Abstract
Seed setting rate is one of the critical factors that determine rice yield. Grain formation is a complex biological process, whose molecular mechanism is yet to be improved. Here we investigated the function of an OVATE family protein, Embryo Sac Development 1 (ESD1), in the regulation of seed setting rate in rice (Oryza sativa) by examining its loss-of-function mutants generated via clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated9 (Cas9) technology. ESD1 was predominantly expressed at Stage 6 of panicle development, especially in the ovules. esd1 mutants displayed reduced seed setting rates with normal stamen development and pollen tube growth but abnormal pistil group. Investigation of embryo sacs revealed that during the mitosis of functional megaspores, some egg cells degraded during differentiation in esd1 mutants, thereby hindering subsequent fertilization process and reducing seed setting rate. In addition, the transcriptional level of O. sativa anaphase-promoting complex 6, a reported embryo sac developing gene, was significantly reduced in esd1 mutants. These results support that ESD1 is an important modulator of ESD and seed setting rate in rice. Together, this finding demonstrates that ESD1 positively regulates the seed setting rate by controlling ESD in rice and has implications for the improvement of rice yield.
EMBRYO SAC DEVELOPMENT 1 regulates the seed setting rate by controlling embryo sac development in rice and has implications for the improvement of rice yield.
Introduction
Rice (Oryza sativa) is one of the most important food crops in the world. At present, the controlling mechanism of rice yield has made great progress, particularly in grain weight and effective panicles, but little is known about the seed setting rate. Seed setting rate is affected by several factors, including biomass–spikelet ratio, spikelet–leaf ratio, specific leaf weight, spikelet-sheath quantity, and spikelet differentiation (Zhou et al., 2011; Toh et al., 2012; Li et al., 2013; Xu et al., 2017; Liu et al., 2018, 2019; He et al., 2019; Xiang et al., 2019; Xu et al., 2019). Moreover, seed setting rate is also easily affected by the external environment, such as drought, high temperature, low temperature, or saline–alkali stress (Fu et al., 2014; Xu et al., 2014; Wang et al., 2016b; Cheabu et al., 2018; Ma et al., 2018; Si et al., 2018). Further study of seed setting rate has important significance both for its mechanism and for the improvement of rice yield.
As an essential part of spikelet, embryo sac development (ESD) can directly affect seed setting rate. The development of rice embryo sacs can be divided into two stages, the first stage involves the formation of functional megaspores, in which the megasporocyte undergoes meiotic division to yield four haploid cells, three of which disappear, while one becomes the functional megaspore, the second stage is the formation of the embryo sac, in which the functional megaspores undergo mitosis 3 times and eventually form a mature embryo sac, there have been some reports about genes related to ESD (Zee 1997; Li et al., 2007; Chang et al., 2009; Wang et al., 2012, 2019; Hou et al., 2019; Khanday et al., 2019; Xu et al., 2020). OsRAD51C, which belongs to the RecA/RAD51 family, is associated with the normal meiotic division of megasporocyte and microsporocyte at an early stage (Kou et al., 2012; Tang et al., 2014). OsMSH4 and OsMSH5, the members of the ZMM family, can form a heterodimer, which plays a critical role in the regulation of homologous chromosome exchange in the meiotic division (Luo et al., 2013; Zhang et al., 2014). Especially, OsMSH4 influences the exchange of homologous chromosomes during meiotic division of male and female (Wang et al., 2016a). The O. sativa anaphase-promoting complex 6 (OsAPC6) gene encodes an APC6 protein, affecting the development of female gametes but not the development of male gametes. OsAPC6 is crucial for the smooth transition from mitosis metaphase to mitosis anaphase. The T-DNA inserted osapc6 mutants are fertile in males and partially sterile in females. The plants exhibit dwarfed, and their seed setting rates are reduced by 45% (Kumar et al., 2010; Awasthi et al., 2012).
The OVATE family proteins (OFPs) are plant-specific proteins with a conserved OVATE domain (Liu et al., 2014). The discovery of OFP protein family originated from the gene OVATE which regulates the shape of tomato (Solanum lycopersicum) fruit. The C-terminal of OVATE protein contains a highly conserved OVATE domain (Liu et al., 2002). The genes of OFP protein family are widely presented in various plant species such as Arabidopsis (Arabidopsis thaliana), rice, tomato, and pepper (Capsicum annuum). OFP family proteins play a key role in regulating the growth and development of plants, including the construction of fruit shape, fruit maturity and quality formation, DNA repair, vascular bundle development, secondary cell wall formation, and ESD (Hackbusch et al., 2005; Pagnussat et al., 2007; Wang et al., 2008, 2010; Li et al., 2011a; Toh et al., 2012; Schmitz et al., 2015; Yang et al., 2016; Ma et al., 2017; Xiao et al., 2017).
In our previous study, microarray data showed that ESD1 protein belongs to the OFP family was highly and abundantly expressed mainly in spikes. In this study, we find that ESD1 positively regulates the rice seed setting rate by controlling ESD. Its loss-of-function esd1 mutants were successfully generated via CRISPR/Cas9 technology. Results showed that esd1 mutants exhibited abnormal development of embryo sac, thereby ultimately decreasing the seed setting rate. Meanwhile, the transcriptional level of OsAPC6, an ESD-related gene, was found to be significantly reduced in esd1 mutants. ESD1 had the highest level of expression at Stage 6 of young panicle development, especially in the ovules, and ESD1–GFP fusion proteins predominantly localized in cytoplasm. Our results support ESD1 as a key modulator of rice seed setting rate. This finding has significance for the improvement of rice yield.
Results
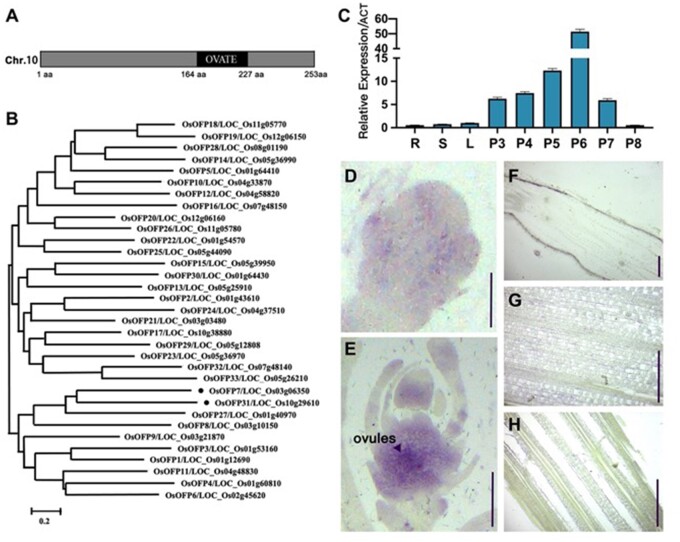
ESD1 belongs to the OFP family
ESD1 contains only one exon without intron, and its predicted protein ESD1 has 253 amino acids (aa). The Simple Modular Architecture Research Tool database (http://smart.embl-heidelberg.de/) showed that ESD1 contains an OVATE domain located at 164–227 aa, and belongs to the OFP family (Hackbusch et al., 2005; Liu et al., 2014; Figure 1A). The OFP family has 33 members in rice (from OsOFP1 to OsOFP33), and ESD1 is numbered as OsOFP31 (Liu et al., 2014). The phylogenetic tree constructed according to the amino acid sequences of 33 members showed that the homological differences between members are relatively significant. ESD1 shares the closest similarity with OsOFP7 and is also close to OsOFP27 and OsOFP8 (Figure 1B). Real-time quantitative polymerase chain reaction (RT-qPCR) showed that the expression levels of ESD1 in roots, stems, and leaves were very low, and there was a parabolic curve at Stages 3–8 of young panicle development. The expression levels at Stages 3–6 of panicle development gradually increased, peaked at Stage 6, followed by a plunge at Stage 7, and reached the lowest point at Stage 8 (Figure 1C). The roots, stems, and leaves at the panicle differentiation stage as well as the young panicles at Stages 3–8 were used for in situ hybridization. The results showed that the strongest hybridization signals of ESD1 presented at Stage 6 of panicle development, which mainly distributed in the ovules (Figure 1, D–H). The results of in situ hybridization were consistent with those of RT-qPCR, indicating that ESD1 functions in ovules at the panicle development process in rice.
Figure 1.
Gene structure and expression patterns of ESD1. A, Structural pattern diagram of ESD1. B, Phylogenetic tree of OFP family members. C, ESD1 expressions in roots, stems, leaves, and young panicles at Stages 3–8; R, roots; S, stems; L, leaves; P3-P8, young panicles at Stages 3–8; Error bars indicate SD (n = 3). D, In situ hybridization of ESD1 at Stage 3 of panicle development; Bar = 150 μm. E, In situ hybridization of ESD1 at Stage 6 of panicle development; Bar = 1 mm. F, In situ hybridization of ESD1 in roots; Bar = 150 μm. G, In situ hybridization of ESD1 in stems; Bar = 150 μm. H, In situ hybridization of ESD1 in leaves; Bar = 150 μm.
ESD1 predominantly localizes in cytoplasm
Some of the OFP family members are generally located in the nuclei and function as transcription factors (Wang et al., 2008; Schmitz et al., 2015). To test whether ESD1 has transcriptional activation activity, the recombinant vector pGBKT7-ESD1 was transferred into yeast (Saccharomyces cerevisiae) strain Y2HGold. The strain expressing ESD1 proteins could not grow on the systematic tri-deficient medium (Figure 2A), indicating that ESD1 was inactive in transcriptional activations. The ESD1 and GFP fusion expression vector was transferred into rice protoplasts. The fluorescent signals of ESD1–GFP fusion proteins were mainly distributed in the cytoplasm (Figure 2B). Therefore, ESD1 might be an atypical transcription factor in rice.
Figure 2.
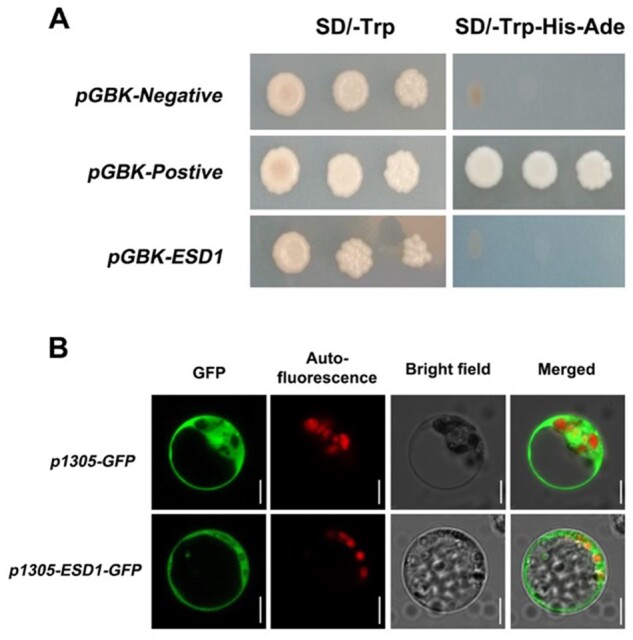
Protein subcellular localization of ESD1. A, Verification of ESD1 transcription activation; SD/-Trp, Tryptophan single-deficient medium; SD/-Trp-His-Ade, Tryptophan, Histidine, and Adenine triple-deficient medium; pGBK-Negative, negative control; pGBK-Postive, positive control. B, Subcellular localization of ESD1 in rice protoplasts; p1305-GFP, the GFP individual expression; p1305-ESD1-GFP, the ESD1 and GFP fusion expression; Bars = 5 μm.
esd1 mutants displayed a phenotype with reduced seed setting rates
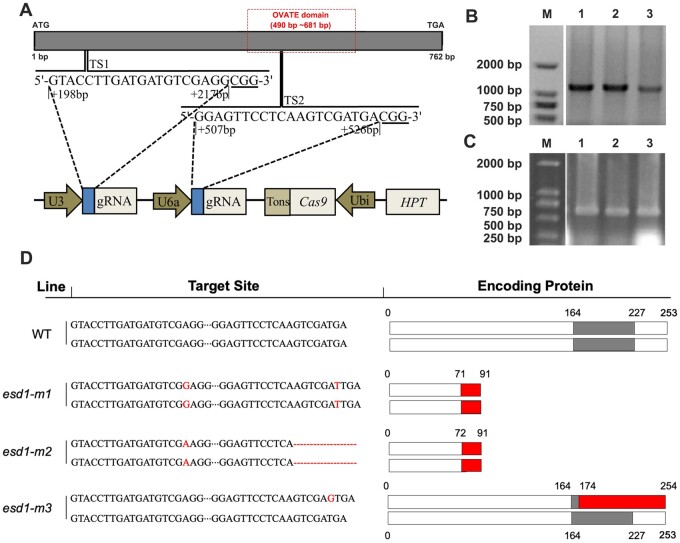
In this study, two targeted editing sites (TSs; TS1 and TS2) of ESD1 were constructed onto the CRISPR/Cas9 based binary expression vector (Figure 3A). The calli induced from a japonica rice 9522 were infected with the Agrobacterium-mediated transformation process and subjected to tissue culturing, resulting in two edited homozygous mutants esd1-m1 and esd1-m2 as well as a heterozygous mutant esd1-m3 (Figure 3, B and C). The mutant esd1-m1 was inserted with G and T at TS1 and TS2, respectively, resulting in an alteration of translated protein sequence after 71 aa, and an early termination after 91 aa, which caused the missing structure of the OVATE domain. The mutant esd1-m2 was inserted with A at TS1 and deleted a sequence of AGTCGATGA at TS2, resulting in a change of translated protein sequence after 72 aa and an early termination after 91 aa, which also caused the missing structure of the OVATE domain. As for the heterozygous mutant esd1-m3, ESD1 of one chromosome was normal, while ESD1 of the other homologous chromosome was inserted with G at TS2, resulting in the translation of two types of protein, one of which was normal ESD1 protein, the other changed after 174 aa, which also caused the missing structure of the OVATE domain (Figure 3D).
Figure 3.
TSs and types of mutations. A, Two TSs, TS1 and TS2, and information on CRISPR/Cas9 recombinant vector element. B, Screening of transgenic plants that integrate gene-edited-related sequence of the recombinant vector into the 9522 genome by PCR. C, Amplification of the TS1 and TS2 sequences of transgenic plants by PCR. D, ESD1’s mutation types and translated protein residues. The gray box represents the OVATE domain, the red box represents the new amino acid sequence.
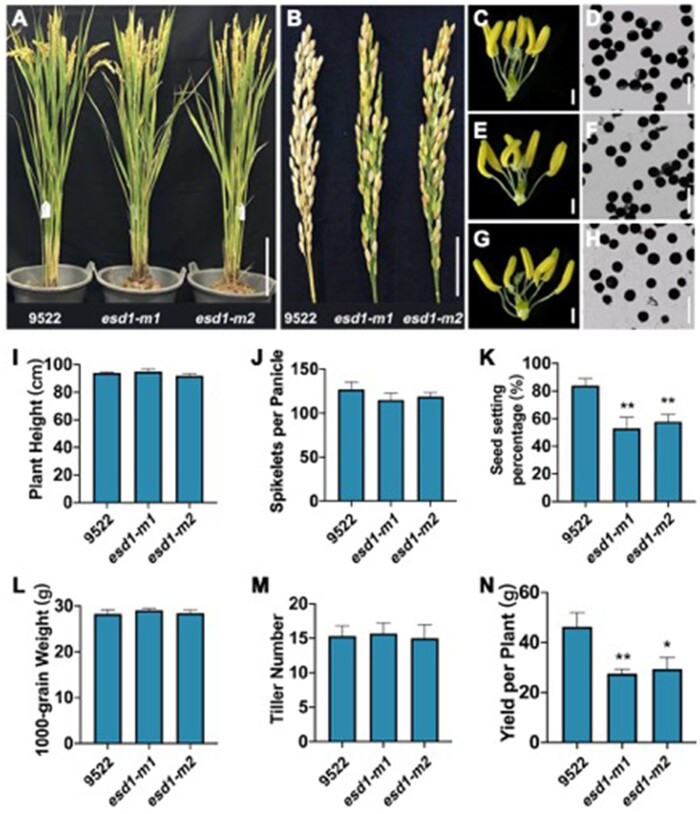
Phenotypic analysis indicated that no obvious difference was observed in the plant morphologies and internal spikelet structure between the wild-type 9522 and the mutants esd1-m1 and esd1-m2 (Figure 4A). Similar to 9552, I2–KI staining assay showed that the anthers of esd1-m1 and esd1-m2 were fertile (Figure 4, C–H). Plant height, spikelets per panicle, 1,000-grain weight, and tiller number of esd1-m1 and esd1-m2 were not significantly different from those of 9522 (Figure 4, I, J, L, and M). However, esd1-m1 and esd1-m2 showed a phenotype with decreased seed setting rates, 53% and 58%, respectively, while that of the 9522 was 84% (Figure 4, B and K), indicating that the deficiency of ESD1 mainly reduced the spikelet fertility in rice. Meanwhile, the grain yield per plant of esd1-m1 and esd1-m2 also markedly decreased compared to that of 9522 (Figure 4N). No significant difference was detected in seed setting rates between esd1-m3 and 9522 (Supplemental Figure S1). The population of T1 generation of esd1-m3 showed a separation with seed setting rate. The result of co-segregation experiment showed that 86 single plants (heterozygous mutants and wild-type plants) had normal seed setting rates, while 28 single plants (homozygous mutants named esd1-m3h) had reduced seed setting rates (Supplemental Figure S2). Therefore, the phenotype with seed setting rate was controlled by ESD1.
Figure 4.
Phenotype identification of esd1 mutants. A, Plant morphology of esd1 mutants; Bar = 20 cm. B, Panicle types of esd1 mutants; Bar = 4 cm. C, Anthers of 9522; Bar = 600 μm. D, Microscopy of 9522 pollen grains; Bar = 100 μm. E, Anthers of esd1-m1; Bar = 600 μm. F, Microscopy of esd1-m1 pollen grains; Bar = 100 μm. G, Anthers of esd1-m2; Bar = 600 μm. H, Microscopy of esd1-m2 pollen grains; Bar = 100 μm. I, Plant height of esd1 mutants. J, Spikelets per panicle of esd1 mutants. K, Seed setting rate of esd1 mutants. L, 1,000-grain weight of esd1 mutants. M, Tiller number of esd1 mutants. N, Yield per plant of esd1 mutants. Error bars indicate sd (n = 15); Asterisks indicate significant difference by Student’s t test (*P < 0.05 and **P < 0.01).
Stamen development and pollen tube growth of esd1 mutants were normal
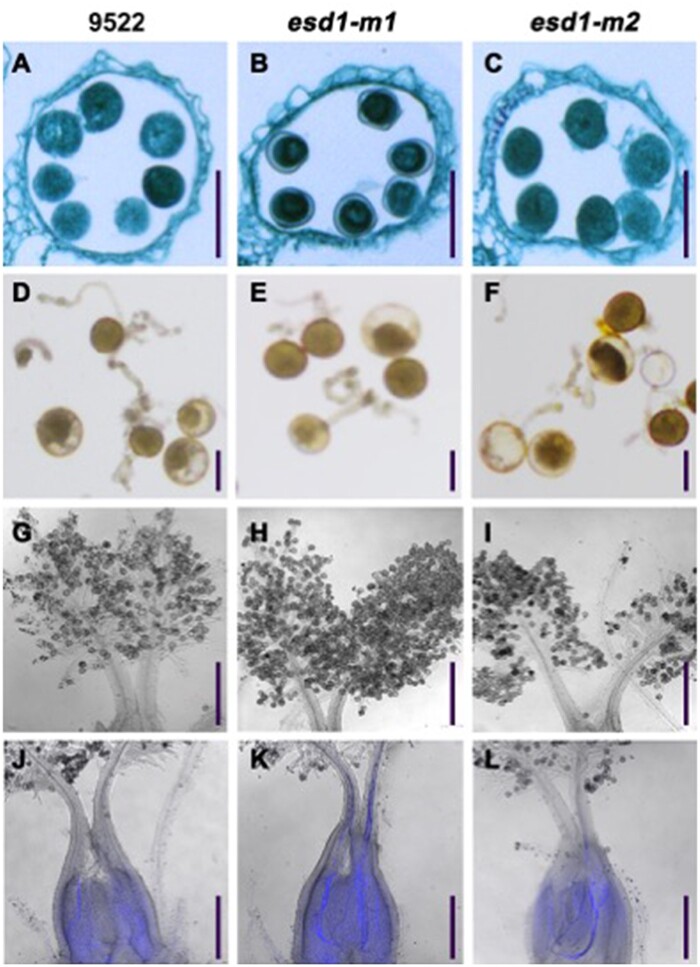
Spikelet samples were taken at Stage 8 of panicle development from 9522, esd1-m1, and esd1-m2, and fixed with FAA fixative to prepare paraffin sections. The results showed that 9522, esd1-m1, and esd1-m2 had no significant differences in the structure of anthers, and all pollen grains were fertile (Figure 5, A–C). Pollen grains were collected from 9522, esd1-m1, and esd1-m2 before flowering and cultured in vitro. The results showed that the germination rate of 9522 was 73.1%, while those of esd1-m1 and esd1-m2 were 72.0% and 71.1%, respectively, indicating that no significant differences were detected in the germination rates among the three genetic lines (Figure 5, D–F). Spikelet samples at different stages after flowering were collected from 9522, esd1-m1, and esd1-m2. The elongation process of pollen tubes was observed using an aniline blue staining. The results showed that the anthers of 9522, esd1-m1, and esd1-m2 could split open normally upon maturity. Their pollen grains could normally adhere to the stigma (Figure 5, G–I), germinate pollen tubes along the styles, and eventually enter the embryo sac through the ovule hole (Figure 5, J–L). These results indicated that the stamen development and pollen tube growth of esd1 mutants were normal.
Figure 5.
Microscopy of pollen grains and pollen tubes of esd1 mutants. A–C, Anther paraffin sections of 9522, esd1-m1, and esd1-m2; Bars = 50 μm. D–F, Pollen grain germinations in vitro of 9522, esd1-m1, and esd1-m2; Bars = 30 μm. G–I, Pollen grains of 9522, esd1-m1, and esd1-m2 attached to the stigmas; Bars = 200 μm. J–L, Pollen tube growth of 9522, esd1-m1, and esd1-m2 (bright field microscopy + fluorescence microscopy); Bars = 250 μm.
The pistil group of esd1 mutants was abnormal
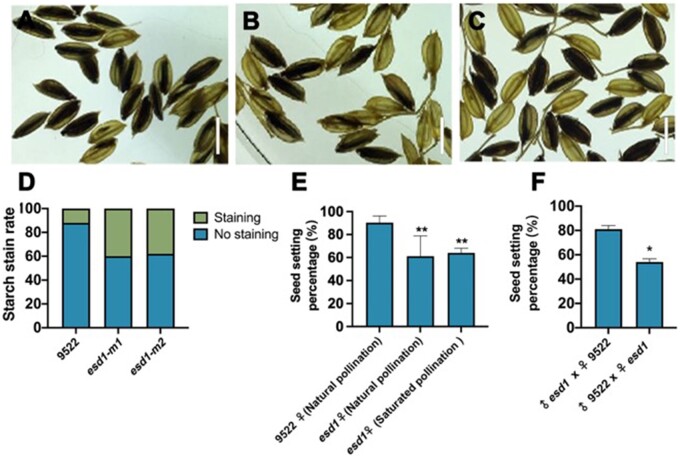
I2–KI staining was performed on the spikelets of 9522, esd1-m1, and esd1-m2 after flowering, and transparent xylene was used to examine the fertilization of spikelets. The results showed that the fertilization rate of 9522 was substantially higher than those of esd1-m1 and esd1-m2 (Figure 6, A–D). The main panicles of esd1-m1 were then subjected to natural pollination and saturated pollination. The results showed that the seed setting rate of esd1-m1 natural and saturated pollinations were 61% and 64%, respectively, which have no significant difference (Figure 6E). When the panicles of de-tasseled 9,522s main panicles were pollinated with esd1-m1 pollens, the resulted seed setting rate of 9,522 was 81%. However, when panicles of de-tasseled esd1-m1’s main panicles were pollinated with 9,522 pollens, the seed setting rate of esd1-m1 was 54% (Figure 6F). These results further indicated that the anther development of esd1 mutants was normal, and the cause of decreased seed setting rate of esd1 mutants was affected by the pistil group and occurred before fertilization.
Figure 6.
Fertilization and hybridization detection of esd1 mutants. A–C, Embryo sac fertilization results of 9522, esd1-m1, and esd1-m2; Bars = 7 mm. D, Starch stain rate of 9522, esd1-m1, and esd1-m2. E, Natural pollination of esd1 mutants and saturation pollination of esd1 mutants with 9522 pollens; Error bars indicate sd (n = 7). F, Reciprocal crosses between 9522 and esd1-m1; Error bars indicate sd (n = 3). Asterisks indicate significant difference by Student’s t test (*P < 0.05 and **P < 0.01).
No egg cells were detected in the partially matured embryo sacs of esd1 mutants
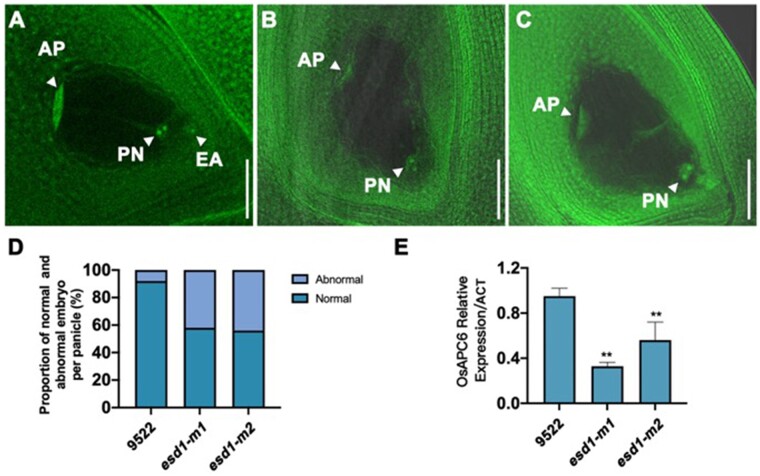
A large number of mature embryo sacs of 9,522, esd1-m1, and esd1-m2 were stained with nuclear-specific fluorescent dyes and hyalinized using hyalinization techniques. Samples were then scanned and photographed at 2 μm/layer under a confocal laser scanning microscope. The mature embryo sacs of 9522 were tested first, and 92% of their embryo sacs were found to develop healthily with a “seven cells and eight nuclei” structure at different levels of definition (Figure 7, A and D). The mature embryo sacs of esd1-m1 were then observed, and 58% of their embryo sacs developed normally, whereas 42% failed to show any egg cells and synergids despite the existence of normal antipodal cells and central polar nuclei (Figure 7, B and D). Similarly, among the embryo sacs of esd1-m2, 56% developed normally, whereas 44% showed no egg cells and synergids (Figure 7, C and D). The transcriptional levels of genes involved in the development of rice embryo sac were detected in the young panicles of 9522 and esd1 mutants by RT-qPCR. The results showed that the deficiency of ESD1 could significantly reduce the expression of OsAPC6 (Figure 7E). These results demonstrated that abnormal development of some embryo sacs led to a reduction in seed setting rates in esd1 mutants.
Figure 7.
Detection of mature embryo sac of esd1 mutants and related RT-qPCR. A, Normal embryo sac of 9522; AP, antipodal cells; PN, polar nuclei; EA, egg cell; Bar = 50 μm. B, Abnormal embryo sac of esd1-m1; Bar = 50 μm. C, Abnormal embryo sac of esd1-m2; Bar = 50 μm. D, Abnormal embryo sac ratio of esd1 mutants. E, Expression of OsAPC6 in the young panicles of 9522 and esd1 mutants. Error bars indicate sd (n = 2); Asterisks indicate significant difference by Student’s t test (**P < 0.01).
The ubiquitin-mediated protein degradation pathway was the most involved metabolic pathway in esd1 mutant differentially expressed genes
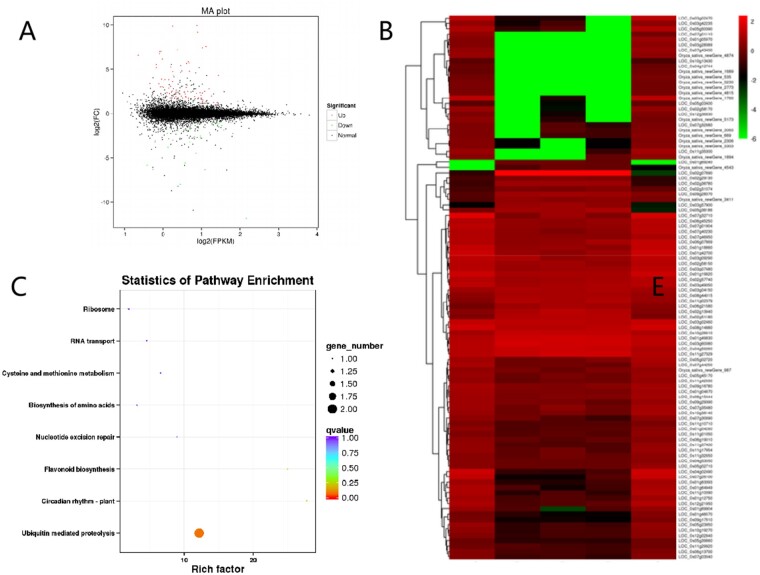
The young panicles at Stages 5 and 6 of panicle development of 9522 and esd1-m1 were taken for transcriptome analysis. A total of 190 significant differentially expressed genes (DEGs) were detected, and 51 common DEGs were presented at two stages. One hundred and two significant DEGs were detected in esd1-m1 with 81 upregulated and 21 downregulated expression genes at Stage 5. A total of 88 significant DEGs, 72 upregulated, and 16 downregulated genes, were detected in esd1-m1 at Stage 6 (Figure 8A). The results of hierarchical cluster analysis showed that DEGs of 9,522 and esd1-m1 could cluster together at Stage 5, indicating a high degree of similarity in genetic background and gene expression patterns between 9,522 and esd1-m1 (Figure 8B). One hundred and ninety DEGs were classified into eight functional categories, namely ubiquitin-mediated proteolysis, circadian rhythm, biosynthesis of amino acids, nucleotide excision repair, flavonoid biosynthesis, cysteine, and methionine metabolism, RNA transport, and ribosome. The ubiquitin-mediated protein degradation pathway is the most DEGs involved metabolic pathway (Figure 8C). Meanwhile, the transcriptome analysis also showed that the transcriptional level of OsAPC6 was significantly reduced in esd1-m1, which was highly consistent with the result of RT-qPCR (Figure 7E). OsAPC6 was identified to encode a cell cycle-related ubiquitin ligase that degrades mitosis-related regulators via the ubiquitin–proteasome pathway (Kumar et al., 2010). These results imply that the ubiquitin-mediated protein degradation pathway might be involved in regulating the development of embryo sacs in rice.
Figure 8.
Transcriptome analysis of ESD1. A, Differential expression volcano map. B, DEG cluster map. C, Scatter plot of enrichment of DEG KEGG pathway.
Discussion
Seed setting rate, grain number, and grain weight are the most important factors that determine rice yield. Seed setting rate is affected by several factors including the pistil and stamen development. It has been reported that OFP family members can regulate pistil development. For example, the OFP family member Arabidopsis OFP 5 (AtOFP5) protein is involved in regulating ovule formation in Arabidopsis (Hackbusch et al., 2005; Pagnussat et al., 2007). In our previous study, microarray data showed that ESD1 protein which contains an OVATE domain located at 164–227 aa and belongs to the OFP family was highly and abundantly expressed mainly in spikes. In this study, we obtained several different types of loss-of-function mutants of ESD1 by CRISPR/Cas9 technology (Ma et al., 2016, 2020; Sun et al., 2016; Zhang et al., 2018, 2019; Selma et al., 2019). The seed setting rates of esd1 mutants were significantly lower compared to that of wild-type 9522, and the co-segregation test showed that the trait was controlled by a single gene. The above research indicated that ESD1 is a key modulator of rice seed setting rate.
Low seed setting rate in rice can be caused by many factors (Zhou et al., 2011; Li et al., 2013; Xu et al., 2019). The pollen fertility I2–KI staining, anther paraffin sections, in vitro culture of pollen grains, and pollen tube germination test in vivo all proved that the stamen development and pollen tube growth of esd1 mutants were normal. These results suggest that the reduced seed setting rate of esd1 mutants has not resulted from the development of stamens. Promoter element prediction showed that ESD1 has elements involved in photoresponse, such as ACE, Box 4, G-box, G-Box, and GA-motif, and so on (Li et al., 2017). To demonstrate whether the fertility of the esd1 mutants was affected by the environment, the esd1 mutants were planted under different photoperiods and temperature treatments. The results showed that anther developments of 9522 and esd1 mutants were normal under different photoperiods and temperature treatments (Supplemental Figures S3 and S4), and no significant difference was detected in the seed setting rates of esd1 mutants under various treatments, indicating that the functions of ESD1 were not affected by light or temperature.
The results of embryo sac fertilization, reciprocal cross test, natural pollination, and saturation pollination further demonstrated that the anther development of esd1 mutants was normal, and the reduced seed setting rates of esd1 mutants have resulted from the development of pistil group before fertilization. Mature embryo sac structure inspection proved that the embryo sac abortion of esd1 mutants was caused by the absence of egg cells. The presence of normal polar nuclei and antipodal cells in all aborted embryo sacs further suggested that ESD was normal during the meiosis from megasporocyte and first two mitotic divisions from functional macrospore. During or after the third mitosis of functional megaspores, egg cells and synergids gradually degenerate, and this process is not influenced by light and temperature. The results of both RT-qPCR and in situ hybridization demonstrated that ESD1 expression was highest at the sixth stage of panicle differentiation. Stage 6 is the time when functional megaspores begin to undergo mitosis (Liu et al., 1997). The alteration of ESD1 expression level coincided with the failure of embryo sacs of esd1 mutants, which further indicated that ESD1 regulates ESD by influencing mitosis of functional megaspores.
Transcriptome data analysis revealed DEGs in esd1 mutant and wild-type material. The ubiquitin-mediated protein degradation pathway was the most involved metabolic pathway in DEGs in esd1 mutants. The ubiquitin–proteasome system is the major pathway of intracellular protein degradation, involved in ˃80% of protein degradation (Tokumaru et al., 2017; Zhang et al., 2017; Majumdar et al., 2020; Xia et al., 2020). The intracellular or extracellular ubiquitin–proteasome system is likely to be involved in regulating the development of the embryo sac by acting on different substrates (Liu et al., 2008; Kumar et al., 2010; Awasthi et al., 2012). Transcriptome analysis showed that the deficiency of ESD1 resulted in the downregulation of OsAPC6 encoding a cell cycle-related ubiquitin ligase involved in the degradation of mitosis-related regulators via the ubiquitin–proteasome pathway, which was consistent with the RT-qPCR result. In the T-DNA insertion mutant of osapc6, the functional megaspores in some of the embryo sacs showed an abnormality during the second mitosis, therefore the seed setting rate of osapc6 mutants was reduced by 45%. Coincidentally, the seed setting rate of the esd1 mutants also decreased by ∼45%. It is hypothesized that ESD1 has a genetic regulatory relationship with OsACP6, both act simultaneously during mitosis of functional macrospores. Consequently, ESD1 might have some regulatory interactions with OsAPC6, which could degrade relevant enzymes involved in the mitosis of functional megaspores through the ubiquitin–proteasome pathway, promote the programmed death of egg cells, and eventually lead to reduction in seed setting rate. However, more data and in-depth studies are needed on how ESD1 affects the ubiquitination pathway and thus regulates the development of the embryo sac.
Subcellular localization analysis revealed that ESD1–GFP fusion proteins are predominantly localized in the cytoplasm (Figure 2B). There are 33 genes that encode the full-length OVATE domain-containing proteins in the rice genome. OFPs are not a typical family of transcription factors. For example, the Arabidopsis AtOFPs have been shown to function as transcriptional repressors (Wang et al., 2016c). In this study, we performed subcellular localization prediction for ESD1 protein with TargetP tools (http://www.cbs.dtu.dk/services/TargetP/), and the result showed that ESD1 does not contain nuclear localization signal sequences (Supplemental Table S1). In addition to the subcellular localization study in rice protoplasts, we also repeated this experiment in Nicotiana benthamiana, and the results showed that the fluorescent signal was distributed throughout the cell, including the nucleus and cytoplasm (Supplementary Figure S5), which indicated that ESD1 does not contain the typical nuclear localization signal peptide. Verification of ESD1 transcription activation showed that ESD1 was inactive in transcriptional activations. Therefore, we speculate that ESD1 might be an atypical transcription factor to regulate the development of embryo sac in rice, but a further investigation should be performed in the future study.
In conclusion, this study demonstrated that ESD1 positively regulates the seed setting rate by controlling ESD in rice, and has implications for the improvement of rice yield. These results not only help to further illustrate the regulatory mechanism of ESD but also have a high theoretical value in revealing the molecular mechanism of rice grain formation.
Materials and methods
Phylogenetic analysis of ESD1
Rice (O. sativa) protein sequence comparisons were conducted using MUSCLE version 3.6 based on the BLAST given in TIGR (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/index.shtml) and Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn) of OFP family members. A phylogenetic tree was constructed with the aligned relative protein sequences using MEGA version 3.0 (http://www.megasoftware.net/index. html) and Neighbor-Joining method with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1,000 replicates; random seed).
Construction and transformation of CRISPR/Cas9 vectors
We used CRISPR/Cas9 target site designing website called E-CRISP (http://www.e-crisp.org/E-CRISP/) to analyze and design the ESD1 sequences. Target sites TS1 and TS2 with high scores were selected to reduce the probability of missing shots. TS1, TS2, and their corresponding reverse complementary sequences were then made into two pairs of primers, TS1-U3-F/R and TS2-U6a-F/R (ggca and aaac were bound at the 5′-ends of the forward primer and reverse primer, respectively; Supplemental Table S2 for primer sequences). Through Golden Gate cloning, DS-TS1 and DS-TS2, the double-stranded sequences obtained by annealing the above two pairs of primers, were inserted into the intermediate vectors pYLgRNA-U3 and pYLgRNA-U6a, respectively. Two expression cassettes U3-TS1-gRNA and U6a-TS2-gRNA were obtained via PCR amplification (Supplemental Table S3 for the amplification primer sequences). Through Golden Gate cloning, two expression cassettes were then inserted into the plant expression vector pYLCRISPR/Cas9-MH, after seedlings were obtained by tissue culture, primers SP1/SP2 were used to detect whether pYLCRISPR/Cas9-MH was introduced into the recipient plants (Supplemental Table S4 for primer sequences), and then primers ESD1-JC-F/R were used to amplify sequences containing the target sites TS1, TS2 (Supplemental Table S4 for the amplification primer sequences), and then sequencing was used to detect the mutations at the target sites.
Microscopy of pollen grains
Spikelet samples of 9522, esd1-m1, and esd1-m2 were collected from their main panicles (in the same position) 1 d before flowering, fixed in Kano solution (absolute ethanol: glacial acetic acid = 3:1), and stored at 4°C. For microscopic examination, each spikelet was stained with 1% (w/v) I2–KI and observed under a microscope with a 10 X ocular. The field of view was switched at least 3 times. The total count of pollen grains was 500. Judging criteria: the round dark brown pollen grains were considered fertile; irregularly shaped, weakly stained, or unstained pollen grains were deemed sterile.
Anther paraffin sections
Spikelet samples at Stage 8 of panicle development were taken from 9522, esd1-m1, and esd1-m2. Samples were dehydrated, hyalinized, immersed, and embedded in paraffin. Samples were then cut into 10 μm-thick sections, which were stained with safranin O-fast green, hyalinized, and sealed. The sections were observed under an Olympus BH2 optical microscope.
Pollen grain culture in vitro
An appropriate amount of liquid culture medium was placed on glass slides. When the plants were about to bloom, the spikelets of 9522, esd1-m1, and esd1-m2 were pinched with tweezers, after which their pollen grains were gently shaken off onto liquid media, respectively. The glass slides were covered with coverslips and placed in petri dishes with moistened filter paper. The petri dishes were sealed and incubated at 28°C in the dark for 60 min. Afterward, a drop of 0.005% (w/v) water-soluble aniline blue solution was added to each liquid culture medium, and the germination of pollens was checked under a fluorescence microscope. The above procedures were repeated 3 times for each group of samples. More than 200 pollen grains were detected each time. The average percentage of germinated pollens was deemed as the germination rate.
Pollen tube germination test in vivo
After heading, the flowering time of 9522, esd1-m1, and esd1-m2 was written down with a marker. Spikelet samples were collected at 0, 10, and 20 min, and 1, 1.5, 2, and 2.5 h after flowering, fixed with FAA fixative for 24 h, immersed in 70% (v/v) ethanol solution, and stored at 4°C. The ovaries were peeled off before observation. The samples were then rehydrated in 50% (v/v), 30% (v/v), and 10% (v/v) ethanol (30 min each), washed with ddH2O 2–3 times, immersed in 2 mol/L NaOH solutions to soften for 12 h, rinsed with ddH2O several times, stained with 0.05% (w/v) aniline blue for 12 h, and then observed under a confocal laser scanning microscope (Zeiss LSM 880) under 405 nm laser of excitation wavelength.
Saturation pollination
One day before flowering, five individual plants sharing the same growth status among the esd1-m1 population were bagged. There were two panicles per bag and two bags per plant, one of which was used for saturation pollination, and the other for self-pollination. For saturation pollination, esd1-m1 plants were pollinated with 9522 pollens at the full-bloom stage. This process was repeated for seven consecutive days. Seeds were tested upon harvest, and the seed setting rate of saturation pollination and self-pollination were compared.
Reciprocal cross tests
The main panicle samples of 9522 and esd1-m1 were collected at the heading stage. Bloomed spikelets and spikelets that could not bloom on that day were cut off. The spikelets that could bloom on that day were retained and de-tasseled before flowering. At the full-bloom stage, de-tasseled spikelets of the esd1-m1 main panicles were pollinated with 9522 pollens, and de-tasseled spikelets of the 9522 main panicles were pollinated with esd1-m1 pollens. Seeds were tested upon harvest, and the seed setting rate of the two hybrids was compared (Li et al., 2011b).
Tests for fertilization
Spikelet samples were taken from 9522, esd1-m1, and esd1-m2 more than one day after flowering, and immersed in 40°C water overnight, followed by 35% (v/v) concentrated hydrochloric acid for 30 min. The residual hydrochloric acid was rinsed off with ddH2O. The samples were then immersed in the I2–KI solution for an additional 30 min. After rinsing off the excess iodine with 70% (v/v) alcohol, the samples were immersed in absolute ethanol for 30 min. Lastly, the spikelets were hyalinized in xylene and examined on a fluorescent box.
Mature embryo sac structure inspection
Spikelet samples at Stage 8 of panicle development were collected from 9522, esd1-m1, and esd1-m2, fixed with FAA fixative, and then immersed in 70% (v/v) ethanol after ovaries were peeled off. In this process, the ovaries of different samples should be, respectively, immersed in ethanol at gradient concentrations (50% (v/v), 30% (v/v), and 15% (v/v)) for 20 min each, and rinsed with ddH2O 2–3 times. After that, the ovaries were hydrolyzed with 1 mol/L HCl in a 60°C water bath for 15 min. Subsequently, the ovaries were stained with 1% (w/v) eosin Y for 8 h. After staining, the ovaries were rinsed with ddH2O until colorless and pre-treated with 0.1 mol/L citric acid–disodium hydrogen phosphate buffer (pH = 5.0) for 8 h. They were then dyed with 20 μg/mL H33342 in the dark at 25°C for 24 h. The ovaries were rinsed with ddH2O 2–3 times and dehydrated in ethanol at gradient concentrations (15% (v/v), 30% (v/v), 50% (v/v), 70% (v/v), 85% (v/v), and 95% (v/v)) for 20 min each. The ovaries were then dehydrated twice with absolute ethanol for 2 h each time and transferred into absolute ethanol overnight. The next day, the dehydrated ovaries were immersed in a mixture of equal amounts of methyl salicylate and absolute ethanol for 1 h as a transition and then hyalinized with methyl salicylate 3 times. The first 2 times took 2 h each, and the last time took 15 h, after which the ovaries could be stored in methyl salicylate. A drop of sesame oil was placed on each concave glass slide before observation, and a treated ovary was then mounted on a slide. After that, the slide was inverted under a confocal laser scanning microscope (Zeiss LSM 880) and scanned at 2 μm/layer under 488 nm laser of excitation wavelength. Meanwhile, photographs were taken.
RT-qPCR
Total RNA was extracted using the TRIzol method, and cDNA was obtained by reverse transcription after digesting the RNA. The RT-qPCR reaction system (ABI QuantStudio 3) included: ESD1-qP-F/R primers (Supplemental Table S5 for primer sequences), 0.5 μL each, 2 μL cDNA, 4.5 μL SYBR, 0.5 μL ROX, and 2.0 μL ddH2O. Three biological replicates and three technical replicates were taken. Settings for the RT-qPCR amplification program: 95°C for 5 min; 95°C for 15 s; 58°C for 35 s; 45 cycles; default settings for dissolution curve and cooling temperature. The internal reference standard was the expression level of OsACTIN1, and the expression of ESD1 was calculated using the 2−ΔΔCT formula (Livak and Schmittge, 2001).
In situ hybridization
Roots, stems, leaves at the stage of panicle differentiation, and young panicles at Stages 3–8 were taken (the whole process was guaranteed to be free of RNase contamination), fixed with FAA fixative, and then made into paraffin sections. ESD1-ish-F/R primers (Supplemental Table S6 for primer sequences) and in situ hybridization probes were designed for in vitro transcription. Kawata’s experimental method was used for reference in the sensitivity test of the in situ probes (Kawata et al., 2010); Kouchi’s experimental method was used for reference in the RNA hybridization and immunoassay (Kouchi and Hata, 1993).
Subcellular localization in rice protoplast
ESD1 and GFP fusion expression vector was constructed. A 10 μL of fusion expression vector plasmid and 110 μL of protoplasts were pipetted, placed in a 2 mL centrifuge tube, and mixed well. A 130 μL of 40% (w/v) PEG4000 prepared on-site was added to the mixture, which was then let sit at room temperature for 15 min. About 500 mL of W5 solution was added to stop the reaction, and the mixture was then centrifuged at 1,188 rpm for 2 min, with the supernatant discarded. Then, 1 mL of W5 solution was added to re-suspend the protoplasts. The mixture was again centrifuged at 1,188 rpm for 2 min, with the supernatant discarded. Finally, 1 mL of W5 solution (containing 1 μL of Kan) was added to re-suspend the protoplasts. The mixture was then transferred onto a 24-well plate, incubated at 28°C, 40 rpm in the dark for 12–16 h, and then observed under a confocal laser scanning microscope (Zeiss LSM 880) under 488 nm laser of excitation wavelength.
Transcription activation verification
The pGBKT7-ESD1 vector was constructed. pGBKT7-CSA vector plasmid (positive control) and pGBKT7 empty vector plasmid (negative control) were used to transform Y2H Gold yeast (S. cerevisiae) competent cells using a heat shock approach. The bacterial solution was spread over the SD/-Trp solid medium, which was then sealed, inverted, and kept in an incubator at 28°C for 3–5 d. Once the colonies reached 2–3 mm, a single colony from each of the three solid media was picked and diluted at 1:10, 1:103, and 1:105. An aliquot of 4 μL of the diluents was transferred onto an SD/-Trp single-deficient plate and also onto an SD/-Trp-His-Ade triple-deficient plate. The plates were sealed, inverted, and kept in an incubator at 30°C for 3–5 d after air-drying the bacterial plaques.
Transcriptome analysis
Extract and isolate mRNA from 9522, esd1-m1 plant young panicles at 5 and 6 stages of development, add a crushing buffer, interrupt the mRNA into short sequences (200–700 nt), and then reverse transcribed to obtain double-stranded cDNA; purified, eluted, with base A added to the end and connected to the sequencing junction; and then enrich the cDNA library by PCR, with Illumina HiSeq 2000 sequenced the library. The sequences obtained after sequencing and filtering were compared to the reference genome of the Japonica variety Nipponbare.
Accession numbers
Sequence data from this article for the cDNA and genomic DNA of ESD1 can be found in the GenBank/EMBL/Gramene data libraries under accession number LOC_Os10g29610.
Supplemental data
The following supplemental materials are available.
Supplemental Figure S1. The seed setting rate of esd1-m3.
Supplemental Figure S2. Phenotypic data of esd1-m3h.
Supplemental Figure S3. Effects of photoperiods on the fertility of esd1 mutants.
Supplemental Figure S4. Effects of temperature treatments on the fertility of esd1 mutants.
Supplemental Figure S5. The subcellular localization of ESD1 in tobacco.
Supplemental Table S1. Subcellular localization prediction for ESD1 protein with TargetP.
Supplemental Table S2. Amplification primers for target sites TS1 and TS2.
Supplemental Table S3. Amplification primers for expression cassettes.
Supplemental Table S4. Detection primers for target sites mutation.
Supplemental Table S5. Primers for RT-qPCR reaction.
Supplemental Table S6. Primers for in situ hybridization.
Supplementary Material
Acknowledgments
We are grateful to Prof. Dabing Zhang, Shanghai Jiao Tong University for kindly providing the microarray data. We are grateful to Prof. Yinghong Yu, Hunan Academy of Agricultural Sciences for revising the manuscript. We are grateful to Prof. Yaoguang Liu, South China Agricultural University for kindly providing the CRISPR/Cas9 editing vectors, pYLgRNA-U3, pYLgRNA-U6a, and pYLCRISPR/Cas9-MH.
Funding
This work was supported by the Hunan Provincial Key Research and Development Program (grant no. 2020NK2041), the Natural Science Foundation of Hunan Province (grant no. 2020JJ2019 and 2020JJ5402), and the National Natural Science Foundation of China (grant no. U19A2031).
Conflict of interest statement. The authors declare that they have no competing interests.
L.L. and J.W. designed the research process, and revised the paper; T.W., Y.L., S.S., M.Q., L.Z., C.L., H.D., and L.L. performed the experiments, and drafted the manuscript.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the instructions for authors (https://academic.oup.com/plphys/pages/general-instructions) is: Li Li (lili@hhrrc.ac.cn).
References
- Awasthi A, Paul P, Kumar S, Verma SK, Prasad R, Dhaliwa HS (2012) Abnormal endosperm development causes female sterility in rice insertional mutant OsAPC6. Plant Sci 183:167–174 [DOI] [PubMed] [Google Scholar]
- Chang YX, Gong L, Yuan WY, Li XW, Chen GX, Li XH, Zhang QF, Wu CY (2009) Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol 151:2162–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheabu S, Moung-Ngam P, Arikit S, Vanavichit A, Malumpong C (2018) Effects of heat stress at vegetative and reproductive stages on spikelet fertility. Rice Sci 25:218–226 [Google Scholar]
- Fu J, Liu J,, Cao ZQ, Wang ZQ, Zhang H, Yang JC (2014) Effects of alternate wetting and drying irrigation during grain filling on the seed-setting rate and grain weight of two super rice cultivars. Acta Agron Sin 40:1056–1065 [Google Scholar]
- Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 102:4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YB, Yan L, Ge CN, Yao XF, Han X, Wang RC, Xiong LZ, Jiang LW, Liu CM, Zhao YD (2019) PINOID is required for formation of the stigma and style in rice. Plant Physiol 180:926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JJ, Cao CH, Ruan YN, Deng YY, Liu YX, Zhang K, Tan LB, Zhu ZF, Cai HW, Liu FX, et al. (2019) ESA1 is involved in embryo sac abortion in interspecific hybrid progeny of rice. Plant Physiol 180:356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata A, Kubota S, Eguchi T, Moritani NH, Shimo T, Kondo S, Nishida T, Minagi S, Takigawa T (2010) Role of the low-density lipoprotein receptor-related protein-1 in regulation of chondrocyte differentiation. J Cell Physiol 222:138–148 [DOI] [PubMed] [Google Scholar]
- Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V (2019) A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565:91–95 [DOI] [PubMed] [Google Scholar]
- Kou YJ, Chang YX, Li XH, Xiao JH, Wang SP (2012) The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J Exp Bot 63:5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238:106–119 [DOI] [PubMed] [Google Scholar]
- Kumar M, Basha PO, Puri A, Rajpurohit D, Randhawa GS, Sharma TR, Dhaliwal HS (2010) A candidate gene OsAPC6 of anaphase-promoting complex of rice identified through T-DNA insertion. Funct Integr Genomic 10:349–358 [DOI] [PubMed] [Google Scholar]
- Li DT, Chen LM, Jiang L, Zhu SS, Zhao ZG, Liu SJ, Su N, Zhai HQ, Ikehashi H, Wan JM (2007) Fine mapping of S32(t), a new gene causing hybrid embryo sac sterility in a Chinese landrace rice (Oryza sativa L). Theor Appl Genet 114:515–524 [DOI] [PubMed] [Google Scholar]
- Li EY, Wang SC, Liu YY, Chen JG, Douglas CJ (2011a) OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 67:328–341 [DOI] [PubMed] [Google Scholar]
- Li M, Yu TF, Xu ZS, Zhang SX, Min DH, Chen M, Ma YZ, Chai SC, Zheng WJ (2017) Soybean Transcription Factor Gene GmNF-YCa Enhances Osmotic Stress Tolerance of Transgenic Arabidopsis. Acta Agron Sin 43:1161–1169 [Google Scholar]
- Li SC, Li WB, Huang B, Cao XM, Zhou XY, Ye SM, Li CB, Gao FY, Zou T, Xie KL, et al. (2013) Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat Commun 4:2793. [DOI] [PubMed] [Google Scholar]
- Li YB, Fan CC, Xing YZ, Jiang YH, Luo LJ, Sun L, Shao D, Xu CJ, Li XH, Xiao JH, et al. (2011b) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269 [DOI] [PubMed] [Google Scholar]
- Liu CZ, Xue ZH, Tang DX, Shen Y, Shi WQ, Ren LJ, Du GJ, Li YF, Cheng ZK (2018) Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J 96:842–854 [DOI] [PubMed] [Google Scholar]
- Liu D, Sun W, Yuan YW, Zhang N, Hayward A, Liu YL, Wang Y (2014) Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann Bot Lond 113:1219–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EB, Zeng SY, Zhu SS, Liu Y, Wu GC, Zhao KM, Liu XL, Liu QM, Dong ZY, Dang XJ, et al. (2019) Favorable alleles of GRAIN-FILLING RATE1 increase the grain-filling rate and yield of rice. Plant Physiol 181:1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Zhang YY, Qin GJ, Tsuge T, Sakaguchi N, Luo G, Sun KT, Shi DQ, Aki S, Zheng NY, et al. (2008) Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-Type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis Gametogenesis. Plant Cell 20:1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Van Eck J, Cong B, Tanksley SD (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA 99:13302–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Xu XB, Lu YG, Xu SX (1997) The process of embryo formation and stages dividing in rice. Chin J Rice Sci 11:141–150 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–△△Ct method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Luo Q, Tang D, Wang M, Luo WX, Zhang L, Qin BX, Shen Y, Wang KJ, Li YF, Cheng ZK (2013) The role of OsMSH5 in crossover formation during rice meiosis. Mol Plant 6:729–742 [DOI] [PubMed] [Google Scholar]
- Ma SQ, Liu XH, Deng KC, Quan HJ, Tong LY, Xi ZX, Chai QR, Yang J (2018) Impact of low temperature in young ear formation stage on rice seed setting. Chin J Appl Ecol 29:125–132 [DOI] [PubMed] [Google Scholar]
- Ma XL, Zhu QL, Chen YL, Liu YG (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant 9:961–974 [DOI] [PubMed] [Google Scholar]
- Ma XN, Zhang XY, Liu HM, Li ZH (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat Plants 6:773–779 [DOI] [PubMed] [Google Scholar]
- Ma YM, Yang C, He Y, Tian ZH, Li JX (2017) Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J Exp Bot 68:4885–4898 [DOI] [PubMed] [Google Scholar]
- Majumdar P, Karidas P, Siddiqi I, Nath U (2020) The ubiquitin-specific protease tni/ubp14 functions in ubiquitin recycling and affects auxin response. Plant Physiol 184:00689.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Sundaresan V (2007) Cell-fate switch of synergid to egg cell in arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19:3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz AJ, Begcy K, Sarath G, Walia H (2015) Rice ovate family protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci 241:177–188 [DOI] [PubMed] [Google Scholar]
- Selma S, Bernabé-Orts JM, Vazquez-Vilar M, Diego-Martin B, Ajenjo M, Garcia-Carpintero V, Granell A, Orzaez D (2019) Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol J 17:1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si CL, Jin ZQ, Zheng JC, Tang RS (2018) Quantitative analysis on the effects of high temperature at meiosis stage on seed-setting rate of rice florets. Acta Agron Sin 34:627–631 [Google Scholar]
- Sun YW, Zhang X, Wu CY, He YB, Ma YZ, Hou H, Guo XP, Du WM, Zhao YD, Xia LQ (2016) Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9:628–631 [DOI] [PubMed] [Google Scholar]
- Tang D, Miao CB, Li YF, Wang HJ, Liu XF, Yu HX, Cheng ZK (2014) OsRAD51C is essential for double-strand break repair in rice meiosis. Front Plant Sci 5:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y (2012) Thermoinhibition uncovers a role for strigolactones in arabidopsis seed germination. Plant Cell Physiol 53:107–117 [DOI] [PubMed] [Google Scholar]
- Tokumaru M, Adachi F, Toda M, Ito-Inaba Y, Yazu F, Hirosawa Y, Sakakibara Y, Suiko M, Kakizaki T, Inaba T (2017) Ubiquitin-Proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol 173:524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Q, Shen Y, Hua YF, Wang JJ, Lin JR, Wu MG, Sun TT, Cheng ZK, Mercier R, et al. (2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol 37:283–286 [DOI] [PubMed] [Google Scholar]
- Wang CL, Wang Y, Cheng ZJ, Zhao ZG, Chen J, Sheng PK, Yu Y, Ma WW, Duan EC, Wu FQ, et al. (2016a) The role of OsMSH4 in male and female gamete development in rice meiosis. J Exp Bot 67:1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LZ, Wang LM, Xiang HT, Luo Y, Li R, Li ZJ, Wang CY, Meng Y (2016b) Relationship of photosynthetic efficiency and seed-setting rate in two contrasting rice cultivars under chilling stress. Photosynthetica 54:581–588 [Google Scholar]
- Wang N, Huang HJ, Ren ST, Li JJ, Sun Y, Sun DY, Zhang SQ (2012) The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol 160:696–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Chang Y, Ellis B (2016c) Overview of OVATE FAMILY PROTEINS, a novel class of plant-specific growth regulators. Front Plant Sci 7:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Diehl A, Wu FN, Vrebalov J, Giovannoni J, Siepel A, Tanksley SD (2008) Sequencing and comparative analysis of a conserved syntenic segment in the solanaceae. Genetics 180:391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YK, Chang WC, Liu PF, Hsiao MK, Lin CT, Lin SM, Pan RL (2010) Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Mol Biol 74:453–466 [DOI] [PubMed] [Google Scholar]
- Xia FN, Zeng BQ, Liu HS, Qi H, Xie LJ, Yu LJ, Chen QF, Li JF, Chen YQ, Jiang LW, et al. (2020) SINAT E3 ubiquitin ligases mediate FREE1 and VPS23A degradation to modulate abscisic acid signalling. Plant Cell 32: tpc.00267.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang XJ, Zhang PP, Yu P, Zhang YX, Yang ZF, Sun LP, Wu WX, Khan RM, Abbas A, Cheng SH, et al. (2019) LSSR1 facilitates seed setting rate by promoting fertilization in rice. Rice 12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YH, Liu DP, Zhang GX, Tong HN, Chu CC (2017) Brassinosteroids regulate OFP1, a DLT interacting protein, to modulate plant architecture and grain morphology in rice. Front Plant Sci 8:1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Tang DX, Cheng XJ, Zhang JX, Tang YJ, Tao QD, Shi WQ, You AQ, Gu MH, Cheng ZK (2019) OsPINOID regulates stigma and ovule initiation through maintenance of the floral meristem by auxin signaling. Plant Physiol 180:952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XJ, Li B, Liu SY, Zhang GL (2014) Effects of high temperature stress at heading stage on flowering habits and seed-setting rate in rice. Hybrid Rice 29:57–62 [Google Scholar]
- Xu Y, Wang FQ, Chen ZH, Wang J, Li WQ, Fan FJ, Tao YJ, Jiang YJ,, Zhu QH, Yang JZ (2020) CRISPR/Cas9‐targeted mutagenesis of the OsROS1 gene induces pollen and embryo sac defects in rice. Plant Biotechnol J 18:1999–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Yang J, Wang YH, Wang JC, Yu Y,, Long Y, Wang YL, Zhang H, Ren YL, Chen J, et al. (2017) OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genet 13:e1006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Shen WJ, He Y, Tian ZH, Li JX (2016) OVATE family protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PLoS Genet 12:e1006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee SY (1997) Changes in the pattern of organization of microtubules in the embryo sac of rice. Plant Physiol 114:248 [Google Scholar]
- Zhang HW, Si XM, Ji X, Fan R, Liu JX, Chen KL, Wang DW, Gao CX (2018) Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol 36:894–898 [DOI] [PubMed] [Google Scholar]
- Zhang L, Tang D, Luo Q, Chen XJ, Wang HJ, Li YF, Cheng ZK (2014) Crossover formation during rice meiosis relies on interaction of OsMSH4 and OsMSH5. Genetics 198:1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang Y, Lu MH, Chai YP, Jiang YY, Zhou Y, Wang XC, Chen QJ (2019) A novel ternary vector system united with morphogenic genes enhances CRISPR/Cas delivery in maize. Plant Physiol 181:1441–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY, Li JH, Li F, Liu HH, Yang WS, Chang K, Xu YY (2017) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell 43:731–743 [DOI] [PubMed] [Google Scholar]
- Zhou SR, Wang Y,, Li WC, Zhao ZG, Ren YL, Wang Y, Gu SH, Lin QB, Wang D, Jiang L, et al. (2011) Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23:111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.