Some of the greatest gains in agricultural yield over the last century have come from the growth of hybrid plants produced by crossing two inbred cultivars. The phenomenon that enables this remarkable crop improvement is termed heterosis or hybrid vigor. Despite its importance, researchers are still working to understand the genetic basis of heterosis, which was first described by Darwin in 1876 (Darwin, 1876).
Through mapping efforts, geneticists have identified heterosis-associated quantitative trait loci, some of which have been further studied to clarify the molecular mechanism by which they contribute to improved performance in hybrid plants. However, studies show that most examples of heterosis are unlikely to be the product of just one or two genes (Botet and Keurentjes, 2020). An accumulating body of evidence suggests that due to a wide range of genetic diversity, from single nucleotide polymorphisms to presence–absence, copy number, and structural variants to epigenetic variants, there are substantial differences in the functional gene content of different crop accessions. These differences can result in the expression of only one parental allele for a given gene, called allele-specific expression, a possible mechanism underlying heterosis (Botet and Keurentjes, 2020).
In this issue of Plant Physiology, Ma et al. address the relationship between epigenetics and allele-specific expression in the elite hybrid rice (Oryza sativa) line SY63 (Ma et al., 2021). Through whole-genome bisulfite sequencing of the shoot and panicles of SY63, its parents MH63 and ZS97, and the reciprocal hybrid MZ, the authors identify differentially methylated regions between lines and tissues. They find an overall increase in methylation throughout development and show differences between the two parental lines in this process, perhaps, unsurprisingly due to their very different phenotypes at the profiled developmental stages (ZS97 is valued as a parent for robust seedling growth, while MH63 contributes a robust panicle phenotype; Xie and Zhang, 2018; Ma et al., 2021).
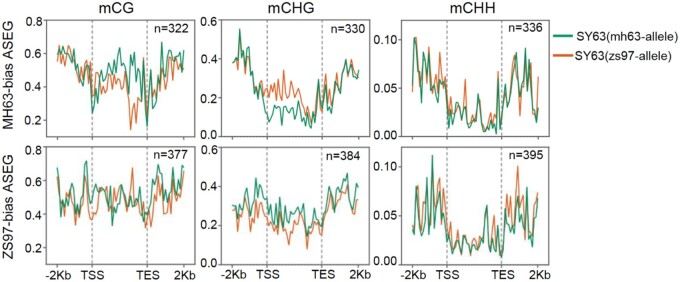
Interestingly, the authors find >60% of the regions differentially methylated at CG and CHG sequence contexts between the two parental genomes are located in gene bodies, despite the more common association of these methylation marks with repetitive sequence and transposable element silencing in the rice genome. By correlating differential methylation with gene expression, the authors demonstrate allele-specific expression is negatively associated with allele-specific differences in genic CHG methylation in SY63, consistent with the role of CHG methylation as a repressor of expression (Figure 1). These methylation differences appear to be maintained from the parents to the F1 hybrid and may explain allele-specific differences in expression of a number of genes associated with agronomically important traits, such as a regulator of plant architecture and flowering time and a Xanthomonas oryzae resistance gene (Sun et al., 2004; Lu et al., 2012, 15).
Figure 1.
Metaplots showing CG, CHG, and CHH methylation relative to the transcription start sites and end sites (TSS and TES, respectively). Plots are shown for MH63 parental alleles (green) and ZS97 parental alleles (orange) of MH63-biased or ZS97-biased allele-specific expressed genes in the SY63 hybrid rice panicle. Notably, there is an inverse correlation between gene body CHG methylation and allele expression. Adapted from Ma et al. (2021).
The findings in this study correspond with published data suggesting the gene DECREASE IN DNA METHYLATION 1 (DDM1), which is required for CHG methylation in Arabidopsis (Arabidopsis thaliana) and rice, is also required for biomass heterosis in Arabidopsis (Kawanabe et al., 2016; Tan et al., 2016; Zhang et al., 2016). While the authors demonstrated treatment with the methylation-blocking drug 5′-Azacytidine decreased allele-specific expression in rice seedlings, it will be interesting to continue studying the genetic mechanism by which parental methylation patterns are maintained in an allele-specific manner in hybrids and the possible role of rice DDM1 in this process. In particular, gene editing of DDM1 in MH63 and ZS97 will be a key to the next steps of understanding the role of epigenetics in hybrid vigor in rice. In the meantime, we can all enjoy the seeds of this work through the estimated 18.8 million tons of increased yield achieved by growing SY63 over other varieties grown from 1984 to 2012 (Xie and Zhang, 2018).
References
- Botet R, Keurentjes JB (2020) The role of transcriptional regulation in hybrid vigor. Front Plant Sci 11: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C ( 1876) The Effects of Cross and Self-Fertilisation in the Vegetable Kingdom, London, Murray. [Google Scholar]
- Kawanabe T, Ishikura S, Miyaji N, Sasaki T, Wu LM, Itabashi E, Takada S, Shimizu M, Yasuda TT, Osabe K, et al. (2016) Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci USA 113: E6704–E6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SJ, Wei H, Wang Y, Wang HM, Yang RF, Zhang XB, Tu JM (2012) Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza Sativa L.). Plant Mol Biol Rep 30: 1461–1469 [Google Scholar]
- Ma X, Xing F, Jia Q, Zhang Q, Hu T, Wu B, Shao L, Zhao Y, Zhang Q, Zhou DX (2021) Parental variation in CHG methylation is associated with allelic-specific expression in elite hybrid rice. Plant Physiol 186: 1025–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas Oryzae Pv. Oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37: 517–527 [DOI] [PubMed] [Google Scholar]
- Tan F, Zhou C, Zhou Q, Zhou S, Yang W, Zhao Y, Li G, Zhou DX (2016) Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol 171: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Zhang J (2018) Shanyou 63: an elite mega rice hybrid in China. Rice 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C, Huang H, Zhang H, et al. (2016) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA 113: E4248–E4256 [DOI] [PMC free article] [PubMed] [Google Scholar]