Parental methylation divergence at CHG sequences is maintained in hybrid rice plants and corresponds to allelic-specific expression that is associated with phenotype variation and hybrid vigor.
Abstract
Heterosis refers to the superior performance of hybrid lines over inbred parental lines. Besides genetic variation, epigenetic differences between parental lines are suggested to contribute to heterosis. However, the precise nature and extent of differences between the parental epigenomes and the reprograming in hybrids that govern heterotic gene expression remain unclear. In this work, we analyzed DNA methylomes and transcriptomes of the widely cultivated and genetically studied elite hybrid rice (Oryza sativa) SY63, the reciprocal hybrid, and the parental varieties ZS97 and MH63, for which high-quality reference genomic sequences are available. We showed that the parental varieties displayed substantial variation in genic methylation at CG and CHG (H = A, C, or T) sequences. Compared with their parents, the hybrids displayed dynamic methylation variation during development. However, many parental differentially methylated regions (DMRs) at CG and CHG sites were maintained in the hybrid. Only a small fraction of the DMRs displayed non-additive DNA methylation variation, which, however, showed no overall correlation relationship with gene expression variation. In contrast, most of the allelic-specific expression (ASE) genes in the hybrid were associated with DNA methylation, and the ASE negatively associated with allelic-specific methylation (ASM) at CHG. These results revealed a specific DNA methylation reprogramming pattern in the hybrid rice and pointed to a role for parental CHG methylation divergence in ASE, which is associated with phenotype variation and hybrid vigor in several plant species.
Introduction
Heterosis or hybrid vigor refers to the phenomenon where hybrids exhibit superior performance in traits of interest relative to their parental inbred lines. This phenomenon has been widely exploited in crop breeding to augment agricultural productivity. Heterosis requires genetic variation between parental lines (Schnable and Springer, 2013). However, the relationship between genetic distance and heterosis is not straightforward. For instance, hybrid vigor can occur in progeny derived from crosses of genetically similar ecotypes in Arabidopsis (Arabidopsis thaliana; Groszmann et al., 2011). Accumulating evidence indicates that epigenetic difference is also involved in heterosis (Chen, 2013; Groszmann et al., 2013; Dapp et al., 2015). For instance, small interfering RNAs (siRNA) and/or DNA cytosine methylation levels in heterotic hybrids of Arabidopsis (Groszmann et al., 2011; Shen et al., 2012; Zhang et al., 2016), rice (Chen et al., 2010; He et al., 2010; Chodavarapu et al., 2012), maize (Zea mays; Barber et al., 2012; He et al., 2013; Seifert et al., 2018), and tomato (Solanum lycopersicum; Shivaprasad et al., 2012) vary from their parental lines. It has been shown that the RNA-directed DNA methylation (RdDM) pathway, which involves siRNAs, is required for differential DNA methylation in Arabidopsis hybrids (Greaves et al., 2012; Greaves et al., 2014; Zhang et al., 2016).
In plants, DNA cytosine methylation occurs in the context of CG, CHG, and CHH (where H is A, C, or T). The major role of DNA cytosine methylation is to silence transposable elements (TEs) and repetitive sequences and repress gene promoter activity. CG methylation is maintained during cell division by DNA METHYLTRANSFERASE 11 (MET1) by recognizing and methylating hemi-methylated CG sites in the newly replicated DNA. Non-CG (i.e., CHG and CHH) methylation in heterochromatin is maintained, respectively, by plant-specific CHROMOMETHYLASE 3 (CMT3 at CHG sites) and CMT2 (at CHH and CHG sites), which are recruited to heterochromatin by interacting with the histone methylation mark H3K9me2, while CHH methylation in euchromatin regions is maintained by the RdDM pathway involving the DOMAINS REARRANGED METHYLTRANSFERASE 2(DRM2) and siRNA (Law and Jacobsen, 2010). DECREASE IN DNA METHYLATION1 (DDM1) is a nucleosome remodeler; the ddm1 mutation led to >70% reduction in DNA methylation, predominantly affecting methylation at CG and CHG contexts in Arabidopsis and rice (Zemach et al., 2013; Tan et al., 2016). It was shown that hybrids between Arabidopsis C24 and Columbia-0 (Col) defective in RNA polymerase IV (Pol IV, required for siRNA production) or MET1 function did not reduce the level of heterosis of biomass. In contrast, hybrids with ddm1 mutation displayed a decreased heterosis level (Kawanabe et al., 2016; Zhang et al., 2016), suggesting that specific DNA methylation patterns and/or levels regulated by DDM1 play a role in heterosis in Arabidopsis.
It has been suggested that epigenetic systems may be involved in the alteration of gene expression in hybrids that, in turn, could contribute to the hybrid phenotype (Greaves et al., 2015). Work in Arabidopsis indicates that locus-specific DNA methylation divergence between the parental lines can directly or indirectly trigger heterosis (Lauss et al., 2018). Some of the changes in DNA methylation in hybrids correspond to changes in transcription levels, but there is no consistent relationship among the changes in DNA methylation, transcription, siRNA, and the generation of the heterotic phenotype (Crisp et al., 2020). It has been shown that gene allelic-specific expression (ASE) can lead to phenotype variation relevant to heterosis (Springer and Stupar, 2007; Paschold et al., 2012; Goff and Zhang, 2013; Shao et al., 2019). Although genetic differences may contribute to ASE, the epigenetic mechanism has been shown to be essential in differential expression of parental alleles of imprinted genes in endosperm in plants (Gehring, 2013). However, whether parental epigenome differences and their interactions in hybrids are involved in ASE is unclear. In addition, the precise nature and extent of parental epigenetic difference involved in heterosis and whether specific epigenetic variation can be considered as indicator to predict heterosis in agriculturally important crops remain unknown.
Rice is one of the most important food crops in the world. Rice cultivated in Asia can be divided into two subspecies: Oryza sativa subsp. Indica (Xian) and O. sativa subsp. Japonica (Geng). Indica/Xian rice can be further genetically subdivided into two major varietal groups, indica I and indica II, which have been independently bred and widely cultivated in China and Southeast Asia, respectively (Xie et al., 2015). Hybrids between these groups usually generate strong heterosis. For example, the elite hybrid Shanyou 63 (SY63) from the cross between female Zhenshan 97 (ZS97, indica I) and male Minghui 63 (MH63, indica II) exhibits superiority for a large array of agronomic traits and has been the most widely cultivated hybrid in China for approximately 3 decades (Zhang et al., 2016; Xie and Zhang, 2018). This particular hybrid rice system has been used as a model system for heterosis study, which has led to identification of a number of genetic loci/genes involved in hybrid vigor relevant to yield and yield-related traits (Yu et al., 1997; Hua et al., 2003; Huang et al., 2010; Zhou et al., 2012; Xie et al., 2015). The availability of high-quality reference genome sequences of ZS97 and MH63 and the detailed comparative annotation and analysis of the two genomes and transcriptomes (Zhang et al., 2016; Shao et al., 2019) provide a unique opportunity to analyze the epigenetic basis of hybrid vigor of the elite intra-subspecific hybrid rice.
In this work, we investigated DNA methylation differences between ZS97 and MH63 and parental methylation interaction in the reciprocal hybrids. Our data reveal a high divergence in the CG and CHG methylation patterns and dynamics between the parental lines, which is associated with the ASE in the hybrids.
Results
DNA methylation differences between shoot and panicle of two parents and hybrids
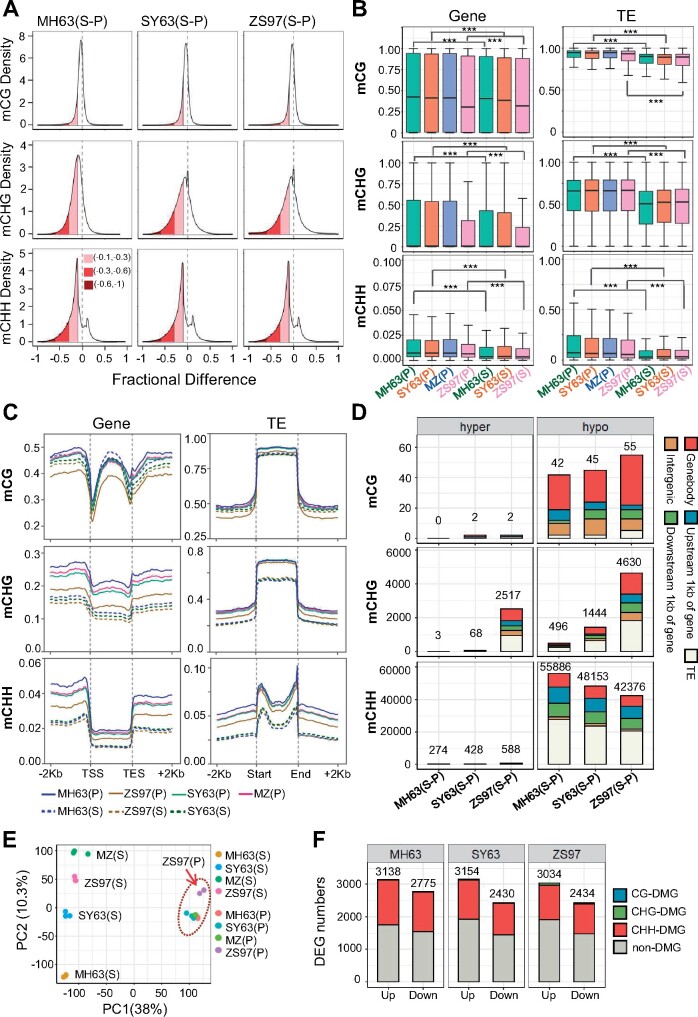
To evaluate DNA methylation differences between parental lines of the elite hybrid rice SY63, we analyzed the DNA methylomes of young panicle (2 mm length, inflorescence meristem) of ZS97 and MH63 and the reciprocal hybrids SY63 and MH63/ZS97 (MZ, with MH63 as female and ZS97 as male). Phenotypically, the reciprocal hybrids showed similar growth vigor and other traits including grain yield (Supplemental Figure S1A). The hybrid SY63 shows a ZS97-like growth vigor at seedling stage and MH63-like panicle growth at mature stage (Xie and Zhang, 2018). To study whether there is any difference in DNA methylation dynamics during shoot to panicle development between the parental lines, we also analyzed seedling DNA methylome of SY63 and the parent lines to compare with panicle methylomes. The sequencing depth of the BS-seq data was about 18–40 × genome coverage and the two biological replicates for each genotype were highly correlated (Pearson’s r was about 0.97–0.99; Supplemental Table S1 and Supplemental Figure S1B). The overall DNA methylation levels in panicle were higher than in shoot of all genotypes (Supplemental Table S1 and Supplemental Figure S1C). The increase of methylation was consistent with the higher expression of DNA methyltransferases CMT3, DRM2, and MET1-2 in panicle (Supplemental Figure S2A). Density plots (see “Materials and Methods” section) indicated that non-CG methylation levels were largely augmented in panicle versus shoot, whereas CG methylation was not changed as much (Figure 1A). The increased level of non-CG methylation in panicle versus shoot was detected in genes and TEs (Figure 1, B and C). Differentially methylated regions (DMRs) were tested using the criteria of methylation difference at CG >0.7, CHG >0.5, and CHH >0.2 within 200 bp bins that showed at least 25 informative sequenced cytosines in both samples (see “Materials and Methods” section). The analysis revealed that at CG context, there were very few hyper (0–2) or hypo (42–55) DMRs between shoot and panicle in the three genotypes (Figure 1D). At CHG context, higher numbers of shoot versus panicle DMRs were found in ZS97 (hypo 4630 and hyper 2517) than MH63 (3 hyper and 496 hypo) and SY63 (68 hyper and 1444 hypo; Figure 1D). At CHH sites, 42,000–55,000 hypo but only 274–588 hyper DMRs were detected in the three genotypes (Figure 1D). The analysis indicated a predominant increase of CHH methylation during shoot to panicle development and a more dynamic (with both hyper and hypo DMRs) CHG methylation in ZS97 than MH63. The data corroborated previous observations that methylation increases during development in rice (Zemach et al., 2010), and revealed substantial differences in CHG methylation dynamics during shoot to panicle development between the two parental lines.
Figure 1.
DNA methylation levels in shoot and panicle of MH63, ZS97 and the hybrids SY63 and MZ. A, Density plots showing the frequency distribution of CG, CHG, and CHH methylation differences at 200 bp windows between shoot and panicle of MH63, ZS97, and SY63. Variations from −0.1 to −0.3, −0.3 to −0.6, and −0.6 to −1.0 are indicated by pink, red, and dark red colors. B, Boxplots showing overall DNA methylation levels (at CG, CHG, and CHH context) in gene and TE of MH63 (green), SY63 (orange), MZ (blue), and ZS97 (pink) in shoot (S) and panicle (P) (*** P < 0.001, Wilcoxon rank-sum test). The y axis represents DNA methylation rate. The horizontal line within the box represents the median, box limits represent upper and lower quartiles, and whiskers represent 1.5 × interquartile range. C, Metaplots showing the average methylation levels within each 50-bp interval plotted in gene and TE of MH63 (blue), ZS97 (dark-orange), SY63 (green), and MZ (deep-pink) in shoot (S) and panicle (P). D, Numbers of DMRs of 200-bp bins between shoot and panicle in MH63, ZS97, and SY63. Different colors show the DMRs distributed in gene body (red), upstream 1 kb of gene (blue), downstream 1 kb of gene (green), TE (white), and intergenic regions (golden). E, Principal component analysis of transcriptomes of MH63, ZS97, SY63, and MZ in shoot (S) and panicle (P). The orange dotted circle indicates the panicle transcriptomes. Red arrow indicates the transcriptome of ZS97 panicle. For each genotype of tissue, three biological replicates are shown. F, DEGs (Q-value <0.01, |log2(Fold Change)| >2) in shoot versus panicle of MH63, ZS97, and SY63. Different colors show the DEGs with or without DMRs of all contexts in gene body or gene flanking regions.
In parallel, we analyzed the shoot and panicle transcriptomes of the three genotypes. Three biological replicates were sequenced and there were high correlations of the transcript numbers between the replicates (r = 0.88–1.00; Supplemental Table S2 and Supplemental Figure S3). Principal component analysis indicated that the shoot transcriptomes were distal from those of panicle of the different genotypes (Figure 1E). The shoot transcriptomes of the hybrids were closer to those of ZS97, while the panicle transcriptomes of the hybrid were closer to those of MH63, which is consistent with the characteristic of phenotype observed in SY63 (Xie and Zhang, 2018). However, there were some differences between SY63 and MZ shoot transcriptomes (Figure 1E), suggesting that there may be some parent-of-origin effects in gene expression. In total, 2,430–3,154 up and downregulated (fold change > 4 ×, Q-value <0.01) genes were detected in shoot versus panicle. About 40% of the DEGs were associated with DMRs (Figure 1F), suggesting that differential DNA methylation is involved in differential expression in shoot and panicle.
DNA methylation difference among the parental lines and hybrids
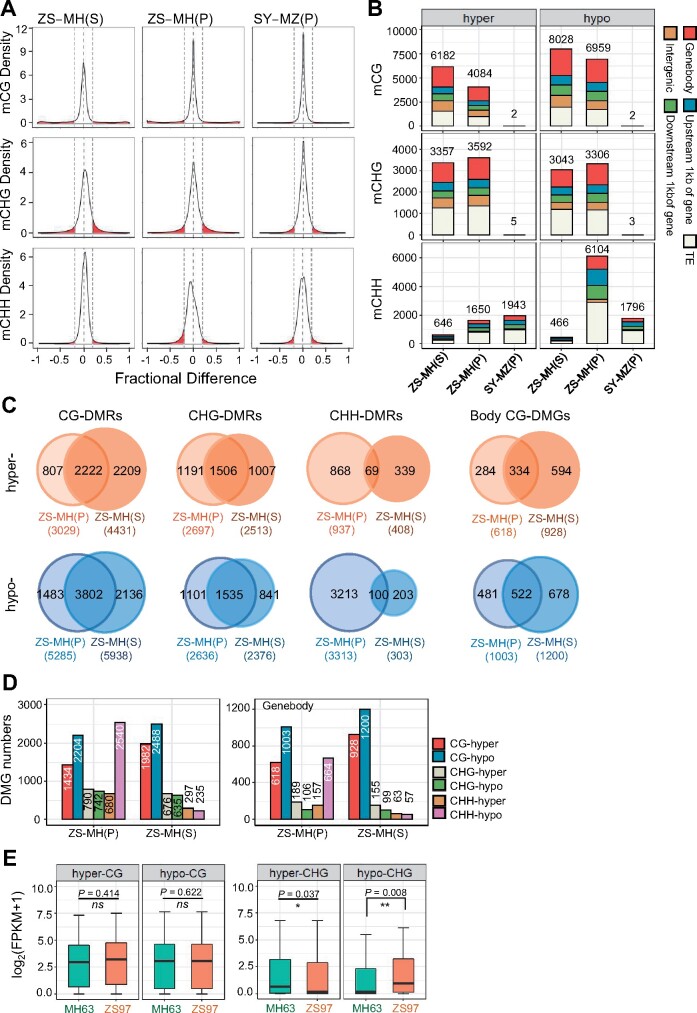
Density plots analysis revealed variation of DNA methylation levels at all sequence contexts between the two parent lines (Figure 2A). In shoot, the CHH methylation variation was relatively low compared with CG and CHG methylation, but became more important in panicle (Figure 2A). Analysis of DMR between ZS97 and MH63 revealed 4,084–6,182 hyper and 6,959–8,028 hypo CG DMRs and 3,357–3,592 hyper and 3,043–3,306 hypo CHG DMRs in shoot and panicle (Figure 2B). More than 72% of the DMR at CG sites and 55% at CHG sites in panicle overlapped with those detected in shoot (Figure 2, C and D). The data indicated that the parental lines had substantial differences in CG and CHG methylation, most of which persisted during shoot to panicle development (Figure 2, C and D; Supplemental Figure S4). In contrast, the difference at CHH methylation between ZS97 and MH63 was relatively low in shoot (646 hyper and 466 hypo DMRs) and increased in panicle (1,650 hyper and 6,104 hypo DMRs in ZS97 versus MH63; Figure 2, A and B). The higher number of hypo than hyper DMRs in ZS97 panicle was consistent with the larger increase of panicle CHH methylation in MH63 (Figure 1D). Nevertheless, methylation variation at CHH sites (especially in shoot) was unexpectedly low between the parental lines. By comparison, the reciprocal hybrids showed 1,796 hypo and 1,943 hyper CHH DMRs in SY63 compared to MZ, with much less or no methylation difference at CG and CHG contexts (Figure 2, A and B). The very low divergence of CG and CHG methylation between the reciprocal hybrids suggests that there is little effect of the parent-of-origin in genome-wide methylation pattern at the sequence contexts. However, larger methylation differences were observed at CHH sites between SY63 and MZ panicle (Figure 2A). This was consistent with the difference of transcriptomes between SY63 and MZ (Figure 1E), suggesting that there may be some parent-of-origin effect on CHH methylation that is associated mainly with genes in the rice genome.
Figure 2.
Divergence of DNA methylation landscape between MH63 and ZS97. A, Density plots showing the frequency distribution of methylation differences at 200-bp windows between MH63 (MH) and ZS97 (ZS) in shoot (S) and panicle (P), and methylation differences between SY63 (SY) and MZ in panicle (P). Variations more than 20% (0.2) are indicated by red color. The gray dotted lines represent −0.2, 0, and 0.2 from left to right. B, Numbers of ZS97 versus MH63 (ZS-MH) shoot (S) and panicle (P) DMRs of 200-bp bins and SY63 versus MZ (SY-MZ) panicle (P) DMRs. Different colors show the DMRs distributed in gene body (red), 1-kb flanking regions of gene (upstream, blue; downstream, green), TE (white), and intergenic regions (golden). C, Venn diagrams showing overlapping DMRs (ZS97-MH63, in CG, CHG, and CHH contexts) between shoot (S) and panicle (P). D, Numbers of DMGs between ZS97 (ZS) and MH63 (MH) in shoot (S) and panicle (P). Left, genes with the flanking regions; right, gene body only. E, Boxplots showing the expression levels of hyper- or hypo-DMGs (ZS97-MH63, at CG or CHG context) with methylation sites in gene body regions in MH63 (green) and ZS97 (orange) panicle. The y axis represents gene expression level log2(FPKM + 1). The horizontal line within the box represents the median, box limits represent upper and lower quartiles, and whiskers represent 1.5 × interquartile range. P < 0.05 indicates statistical significance (* P < 0.05, ** P < 0.01, NS, not significant, two-tailed t test).
Although CG and CHG methylation mainly target TEs and repetitive sequences in the rice genome (Tan et al., 2016), more than 60% of the CG and CHG DMRs between ZS97 and MH63 were detected in genes (body and flanking regions; Figure 2, B and D; Supplemental Data set S1 and S2). In panicle, there were 1,434 and 2,204 genes with, respectively, hyper and hypo CG methylation and 790 and 742 genes with, respectively, hyper and hypo CHG methylation detected in ZS97 versus MH63 (Figure 2D). The data highlighted that substantial variations of genic CG and CHG methylations exist between ZS97 and MH63, which are maintained in most cases during shoot to panicle development.
Many CG and, to a lesser extent, CHG DMRs were located within the gene body (Figure 2B). In fact, nearly 45% of the CG differentially methylated genes (DMGs) were gene body CG DMGs (Figure 2, C and D; Supplemental Figure S4). To study whether differential gene body CG methylation is associated with differential gene expression, we plotted transcript levels of the DMGs between ZS97 and MH63. The analysis revealed no significant difference of expression (Figure 2E). In contrast, gene body CHG DMGs displayed a reverse relationship with expression levels between ZS97 and MH63 (Figure 2E).
There are 1,284,423 single nucleotide polymorphisms (SNPs, not including InDels) between MH63 and ZS97 genomes (Zhang et al., 2016). Bins with both SNP and non-SNP showed methylation differences between ZS97 and MH63 (Supplemental Figure S5A), but percentages of DMR in SNP bins are higher than those in non-SNP bins (Supplemental Figure S5B).
Differential methylation in the hybrids relative to the parents
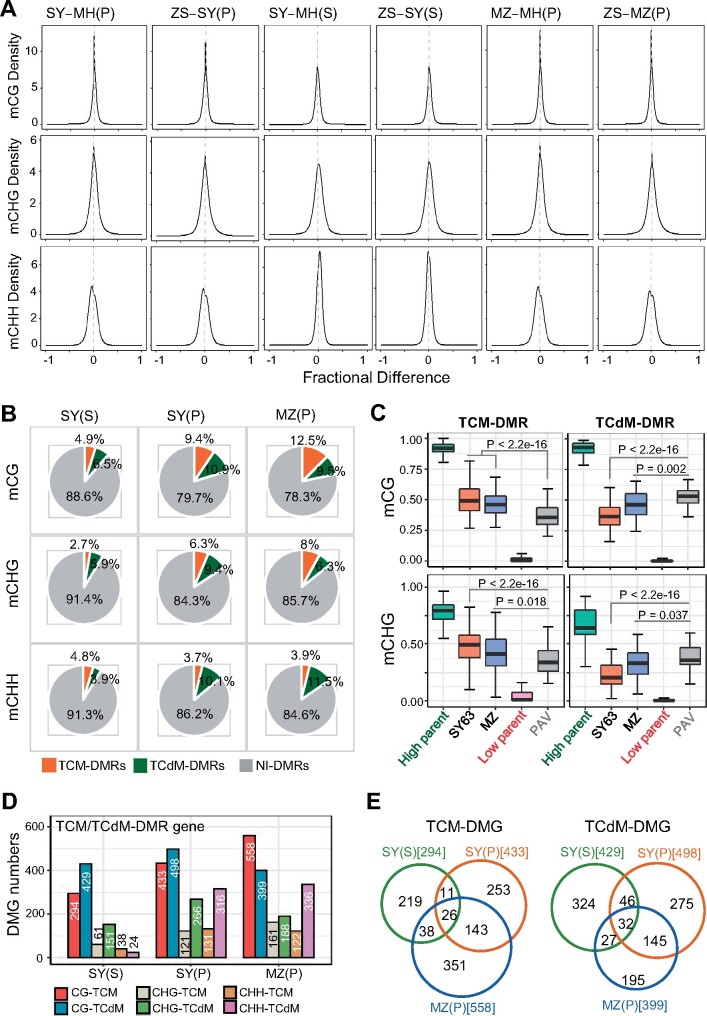
Overall methylation levels in the reciprocal hybrids SY63 and MZ were intermediate between the parents (Figure 1B; Supplemental Figure S1C; Supplemental Table S1). Density plots revealed that in shoot, CG and CHG methylations show clear variations between SY63 and the parents, but the differences became less pronounced in panicle (Figure 3A). Conversely, the variation of CHH methylation in shoot was lower than that of CG or CHG methylations, but became more pronounced in panicle (Figure 3A). This likely reflects the dynamic change of the methylation divergences from shoot to panicle between the parents (Figure 2, A and B). Similar profiles of density plots are observed in panicle methylation of the reciprocal hybrid MZ relative to the parents (Figure 3A), consistent with the low methylation divergences between SY63 and MZ (Figure 2, A and B). The analysis revealed substantial DNA methylation differences in the hybrids compared to either of the parents, which became more or less pronounced in panicle depending on the sequence context.
Figure 3.
Methylation variation and interaction of parental DMRs in the hybrids. A, Density plots showing the distribution of methylation difference frequency at 200-bp windows between the hybrids SY63 (SY) and MZ and either of the parents (MH63, MH; ZS97, ZS) in shoot (S) and/or panicle (P). B, Percentage of the TCM-DMR (orange), TCdM-DMR (green), and NI-DMR (gray) regions in SY63 shoot [SY(S)] and panicle [SY(P)] and MZ panicle [MZ(P)]. C, Boxplots showing CG and CHG methylation levels of TCM-DMR, TCdM-DMR in SY63 panicle, MZ panicle and the parents (parent with high methylation level, green; parent with low methylation level, pink). The gray box represents the PAV. The y axis represents methylation rate. The horizontal line within the box represents the median, box limits represent upper and lower quartiles, and whiskers represent 1.5 × interquartile range. P < 0.05 indicates statistical significance (Wilcoxon rank-sum test). D, Numbers of TCM-DMGs, TCdM-DMG and NI-DMG in SY63 shoot [SY (S)], SY63 panicle [SY (P)], and MZ panicle [MZ (P)]. E, Venn diagrams of overlapping TCM-DMGs or TCdM-DMGs in CG context between SY63 shoot [SY (S)], SY63 panicle [SY (P)], and MZ panicle [MZ (P)].
To identify differential methylation between hybrids and the parents, we calculated the predicted additive value (PAV) of the DMRs identified between the parents as previously described (Schultz et al., 2012; Zhang et al., 2016; see “Materials and Methods” section), and compared the methylation levels of hybrid with the PAVs in corresponding regions (within 200 bp bins). The hybrid bins (F1) that show >PAV and FDR <0.01 are known as trans-chromosomal methylation regions (TCMs), whereas bins with F1 < PAV and FDR <0.01 are considered as trans-chromosomal demethylation regions (TCdMs; Zhang et al., 2016; Supplemental Figure S6A). These differential methylations are supposed to be from nonadditive interaction, while the remaining bins resulted from additive interaction between the parental methylation (Zhang et al., 2016). In shoot, only a small portion of the parental DMRs display TCM and TCdM (i.e., nonadditive interactions), and more than 88% of them display additive methylation interaction (NI-DMRs) in the hybrid (Figure 3, B and D). The percentages of TCM and TCdM increased in panicle (Figure 3B). There were 294–558 genes with TCM or TCdM at CG sites (Figure 3D), while and relatively few genes with TCM or TCdM were found at non-CG sites (Figure 3D; Supplemental Figure S6B). However, the TCM/TCdM showed little overlapped between shoot and panicle (Figure 3E). The reciprocal hybrid MZ exhibited similar numbers of TCM and TCdM in panicle, about 40% of which overlapped with those detected in SY63 (Figure 3E). The analysis indicated that in the regions where the parental genomes display differential methylation, mainly additive interaction between alleles takes places in the hybrid.
About 2% of the parental similarly methylated regions (SMRs) showed nonadditive methylation, which, however, correspond to >86% of the total nonadditive methylation in the hybrid (Supplemental Figure S6B and Supplemental Figure S7A). The remaining SMRs showed additive methylation in the hybrid. The nonadditive methylation detected in the SMR principally concerns CHH and CG sequences (Supplemental Figures S6B and S7B). However, in shoot, only about 16% of nonadditive CHH methylation bins were associated with 21- and 24-nt siRNAs in SY63 (Supplemental Figure S7D). The percentage increased to 30% in panicle, which is consistent with the higher portions of 21- and 24-nt siRNAs among the total siRNAs in panicle compared with shoot (Supplemental Figures S7C and S7D). The analysis suggests that besides the RdDM pathway that is suggested to regulate nonadditive methylation interaction in the hybrid (Zhang et al., 2016), additional mechanisms are likely to be involved. Analysis of expression levels of the genes with nonadditive methylation revealed no differences between the hybrid SY63 and either of the parents or the mid-parent value (MPV), although a small percentage of the genes showed up and down-regulation in the hybrid (Supplemental Figure S8).
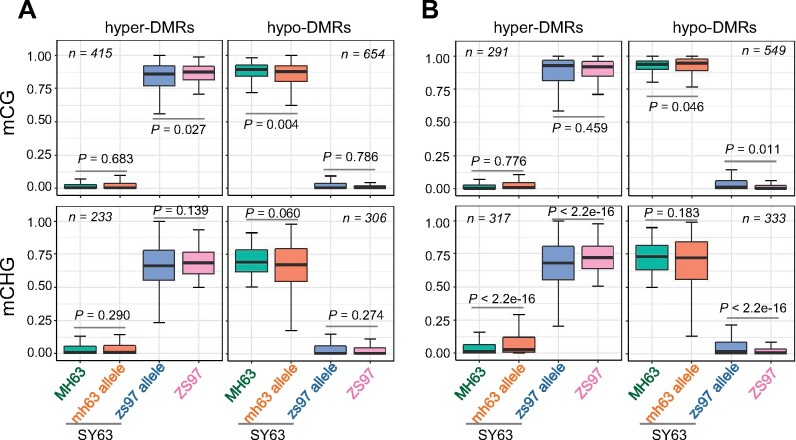
To get information on how the parental CG and CHG DMRs are remodeled in the hybrid, we investigated allelic methylation levels of the SNP-bearing DMRs between ZS97 and MH63 in the hybrid (Supplemental Figure S5). The analysis showed that parental CG and CHG DMRs were essentially conserved in the respective allelic bins in the hybrid (Figure 4), indicating that parental CG and CHG methylation variations are essentially maintained in the parental alleles in the hybrid.
Figure 4.
Parental methylation variations are maintained in the parental alleles in the hybrid SY63. Boxplots showing the methylation levels of DMRs (with SNPs) identified between the parental lines (MH63 and ZS97) and ASM in hybrid [SY63 (mh63-allele, orange; zs97-allele, blue) shoot (A) and panicle (B). The y axis represents methylation rate. The horizontal line within the box represents the median, box limits represent upper and lower quartiles, and whiskers represent 1.5 × interquartile range. P < 0.05 indicates statistical significance (Wilcoxon rank-sum test).
ASE is associated with DNA methylation
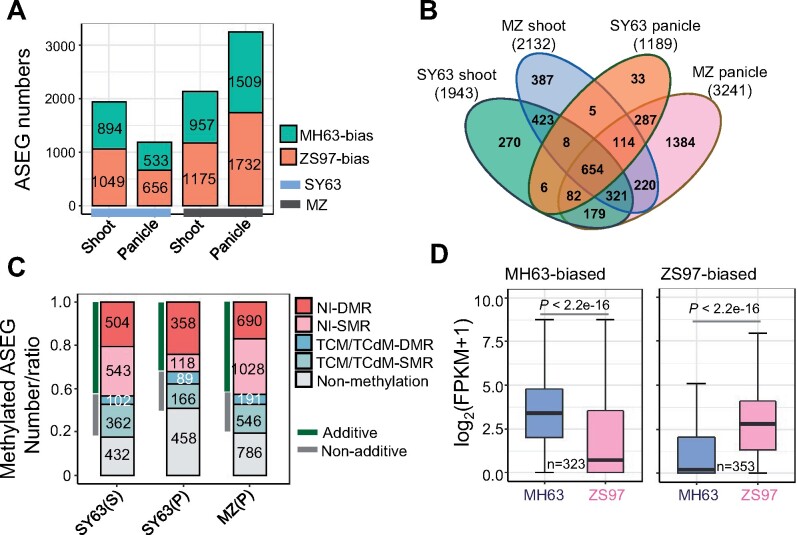
To investigate whether parental DNA methylation variations are associated with ASE in the hybrids, we analyzed shoot and panicle transcriptomes of the ZS97 and MH63 and their reciprocal hybrids SY63 and MZ. We used the 1,284,423 SNPs between MH63 reference sequence2 (MH63RS2) and ZS97 reference sequence2 (ZS97RS2), which were identified previously for ASE calling to identify allele-specific expression genes (ASEGs; see “Materials and Methods” section; Shao et al., 2019). A total of 1,943 ASEGs are identified in SY63 shoot, 894 with MH63 allele-biased, and 1,049 with ZS97 allele-biased expression (Figure 5A, Supplemental Data set S3). In SY63 panicle, 533 genes exhibited MH63 allele-biased expression, and 656 genes exhibited ZS97 allele-biased expression (Figure 5A). About 35%–64% of the MH63 or ZS97-biased ASEG were maintained in shoot and panicle with the same biased direction (Supplemental Figure S9A). In the reciprocal hybrid MZ shoot, the ASEG numbers were comparable with those in SY63, but in panicle, higher numbers of ASEG were detected (Figure 5A; Supplemental Data set S4). Similarly, 38%–68% of the same parent-biased ASEG were persistently expressed in MZ shoot and panicle (Supplemental Figure S9A). We noticed that most of the ASEG identified in SY63 shoot (651/894 for MH63-biased and 755/1,049 for ZS97-biased) and panicle (518/533 for MH63-biased and 621/656 for ZS97-biased) were also detected in the reciprocal hybrid MZ (Supplemental Figure S9B), suggesting that a majority of the ASEGs were independent of the parent-of-origin. Comparison of all ASEGs (a total of 4,373 genes) identified in shoot and panicle of SY63 and MZ revealed 654 ASEGs that were consistently present in the reciprocal hybrids and in shoot and panicle, which includes 283 MH63-biased and 366 ZS97-biased ASEGs (Figure 5B; Supplemental Data set S5). The remaining 3,719 genes exhibited inconsistent ASE directions among the reciprocal hybrids and analyzed tissues. The results were consistent with previously reported results (Shao et al., 2019). Among the identified ASEGs, many were previously shown to be associated with important agronomic traits and/or related to heterosis, such as Grain number, plant height, and heading date7.1 (Ghd7.1), Ghd8, MONOCULM1 (MOC1), Oryza sativa SQUAMOSA PROMOTER BINDING PROTEIN-LIKE13 (OsSPL13), OsMADS56 (Oryza sativa MCM1/AGAMOUS/DEFICIENS/SFR56), Xa1 (against Xanthomonas oryzae pv. Oryzae), etc. (Table 1; Yan et al., 2011, 2013; Si et al., 2016; Shao et al., 2019). The majority (60%–77%) of the ASEGs in the shoot and panicle of the reciprocal hybrids were associated with DNA methylation (Figure 5C), which were several folds higher than the genomic average (Tan et al., 2016). Among the methylation-associated ASEG, more than two-thirds showed additive methylation in the hybrids (Figure 5C). The remaining ASEGs were associated with TCM/TCdM-DMR (Supplemental Data set S6). Among the genes with TCM-DMR, several were previously characterized to be related to important agronomical traits, including Heading date1 (Hd1; OsMH_06G0155400), OsMADS51 (OsMH_01G0669400), O. sativa SQUAMOSA PROMOTER BINDING PROTEIN-LIKE1 (OsSPL1; OsMH_01G0000400), Xa26 (OsMH_11G0448200), and several LRR receptor-like serine/threonine–protein kinase genes (OsMH_01G0398900, OsMH_11G0447100, OsMH_0447800). Among the genes with TCdM-DMR, several were O. sativa Gibberellin 2-oxidase1 (OsGA2ox1; OsMH_05G0050000), O. sativa VERNALIZATION INSENSITIVE3-LIKE2 (OsVIL2; OsMH_12G0279200), GLUTAMINE SYNTHETASE1.2 (GS1.2; OsMH_03G0107700), OsYUCCA4 (OsMH_01G0112300), and OsMADS22 (OsMH_02G0527200) that are also important growth regulatory genes characterized in rice (Supplemental Table S3). The ASEGs in the parental lines exhibited similar difference of expression, suggesting that ASE of the genes may be inherited from the parental expression pattern (Figure 5D).
Figure 5.
Identification of ASEGs in the reciprocal hybrids shoot and panicle. A, Numbers of ASEGs identified in SY63 and MZ shoot or panicle. B, Venn diagram showing the numbers of consistent ASEGs in shoot and panicle of SY63 and MZ. C, Methylated ASEG numbers or ratio in SY63 shoot (S) or panicle (P) and in MZ panicle (P). D, Boxplot showing the difference of expression between the parents of MH63-biased or ZS97-biased ASEGs with DNA methylation identified in SY63 panicle. The y axis represents gene expression level log2(FPKM + 1). The horizontal line within the box represents the median, box limits represent upper and lower quartiles, and whiskers represent 1.5 × interquartile range. P < 0.05 indicates statistical significance (two-tailed t test).
Table 1.
The previously identified genes in the identified ASEGs in SY63 or MZ that is important for agronomic traits or related to heterosis
| Gene Symbol | MH63 Locus | Allele Type(S) | Allele Type(P) | ZS97 Locus | ASEG Type |
|---|---|---|---|---|---|
| Ghd7.1/DTH7 | OsMH_07G0481500 | ZS97 | ZS97 | OsZS_07G0493800 | Consistent |
| OsMADS30 | OsMH_06G0437500 | ZS97 | ZS97 | OsZS_06G0417800 | Consistent |
| Xa1 | OsMH_04G0491400 | ZS97 | ZS97 | OsZS_04G0493900 | Consistent |
| Rf1a/Rf5 | OsMH_10G0314300 | ZS97 | ZS97 | OsZS_10G0336200 | Consistent |
| OsVIL1 | OsMH_08G0115500 | MH63 | MH63 | OsZS_08G0106100 | Consistent |
| Rf4/PPR782a | OsMH_10G0311800 | MH63 | MH63 | OsZS_10G0333900 | Consistent |
| MOC1 | OsMH_06G0494800 | ZS97 | NA | OsZS_06G0364100 | Inconsistent |
| LAZY1 | OsMH_11G0270000 | ZS97 | NA | OsZS_11G0283700 | Inconsistent |
| Ghd8/DTH8 | OsMH_08G0066000 | MH63 | NA | OsZS_08G0071600 | Inconsistent |
| OsMADS22 | OsMH_02G0527200 | MH63 | MH63 | OsZS_02G0620600 | Inconsistent |
| 14-3-3/GF14f | OsMH_03G0497400 | MH63 | NA | OsZS_03G0494600 | Inconsistent |
| Wx | OsMH_06G0027500 | MH63 | NA | OsZS_06G0028900 | Inconsistent |
| Xa26 | OsMH_11G0447800 | MH63 | MH63 | OsZS_11G0447800 | Inconsistent |
| TAW1 | OsMH_10G0384000 | MH63 | NA | OsZS_10G0317200 | Inconsistent |
| OsGID1 | OsMH_05G0309000 | NA | MH63 | OsZS_05G0316500 | Inconsistent |
| OsSPL13 | OsMH_07G0316100 | NA | MH63 | OsZS_07G0327600 | Inconsistent |
| OsYUCCA3 | OsMH_01G0511100 | NA | MH63 | OsZS_01G0717200 | Inconsistent |
| OsGA2OX3 | OsMH_01G0531100 | NA | MH63 | OsZS_01G0526400 | Inconsistent |
| Hd1 | OsMH_06G0155400 | NA | MH63 | OsZS_06G0152400 | Inconsistent |
| OsMADS15 | OsMH_07G0007900 | NA | MH63 | OsZS_07G0006000 | Inconsistent |
S, shoot; P, panicle.
Allelic-specific CHG methylation is associated with allelic gene expression
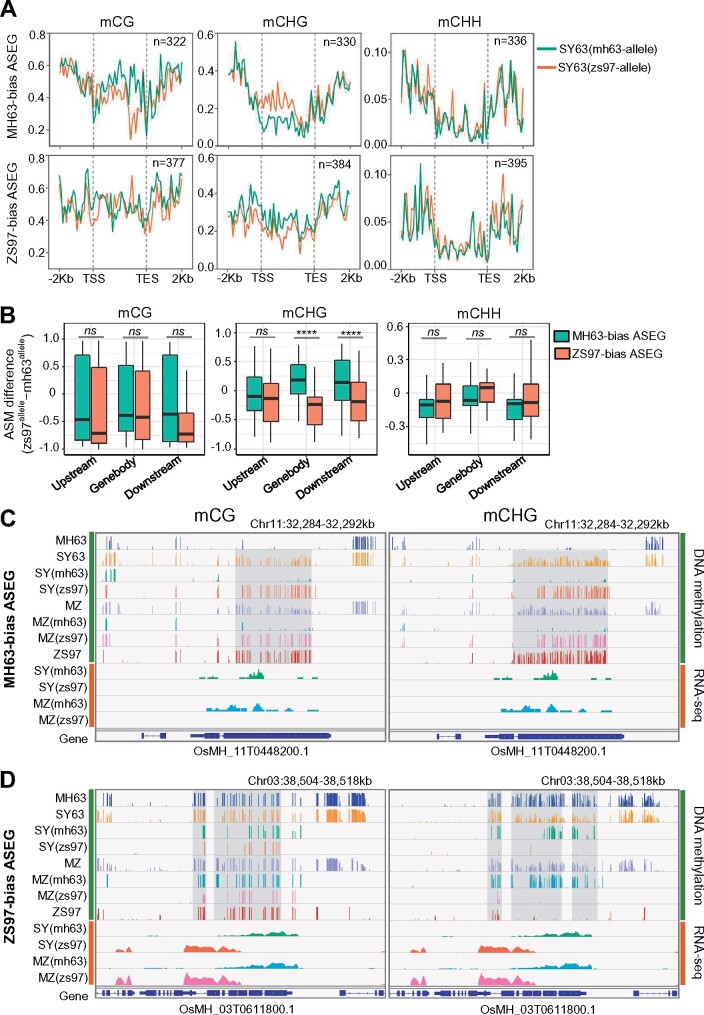
To study whether the methylated ASEG is associated with allelic-specific methylation (ASM), we analyzed methylation levels of ASEGs (gene body and 2-kb 5′- and 3′-flanking regions) in the parents and the hybrid parental alleles at CG, CHG, and CHH contexts. For the 533 panicle MH63-biased ASEGs, 322–336 have CG, CHG, and/or CHH methylation, and for the 656 panicle ZS97-biased ASEGs, 377–395 have CG, CHG, and/or CHH methylation (Figure 6A; Supplemental Data set S7). Meta-plot analysis revealed that MH63-biased ASEGs exhibited lower gene body CHG methylation in the MH63 alleles than the ZS97 alleles in the hybrid (Figure 6A). Conversely, in the 384 (out of 656) ZS97-biased ASEGs that show CHG methylation (Supplemental Data set S7), the ZS97 alleles display lower CHG methylation than the MH63 alleles in the hybrid (Figure 6A). Quantitative analysis indicated that the body region of the ZS97 alleles showed significantly higher CHG methylation than the MH63 alleles in the MH63-biased ASEGs (green in Figure 6B), and lower CHG methylation in the ZS97-biased ASEGs (Figure 6B), indicating that ASM at CHG sites was negatively associated with ASE, consistent with the repressive role of CHG methylation in gene expression. This pattern of ASM was detected in several agronomically important genes such as MOC1, GDP-D-mannose-3,5-epimerase (OsGME), Xa26, and OsMADS15 (Figure 6, C and D, Supplemental Figure S10; Supplemental Data set S7; Shao et al., 2019). At CG and CHH contexts, no clear ASM could be discerned, except that MH63-biased ASEGs exhibited slightly higher gene body CG methylation in the MH63 alleles than the ZS97, and that CHH methylation peaks could be observed mainly in the flanking regions in both alleles of MH63-biased or ZS97-biased ASEGs (Figure 6A). Similar trends were observed for the SY63 shoot ASEGs and the reciprocal hybrid (MZ) panicle ASEGs (Supplemental Figures S11 and S12; Supplemental Data set S8 and S9). However, the ASM differences in SY63 shoot or MZ panicle were weaker, probably because larger numbers of ASEGs were taken into consideration. Taken together, the analysis identifies a role of ASM at gene body CHG sites in ASE.
Figure 6.
Differential ASM of the ASEGs identified in the hybrid SY63 panicle. A, Metaplots showing CG or CHG methylation difference between MH63-alleles (green) and ZS97 alleles (orange) of MH63-biased or ZS97-biased ASEG in SY63 panicle. B, Boxplots showing ASM differences (ZS97 allele minus MH63 allele) of MH63-biased ASEG (green) and ZS97-biased ASEG (orange) in SY63 panicle. The y axis represents ASM difference values. The horizontal line within the box represents the median, box limits represent upper and lower quartiles, and whiskers represent 1.5 × interquartile range. ****P < 0.0001; NS, P > 0.05 (Wilcoxon rank-sum test). C, Genome browser screenshots showing ASM (upper, first to eighth lines) at CG (left) or CHG context (right) and allele-specific RNA-seq reads (lower, 9th–12th lines) of a MH63-biased ASEG (Xa26, OsMH_11T0448200) in the reciprocal hybrids SY63 and MZ panicle, and ASM comparison with the hybrid average methylation (not distinct allele-specific) and the parental methylation levels. SY (mh63 or zs97) and MZ (mh63 or zs97) indicate the MH63 or ZS97 allele in SY63 or MZ. For DNA methylation, different colors indicate the methylation of MH63 (blue), ZS97 (dark red), SY63 (average, yellow), MH63-allele in SY63 [SY (mh63), green], ZS97-allele in SY63 [SY (zs97), orange], MZ (average, slate blue), MH63-allele in MZ [MZ (mh63), turquoise], ZS97-allele in MZ [MZ (zs97), pink]. The gray shading indicates regions of AMS with clear difference between parent alleles. For allele-specific RNA-seq reads, green stands for MH63-allele in SY63 [SY (mh63)], orange stand for ZS97-allele in SY63 [SY (zs97)], turquoise stand for MH63-allele in MZ [MZ (mh63)] and pink stand for ZS97-allele in MZ [MZ (zs97)]. D, Genome browser screenshots showing allele-specific CG (left) or CHG (right) methylation (upper, first–eighth lines) and RNA-seq reads (lower, 9th–12th lines) of a ZS97-biased ASEG (OsMH_03T0611800) between the parent alleles in the hybrid panicles.
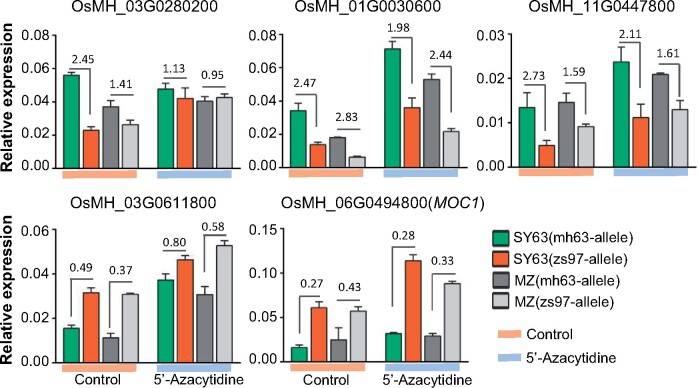
To confirm involvement of DNA methylation in ASE, we treated SY63 and MZ seedlings with DNA methylation inhibitor 5′-azacytidine (200 µM) and analyzed ASE of five genes with allele-specific primer sets (Supplemental Figure S13 and Supplemental Table S4). The treatment reduced DNA methylation (Supplemental Figure S14). In four of the five tested genes, we observed a reduction of ASE between the parental alleles in treated plants compared with untreated seedlings (Figure 7).
Figure 7.
Effects of DNA methylation inhibitor 5′-azacytidine on ASE. RT-qPCR analysis of the expression levels of MH63-alleles or ZS97-alleles of 5 ASEG [3 MH63-biased ASEG (OsMH_03G0280200, OsMH_01G0030600, OsMH_11G0447800) and 2 ZS97-biased ASEG (OsMH_03G0611800, OsMH_06G0494800 (MOC1))] in SY63 or MZ seedlings treated with or without DNA methylation inhibitor 5′-azacytidine (200 µM). Allele-specific primer sets were used. Relative expression levels (relative to Actin transcript) are shown. Bars indicate means ± sd from three biological replicates. The values on the bars are ratios of mh63-allele/zs97-allele expression level.
Discussion
DNA methylation divergence between the parental lines ZS97 and MH63
This work reveals higher methylation variations at CG and CHG than CHH sites between the parental lines of the elite hybrid rice SY63, although the CHH methylation variation increases in panicle. This is unexpected, as CHH methylation appears to be more stochastic and tends to be more variable in populations and during development (as shown in Figure 1D). Despite that CG and CHG methylation mainly target TEs and repetitive sequences in the rice genome (Tan et al., 2016), a majority of the CG and CHG DMRs between ZS97 and MH63 are located in genes (Figure 2B). Since only the syntenic regions (with or without SNP, but excluding InDels) between the parental genomes were taken into consideration, it is unlikely that the CG and CHG methylation variations resulted solely from the parental DNA sequence divergence, although higher percentages of DMRs were detected in SNP bins (Supplemental Figure S5). In addition, this work reveals differences of overall methylation levels and variation during development between MH63 than ZS97. The present results suggest that methylation differences between the parental lines can be evaluated to allow predictions of heterotic performance between different varieties.
The difference in gene body CG and CHG methylation between MH63 and ZS97 may have arisen during their breeding and adaptation to different subcontinental regions. It has been shown that gene body CG methylation is related to activity of CMT3, which catalyzes CHG methylation (Bewick et al., 2016; Wendte et al., 2019). This would suggest that differential CMT3 activities between the parental lines could be related to the CG and CHG methylation divergence. However, no difference of CMT3 amino acid sequence is detected between ZS97 and MH63 (Supplemental Figure S2C). In contrast, CMT2 presents a large deletion in ZS97 (Supplemental Figure S2C). As CMT2 is involved in both CHG and CHH methylation, CMT2 polymorphism may be related to the difference in CG and CHG methylation between MH63 and ZS97. Alternatively, differences in chromatin modification, such as H3K9me2, may also contribute to the differences in CG and CHG methylation between the two parental lines.
Interaction of parental DNA methylation epigenomes in hybrid rice
Processes such as the nonadditive interactions between the parental alleles leading to either gain (TCM) or loss (TCdM) of allelic methylation have been found to contribute to the observed methylation changes in hybrids (Greaves et al., 2012; Shivaprasad et al., 2012; Groszmann et al., 2013; Chen et al., 2014; Zhang et al., 2016). The present data show that in the hybrid SY63, only a small percentage of bins show nonadditive methylation, most of which is detected in the genomic regions where parental DNA methylations are similar (Supplemental Figures S6 and S7B). Due to the presence of siRNAs at regions that undergo TCM/TCdM, it has been suggested that siRNAs are the initiating molecules that establish these TCM/TCdM events (Greaves et al., 2015). However, in the rice hybrids, only about 15% (in shoot) and 30% (in panicle) of the nonadditive (both hyper and hypo) DNA methylation regions are associated with 21- and 24-nt siRNA (Supplemental Figure S7D). In addition, besides CHH methylation, nonadditive interaction also involves CG methylation in SY63. Together, these observations suggest that besides RdDM, which is suggested to be important for communication between alleles (Zhang et al., 2016), additional mechanisms are likely involved in the nonadditive methylation interaction between alleles in the hybrid rice. That no overall expression difference of the nonadditive methylation genes is observed between the hybrid and either of the parents or the MPVs (Supplemental Figure S8) supports the hypothesis that nonadditive methylation variation has principal impacts at specific loci rather than on a global scale.
ASM at CHG sites represses ASE
It is suggested that ASE can lead to phenotypic variation and play a role in heterosis (Springer and Stupar, 2007; Paschold et al., 2012; Goff and Zhang, 2013; Shao et al., 2019). This notion is supported by the identification of several ASE genes involved in key agronomic traits and/or hybrid vigor in this and a previous study (Supplemental Data set S3 and S5; Table 1; Shao et al., 2019). Although genetic variations may cause gene expression differences between alleles, it has been shown that epigenetic mechanisms are involved in ASE. Unlike the parentally imprinted genes, nearly all of the ASEG in the SY63 are detected in the reciprocal hybrid MZ (Supplemental Figure S9B). This suggests that effect of the parent-of-origin is not generally involved in ASE. As most of the identified ASEGs in the rice hybrids are methylated, DNA methylation may be an important factor in the ASE. The reverse association between ASE and ASM at CHG sites suggests that CHG methylation maintains or reinforces repression of the parental alleles in the hybrids, consistent with the observation that genes with variation in CHG methylation show difference of expression between the parental lines (Figure 2E). This is consistent with the finding that the silent maternal allele of paternally expressed genes in endosperm of Arabidopsis lyrata is marked by hyper CHG methylation (Klosinska et al., 2016). The present results indicate that allelic-specific gene body CHG methylation is likely to be inherited from the parental epigenomes and is maintained or reinforced in an allelic-specific manner in the hybrid and during development (Figure 4; Supplemental Figure S10), possibly through the reinforcing feedback loop with H3K9me2 (Law and Jacobsen, 2010). As nearly all of the genes with ASMs at CHG also show CG methylation, retention of allelic-specific CG gene body methylation could possibly protect them from gain of CHG methylation. This is consistent with previous results showing that in hybrids of Arabidopsis Ler and C24 ecotypes, both cis- and trans-regulated DNA methylation play roles in ASM, with cis-regulation playing a major role in CG methylation and trans-regulation (e.g., involving siRNA, DNA methylation regulators, etc.) playing major roles in CHG and CHH methylation (Chen et al., 2010). It was shown that in Arabidopsis the mutation of DDM1 reduces heterosis level of biomass (Kawanabe et al., 2016; Zhang et al., 2016), while mutations in RNA polymerase IV (involved in siRNA production and RdDM) or MET1 (required for CG methylation) had no effect on heterosis (Kawanabe et al., 2016). As DDM1 is required mainly for CHG methylation in rice (Tan et al., 2016; Tan et al., 2018), the effect of DDM1 mutation on heterosis supports the hypothesis that CHG methylation variation may play a role in epigenetic mechanism involved in heterosis.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa spp. Indica/Xian) varieties Zhenshan 97 (ZS97) and Minghui 63 (MH63) and their reciprocal hybrids [Shanyou 63 (SY63) and MH63/ZS97 (MZ)] were used in this study. Germinated seeds were planted under growth chambers with conditions of 14-h light and 10-h darkness and with temperatures of 32°C/28°C. Seedling shoots at 4-leaf stage were collected for total RNA or genomic DNA extraction. For each RNA-seq sample, three biological replicates were performed with nine plants in each replicate. For each bisulfite-seq sample, two biological replicates were performed with six plants in each replicate. The plants used for collection of young panicle were grown in field and managed under normal agricultural conditions on the Huazhong Agricultural University rice experimental field, Wuhan, China. Young panicles with 2-mm length at the early stage of floral organs differentiation were collected and immediately placed in liquid N2 and stored at −80°C until RNA and genomic DNA extraction.
RNA-seq data analysis and ASE identification
Total RNA was extracted from the indicated tissues using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. RNA concentrations were measured using Nanodrop 2000 (Thermo Scientific, USA) and further quantified using Qubit 2.0 Fluorometer (Invitrogen, USA). A total of 2-µg RNA was used for mRNA isolated and RNA-seq library construction using the TruSeq RNA Library Preparation Kit (Illumina, USA), according to the manufacturer’s recommendations. The libraries were sequenced using Illumina HiSeq 2000 platform.
RNA-seq raw reads were filtered to remove adapter and low-quality bases by Trimmomatic (version 0.35). Allele-specific expression (ASE) reads separation and identification were carried out as previously described (Shao et al., 2019). Briefly, the cleaned reads of 24 libraries (4 genotypes × 2 tissues × 3 replicates) were aligned to two parental reference genomes (MH63RS2 and ZS97RS2 from Rice Information GateWay [RIGW], http://rice.hzau.edu.cn/rice/) by HISAT2 (version 2.1.0), using default parameters (http://ccb.jhu.edu/software/hisat2/index.shtml). For the mapping reads, a stringent filtration was conducted using a customized perl script. It was retained when the filtered reads could be perfectly mapped to one parental genome or with SNPs mapped to the other parental genome. The trimmed high-quality reads from the hybrid were divided into six sets, according to the reads of each replicate aligned to MH63RS2 and ZS97RS2, to separate MH63- or ZS97-specific reads in the SNP calling. A total of 1,284,423 SNPs between MH63RS2 and ZS97RS2 were used as the reference for ASE calling. The identification of ASE for each gene was based on the SNPs between two parental genomes, and it needed to meet the following three criteria: (1) every SNP was covered by no <5 reads; (2) the ratio of read counts of SNPs from the two parental alleles differed significantly from 1:1 (Q < 0.01); and (3) the significant bias of different SNPs of the same gene was not in different directions. A gene showing ASE of no less than one SNP was referred to as an ASEG.
Gene expression levels were calculated by StringTie (version 1.2.1) with parameters for strand-specific RNA-seq (Pertea et al., 2016). Differentially expressed gene of statistical significance (adjusted P < 0.01 and abs [log2 (fold change)] > 1) were identified between ZS97 and MH63 by the nbinomTest function of the DESeq package (Anders and Huber, 2010).
Bisulfite-seq library construction and whole-genome bisulfite sequencing (WGBS)-seq data analysis
Genomic DNA was extracted using DNeasy plant mini kit (QIAGEN, Germany) according to the manufacturer’s instructions. Bisulfite conversion of DNA, library construction, and sequencing were performed at the Beijing Genomics Institute.
Sequence quality of the WGBS data was evaluated by FastQC (version 0.11.5). The adapter and low-quality reads were removed by Trimmomatic (version 0.35), and the clean data were mapped to the MH63RS2 reference genome by Bismark (version 0.22.3) using default parameters (Krueger and Andrews, 2011). The uniquely mapped reads were retained for further analysis. Individual cytosines with more than four reads were considered for DNA methylation-level calculation.
Kernel density plots were generated by comparing the average cytosine methylation level within 200-bp window between two samples; the window contained at least five cytosines, and every cytosine that was covered by at least five reads was used.
Identification of DMRs and analysis of methylation interaction in F1 hybrids
To identify the DMRs, the whole genome was divided into 200-bp bins. Bins that contained at least five cytosines each and every cytosine with at least a five-fold coverage were retained. Bins with absolute methylation difference of 0.7, 0.5, 0.2 for CG, CHG, and CHH sites, respectively, and P <0.01 (Fisher’s exact test) between comparisons were considered as DMRs. After excluding the DMRs identified between MH63 and ZS97, the remaining bins in the syntenic regions (with or without SNP, but excluding InDels) between MH63 and ZS97 genome were considered as SMRs.
Identification of methylation interaction in F1 hybrid was conducted according to a previous study (Schultz et al., 2012; Zhang et al., 2016). The weighted methylation level of F1 hybrid for each DMR between MH63 and ZS97 was calculated as the PAV. In brief, the combined parental methylated reads were divided by the combined parental total reads inside parental DMR by the following formula:
i = position of cytosine, n = total number of cytosine positions in the DMR, MC = maternal methylated reads, PC = paternal methylated reads, MT = maternal methylated and unmethylated reads, PT = paternal methylated and unmethylated reads.
To compare the methylation level in F1 hybrids with parental DMRs, Fisher’s exact test was used to validate the methylation difference between F1 hybrid and corresponding PAV. The FDR was generated from an adjusted P-value using the Benjamini–Hochberg method. The F1 hybrid bins whose methylation levels were higher than the corresponding PAV with FDR <0.01 were considered as TCM-DMRs, whereas bins with F1 methylation levels lower than the PAV and FDR <0.01 were considered as TCdM-DMRs, and the remaining bins with FDR >0.01 were considered as non-interaction methylation regions (NI-DMRs). For the comparison of methylation differences between F1 hybrid and parental SMRs, the calculation method was the same as described above.
sRNA-seq data analysis
Analysis of small RNA-seq data was performed as previously described (Tan et al., 2016). Briefly, sRNA-seq raw data were cleaned by removing adaptor and low-quality reads by Trimmomatic (version 0.35). MicroRNA (http://www.mirbase.org/ftp.shtml); pre-miRNA, tRNA, and rRNA were removed. The cleaned reads were aligned to the MH63RS2 reference genome by Bowtie (version 1.3.0, https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.3.0/), allowing zero mismatches and unique mapping. Two types of small RNA abundance were calculated by 21- or 24- nt read numbers falling into 1-kb windows/regions throughout the whole genomes.
ASM analysis
To separate the MH63-allele or ZS97-allele reads from the hybrids BS-seq data, SNPs in the syntenic block regions of the parental genomes were used to filtrate the BS-seq reads. A total of 1,284,423 SNPs between two parents were identified by comparing the high-quality genome sequences of MH63 and ZS97 using Mummer3.23 (https://sourceforge.net/projects/mummer/files/). First, the filtered reads of BS-seq libraries were aligned to the parental reference genomes (MH63RS2 and ZS97RS2) by Bismark (version 0.22.3) using default parameters. Then the mapped reads of F1 hybrids were separated into MH63-allele or ZS97-allele reads, according to the identified SNPs between MH63RS2 and ZS97RS2 and their locations in the synteny block, using a customized perl script. To assess the accuracy of the separated reads, the parental reads were also mapped to two reference genomes by the same process. Finally, the allele-specific-methylation level of hybrid F1s were calculated based on the separated MH63-allele or ZS97-allele reads independently by BatMeth2 (version 2.01) using default parameters (Zhou et al., 2019). Fisher’s exact tests were used to examine the methylation difference between MH63-allele and ZS97-allele reads within 200-bp windows.
Reverse-transcription quantitative PCR and 5′-azacytidine treatment
Seeds of SY63, MZ and their parents were germinated and grown on hormone-free, one-half-strength Murashige and Skoog (1/2 MS) medium under 16/8 h of light/dark at 30°C/25°C. Eight-day-old seedlings were carefully transferred to the 1/2 MS medium containing 200 µM of DNA methylation inhibitor 5′-azacytidine (Sigma, A2385). The control seedlings were transferred to the medium without 5′-azacytidine. Other conditions were maintained as normal. After 5 d, shoots of the control or treatment seedlings were collected and immediately placed in liquid N2 for RNA and genomic DNA extraction.
Total RNA was isolated from the shoot of seedlings using TRIzol reagent (Invitrogen, USA). For each genotype, three biological replicates of RNA isolation were performed with six plants in each replicate. Four micrograms of total RNA was used to synthesize first-strand cDNA with a reverse transcription kit (Invitrogen, USA). Reverse transcription quantitative PCR (RT-qPCR) was performed using SYBR Premix ExTaq (TaKaRa) in a real-time system. The transcript level of ACTIN was used as the internal control. Three biological replicates were performed for each RT-qPCR analysis. Primer sequences are listed in Supplemental Table S4. Genomic DNA (1 µg) isolated from the control or treatment seedlings was digested with 40 units of the methylation-sensitive restriction enzyme McrBC (New England Biolabs; M0272) for 6 h, which cuts methylated DNA, followed by PCR with specific primers (Supplemental Table S4).
Accession numbers
The BS-seq and RNA-seq data produced in this study are deposited to NCBI Sequence Read Archive (BioProject ID: PRJNA664649; https://submit.ncbi.nlm.nih.gov/subs/sra/SUB8163360/ overview).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. BS-seq data and DNA methylation levels of MH63, ZS97 and their reciprocal hybrids.
Supplemental Figure S2. Expression levels and amino acid sequence comparison of DNA methylation maintenance genes in rice.
Supplemental Figure S3. Heatmap and Pearson correlation coefficients of RNA-seq replicates of MH63 (MH), ZS97 (ZS), SY63 (SY) and MH63/ZS97 (MZ) in shoot (S) and panicle (P).
Supplemental Figure S4. Genome browser screenshots of DNA methylation in MH63 and ZS97 shoot or panicle.
Supplemental Figure S5. DMRs with SNPs or without SNPs between MH63 and ZS97 in shoot and panicle.
Supplemental Figure S6 . Identification of methylation interaction in hybrids panicle and shoot.
Supplemental Figure S7. Methylation interaction in hybrid panicle and shoot.
Supplemental Figure S8. The expression levels of genes with SMR-TCM or SMR-TCdM in SY63 panicle.
Supplemental Figure S9. Identification of allele-specific expression genes (ASEGs) in the hybrids shoot and panicle.
Supplemental Figure S10. Genome browser screenshots of methylated ASEGs in hybrids.
Supplemental Figure S11. Differential allele-specific methylation of the ASEGs identified in SY63 shoot.
Supplemental Figure S12. Differential allele-specific methylation of the ASEGs identified in MZ panicle.
Supplemental Figure S13. Parental allele structure and sequence variation of 5 ASEGs and location of allele-specific primers used for RT-qPCR.
Supplemental Figure S14. McrBC-digestion and PCR test of ASEGs in F1 seedling treated with or without 5′-azacytidine (5-Aza; 200 µM).
Supplemental Table S1. Information of BS-seq data of MH63, ZS97 and their reciprocal hybrids F1 in shoot and panicle.
Supplemental Table S2. Information of RNA-seq data of MH63, ZS97 and their reciprocal hybrids F1 in shoot and panicle.
Supplemental Table S3. List of ASEGs with TCM/TCdM-DMRs in SY63 or MZ.
Supplemental Table S4. Primers used for RT-qPCR and McrBC-PCR and genotyping of F1 hybrids.
Supplemental Data set S1. Differentially methylated genes identified between ZS97 and MH63 in CG, CHG, and CHH contexts in panicle.
Supplemental Data set S2. Differentially methylated genes identified between ZS97 and MH63 in CG, CHG, and CHH contexts in shoot.
Supplemental Data set S3. Information of the ASEGs identified in SY63 shoot and panicle.
Supplemental Data set S4. Information of the ASEGs identified in MZ shoot and panicle.
Supplemental Data set S5. Information of the consistent ASEGs identified in hybrid F1s.
Supplemental Data set S6 . List and information of genes with CG or CHG TCM/TCdM-DMRs identified in F1 hybrids.
Supplemental Data set S7. Allele-specific methylated ASEGs of MH63-biased or ZS97-biased identified in SY63 panicle.
Supplemental Data set S8. Allele-specific methylated ASEGs of MH63-biased or ZS97-biased identified in SY63 shoot.
Supplemental Data set S9. Allele-specific methylated ASEGs of MH63-biased or ZS97-biased identified in MZ panicle.
Supplementary Material
Acknowledgments
We thank Drs Xianghua Li and Jinghua Xiao for assistance.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31821005; 31730049; 31900480); the National Key Research and Development Program of China (2016YFD0100802); Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Program No. 2016RC003), and the Fundamental Research Funds for the Central Universities (2662015PY228).
Conflict of interest statement. The authors declare that they have no competing interests.
X.M. did most of the experimental work and data analysis; F.X. did bioinformatics analysis; Q.J. and Q.Z. did field work; T.H., B.W., L.S., and Y.Z. participated in experimental work, Q.Z. and D.X.Z. designed and supervised the work; D.X.Z. wrote the article with inputs from X.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Dao-Xiu Zhou (dao-xiu.zhou@universite-paris-saclay.fr).
References
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber WT, Zhang W, Win H, Varala KK, Dorweiler JE, Hudson ME, Moose SP (2012) Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci U S A 109: 10444–10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Ji LX, Niederhuth CE, Willing EM, Hofmeister BT, Shi XL, Wang L, Lu ZF, Rohr NA, Hartwig B, et al. (2016) On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A 113: 9111–9116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Schmitz RJ (2017) Gene body DNA methylation in plants. Curr Opin Plant Biol 36: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, He G, He H, Chen W, Zhu X, Liang M, Chen L, Deng XW (2010) Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J Integr Plant Biol 52: 971–980 [DOI] [PubMed] [Google Scholar]
- Chen S, He H, Deng X (2014) Allele-specific DNA methylation analyses associated with siRNAs in Arabidopsis hybrids. Sci China Life Sci 57: 519–525 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14: 471–482 [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang GL, Meyers BC, Jacobsen SE, Pellegrini M (2012) Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci U S A 109: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Hammond R, Zhou P, Vaillancourt B, Lipzen A, Daum C, Barry K, de Leon N, Buell CR, Kaeppler SM et al. (2020) Variation and inheritance of small RNAs in maize inbreds and F1 hybrids. Plant Physiol 182: 318–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapp M, Reinders J, Bediee A, Balsera C, Bucher E, Theiler G, Granier C, Paszkowski J (2015) Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants 1: 15092. [DOI] [PubMed] [Google Scholar]
- Gehring M (2013) Genomic imprinting: insights from plants. Annu Rev Genet 47: 187–208 [DOI] [PubMed] [Google Scholar]
- Goff SA, Zhang Q (2013) Heterosis in elite hybrid rice: speculation on the genetic and biochemical mechanisms. Curr Opin Plant Biol 16: 221–227 [DOI] [PubMed] [Google Scholar]
- Greaves IK, Gonzalez-Bayon R, Wang L, Zhu A, Liu PC, Groszmann M, Peacock WJ, Dennis ES (2015) Epigenetic changes in hybrids. Plant Physiol 168: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES (2014) Inheritance of Trans Chromosomal Methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci U S A 111: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES (2012) Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci U S A 109: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci U S A 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 29: 684–690 [DOI] [PubMed] [Google Scholar]
- He G, Chen B, Wang X, Li X, Li J, He H, Yang M, Lu L, Qi Y, Wang X et al. (2013) Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol 14: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Xing Y, Wu W, Xu C, Sun X, Yu S, Zhang Q (2003) Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci U S A 100: 2574–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q,, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Kawanabe T, Ishikura S, Miyaji N, Sasaki T, Wu LM, Itabashi E, Takada S, Shimizu M, Takasaki-Yasuda T, Osabe K, et al. (2016) Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci U S A 113: E6704–E6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska M, Picard CL, Gehring M (2016) Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat Plants 2: 16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC (2004) Allele-specific gene expression uncovered. Trends Genet 20: 113–116 [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauss K, Wardenaar R, Oka R, van Hulten MHA, Guryev V, Keurentjes JJB, Stam M, Johannes F (2018) Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol 176: 1627–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J, Jiang H, Santos-Gonzalez J, Kohler C (2016) Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J 35: 1298–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Jia Y, Marcon C, Lund S, Larson NB, Yeh CT, Ossowski S, Lanz C, Nettleton D, Schnable PS, et al. (2012) Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res 22: 2445–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Springer NM (2013) Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64: 71–88 [DOI] [PubMed] [Google Scholar]
- Schultz MD, Schmitz RJ, Ecker JR (2012) ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet 28: 583–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert F, Thiemann A, Grant-Downton R, Edelmann S, Rybka D, Schrag TA, Frisch M, Dickinson HG, Melchinger AE, Scholten S (2018) Parental expression variation of small RNAs is negatively correlated with grain yield heterosis in a maize breeding population. Front Plant Sci 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao GN, Lu ZF, Xiong JS, Wang B, Jing YH, Meng XB, Liu GF, Ma HY, Liang Y, Chen F, et al. (2019) Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol Plant 12: 1090–1102 [DOI] [PubMed] [Google Scholar]
- Shao L, Xing F, Xu C, Zhang Q, Che J, Wang X, Song J, Li X, Xiao J, Chen LL, et al. (2019) Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc Natl Acad Sci U S A 116: 5653–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, He H, Li J, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, et al. (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24: 875–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, et al. (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48: 447–456 [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007) Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 17: 264–275 [DOI] [PubMed] [Google Scholar]
- Tan F, Lu Y, Jiang W, Wu T, Zhang RY, Zhao Y, Zhou DX (2018) DDM1 represses noncoding RNA expression and RNA-directed DNA methylation in heterochromatin. Plant Physiol 177: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Zhou C, Zhou Q, Zhou S, Yang W, Zhao Y, Li G, Zhou DX (2016) Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol 171: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendte JM, Zhang Y, Ji L, Shi X, Hazarika RR, Shahryary Y, Johannes F, Schmitz RJ (2019) Epimutations are associated with CHROMOMETHYLASE 3-induced de novo DNA methylation. Elife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Zhang J (2018) Shanyou 63: an elite mega rice hybrid in China. Rice (N Y) 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Wang G, Yuan M, Yao W, Lyu K, Zhao H, Yang M, Li P, Zhang X, Yuan J, et al. (2015) Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc Natl Acad Sci U S A 112: E5411–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, Liu T, Liu H, Zhang C, Zhang Z, et al. (2013) Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res 23: 969–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, et al. (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4: 319–330 [DOI] [PubMed] [Google Scholar]
- Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Saghai Maroof MA (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci U S A 94: 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci U S A 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen LL, Xing F, Kudrna DA, Yao W, Copetti D, Mu T, Li W, Song JM, Xie W, et al. (2016) Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc Natl Acad Sci U S A 113: E5163–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Xu T, Srivastava AK, Wang D, Zeng L, Yang L, He L, Zhang H, Zheng Z, et al. (2016) The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov 2: 16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C, Huang H, Zhang H, et al. (2016) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci U S A 113: E4248–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Chen Y, Yao W, Zhang C, Xie W, Hua J, Xing Y, Xiao J, Zhang Q (2012) Genetic composition of yield heterosis in an elite rice hybrid. Proc Natl Acad Sci U S A 109: 15847–15852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Lim JQ, Sung WK, Li G (2019) An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinformatics 20: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.