levels many times those of the lower part of the leaf (Weinstein, 1977; Cooke et al., 1976; Weinstein and Davison, 2004). Fluoride sensitive plants typically exhibit leaf damage or leaf burn due to fluoride concentration in leaf tips and margins (Posthumus, 1982; Coulter et al., 1985). In general, higher concentrations of environmental fluoride result in higher fluoride concentrations in the plant. Most plants have some tolerance to fluoride, but the degree of tolerance varies. Sorghum (Sorghum bicolor) is more fluoride tolerant at 100–425 µM than maize (Zea mays) or soybean (Glycine max), despite also accumulating higher levels of fluoride (Fina et al., 2016). Plants able to grow in fluorspar mine waste include species such as white clover (Trifolium repens) and red fescue (Festuca rubra), which were able to thrive at the extremely high levels of soil fluoride that would damage most other plants (Cooke et al., 1976). At the other extreme, many house plants are highly sensitive to fluoride. Even 50µM in tap water can cause foliar damage (Pscheidt, 2015). For comparison, Gladiolus leaves are injured at a concentration of 1 mM while concentrations greater than 63 mM do not affect fluoride-tolerant tea (Camellia sinensis; Jacobson et al., 1966; Ruan and Wong, 2001; Shu et al., 2003). Tea is known to both accumulate and tolerate fluoride, resulting in release of fluoride during steeping of the leaves (Zimmerman et al., 1957; Davison, 1983). Although the level varies, all plants have some ability to avoid toxic fluoride build-up, but the mechanism of tolerance is unknown.
Studies have identified a fluoride-specific channel that is responsible for tolerance of fluoride in bacteria and yeast. Acidic environments favor the formation of HF (pKa = 3.4), which can pass readily through biological membranes. Inside a cell the pH is higher and HF dissociates to H+ and F− ions. F− is no longer membrane permeable and concentrates inside the cell. It is possible that F− could also enter cells via transport proteins that favor Cl− and OH− due to the charge and size similarity. In single-celled organisms, the intracellular concentration of F− is reduced to non-toxic levels by rapid efflux through a fluoride-specific channel called Fluc for Fluoride channel in bacteria and FEX for Fluoride EXporter in yeast (Li et al., 2013; Stockbridge et al., 2013). Fluc is situated in the membrane as an antiparallel dimer that forms two pores. Saccharomyces cerevisiae FEX is encoded in a single polypeptide, also in an antiparallel conformation that results in two pores, though only the second one is active for fluoride transport (Li et al., 2013; Smith et al., 2015; Berbasova et al., 2017). Yeast cells lacking FEX loci are over 1,000-fold more sensitive to fluoride. The FEX knock-out strain has an IC50 of 60µM, compared with an IC50 of 70 mM for wild-type yeast (Li et al., 2013). A plasmid containing a single FEX gene rescues yeast growth to wild-type levels, confirming the role of the fluoride transport protein in preventing toxic intracellular build-up of fluoride ions. Using the yeast knockout, we have shown that heterologous expression of a putative FEX homolog from Arabidopsis thaliana rescues the fluoride growth phenotype in yeast (Berbasova et al., 2017). It is unknown whether plant FEX (FLUORIDE EXPORTER) functions as a channel or a transporter. The identification of FEX homologs in plants has provided insight into one possible mechanism of tolerance, though the role of FEX in plant fluoride tolerance has not been studied (Li et al., 2013; Berbasova et al., 2017; Zhu et al., 2019).
It is possible that FEX in plants is responsible for the majority of fluoride tolerance. Complex multicellular organisms, like plants, might detoxify fluoride in the same way as a single-celled yeast, by moving ions out of the cytosol of all cells and into the extracellular space. However, this just passes the toxin into the apoplast. Alternatively, there might be a coordinated mechanism to sequester or transfer toxic fluoride ions to a less vulnerable organelle or cell type. To elucidate the role FEX plays in this process and whether it is redundant with other functions, both FEX knock out and overexpression lines were generated in Arabidopsis.
Here, we report that FEX homologs are found in all plant genomes examined to date and that all those tested could function as a F− efflux protein. Further, FEX is required for tolerance of fluoride during all stages of plant development. Determination of the role of FEX in plant fluoride tolerance will aid in the understanding of how fluoride found in water, growth substrates, and fertilizers affects plant survival.
Results
Plant genomes contain FEX homologs that rescue yeast lacking FEX
The yeast FEX protein sequence was used to search for FEX homologs in plants. Using the consensus sequence it was possible to identify a FEX homolog in every available plant genome (Phytozome V12.1; Goodstein et al., 2012; Li et al., 2013; Smith et al., 2015). Unlike many plant transporter genes, the putative FEX gene is usually present in only one or two copies. The predicted protein sequences of the plant FEX candidates used in this study were aligned with yeast FEX1 (Supplemental Figure S1). The most strongly conserved regions between yeast and the plant FEX predicted proteins include the nine transmembrane (TM) domains. The N-termini vary in sequence and length, and plant genes encode a longer segment compared with yeast. Yeast, however, encodes longer loops between TM3 and 4 and TM4 and 5. Although amino acid sequence identity is relatively low, ∼20% between yeast and plant, the predicted secondary structures are similar and conserved sequence elements important for function are preserved (Stockbridge et al., 2015; Berbasova et al., 2017). These include the amino acid sequence PxGTxxxN found in TM7 and the sequence LT/STxSTFxxE in TM8, which includes a critical phenylalanine residue involved in fluoride efflux (Supplemental Figure S1; Berbasova et al., 2017).
To test the function of putative plant FEX proteins, cDNAs corresponding to expressed versions of various plant FEX loci were cloned and evaluated for their ability to rescue the growth of double knock-out (DKO) yeast on fluoride-containing media (fex1Δfex2Δ yeast strain SSY3; Smith et al., 2015). Several cloned forms of the A. thaliana (AtFEX) cDNA were isolated and inserted downstream of the glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter in the low copy yeast shuttle vector pRS416GPD (Mumberg et al., 1995). Sequence-verified clones were transformed into DKO yeast to test activity both on solid fluoride-containing media and in liquid growth assays. Previously, the fully spliced A. thaliana cDNA has been shown to rescue the yeast DKO strain to the same extent as native ScFEX1 (Berbasova et al., 2017; Figure 1, A andTable 1). However, we also identified differentially spliced versions in Arabidopsis mRNA samples. RNAseq data obtained by Yu et al. (2015) confirmed the inclusion of intron 2 about half the time in seedling tissue mRNA. However, all the transcripts found associated with ribosomes were spliced, perhaps indicating that splicing has a role in FEX regulation (Yu et al., 2015). We found that the alternatively spliced cDNA that retained intron 2 also rescued the yeast DKO, but to a lesser degree (Table 1). The likely open-reading frame resulting from the retention of intron 2 eliminates the entire N-terminus (PlusIntron2; Supplemental Figure S1, B). Deletion of 17 or 129 AA from the AtFEX N-terminus has confirmed that the plant N-terminal region is unnecessary for FEX function in yeast (Table 1; Del17 and Del129 in Supplemental Figure S1, B). A genomic clone retaining all introns did not rescue the yeast DKO (Table 1).
Figure 1.
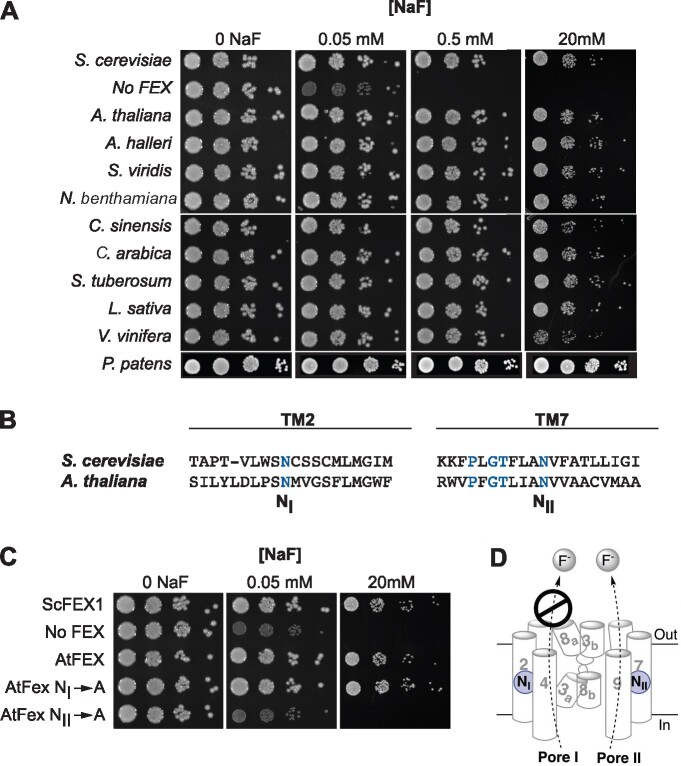
Any plant FEX rescues the yeast FEX DKO. A, DKO yeast were transformed with cDNA sequences of FEX homologs from the indicated plants and 10-fold dilutions of yeast cultures at OD600 of 1 were tested on plates with increasing amounts of NaF. B, Relative positions of the conserved N in yeast and A. thaliana sequences. Blue lettering highlights conserved amino acid residues. C, Growth assay of yeast DKO transformed with N to A substitutions. Rescue of yeast DKO fails only when A is substituted for N in TM7/Pore II. D, Model of FEX in cellular membrane based on bacterial crystal structure and yeast modeling. Cylinders and numbers denote transmembrane domains. A conserved N (blue circle) is positioned in both Pore I and II. In yeast, Pore II is a functional conduit for fluoride ions and Pore I is not.
Table 1.
Quantitative growth tolerance of plant FEX sequences compared with native yeast FEX1 and yeast DKO
| Gene | Organism | IC50 (mM) | Fold rescue |
|---|---|---|---|
| p416GPD ScFEX1 | Saccharomyces cerevisiae | 61 ± 7 | 924 |
| p416GPD (empty vector) | None | 0.066 ± 0.003 | |
| At2g41705.1 fully spliced | Arabidopsis thaliana | 47 ± 1 | 712 |
| At2g41705.1 plus intron 2 | Arabidopsis thaliana | 17 ± 2 | 257 |
| At2g41705.1 N-terminal deletion (−17 AA) | Arabidopsis thaliana | 42 ± 9 | 636 |
| At2g41705.1 N-terminal deletion (−129 AA) | Arabidopsis thaliana | 37 ± 5 | 560 |
| At2g41705.1 N186A | Arabidopsis thaliana | 62 ± 16 | 939 |
| At2g41705.1 N373A | Arabidopsis thaliana | 0.018 ± 0.011 | |
| MT248988 | Arabidopsis halleri | 69 ± 9 | 1,045 |
| Sevir.9G321900.1 | Setaria viridis | 58 ± 5 | 879 |
| Niben101Scf02429 | Nicotiana benthamiana | 56 ± 5 | 848 |
| GEFQ01081211.1 | Camellia sinensis | 25 ± 1 | 379 |
| Scaffold_631.172 | Coffea arabica | 71 ± 3 | 1,076 |
| PGSC0003DMT400031487 | Solanum tuberosum | 65 ± 6 | 985 |
| Lsat_1_v5_gn_3_20261.1 | Lactuca sativa | 74 ± 6 | 1,121 |
| GSVIVT01008915001 | Vitis vinifera | 25 ± 7 | 379 |
| Pp3c27_680V3.1 | Physcomitrium patens | 33 ± 1 | 500 |
Putative full-length FEX cDNA sequences derived from nine crop and model plants were tested for the ability to confer fluoride tolerance in the heterologous yeast system. All nine plant cDNAs rescued the yeast DKO to the same extent as ScFEX1 (Figure 1, A andTable 1). These included plant models Setaria viridis, a C4 monocot, and Arabidopsis halleri, an Arabidopsis species known to hyper-accumulate zinc and cadmium (Briskine et al., 2017). Sequences from crop plants from which different tissues are harvested, including grape (Vitis vinifera, fruit and leaves), potato (Solanum tuberosum, stem), lettuce (Lactuca sativa, leaf), tobacco (Nicotiana benthamiana, leaf), and coffee (Coffea arabica, seed), all restored fluoride tolerance when heterologously expressed in yeast. Tea (Cameilla sinensis) is of particular interest as this plant is a known fluoride accumulator that is also fluoride tolerant. Finally, FEX from the moss Physcomitrium patens, which represents the basal lineage of nonvascular land plants, also complemented the yeast FEX deficiency (Figure 1, A andSupplemental Figure S2).
Site-directed mutagenesis of the yeast FEX protein revealed that the two pores do not function equivalently. In ScFEX, only Pore II is functional for fluoride transport, while Pore I is vestigial and no longer able to expel F− (Berbasova et al., 2017). To test if there is differential activity of two pores in plant FEX, an alanine (A) substitution was made in AtFEX at a conserved asparagine (N) in each pore individually (Figure 1, B). The substitution in Pore I (N186A) had no effect on fluoride sensitivity. The IC50 was 62 ± 16 mM, which is equivalent to wild-type yeast (Table 1). In contrast, the N373A substitution in Pore II resulted in total loss of rescue, resulting in an IC50 of 18 ± 11 µM. This result was also evident in the serial dilution spot growth assay (Figure 1, C). Thus, AtFEX, like its homolog in yeast, has only one functional pore for fluoride efflux. The conservation of Pore II and the lack of conservation in Pore I are probably a common characteristic of eukaryotic FEX (Figure 1, D).
If the yeast DKO survival was due to efflux of F− by plant FEX, the amount of fluoride retained within the rescued yeast cells should be lower than the yeast DKO. To determine if this was the case, DKO yeast with plasmids containing different FEX constructs were grown in 50-µM NaF for 14 h and the intracellular fluoride content measured with a fluoride ion probe. The intracellular fluoride concentration was approximately five-fold lower in the rescued yeast strains (Figure 2). Yeast that were not rescued accumulated equal or more fluoride than that of the surrounding media. Thus, all putative FEX sequences from a wide variety of plant sources rescued the yeast knock-out, suggesting that this gene plays an important role in fluoride efflux in plants.
Figure 2.
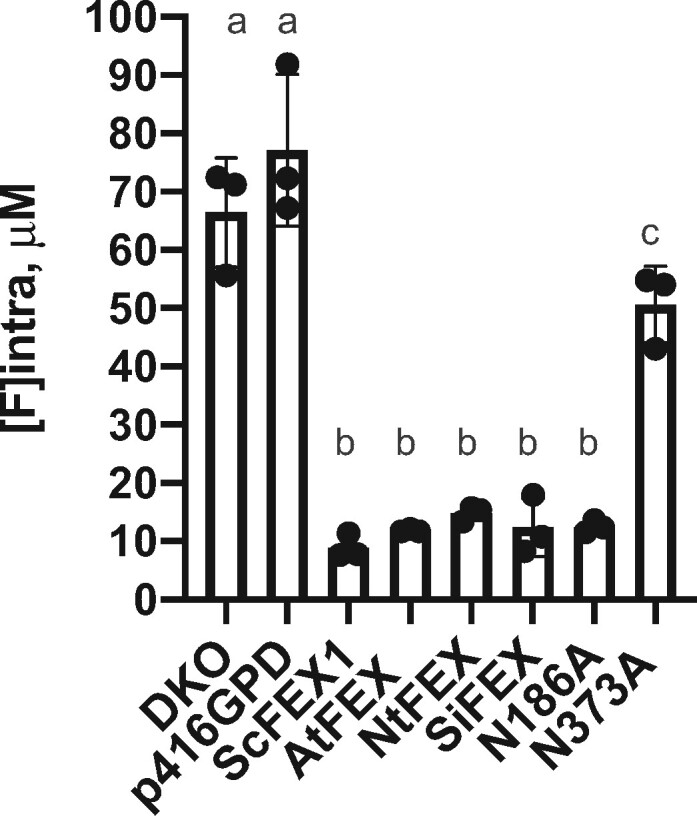
Intracellular fluoride measurements show higher accumulation in yeast cells without a functioning FEX. DKO yeast and DKO with empty vector grown in 50-μM NaF for 14 h accumulated fluoride. DKO transformed with yeast FEX (ScFEX1) or plant FEX (AtFEX, NtFEX, SiFEX) recovered the ability to export fluoride. Substitution of a conserved N in Pore II (N373A), but not Pore I (N186A) impaired fluoride efflux. Points represent biological replicates, bars are the mean, and error bars are SD. One-way ANOVA (Dunnett) results between a and b are P < 0.0001, and P = 0.0428 between a and c.
Plant FEX exports fluoride ions in vitro
Given that plant FEX rescues the yeast DKO and that the intracellular fluoride was also lower in the rescued yeast strains, we predicted that plant FEX would efficiently export fluoride ions. The flux capacities of FEX from yeast (ScFEX1), tea (CsFEX), and Arabidopsis (AtFEX) were determined using an established liposome-based ion flux assay developed to examine the bacterial fluoride channel Fluc (Figure 3, A;Stockbridge et al., 2013). Proteoliposomes with an average of less than one FEX protein molecule were internally loaded with a high concentration (150-mM KF) of F− and added to assay buffer containing low F− (1-mM KF), thereby creating a fluoride ion gradient. The movement of F− was initiated by the addition of the K+-specific ionophore valinomycin, which moves K+ through the lipid bilayer. Fluoride ion flux by FEX was evident by the immediate increase in F− as monitored by a fluoride selective electrode-based probe (Figure 3, B). Once the reading plateaued, the detergent β-OG was added to dissolve proteoliposomes, releasing the F− ions still trapped in liposomes that were devoid of an active membrane protein. Yeast FEX, as well as both plant FEX proteins, displayed flux of F− as demonstrated by the increase in F− efflux following valinomycin addition. We calculated a flux rate of 18,000 and 4,000 F− ions per second per FEX molecule for AtFEX and CsFEX, respectively. This is substantially slower than the bacterial protein Fluc (>30,000 F−/s), but it is within the same range as observed for yeast FEX1 (6,000 F−/s; Figure 3, B) under the same conditions.
Figure 3.
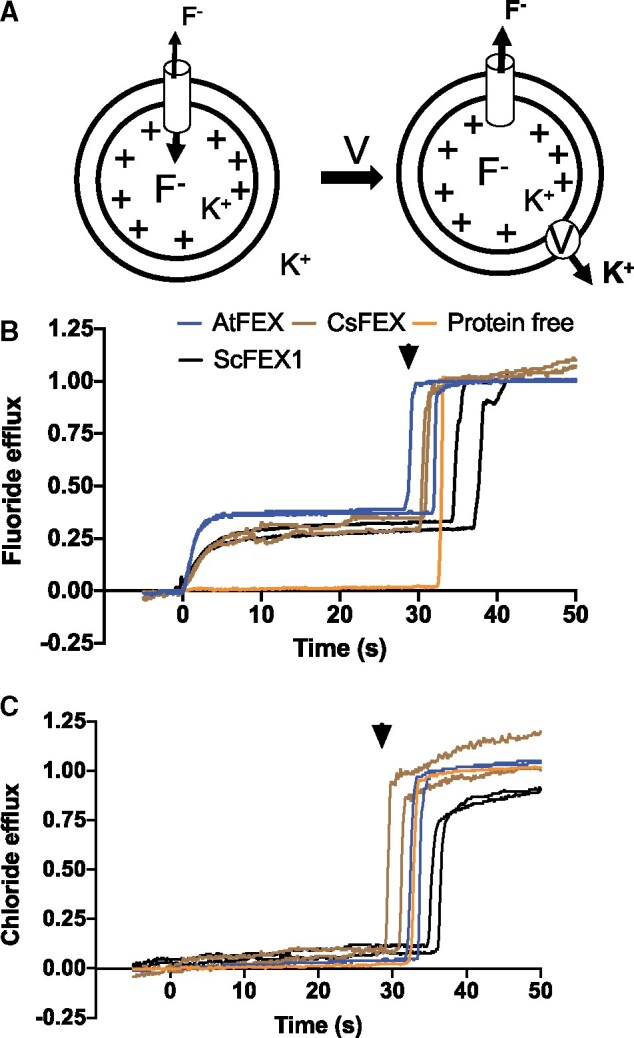
AtFEX, CsFEX, and ScFEX1 efflux F− but not Cl−. A, Model of in vitro assay using proteoliposomes with an inside concentration of 150-mM KF (pH 7) and an outside concentration of 1-mM KF (pH 7). Valinomycin (V) was added at time 0 (arrow), creating a chemical gradient and efflux of ions. B, Relative fluoride efflux by ScFEX1 (black), AtFEX (blue), CsFEX (brown), or no FEX (orange), as detected with a fluoride ion selective electrode-based probe. The Y-axis represents negative voltage versus time, where the negative voltage has been transformed into nanomoles of ions based on a calibration with an ion standard and then compared with how many nanomoles of fluoride ions there were in all liposomes. C, Instead of F−, Cl− ions were loaded into proteoliposomes and efflux was detected with a chloride specific electrode–base probe. Arrowheads indicate the approximate time of β-OG addition to break open proteoliposomes and release trapped ions.
To test the selectivity of the efflux for fluoride over chloride, the ion flux assay was repeated with KCl in place of KF using a Cl− electrode-based probe. After addition of valinomycin, there was zero net movement of Cl− (Figure 3, C). Therefore, yeast and plant FEX fluoride transport proteins, like their bacterial homologs, have a high degree of specificity for F− over Cl−.
The distribution of plant FEX expression is broad especially in young tissues
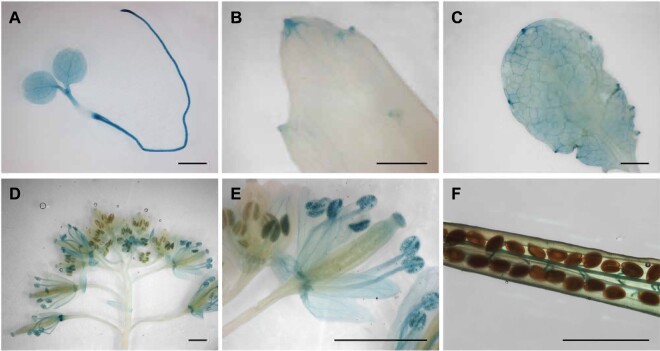
Available RNA expression data indicated that AtFEX is expressed at a low level in most tissues (BAR-Arabidopsis eFP browser; Schmid et al., 2005; Winter et al., 2007). To more precisely analyze expression, glucuronidase (GUS) reporter constructs were made including 1,968 bp of the sequence upstream of the AtFEX start codon, which preceded the start site of the adjacent gene. In transformed plants, GUS staining was somewhat variable in mature leaves between and within different lines, but appeared consistently in young tissues, veins, and mature flowers (Figure 4). Staining was strong throughout very young seedlings and the meristematic region (Figure 4, A). Intriguingly, hydathodes of mature leaves also stained blue, but other mature leaf cells often remained largely unstained (Figure 4, B and C). In mature flowers, sepals, nectaries, and stamen stained blue. Mature, but not immature, pollen also stained blue (Figure 4, D and E). In maturing siliques, funiculi and the abscission zone stained blue (Figure 4, D and F). This suggests that FEX is expressed broadly throughout the plant.
Figure 4.
Promoter GUS expression indicates AtFEX is expressed in young tissues, veins, and hydathodes. Promoter constructs in two different vectors (pKGWFS7, 8 lines and pGWB3, 7 lines) were analyzed with similar results. Pictures are from the pGWB3 lines. A, Widespread staining in seedling. B and C, Staining in older leaves reveals expression in hydathodes and veins. D, Flowers exhibit differential staining depending on the developmental stage. E, Magnification of flowers in (D) showing staining in pollen and sepals in more mature flowers. F, Silique from heterozygous parent with staining in funiculi. Scale bars are 1 mm.
fex/fex mutants do not produce viable seed even at low concentrations of fluoride
The work to date on plant FEX function has been done using rescue experiments in yeast. Experiments to measure the fluoride sensitivity in plants lacking a functional FEX is needed to determine if this gene plays a critical or redundant role in mitigating fluoride toxicity. Unfortunately, there are no DNA insertion lines available within the coding region of AtFEX and insertion lines upstream and downstream of AtFEX did not affect expression. Therefore, to create a plant FEX knock out, we used Crispr/Cas9 to mutate AtFEX and generated a frameshift mutation. The mutation consisted of the insertion of a single T (thymine) in the coding region of exon 2, resulting in a frameshift and a predicted truncated protein of just 60 amino acids (Supplemental Figure S1, B). To identify KO (fex/fex) mutants in a population, a set of derived cleaved amplified polymorphic sequence (dCAPS) primers was designed to introduce a HinfI site in WT (FEX/FEX) genomic sequences only (Supplemental Table S1 and Supplemental Figure S3, A; Neff et al., 2002). Seed from heterozygous plants from all generations reliably segregated 1:2:1 (FEX/FEX: FEX/fex: fex/fex). These plants were used as a source of fex/fex plants for the experiments described in this paper, though it required that each plant be genotyped. At the lowest achievable fluoride level (Fafard Nursery mix, distilled water, no fertilizer, ∼0.6-µM fluoride), seeds from homozygous fex/fex plants were not viable and appeared light in color and empty (Supplemental Figure S3, B).
Different levels of FEX exhibit corresponding differences in fluoride tolerance
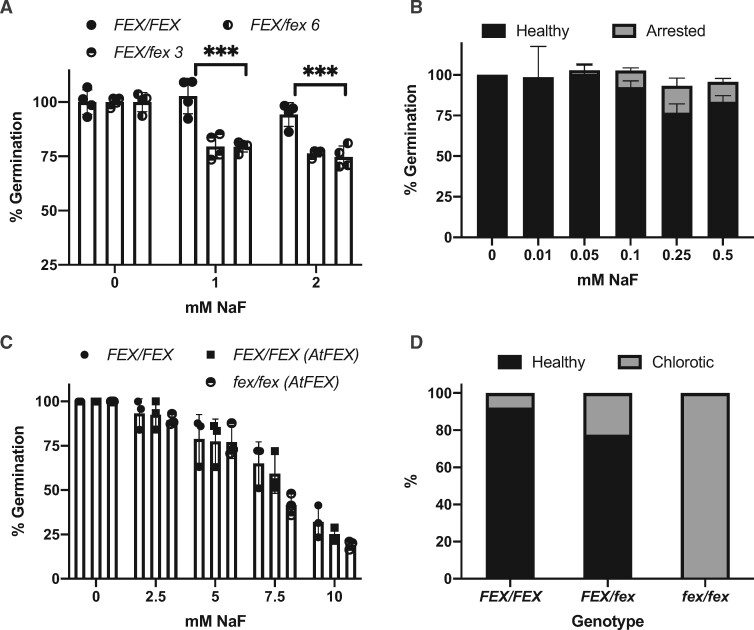
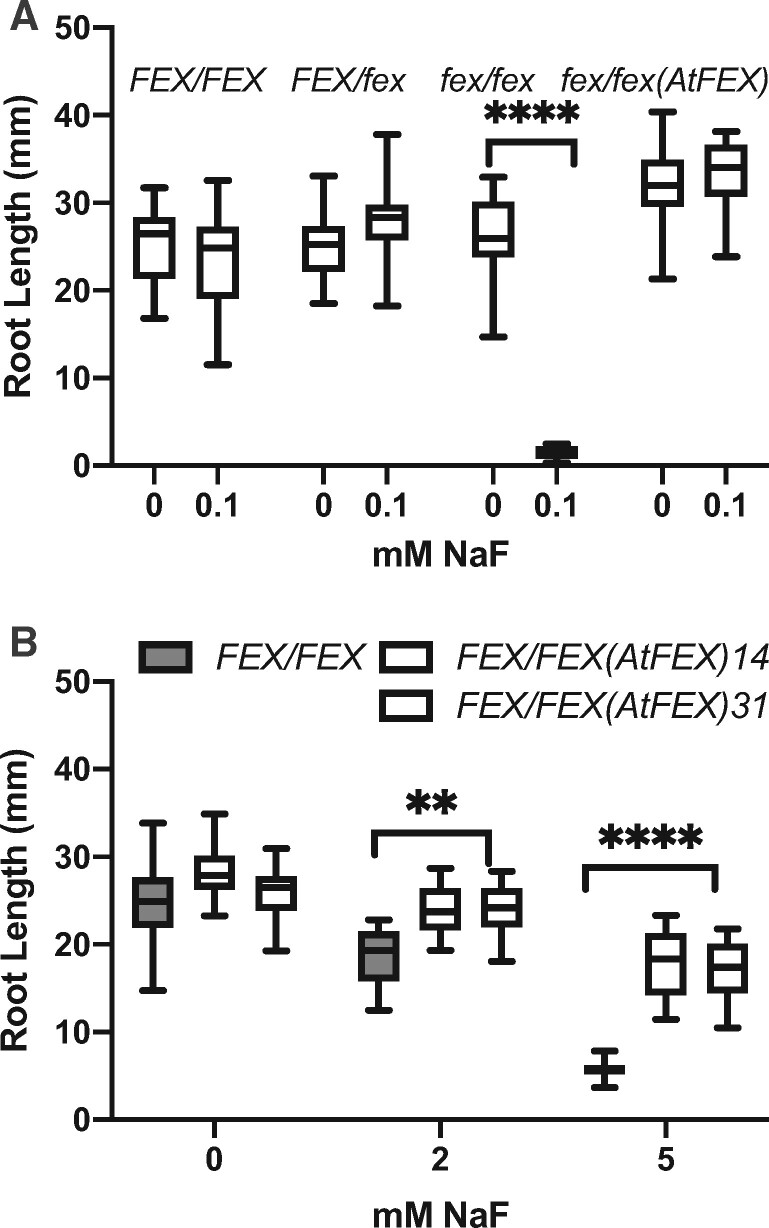
The FEX frameshift enabled testing of homozygotic and heterozygotic mutant plants at various developmental stages. First, fluoride sensitivity of mutants during germination was tested. Seeds from heterozygote mutant plants were germinated on media containing 0-, 1-, or 2-mM NaF (Figure 5, A). In the absence of fluoride, all the seeds germinated and produced green seedlings. However, when fluoride was present, only about 75% of the seeds produced seedlings. We predicted the quarter that failed were homozygous mutants. Genotyping the surviving plants confirmed that only FEX/FEX and FEX/fex seedlings grew in the presence of 1- or 2-mM fluoride. To test the fluoride sensitivity limit of the fex/fex mutants, seeds from heterozygous plants were germinated on media with lower concentrations of fluoride (10–500 µM; Figure 5, B). Overall higher germination rates were observed. Closer examination of the seeds revealed that some had arrested during germination with just the root tip emerging from the seed coat. Genotyping of the arrested seeds revealed that these were homozygous (fex/fex) mutants. The arrested plants did not progress beyond root tip emergence and eventually died if left on fluoride-containing media. Thus, at high concentration (1 mM), the fex/fex seeds failed to germinate and at lower concentration (100–500 µM) they germinated, but they were developmentally arrested. However, if transferred within 5 d to media without fluoride, the fex/fex plants could be rescued and resumed growth. Germination of WT seed was not inhibited until the fluoride concentration reached 4 mM or about 40 times the concentration that inhibited fex/fex seed germination (Figure 5, C).
Figure 5.
Germination and fluoride sensitivity of the AtFEX mutant. A, Germination of seeds from either FEX/FEX (WT) (filled circle) or FEX/fex heterozygous (half-filled circle) plants on 0 NaF or NaF-containing plates. Germination on 0 NaF was set to 100% for each genotype. Each data point represents 40–50 seeds. ***P < 0.001 as determined by one-way ANOVA (Dunnett’s) between WT and heterozygous plants. B, Germination of seeds from a FEX/fex plant on lower concentrations of NaF. Arrested represents seeds where the seed coat was broken and the root emerged, but further growth stalled. Sample size was at least 200 per fluoride treatment. C, Germination of seeds from plants with either WT (closed circle) or OX (closed square) levels of FEX and mutant seeds rescued with AtFEX (half-filled circle). Data points represent replicates of 40–70 seeds each. There was no difference among genotypes at each fluoride concentration as determined by ANOVA (Dunnett’s). D, Germinated seedlings from FEX/fex plants transferred to plates containing 0.25-mM NaF were phenotyped after 10 d and then genotyped. A total of 230 plants were analyzed and the percentage of healthy versus chlorotic for each genotype is represented. In all cases, error bars are SD.
Next, we tested if overexpression of FEX, either in expression levels or by distribution of expression, might enhance germination on media with high fluoride concentrations. WT and heterozygous plants were transformed with AtFEX under the control of the constitutive 35S promoter (p35S:AtFEX). Lines with overexpression (OX) of AtFEX contained about 10× the WT level of FEX as determined by qPCR (Supplemental Figure S4, A). All genotypes with constitutive AtFEX expression germinated as well as WT at 5-mM NaF including the rescued fex/fex mutant, which did not survive at concentrations above 100-µM (Figure 5, C). This establishes that the phenotype in the knockout line was due to loss of FEX function. Germination assays substituting NaCl for NaF at the same concentration had no effect on germination of any genotype. This was expected since this is below the concentration of NaCl known to be inhibitory (>100 mM; Supplemental Figure S4, B; Li et al., 2016), though it establishes that the phenotype of the fex/fex mutant is specific to fluoride. Thus, extreme germination sensitivity due to fluoride was exhibited only by the homozygous mutant and tolerance of fluoride was recovered when FEX was present.
During vegetative growth, fluoride concentrates at the tips and margins of leaves, which causes chlorosis, tip burn, and size reduction. A plant with no ability to detoxify fluoride might be more susceptible to fluoride and exhibit growth phenotypes at lower fluoride concentrations. To test post-germination sensitivity of mutant plants to fluoride, seeds from heterozygous plants were sown on sterile polypropylene filters on media with no fluoride. When the filter with seedlings was transferred to either 0 or 250-µM NaF, some plants on the fluoride plate stopped growing and became chlorotic (Supplemental Figure S3, C). By 10 d after transfer, a subset of plants turned yellow to white, while others remained vigorous and green, identical to those growing on no fluoride. Genotyping revealed that all homozygous mutants were smaller and chlorotic (Figure 5, D). Heterozygous plants were mostly green and healthy, but just under a quarter (22%) were also compromised. This was surprising because heterozygous plants grown to flowering were phenotypically indistinguishable from wild-type.
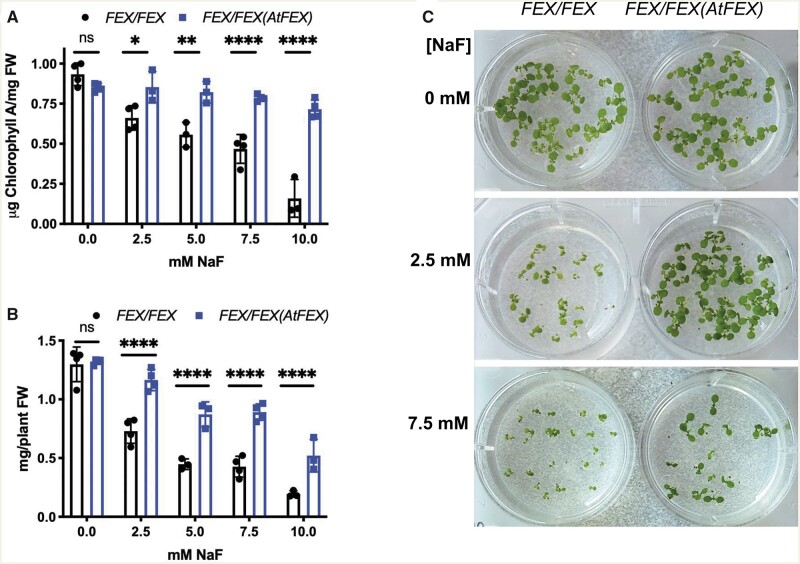
The absence of FEX makes the seedling more susceptible to fluoride, so we tested whether more FEX would make the seedling more tolerant. WT seedlings grown for 10 d on media containing increasing amounts of NaF exhibited a decrease in size and fresh weight and appeared less green, roughly in proportion to the NaF concentration (Figure 6). OX plants grown under the same conditions showed higher tolerance of all the fluoride concentrations tested, starting at 2.5 mM. Mutant plants cannot germinate or grow at this fluoride concentration. Chlorophyll a and fresh weight decreased more quickly with increasing fluoride in the WT plants compared with OX plants (Figure 6). During vegetative growth FEX overexpression protected leaves from fluoride toxicity while lack of FEX increased fluoride sensitivity in leaves.
Figure 6.
FEX overexpression protects from fluoride toxicity. A, Chlorophyll a concentrations from either WT (FEX/FEX) or OX (FEX/FEX (AtFEX)) 10 d plants grown on media with increasing amounts of NaF. Data points represent a replicate of at least 10 seedlings. One-way ANOVA (Tukey) analysis P-value results between the two genotypes are indicated. Error bars are SD. B, Fresh weights of the WT and OX seedlings. Statistical analysis as in A. C, WT and OX seedlings on media with or without NaF.
Leaf damage is the common diagnostic of high fluoride content. Damage to roots, because they are underground, is less documented. We examined root growth to see if different levels of FEX affected elongation in fluoride-containing media. Growth of roots with and without fluoride was tested in plants with WT, fex/fex, or OX levels of FEX expression (Supplemental Figure S3, D). When FEX was absent, roots were more sensitive to fluoride in the media as indicated by the length of the primary root. When grown on 100-µM NaF, the primary roots of the fex/fex plants were almost 20 times shorter than the roots of WT plants grown under the same conditions (Figure 7, A). However, roots of mutants carrying the p35S:AtFEX construct were restored to WT lengths. The overexpressing lines grown on 2- and 5-mM NaF exhibited more robust root growth compared with WT (Figure 7, B). Fluoride tolerance in roots may be due either to higher FEX expression levels or to expression in additional root cell types.
Figure 7.
Root growth in fluoride is impaired in mutants and restored by AtFEX. A, Seeds from FEX/fex plants, mutant plants (fex/fex), and rescued mutant plants (fex/fex (AtFEX)) were plated on no and low fluoride. The root lengths of 7-d seedlings were measured and then genotyped. B, Root lengths of plants overexpressing AtFEX (line 14 data are the first open box followed by line 31) were compared with those of FEX/FEX (WT) plants. In both panels, one-way ANOVA (Tukey: calculated P-values are **** and ** indicating <0.0001 and <0.01, respectively). Both graphs represent one biological replicate of three total replicates with N ≥ 10. Bars on box plots indicate min and max measurements and the horizontal line indicates the mean.
Fluoride accumulates in flowers of FEX mutants and causes male sterility
Important phenotypic differences were observed even at the lowest fluoride concentrations that could be tested in soil substrates. Homozygous mutants grown in soil substrates at very low fluoride (∼2-µM; Fafard Growing mix #2, distilled water, no fertilizer) were initially phenotypically identical to WT and heterozygote plants; however, after the initiation of floral bolting a difference was observed (Figure 8, A). The flowers on heterozygote plants were indistinguishable from WT and produced full seed pods (Figure 8, B). All the flowers of the mutant appeared yellow due to chlorosis of the sepals, did not open, and eventually died (Figure 8, C and D). Some cauline leaves near the flower buds displayed the signature tip death and chlorosis characteristic for fluoride toxicity in plants (Figure 8, E).
Figure 8.
FEX mutant flowers are sensitive to fluoride and cannot produce viable seed. A, Flowering plants grown on regular soilless substrate Fafard Growing mix #2. Heterozygote FEX/fex plants appear WT and homozygote fex/fex plants have shorter flower stalks. B, Heterozygote flowers were phenotypically identical to WT. C, fex/fex flowers at a later time point were dead. D, Flower sepals on fex/fex mutant were chlorotic. E, Chlorosis in fex/fex cauline leaf tip. F, Stage 13 FEX/fex flower grown under low fluoride conditions (∼2-μM) opened to reveal pollen-shedding stamen. G, Stage 13 fex/fex flower opened to reveal stunted stamen with no pollen shed. Scale bars are 1 mm.
Close examination of flowers from the homozygous mutant plants grown on media with the lowest free fluoride content (∼0.6-µM; Fafard Nursery Mix) revealed defects in the stamen and pollen (Figure 8, G) when compared with heterozygous mutant flowers (Figure 8, F). In the fex/fex mutant flowers, filaments did not elongate and the anthers did not release pollen. Both Alexander and KI staining of anthers and pollen revealed that only the pollen grains from the homozygote, but not those from the heterozygote, were inviable (Figure 9). One half of the pollen grains in the stamen of the heterozygote plant should carry only the mutant FEX gene, but all appear unaffected. The level of FEX produced by the heterozygote appears sufficient to protect the anthers from fluoride accumulation and shields the developing pollen grains. Pollen is particularly affected, failing to develop even at extremely low levels of fluoride in the growth medium.
Figure 9.
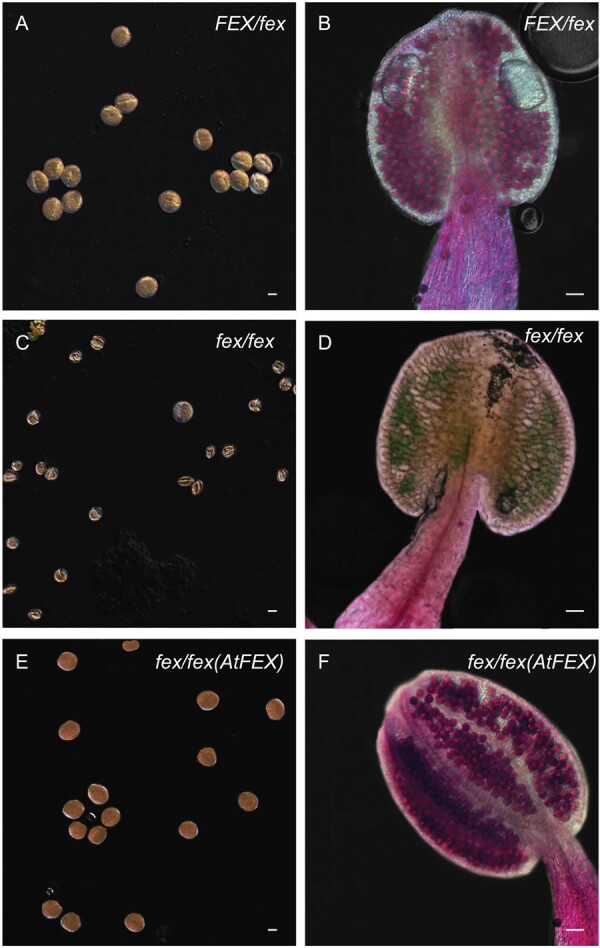
Mutant stamen and pollen show defects from fluoride. A, C, and E show KI-stained pollen from heterozygote, mutant, and rescued plants, respectively. Scale bar is 10-μm. B, D, and F show stamen stained with Alexander’s stain. Viable pollen stains pink and inviable pollen stains blue/green. Scale bar is 50-μm. Each panel (A)–(F) was adjusted to give a uniform black background.
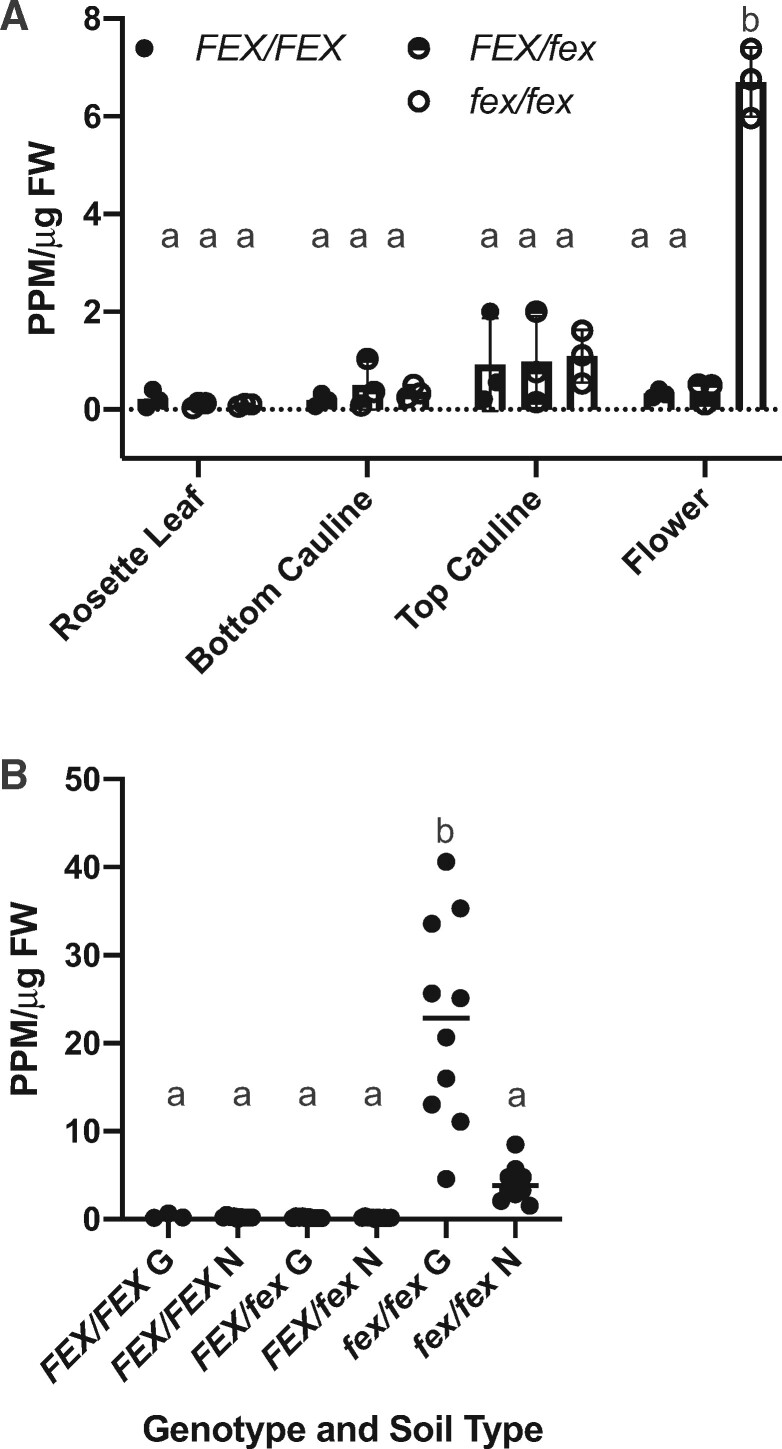
The defective flower phenotype was likely due to the inability of the mutant to reduce the intracellular fluoride content in the cells at the apex of the plant. The fluoride content of leaves and flowers from WT, homozygote, and heterozygote mutant plants grown on regular plant growth media (Fafard Growing mix #2, distilled water, no fertilizer) was determined. Some fluoride (0.1- to 7-ppm/µg FW) was detected in all three genotypes in rosette leaves, cauline leaves, and flowers. In the green tissues, the levels of fluoride were similar. However, it was clear that the flowers from the homozygote mutant retained more fluoride than any other tissue or genotype (Figure 10, A). In general, more distal tissues accumulated more fluoride. The cauline leaves closer to the floral buds had more fluoride than those located lower on the floral stem, which matched with the chlorotic phenotype seen primarily in these leaves (Figure 8, E).
Figure 10.
FEX mutant plants accumulate fluoride in the flowers. A, Samples from plants growing on growth substrate with low fluoride (∼2-μM) exhibited continued fluoride accumulation in the flowers of mutants. The top of the bar indicates the mean and error bars are SD. B, A comparison of flowers of plants on two growth substrates Growing mix #2 (∼2-μM fluoride; G) and Nursery mix (∼0.6-μM fluoride; N) demonstrated accumulation in mutants in proportion to the fluoride in the substrate. The horizontal bar indicates the mean. A and B, Each point represents one biological replicate. Lower case letters above data points indicate statistically significant differences P < 0.0001 as calculated by one-way ANOVA (multiple comparisons, Tukey).
To explore further the relationship between fluoride availability and tissue accumulation, flowers from plants grown on the two different growth substrates Fafard Growing Mix #2 (∼2-µM) and Fafard Nursery Mix (at ∼0.6-µM) were tested for fluoride content. Even the smallest amounts of fluoride present in the growth substrates adversely affected reproduction. Homozygous flowers contained more fluoride than either the heterozygous or WT flowers in either soil type, corresponding to the amount of fluoride available from the growth substrate (Figure 10, B). Clearly, a functional FEX is necessary for fluoride tolerance. Although fluoride is known to accumulate in leaves, especially the tips, the FEX mutant demonstrates the toxic effect of accumulation in the reproductive organs at the apex of the plant. Even the smallest amounts of fluoride in the growth media were sufficient to cause male sterility in the absence of a functional FEX gene.
Discussion
We have shown that FEX is necessary and sufficient for fluoride tolerance in Arabidopsis. Without an active FEX, Arabidopsis plants exhibit elevated sensitivity to concentrations of fluoride equivalent to that found in municipal tap water (∼40-µM) and a concentration commonly found in naturally occurring water sources for irrigation. Mutant FEX plants demonstrated fluoride sensitivity at all stages of development from germination to flowering, which indicates that the fluoride transport protein is the primary defense against toxic fluoride build-up. Our analysis of available genomic or transcriptomic data indicates that all plants have a FEX homolog. The ability of FEX genes from numerous plant sources to rescue the yeast DKO by heterologous expression established some important observations. First, plant FEX permits the efflux of F− out of the yeast cell. Second, sufficient amounts of the plant FEX protein have been properly inserted and are functional within the yeast cell membrane. Third, expression provides levels of fluoride tolerance (IC50 levels) comparable to those of native yeast FEX. Fourth, no plant-specific factors are necessary for function of any of these proteins within yeast, or the factor is able to functionally interact with a wide variety of different proteins. Fifth, FEX function is conserved from single-celled yeast to an early multicellular plant to more complex plants. In summary, plant FEX is a confirmed candidate for fluoride detoxification in plants and is an example of evolutionary conservation of function. This work established that the fluoride transport protein is an essential component of fluoride tolerance.
Without a functioning FEX, every aspect of plant growth was severely impaired or eliminated in the presence of fluoride concentrations well below those found naturally in the environment. At the lowest fluoride concentration achievable in soil substrates (<1-µM), FEX mutants accumulated fluoride to lethal levels in the flower buds. Although the diagnostic phenotype of plant fluoride toxicity is leaf chlorosis and necrosis, we found the flower, and pollen in particular, were most vulnerable in the Arabidopsis AtFEX mutant. This is consistent with studies done in corn, which showed that fluoride was concentrated in the tassels (Kornberger, 1978), and in wheat and sorghum, in which anthesis was found to be the most fluoride-sensitive stage of development (MacLean et al., 1984). Fluoride travels with the transpiration stream to the most distal sites of the plant, which are leaf tips in the vegetative stage, but flowers in the reproductive stage. Leaves could be replaced during growth and, thus, may be somewhat dispensable, but flowers are essential to the survival of future generations of the plant. The lack of elongation of the stamen and the defective appearance of the pollen grains in the AtFEX mutant indicates the adverse effect of fluoride during pollen maturation. Stamen filaments are among the few plant organs that lack stomata and the anther has only a few, reducing the possibility of eliminating fluoride by evapotranspiration. Although available transcriptomic data indicated Arabidopsis pollen did not express AtFEX, our GUS expression data suggest that older pollen grains do express AtFEX and that there is developmental control of FEX expression. There are also data in tea that indicate pollen expresses FEX (Zhu et al., 2019). The more fluoride the plant accumulates, the more flowers and flower organs are affected. At the lowest possible fluoride level the stamen were affected, but as levels increased, sepals became chlorotic and petals shriveled.
A number of other mutations have caused a similar floral phenotype to that described for the fex/fex mutant. For example, VACUOLAR PHOSPHATE TRANSPORTER1 (VPT1) and VPT3 localize to the vacuolar membrane and influx Pi for storage (Luan et al., 2019). A vpt1vpt3 mutant, unable to shunt Pi into the vacuole, accumulates excess Pi in floral organs, causing infertility and notably affecting the pollen. Similarly, a DKO of two K+/H+ antiporters, Na+/H+ ANTIPORTER1 (NHX1) and NHX2, located in the vacuolar membrane had a similar floral phenotype consisting of short filaments and stamen that could not dehisce (Bassil et al., 2011). In this case, infertility was due to the inability to store K+ in the vacuole resulting in a disruption of K+ homeostasis and hydration control (Bassil et al., 2011). In each case, the inability to sequester ions in the vacuole caused the ions to accumulate in the flower. Although it is clear for the plant FEX rescue in yeast that FEX functions in the yeast cell membrane, the membrane location in the plant cell is currently unknown. It may be that FEX can function in the cell membrane and/or the tonoplast. There is evidence that tea plants, which are fluoride-tolerant, accumulate fluoride in vacuoles (Gao et al., 2014). If fluoride ions are sequestered by FEX in root vacuoles, there would be fewer in the transpiration stream. One interesting dilemma is the lower pH in the vacuole allows for the formation of membrane permeable HF, which would be counterproductive for storage. Fluoride ions in the vacuole might be chelated, perhaps by reaction with cations like Ca2+, to halt the return to the cytoplasm.
Changing the levels of FEX expression had different effects on the phenotype exhibited in response to fluoride. The FEX/fex heterozygote of Arabidopsis appeared to be as tolerant to fluoride as the wild type, indicating that FEX is haplosufficient under most conditions. However, the overexpression of AtFEX increased fluoride tolerance. FEX overexpression in roots, either through higher transcription or through expression in additional cell types, could aid in keeping fluoride out of the transpiration stream when fluoride is present in soil and water. Additional FEX in the vegetative tissues might have the opposite beneficial function of keeping fluoride out of the cells and in the transpiration stream. Under our conditions, FEX overexpression did not affect seed germination in fluoride-containing media. This result was unexpected because it is known that phytases are inhibited by fluoride and are required for release of phosphate stores during germination (Loewus and Murthy, 2000). When a different CsFEX construct was overexpressed in A. thaliana under control of the 35S promoter, Zhu et al. (2019) reported an increase in germination rate on fluoride in some lines. Perhaps this reflects an optimal concentration or localization of FEX within the plant.
Little is known about how a plant detoxifies and/or eliminates fluoride (Weinstein and Davison, 2004). Fluoride accumulates at the margins and the tips of leaves where water loss also occurs. Greater than 90% of the water taken up by roots is transpired through the stomata. Fluoride ions and HF are water soluble and could be released along with the water vapor, although this has yet to be shown. Alternatively, hydathodes at the xylem ends at the leaf margins cause water flux when transpiration is curtailed and have been shown to exude dissolved substances like the metals Ni2+, Cu2+, Zn2+, and ions like Cl− (Neumann et al., 1997; Mizuno et al., 2002; Nagai et al., 2013). Transcripts of a boron transporter (Bot1) identified in a boron-tolerant strain of barley were found to accumulate in leaf tips and were proposed to cause removal of excess boron through guttation (Sutton et al., 2007). Our finding of increased transcription of AtFEX in the hydathodes may also indicate a mechanism for fluoride elimination.
Fluoride toxicity in crop plants is an important issue not only in areas of high fluoride in water and soil, but also for plants grown with high phosphate fertilizer. Phosphate is crucial for crop growth, yet common phosphate resources are limited and are often contaminated with fluoride. Fluoride from fertilizers builds up in soil and is subsequently taken up by plants (Anbuvel et al., 2014). Understanding how plants tolerate fluoride toxicity and where fluoride is accumulated in the whole plant is crucial to future food production. It may be possible to manipulate FEX expression to enhance fluoride tolerance in crop plants, resulting in higher yields especially in areas of high fluoride. Now we can test directly if FEX expression is coordinated with fluoride tolerance or if sequence differences result in distinct tolerance levels. Studying FEX expression in a cell-specific manner might reveal which plant cells are particularly vulnerable to or most efficient in reducing fluoride toxicity. Manipulation of FEX expression could also direct fluoride accumulation to plant tissues that are neither crucial to plant survival nor harvested for consumption.
Materials and methods
Plant FEX cloning and yeast rescue
Plant homologs to S. cerevisiae FEX were identified in Phytozome (V12.1). Putative FEX cDNAs from A. thaliana, A. halleri var. Langelsheim (TAIR stock CS9852; Ernst, 1974), S. viridis—foxtail millet, C. sinensis—tea, and N. benthamiana—tobacco (Notredame et al., 2000; Möller et al., 2001). Note that the tea sequence is the same as published by Zhu et al. (2019) except that we predicted an extra 17 amino acids at the N terminus. No A. halleri FEX homolog was predicted (Phytozome V12.1), but we noted a gap in the syntenic region to A. thaliana between two contigs. Primers designed to replicate genomic DNA within the gap resulted in the genomic sequence for AhFEX. AtFEX and AhFEX are 94% identical at the amino acid level. RNA was isolated using TRIzol (Thermo Fisher Scientific) and converted to cDNA using the RETROscript kit (Thermo Fisher Scientific) according to the manufacturer’s directions. The resulting cDNA was cloned into TA vector 2.1 (Thermo Fisher Scientific), sequence-verified, and re-amplified using Phusion (NEB) with primers that included 45 bp of homology to the pRS416GPD vector (Mumberg et al., 1995; Supplemental Table S1). Sequences of cDNAs for L. sativa—lettuce, C. arabica—coffee, V. vinifera—grape, S. tuberosum—potato, and P. patens—moss were synthesized (GeneArt, Thermo Fisher Scientific) and amplified as above. In all cases, the PCR-amplified cDNAs and the HindIII-digested pRS416GPD vector were transformed into the yeast DKO (fex1Δfex2Δ yeast strain SSY3; Li et al., 2013) using the standard lithium acetate procedure (Gietz et al., 1995). Successfully transformed yeast were selected on SD-ura media, the DNA isolated and used to transform DH10B Escherichia coli cells for re-isolation and sequence verification. Yeast DKO rescue was determined with both spot and growth assays performed as described (Li et al., 2013; Berbasova et al., 2017). A positive control consisting of pRS416-FEX1-Sc and a negative control of empty pRS416GPD were used in both assays.
Measurement of intracellular fluoride
A 1–10 dilution of overnight yeast cultures in YPD were grown at 30°C for 2 h at which point NaF was added to 50-μM. Cells were incubated with shaking for another 14 h. Cells were collected by centrifugation at 6,000g for 30 s and washed twice with sterile water. After resuspension in 0.3 mL of 10-mM Tris pH 6.8 with 2% Triton X100 (Sigma) and 1% SDS (American Bioanalytical) and the addition of ∼200-μL of glass beads, cells were vortexed 15 min at 4°C, sonicated in a water bath (Branson ultrasonic cleaner) for 15 min followed by another round of vortexing. Measurement of fluoride was as described below. Intracellular fluoride concentration was calculated based on cell count and the corresponding dilution factor.
Protein isolation and anion efflux assay
ScFEX, AtFEX, and CsFEX were cloned into yeast vector pEGH by homologous recombination in yeast DKO cells. After sequence verification of the GST–FEX fusion, yeast cells containing the constructs were also tested for growth in fluoride to confirm functionality. For protein isolation, cell pellets were resuspended in lysis buffer (50-mM Na2HPO4/NaH2PO4, 150-mM NaCl, 1 protease inhibitor tablet, 1-mM EDTA, 1-mM PMSF, pH 7.0), lysed via eight passes through a microfluidizer set to 30,000 PSI on ice, and clarified by centrifugation at 30,000 g for 10 min at 4°C. Supernatant was then re-centrifuged at 250,000 g, the membrane pellet resuspended in 50 mL Buffer A (50-mM Na2HPO4/NaH2PO4, 20-mM NaCl, 1-mM EDTA, 0.2% DM, pH 7.0) with a tissue grinder, and filtered through a 0.45-μm PES membrane (Millipore) on ice. Resuspended sample was combined with glutathione Sepharose 4B (GE) resin equilibrated with Buffer A resin and rocked 1 h at 4°C. Sample was passed through the resin using and washed with Buffer A. Buffer B (20-mM Tris, 150-mM NaCl, 40-mM glutathione reduced, and 0.2% DM, pH 8.0) was incubated with standing resin for 1 h at room temperature to elute protein.
Proteoliposomes containing 0.5-µg protein per mg of E. coli polar lipids were created as described in Stockbridge et al. (2013). The assay buffer consisted of 150-mM potassium gluconate, 25-mM HEPES, and 1-mM KF at pH 7.0. The buffer inside the liposomes consisted of 25-mM HEPES and 150-mM KF, pH 7.0 which resulted in a fluoride gradient. Anion movement was continuously monitored with either a F− or Cl− ion selective electrode-based probe (Cole-Parmer) attached to an Orion 710A pH meter outputting voltage measurements to a DI-245 datalogger (MicroDAQ). Valinomycin was added to initiate flux and n-octyl-β-D-glucopyranoside (β-OG) was added to destroy liposomes and release intraliposomal ions.
Plant materials and growth
The Columbia ecotype of A. thaliana (A. thaliana var. Col-0) was used for all A. thaliana experiments. Plants were grown on either Growing Mix #2 or Nursery Mix (Fafard Inc., Apopka, Fla.), which were sometimes amended with garden lime. Plants were grown in chambers (16 h light/8 h dark, 22 ± 3°C, 60% RH, 150 μmol m−2 s−1) or on a light cart (16 h light/8 h dark, 22 ± 3°C, ∼15% RH, 50 μmol m−2 s−1). For solid media growth, A. thaliana seeds were sterilized with 30% bleach and 0.05% Tween20, followed by incubation in the dark for 3 d at 4°C before being plated on 0.5× MS (PhytoTechnology Laboratories) pH 5.2 plus 1% sucrose with varying amounts of NaF (Sigma).
Promoter GUS constructs and activity localization
The entire region upstream of the AtFEX start ATG to the start of the adjacent gene (1,968 bp) including the 5′-UTR and first intron was cloned by PCR into pENTR/D-Topo (Thermo Fisher Scientific; Supplemental Table S1). After sequence confirmation, GUS reporter binary plasmids were generated using Gateway LR Clonase II (Thermo Fisher Scientific) with the vectors pKGWFS7 (eight lines examined: Karimi et al., 2002) and pGWB3 (seven lines examined; Nakagawa et al., 2009). GUS staining was performed as in Carland and Nelson (2009) except 3-mM K3/K4(Fe(CN)6) was used and chloramphenicol was not. Staining was routinely done overnight at 37°C after 10 min of vacuum infiltration. Photography was performed using an AxioCam HRc attached to a Zeiss Stemi 2000‐C dissecting microscope or Nikon Ti-E inverted microscope with NIS Elements software.
Generation and identification of Crispr/Cas9 mutant Arabidopsis line
No available T-DNA insertions disrupted AtFEX transcription so a mutation was generated using CRISPR/Cas9 as described in LeBlanc et al. (2018) using the vector pYAO:hSpCas9 (Yan et al., 2015). A frameshift mutation in the second exon was generated and analyzed in depth. The mutant sequence was distinguished from the wild-type sequence using dCAPSs (Neff et al., 1998, 2002). A forward PCR primer was designed to introduce a HinfI site by replacing a T with an A so WT would be cut with HinfI, but mutant DNA would not (Supplemental Table S1 and Supplemental Figure S2, A).
Generation and identification of transgenic Arabidopsis lines
An OX line in A. thaliana was made by transforming WT Arabidopsis plants with a clone containing the AtFEX cDNA with an additional 46 bp immediately preceding the ATG start codon under the control of the 35S promoter in pEarleyGate100 (p35S:AtFEX; Earley et al., 2006). For rescue of the FEX mutant, FEX/fex plants were transformed with the same construct. The resulting T2 and T3 plants were genotyped with a different dCAPS primer complimentary to the intron immediately following the T insertion site, allowing determination of the parental genotype (Supplemental Table S1). All plant transformations were performed using agrobacterium GV3101 with selection on appropriate antibiotics according to established methods (Clough and Bent, 1998; Davis et al., 2009).
Quantitative reverse transcription polymerase chain reaction
RNA was isolated using TRIzol followed by treatment with TURBO DNAse I according to the manufacturer’s instructions (Thermo Fisher Scientific). The Luna Universal One-Step RT-qPCR Kit (New England Biolabs) was used, reactions run in a CFX384 Detection System (Bio-Rad) and transcript levels normalized to At4g33380 (adenosine tRNA methylthiotransferase) and At4g34270 (TIP41-like; Czechowski et al., 2005). Quantification was calculated following the ΔΔCt method (Livak and Schmittgen, 2001).
Germination, seedling, root fluoride tolerance assays
All growth assays were repeated at least three times. Germinability was determined both by root emergence and cotyledon appearance and percentages calculated by comparing to 0 NaF plates. The total number of seeds tested was about 100 per trial. Whole plate transfer of seedlings utilized sterilized polypropylene mesh (all-purpose fabric- point-bonded polypropylene, Gardeners Supply) on top of solid media. For root assays, seeds were vernalized in water for 3 d before plating on 0.5× MS plus 1% sucrose media either with 0 or a defined concentration of NaF and grown vertically. After 7 or 10 d, root length was measured using a ChemiDoc MP (Bio-Rad) and the line tool in FIJI (Schindelin et al., 2012), and each plant was genotyped. P-values were determined using one-way ANOVA with multiple comparisons and either Tukey or Dunnett test in GraphPad Prism 9. P < 0.0001 is ****, P =0.0001–0.001 is ***, P =0.001–0.01 is **, P =0.01–0.05 is *, and ns is not significant. In some graphs, small letters were used to denote significant differences among samples.
Fluoride measurements
The direct method of measuring fluoride with a fluoride ion selective electrode-based probe (Cole-Parmer) attached to an Orion 710A pH meter was used. Fresh tissue was weighed, placed in 500-µL sterile ultrapure water, and ground in an Eppendorf tube with a plastic pestle until the tissue was totally macerated (broken cells >90% by visual inspection). An equal volume of total ionic strength adjustment buffer (TISABII: NaCl (1M), glacial acetic acid (1 M), CDTA (100 mM), NaOH to adjust to pH 5.3) was added to standardize pH and ionic strength (Agarwal et al., 2002). Standard fluoride solutions of NaF were used for calibration. We compared our results using this stream-lined approach to drying and digesting with nitric acid in a high-pressure digestion bomb for approximately 18 h at 105°C. In each trial, the fluoride determinations were on the same order of magnitude. For example, a sample was split in half. One half was measured after maceration in water and contained 34-μg free fluoride. The second half was dried, digested, and found to contain 45-μg of total fluoride.
Fluoride analysis of growth substrates was done by drying overnight at 80°C, putting 1 g in 5 mL of sterile ultrapure water, incubating for at least 1 h with mixing, and then measuring 500 µL of the liquid with TISABII.
Chlorophyll content
Tissue from 10-d seedlings was incubated in 100% DMSO at 65°C, which efficiently and stably extracted the chlorophyll (Hiscox and Israelstam, 1979; Barnes et al., 1992). Readings at ODs 647, 663, and 470 were taken in a plate reader (Biotek Synergy 4) and adjusted for path length as described in Warren (2008). The equations of Barnes et al. (1992) were used for concentration calculation.
Pollen staining
Slides prepared with Alexander’s stain (Peterson et al., 2010) were laid on a 70°C heat block, covered with foil, and incubated for 10 min instead of heating using a flame. Images were captured on a Nikon Ti-E inverted microscope and processed with Adobe Photoshop.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers found in Table 1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Predicted protein alignment of putative plant and yeast FEX1 sequences used in this study.
Supplemental Figure S2. An example of the serial dilution spot growth assay with plates containing different concentrations of NaF for DKO yeast rescued with either P. patens FEX or S. cerevisiae FEX1.
Supplemental Figure S3. Examples of genotypic and phenotypic data.
Supplemental Figure S4. Overexpression level of AtFEX and germination on NaCl.
Supplemental Table S1. Primer sequences.
Supplementary Material
Acknowledgments
The authors thank Padma Mamillipalli and Sunitha Nallur for excellent technical assistance, Steven Ertle and Adam Saffer for timely advice, and the ongoing support of and discussions with members of the Strobel Laboratory.
Funding
This work was supported by the National Science Foundation [grant no. GR108639].
Conflict of interest statement. There are no conflicts of interest.
S.L.T. conceived the research plans, designed and performed the experiments, and analyzed the data. S.S. supervised the experiments. T.B. helped perform and document the yeast rescue experiments. M.P. performed the in vitro experiments. S.L.T. wrote the article with contributions of all the authors. S.A.S. supervised, completed the writing, and agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Scott A. Strobel (scott.strobel@yale.edu).
References
- Agarwal M, Rai K, Shrivastav R, Dass S (2002) A study on fluoride sorption by montmorillonite and kaolinite. Water Air Soil Pollut 141: 247–261 [Google Scholar]
- Anbuvel D, Kumaresan S, Margret RJ (2014) Fluoride analysis of soil in cultivated areas of Thovalai channel in Kanyakumari district, Tamil Nadu, India: correlation with physico-chemical parameters. Int J Basic Appl Chem Sci 4: 20–29 [Google Scholar]
- Barnes JD, Balaguer L, Manrique E, Elvira S, Davison AW (1992) A reappraisal of the use of DMSO for the extraction and determination of chlorophylls a and b in lichens and higher plants. Environ Exp Bot 32: 85–100 [Google Scholar]
- Bassil E, Tajima H, Liang Y-C, Ohto M, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E (2011) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23: 3482–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbasova T, Nallur S, Sells T, Smith KD, Gordon PB, Tausta SL, Strobel SA (2017) Fluoride export (FEX) proteins from fungi, plants and animals are “single barreled” channels containing one functional and one vestigial ion pore. PLoS ONE 12: e0177096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briskine RV, Paape T, Shimizu‐Inatsugi R, Nishiyama T, Akama S, Sese J, Shimizu KK (2017) Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol Ecol Resour 17: 1025. [DOI] [PubMed] [Google Scholar]
- Carland F, Nelson T (2009) CVP2- and CVL1-mediated phosphoinositide signaling as a regulator of the ARF GAP SFC/VAN3 in establishment of foliar vein patterns. Plant J 59: 895–907 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooke JA, Johnson MS, Davison A, Bradshaw A (1976) Fluoride in plants colonising fluorspar mine waste in the Peak District and Weardale. Environ Pollut 11: 9–23 [Google Scholar]
- Coulter CT, Pack MR, Sulzbach CW (1985) An evaluation of the dose–response relationship of fluoride injury to Gladiolus. Atmos Environ 19: 1001–1007 [Google Scholar]
- Cronin SJ, Manoharan V, Hedley MJ, Loganathan P (2000) Fluoride: a review of its fate, bioavailability, and risks of fluorosis in grazed‐pasture systems in New Zealand. NZ J Agric Res 43: 295 [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AM, Hall A, Millar AJ, Darrah C, Davis SJ (2009) Protocol: streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AW (1983) Uptake, translocation and accumulation of soil and airborne fluorides by vegetation. InShupe JL, Peterson HB, Leone NC, eds, Fluorides: Effects on Vegetation, Animals and Humans. Paragon Press, Salt Lake City, UT, pp 62–82 [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fina BL, Lupo M, Dri N, Lombarte M, Rigalli A (2016) Comparison of fluoride effects on germination and growth of Zea mays, Glycine max and Sorghum vulgare. J Sci Food Agric 96: 3679–3687 [DOI] [PubMed] [Google Scholar]
- Gao H, Zhao Q, Zhang X, Wan X, Mao J (2014) Localization of fluoride and aluminum in subcellular fractions of tea leaves and roots. J Agric Food Chem 62: 2313–2319 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57: 1332–1334 [Google Scholar]
- Jacobson JS, Weinstein LH, Mccune DC, Hitchcock AE (1966) The accumulation of fluorine by plants. J Air Pollut Control Assoc 16: 412–417 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kornberger W, Halbwachs G, Richter H (1978) Distribution of fluoride in Zea mays grown near an aluminum plant. Fluoride 12: 129–135 [Google Scholar]
- LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y (2018) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93: 377–386 [DOI] [PubMed] [Google Scholar]
- Li S, Smith KD, Davis JH, Gordon PB, Breaker RR, Strobel SA (2013) Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc Natl Acad Sci U S A 110: 19018–19023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pan Y, Chang B, Wang Y, Tang Z (2016) NO promotes seed germination and seedling growth under high salt may depend on EIN3 protein in Arabidopsis. Front Plant Sci 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN (2000) myo-Inositol metabolism in plants. Plant Sci 150: 1–19 [Google Scholar]
- Luan M, Zhao F, Han X, Sun G, Yang Y, Liu J, Shi J, Fu A, Lan W, Luan S (2019) Vacuolar phosphate transporters contribute to systemic phosphate homeostasis vital for reproductive development in Arabidopsis. Plant Physiol 179: 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean DC, McCune DC, Schneider RE (1984) Growth and yield of wheat and sorghum after sequential exposures to hydrogen fluoride. Environ Pollut Ser A 36: 351–365 [Google Scholar]
- Mclaughlin MJ, Tiller KG, Naidu R, Stevens DP (1996) Review: the behaviour and environmental impact of contaminants in fertilizers. Soil Res 34: 1–54 [Google Scholar]
- Mizuno N, Takahashi A, Wagatsuma T, Mizuno T, Obata H (2002) Chemical composition of guttation fluid and leaves of Petasites japonicus v. giganteus and Polygonum cuspidatum growing on ultramafic soil. Soil Sci Plant Nutr 48: 451–453 [Google Scholar]
- Möller S, Croning MD, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17: 646–653 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Nagai M, Ohnishi M, Uehara T, Yamagami M, Miura E, Kamakura M, Kitamura A, Sakaguchi S-I, Sakamoto W, Shimmen T, et al. (2013) Ion gradients in xylem exudate and guttation fluid related to tissue ion levels along primary leaves of barley. Plant Cell Environ 36: 1826–1837 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Ishiguro S, Kimura T (2009) Gateway vectors for plant transformation. Plant Biotechnol 26: 275–284 [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Neumann D, zur Nieden U, Schwieger W, Leopold I, Lichtenberger O (1997) Heavy metal tolerance of Minuartia verna. J Plant Physiol 151: 101–108 [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Oelschläger W (1971) Fluoride uptake in soil and its depletion. Fluoride Official Quart J Int Soc Fluoride Res 4: 80–84 [Google Scholar]
- Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1: e13–e13 [Google Scholar]
- Posthumus AC, Unsworth MH, Ormrod DP (1982) Biological indicators of air pollution. In MH Unsworth, DP Ormrod, eds, Effects of Gaseous Air Pollution in Agriculture and Horticulture. Butterworths, London, pp 27–42 [Google Scholar]
- Pscheidt JW (2015) Fluorine toxicity in plants. In JW Pscheidt, CM Ocamb, eds, Pacific Northwest Pest Management Handbooks, Oregon State University, Corvallis, OR.
- Ramteke LP, Sahayam AC, Ghosh A, Rambabu U, Reddy MRP, Popat KM, Rebary B, Kubavat D, Marathe KV, Ghosh PK (2018) Study of fluoride content in some commercial phosphate fertilizers. J Fluorine Chem 210: 149–155 [Google Scholar]
- Ruan J, Wong MH (2001) Accumulation of fluoride and aluminium related to different varieties of tea plant. Environ Geochem Health 23: 53–63 [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shu WS, Zhang ZQ, Lan CY, Wong MH (2003) Fluoride and aluminium concentrations of tea plants and tea products from Sichuan Province, PR China. Chemosphere 52: 1475–1482 [DOI] [PubMed] [Google Scholar]
- Smith KD, Gordon PB, Rivetta A, Allen KE, Berbasova T, Slayman C, Strobel SA (2015) Yeast Fex1p is a constitutively expressed fluoride channel with functional asymmetry of its two homologous domains. J Biol Chem 290: 19874–19887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Kolmakova-Partensky L, Shane T, Koide A, Koide S, Miller C, Newstead S (2015) Crystal structures of a double-barrelled fluoride ion channel. Nature 525: 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Robertson JL, Kolmakova-Partensky L, Miller C (2013) A family of fluoride-specific ion channels with dual-topology architecture. eLife 2: e01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton T, Baumann U, Hayes J, Collins NC, Shi B-J, Schnurbusch T, Hay A, Mayo G, Pallotta M, Tester M, et al. (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449 [DOI] [PubMed] [Google Scholar]
- Symonds RB, Rose WI, Reed MH (1988) Contribution of C1- and F-bearing gases to the atmosphere by volcanoes. Nature 334: 415–418 [Google Scholar]
- Wang GX, Cheng GD (2001) Fluoride distribution in water and the governing factors of environment in arid north-west China. J Arid Environ 49: 601–614 [Google Scholar]
- Warren CR (2008) Rapid measurement of chlorophylls with a microplate reader. J Plant Nutr 31: 1321–1332 [Google Scholar]
- Weinstein LH (1977) Fluoride and plant life. J Occup Med 19: 49–78 [DOI] [PubMed] [Google Scholar]
- Weinstein LH, Davison A (2004) Fluorides in the Environment: Effects on Plants and Animals. CABI Publishing, Cambridge, MA [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yu H, Tian C, Yu Y, Jiao Y (2015) Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol Plant 9: 749–752 [DOI] [PubMed] [Google Scholar]
- Zhu J, Xing A, Wu Z, Tao J, Ma Y, Wen B, Zhu X, Fang W, Wang Y (2019) CsFEX, a fluoride export protein gene from Camellia sinensis, alleviates fluoride toxicity in transgenic Escherichia coli and Arabidopsis thaliana. J Agric Food Chem 67: 5997–6006 [DOI] [PubMed] [Google Scholar]
- Zimmerman PW, Hitchcock AE, Gwirtsman J (1957) Fluorine in food with special reference to tea. Contrib Boyce Thompson Inst 19: 610 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.